Abstract
We have previously demonstrated that the KH-domain protein αCP binds to a 3′ untranslated region (3′UTR) C-rich motif of the nascent human alpha-globin (hα-globin) transcript and enhances the efficiency of 3′ processing. Here we assess the genome-wide impact of αCP RNA-protein (RNP) complexes on 3′ processing with a specific focus on its role in alternative polyadenylation (APA) site utilization. The major isoforms of αCP were acutely depleted from a human hematopoietic cell line, and the impact on mRNA representation and poly(A) site utilization was determined by direct RNA sequencing (DRS). Bioinformatic analysis revealed 357 significant alterations in poly(A) site utilization that could be specifically linked to the αCP depletion. These APA events correlated strongly with the presence of C-rich sequences in close proximity to the impacted poly(A) addition sites. The most significant linkage was the presence of a C-rich motif within a window 30 to 40 bases 5′ to poly(A) signals (AAUAAA) that were repressed upon αCP depletion. This linkage is consistent with a general role for αCPs as enhancers of 3′ processing. These findings predict a role for αCPs in posttranscriptional control pathways that can alter the coding potential and/or levels of expression of subsets of mRNAs in the mammalian transcriptome.
INTRODUCTION
Posttranscriptional controls play a major role in the regulation of eukaryotic gene expression (1, 2). These controls reflect interactions of RNA-binding proteins and/or noncoding RNAs with sequences and structures on target RNAs (3). Significant efforts have focused on the role(s) of 3′ untranslated region (3′UTR) determinants in these processes. Regulatory roles of 3′UTRs have been defined in nuclear RNA processing and mRNA transport as well as in the localization, translation, and decay of cytoplasmic mRNAs (4, 5).
Within the nucleus, the efficiency and specificity of 3′ processing of nascent polymerase II (Pol II) transcripts can have an impact both on the level of mRNA synthesis and on the structure of the final mature mRNA product (6–10). A widespread impact of 3′ processing on gene expression was predicted by initial studies of expressed sequence tags (11). These and subsequent studies revealed that the majority of gene transcripts have multiple functional polyadenylation sites. Alternative polyadenylation (APA) can impact on a spectrum of regulatory functions (12). The cellular transcriptome can undergo alterations in 3′ processing in response to physiological and developmental cues. For example, cells that are rapidly proliferating and/or have undergone malignant transformation have been observed to generate mRNAs with a shorter 3′UTR (13, 14). In a reciprocal manner, embryonic cells that are committing to specific differentiation pathways undergo 3′ processing that generates mRNAs with longer 3′UTRs (15). While the impact of these global changes remains to be rigorously determined, a model proposes that the global shortening of 3′UTRs releases mRNAs from the negative effects of endogenous miRNAs whereas the reciprocal lengthening of 3′UTRs facilitates cell differentiation and functional specification (13–15).
3′ processing of Pol II transcripts is mediated by concerted actions of multiple macromolecular complexes that have been functionally defined using in vitro processing systems (16, 17). These complexes include the cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CFIm), cleavage factor II (CFIIm), poly(A) polymerase (PAP), and symplekin, the scaffold protein (18). Assembly of a subset of these complexes on the nascent transcript may be directed and facilitated by their interactions with the phosphorylated C-terminal domain of the elongating Pol II (8, 9, 19–21).
The accuracy and efficiency of 3′ processing are determined by two major cis elements: the polyadenylation “signal,” AAUAAA, positioned 10 to 30 nucleotides (nt) upstream of the cleavage site, and a GU/U-rich region, or “downstream sequence element” (DSE), within a window located 30 nt 3′ of the cleavage site. The CPSF complex associates with the AAUAAA motif via its CPSF-160 subunit and determines the positioning of the cleavage reaction (18, 22). The CstF complex interacts with the GU/U DSE via binding of its CstF-64 subunit and functions to regulate the efficiency of 3′ processing (18, 22, 23).
Additional cis elements, working together with their interacting factors, can modulate, sometimes dramatically, the efficiency of the 3′-end processing reaction at a particular site. Two such determinants are “upstream sequence elements” (USEs) characteristically located 5′ of the AAUAAA signal and “auxiliary downstream sequence elements” (AuxDSEs) positioned 3′ of the GU/U-rich element (24). The USEs are generally U rich, but a consensus sequence has not been established (18, 25). The few AuxDSEs that have been identified are G rich, but they appear to lack a conserved sequence or distance from the cleavage site (18). These and other less-well-defined auxiliary elements (24) can enhance, either directly or indirectly, the efficiency of 3′ processing by recruiting the basal 3′ processing complexes to the RNA (18, 26). Thus, the net level and efficiency of 3′ processing of a Pol II transcript are determined by the recruitment of a complex set of macromolecular complexes to an array of cis-acting regulatory elements.
Prior studies in our laboratory have identified a novel RNA-protein (RNP) complex that assembles on the 3′UTR of the human alpha-globin (hα-globin) mRNA. This complex, initially identified based on its ability to enhance stability of hα-globin mRNA in the cytoplasm of erythroid cells (27–32), is comprised of the KH-domain RNA-binding protein, αCP [also known as poly(C)-binding protein (PCBP) and heterogeneous nuclear ribonucleoprotein (hnRNP) E (reviewed in reference 33)], bound to a repeated C-rich motif within the 3′UTR (34, 35). This αCP/poly(C) RNP complex plays a role in stability control of multiple mRNAs, in both erythroid and nonerythroid cells, and is likely to constitute a widely distributed cytoplasmic determinant of gene regulation (36–39). The sequences and structures of these native C-rich elements parallel the C-rich motifs in single-stranded configurations that have been identified by in vitro systematic evolution of ligands by exponential enrichment (SELEX) as the optimal binding site for αCP2 (35).
In addition to their mRNA-stabilizing role, αCP/poly(C) complexes also function in the nucleus during transcript processing (40). For example, αCP has been demonstrated to initially bind to the nascent hα-globin transcript in the nucleus (40), where it acts in vivo as a splicing regulator (40). Our recent study indicated that αCP also enhances mRNA 3′ processing (4). These studies demonstrate that αCP bound to the C-rich USE enhances both steps in 3′-end processing, cleavage and polyadenylation (4). The idea of the ability of the αCP complex to enhance 3′-end processing is further supported by the in vivo interaction of αCP with core components of the 3′-end processing complex (4). These observations support a model in which αCP assembles cotranscriptionally on the 3′UTR, setting the stage for a coordinated set of nuclear and cytoplasmic controls.
In the current study, we extended these observations by exploring a wider role for the αCP/poly(C) complex in the control of the mammalian transcriptome. The results demonstrate that αCPs, in conjunction with their cognate C-rich binding sites, control the utilization of poly(A) processing sites in a defined subset of the mRNAs. Thus, the αCP RNP complex has the capacity to play a pivotal and global role in determining the structure and expression of specific transcripts via its impact on the 3′ processing pathway.
MATERIALS AND METHODS
Cell culture and siRNA transfection.
K562 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone) and antibiotic/antimycotic at 37°C in a 5% CO2 incubator. Cells were transfected with a total of 2.0 μg of small interfering RNA (siRNA) using Nucleofector V (Amaxa) according to the manufacturer's instructions. All siRNAs were from Dharmacon. The siRNA sequences are as follows: for αCP(1/2)-1, GUG AAA GGC UAU UGG GCA A; for αCP(1/2)-2, UGU AAG AGU GGA AUG UUA A; for GLD2-1, GUG AUU AAG AAG UGG GCA A; and for GLD2-2, CCA AAG AUA AGU UGA GUC A.
Standard control siRNA for cyclophilin was directly purchased from Dharmacon. At 24 h after the initial siRNA transfection, these cells were retransfected with same siRNA. Cells were harvested 72 h after the initial transfection, and RNAs were purified using an Absolutely RNA Miniprep kit (Stratagene) according to manufacturer's instructions. Western blot analysis was performed as described previously (27, 34).
Direct RNA sequencing (DRS).
Massively parallel sequencing of the RNA 3′ termini was carried out by the Helicos Bioscience Corporation (Cambridge, MA) according to established protocols (41, 42).
Mapping and APA analysis of DRS data.
The DRS reads were aligned to human genome assembly 19 (hg19) using the indexDP genomic tool provided by Helicos Biosciences. The uniquely mapped reads with a minimal mapped length of 25 nt and an alignment score of 4.0 were retained for further analysis. The replicate samples for control and αCP knockdown experiments were pooled for differential expression analysis and APA studies. All mapped reads were initially screened and filtered for those arising from internal poly(A) priming using a previously described approach (43). Individual poly(A) sites were identified by reversing 5′ ends of the non-internal-priming reads. Pooled data from both pooled control and αCP experiments were used to construct a consensus poly(A) annotation for downstream analysis and to iteratively cluster all individual poly(A) sites within 40 nt of its nearest poly(A) site on the same chromosome strand. The weighted coordinate, which was calculated as the sum of the product of the coordinate of an individual poly(A) and its percentage of usage in the whole cluster, was taken as the representative coordinate of the corresponding poly(A) cluster. The frequencies of poly(A) clusters in the different samples were calculated according to the consensus coordinates of poly(A) clusters in the pooled data as described above. Next, the poly(A)s residing in the whole gene region, including exons, introns, and the downstream 100-nt region of the terminal exon, were collected as possible poly(A)s of a defined gene (UCSC genes [hg19] and Ensembl genes [release 61]).
To test whether there was a change of usage for any single poly(A) cluster of a particular gene, Fisher's exact test was conducted to compare the ratio of DRS counts of a single poly(A) cluster to the sum of those of all the other poly(A) clusters between pooled control and αCP knockdown samples. The P values were adjusted by Benjamini-Hochberg method for calculating the false discovery rate (FDR). Finally, the poly(A)s with FDRs of less than 0.05 and a percentage change of total poly(A) usage greater than 10% (|ΔΨ| > 10%) were defined as significantly changed poly(A)s.
Detection of differential gene expression.
The expression level of a gene is represented by the sum of DRS read counts of all the overlapping poly(A) reads. DEGSeq (44) was run to detect differential gene expression between the control and αCP knockdown samples. The genes with FDRs of less than 0.05 and normalized fold change values greater than 1.5 (as determined by number of mapped DRS reads) were defined as significantly differentially expressed genes (DEG).
Motif enrichment analysis.
Significantly changed poly(A)s in APA analyses were divided into upregulated sets (FDR < 0.05 and ΔΨ > 0.1) and downregulated sets (FDR < 0.05 and ΔΨ < −0.1). Motif enrichment and occurrence analyses were conducted on these two subsets separately. The upstream 200-bp sequences of poly(A)s were first scanned by MEME (45). To control for the poly(A) abundance, significantly changed poly(A)s and unchanged poly(A)s (FDR > 0.5) were grouped into bins [the borders of the bins were defined by 20, 21, …, 2n, where n is determined by the most highly abundant poly(A) in the data set] according to DRS read counts. The background poly(A)s were next randomly sampled from unchanged poly(A)s (FDR > 0.5) with bin sizes 10 times bigger than those in the significant poly(A) sets. To draw an RNA map, the motif score of a sequence position [in the upstream 200-nt region of a poly(A) site] is calculated as the average number of occurrences of overlapped nucleotides in a 31-nt window (upstream and downstream 15 nt) for both significant and background poly(A)s. A Wilcoxon rank sum test was performed to measure the significance of differences in average motif scores for a specific position. The P value of Wilcoxon rank sum test was adjusted using the Benjamin-Hochberg algorithm to calculate an FDR. The dominant poly(A)s of the significantly upregulated and downregulated genes were retrieved for motif analysis using a method similar to that used in the APA study. The background data set was generated by controlling for the gene expression level of significantly differentially expressed genes, which used the same binning and random sampling method as that described for the APA study (see above). All the subsequent analyses were also the same as those described for the APA study.
GO analysis.
DAVID software (using the PANTHER classification system) was used to predict gene ontology (GO) groupings for mRNAs impacted in their expression or in their patterns of poly(A) site selection and utilization by αCP depletion (46). The background gene sets were controlled for the distribution of gene expression level in the foreground gene sets using a binning and random sampling method similar to that described for the motif enrichment analysis.
RT-qPCR.
RNAs were treated with DNase I (Invitrogen) and then reverse transcribed (RT) using a first-strand cDNA synthesis kit (GE). Quantitative (qPCR) procedures were performed using a Fast SYBR green Master Mix kit (Applied Biosystems) and a 7900HT Fast qPCR thermocycler (Applied Biosystems) according to the manufacturer's instructions. Primers used in the differential gene expression (DGE) and APA studies are listed in Table S3 in the supplemental material.
3′ RACE.
3′ rapid amplification of cDNA ends (RACE) was performed according to an established protocol (3′ RACE system for rapid amplification of cDNA ends; Invitrogen).
RNA UV-cross-linking and electrophoretic mobility shift assay (EMSA).
UV-cross-linking assays were performed as described previously (4, 40).
In vitro polyadenylation.
In vitro polyadenylation assays were performed as described previously (4). Recombinant αCP2 protein was expressed and purified as described previously (34). 32P-labeled PRG2 and AES were generated by in vitro transcription from PCR-generated DNA templates with an sp6 sequence added to the 5′ end. The probe sequences are as follows: for PRG2-wt, GCTGGTCCCAGCCAGCAGTTCAGAGCTGCCCTCTCCTGGGCAGCTGCCTCCCCTCCTCTGCTTGCCATCCCTCCCTCCACCTCCCTGCAATAAAATGGGTTTTACTGAAATGGA; for PRG2-mut, GCTGGTCCCAGCCAGCAGTTCAGAGCTGCCGTCTCCTGGGCAGCTGCCTGCCGTCCTCTGCTTGCCATCGCTCGCTGCACCTCGCTGCAATAAAATGGGTTTTACTGAAATGGA; for AES-WT, GCTGGGAGGAGCAGGGTGAGGGTGGGCGACCCAGGATTCCCCCTCCCCTTCCCAAATAAAGATGAGGGTACTA; and for AES-Mut, GCTGGGAGGAGCAGGGTGAGGGTGGGCGACCCAGGATTCCGCCTCGCCTTGCCAAATAAAGATGAGGGTACTA.
RESULTS
Direct RNA 3′ sequencing of the transcriptome in cells acutely depleted of αCP.
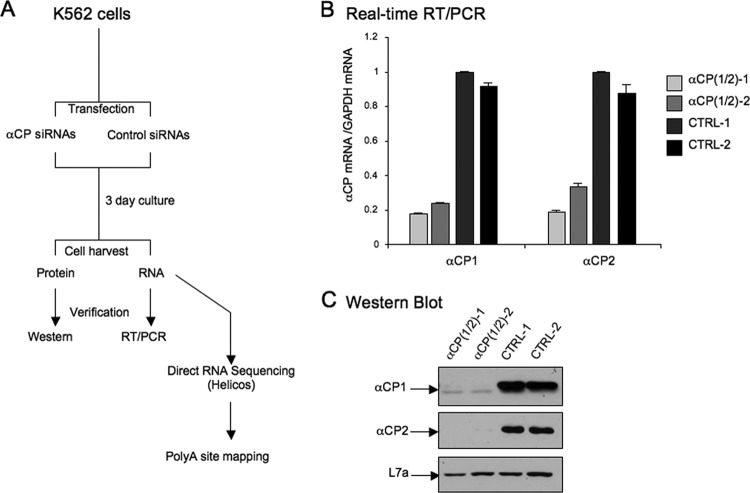
We have previously demonstrated that the RNA-binding proteins (αCPs) markedly enhance 3′ processing of the hα-globin transcript via a sequence-specific association of the αCP proteins with a C-rich motif within the 3′UTR (4). These studies led us to ask whether αCPs play a global role in 3′ processing in erythroid cells. To address this question, we assessed the impact of αCP depletion on the K562 transcriptome. K562 cells are a human Tier I ENCODE cell line with hematopoietic properties. We separately transfected the K562 cells with two distinct siRNAs, each of which cotargets the two major αCP transcripts, αCP1 and αCP2 (33). Parallel control transfections were carried out with siRNAs against an unrelated protein (GLD-2) (Fig. 1A). Effective and specific codepletion of αCP1 and αCP2 from the siRNA-treated cells was demonstrated by mRNA and protein analyses at 3 days posttransfection (Fig. 1B and C).
Fig 1.
siRNA-mediated codepletion of αCP1 and αCP2 from K562 cells. (A) Experimental procedure. K562 cells were separately transfected with two distinct siRNAs, each cotargeting αCP1 and αCP2 mRNAs [αCP(1/2)-1 and αCP(1/2)-2]. Parallel transfections were carried out with 2 distinct control siRNAs targeting an unrelated protein (GLD-2 mRNA; CTRL-1 and CTRL-2). At 24 h posttransfection, cells were retransfected with same siRNAs, cultured an additional 2 days (total, 3 days of culture), and assessed for effective siRNA-mediated depletion by protein and RNA analyses. RNA isolated from each culture was subjected to DRS analysis for mapping and quantification of 3′ processing sites. (B) Assessment of αCP depletion by real-time RT-PCR. Levels of mRNAs encoding the two αCP isoforms, αCP1 and αCP2, are displayed. The values on the y axis represent the αCP mRNA levels normalized to levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA in the respective samples. The ratio of αCP to GAPDH for CTRL-1 is arbitrarily defined as 1.0. The standard deviation for each sample is shown (n = 2). (C) Assessment of αCP depletion by Western blotting. Affinity-purified antibodies specific for either αCP1 or αCP2 (34) were used for detection in the top and middle panels. Detection of the large ribosomal subunit, L7a (27), controlled for sample loading, is shown in the bottom panel.
Total RNA isolated from each set of siRNA-transfected cells was subjected to direct RNA sequencing (DRS; Helicos BioSciences, Cambridge, MA). DRS isolates individual tethered poly(A) RNAs for massively parallel sequencing of 3′ termini. This direct approach eliminates the need for generating cDNA intermediates, amplification steps, or ligation reactions, any of which has the capacity to introduce bias in the final quantification of mRNA species (41, 42).
Total cellular RNAs from cells individually treated with each of the two αCP siRNAs and with each of the two control siRNAs were sequenced on four separate channels. The sequenced DRS reads had a mean read length of 32 nt (24 nt to 70 nt; see Fig. S1A in the supplemental material). Three of the channels generated 16 to 18 million reads, while the yield in the fourth was somewhat lower (10 million) (see Table S1 in the supplemental material). The raw reads were mapped back to the hg19 genome assembly and were filtered for internal priming to generate a final data set of positions and numbers of poly(A) termini (see Table S1). Approximately one-third (28% to 35%) of the sequenced reads were retained for poly(A) site quantification. A total of 55.7% to 61.4% of the retained DRS reads are within 40 nt of the ends of UCSC and Ensembl genes or poly(A) sites in polyA_DB2 (47). The DRS data were highly reproducible, with Pearson correlation coefficients higher than 0.92 and 0.94 for two siRNA control samples and two αCP siRNA samples, respectively (see Fig. S1B in the supplemental material). Based on this level of reproducibility, we pooled siRNA control data and αCP siRNA data, respectively, for the subsequent computational analysis.
Identification of mRNAs impacted by αCP depletion.
The DRS data were evaluated for the impact of the αCP depletion on overall gene expression levels and on the relative abundances of alternative poly(A) (APA) sites. The steady-state expression from each locus was determined by summing the total number of overlapping poly(A) site reads. This sum is referred to as the digital gene expression (DGE) value. We applied DEGseq (44) to identify differentially expressed genes. Using a false discovery rate (FDR) of less than 0.05 and a minimal normalized fold change value of 1.5, the data revealed that acute depletion of αCPs altered the expression of 586 genes; 231 were increased in expression and 355 were decreased in expression relative to cells transfected with either of the two control siRNAs (Table 1). Increasing the cutoff to a 2-fold change in transcript abundance revealed a significant impact on the expression of 117 genes; 42 were increased in expression and 75 were decreased in expression relative to the two controls (Table 1). Heat map profiling of the comparative DGE values for the 117 most significantly impacted genes (>2-fold change) revealed excellent concordance between the analyses of RNAs isolated from cells treated with the two distinct αCP siRNAs and those with the two distinct control siRNAs (Fig. 2A).
Table 1.
Genes impacted by αCP depletion as detected by DGE analysis from direct 3′-terminal RNA sequencing data
| Change (FDR < 0.05) | No. of genes |
||
|---|---|---|---|
| Upregulated | Downregulated | Total | |
| 2-fold | 42 | 75 | 117 |
| 1.5-fold | 231 | 355 | 586 |
Fig 2.

Impact of αCP depletion on the K562 transcriptome. (A) Heat map. Direct RNA sequencing (DRS) analyses were carried out on poly(A) RNAs isolated from cell cultures treated with the αCP siRNAs or the control siRNAs (as described for Fig. 1). The heat map represents all 117 mRNA species that showed a >2-fold change in expression (increased or decreased) subsequent to the αCP depletion. The color gradient (log scale) for the heat map represents the change in the overall representation of each mRNA normalized to the corresponding level in the RNA isolated from the cells treated in parallel with the control siRNAs. The positions of the direct siRNA targets, αCP1 and αCP2 mRNAs, are indicated by the arrows to the left of the heat map. (B) GO analysis of mRNAs altered in overall expression (DGE levels) by αCP depletion. GO analyses (DAVID algorithm) of mRNAs that undergo an alteration in steady-state levels (1.5-fold or greater change) subsequent to αCP depletion were included in the analysis. The data were assessed with Fisher's exact test with the FDR adjustment. The asterisks indicate the level of significance of the effect. *, 0.01 < FDR < 0.05; **, 0.001 < FDR < 0.01; ***, FDR < 0.001.
Gene ontology (GO) analysis revealed that the 586 genes with a 1.5-fold or greater change in expression subsequent to αCP depletion were enriched in genes related to amino acid metabolism, amino acid biosynthesis, oxidation-reduction reactions, cholesterol metabolism, lyase reactions, and immunity and defense (Fig. 2B and Table 2). The impact of αCP depletion on these gene categories is consistent with a role in the modulation of pathways controlling basic metabolism and cell stress responses (48).
Table 2.
GO analysis of genes with a >1.5-fold change in expression subsequent to αCP depletion
| GO term | No. of genes | Fold enrichment | FDR |
|---|---|---|---|
| BP00013:amino acid metabolism | 24 | 3.377374 | 1.19E-06 |
| BP00148:immunity and defense | 51 | 1.870906 | 8.57E-05 |
| MF00157:lyase | 14 | 2.864161 | 0.008 |
| BP00026:cholesterol metabolism | 10 | 3.732243 | 0.008 |
| MF00123:oxidoreductase | 33 | 1.668409 | 0.039 |
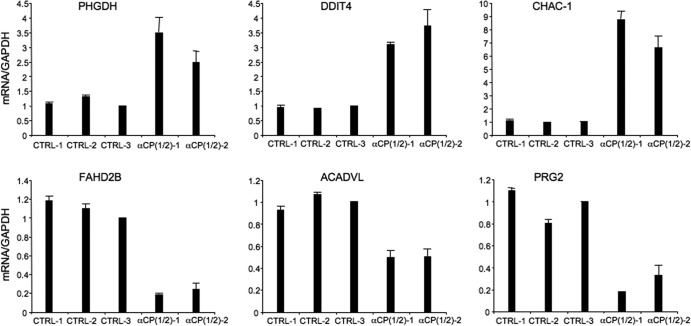
A subset of these transcripts with changes in expression greater than 1.5-fold as shown by the DGE analysis was subjected to verification by real-time RT-PCR. Each amplimer set corresponded to an internal region of the target mRNA so as to detect all mRNA isoforms, irrespective of their 3′-end processing patterns. These analyses, carried out on the same RNA samples that were assessed in the original DRS study, confirmed the DGE results (an increase or decrease of more than 1.5-fold in steady-state mRNA representation) (Fig. 3; see also Table S2 in the supplemental material).
Fig 3.
Confirmation of DRS results by targeted real-time RT-PCR analysis. Results of real-time analyses of three mRNAs that increased and three mRNAs that decreased in overall abundance relative to controls subsequent to αCP depletion (see Table S2 in the supplemental material) are shown. These studies were carried out on the same RNA preparations as were used in the original DRS analysis. To further validate these results, analyses of mRNA levels in K562 cells were additionally carried out with cells treated with a third distinct control siRNA corresponding to an unrelated mRNA (CTRL-3; cyclophilin siRNA). All values shown were normalized to the corresponding levels of GAPDH mRNA. The data are represented as ratios, with the ratio for the CTRL-3 siRNA sample defined as 1.0. The standard deviation for each sample is shown (n = 3).
Motif analysis reveals C-rich determinants in the 3′UTRs of mRNAs impacted by αCP depletion.
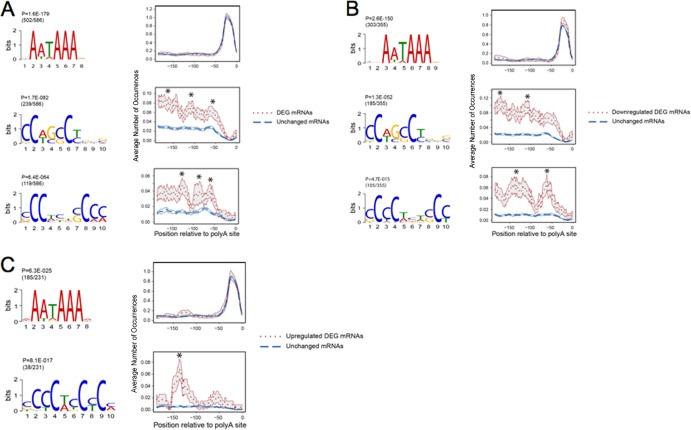
We applied MEME (45) software to infer motifs in the differentially expressed genes (DEG) associated with αCP depletion (“αCP knockdown”). The search was initiated on the full set of transcripts that underwent a >1.5-fold change in expression subsequent to αCP depletion. This MEME analysis was limited to the 200-nt segment immediately 5′ to the functional poly(A) cleavage site. By setting a rigorous P value cutoff of 1.0E–10, we found 3 motifs that were significantly enriched in the 200-nt segments upstream of the major poly(A) sites of the significantly changed genes. As expected, the most strongly conserved elements were the canonical poly(A) signal, AAUAAA, and its variants, peaking at 15 to 20 nt 5′ to the poly(A) site. These data corroborate the quality of DRS in recovering functional poly(A) sites. As expected, this poly(A) signal was observed in the mRNAs irrespective of whether or not they were impacted by the αCP depletion. Both of the next two most prominent motifs contained several prominent C's (Fig. 4A). An RNA-Map and Wilcoxon rank sum test were employed to identify the positioning of motifs relative to the respectively utilized poly(A) sites. The two C-rich motifs were significantly enriched in the αCP-impacted transcripts at multiple positions relative to the poly(A) site (nt −150, −100, and −50; FDR < 0.05).
Fig 4.
Motif analysis within the 3′UTRs of mRNAs impacted by αCP depletion. (A) MEME analyses of the sequences 5′ to the dominant poly(A) sites of all mRNAs that underwent a 1.5-fold or greater change (up or down) in their representation subsequent to αCP depletion (differentially expressed gene [DEG] mRNAs). The RNA map encompassed the 200-nt segments immediately 5′ to the sites of poly(A) addition. The top 3 motifs as detected by MEME are shown. For each motif, the P value and number of mRNAs containing corresponding motifs among the total number of mRNAs being studied are shown. The distance distributions [the poly(A) cleavage site is defined as base 0] are shown below each motif (x axis). The y axis indicates the percentage of nucleotides at each indicated site. An asterisk symbolizes a significant peak detected by the Wilcoxon rank sum test (FDR < 0.05). The P value measures the significance of a motif, and the ratio measures the fraction of mRNAs harboring corresponding motifs in the whole set of mRNAs. DEG mRNAs, 1.5-fold or greater change in expression as determined in comparisons of the αCP-depleted cells with control siRNA-treated cells. Unchanged mRNAs represent mRNAs whose expression was not changed subsequent to αCP depletion in comparison with mRNAs from control siRNA-treated samples. (B) Summary of MEME analyses of all mRNAs downregulated by greater than 1.5-fold subsequent to αCP depletion. Data are displayed as described for panel A. (C) Summary of MEME analyses of all mRNAs upregulated by greater than 1.5-fold subsequent to αCP depletion. Data are displayed as described for panel A.
MEME analysis was next separately applied to 355 and 231 transcripts that were downregulated and upregulated, respectively, in response to the acute αCP depletion (Fig. 4B and C). Analysis of the downregulated genes revealed one C-rich motif 5′ to sequences of poly(A) sites in approximately 30% of these transcripts (105 of 355 genes). Another motif, containing several Cs, occurred in 50% of these transcripts (185 of 355 genes) (Fig. 4B). These C-rich motifs in the downregulated genes were enriched at multiple locations relative to the poly(A) site (Fig. 4B). In contrast, only 38 of 231 (16%) of the upregulated genes harbored a C-rich motif; this motif was centered at a mean distance of 125 nt 5′ to the poly(A) sites (Fig. 4C).
In summary, the DGE analysis points to a significant impact of αCP on the overall expression level of a defined subset of mRNAs. The markedly greater number of mRNAs that were downregulated following the αCP depletion was consistent with an overall enhancing action of this αCP complex on steady-state mRNA levels and with the enrichment for C-rich motifs in the mRNAs impacted negatively by αCP depletion transcripts. These data thus support a direct role for the poly(C)-binding proteins in one or more posttranscriptional control pathways that impact on steady-state mRNA representation.
αCP impacts on patterns of alternative poly(A) selection.
The αCP/poly(C) complex within the hα-globin 3′UTR enhances 3′ cleavage and polyadenylation (4). Based on these studies, we proposed that C-rich motifs might act as upstream sequence element (USE) enhancers of 3′ processing in a subset of cellular transcripts. This activity could alter overall production of mature mRNAs by enhancing the use of a unique poly(A) site and/or have its impact via modulation of alternative poly(A) (APA) utilization. The preceding DGE analysis is consistent with a positive impact of the 3′UTR αCP/poly(C) complex on steady-state levels of a subset of mRNAs. To assess the impact of this complex on APA, we screened the DRS data set for shifts in poly(A) site utilization. For each poly(A) site, we applied Fisher's exact test to compare its DRS count to the sum of DRS counts of all the other poly(A)s within the same gene between two cell conditions (cells transfected with αCP siRNAs and with control siRNAs). This comparison revealed a total of 357 significant changes in poly(A) site utilization [198 downregulated poly(A) sites and 159 upregulated poly(A) sites] subsequent to αCP depletion, corresponding to a total 264 gene transcripts (FDR < 0.05). These data further revealed multiple APA events occurring in a subset of these 264 genes (Table 3). Of these APA events, 102 poly(A) sites occurred within the same terminal exon (“SE-APA”). This SE-APA subset of APA events should be particularly informative regarding the identification of 3′UTR motifs functional in APA, as they should be independent of alterations in transcript splicing. Another 122 genes with APA events linked to alterations in splicing patterns and occurred in different terminal exons (“DE-APA”). The remaining 133 APA events could not be simply assigned to either SE-APA or DE-APA categories and are termed “complex-APA” events. Examples of several complex-APA events are shown in Fig. S2 in the supplemental material. The pathways controlling these last two sets of APA events are likely to be complex and difficult to attribute to defined 3′UTR motifs.
Table 3.
Number of alternative poly(A) events impacted by depletion of αCPs
| Regulation change | No. of sites with indicated change |
|||
|---|---|---|---|---|
| SE-APA | DE-APA | Complex-APA | Total | |
| Upregulation | 44 | 58 | 57 | 159 |
| Downregulation | 58 | 64 | 76 | 198 |
| Total | 102 | 122 | 133 | 357 |
Motif analysis of APA events.
We searched for sequence motifs that could establish a direct mechanistic link(s) between αCP depletion and APA events. As with the DGE gene analysis, we examined the 200-nt regions upstream of poly(A) sites that underwent significant alteration in utilization for the structure and positioning of enriched motifs. A set of unchanged poly(A)s (FDR > 0.8), with DRS count distributions that were similar to those of the group with significantly changed poly(A)s, was randomly selected to serve as a background set for the analysis.
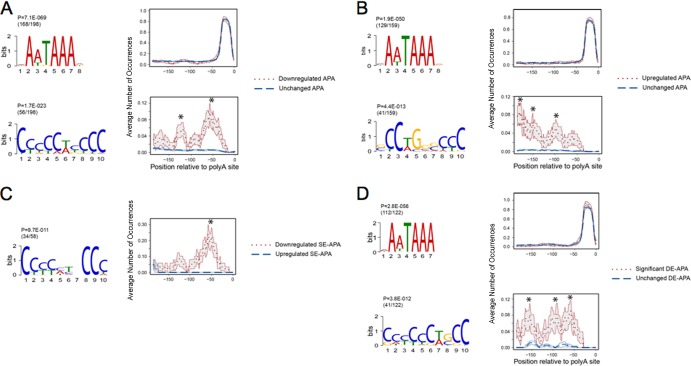
The initial analysis was carried out on the entire set of 198 poly(A) sites that were selectively downregulated upon αCP depletion. As expected, the canonical poly(A) signals, AATAAA and its variants, were consistently identified 15 to 20 bp 5′ to each of the utilized poly(A) sites (168/198 mRNAs) and their representation in αCP-impacted APA events was equal to that seen in the control group. A motif strongly enriched for C's was identified in 56 of these 198 APA sites (Fig. 5A). This motif was pyrimidine pure, with C's the predominant base at 9 of the 10 positions, and was not observed in the control group. This C-rich motif was highly represented 5′ of the poly(A) sites that were downregulated subsequent to αCP depletion and peaked at a position 35 to 45 bp 5′ of the site of poly(A) addition.
Fig 5.
Motif analysis of transcripts undergoing APA in response to αCP depletion. (A) Analyses of motifs 5′ to poly(A) sites that are involved in APA [DE-APA and SE-APA categories combined; 198 poly(A) sites] and are repressed in their representation subsequent to αCP depletion. The distance distribution plot is shown below each corresponding motif. For each motif, the P value and number of mRNAs containing corresponding motifs among the total number of mRNAs being studied are shown. The y axis indicates the percentage of each nucleotide at each indicated site at the indicated distance to the poly(A) cleavage site location (defined as base 0). The analyses from the cells treated with the control or the αCP siRNAs are directly compared in each setting. Asterisks illustrate positions with FDR (Benjamini-Hochberg algorithm) of less than 0.05 as determined by the Wilcoxon rank sum test. (B) Analyses of motifs 5′ to poly(A) sites that are involved in APA [DE-APA and SE-APA categories combined; 159 poly(A) sites] and are enhanced in their representation by αCP depletion (figure organized as described for panel A). (C) Analysis of motifs 5′ to poly(A) sites that are involved specifically in APA between sites in the same terminal exon (SE)-APA category. A discriminative motif discovery analysis by MEME was executed to specifically identify motifs overrepresented in the downregulated SE-APA [58 poly(A)s] and underrepresented in the upregulated SE-APA [44 poly(A)s]. The repressed SE-APA was defined in this comparison as the positive set, and the enhanced SE-APA was defined as the negative/background set. The position-specific prior probabilities were first estimated for the background set. Next, a normal motif search was done in downregulated SE-APA based on the position-specific prior probabilities. Note that the nucleotide at position 7 can be any of the four nucleotides. (D) Motifs 5′ to poly(A) sites that are involved specifically in APA between sites in different exons [DE-APA poly(A) sites] [122 poly(A)s]. The analysis was carried out as described for panel A.
The complementing analysis of the set of poly(A) sites that were upregulated following αCP depletion revealed a complex motif in 41 of 159 poly(A) sites. This motif contained central purines, lacked a significant poly(C) tract, and lacked a specific or predominant localization relative to the affected poly(A) site (Fig. 5B).
To further refine the analysis of the APA events, we limited the motif search to the 102 APA events at the same terminal exon (SE-APA events). This eliminated complicating influences of coexisting alterations in splicing events (Fig. 5C). To more directly link the C-rich motifs with the proposed USE function, we established a discriminative MEME motif approach that directly compared the sequence environment of 58 downregulated SE-APA (positive set) to that of 44 upregulated SE-APA (negative set). In this manner, the analysis was specifically configured to identify motifs associated with the downregulated poly(A) sites that were underrepresented in the environment of the upregulated poly(A) sites. The top-ranking motif in this discriminative analysis was a pyrimidine-pure and C-rich motif (Fig. 5C). This motif was present in 34 of the 58 downregulated poly(A) sites (Fig. 5C) and was positioned approximately 50 bp 5′ to the downregulated poly(A) site. When this same motif search was extended to the DE-APA events [122 poly(A) sites] (Fig. 5D), we again identified a C-rich motif (41/122 sites; 21 enhanced APA and 20 repressed APA), although in this case the positioning was somewhat less focused and had a mean distance of 80 bp upstream from the poly(A) site. These studies thus reveal a strong correlation between repression of a poly(A) site utilization subsequent to αCP depletion and the presence of a pyrimidine-pure and C-rich motif in close proximity to the site of 3′ processing.
αCP2 controls the 3′ processing of its own transcript.
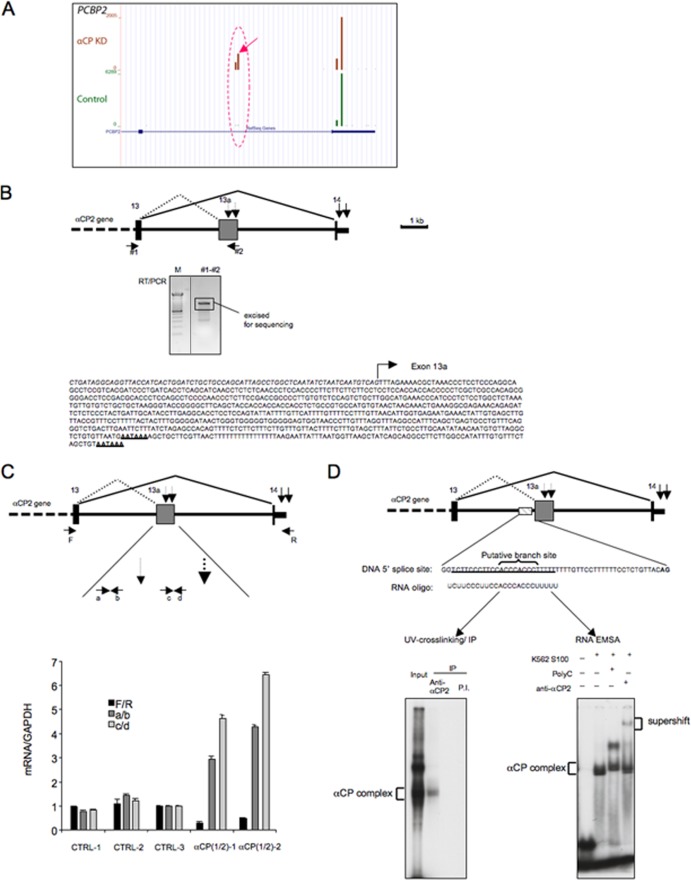
An unexpected observation from the APA analyses was that αCPs autoregulate the poly(A) selection of the αCP2 transcript. The codepletion of the two major αCP transcripts activated a set of two adjacent cryptic poly(A) sites within the last intron (intron 13) of the αCP2 RNA (Fig. 6A and B; dotted oval in the genome browser diagram). Both of these poly(A) sites are located immediately 3′ to a cryptic poly(A) signal, AATAAA. The use of these two sites was linked to the activation of a cryptic splice acceptor site upstream of these poly(A) sites, thus generating an mRNA with a unique 3′-terminal exon (exon 13a). Targeted RT-PCR analysis and 3′ RACE both confirmed the positioning of the novel 3′ processing sites within intron 13 and the generation of the new exon 13a (Fig. 6B). The generation of exon 13a subsequent to αCP depletion was accompanied by a decrease in the use of the poly(A) site in exon 14. This reciprocal relationship was validated by targeted real-time RT-PCR (Fig. 6C). The presence of a C-rich sequence approximately 40 nt upstream of the splicing acceptor site and overlapping the likely lariat branch site for this new intronic exon (13a) (Fig. 6D) may play a role in this alternative processing event. The absence of a C-rich motif near these new poly(A) sites and exon 14 poly(A) site supports the idea of a possible direct effect on alternative splicing rather than on poly(A) site utilization change at these two polyadenylation sites (40). The idea of a primary splicing mechanism is further supported by the interaction between this upstream C-rich element and αCP proteins, as evidenced by the RNA EMSA and UV-cross-linking assay (Fig. 6D). Thus, under normal conditions, the usage of the branch site encompassed by the C-rich motif may be repressed by bound αCPs. When αCP levels or activities are depleted from the cell, a new set of αCP2 mRNA isoforms is generated.
Fig 6.
αCP depletion alters 3′ processing of the αCP2 transcript. (A) Genome browser view of the DRS reads at the αCP2 locus. Comparison of 3′ processing site utilization in cells treated with αCP siRNAs (pooled, upper panels) and control siRNAs (pooled, lower panels). The y axis represents the number of read counts corresponding to each of the poly(A) sites. The two novel poly(A) sites observed in the αCP-depleted cells are encompassed in the dotted oval. (B) Activation of the two novel poly(A) sites in αCP mRNA subsequent to αCP depletion reflects linked alterations in splicing and 3′ processing. Exons 13 and 14 (terminal exon) of the αCP gene are shown in the diagram. The splicing patterns and the positions of the corresponding 3′ poly(A) termini detected in the control cells are indicated by the solid lines and the solid vertical arrows, respectively. (Upper panel) The alternative splicing and 3′ processing events that occur subsequent to αCP depletion are indicated by the corresponding set of dotted lines and dotted vertical arrows, respectively. (Middle panel) The RT-PCR analysis, shown in the gel image, represents amplification between primers 1 and 2. The amplified fragment (boxed) was excised and sequenced. The sequence confirmed the use of the novel splice acceptor site in exon 13a, with the two poly(A) signals highlighted (lower panel: the partial exon 13 sequence is shown in italics; the arrow indicates the starting point of new novel exon 13). (C) Real-time PCR analysis confirms the switch in terminal intron splicing and 3′ poly(A) site selection within the αCP2 transcript subsequent to αCP depletion. The amplimer generated between primers F and R assesses the canonical poly(A) usage, the amplimer generated between primers c and d assesses use of the distal novel poly(A) site within exon 13a, and the amplimer generated by primers a and b assesses overall levels of alternative splicing and 3′ processing with exon 13a. The assays were carried out on RNAs isolated from cells treated with each of the 3 distinct control siRNAs and with each of the two distinct αCP2-targeting siRNAs. Each real-time assay was normalized to the GAPDH amplicon. The ratio in the CTRL-3 sample is defined as 1.0. The standard deviation for each sample is shown (n = 3). (D) In vitro RNA-protein interaction assay. A 24-nt RNA oligonucleotide (shown below the diagram), encompassing the region immediately 5′ to exon 13a (dashed rectangle), was synthesized and 32P labeled. The DNA sequence 5′ to the splice site is also shown. (Left panel) The labeled RNA oligonucleotide was incubated with HeLa cell nuclear extract, UV-cross-linked, subjected to immunoprecipitation (IP) with anti-αCP2/KL, and resolved on an SDS-PAGE gel (4). The position of the αCP complex is defined by IP using anti-αCP2/KL antibody. (Right panel) The same 32P-labeled probe was subjected to RNA EMSA with poly(C) competition and anti-αCP supershift analysis using K562 cell S100 extract as described previously (40).
PA pattern changes impacted by αCPs.
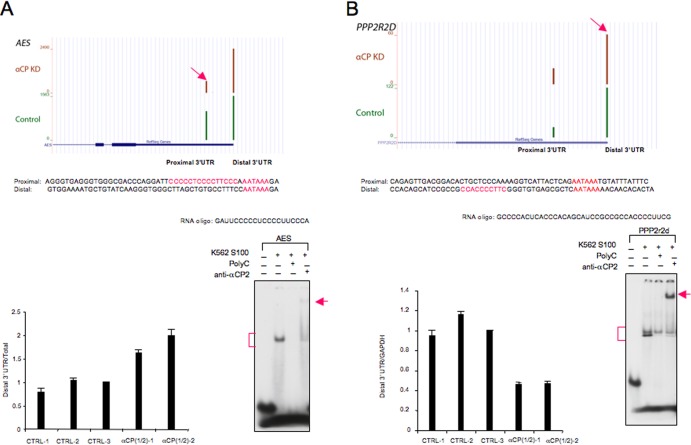
The preceding DRS analysis identified an enrichment for C-rich motifs 5′ to poly(A) sites that were downregulated subsequent to αCP depletion. These data were next validated by targeted real-time RT-PCR analyses. Six examples of mRNAs with an SE-APA pattern were assessed (Fig. 7; see also Fig. S3A to D in the supplemental material). In four of these transcripts, a C-rich motif preceded the more proximal of the competing alternative poly(A) sites, and for the remaining two transcripts, it preceded the more distal site of the competing sites. In each case, the poly(A) site located directly 3′ to the C-rich motif was repressed subsequent to αCP depletion. RNA EMSA and UV-cross-linking assays demonstrated that in each of these cases the C-rich motif in question binds αCPs (Fig. 7; see also Fig. S4 in the supplemental material). Direct confirmation of APA was also carried out on two mRNAs that undergo DE-APA. The real-time RT-PCR analysis confirmed the DE-APA events for the Ssu72 gene and NPM1 gene following αCP depletion (see Fig. S3E and F). Of note, both of these genes have been implicated in mRNA 3′-end processing regulation (49–51) (see also Discussion).
Fig 7.
RT-qPCR validations of APA events. A subset of APA events identified in the DRS analysis was independently assessed by targeted RT-qPCR. Each set of DRS data is shown in the context of the genome browser diagram of the respective locus. The positions of the proximal 3′UTR and the distal 3′UTR are marked below the browser view. The arrow indicates the position of the site of reduced polyadenylation site usage triggered by αCP depletion. All the real-time RT-PCR quantifications were normalized to GAPDH mRNA and are presented as ratios versus CTRL-3 (cyclophilin siRNA) defined as 1. The standard deviation for each sample is shown (n = 3). (A and B) Examples of APA involving competing PA sites within the same terminal exon (SE-APA). KD, knockdown. (A) Amino-terminal enhancer of split (AES) gene. (B) Protein phosphatase 2, subunit B, isoform delta (PPP2r2d) gene. The sequences encompassing and 5′ to each APA sites are shown, and the C-rich sequences (putative αCP binding sites) and the alternative poly(A) signals (AAUAAA) are highlighted in red. These two examples of mRNAs with the SE-APA pattern were confirmed by real-time RT-PCR. Panel A shows the analysis of a transcript with a C-rich motif preceding the proximal alternative poly(A) site; the decreased usage of the proximal poly(A) site subsequent to αCP depletion was confirmed and demonstrated in the individual histogram and is presented as the elevated ratio of the distal 3′UTR isoform [i.e., use of the distal poly(A) site] relative to the total poly(A) site usage (as described in reference 54). Panel B shows a transcript with a C-rich motif preceding the distal site of the alternative poly(A) sites; the decreased usage of the distal poly(A) site was confirmed and is shown in the individual histogram, simply presented as the ratio of the distal 3′UTR isoform [i.e., use of the distal poly(A) site] to control GAPDH mRNA. All the real-time RT-PCR quantifications were normalized to GAPDH mRNA and are presented as a ratio versus CTRL-3 (cyclophilin siRNA) defined as 1.0. The standard deviation for each sample is shown (n = 3). RNA EMSAs were performed on 32P-labeled RNA probes corresponding to each C-rich segment in the tested genes to confirm αCP complex assembly. RNA oligonucleotide sequences are shown above the EMSA data. Brackets indicate the αCP complex, and the arrows indicate the supershift that occurred when anti-αCP antibody was included in the reaction.
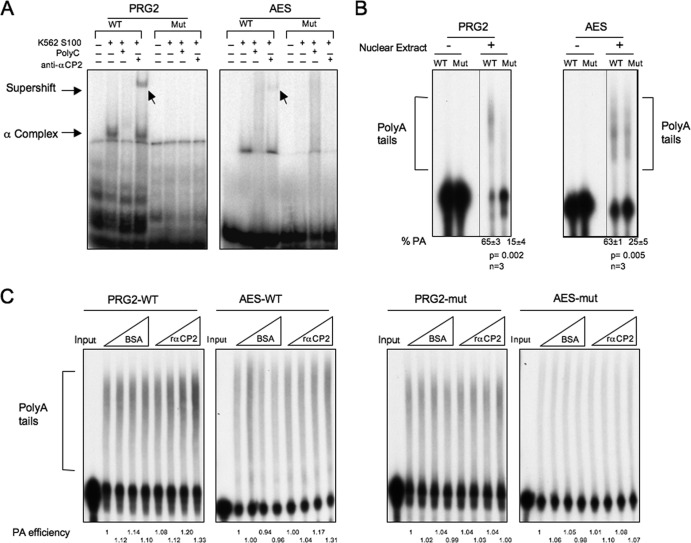
Our prior studies of hα-globin transcript processing demonstrated a direct impact of the 3′UTR C-rich domain and its bound αCP on the recruitment and activity of the 3′ processing machinery. In that setting, αCPs assemble on the nascent transcript at the hα-globin locus and enhance transcript processing, and bound αCP is then coexported to cytoplasm where it regulates cytoplasmic hα-globin mRNA stability. The current data support the idea of an enhancement of mRNA expression by the 3′UTR C-rich determinant of a broad array of transcripts and directly support the idea of a role for this complex as a USE of 3′ processing. To further support these findings and conclusions, we assessed the impact of the C-rich element on in vitro polyadenlyation. We chose to test the function of C-rich determinants in this assay in two representative transcripts: the PRG2 (3′UTR) as an example of an mRNA whose steady-state levels are repressed in cells depleted of αCP (Fig. 3) and the AES 3′UTR as an example of an mRNA that undergoes SE APA impacted by αCP depletion (Fig. 7). In both cases, we observed that the C-rich determinant immediately 5′ to the αCP-impacted poly(A) site binds αCP (Fig. 8A; EMSA) and supports polyadenylation [Fig. 8B; in vitro poly(A) assay]. In both cases, the mutation of the C-rich determinants ablates αCP binding and diminishes the conversion of corresponding RNA template to a polyadenylated form (Fig. 8A and B). Furthermore, the addition of recombinant αCP2 to the reaction increases the level of polyadenylation of wild-type (WT) substrates but not that of the mutants in which the C-rich motif was disrupted (Fig. 8C). As an additional control, the addition of a comparable amount of bovine serum albumin (BSA) to the in vitro reaction had no effect on poly(A) addition. These two examples further support the idea of the activity of the C-rich determinant as a USE of 3′ processing and as a mediator of APA.
Fig 8.
In vitro polyadenylation assay. 32P-labeled RNA substrates representing the poly(A) addition sites for the PRG2 and AES mRNAs were synthesized by in vitro transcription. In each case, the wild-type (WT) sequence or a sequence of a corresponding substrate containing a mutation of the C-rich region (Mut) was synthesized. (A) EMSA was performed to determine the efficiency of αCP assembly on the WT and Mut templates (as described for Fig. 7). (B) In vitro polyadenylation assay. The 32P-labeled templates were added to an in vitro polyadenylation reaction mixture (HeLa nuclear extract), and the products were resolved on a denaturing polyacrylamide gel. The poly(A) tails were added to the template (bracketed) in the presence (+) but not the absence (−) of the nuclear extract. The polyadenylation efficiency was calculated as the ratio of labeled RNA in the poly(A) tail region divided by total RNA activity (polyadenylated plus remaining substrate). These values (% PA) are shown at the bottom of the respective lanes. Standard deviations, P values, and numbers of repeats (n) are shown below the respective gels. (C) αCP proteins increase the efficiency of polyadenylation in vitro. The WT (left) and mutant (right) PRG2 and AES RNA substrates were incubated with HeLa cell nuclear extract (as described for panel B). Increasing equivalent amounts (0 μg, 0.3 μg, 0.6 μg, and 1.2 μg) of BSA or recombinant αCP2 were added to the indicated reaction mixtures. The reactions were terminated after a 90-min incubation, and the RNAs were analyzed on 6% denaturing PAGE for the addition of poly(A) tails. The PA efficiency, the ratio of labeled RNA in the poly(A) tail region divided by total RNA activity (polyadenylated plus remaining substrate), was quantified as indicated below the respective lanes (the activity at the lowest level of BSA for each set of reactions was defined as 1.0).
Taken together, these studies demonstrated that αCP proteins, via interactions with C-rich RNA elements, impact on alternative poly(A) site choices. These observations are summarized in a model in Fig. 9.
Fig 9.
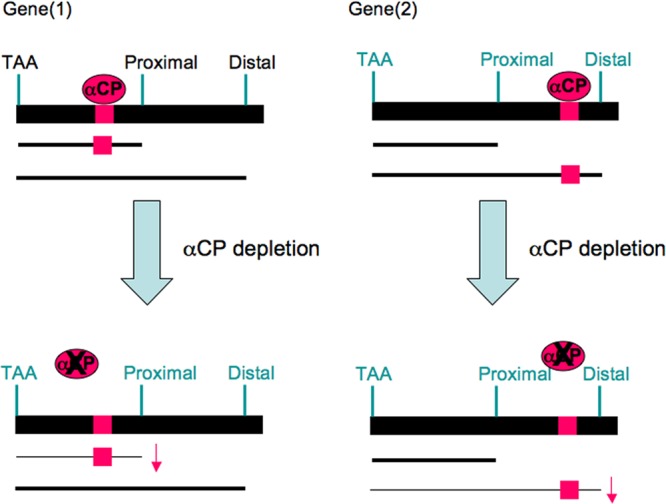
Impact of αCPs on expression and alternative polyadenylation of Pol II transcripts. Based on our analysis of 3′ processing of the hα-globin transcript (4) and on the present genome-wide analysis, we propose that αCPs can act as general regulators of 3′ processing. The αCP RNP complex recruits core components of the 3′-end processing machinery to a defined subset of human transcripts containing cognate C-rich binding motifs situated in proximity to poly(A) signals. This RNP complex serves as an upstream sequence element (USE) enhancer of 3′ cleavage and polyadenylation (4). This enhancement of 3′ processing can increase the net levels of steady-state mRNA and/or alter the pattern of poly(A) site selection. Physiological (59) and/or pathological (60) shifts in the levels or biological activities of αCP can result in major alterations in the transcriptome by altering steady-state levels of subsets of mRNAs and/or by shifts in the relative utilization of alternative poly(A) sites.
DISCUSSION
We previously demonstrated that αCPs enhance 3′ processing of the hα-globin transcript via binding to a C-rich motif in the 3′UTR. These findings led us to conclude that the poly(C) motif in the hα-globin 3′UTR acted as a USE that enhances 3′ processing of the hα-globin transcript (4). The current findings support and extend the idea of a role of αCPs in the control of 3′-end processing by documenting their broad impact on steady-state levels and poly(A) site utilization of mRNAs within the human transcriptome. These data specifically identify a subset of mRNA transcripts in which the enhancement of 3′ processing is tightly linked to the presence of the cognate C-rich binding sites in close proximity to a poly(A) signal.
A global relationship of 3′ processing to gene regulation has been highlighted by a number of recent studies (13, 52). Alterations in levels of general factors and complexes involved in 3′ processing, such as CPSF, CSTF, and the nuclear poly(A)-binding protein PABPN1, can impact on poly(A) site selection and the efficiency of poly(A) addition (53). In addition, particular RNA-binding proteins have the capacity to impact on the 3′ processing of specific transcripts or groups of transcripts. For example, the KH-domain-binding protein Nova2, a protein closely related to αCP, exerts controls over poly(A) site choices in a position-dependent manner (54). Polypyrimidine tract binding (PTB) has been implicated in the enhancement of 3′-end processing of several genes (55, 56) via stimulating hnRNP H binding to a G-rich binding sites (24). This pathway appears to have a global role in alternative poly(A) site selection (57). Likewise, recent genome-wide surveys have revealed that alterations in the levels of the epithelium-specific splicing regulatory proteins (ESRPs) can trigger widespread shifts in polyadenylation patterns (58).
In the current study, we demonstrated that αCP proteins are actively involved in the determination of mRNA expression levels and alternative polyadenylation. We observed that αCP depletion from the cell represses the steady-state levels of substantially more mRNA than are increased. This result was consistent with the known enhancing action of the αCP complex for a number of regulatory steps that determine steady-state mRNA levels (27, 30, 38). The impact of αCPs on nuclear functions, and in particular on splicing and poly(A) activity, is likely to also include roles in influencing how much of the mRNA is generated and exported to the cytoplasm. Importantly, these control pathways are likely to be mechanistically interrelated. We have demonstrated in the case of the hα-globin gene expression that the αCP complex assembles on the nascent hα-globin transcript in the nucleus and appears to travel on the mRNA to the cytoplasm, where it stabilizes the mRNA (46). Thus, the nuclear and cytoplasmic pathways are linked and may coordinate overall levels of gene expression and protein production. Future studies will determine whether αCPs regulate mRNA steady-state level through other defined mechanisms as well. For example, APA may have an impact on mRNA steady-state levels by inclusion or exclusion of miRNA target sites and/or additional RNA-binding protein-binding sites in the mRNA products.
The current data directly support a model in which alterations in αCP protein availability can regulate alternative poly(A) site utilization choices in a subset of PolII transcripts by interacting with C-rich sequences. The idea of this involvement of αCP in the global control of 3′ processing is supported by the results of a recent general screen for proteins involved in 3′ processing (53). We observed that the formation of an αCP RNP complex near the poly(A) sites (either proximal or distal) enhances useage of the corresponding poly(A) site (reference 4 and current data). Following αCP depletion, the AES, Get 4, CDK16, and SHMT2 transcripts undergo a decrease in their usage of proximal poly(A) sites and shift to the distal poly(A) sites (Fig. 7; see also Fig. S3 in the supplemental material). These four genes all have C-rich motifs closely located upstream of their proximal poly(A) site. In a reciprocal fashion, the depletion of αCP results in decreased usage of the distal poly(A) sites and a shift to the proximal poly(A) site in the CSTF1 and PPP2r2d transcripts. In agreement with the model that the C-rich USE enhances poly(A) site activity (Fig. 9), we identified C-rich motifs 5′ to the distal poly(A) sites in both of those transcripts. The identification of the C-rich motif 5′ of the repressed sites by MEME analysis is in accord with the definition of what constitutes an αCP binding site as defined in prior analyses of mRNAs targeted by αCP (4, 28, 37, 40, 59, 60) and with αCP binding features as determined by in vitro SELEX (35). Consistent with this role, we show that these C-rich motifs interact with αCP proteins (Fig. 7; see also Fig. S4 in the supplemental material) and that the addition of recombinant αCP to an in vitro polyadenylation reaction enhances poly(A) addition to substrates containing the C-rich binding site motif (Fig. 8). From these complementing lines of evidence, we conclude that the binding of αCP to a C-rich motif acts as a potent USE enhancer of 3′ processing.
It should be noted that a significant number of mRNAs have altered poly(A) site utilization in the absence of the C-rich motif. This may represent secondary effects of αCP depletion. As revealed in the current study, αCP depletion can result in changes in the expression levels and/or structures of mRNAs that encode RNA-binding proteins and components of RNA processing machinery, such as NPM1, Ssu72, and CSTF1 (this study) and CPSF1, SF3A2, CSTF3, hnRNPC, and hnRNP LL (our unpublished data). One can imagine that these changes will indirectly alter APA patterns of a subset of genes, although elucidation of the exact mechanism must await future studies.
It is interesting that there was only a small overlap between the mRNAs that changed significantly in their overall steady-state levels (DGE values) and those that were impacted by significant alterations in their poly(A) site utilization (APA patterns). When the DEG mRNAs with greater than 2-fold changes and APA data sets were compared, only 7 (the αCP2, ACOT2, ACSM3, SLC6A6, PRG2, C3orf75, and C1orf86 genes) of 117 genes were present in both categories. When the more inclusive 1.5-fold change in DGE was used for the comparison, we observed only 29 of the 586 genes in both categories. It is clear that a reciprocal switch between two sets of alternative poly(A) sites need not result in a significant net change in mRNA abundance. The main impact of such a switch may instead reflect alterations in translational activity and/or protein coding content. Thus, the data suggest that the impact of αCP depletion on steady-state levels for many of the genes studied is likely to reflect substantial changes in the efficiency of 3′ processing at a unique poly(A) site with consequent alterations in mRNA production rather than triggering of an APA event.
How might the role of an αCP/poly(C) complex as a USE enhancer of 3′ processing relate to known physiological and pathophysiological processes? The current study revealed that αCPs can autoregulate poly(A) site selection on the αCP2 transcript. This finding is in general agreement with multiple observations of autoregulatory control over expression of RNA-binding proteins (61). These new αCP2 poly(A) sites were generated secondary to an alternative splicing event mediated by the shift in αCP levels (40). The newly generated αCP2 mRNA is predicted to encode a protein that is structurally similar to the αCP4 protein, which has been implicated in apoptosis regulation (62). Future work will determine whether this novel αCP2 isoform plays a similar role under some circumstances.
αCPs are considered to be ubiquitously expressed and are linked to a variety of activities (33). αCP expression and function (RNA-binding activity [52, 63, 64]) can be significantly impacted by defined physiological and pathological events. These include environmental stress (48), cell transformation (65), differentiation (59), chronic myeloid leukemia (60), and epithelial-mesenchymal transdifferentiation (EMT) during the development and metastatic progression of tumors (63). The changes can be in the overall levels of αCPs or in their RNA-binding activities. It is well established, for example, that phosphorylation of αCP can have a dramatic impact on its RNA-binding activity and biological functions (59, 63, 66). On the basis of our current study, it is likely that these changes of levels and/or RNA-binding activity of αCP proteins will have a broad effect on the 3′ processing efficiency and choice of poly(A) signals, with a consequent global impact on the cellular transcriptome.
In summary, the present report reveals that the αCP RNA-binding proteins play an important role in the 3′-end processing of a subset of genes and that this effect is mediated by the USE function of the αCP RNP complex. Combining our recent studies on human α-globin mRNA (4) with the current work, we propose that αCP complexes assemble on target RNAs cotranscriptionally and that the nucleus-assembled α-complexes impact on nuclear processing of the transcript and are subsequently retained on the mature mRNAs. The assembled mRNP is then exported to the cytoplasm, where αCP impacts on cytoplasmic events. In this way, αCPs effectively link nuclear transcript processing and cytoplasmic mRNA metabolism and impact on gene expression in a global and multifaceted manner. The current report defines 3′ processing as an integral step in this pathway of αCP-mediated gene regulation.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the generosity of laboratory members for sharing various reagents and thoughts.
This work was supported by NIH MERIT HL 65449 and CA72765 (S.A.L.), NIDDK T32-DK007780 Hematopoiesis Training Grant (M.V.), and a junior faculty grant from the Edward Mallinckrodt, Jr., Foundation to Y.X.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 29 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01380-12.
REFERENCES
- 1. Keene JD. 2010. Minireview: global regulation and dynamics of ribonucleic acid. Endocrinology 151:1391–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore MJ. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514–1518 [DOI] [PubMed] [Google Scholar]
- 3. Glisovic T, Bachorik JL, Yong J, Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582:1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ji X, Kong J, Liebhaber SA. 2011. An RNA-protein complex links enhanced nuclear 3′ processing with cytoplasmic mRNA stabilization. EMBO J. 30:2622–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao W, Blagev D, Pollack JL, Erle DJ. 2011. Toward a systematic understanding of mRNA 3′ untranslated regions. Proc. Am. Thorac. Soc. 8:163–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755–2766 [DOI] [PubMed] [Google Scholar]
- 7. Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43:853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsin JP, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26:2119–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsin JP, Sheth A, Manley JL. 2011. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 334:683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richard P, Manley JL. 2009. Transcription termination by nuclear RNA polymerases. Genes Dev. 23:1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tian B, Hu J, Zhang H, Lutz CS. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33:201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi Y. 2012. Alternative polyadenylation: new insights from global analyses. RNA 18:2105–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320:1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U. S. A. 106:7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilmartin GM. 1997. In vitro analysis of mammalian cell mRNA 3′ processing, p79–98 In Richter JD. (ed), mRNA formation and function. Academic Press, New York, NY [Google Scholar]
- 17. Moore CL, Sharp PA. 1985. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell 41:845–855 [DOI] [PubMed] [Google Scholar]
- 18. Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65:1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. 2011. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol. Cell 41:409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33:365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Millevoi S, Vagner S. 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38:2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. 2012. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc. Natl. Acad. Sci. U. S. A. 109:18773–18778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Millevoi S, Decorsiere A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, Vagner S. 2009. A physical and functional link between splicing factors promotes pre-mRNA 3′ end processing. Nucleic Acids Res. 37:4672–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Danckwardt S, Hentze MW, Kulozik AE. 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 27:482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brackenridge S, Proudfoot NJ. 2000. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol. Cell. Biol. 20:2660–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ji X, Kong J, Liebhaber SA. 2003. In vivo association of the stability control protein alphaCP with actively translating mRNAs. Mol. Cell. Biol. 23:899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiledjian M, Wang X, Liebhaber SA. 1995. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 14:4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kong J, Ji X, Liebhaber SA. 2003. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell. Biol. 23:1125–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kong J, Liebhaber SA. 2007. A cell type-restricted mRNA surveillance pathway triggered by ribosome extension into the 3′ untranslated region. Nat. Struct. Mol. Biol. 14:670–676 [DOI] [PubMed] [Google Scholar]
- 31. Weiss IM, Liebhaber SA. 1994. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol. Cell. Biol. 14:8123–8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiss IM, Liebhaber SA. 1995. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3′ nontranslated region. Mol. Cell. Biol. 15:2457–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makeyev AV, Liebhaber SA. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. 1999. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol. Cell. Biol. 19:4572–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thisted T, Lyakhov DL, Liebhaber SA. 2001. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest distinct modes of RNA recognition. J. Biol. Chem. 276:17484–17496 [DOI] [PubMed] [Google Scholar]
- 36. Chaudhury A, Chander P, Howe PH. 2010. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16:1449–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holcik M, Liebhaber SA. 1997. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. U. S. A. 94:2410–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Waggoner SA, Liebhaber SA. 2003. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol. Cell. Biol. 23:7055–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waggoner SA, Liebhaber SA. 2003. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. (Maywood) 228:387–395 [DOI] [PubMed] [Google Scholar]
- 40. Ji X, Kong J, Carstens RP, Liebhaber SA. 2007. The 3′ untranslated region complex involved in stabilization of human alpha-globin mRNA assembles in the nucleus and serves an independent role as a splice enhancer. Mol. Cell. Biol. 27:3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143:1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. 2009. Direct RNA sequencing. Nature 461:814–818 [DOI] [PubMed] [Google Scholar]
- 43. Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 21:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang L, Feng Z, Wang X, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138 [DOI] [PubMed] [Google Scholar]
- 45. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee JY, Yeh I, Park JY, Tian B. 2007. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 35:D165–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ghosh D, Srivastava GP, Xu D, Schulz LC, Roberts RM. 2008. A link between SIN1 (MAPKAP1) and poly(rC) binding protein 2 (PCBP2) in counteracting environmental stress. Proc. Natl. Acad. Sci. U. S. A. 105:11673–11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sagawa F, Ibrahim H, Morrison AL, Wilusz CJ, Wilusz J. 2011. Nucleophosmin deposition during mRNA 3′ end processing influences poly(A) tail length. EMBO J. 30:3994–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xiang K, Manley JL, Tong L. 2012. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev. 26:2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. 2010. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467:729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perrotti D, Calabretta B. 2004. Translational regulation by the p210 BCR/ABL oncoprotein. Oncogene 23:3222–3229 [DOI] [PubMed] [Google Scholar]
- 53. Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, Rubinsztein DC, Agami R. 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149:538–553 [DOI] [PubMed] [Google Scholar]
- 54. Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456:464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hall-Pogar T, Liang S, Hague LK, Lutz CS. 2007. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA 13:1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. 1998. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 12:2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7:1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. 2012. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell. Biol. 32:1468–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Naarmann IS, Harnisch C, Flach N, Kremmer E, Kuhn H, Ostareck DH, Ostareck-Lederer A. 2008. mRNA silencing in human erythroid cell maturation: heterogeneous nuclear ribonucleoprotein K controls the expression of its regulator c-Src. J. Biol. Chem. 283:18461–18472 [DOI] [PubMed] [Google Scholar]
- 60. Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, Iervolino A, Condorelli F, Gambacorti-Passerini C, Caligiuri MA, Calabretta B. 2002. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat. Genet. 30:48–58 [DOI] [PubMed] [Google Scholar]
- 61. Buratti E, Baralle FE. 2011. TDP-43: new aspects of autoregulation mechanisms in RNA binding proteins and their connection with human disease. FEBS J. 278:3530–3538 [DOI] [PubMed] [Google Scholar]
- 62. Zhu J, Chen X. 2000. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M. Mol. Cell. Biol. 20:5602–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. 2010. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12:286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Perrotti D, Calabretta B. 2002. Post-transcriptional mechanisms in BCR/ABL leukemogenesis: role of shuttling RNA-binding proteins. Oncogene 21:8577–8583 [DOI] [PubMed] [Google Scholar]
- 65. Molinaro RJ, Jha BK, Malathi K, Varambally S, Chinnaiyan AM, Silverman RH. 2006. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucleic Acids Res. 34:6684–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chang JS, Santhanam R, Trotta R, Neviani P, Eiring AM, Briercheck E, Ronchetti M, Roy DC, Calabretta B, Caligiuri MA, Perrotti D. 2007. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP-E2 dependent suppression of C/EBPalpha-driven myeloid differentiation. Blood 110:994–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.