Abstract
Significance: The maintenance of mitochondrial genome integrity is a major challenge for cells to sustain energy production by respiration. Recent Advances: Recently, mitochondrial membrane dynamics emerged as a key process contributing to prevent mitochondrial DNA (mtDNA) alterations. Indeed, both fundamental and clinical data suggest that disruption of mitochondrial fusion, related to mutations in the OPA1, MFN2, PINK1, and PARK2 genes, leads to the accumulation of mutations in the mitochondrial genome. Critical Issues: We discuss here the possibility that mitochondrial fusion acts as a direct mechanism to prevent the generation of altered mtDNA and to eliminate mutated deleterious genomes either by trans-complementation or by mitophagy. Future Directions: Finally, we conclude this review with a short evolutionary comparison between the mechanisms involved in mitochondrial and bacterial modes of genome distribution and plasticity, highlighting possible common conserved processes required for the maintenance of their genome integrity, which should inspire our future investigations. Antioxid. Redox Signal. 19, 379–388.
Introduction
The concept of mitochondrial network dynamics was undermined originally when the name of “mitochondria,” the etymological assembly of mitos (wire, string) and chondrios (bead, grain, seed) was given to these cytoplasmic organelles, according to their distinct morphological structures when observed by electron microscopy (36). The reality of the dynamic features of the mitochondrial reticulum was subsequently revealed when the processes of fusion and fission of this double membrane network was discovered in living cells (5). Since these early descriptions, it is now well accepted that fusion and fission events contribute to the mitochondrial life cycle, being intimately interconnected to mitochondrial biogenesis, turnover, and degradation (52). Fusion and fission of mitochondrial membranes require the combination of dynamin-related mechano-enzymes: the dynamin-related-protein (dynamin related protein 1 [DRP1]) for the fission of the outer and inner membranes (6), the mitofusins (MFN1 and MFN2) (8, 42), and the optic atrophy (optic atrophy 1 [OPA1]) (13, 35) for the fusion of the outer and the inner membrane, respectively. So far, a single mutation in DRP1 gene has been reported in a young patient with severe neonatal developmental neurological presentation (54), whereas tens of mutations in both OPA1 and MFN2 genes were identified in patients with dominant optic atrophy (DOA) (17) and Charcot-Marie-Tooth (30) neurodegenerative diseases; importantly among them, some are leading to syndromic presentations with disruption of the mitochondrial genome integrity (2, 20, 44).
Understanding the central roles of mitochondrial membrane dynamics remains an ongoing fundamental challenge, but increasing evidence, first gathered from studies in yeast and now ultimately collected from investigations of patient biopsies affected by mitochondrial disease, suggests that it is intimately associated to the maintenance of the mitochondrial genome. How the network dynamics are related to the integrity of mitochondrial DNA (mtDNA) remains elusive, although emerging novel data linking these two processes deserves consideration, opening novel routes of investigations.
In this review we will address this question by describing first the main alterations of the mitochondrial genome and how they are generated, with major emphasis on those induced by impairment of the mitochondrial dynamics. Subsequently we will illustrate how mitochondria with altered genomes require mitochondrial dynamics to circumvent the consequences of the loss of mtDNA integrity. Finally, we will discuss a possible evolutionary parallel between bacterial mechanisms involved in genome dynamics and plasticity, and mitochondrial mechanisms linking the membrane fusion to the maintenance of mtDNA integrity.
Generation of Altered Mitochondrial Genome
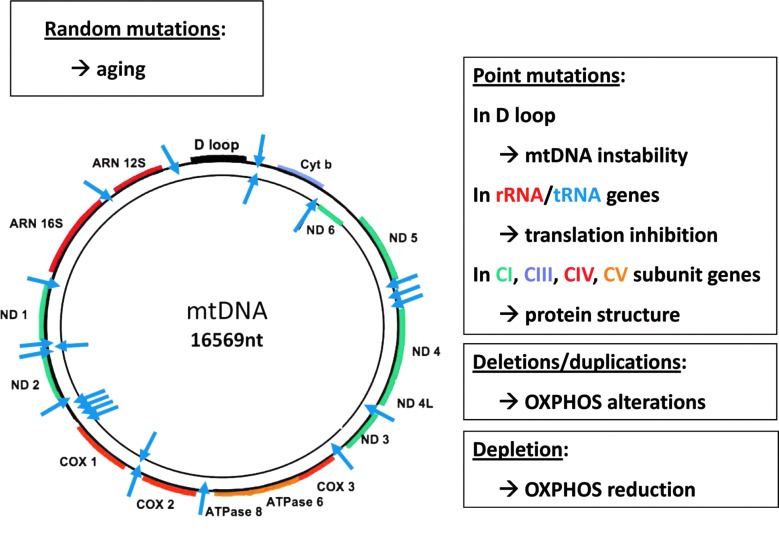
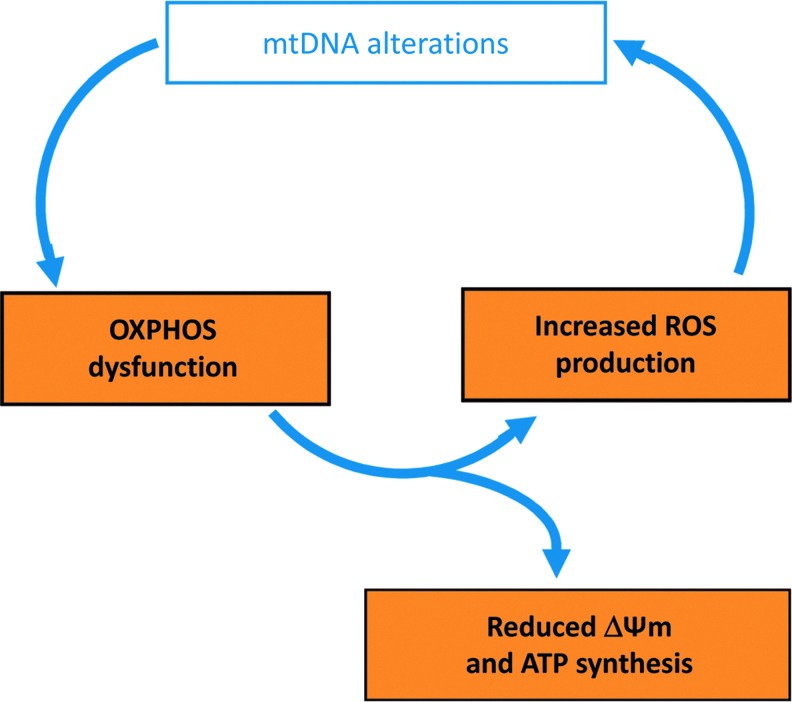
As a relic of their bacterial origin, mitochondria possess a small genome composed of a circular DNA molecule present in high copy number, ranging from tens to hundred thousand copies per cell. The mtDNA are arranged in nucleoprotein complexes called nucleoids, evenly distributed along the mitochondrial network (28). In mammals, the mitochondrial genome encodes 13 proteins embedded in the oxidative phosphorylation complexes I, III, IV, and V, two of the three ribosomal RNAs and all tRNAs required for translation (22). The mtDNA sequence bears many prokaryote features, as an AT rich composition, the organization in two polycistronic units of transcription, and the absence of introns and intervening sequences among and inbetween genes (31). Unlike the nuclear genome, mtDNA is strictly maternally inherited, a process that has key consequences for the genetic diagnosis of mitochondrial diseases and for mtDNA transmission throughout generations. Since the discovery of the first pathogenic mutation in the mtDNA in 1988, it is now well established that many alterations of the 16.569 nt long sequence of the human mitochondrial genome lead to mitochondrial diseases, with an amazingly broad range of clinical presentations. These mtDNA alterations can be uniformly integrated in all mtDNA copies, a situation termed homoplasmy, or only present in a restricted proportion of the mtDNA population, a situation referred to as heteroplasmy. Summarizing briefly genetic and molecular investigations of mtDNA from the last 20 years, we can enumerate four types of alterations that can affect the mitochondrial genome (Fig. 1). The first is the accumulation of random mutations spread all over the mtDNA sequence, which is physiologically correlated to the ageing process (53). The second is the presence of discrete point mutations in the mtDNA sequence that can affect either the genome stability when present in the D-loop, the translation process when localized in the rRNA or tRNA genes, or the protein sequence of one of the 13 peptides encoded by this genome (16). The third type is due to partial deletions or duplications of mtDNA fragments, leading to the loss or to the uneven synthesis of some oxidative phosphorylation complex subunits (47). Finally, the last alteration is the depletion of the mtDNA, which becomes pathogenic in humans when reaching a loss of 60%–70% of the normal mtDNA abundance (45). Altogether, these deleterious events affecting the integrity of the mitochondrial genome have dramatic consequences on the efficiency of the mitochondrial oxidative phosphorylation process, leading to reduced ATP production, and to increased generation of reactive oxygen species (ROS) (23). Importantly, as illustrated in Figure 2, this enhanced ROS production will lead to a vicious cycle further damaging the mtDNA, with a rate of mutagenesis that can be 50 times higher than the one reported for the nuclear genome (55).
FIG. 1.
The main alterations of the mtDNA. The circular mtDNA is presented with the D-Loop (in black) responsible for mtDNA stability, and the genes coding respiratory chain complex I (CI, ND1, 2, 3, 4, 4b, 5, and 6 in green), III (CIII, Cytb in purple), IV (CIV, COX1, 2, and 3 in red) and V (CV, ATPase 6, and 8 in orange) subunits, together with the genes coding the ribosomal RNA 12S and 16S (rRNA 12S and 16S) and the 22 tRNAs (blue arrows). Alterations of the mtDNA are categorized in four items with their consequences on mitochondrial physiology. ATPase, adenosine triphosphate synthase; COX, cytochrome oxidase; mtDNA, mitochondrial DNA; ND, NADH dehydrogenase. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 2.
The vicious cycle leading to progressive alterations of the mtDNA. mtDNA alterations affect the efficiency of the OXPHOS, which consequently leads to reduced membrane potential (ΔΨm) and ATP synthesis on one hand, and to increased ROS production on the other hand. The latter will further affect the integrity of the mitochondrial genome, and contribute to the increment of the vicious cycle. OXPHOS, oxidative phosphorylation reaction; ROS, reactive oxygen species; ΔΨm, mitochondrial membrane potential. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
How alterations of mtDNA occur is a hot topic of investigations involving a broad range of studies from yeast genetics to clinical investigations on patients. It is now well established that mtDNA alterations can occur randomly, for example as a consequence of mistakes during mtDNA replication by the mtDNA-polymerase γ enzyme (26), or can be induced by drugs as antiretrovirals (37). In addition, in humans these alterations can be innate and transmitted directly by the pool of mtDNA from the oocyte, or acquired resulting from autosomal mutations in genes encoding mitochondrial proteins. In the latter situation, genetically inherited diseases involving instability of the mitochondrial genome are mostly due to the alteration of genes encoding proteins involved in mtDNA metabolism, as those responsible for replicating the mtDNA (POLG1/2, PEO) or for generating the pool of mitochondrial nucleotides (SUCLG1, SUCLA2, RRM2B, TK2, DGUOK, TYMP, ANT1) (Table 1) (11, 46). Intuitively, it was not surprising to find that when these genes are mutated the process of mtDNA replication is affected, generating and accumulating mutations in the mitochondrial genome.
Table 1.
Genes Involved in Mitochondrial DNA Instability
| Function | Gene | Protein |
|---|---|---|
| mtDNA replication | POLG1 | mtDNA polymerase γ |
| POLG2 | Polymerase γ accessory subunit | |
| PEO1 | Twinkle mtDNA helicase | |
| Mitochondrial nucleotide metabolism | SUCLG1 | Succinate CoA ligase, α subunit |
| SUCLA2 | Succinate CoA ligase, β subunit | |
| RRM2B | Ribonucleotide reductase, small subunit P53-inducible | |
| TK2 | Thymidine kinase 2 | |
| DGUOK | Deoxyguanosine kinase | |
| TYMP | Thymidine phosphorylase | |
| ANT1 | Adenine nucleotide translocator | |
| Mitochondrial fusion | OPA1 | Optic atrophy 1 |
| MFN2 | Mitofusin 2 | |
| Mitophagy | PINK1 | PTEN-induced kinase 1 |
| PARK2 | E3-ubiquitin ligase or Parkin | |
| Unknown | MPV17 | Mitochondrial inner membrane protein |
mtDNA, mitochondrial DNA.
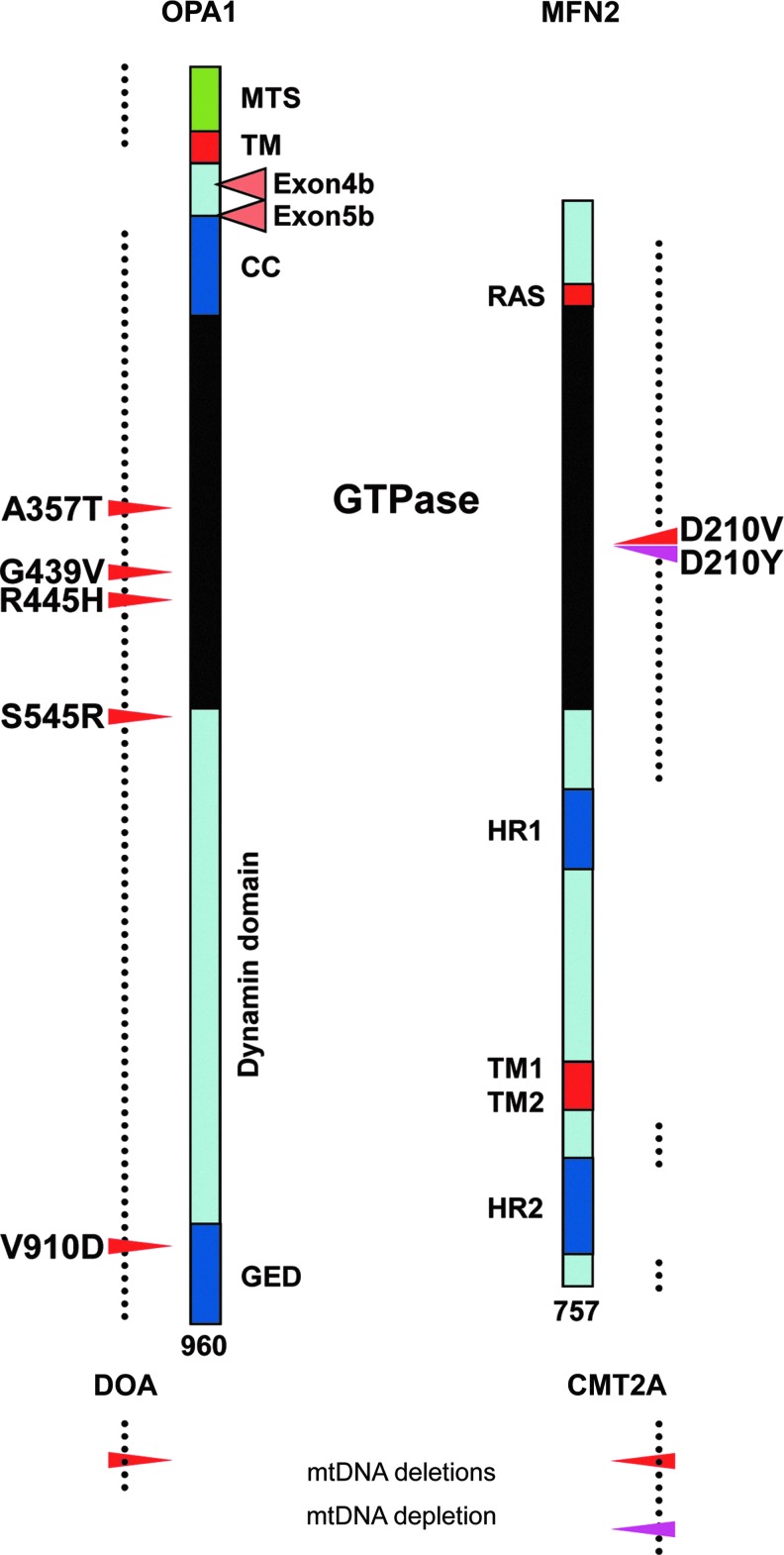
Conversely, it was quite surprising when two European consortia simultaneously identified in patients a form of mtDNA instability associated to alteration of the mitochondrial network dynamics. Indeed, mtDNA deletions were found in the calf muscle of patients with dominant negative mutations in the OPA1 gene (Fig. 3, left) responsible for syndromic dominant optic atrophy (DOAplus), a clinical presentation combining severe neurosensorial visual and auditory symptoms to peripheral neuropathy and myopathies (2, 20). This observation has now been further extended to a single family with a similar DOAplus clinical presentation associated to a novel dominant mutation in the MFN2 gene (Fig. 3, right); thus, suggesting that severe alterations of the mitochondrial fusion actors, and, in particular, of their guanosine tri phosphatase domain, are responsible for the accumulation of mtDNA deletions (44). These clinical data can be further related to the aetiology of Parkinson disease (PD), as mtDNA deletions were identified at autopsy in dopaminergic midbrain neurons of patients with inherited form of this disease (4). Some familial forms of PD are provoked by mutations in the PINK1 and PARK2 genes, encoding the PTEN Induced Kinase 1 (PINK1) and the E3 ubiquitin ligase (Parkin) proteins, respectively, regulating mitochondrial dynamics (39, 56) and mitophagy (29). Thus, mtDNA deletions seem to be frequent in diseases associated to impaired fusion (Fig. 4, left column), whereas mtDNA depletion has yet never been reported in cohorts of patients with mutations in OPA1, MFN2, PINK1, or PARK2 gene. A single exception concerns a patient presenting a mitochondrial cytopathy due to mtDNA depletion, associated to a mutation in the MFN2 gene, that remarkably affects the same amino-acid as the one reported to generate deletions in the mtDNA (41) (Fig. 3, right). Notably, the level of random point mutations has not yet been considered in patient biopsies, owing to the requirement of next generation sequencing of mtDNA, a technology that only recently became broadly accessible. It should further be considered that no severe mtDNA alteration has yet been characterized so far in fibroblasts from patients with mutation in OPA1, MFN2, PINK1, or PARK2 gene.
FIG. 3.
Mutations in OPA1 and MFN2 proteins leading to mtDNA instability. The linear structure of the OPA1 (left) and MFN2 (right) proteins are schematically represented. In addition, for OPA1, the domains corresponding to the alternate spliced exons 4b and 5b are shown. The regions mutated in OPA1 in pure DOA (left) and in MFN2 in pure CMT2A (right) patients are indicated by dotted lines, whereas mutations responsible for mtDNA deletions are represented by red arrow heads and the mutation responsible for mtDNA depletion by a purple arrow head, together with the amino-acid position and change. CC/HR, coiled coil domains/heptad repeat; CMT2A, Charcot Marie Tooth type 2A; DOA, Dominant optic atrophy; GED, GTPase effector domain; GTPase, guanosine tri-phosphate hydrolase domain; MFN1/MFN2: mitofusin 1/2; MTS, mitochondrial targeting sequence; OPA1, optic atrophy 1; RAS, Ras binding domain; TM, transmembrane domain. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
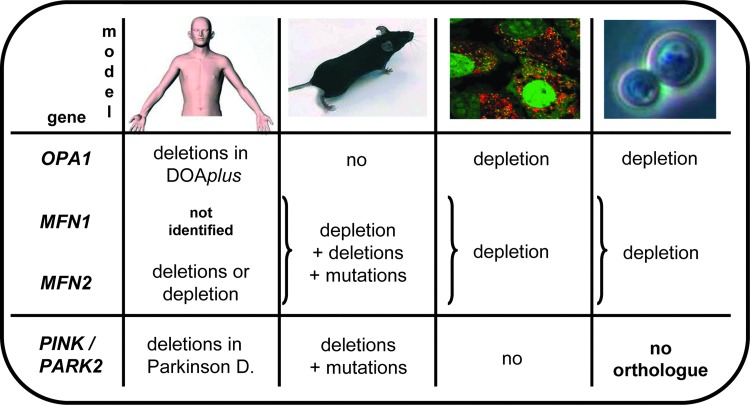
FIG. 4.
Alterations of the mtDNA in different models with disruption of genes involved in mitochondrial fusion. Data are collected from patients with DOAplus (OPA1 and MFN2), mitochondrial cytopathy (MFN2), or Parkinson disease (PINK1/PARK2) (left column), mouse models with Opa1 mutation, Mfn1, and Mfn2 or both knockouts, Pink1 and PARK2 mutations (middle left column), mammalian cells manipulated for the OPA1, MFN1/MFN2, and PINK1/PARK2 genes (middle right column), and from yeasts invalidated for the MGM1/Msp1 or Fzo genes (right column). In yeasts, MFNs have a single orthologue, whereas PINK1 and PARK2 have none. No mutation in the MFN1 gene has yet been found in humans. DOAplus, syndromic dominant optic atrophy; Fzo, Fuzzy onions; MGM1, mitochondrial genome maintenance 1; Msp1, MGM1 in Schizosaccharomyces pombe; PINK1, PTEN induced kinase 1. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
No mtDNA alteration has yet been reported in mice invalidated for Pink1 or Park2 genes (12) or carrying pathogenic mutations in Opa1 (1, 57) or Mfn2 gene (7). Only in the case of mice with targeted deletions of both Mfn1 and Mfn2 genes in skeletal muscles, the presence of mtDNA deletions, depletion, and point mutations was reported (9) (Fig. 4, middle left column).
In human cellular models, the combined deletion of MFN1 and MFN2 genes (9) and the silencing of the single OPA1 isoform mandatory for mitochondrial genome maintenance and distribution (15), lead to mtDNA depletion (Fig. 4, middle right column), while the coupling of PINK1/Parkin overexpression with pharmacological dissipation of the mitochondrial membrane potential induces a drastic elimination of mitochondria with their genome (32).
In yeast models, disruption of the mitochondrial genome maintenance 1 (MGM1)/MGM1 in Schizosaccharomyces pombe (Msp1) or Fuzzy onions (Fzo) gene, the orthologues of OPA1 and MFNs genes, respectively, leads to the total loss of mitochondrial genome (Fig. 4, right column), and consequently to the complete inhibition of the respiratory capacity (21, 38, 40), while orthologues of the PINK and PARK2 genes have not been identified in lower eukaryotes.
Thus, summing up these data, in human tissues with disrupted mitochondrial fusion, we observed mainly the accumulation of mtDNA deletions, whereas in cultured cell, mtDNA depletion was evidenced in cells with altered mitochondrial fusion, an intriguing discrepancy that confers complexity to the study of the links associating mitochondrial fusion to the maintenance of mtDNA integrity.
How Alterations of Mitochondrial Fusion Might Primarily Impact on mtDNA Integrity
When considering the different hypotheses that could link mitochondrial dynamics to mtDNA alterations, the first to take into account is that defective mitochondrial fusion could be primarily responsible for generating different types of mtDNA alterations.
Generation of point mutations in relation to mitochondrial fusion alteration
It is well established that the mitochondrial genome is a direct target of ROS that induce nucleotide oxidation and base damage, leading to the accumulation of point mutations (55). Therefore, because OPA1 and MFNs mutations alter directly membrane dynamics and affect the mitochondrial respiration efficiency (10, 27), ROS production may increase and consequently induce the accumulation of mtDNA mutations. Although never addressed in human or mouse models reproducing pathological conditions, it will soon become clear if this hypothesis is relevant, and if point mutations accumulate and contribute to the pathophysiology of OPA1 and MFN2 associated diseases.
Generation of mtDNA deletions in relation to mitochondrial fusion alteration
How mtDNA deletions accumulate in organs from OPA1 and MFN2 patients remains an open question. In cells with highly fragmented mitochondria, due to severe mutations in MFN2 or OPA1 gene, the distribution of the mitochondrial genome might be deeply affected, resulting in the accumulation of many mitochondria devoid of mtDNA. In the remaining mitochondria, where mtDNA replication occurs, the segregation of nucleoids could be impaired, creating superstructures, including multiple copies of mtDNA in aggregated nucleoids. The physical constraints on these catenated mtDNA copies could favor recombination between homologous sequences, leading to the deletion or duplication of some mtDNA regions. In this respect, the analysis of the sequences at the edges of the mtDNA deletions in the calf muscle of DOAplus patients revealed short stretches of sequences, around 10 nucleotides long, which were repeatedly found in many other diseases associated to mtDNA instability, suggesting that a common mechanism is responsible for generating these deletions during mtDNA replication and segregation (2). Thus, fusion of mitochondria and proper nucleoid distribution could be critical events required to untangle copies of mtDNA to evenly segregate nucleoids along the reticulum and prevent situations where illegitimate mtDNA recombinations occur. Although never addressed in terms of pathophysiological mechanism, this hypothesis could explain how mutations in OPA1 or MFN2 genes generate deletions in the mtDNA from the calf muscle of syndromic patients with myopathy.
Generation of mtDNA depletion in relation to mitochondrial fusion alteration
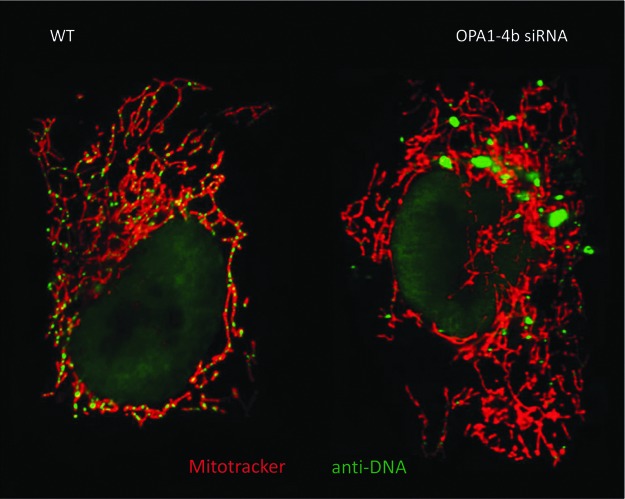
In cellular models as well as in yeasts, altered fusion can lead to mtDNA depletion. We have recently shown that in HeLa cells a small peptide inferred from the cleavage of a specific OPA1 isoform is involved in anchoring the nucleoid to the inner mitochondrial membrane, and that its silencing induces a major mtDNA depletion and the alteration of the distribution of the remaining mtDNA (15) (Fig. 5). Importantly, the biochemical structure of this peptide, including two transmembrane domains, is conserved throughout the eukaryote kingdom, suggesting that this mechanism could be responsible for mitochondrial genome maintenance whatever the model considered (14). In this respect, in yeasts, MGM1/Msp1 gene knockout or alteration of the structure of the anchoring peptide causes complete mtDNA loss (21, 38). Thus, alteration of any OPA1 orthologues in cellular models results in mtDNA depletion that eventually can be dissociated from their involvement in the network fusion. Now, how MFNs/Fzo disruption leads to a similar phenotype remains unclear, although the physical interaction between MFNs/Fzo and OPA1/MGM1/Msp1, which contributes to synchronize outer and inner membrane fusion, might affect the genome maintenance in relation to the function of the latter protein. Altogether, these hypotheses might explain how the disruption of the function of mitochondrial fusion actors can generate different types of mtDNA alterations.
FIG. 5.
Alterations of the mtDNA abundance and distribution in HeLa cells silenced for a specific OPA1 isoform. Fluorescent pictures of a control HeLa cell (WT, left) or a cell silenced for OPA1-exon4b (OPA1-4b siRNA, right), after labeling the mitochondrial network with Mitotracker (in red) and the mtDNA with anti-DNA antibodies (in green), which mainly label the mitochondrial nucleoids. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
How Alterations of Mitochondrial Fusion Might Influence the Recycling of Altered mtDNA
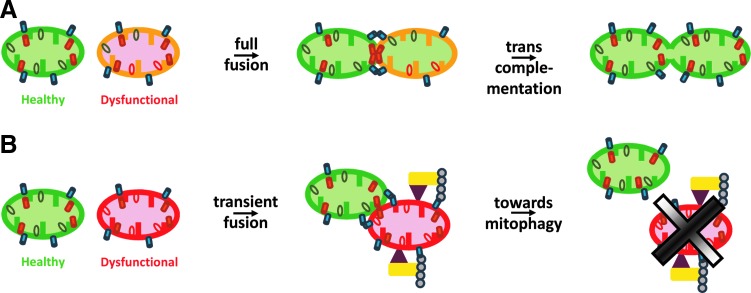
An alternative to the previous hypotheses concerns the involvement of mitochondrial fusion in the elimination of altered mitochondrial genome. Although not exclusive with the previous ones, this concept undermines that mtDNA alterations are generated throughout life time, but are not properly corrected, or counterselected and eliminated, due to the impairment of the mitochondrial fusion process. Two mechanisms may contribute to this hypothesis. The first one is based on transcomplementation between mitochondria (49) (Fig. 6A). It requires the complete fusion of the outer and inner membranes, and consequently the function of MFNs and OPA1 proteins. In this process, fused mitochondria are able to share and homogenize the content of all their compartments: both the soluble ones from the inter membrane space and the matrix, and the insoluble ones from the outer and inner membranes. Thus, the lack of a functional component in one mitochondrion might be complemented by the presence of the component in the sister mitochondrion. Similarly, in cells with a heteroplasmic mtDNA mutation, malfunctioning mitochondria can benefit from transcomplementation to recover a wild-type mtDNA copy (18). This mutual exchange would allow either for removal of the defective copy of the mtDNA as a consequence of mitochondrial turnover, or for its repair by using the subset of enzymes dedicated to this process and the wild-type mtDNA copy (24).
FIG. 6.
Mitochondrial fusion allows for trans-complementation or mitophagy of dysfunctional mitochondria. Cartoons illustrating the two possible modes of processing of dysfunctional mitochondria. In (A), one mitochondrion with defective genome (red ellipses) will fully fuse with a functional mitochondria to restore the wild-type mtDNA (green ellipses), allowing for transcomplementation of the initial defect. This process requires the mitofusins MFN1/2 (blue cylinders) on the outer mitochondrial membrane and OPA1 (orange cylinders) on the inner mitochondrial membrane to perform complete fusion of all mitochondrial compartments. In (B), one mitochondrion with defective genome and low membrane potential will transiently fuse with healthy mitochondria to assess its ability to be rescued. If too deficient, it will be directed to mitophagy by the PINK1 (brown triangle) and Parkin (yellow rectangle) combined activities, leading to MFNs ubiquitinylation (blue circles), followed by the autophagy and elimination (black cross) of the defective mitochondria. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The second mechanism is associated to the recently reported mitochondrial transient fusion of mitochondria, referred to as “kiss-and-run” (Fig. 6B) (25, 51). In this process, mitochondria come into close apposition, transiently fuse to exchange soluble intermembrane space and matrix proteins, and re-separate, preserving their original morphology (51). “Kiss-and-run” has been shown to require the coordinate activity of OPA1, MFN2, and DRP1, and represents an important element of the mitochondrial quality control, working as a “pit stop” for mitochondria and providing an efficient recharging mechanism for quick equilibration of small solutes, mRNA, and proteins, but not integral membrane proteins or mtDNA (25). Thus, when mitochondrial function is not hampered by severe alterations of the mtDNA, complete and transient fusion of mitochondria can provide temporary solution to complement the defective activities and eventually provide the enzyme required for repairing the damaged mtDNA. Conversely, when mitochondrial functions are severely compromised, that is, when the membrane potential is collapsed, the fate of malfunctioning mitochondria consists in their delivery to lysosomes for autophagic elimination (52). The process responsible for this mitochondrial quality control mechanism, known as mitophagy, involves PINK1 and Parkin proteins (29). In the presence of mitochondrial membrane potential, PINK1 is continuously degraded in a proteasome-dependent manner, whereas under depolarizing conditions, PINK1 accumulates on mitochondrial outer membrane, where it recruits Parkin that ubiquitinylates specific proteins, among which MFN2. This process triggers an irreversible signal that targets mitochondria for autophagic elimination (33). In this regard, it has been recently reported that long-term overexpression of Parkin can eliminate mitochondria with deleterious mtDNA mutation affecting cytochrome oxidase I subunits of complex IV, enriching cells with wild-type mtDNA, suggesting that increasing levels of Parkin expression might contribute to ameliorate certain mitochondrial diseases related to mtDNA instability (48). In agreement with this, a direct association of endogenous Parkin with mtDNA and Mitochondrial Transcription Factor A has been reported in SH-SY5Y cells, and most importantly, Parkin overexpression has been shown to enhance mtDNA replication and transcription to protect this genome from oxidative damage (43).
On the other hand, inhibition of mitophagy can impair the efficient elimination of dysfunctional mitochondria; thus, allowing for the accumulation of mutated genomes, as observed in patients with mutations in the OPA1, MFN2, PINK1, and PARK2 genes. Although mechanisms of DNA repair mostly similar to those found in the nucleus have been characterized recently in mitochondria (24), their efficiency, except for the base excision repair counteracting the direct effects of ROS, seems rather limited and probably cannot comply with the multiple harmful mtDNA alterations generated permanently. Thus, mitophagy remains an efficient solution to eliminate mutated mtDNA and generate sustained mtDNA turnover, even in nonproliferative cells. A critical question is how mitochondria are selected to be degraded, as no intramitochondrial checkpoint sensing mtDNA alterations is yet suspected to exist. The mechanism that was retained is based on the most prominent mitochondrial bioenergetic parameter: the membrane potential, from which depends ATP production. As membrane fusion and the switch from long fusion competent to short fusion incompetent OPA1 isoforms are governed by the membrane potential and ATP concentration (3, 19), the capacity of an individual mitochondrion to fuse or “kiss-and-run,” is an excellent marker of its functional integrity, and can be monitored extrinsically by cytoplasmic sensors, as PINK1 does. Thus, by selecting mitochondria that have not retained an adequate capacity to fuse, that is, that are fragmented, cells can readily identify and eliminate by mitophagy the pool of defective mitochondria with potentially altered mtDNA sequences. Thus, facing the large pool of mtDNA per cells, the most proficient mechanism that was selected to favor the perenniality of the mitochondrial genome consists in the elimination rather than the reparation of damaged mtDNA.
Evolutionary Considerations Linking Mitochondrial mtDNA Integrity and Network Dynamics
The data presented here on mitochondrial dynamics and genome stability prompt an evolutionary parallel with the mechanisms contributing to genome maintenance and plasticity in bacteria. During prokaryote proliferation, it is well documented that while genomic or large plasmid DNA is replicated, the neosynthesized strands are segregated to the distal cell extremities by duplicated structures embedded in the plasma membrane, so that the replicated genetic contents are evenly distributed in the two future daughter cells (50). This mechanism establishes a tight coupling between bacterial genome replication and membrane dynamics which, when deficient, leads to chromosome instability and recombination. In this respect, we can draw a parallel with mitochondria, in the way that alterations in mtDNA anchoring to the inner membrane or in its distribution along the network, by selective disruption of OPA1, MFNs, or DRP1 functions, induce mtDNA depletion or deletions. Thus, although no dynamin-related protein exists in bacteria, it is nonetheless meaningful that certain aspects of the coupling between genome replication, distribution and integrity have been conserved after 3 billion years of evolutionary divergence between bacteria and mitochondria.
A second parallel can be drawn from what is known on bacterial conjugation. In this process, bacteria from opposite mating type fuse to allow the transfer of genetic material from the donor to the acceptor cell. The transferred genetic material can be kept as episomal, bringing a novel set of genes, or used to recombine with the acceptor chromosome, thus allowing for novel genetic combinations (34). Ultimately, facing stressing conditions, this process can lead to the selection of the genomes most able to respond to novel environmental challenges. Consequently, membrane fusion of two cells with different genetic patterns can generate a third cell with a novel genetic program. This mechanism is reminiscent to the transcomplementation process that has been demonstrated after fusion of mitochondria bearing two different point mutations affecting different respiratory chain subunits (49). Thus, both in mitochondria and in bacteria, the transfer of genetic material associated to membrane fusion allows first to transcomplement possible genetic defects, and second to select eventually for novel genetic programs by homology recombination. Nevertheless, in mammalian cells the selective constraints acting on the mitochondrial genome are not subject to sudden environmental changes; thus, the need for frequent recombination of the mitochondrial genome remained low. Altogether, it sounds clear that mitochondria have maintained throughout evolution some bacterial processes that link the membrane dynamics to the plasticity and motility of its genome, that ultimately are required for the maintenance of mtDNA integrity.
Open Questions
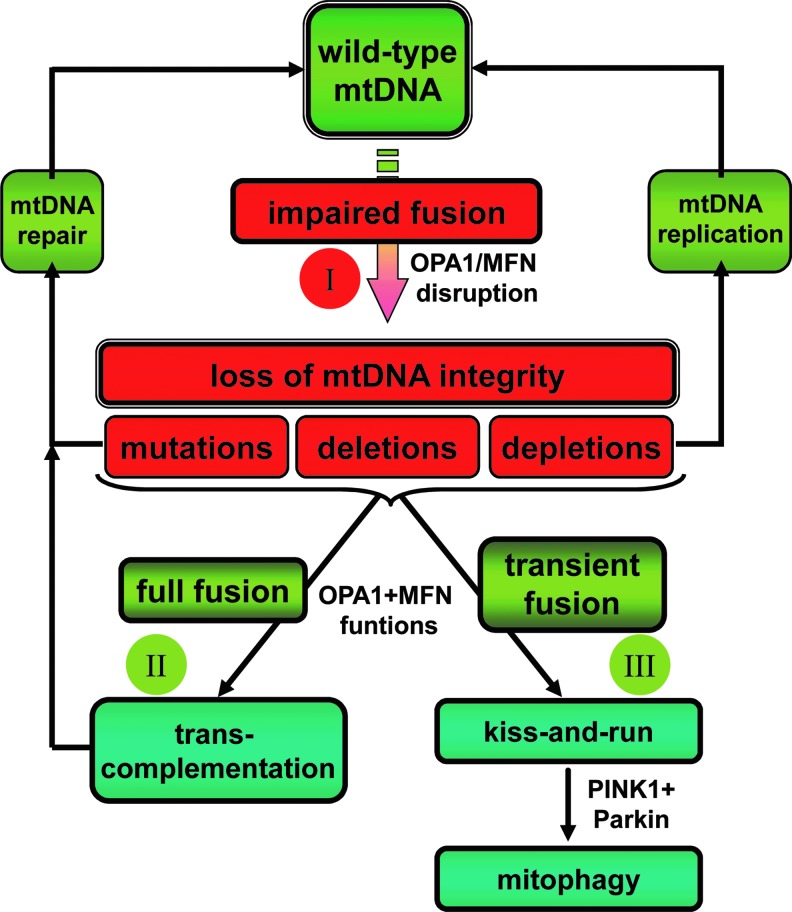
Summarizing the topic addressed in this review (Fig. 7), we should recognize that the number of facts supporting a direct link between mitochondrial fusion and genome maintenance are still scarce, leaving tremendous space for open questions. The limited relevant data that have been gathered to date consist in fundamental observations obtained from cell and genetic models suggesting that mitochondrial fusion dynamins are required for the quantitative maintenance of the mtDNA, whereas clinical analysis of human biopsies bearing mutations in the OPA1, MFN2, PINK, and PARK2 genes revealed qualitative alterations of the mitochondrial genome, mainly multiple deletions in the mtDNA, as reported in samples from old human beings.
FIG. 7.
Summary of the roles of mitochondrial fusion in the maintenance of the mitochondrial genome integrity. Schematic cartoon of the three possible roles of mitochondrial fusion: I in generating mtDNA alterations when OPA1 or MFNs functions are disrupted; II in promoting mitochondrial trans-complementation; III in triggering the “kiss-and-run” process before mitophagy. PINK1 and Parkin functions lie in-between the “kiss-and-run” process and the removal of depolarized mitochondria by mitophagy. Green colors illustrate physiological processes contributing to the maintenance of mitochondrial genome integrity, whereas red colors highlight deleterious situations affecting the mtDNA. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
So questions are open! Does disruption of mitochondrial fusion cause by itself mtDNA depletion and deletions? Or is the process of mitochondrial fusion only required for the elimination of pre-existing mtDNA alterations? Why only restricted organs do present mtDNA alterations and become pathogenic, whereas the mutated alleles of these genes are ubiquitously expressed? Could the answer of this latter question be linked to a difference of efficiency of the transcomplementation and the mitophagy processes between organs? Have organs differential capacities for oxidative phosphorylation and preventing ROS production? Is the accumulation of mtDNA alterations just a matter of time, which would be accelerated by decreasing the efficiency of mitochondrial fusion? Consequently, could we relate the accumulation of mtDNA alterations in elders to a decrease of fusion efficiency and conversely imagine that increased fusion would prevent aging?
To answer these questions, we need to investigate deeper the relevant models that are available now, and generate new ones that reproduce what has been observed in patients with altered fusion. Ultimately, we should find relevant evidence proving that direct links exist between the different processes of mitochondrial fusion, mitophagy, and maintenance of the mitochondrial genome integrity.
Abbreviations Used
- ΔΨm
mitochondrial membrane potential
- ATPase
adenosine triphosphate synthase
- CC
coiled coil
- CMT2A
Charcot Marie Tooth type 2A
- COX
cytochrome oxidase
- DOA
dominant optic atrophy
- DOAplus
syndromic dominant optic atrophy
- DRP1
dynamin related protein 1
- Fzo
fuzzy onions
- GED
GTPase effector domain
- GTPase
guanosine tri phosphatase
- HR
heptad repeat
- MFN1/MFN2
mitofusin 1/2
- MGM1
mitochondrial genome maintenance 1
- Msp1
MGM1 in Schizosaccharomyces pombe
- mtDNA
mitochondrial DNA
- MTS
mitochondrial targeting sequence
- ND
NADH dehydrogenase
- OPA1
optic atrophy 1
- OXPHOS
oxidative phosphorylation reaction
- PD
Parkinson disease
- PINK1
PTEN induced kinase 1
- RAS
Ras binding domain
- ROS
reactive oxygen species
- TM
trans membrane domain
Acknowledgments
We are indebted to the INSERM, CNRS and Université Montpellier I and II for providing institutional supports and to the patient associations Retina France, Union Nationale des Aveugles et Déficients Visuels and the Association Française contre les Myopathies, for their financial support. This work was supported by an ERA-Net Research Program on Rare Diseases to G.L. and M.R., and by a NIH grant GM 38237 to M.R. We acknowledge Dr. Mourties (Montpellier, France) and Dr. Barbera (Bologna, Italy) for generating helpful discussions.
References
- 1.Alavi MV. Bette S. Schimpf S. Schuettauf F. Schraermeyer U. Wehrl HF. Ruttiger L. Beck SC. Tonagel F. Pichler BJ, et al. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy. Brain. 2007;130:1029–1042. doi: 10.1093/brain/awm005. [DOI] [PubMed] [Google Scholar]
- 2.Amati-Bonneau P. Valentino ML. Reynier P. Gallardo ME. Bornstein B. Boissiere A. Campos Y. Rivera H. de la Aleja JG. Carroccia R, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus' phenotypes. Brain. 2008;131:338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- 3.Baricault L. Segui B. Guegand L. Olichon A. Valette A. Larminat F. Lenaers G. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res. 2007;313:3800–3808. doi: 10.1016/j.yexcr.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Bender A. Krishnan KJ. Morris CM. Taylor GA. Reeve AK. Perry RH. Jaros E. Hersheson JS. Betts J. Klopstock T, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 5.Bereiter-Hahn J. Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 6.Bleazard W. McCaffery JM. King EJ. Bale S. Mozdy A. Tieu Q. Nunnari J. Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartoni R. Arnaud E. Medard JJ. Poirot O. Courvoisier DS. Chrast R. Martinou JC. Expression of mitofusin 2(R94Q) in a transgenic mouse leads to Charcot-Marie-Tooth neuropathy type 2A. Brain. 2010;133:1460–1469. doi: 10.1093/brain/awq082. [DOI] [PubMed] [Google Scholar]
- 8.Chen H. Detmer SA. Ewald AJ. Griffin EE. Fraser SE. Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H. Vermulst M. Wang YE. Chomyn A. Prolla TA. McCaffery JM. Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevrollier A. Guillet V. Loiseau D. Gueguen N. de Crescenzo MA. Verny C. Eng MF. Dollfus H. Odent S. Milea D, et al. Hereditary optic neuropathies share a common mitochondrial coupling defect. Ann Neurol. 2008;63:794–798. doi: 10.1002/ana.21385. [DOI] [PubMed] [Google Scholar]
- 11.Copeland WC. Defects in mitochondrial DNA replication and human disease. Crit Rev Biochem Mol Biol. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson TM. Ko HS. Dawson VL. Genetic animal models of Parkinson's disease. Neuron. 2010;66:646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delettre C. Lenaers G. Griffoin JM. Gigarel N. Lorenzo C. Belenguer P. Pelloquin L. Grosgeorge J. Turc-Carel C. Perret E, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 14.Diot A. Guillou E. Daloyau M. Arnaune-Pelloquin L. Emorine LJ. Belenguer P. Transmembrane segments of the dynamin Msp1p uncouple its functions in the control of mitochondrial morphology and genome maintenance. J Cell Sci. 2009;122:2632–2639. doi: 10.1242/jcs.040139. [DOI] [PubMed] [Google Scholar]
- 15.Elachouri G. Vidoni S. Zanna C. Pattyn A. Boukhaddaoui H. Gaget K. Yu-Wai-Man P. Gasparre G. Sarzi E. Delettre C, et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21:12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott HR. Samuels DC. Eden JA. Relton CL. Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferre M. Bonneau D. Milea D. Chevrollier A. Verny C. Dollfus H. Ayuso C. Defoort S. Vignal C. Zanlonghi X, et al. Molecular screening of 980 cases of suspected hereditary optic neuropathy with a report on 77 Novel OPA1 mutations. Hum Mutat. 2009;30:E692–E705. doi: 10.1002/humu.21025. [DOI] [PubMed] [Google Scholar]
- 18.Gilkerson RW. Schon EA. Hernandez E. Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griparic L. Kanazawa T. van der Bliek AM. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson G. Amati-Bonneau P. Blakely EL. Stewart JD. He L. Schaefer AM. Griffiths PG. Ahlqvist K. Suomalainen A. Reynier P, et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131:329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- 21.Jones BA. Fangman WL. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- 22.Kogelnik AM. Lott MT. Brown MD. Navathe SB. Wallace DC. MITOMAP: a human mitochondrial genome database—1998 update. Nucleic Acids Res. 1998;26:112–115. doi: 10.1093/nar/26.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenaz G. Baracca A. Carelli V. D'Aurelio M. Sgarbi G. Solaini G. Bioenergetics of mitochondrial diseases associated with mtDNA mutations. Biochim Biophys Acta. 2004;1658:89–94. doi: 10.1016/j.bbabio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Liu P. Demple B. DNA repair in mammalian mitochondria: much more than we thought? Environ Mol Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 25.Liu X. Weaver D. Shirihai O. Hajnoczky G. Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeb LA. Wallace DC. Martin GM. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc Natl Acad Sci U S A. 2005;102:18769–18770. doi: 10.1073/pnas.0509776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loiseau D. Chevrollier A. Verny C. Guillet V. Gueguen N. Pou de Crescenzo MA. Ferre M. Malinge MC. Guichet A. Nicolas G, et al. Mitochondrial coupling defect in Charcot-Marie-Tooth type 2A disease. Ann Neurol. 2007;61:315–323. doi: 10.1002/ana.21086. [DOI] [PubMed] [Google Scholar]
- 28.Malka F. Lombes A. Rojo M. Organization, dynamics and transmission of mitochondrial DNA: focus on vertebrate nucleoids. Biochim Biophys Acta. 2006;1763:463–472. doi: 10.1016/j.bbamcr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda N. Tanaka K. Uncovering the roles of PINK1 and parkin in mitophagy. Autophagy. 2010;6:952–954. doi: 10.4161/auto.6.7.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCorquodale DS., 3rd Montenegro G. Peguero A. Carlson N. Speziani F. Price J. Taylor SW. Melanson M. Vance JM. Zuchner S. Mutation screening of mitofusin 2 in Charcot-Marie-Tooth disease type 2. J Neurol. 2011;258:1234–1239. doi: 10.1007/s00415-011-5910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montoya J. Gaines GL. Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 32.Narendra D. Tanaka A. Suen DF. Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narendra DP. Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narra HP. Ochman H. Of what use is sex to bacteria? Curr Biol. 2006;16:R705–R710. doi: 10.1016/j.cub.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 35.Olichon A. Baricault L. Gas N. Guillou E. Valette A. Belenguer P. Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 36.Palade GE. An electron microscope study of the mitochondrial structure. J Histochem Cytochem. 1953;1:188–211. doi: 10.1177/1.4.188. [DOI] [PubMed] [Google Scholar]
- 37.Payne BA. Wilson IJ. Hateley CA. Horvath R. Santibanez-Koref M. Samuels DC. Price DA. Chinnery PF. Mitochondrial aging is accelerated by anti-retroviral therapy through the clonal expansion of mtDNA mutations. Nat Genet. 2011;43:806–810. doi: 10.1038/ng.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelloquin L. Belenguer P. Menon Y. Ducommun B. Identification of a fission yeast dynamin-related protein involved in mitochondrial DNA maintenance. Biochem Biophys Res Commun. 1998;251:720–726. doi: 10.1006/bbrc.1998.9539. [DOI] [PubMed] [Google Scholar]
- 39.Poole AC. Thomas RE. Andrews LA. McBride HM. Whitworth AJ. Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rapaport D. Brunner M. Neupert W. Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- 41.Renaldo F. Amati-Bonneau P. Slama A. Romana C. Forin V. Doummar D. Barnerias C. Bursztyn J. Mayer M. Khouri N, et al. MFN2, a new gene responsible for mitochondrial DNA depletion. Brain. 2012;135:e223. doi: 10.1093/brain/aws111. [DOI] [PubMed] [Google Scholar]
- 42.Rojo M. Legros F. Chateau D. Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- 43.Rothfuss O. Fischer H. Hasegawa T. Maisel M. Leitner P. Miesel F. Sharma M. Bornemann A. Berg D. Gasser T, et al. Parkin protects mitochondrial genome integrity and supports mitochondrial DNA repair. Hum Mol Genet. 2009;18:3832–3850. doi: 10.1093/hmg/ddp327. [DOI] [PubMed] [Google Scholar]
- 44.Rouzier C. Bannwarth S. Chaussenot A. Chevrollier A. Verschueren A. Bonello-Palot N. Fragaki K. Cano A. Pouget J. Pellissier JF, et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus' phenotype. Brain. 2012;135:23–34. doi: 10.1093/brain/awr323. [DOI] [PubMed] [Google Scholar]
- 45.Sarzi E. Bourdon A. Chretien D. Zarhrate M. Corcos J. Slama A. Cormier-Daire V. de Lonlay P. Munnich A. Rotig A. Mitochondrial DNA depletion is a prevalent cause of multiple respiratory chain deficiency in childhood. J Pediatr. 2007;150:531–534. doi: 10.1016/j.jpeds.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 46.Sarzi E. Goffart S. Serre V. Chretien D. Slama A. Munnich A. Spelbrink JN. Rotig A. Twinkle helicase (PEO1) gene mutation causes mitochondrial DNA depletion. Ann Neurol. 2007;62:579–587. doi: 10.1002/ana.21207. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava S. Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suen DF. Narendra DP. Tanaka A. Manfredi G. Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takai D. Isobe K. Hayashi J. Transcomplementation between different types of respiration-deficient mitochondria with different pathogenic mutant mitochondrial DNAs. J Biol Chem. 1999;274:11199–11202. doi: 10.1074/jbc.274.16.11199. [DOI] [PubMed] [Google Scholar]
- 50.Thanbichler M. Shapiro L. Chromosome organization and segregation in bacteria. J Struct Biol. 2006;156:292–303. doi: 10.1016/j.jsb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Twig G. Elorza A. Molina AJ. Mohamed H. Wikstrom JD. Walzer G. Stiles L. Haigh SE. Katz S. Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Twig G. Hyde B. Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 54.Waterham HR. Koster J. van Roermund CW. Mooyer PA. Wanders RJ. Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 55.Wei YH. Lee HC. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp Biol Med (Maywood) 2002;227:671–682. doi: 10.1177/153537020222700901. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y. Ouyang Y. Yang L. Beal MF. McQuibban A. Vogel H. Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu-Wai-Man P. Davies VJ. Piechota MJ. Cree LM. Votruba M. Chinnery PF. Secondary mtDNA defects do not cause optic nerve dysfunction in a mouse model of dominant optic atrophy. Invest Ophthalmol Vis Sci. 2009;50:4561–4566. doi: 10.1167/iovs.09-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]