Abstract
Nuclear magnetic resonance (NMR) spectroscopy has evolved into a powerful tool for fragment-based drug discovery over the last two decades. While NMR has been traditionally used to elucidate the three-dimensional structures and dynamics of biomacromolecules and their interactions, it can also be a very valuable tool for the reliable identification of small molecules that bind to proteins and for hit-to-lead optimization. Here, we describe the use of NMR spectroscopy as a method for fragment-based drug discovery and how to most effectively utilize this approach for discovering novel therapeutics based on our experience.
Keywords: NMR spectroscopy, fragment-based drug discovery, fragment-based screening, hit identification, fragment libraries
1. Introduction
Fragment-based drug discovery (FBDD) is a powerful method for discovering high-affinity ligands for target proteins. Vemurafenib, the first small molecule inhibitor originating from a fragment-based screen, was approved by the FDA in 2011 (Bollag et al. 2010), demonstrating that fragment-based methods can be useful for drug discovery. Numerous other success stories illustrate the use of fragment-based approaches for the discovery of clinical candidates (Hajduk and Greer 2007, Chessari and Woodhead 2009, Murray and Blundell 2010). Generally speaking, there is no strict size requirement in order for a compound to be designated as a “fragment”, but the term is customarily used for small organic molecules with less than ~ 25 heavy atoms or a molecular weight of less than 250 Da. Fragment-based screening has several advantages over conventional high-throughput screening (HTS). First, structure-activity relationship (SAR) studies of the attained fragment hits proceed quickly because analogs are simpler to prepare synthetically or can be purchased commercially due to their smaller size and lower complexity. Another advantage stemming from the small size of the fragments screened is that a greater chemical space can be covered. In addition, fragment binding to a target protein is not constrained by the rest of the molecule, allowing the small molecules to optimally bind to proteins. Also, the stringency in fragment-based assays is less (up to millimolar binding affinities can be detected) allowing chemical starting points to be more easily obtained. Finally, the fragment hits identified have improved ligand efficiencies over HTS hits and form fewer but higher-quality intermolecular interactions with the target protein (Kuntz et al. 1999). FBDD is frequently used to find lead molecules for proteins lacking deep or well-defined ligand binding pockets, so-called “undruggable” targets (Hopkins and Groom 2002). Because fragment screens often identify hits within multiple chemical classes, this methodology is also a great way to circumvent patents and discover novel chemical matter against any target.
The detection of weak fragment binding by biochemical approaches (e.g. spectrophotometric and fluorescence-based experiments) is challenging, as a change in signal above the baseline of the assay is difficult to observe. In contrast, biophysical techniques (e.g. NMR spectroscopy, isothermal titration calorimetry (ITC), thermal denaturation, surface plasmon resonance (SPR), and X-ray crystallography, among others) are more robust in detecting such weak interactions (Hoffer et al. 2011). In particular, NMR spectroscopy is ideally suited for fragment-based screening because it can reliably detect binding up to single digit millimolar Kd values, often the only hits found for challenging targets (Carr et al. 2005, Jhoti et al. 2007). Additionally, NMR can be used to quantify binding affinities in order to establish the structure activity relationships (SAR). Furthermore, unlike with other techniques, ex apt X-ray crystallography, the binding sites of fragment hits and modes of binding can be ascertained from NMR experiments. Another disadvantage of plate-based spectrophotometric or fluorescence-based experiments is that many false-positive hits are often found as a result of assay artifacts (Wu et al. 2013). Because binding events are directly observed by NMR, the technique does not usually suffer from false-positive hit identification that riddles other screening techniques. Furthermore, the unbiased binding events observed by NMR can be used to identify novel ligand-binding “hot spots” not previously known, such as sites allosteric to the known binding site and sites that result from protein conformational changes. The use of NMR for FBDD also does not require a priori knowledge of protein function or endogenous binding partners (i.e. for design of a reference molecule) as needed in enzymatic or displacement-based assays.
From our experience in using NMR in FBDD, we have implemented a preferred research plan that spans from hit identification to lead optimization. Carrying out a successful fragment-based screen by NMR relies on implementation of the following steps: obtaining a suitable fragment library, performing the screen by either monitoring changes in the spectra of the protein or small molecule, determining structures of protein-fragment complexes, and aiding in the generation of lead compounds.
2. Designing a Fragment Library
FBDD requires a collection of small organic molecules (fragments). The size and quality of a fragment library are of utmost importance to the screening process.
2.1 Size of a Fragment Library
As the goal of any screening campaign is to identify hit molecules against a particular target, large libraries provide a greater chance of finding starting points for further elaboration. Fragment screens have been conducted using libraries of anywhere from several hundred to several thousand molecules (500-10,000) (Hajduk et al. 2000). Although a follow-up screen to explore the SAR of hit analogs can be performed when starting with a library of small size, we prefer to start with a library on the upper end of this range that already contains multiple examples of each chemotype. Building a library with multiple related compounds within each specific chemical class seems redundant; however, the presence of these analogs could be the difference between identifying a hit or not. An excellent example of this principle comes from our recent work on the discovery of Mcl-1 inhibitors (Friberg et al. 2013). The initial screen did not identify unsubstituted benzothiophene-, benzofuran-, or indole-2-carboxylic acids, yet analogs containing methyl or chlorine substitutions were found to be low micromolar affinity ligands for Mcl-1 and serve as excellent starting points for further medicinal chemistry modifications. Therefore, the standard procedure of first screening a small library and then following-up with a screen of closely related analogs could preclude the identification of additional and perhaps novel chemotypes as starting points.
2.2 Overall Guidelines for Compound Selection
In the literature, several rules or recommendations have been suggested for the selection of compounds when building a fragment library. The most commonly applied strategy follows the rule-of-three (molecular weight ≤ 300 Da, ClogP ≤ 3, hydrogen bond donors and acceptors each ≤ 3) (Congreve et al. 2003).
Our fragment library which currently consists of approximately 14,000 fragments was constructed by close consideration of the rule-of-three, but with slight modifications. Fragments between a molecular weight of 100-250 Da, having up to 4 hydrogen bond donors, and with a ClogP of up to 3.5 were selected in the initial search of commercial collections of compounds. Additional privileged fragments were selected by searching for compounds with substructures known to bind frequently to proteins (Hajduk et al. 2000), including carboxylic acid-, biphenyl-, diphenylmethyl-containing compounds, and many different heterocycles. Looser definitions of what constitutes a fragment were allowed for these privileged compounds, including having a molecular weight up to 275 Da, up to 5 hydrogen bond donors, and up to a ClogP of 4.
2.3 Removing “Bad Actors”
Before beginning a fragment-based screen, it is important to remove fragments that may misbehave, as the presence of such molecules can partially or fully confound the screen. Misbehaving molecules, so called “bad actors”, include nonspecific binders, reactive covalent modifiers, chelators, or aggregators. These can include molecules with functional groups such as Michael acceptors, hydrazines, hydroxyl amines, and free thiols, among others. When a fragment library is screened in mixtures, it is especially imperative to carefully analyze all molecules in the library. For example, if 10% of a library consists of bad actors, and the library is screened in mixtures of 10, every mixture is, on average, flawed. Therefore, the results for all samples are not to be trusted without further experimentation. Sorting out all of the misbehaving molecules from a fragment library is a daunting task since not all nuisance compounds are known a priori. Whereas the aforementioned rules and guidelines ensure that fragments are small, neither too hydrophilic nor hydrophobic and have higher chances to interact with the targets of interests, they do not assist in the removal of bad actors. However, compounds that do not behave well in screens have been identified and reported. For example, several classes of molecules have been described in the literature that should be removed from screening libraries (Fig. 1) (e.g., ALARM compounds (Huth et al. 2005), PAINS molecules (Baell 2010, Baell and Holloway 2010), and “Shoichet” aggregators (Seidler et al. 2003, Feng et al. 2005)).
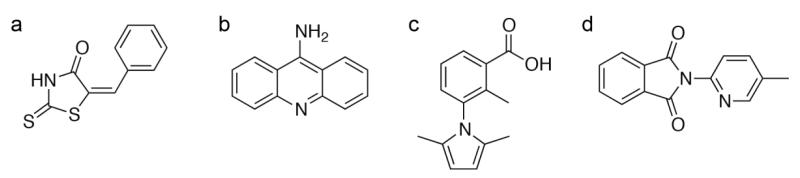
Fig. 1.
Examples of undesirable molecules in a fragment library. (a) Rhodanines are known to bind to a plethora of different protein targets. (b) 9-Aminoacridines can form soluble aggregates in aqueous solutions and can nonspecifically interact with hydrophobic residues on protein surfaces. (c) Phenyl dimethylpyrroles have been shown to covalently bind to proteins. (d) Maleimides can be hydrolyzed and are reactive.
Beyond these suggestions, further care must still be taken to filter out additional compounds such as aggregation-prone and insoluble fragments. Towards this end, NMR spectroscopy itself can help to build and maintain a clean fragment library. Fragment aggregates that can cause promiscuous inhibition are often soluble in aqueous solutions or are very small and therefore not detectable by visual inspection. NMR can be used to identify aggregators by observing a broadened water resonance or poor water suppression along with broadening of ligand resonances as the concentration of the fragment is increased. Solubility-limited fragments are also problematic in FBDD, as high fragment concentrations are necessary for binding detection, for X-ray studies (see section 4), and for second-site screening (see section 5). Insolubility can also result in false-negatives and errors in the quantification of the hits. NMR can be used to identify insoluble compounds by observing concentration-dependent signal intensities of the ligands. Compared to aggregation-prone and insoluble fragments, it is more difficult to eliminate reactive molecules from a library a priori since covalent binding can be target dependent. It is advisable to reexamine the library after several, unrelated targets have been screened to remove fragments that are found to be hits in all screens. Using this approach, we removed 450 fragments from our in-house library.
3. NMR Fragment-based Screening Methods
Several experimental NMR approaches for screening have been developed allowing hits to be identified for targets that are small, medium, and large in size (Fernandez and Jahnke 2004, Dalvit 2009, Campos-Olivas 2011, Shortridge and Powers 2011). Two general methodologies can be implemented for an NMR-based screen: monitoring differences in the spectra of the small molecules or the protein.
3.1 Screening by Observing the Small Molecule
Fragment-based screening methods that monitor changes in the spectra of the small molecules are popular approaches. The two most commonly applied techniques are saturation transfer difference spectroscopy (STD) (Mayer and Meyer 1999) and Water-LOGSY (Dalvit et al. 2000), both being transfer-NOE-type experiments. Both methods are well suited to detect weak binding, and the larger the target protein, the better they work. Other NMR methods where changes in the small molecule spectra are monitored include diffusion editing, relaxation-based experiments (Hajduk et al. 1997), approaches using target-attached paramagnetic probes (SLAPSTIC) (Jahnke 2002), heteronuclear detection schemes (19F- or 31P-based screening) (Dalvit et al. 2002, Tengel et al. 2004, Manzenrieder et al. 2008), and target-immobilized NMR screening (TINS) (Vanwetswinkel et al. 2005). For more details about ligand-based screening techniques, the interested reader is referred to the recent literature (e.g., (Meyer and Peters 2003, Klages et al. 2007, Pellecchia et al. 2008, Ludwig and Guenther 2009)). Ligand-based methods are also often used for medium-sized proteins (~15-100 kDa), as no isotope labeling is necessary and the quantity of protein needed is small (0.5-5 μM is required vs. 20-50 μM concentrations needed for protein-observed methods). When ligand-observed mixture screens are conducted (e.g., by STD or Water-LOGSY experiments) knowledge of the chemical shift pattern for each ligand makes time-consuming deconvolution of mixture hits unnecessary. However, ligand-based screening does not provide information on binding sites and, in our hands, is not as robust as observing protein chemical shift perturbations (greater number of false-positive and false-negative hits) (Lepre 2011). There are several reasons for the occurrence of the higher error rates, one being that it is usually difficult to discriminate between promiscuous binding due to compound aggregation and the desired site-specific binding by STD, WaterLOGSY, or fluorine-based experiments. In addition, transfer-NOE-based techniques are prone to artifacts arising from aberrant excitation of ligand resonances or spillover of radio-frequency power. Furthermore, since screening by observing the small molecule does not reveal atom-resolved information about the target, fragment induced protein precipitation can result in both false-positives and false-negatives, especially when ligands are screened as mixtures. If the protein fully precipitates, all members of a mixture may mistakenly be classified as non-binders (potential false-negatives). Partial unfolding of the target can lead to the appearance of transient binding sites which can attract hydrophobic fragments, and cause them to be considered as hits. Finally, ligand-based screening becomes challenging when binding is too strong (Kex ≪ | Δν|). To overcome the high-affinity limit, so-called reporter screening is of great value (Jahnke 2002, Zhang et al. 2009); instead of detecting direct binding, the concentration-dependent replacement of a weaker ligand by a stronger one is followed by ligand-observed NMR.
3.2 Screening by Chemical Shift Changes of Isotopically Labeled Proteins
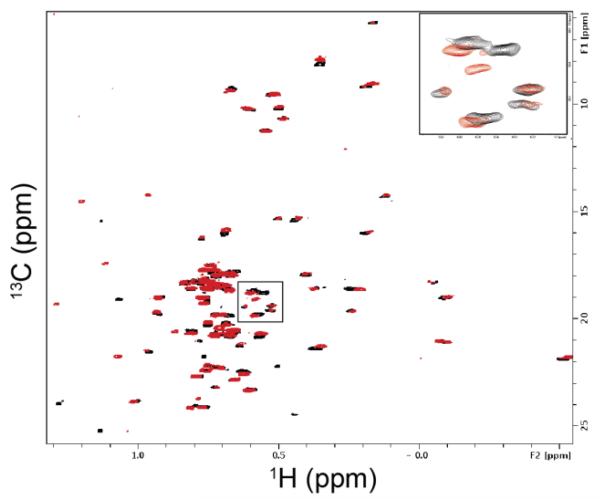
The most robust (and our preferred) fragment screening technique is based on monitoring chemical shift changes of an isotopically labeled protein upon the addition of fragments (Shuker et al. 1996). We typically screen targets up to 30-40 kDa by recording 1H-15N-HSQC spectra if uniformly 15N-labeled protein can be obtained, providing that the spectra show resonances with good lineshapes and only moderate peak overlap. Fast pulsing techniques such as the SOFAST-HMQC (Schanda et al. 2005) experiment allow for the recording of more than 200 2D heteronuclear correlation spectra in 24 hours (500 MHz with cryoprobe and automatic sample changer, 40 μM protein concentration, globular 15 kDa protein). Other techniques that can speed up screening by monitoring protein chemical shift perturbations involve sparse or non-uniform sampling (Felli and Brutscher 2009). Another advantage of using the protein signals is that by following the chemical shift perturbations upon the addition of small molecules, not only can hits be identified, but binding strengths (Kd values) can be determined and binding sites (if the protein signals have been assigned) can be identified. For proteins larger than 40 kDa, either TROSY-type experiments (Pervushin et al. 1997, Lescop et al. 2010), e.g., 1H-15N-TROSY-HSQC, or 1H-13C-HSQC measurements of selectively 13C-labeled (valine, leucine, isoleucine) proteins can be used (Hajduk et al. 2000). TROSY-based experiments work best at higher magnetic field strengths and when perdeuterated protein samples are used, making these experiments less attractive than 13C(methyl)-based experiments in our opinion. Protein binding sites almost always contain hydrophobic pockets, and thus side-chain methyl groups are perfectly suited as reporters for ligand binding events (Fig. 2). Compared to TROSY experiments, 13C(methyl)-based screening has the advantage of being very efficient even at lower magnetic fields (500 / 600 MHz spectrometer which are often equipped with automatic sample changer systems) and also has a high sensitivity due to the fact that three protons contribute to the NMR signal and the long relaxation of methyl resonances. While greater amounts of isotopically labeled protein are required for these methods that monitor the changes in chemical shift of the protein, it is straightforward to recycle and reuse protein with good yields (50-80%) throughout the screen.
Fig. 2.
1H-13C-HSQC of a 30 kDa protein selectively 13C-labeled on Ile, Leu, and Val methyl groups in the presence (red) and absence (black) of a twelve-fragment mixture (75 μM protein, 600 MHz spectrometer, 12 minute collection time). The expansion in the top right corner is of the boxed portion of the spectrum.
3.3 Measuring Binding Constants and Determining Preliminary SAR
NMR spectroscopy is not only well-suited to detect but also to quantify protein-fragment interactions. The great advantage of using NMR for binding affinity determination is that quantification is direct and a reference molecule is not required as in other techniques (e.g. fluorescence polarization anisotropy; FPA). Because fragments usually have target affinities weaker than ~50 micromolar, the fast exchange regime almost always applies, and Kd values can be directly determined from changes in chemical shifts. Direct binding can be measured by increasing target or fragment concentrations in a stepwise manner followed by the recording of protein spectra (Fielding 2007). Functional data and IC50 values can be obtained by replacing a known binder, e.g., a partner protein or a peptide derived thereof, with increasing concentrations of screening hits. When fragment binding affinity approaches the low micromolar range (5-50 μM), intermediate exchange of resonances becomes an issue (resonances broaden and disappear under these conditions) and NMR is no longer useful for Kd determinations. In addition, NMR titration measurements are slow, laborious, and demand larger amounts of protein compared to other biophysical methods (e.g. SPR and ITC) or biochemical assays (e.g. FPA). Hence, we recommend switching from NMR-based quantification of fragment binding to alternative methods when expected affinities have a Kd ≲ 500 μM.
4. Structural Elucidation of Intermolecular Interactions
Success or failure of a FBDD campaign often depends on the availability of high-resolution structural information for protein-ligand complexes. Extensive medicinal chemistry efforts usually begin only when structures of fragments bound to a target are known. NMR spectroscopy and X-ray crystallography can be used to obtain this structural information.
4.1 Structure Determination by NMR
NMR is a powerful tool for the structure determination of protein-small molecule complexes. Its use is limited to proteins of small and medium sizes and requires the assignment of backbone and side-chain resonances to elucidate the complete protein structure. Despite developments toward semi-automatic processes, the determination of solution structures is still a time-consuming procedure (Guntert 2009). As an alternative to solving the full NMR solution structure of a target-fragment complex, there are several less time-consuming ways for obtaining structural information on protein-ligand complexes for the purpose of driving medicinal chemistry and lead optimization. For example, if the size of the target allows, an X-ray structure of the protein is available, and amino acid backbone assignments are known, then ligand induced changes in chemical shift, e.g., from 15N- or 13C-HSQC spectra, can be used in both a qualitative and a quantitative way to map binding sites. The quantitative mapping of chemical shift perturbations on the protein surface is an especially powerful way to identify binding sites, modes, and stoichiometries (McCoy and Wyss 2002, Cioffi et al. 2008, Krishnamoorthy et al. 2010). In addition to this method, ligand-induced line broadening of protein resonances can be used to characterize binding modes (Reibarkh et al. 2006). However, most of the methods developed to obtain structural information on protein-ligand complexes are based on nuclear Overhauser effects (NOEs) (Meyer and Peters 2003). In particular, if a few intermolecular NOEs can be identified, a good model can usually be obtained of the protein-ligand complex that is suitable for drug design. In cases where no sequence-specific protein resonance assignments are available, a method called NOE Matching can be useful (Constantine et al. 2006). This concept takes advantage of the fact that 1H-13C groups in amino acids have characteristic chemical shifts. Experimental intermolecular NOEs from filtered 3D NOESY spectra are compared to a predicted NOE pattern based on simulated protein-ligand binding poses. A prerequisite of the method is that a sufficient number of target-ligand NOEs can be measured. This may be problematic if fragments are small and binding is weak. Another approach involves the use of STD signal intensities to map fragment epitopes and hence guide chemistry efforts (Mayer and Meyer 2001); protons which do not have close contacts to the binding interface typically show weaker STD signals and can serve as fragment growing or linking sites. In addition, WaterLOGSY experiments have been proven to aid in identifying the orientation of a ligand bound to a target (SALMON) (Ludwig et al. 2008). This is achieved by mapping the solvent accessibility of a ligand using protein-dependent signal sign changes in WaterLOGSY spectra. Information on the orientation of a molecule bound to a receptor can also be obtained by the INPHARMA inter-ligand NOE (Sanchez-Pedregal et al. 2005, Krimm 2012). Spin diffusion (for mixing times between ca. 100-800 ms) transfers polarization from a weak, known binder to the protein and back to a fragment that binds competitively. Atoms from two ligands that interact with the same target residues show the strongest INPHARMA inter-ligand NOEs and consequently, relative binding orientations can be derived. Another approach to map fragment binding sites is SOS-NMR (Hajduk et al. 2004). Using this method, a series of STD spectra is recorded on different protein samples that are uniformly deuterium-labeled except for selective amino acid types (e.g., valine, leucine, isoleucine, and methionine that form hydrophobic binding pockets). Ambiguous restraints localizing the binding site of a ligand are gained by detecting the amino acid-type dependent occurrence of STD signals.
4.2 Structure Determination by X-ray Crystallography
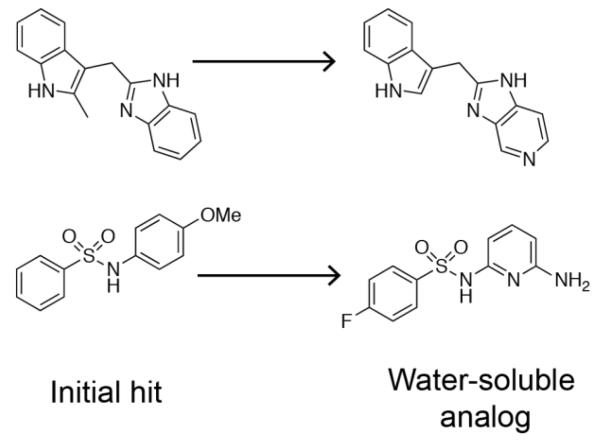
Because NMR structure determination is time intensive and NMR-derived structures typically have low resolution, X-ray crystallography is the preferred method to elucidate the three-dimensional structures of protein-ligand complexes. Once suitable conditions for crystal growth are identified, X-ray crystallography is commonly used as a rapid means of producing 3D structures (Bottcher et al. 2011). The most efficient approach is to identify conditions under which the apo-protein forms crystals that are amenable to fragment soaking experiments for obtaining structures of protein-fragment complexes in a reproducible and rapid manner. If ligand binding is associated with large conformational changes or required for protein stability, cocrystallization may be necessary. In any case, obtaining a protein/fragment complex structure with well-resolved electron density for small weak binding molecules is not always straightforward. Problems arise when proteins are too flexible to crystallize. In addition, weak, partial, or no electron density is often observed for fragments when the solubility of the ligand falls below 3-10 fold the protein concentration. Fragment binding can also be problematic when protein crystal lattices block the ligand binding site or when solvent channels are small. Deletion of flexible protein regions, the synthesis of more soluble analogs of the screening hits, and the introduction of amino acid point mutations into proteins that alter the crystal packing are common ways to overcome these issues (Fig. 3) (Sun et al. 2012). For example, both protein engineering and the synthesis of more soluble fragment analogs were necessary for the cocrystallization of K-Ras-GDP with hits identified from our primary screen (Fig. 3) (Sun et al. 2012). At times, the necessary modifications to be made to the protein or fragment are not intuitive. However, successful crystallization often results when many protein constructs are screened against several fragment analogs using a broad range of crystallization conditions. If no X-ray structures are obtained despite great efforts in optimizing crystallization conditions, then the NMR methods described above can be used to obtain the structural information.
Fig. 3.
Structures of K-Ras-GDP bound to fragment hits were obtained by improving fragment aqueous solubility through chemical modifications.
5. Generation of Lead Compounds
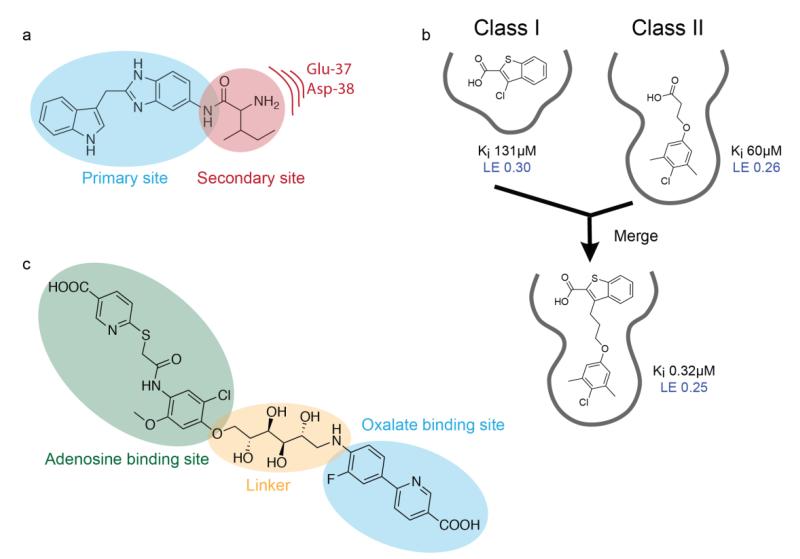
Since fragments are typically weak binders, the hits obtained in a screen must be developed into lead molecules. Three ways to achieve this goal are fragment growing, merging, and linking (Fig. 4). Which strategy to apply for a particular target depends on the data available from the first- and second-site NMR screens along with structural data from crystallography and NMR.
Fig. 4.
Lead generation by fragment growing, merging, and linking. (a) K-Ras inhibitors were designed by growing the first-site ligand into a neighboring negatively charged secondary pocket. (b) Inhibitors of Mcl-1 were prepared by merging two scaffolds identified in the primary screen that had overlapping binding sites (Friberg et al. 2013). (c) Fragment linking of adenosine- and oxalate-binding fragments via a flexible linker resulted in potent inhibitors of lactate dehydrogenase.
5.1 Fragment Growing
NMR-based screening for first-site binders is generally straightforward, resulting in the identification of several hit series (Warr 2011). Such first-site binders are often used as anchor moieties for the addition of small atomic groups or bigger building blocks to improve binding affinities. In fragment growing, the chemical addition of small groups is based on structures obtained of the target bound to first-site fragments. The goal of this strategy is to gain access to nearby empty pockets or to increase affinity by efficiently filling gaps in the current binding site. Fragment growing was recently used by our group in the discovery of K-Ras inhibitors (Fig. 4a) (Sun et al. 2012). In a primary screen against K-Ras, indole-containing compounds were found to bind with Kd values around 1 mM. Based on X-ray structures of K-Ras bound to these hits, a secondary pocket of electronegative character was observed proximal to the first site. To grow into this pocket, an indole-benzimidazole fragment was functionalized with amino acids. A near 10-fold improvement in potency resulted from the addition of Ile, with the positively charged amine group directed towards the negatively charged secondary pocket. While the fragment growing method is the most common strategy found in the literature, it often does not yield very large gains in potency. Also, the growing of fragments can be accompanied by a substantial decrease in ligand efficiency if the added atoms do not optimally interact with the target (Hopkins et al. 2004). This limits the successful use of this approach when applied to challenging targets where first-site hits are weak (Kd ≳ 1 mM) and have low ligand efficiencies (LE ≲ 0.20).
5.2 Fragment Merging / Hopping
An alternative approach to develop fragment hits into leads is fragment merging or scaffold hopping of fragments. If the binding modes for several first-site ligands which have overlapping binding sites are known, larger and more potent molecules can be synthesized that combine chemical features important for binding from two or more ligands. Merging is accomplished by attaching a piece of one fragment to another one, as illustrated by our recent discovery of Mcl-1 inhibitors (Fig. 4b) (Friberg et al. 2013). In this work, two classes of fragments identified in the primary screen were found to bind with overlapping poses in the binding site of Mcl-1. Based on NOE-derived distance restraints and molecular modeling, benzothiophene-2-carboxylic acid was merged with 4-chloro-3,5-dimethylphenyl tethered carboxylic acid to produce a molecule with improved potency and ligand efficiency. Scaffold hopping is accomplished by combining molecules with dissimilar fragment cores but sharing physically similar functional groups (Bohm et al. 2004). Fragment hopping is frequently used if fragment cores are not drug-like or already protected by patents.
5.3 Fragment Linking
In our opinion, the most powerful way to develop a fragment hit into a lead compound is to link two or more fragments together. The concept is based on the additivity of binding free energies when compounds are linked in an optimal way (Jencks 1981, Ichihara et al. 2011). Based on this principle, the Kd of the linked molecule is equal to the Kd of one fragment times the Kd of another fragment. Thus, linked molecules with micromolar affinities can be obtained from fragments that bind in the millimolar range. However, the linking requires that two fragments bind within close proximity of one another. In many cases, only one hotspot is present on a protein surface, and all hits found in a primary screen prefer to bind at this site. Thus, a second-site screen conducted under conditions in which the first site is occupied or occluded may be necessary to identify fragments that bind proximal to one another for linking purposes. Second-site binders are almost always much weaker in affinity (Kd ≳ 1 mM) for the target than first-site ligands, making their detection difficult. NMR spectroscopy is the only method that can robustly detect such weak interactions, and several NMR-based techniques have been developed which can be used.
One option to identify second site ligands is to saturate the first-site hotspot with a known binder. The ligand of choice should be as small and potent as possible, with aqueous solubility to allow for full occupancy of the pocket under the screening conditions. Then, in the presence of the first-site ligand, another NMR-based fragment screen is conducted to identify second-site ligands. Because interactions are expected to be very weak, fragment concentrations used in the second site screen must be high. It is advisable to screen a subset of the original library containing only fragments with a water solubility greater than ~5 mM (second-site screening library). This may also mean reorganization of the fragment library into mixtures of a handful of compounds as opposed to mixtures of ten or more that were used in the primary screen.
Another way to identify second-site binders by NMR spectroscopy is to use paramagnetic labeling (Jahnke 2002, Jahnke et al. 2003), taking advantage of paramagnetic relaxation enhancement (PRE). The relaxation rate of a resonance is increased when the spin is in proximity to a paramagnetic center, affecting its line width in an NMR experiment. Since the effect is distance dependent (line broadening is commonly observed for atoms which are closer than ~20 Å from the paramagnetic center), spatial information can be obtained by analyzing ligand signal widths. A first-site ligand can be labeled with a paramagnetic agent (e.g., TEMPO) and used as a probe in a second-site screen. Molecules that bind close to the first-site ligand can be identified by the broadened NMR signals. The method is very robust because the PRE effect is detectable only if first- and second-site ligands bind to the target simultaneously and close to each other. Successful use of this labeling technique is illustrated in the recent design of protein kinase and tyrosine phosphatase inhibitors (Vazquez et al. 2007, Vazquez et al. 2008).
Another method often used to identify second-site binders is based on the so-called inter-ligand NOE (ILOE experiments) (Li et al. 1999, Leone et al. 2006). Using this method, a library is screened in the presence of high concentrations of first-site binders and intermolecular ligand-ligand NOEs are detected in NOESY-type experiments. ILOEs arise only if first- and second-site fragments bind to the target simultaneously and are in close proximity (~5 Å). The strength of the inter-ligand NOEs depends on the occupancy of ligands at their corresponding sites. Since first-site fragments are often not soluble enough to fully saturate their binding site, and second-site ligands are expected to bind with very low affinity, the ILOE effect is almost always very weak. To overcome this obstacle, it is recommended to screen for second-site binders at very high ligand concentrations (e.g., by using a highly soluble subset of fragment library members), to apply longer NOESY mixing times (200-600 ms), and to use first-site binders containing a methyl group (better signal-to-noise ratio) which points toward the potential second site. In recent work aimed at the discovery of protein-protein interaction inhibitors, ILOEs have been used to identify both first- and second-site binders from fragment mixtures, thus eliminating the traditional first-site screen and the necessity for isotopically labeled protein (Becattini et al. 2006, Becattini and Pellecchia 2006, Rega et al. 2011). Using this “SAR by ILOE” methodology, one can obtain both pairs of fragments that bind together and information on which parts of the fragments bind proximal to one another from a single experiment. In this approach, even longer NOESY mixing times are used to maximize ILOE detection (300-800 ms). A potential source of errors is introduced by the INPHARMA inter-ligand NOE effect, in which magnetization is transferred by spin diffusion from the first-site ligand to target protein protons and back to ligands that compete with the first-site ligand for the same binding site (see section 4). The utilization of perdeuterated protein samples for ILOE measurements helps to eliminate the INPHARMA effect (Krimm 2012). False positive ILOE signals can also arise if fragments from the second-site library form aggregates with first-site binders. In such a case, stronger ILOEs between (almost) all protons of the first- and second-site ligands arise, thereby suggesting compound misbehavior (Sledz et al. 2010). Experiments should always be repeated in the absence of protein to test that the detected ILOEs or PREs are target dependent and therefore indicators of true second-site binding.
One problem with detecting fragments that bind to a second site is that the first-site fragments are often not soluble enough to fully saturate a hotspot due to their weak binding affinities. In these cases, the results from a second-site screen are difficult to interpret. In such a situation, alternative NMR-based approaches can be utilized. One concept is to covalently tether the first-site fragment via engineering cysteine residues in the target’s binding site, similar to the tethering technology pioneered by the Wells’ group (Erlanson et al. 2004). If structural information is available, a residue that is adjacent to the first binding site is mutated to a cysteine. This cysteine can be connected via a covalent bond to a first-site fragment modified with a reactive, flexible linker. The flexible linker allows the fragment to bind in its native protein-binding mode. If no structures of the target bound to first-site fragments are available, NOE or STD measurements can help to determine which atoms of the first-site binder are not directly involved in binding to the protein, and therefore could be used as linker attachment sites. As a result of the target-fragment covalent linking, the binding site is occluded and a second-site screen can be conducted. However, insertion of amino acid mutations is often not only accompanied by local changes, but also by global perturbations of the target surface topology. Consequently, the identified second-site binders might not bind to the native protein. In addition, the tethered first-site ligand may not achieve its preferred binding pose, and the second-site screen may identify fragments that would, again, not bind to the wild-type protein. Hence, the covalent attachment of first-site fragments must be carefully designed, and it is good practice to explore and thoroughly test more than one attachment site for the second-site screen. It is also advisable to determine the structures of the cysteine-engineered mutants tethered to first-site fragments before the second-site screen is attempted.
The chemical linkage of fragments can begin once first- and second-site ligands have been identified by the methods described above. The power of the fragment linking approach becomes obvious by the following example: linking together two fragments both having Kd values of 1 mM may result in a compound with an affinity of 1 μM or less. If the linker contributes to the binding and/or if the binding entropy of the linked compound is smaller than the binding entropies of the two fragments, super-additivity can be reached (Hajduk et al. 1997, Borsi et al. 2010, Ichihara et al. 2011). However, linking fragments in an optimal way is extremely difficult, and in most cases, the expected affinity boost from linking two fragments is not achieved (Erlanson 2006, Chung et al. 2009, Hung et al. 2009). First, it is usually complicated to identify so called second-site fragments binding close to a first site molecule. In addition, even if second-site binders can be identified and structural information on their binding modes is available, it can be challenging to discover a suitable linker with the proper geometry that allows the individual pieces of the linked molecule to bind in the exact same manner as the fragments. To help solve these issues, we suggest a combinatorial approach to fragment linking in which multiple first- and second-site ligands are linked using different flexible linkers. Attempting to link several different second-site ligands to only one first-site ligand may never generate the proper geometries or poses needed for achieving cooperativity in binding affinity. However, assembly of different first- and second-site combinations provides a better probability of the fragments binding in their optimal poses for additivity of affinities. In this process, we cannot stress enough the importance of conformationally flexible linkers to allow fragments to sample their desired poses. Successful fragment linking was recently demonstrated in the discovery of inhibitors of lactate dehydrogenase (Fig. 4c) (Kohlmann et al. 2013). Fragments that bind in the adenosine-binding pocket were linked to those that bind in the oxalate-binding pocket via flexible ether, amide, or poly-alcohol linkers, with the poly-alcohol linkers providing the best improvements in binding affinity.
6. Conclusions and Perspective
NMR spectroscopy has evolved into a powerful tool for FBDD. In addition to structure determination, NMR is useful for identifying small molecules that bind with weak affinity to protein targets, elucidating ligand binding sites, determining ligand binding affinities, and driving initial SAR studies. Because it can be used to detect very weak binding events, NMR is also useful in second-site screening and fragment linking strategies. The technique, especially when conducted using protein-observed methods, has few artifacts relative to other biophysical techniques.
In the future, we foresee NMR becoming an even more valuable technique in FBDD. We envision larger fragment libraries being screened to cover more chemical space. One approach for accomplishing this is the recently described “HTS by NMR” methodology that combines the power of combinatorial chemistry with NMR-based screening (Wu et al. 2013). In addition, to screen larger libraries by NMR, we can apply new technological advances, such as non-linear sampling and new hardware that facilitate shorter data acquisition times and the use of less sample material. Information from NMR experiments could also be applied to drive the development of improved computational tools for docking and scoring of fragment binding. For example, it would be very beneficial to have improved computational tools that can predict which fragments can bind and define how they bind using only a small number of restraints derived from NMR. Improved computational tools could also be useful to guide fragment linking.
Although an important application of NMR-based screening is for the design of challenging protein-protein interaction inhibitors that are often deemed “undruggable,” NMR-based screening can be useful in other applications as well. FBDD using NMR can be used to identify inhibitors against any target by discovering novel chemical matter that can circumvent existing patents. We also foresee NMR as a way to identify novel allosteric ‘hot spots’ on traditionally targeted proteins such as those present in protein kinases as well as for attempting to find hits against intrinsically disordered proteins. NMR seems uniquely suited for screening this class of targets. FBDD using NMR clearly represents an important tool in the discovery of drug molecules against targets important to human disease.
Acknowledgments
This research was supported in part by grants from the National Institutes of Health (NIH Director’s Pioneer Award 5DP1OD006933/8DP1CA174419 to S.W.F. and ARRA stimulus grant 5RC2CA148375 to L.J. Marnett). M.J.H. was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation, and A.O.F. was supported by a postdoctoral fellowship from the German Academic Exchange Service (DAAD).
References
- Baell JB. Observations on screening-based research and some concerning trends in the literature. Future Med Chem. 2010;2:1529–1546. doi: 10.4155/fmc.10.237. [DOI] [PubMed] [Google Scholar]
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Becattini B, Culmsee C, Leone M, Zhai D, Zhang X, Crowell KJ, Rega MF, Landshamer S, Reed JC, Plesnila N, et al. Structure-activity relationships by interligand NOE-based design and synthesis of antiapoptotic compounds targeting Bid. Proc Natl Acad Sci U S A. 2006;103:12602–12606. doi: 10.1073/pnas.0603460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becattini B, Pellecchia M. SAR by ILOEs: an NMR-based approach to reverse chemical genetics. Chemistry. 2006;12:2658–2662. doi: 10.1002/chem.200500636. [DOI] [PubMed] [Google Scholar]
- Bohm HJ, Florh A, Stahl M. Scaffold hopping. Drug Discov Today. 2004;1:217–224. doi: 10.1016/j.ddtec.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsi V, Calderone V, Fragai M, Luchinat C, Sarti N. Entropic contribution to the linking coefficient in fragment based drug design: a case study. J Med Chem. 2010;53:4285–4289. doi: 10.1021/jm901723z. [DOI] [PubMed] [Google Scholar]
- Bottcher J, Jestel A, Kiefersauer R, Krapp S, Nagel S, Steinbacher S, Steuber H. Key factors for successful generation of protein-fragment structures requirement on protein, crystals, and technology. Methods Enzymol. 2011;493:61–89. doi: 10.1016/B978-0-12-381274-2.00003-0. [DOI] [PubMed] [Google Scholar]
- Campos-Olivas R. NMR screening and hit validation in fragment based drug discovery. Curr Top Med Chem. 2011;11:43–67. doi: 10.2174/156802611793611887. [DOI] [PubMed] [Google Scholar]
- Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery: leads by design. Drug Discov Today. 2005;10:987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- Chessari G, Woodhead AJ. From fragment to clinical candidate--a historical perspective. Drug Discov Today. 2009;14:668–675. doi: 10.1016/j.drudis.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Chung S, Parker JB, Bianchet M, Amzel LM, Stivers JT. Impact of linker strain and flexibility in the design of a fragment-based inhibitor. Nat Chem Biol. 2009;5:407–413. doi: 10.1038/nchembio.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi M, Hunter CA, Packer MJ, Spitaleri A. Determination of protein-ligand binding modes using complexation-induced changes in (1)h NMR chemical shift. J Med Chem. 2008;51:2512–2517. doi: 10.1021/jm701194r. [DOI] [PubMed] [Google Scholar]
- Congreve M, Carr R, Murray C, Jhoti H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- Constantine KL, Davis ME, Metzler WJ, Mueller L, Claus BL. Protein-ligand NOE matching: a high-throughput method for binding pose evaluation that does not require protein NMR resonance assignments. J Am Chem Soc. 2006;128:7252–7263. doi: 10.1021/ja060356w. [DOI] [PubMed] [Google Scholar]
- Dalvit C. NMR methods in fragment screening: theory and a comparison with other biophysical techniques. Drug Discov Today. 2009;14:1051–1057. doi: 10.1016/j.drudis.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Dalvit C, Flocco M, Veronesi M, Stockman BJ. Fluorine-NMR competition binding experiments for high-throughput screening of large compound mixtures. Comb Chem High Throughput Screen. 2002;5:605–611. doi: 10.2174/1386207023329923. [DOI] [PubMed] [Google Scholar]
- Dalvit C, Pevarello P, Tato M, Veronesi M, Vulpetti A, Sundstrom M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J Biomol NMR. 2000;18:65–68. doi: 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- Erlanson DA. Fragment-based lead discovery: a chemical update. Curr Opin Biotechnol. 2006;17:643–652. doi: 10.1016/j.copbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- Felli IC, Brutscher B. Recent advances in solution NMR: fast methods and heteronuclear direct detection. Chemphyschem. 2009;10:1356–1368. doi: 10.1002/cphc.200900133. [DOI] [PubMed] [Google Scholar]
- Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Jahnke W. New Approaches for NMR screening in drug discovery. Drg Discov Today. 2004;1:277–283. doi: 10.1016/j.ddtec.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Prog NMR Spec. 2007;51:219–242. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- Friberg A, Vigil D, Zhao B, Daniels RN, Burke JP, Garcia-Barrantes PM, Camper D, Chauder BA, Lee T, Olejniczak ET, et al. Discovery of potent myeloid cell leukemia 1 (mcl-1) inhibitors using fragment-based methods and structure-based design. J Med Chem. 2013;56:15–30. doi: 10.1021/jm301448p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntert P. Automated structure determination from NMR spectra. Eur Biophys J. 2009;38:129–143. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- Hajduk PJ, Augeri DJ, Mack J, Mendoza R, Yang J, Betz SF, Fesik SW. NMR-Based Screening of Proteins Containing 13C-Labeled Methyl Groups. J Am Chem Soc. 2000;122:7898–7904. [Google Scholar]
- Hajduk PJ, Bures M, Praestgaard J, Fesik SW. Privileged molecules for protein binding identified from NMR-based screening. J Med Chem. 2000;43:3443–3447. doi: 10.1021/jm000164q. [DOI] [PubMed] [Google Scholar]
- Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- Hajduk PJ, Mack JC, Olejniczak ET, Park C, Dandliker PJ, Beutel BA. SOS-NMR: a saturation transfer NMR-based method for determining the structures of protein-ligand complexes. J Am Chem Soc. 2004;126:2390–2398. doi: 10.1021/ja039480v. [DOI] [PubMed] [Google Scholar]
- Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins. Science. 1997;278:497–499. doi: 10.1126/science.278.5337.497. [DOI] [PubMed] [Google Scholar]
- Hoffer L, Renaud JP, Horvath D. Fragment-based drug design: computational & experimental state of the art. Comb Chem High Throughput Screen. 2011;14:500–520. doi: 10.2174/138620711795767884. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- Hung AW, Silvestre HL, Wen S, Ciulli A, Blundell TL, Abell C. Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase. Angew Chem Int Ed Engl. 2009;48:8452–8456. doi: 10.1002/anie.200903821. [DOI] [PubMed] [Google Scholar]
- Huth JR, Mendoza R, Olejniczak ET, Johnson RW, Cothron DA, Liu Y, Lerner CG, Chen J, Hajduk PJ. ALARM NMR: a rapid and robust experimental method to detect reactive false positives in biochemical screens. J Am Chem Soc. 2005;127:217–224. doi: 10.1021/ja0455547. [DOI] [PubMed] [Google Scholar]
- Ichihara O, Barker J, Law RJ, Whittaker M. Compound Design by Fragment-Linking. Mol Inf. 2011;30:298–306. doi: 10.1002/minf.201000174. [DOI] [PubMed] [Google Scholar]
- Jahnke W. Spin labels as a tool to identify and characterize protein-ligand interactions by NMR spectroscopy. Chembiochem. 2002;3:167–173. doi: 10.1002/1439-7633(20020301)3:2/3<167::AID-CBIC167>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Jahnke W, Florsheimer A, Blommers MJ, Paris CG, Heim J, Nalin CM, Perez LB. Second-site NMR screening and linker design. Curr Top Med Chem. 2003;3:69–80. doi: 10.2174/1568026033392778. [DOI] [PubMed] [Google Scholar]
- Jencks WP. On the attribution and additivity of binding energies. Proc Natl Acad Sci U S A. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhoti H, Cleasby A, Verdonk M, Williams G. Fragment-based screening using X-ray crystallography and NMR spectroscopy. Curr Opin Chem Biol. 2007;11:485–493. doi: 10.1016/j.cbpa.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Klages J, Coles M, Kessler H. NMR-based screening: a powerful tool in fragment-based drug discovery. Analyst. 2007;132:693–705. doi: 10.1039/b709658p. [DOI] [PubMed] [Google Scholar]
- Kohlmann A, Zech SG, Li F, Zhou T, Squillace RM, Commodore L, Greenfield MT, Lu X, Miller DP, Huang WS, et al. Fragment Growing and Linking Lead to Novel Nanomolar Lactate Dehydrogenase Inhibitors. J Med Chem. 2013 doi: 10.1021/jm3014844. [DOI] [PubMed] [Google Scholar]
- Krimm I. INPHARMA-based identification of ligand binding site in fragment-based drug design. MedChemComm. 2012;3:605–610. [Google Scholar]
- Krishnamoorthy J, Yu VC, Mok YK. Auto-FACE: an NMR based binding site mapping program for fast chemical exchange protein-ligand systems. PLoS One. 2010;5:e8943. doi: 10.1371/journal.pone.0008943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc Natl Acad Sci U S A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Freeze HH, Chan CS, Pellecchia M. The Nuclear Overhauser Effect in the lead identification process. Curr Drug Discov Technol. 2006;3:91–100. doi: 10.2174/157016306778108884. [DOI] [PubMed] [Google Scholar]
- Lepre CA. Practical aspects of NMR-based fragment screening. Methods Enzymol. 2011;493:219–239. doi: 10.1016/B978-0-12-381274-2.00009-1. [DOI] [PubMed] [Google Scholar]
- Lescop E, Kern T, Brutscher B. Guidelines for the use of band-selective radiofrequency pulses in hetero-nuclear NMR: example of longitudinal-relaxation-enhanced BEST-type 1H-15N correlation experiments. J Magn Reson. 2010;203:190–198. doi: 10.1016/j.jmr.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Li D, DeRose EF, London RE. The inter-ligand Overhauser effect: a powerful new NMR approach for mapping structural relationships of macromolecular ligands. J Biomol NMR. 1999;15:71–76. doi: 10.1023/a:1008360208627. [DOI] [PubMed] [Google Scholar]
- Ludwig C, Guenther UL. Ligand based NMR methods for drug discovery. Front Biosci. 2009;14:4565–4574. doi: 10.2741/3549. [DOI] [PubMed] [Google Scholar]
- Ludwig C, Michiels PJ, Wu X, Kavanagh KL, Pilka E, Jansson A, Oppermann U, Gunther UL. SALMON: solvent accessibility, ligand binding, and mapping of ligand orientation by NMR spectroscopy. J Med Chem. 2008;51:1–3. doi: 10.1021/jm701020f. [DOI] [PubMed] [Google Scholar]
- Manzenrieder F, Frank AO, Kessler H. Phosphorus NMR spectroscopy as a versatile tool for compound library screening. Angew Chem Int Ed Engl. 2008;47:2608–2611. doi: 10.1002/anie.200705256. [DOI] [PubMed] [Google Scholar]
- Mayer M, Meyer B. Characterization of Ligand Binding of Saturation Transfer Difference NMR Spectroscopy. Angew Chem Int Ed Engl. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- McCoy MA, Wyss DF. Spatial localization of ligand binding sites from electron current density surfaces calculated from NMR chemical shift perturbations. J Am Chem Soc. 2002;124:11758–11763. doi: 10.1021/ja026166c. [DOI] [PubMed] [Google Scholar]
- Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed Engl. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- Murray CW, Blundell TL. Structural biology in fragment-based drug design. Curr Opin Struct Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Pellecchia M, Bertini I, Cowburn D, Dalvit C, Giralt E, Jahnke W, James TL, Homans SW, Kessler H, Luchinat C, et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat Rev Drug Discov. 2008;7:738–745. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rega MF, Wu B, Wei J, Zhang Z, Cellitti JF, Pellecchia M. SAR by interligand nuclear overhauser effects (ILOEs) based discovery of acylsulfonamide compounds active against Bcl-x(L) and Mcl-1. J Med Chem. 2011;54:6000–6013. doi: 10.1021/jm200826s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibarkh M, Malia TJ, Wagner G. NMR distinction of single- and multiple-mode binding of small-molecule protein ligands. J Am Chem Soc. 2006;128:2160–2161. doi: 10.1021/ja055971z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pedregal VM, Reese M, Meiler J, Blommers MJ, Griesinger C, Carlomagno T. The INPHARMA method: protein-mediated interligand NOEs for pharmacophore mapping. Angew Chem Int Ed Engl. 2005;44:4172–4175. doi: 10.1002/anie.200500503. [DOI] [PubMed] [Google Scholar]
- Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J Med Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- Shortridge MD, Powers R. Biomolecular NMR Spectroscopy. IOS Press BV; Netherlands: 2011. NMR Screening Methods for Drug Discovery; p. 381. [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- Sledz P, Silvestre HL, Hung AW, Ciulli A, Blundell TL, Abell C. Optimization of the interligand Overhauser effect for fragment linking: application to inhibitor discovery against Mycobacterium tuberculosis pantothenate synthetase. J Am Chem Soc. 2010;132:4544–4545. doi: 10.1021/ja100595u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tengel T, Fex T, Emtenas H, Almqvist F, Sethson I, Kihlberg J. Use of 19F NMR spectroscopy to screen chemical libraries for ligands that bind to proteins. Org Biomol Chem. 2004;2:725–731. doi: 10.1039/b313166a. [DOI] [PubMed] [Google Scholar]
- Vanwetswinkel S, Heetebrij RJ, van Duynhoven J, Hollander JG, Filippov DV, Hajduk PJ, Siegal G. TINS, target immobilized NMR screening: an efficient and sensitive method for ligand discovery. Chem Biol. 2005;12:207–216. doi: 10.1016/j.chembiol.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Vazquez J, De SK, Chen LH, Riel-Mehan M, Emdadi A, Cellitti J, Stebbins JL, Rega MF, Pellecchia M. Development of paramagnetic probes for molecular recognition studies in protein kinases. J Med Chem. 2008;51:3460–3465. doi: 10.1021/jm800068w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Tautz L, Ryan JJ, Vuori K, Mustelin T, Pellecchia M. Development of molecular probes for second-site screening and design of protein tyrosine phosphatase inhibitors. J Med Chem. 2007;50:2137–2143. doi: 10.1021/jm061481l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr WA. Some Trends in Chem(o)informatics. Methods Mol Biol. 2011;672:1–37. doi: 10.1007/978-1-60761-839-3_1. [DOI] [PubMed] [Google Scholar]
- Wu B, Zhang Z, Noberini R, Barile E, Giulianotti M, Pinilla C, Houghten RA, Pasquale EB, Pellecchia M. HTS by NMR of combinatorial libraries: a fragment-based approach to ligand discovery. Chem Biol. 2013;20:19–33. doi: 10.1016/j.chembiol.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sanger A, Hemmig R, Jahnke W. Ranking of high-affinity ligands by NMR spectroscopy. Angew Chem Int Ed Engl. 2009;48:6691–6694. doi: 10.1002/anie.200902591. [DOI] [PubMed] [Google Scholar]