Abstract
The mitogen-activated protein kinase (MAPK)-activated protein kinases 2 and 3 (MK2/3) represent protein kinases downstream of the p38 MAPK. Using MK2/3 double-knockout (MK2/3−/−) mice, we analyzed the role of MK2/3 in cross-striated muscle by transcriptome and proteome analyses and by histology. We demonstrated enhanced expression of the slow oxidative skeletal muscle myofiber gene program, including the peroxisome proliferator-activated receptor gamma (PPARγ) coactivator 1α (PGC-1α). Using reporter gene and electrophoretic gel mobility shift assays, we demonstrated that MK2 catalytic activity directly regulated the promoters of the fast fiber-specific myosin heavy-chain IId/x and the slow fiber-specific sarco/endoplasmic reticulum Ca2+-ATPase 2 (SERCA2) gene. Elevated SERCA2a gene expression caused by a decreased ratio of transcription factor Egr-1 to Sp1 was associated with accelerated relaxation and enhanced contractility in MK2/3−/− cardiomyocytes, concomitant with improved force parameters in MK2/3−/− soleus muscle. These results link MK2/3 to the regulation of calcium dynamics and identify enzymatic activity of MK2/3 as a critical factor for modulating cross-striated muscle function by generating a unique muscle phenotype exhibiting both reduced fatigability and enhanced force in MK2/3−/− mice. Hence, the p38-MK2/3 axis may represent a novel target for the design of therapeutic strategies for diseases related to fiber type changes or impaired SERCA2 function.
INTRODUCTION
Cross-striated heart and skeletal muscles comprise on average almost half of mammalian body mass. Each muscle cell is characterized by a regular arrangement of contractile proteins in an extraordinary degree of order termed sarcomeres. These structures are optimized for both constant and intermittent movement. Proteins must be selectively replaced in response to altered physiological demands (1, 2). Hence, skeletal muscle creates a highly adaptive tissue that is classified on the basis of the expression of myosin heavy-chain (MyHC) isoforms and speed of contraction (3). Slow type I fibers are rich in MyHC isoform I/β (MyHCI/β) and mitochondria, their metabolism is oxidative leading to fatigue resistance, while fast type IIb fibers are glycolytic with few mitochondria and rich in MyHCIIb. Type IIa and IId/x fibers comprise an intermediate oxidative/glycolytic fast phenotype. Adaptive changes by endurance exercise shifts fiber composition to increased oxidative and endurance capacity (4), while muscle inactivity switches fibers to more glycolytic types (5). Fiber type shifts arise further during ageing and diseases, such as type 2 diabetes, and even obesity (1, 6). The cardiac muscle also undergoes adaptive changes in gene expression and performance in response to physiological and pathological stimuli, and several protein kinases, such as extracellular signal-regulated kinases (ERKs) and p38 mitogen-activated protein kinases (MAPKs), have been shown to be involved in these processes (7, 8).
p38 MAPK and the downstream MAPK-activated protein kinases 2 and 3 (MAPKAPK2/3 [MK2/3]) are highly abundant in skeletal muscle and in the heart (9–12). Persistent activation of the p38 pathway occurs early during skeletal muscle differentiation (13), and a key role for p38 in myogenesis and regeneration has been demonstrated (14, 15). Several studies have demonstrated an activation of the p38 signaling pathway by muscle contractile activity (16). Interestingly, total and activated p38 is decreased in endurance-trained versus untrained skeletal muscle (16). Whether the downregulation of p38 signaling is correlated with the maintenance of adaptive processes and improved endurance work capacity is not known, and a possible role of MK2/3 remains to be elucidated. In the heart, chronically activated p38 has been implicated in a wide spectrum of cardiac pathologies (8).
MK2 and MK3 share activators and substrates. They have similar physiological functions possibly reflecting a certain functional congruence (17). Apart from their role in substrate phosphorylation, MK2/3 bind to and stabilize p38α (18). In most cells and tissues, MK3 expression is minor compared to MK2 expression. The phenotype of MK2-deficient mice indicates an essential role of MK2 in posttranscriptional regulation of the biosynthesis of cytokines (19). In contrast to p38, MK2 and MK3 basal activity levels and functions in cross-striated muscles remain unclear. Some aspects of p38-induced cardiomyopathy are mediated by MK2, including the posttranscriptional regulation of the proinflammatory protein cyclooxygenase 2 (COX-2) (20). However, a direct role of MK2/3 activity and precise molecular mechanisms underlying the reported effects in cardiac muscle are not clearly defined. The role of MK2/3 in skeletal muscle function is yet unknown.
The sarco-endoplasmic reticulum Ca2+-ATPase 2a (SERCA2a) is responsible for Ca2+ reuptake into the sarcoplasmic reticulum (SR) (21) and directly linked to contractility and alterations in excitation-contraction coupling. Thus, impaired Ca2+ reuptake resulting from decreased abundance and reduced activity of SERCA2a is a hallmark of heart failure (22). An associated regulatory protein of SERCA2a is phospholamban (PLB), expressed in slow-twitch fibers and cardiomyocytes. SERCA2a is inhibited by dephosphorylated PLB, while phosphorylation of PLB by protein kinase A (PKA) or by Ca2+/calmodulin-dependent protein kinase II (CaMKII) relieves inhibition and increases Ca2+ reuptake (23).
Here, we utilized an MK2/MK3 double-knockout (DKO or MK2/3−/−) mouse model (18) to examine functional roles of MK2/3 in muscle. DKO soleus muscles displayed a shift toward the slow skeletal muscle myofiber gene program. This included increased expression of peroxisome proliferator-activated receptor gamma (PPARγ) coactivator 1α (PGC-1α), a master regulator of oxidative energy metabolism (24, 25). Furthermore, we identified SERCA2 and MyHCIId/x as novel MK2/3 target genes. The increased SERCA2a gene expression in MK2/3−/− cells can be directly attributed to MK2 catalytic activity via reduced binding of the transcription factor Egr-1 and is associated with faster relaxation and increased contractility of MK2/3-deficient cardiomyocytes. Thus, results link MK2/3 to the regulation of calcium dynamics generating a unique phenotype exhibiting both reduced fatigability and enhanced force. Hence, this study indicates an essential role of the p38-MK2/3 pathway in the regulation of cross-striated muscle performance.
MATERIALS AND METHODS
MK2−/− and MK2/3−/− mice.
The generation of mice deficient for MK2 and for MK2/3 has been described previously (18, 19). MK2/3−/− mice have a mixed genetic background (129Sv × C57BL/6).
Cell culture and transfection assays.
Mouse C2C12 cells were cultured and transfected as described previously (26). MK2/3-deficient immortalized mouse embryonic fibroblasts (MEFs) were generated as reported earlier (18). MEFs were transiently transfected at 50 to 80% confluence. Nonmodified branched polyethylenimine (Sigma-Aldrich, Taufkirchen, Germany) (4.5 μl of a 1-mg/ml solution) was added to a mixture of 1.5 μg DNA and 90 μl serum- and antibiotic-free Dulbecco modified Eagle medium (DMEM), with the subsequent addition of 900 μl serum- and antibiotic-free DMEM (27). Cells were transfected using 1,000 ng of one of the following promoter firefly luciferase reporter gene constructs: a human SERCA2 promoter fragment extending from bp −2577 to +321 (−2577 fragment; +1 indicates the transcription start site), one of several SERCA2 promoter deletion mutants, a porcine MyHCIId/x promoter fragment extending from bp −2742 to +9 (−2.8 fragment) (26), a rabbit MyHCI/β promoter fragment extending from bp −2345 to +99 (−2.4 fragment) (26), or a murine PGC-1α promoter fragment extending from bp −3036 to +119 (−3.0 fragment). In some experiments cells were also cotransfected with one or several of the following plasmids, as indicated in the figures: 750 ng pcDNA3-myc-MK2, 750 ng MK2K76R, 250 ng pcDNA3-MKK6EE, 500 ng pcDNA3.1(-)myc-His-Erg-1, and/or an empty vector(s). Promoter reporter constructs containing deletion mutants of the human SERCA2 promoter fragments (extending from bp −2577, −1741, −412, −263 [−263 fragment], or −68 [−68 fragment] to +321) inserted into pGL2 basic were described previously (28, 29). The expression vectors for wild-type MK2 (pcDNA3-myc-MK2), mutant catalytic-dead, p38α stabilizing MK2, MK2K76R (pcDNA3-myc-MK2K76R), and constitutively active MK2, MK2EE (pcDNA3.1-MK2EE), were reported earlier (30, 31). Constitutively active MKK6, MKK6EE (pcDNA3-MKK6EE) (32), was a gift from R. Davis. In a dual-luciferase reporter gene assay, firefly luciferase activities were normalized for transfection efficiency using Renilla activity. Firefly luciferase reporter gene assays were performed as described previously (27). Renilla activity was measured by using a Glomax microplate reader (Promega, Mannheim, Germany) and 20 μl of lysate, injecting 100 μl of 1× Renilla substrate buffer (100 mM phosphate buffer [pH 7.8], 1 mM EDTA, 0.5 M NaCl, 4 μM coelenterazine [Promega]) into the microplate reader, and reading for 10 s after a 2-s delay. In other experiments, cells were cotransfected with pCMV-Gal (CMV stands for cytomegalovirus, and Gal stands for β-galactosidase) as an internal reference. The β-galactosidase activity was determined as described previously (26). A c-Myc-tagged Egr-1 expression vector was generated by cloning PCR-generated murine Egr-1 cDNA into pcDNA3.1(-)-myc-His (Invitrogen, Karlsruhe, Germany) using the forward (F) 5′-CTG TCA GAA TTC ATG GCA GCG GCC AAG GCC GAG-3′ and reverse (R) 5′-CTG TCA AAG CTT GCA AAT TTC AAT TGT CCT GGG-3′ primers. To generate a −263 SERCA2 promoter construct mutated at putative Sp1 and Egr-1 binding sites A, B, and/or C, nucleotides GAGG were changed to TATA (A and B) and nucleotides CGCC were changed to TATA (C) as indicated in Fig. 5F, by using the Phusion site-directed mutagenesis kit (New England BioLabs GmbH, Frankfurt, Germany) according to the manufacturer's instructions. A −3.0 promoter reporter construct of the murine PGC-1α was generated with the F 5′-CTG TCA ACG CGT GTC TGT CAG CCC TTG TAA TGT G-3′ and R 5′-CTG TCA AGA TCT CCA GCT CCC GAA TGA CGC CAG TCA A-3′ primer pair.
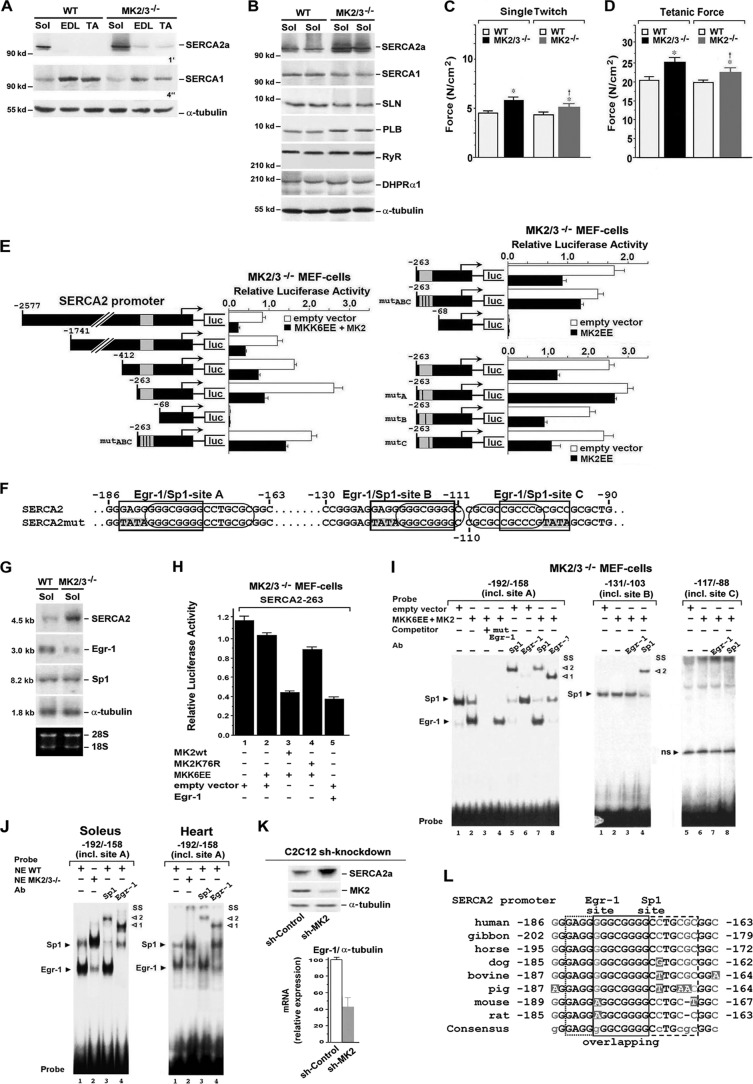
Fig 5.
MK2/3 ablation and shMK2 knockdown increased SERCA gene expression via reduced MK2 activity-dependent Egr-1 function. (A and B) Western blot analysis of proteins involved in Ca2+ homeostasis as indicated, from whole-cell lysates of soleus (Sol), EDL, and TA muscles from WT and MK2/3−/− mice. The exposure times (4 s and 1 min) for enhanced chemiluminescence detection are indicated. α-Tubulin was used as a loading control. (C and D) Single twitch (C) and tetanic (D) force parameter in MK2/3−/− soleus. Soleus muscles were stimulated under isometric conditions. The length and weight (wet weight) of each muscle were determined to estimate specific force (N/cm2). In panels C and D, from left to right, the numbers of WT and mutant mice were as follows: for WT, n = 30; for MK2/3−/−, n = 25; for WT, n = 14; for MK2−/−, n = 12. Values that were significantly different (P < 0.05) from the values for WT mice are indicated by an asterisk, while values that were significantly different (P < 0.05) from the values that were significantly different from the values for MK2/3−/− mice are indicated by a dagger. (E) Reporter gene analysis of a −2577 SERCA2 promoter firefly luciferase (luc) construct, of deletion constructs as indicated, and of −263 SERCA2 constructs mutated in the putative Egr-1 sites of the overlapping Egr-1/Sp1 region A, and/or B, and/or C (mutA, mutB, mutC, and mutABC [see panel F]), in MK2/3−/− MEFs. The cells were cotransfected as indicated. (F) Putative overlapping binding regions A, B, and C for Egr-1 (rectangles) and Sp1 (ovals) within the −263 SERCA2 gene sequence (mutated sequences indicated by a gray background in SERCA2mut). (G) Northern blot analysis of SERCA2a, Egr-1, and Sp1 mRNA expression in soleus (Sol) muscles from WT and MK2/3−/− mice. α-Tubulin was used as a loading control. Ethidium bromide-stained 18S and 28S rRNA is shown in the bottom blot. (H) Reporter gene analysis of a −263 SERCA2 promoter firefly luciferase construct in MK2/3−/− MEFs. The cells were cotransfected as indicated below the bars. In addition, cells were cotransfected with a Renilla luciferase-encoding plasmid. The promoter activities determined on the basis of firefly luciferase activities (E and H) were normalized for transfection efficiency with Renilla luciferase activity (mean ± SD of triplicate data points). (I and J) EMSAs with radiolabeled oligonucleotide probes as indicated containing the bp −184 to −166, the bp −123 to −111, or the bp −110 to −96 putative overlapping Egr-1/Sp1 binding regions (sites A, B, and C). Nuclear extracts from MK2/3−/− MEFs transfected as indicated (I) or from WT or MK2/3−/− soleus or heart (J). SS, supershift, with the antibody (Ab) indicated. Arrowheads to the right of the blots indicate complex formation (1, with Egr-1; 2, with Sp1); ns, nonspecific. (K) Western blot analysis of SERCA2a and MK2 abundance (α-tubulin was used as the loading control) and qPCR analysis of Egr-1 mRNA expression, normalized to α-tubulin transcript levels, in shRNA-based MK2 knockdown C2C12 muscle cells. The Egr-1/α-tubulin mRNA ratio in cells transduced with sh-control particles was set at 100 (arbitrary units) (mean ± SEM) (n = 4). (L) Sequence alignment of SERCA2 gene upstream regulatory regions from several vertebrate species to the −186 to −163 region of the human SERCA2 promoter including the functional Erg-1 site of the Egr-1/Sp1 overlapping site A. Black letters, nucleotides conserved in all vertebrates; grey letters, nucleotides conserved in most of the vertebrates; white letters, nonconserved nucleotides. In the resulting consensus sequence, uppercase letters indicate nucleotides conserved in all species and lowercase letters indicate nucleotides not conserved in all species.
Fiber type analysis and histology.
Soleus muscle from wild-type (WT) and MK2/3−/− mice was isolated, either flash frozen in embedding medium containing a 3:1 mixture of TFM tissue freezing medium (Triangle Biomedical Sciences, Prolab GmbH, Lollar, Germany) and gum tragacanth (Sigma-Aldrich) or fixed in 4% paraformaldehyde, and processed for routine paraffin histology. Frozen sections were cut on a cryotome, and staining was performed by the metachromatic dye-ATPase method (33). Furthermore, staining of soleus fibers at different pHs (4.3 and 9.4) for ATPase enzyme activity was performed as described by Brooke and Kaiser (34). Some of the transverse cryostat sections from the same soleus region were stained with hematoxylin and eosin (HE) solution. Staining for NAD (NADH) dehydrogenase activity, a marker of oxidative fibers, was performed as described previously (33). Briefly, sections were washed in phosphate-buffered saline (PBS) and then incubated in a solution containing 100 mg/ml NADH and 0.1 g/ml nitroblue tetrazolium (NBT) for 30 min at 37°C. The slides were washed three times with deionized water. Unbound NBT was removed from the sections by washing the slides three times each with 30%, 60%, and 90% acetone. The sections were then washed with deionized water and mounted. Dark blue staining of fibers indicates high levels of NADH dehydrogenase activity. Fibers with lower level of oxidative but predominantly glycolytic energy metabolism stain lighter blue.
Light and transmission electron microscopy.
The soleus muscles from WT or MK2/3−/− mice were isolated, immediately immersed in a fixative solution composed of 2.5% glutaraldehyde, 2% formaldehyde that had been freshly prepared from paraformaldehyde, 1.7 mM CaCl2, and 0.1 M sodium cacodylate (Na cacodylate)-HCl buffer (pH 7.3), and at once minced into blocks of about 1 mm (length of the side). Subsequently, the samples were transferred to fresh fixative solution and stored for at least 4 h at 4°C. After the specimens were washed in 0.1 M Na cacodylate-HCl buffer to which 0.22 M sucrose was added, they were postfixed in Na cacodylate-buffered 2% OsO4 for 90 min at room temperature, dehydrated in ascending concentrations of ethanol, and embedded in epoxy resin (Serva, Heidelberg, Germany). Semithin sections (about 1 μm thick) and thin sections (about 65 nm thick) were cut with a Reichert Ultracut E ultramicrotome (Leica, Wetzlar, Germany). Semithin sections were stained with a solution of 1% alkalinized toluidine blue and observed with a Leitz Orthoplan light microscope (Leica). Thin sections were collected on Formvar-coated copper slot grids, stained with uranyl acetate and lead citrate, and investigated with a Zeiss EM 10 CR transmission electron microscope (Zeiss, Oberkochen, Germany) at an acceleration voltage of 80 kV.
Western blot and pulldown analyses.
For Western blot analysis, soluble protein extract from soleus, extensor digitorum longus (EDL), and tibialis anterior (TA) muscle was run on sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gels (for MyHC) or SDS–10% polyacrylamide gels and transferred to Hybond enhanced chemiluminescence (ECL) membranes (Amersham Pharmacia Biotech GmbH, Freiburg, Germany). The blots were incubated for 2 h in PBS–1% Tween 20 containing 5% powdered skim milk. After three washes with PBS–1% Tween 20, the membranes were incubated for 16 h with the primary antibody at 4°C and for 1 h with horseradish peroxidase-conjugated secondary antibodies (diluted 2,000-fold) at room temperature. The blots were developed with an ECL detection kit (Santa Cruz Biotechnology, Inc., Heidelberg, Germany), and the digital chemiluminescence images were taken by a LAS-3000 luminescent image analyzer (Fujifilm, Europe GmbH, Düsseldorf, Germany). Positive controls for p38β and p38δ were from brain and kidney (longest exposure time, 8 min). In additional experiments, hearts were homogenized in Tris buffer containing 20 mM Tris-HCl, 200 mM NaCl, 20 mM NaF, 1 mM Na3VO4, 1 mM dithiothreitol (DTT), 1% Triton X-100 (pH 7.4), and Complete protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). Protein concentration was determined by BCA (bicinchoninic acid) assay (Pierce Biotechnology, Rockford, IL, USA). Denatured tissue homogenates were subjected to Western blotting (4 to 15% gradient and 10% SDS–polyacrylamide gels). Chemiluminescence detection of cardiac Western blots was done with SuperSignal West Pico substrate (Pierce Biotechnology). For glutathione S-transferase (GST)-p38α pulldown assays, fibers from 2 or 3 animals were pooled, and 1 mg of lysate protein was incubated with 0.1 nmol of recombinant catalytic-dead mutant GST-p38α (TGY/AFG) bound to glutathione-Sepharose 4B (Amersham Pharmacia). Affinity-selected proteins were eluted from the washed beads with SDS sample buffer and detected by immunoblotting as described previously (18).
For Western blot analysis of skeletal muscles, the antibodies used were anti-slow MyHCI/β, anti-fast MyHC (MY-32; detects all fast isoforms), anti-α-tubulin (Sigma-Aldrich, Taufkirchen, Germany), anti-slow fiber-specific troponin I (anti-TnIs), anti-myocyte enhancer factor 2C (anti-MEF2C; C-17), anti-phosphorylated MEF2C (anti-p-MEF2C) (Thr300), anti-p-MKK6 (Ser202), anti-ryanodine receptor (anti-RyR), anti-p38α, anti-p38β, and antiserum response factor (anti-SRF) antibodies (Santa Cruz Biotechnology). The anti-dihydropyrimidine receptor antibody (anti-DHPR) was from Abcam (Cambridge, United Kingdom). Anti-Na+/Ca2+ exchanger (NCX) antibody, detecting NCX isoforms 1 and 2 (NCX1 is predominantly found in oxidative type 1 and 2A, and NCX3 is found in fast glycolytic 2B fibers [35]) was from Swant (Marly, Switzerland). Antiphospholamban (anti-PLB) antibody was from Upstate Biotechnologies Millipore (Schwalbach am Taunus, Germany), anti-SERCA2a antibody was from Affinity Bioreagents (Rockford, IL, USA) and from F. Wuytack and P. Vangheluwe (K. U. Leuven, Belgium), and anti-sarcolipin (anti-SLN) antibody was from Acris Antibodies GmbH (Germany). Antibodies that recognize p38, phosphorylated p38 (p-p38), p38γ, MAPKAPK-2 (MK2), and p-MAPKAPK-2 (Thr222; p-MK2) were from Cell Signaling Technology (Beverly, MA, USA).
For Western blot analysis of the heart, additional antibodies used were anti-phospho-Thr286 CaMKII (PT-286) (Thermo Fisher Scientific, Schwerte, Germany), anti-RyR2 (Sigma-Aldrich), anti-p-Ser2809 (PS-2809) RyR2, anti-p-Ser (PS-2815) RyR2, anti-p-Ser16 (PS-16) p-PLB, and anti-p-Thr17 (PT-17) p-PLB were from Badrilla Ltd. (Leeds, United Kingdom). The anti-SERCA2a antibody was purchased from Affinity Bioreagents (Rockford, IL, USA), anti-PLB antibody was from Millipore (Schwalbach am Taunus, Germany), anti-L-type Ca2+-channel (CaV1.2a) antibody from Alomone Labs (Jerusalem, Israel), and anti-GAPDH antibody (GAPDH stands for glyceraldehyde-3-phosphate dehydrogenase) was from Biotrend (Köln, Germany). The anti-MK3 antibody has been described previously (18). The rabbit polyclonal anti-p38δ antibody was a gift from R. Ricci (ETH Zurich, Switzerland), and the anti-CaMKII antibody was a gift from D. M. Bers (Loyola University, Chicago, IL, USA).
High-resolution glycerol gels.
MyHC isoforms were separated by 7.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Total proteins were extracted from freshly frozen muscle samples in extraction buffer (0.3 M KCl, 0.1 M KH2PO4, 50 mM K2HPO4, 10 mM EDTA [pH 6.5]) with the addition of Complete protease inhibitor cocktail (Roche Applied Science) by using a method of Kubis and Gros (36) with minor modifications. In brief, myosin extracts were diluted (1:7) with SDS-PAGE sample buffer (37) and heated for 8 min at 95°C. Heated protein samples (0.2 μg of protein/lane) were loaded onto a slab gel (15 by 22 cm) with two stacking gels (3.5 and 6.5% polyacrylamide) and two separating gels (6.5 and 8.5% polyacrylamide) containing glycerol increasing from 3 to 35%. The gels were run at 4°C under constant current conditions (70 V) for 42 h. Following electrophoresis, the gels were silver stained with Silver Stain Plus (Bio-Rad Laboratories GmbH, Munich, Germany).
Muscle force and fatigue measurements.
Measurements of isometric force or muscle fatigue of isolated soleus muscle from WT, MK2−/−, or MK2/3−/− mice were performed as described previously (38) with some modifications. After cervical dislocation of wild-type or transgenic mice, soleus muscle was immediately dissected and mounted between a fixed clamp and a force transducer (catalog no. 529503; Harvard Bioscience, Holliston, MA, USA) in a waterjacketed bath. The muscles were equilibrated for 60 min in an oxygenated (95% O2/5% CO2) physiological buffer solution containing 120 mM NaCl, 4 mM KCl, 1 mM KH2PO4, 1.2 mM MgSO4, 25 mM NaHCO3, 5.5 mM glucose, and 1 mM CaCl2 at 25°C and pH 7.4. Each muscle was stimulated directly via platinum electrodes placed on either side of the muscle with supramaximal voltage (pulse duration, 1 ms) under isometric conditions. Tensions were recorded using a storage oscilloscope (catalog no. 3091; Nicolet, Hodenhagen, Germany) and a recorder (model 2200; Gould Electronics, Eichstetten, Germany). After adjustment of the optimal length to give maximum isometric twitch tension, the following stimulation protocol was performed: 20 to 30 pairs of single twitches, a short submaximal tetanus at 40 Hz (0.5 s), followed by a maximal tetanus at 50 Hz (1 s). For fatigue experiments, the muscles were stimulated for submaximal tetanus with a train of stimulations of 40 Hz (0.5 s) every 5 s over a 30-min period, followed by one maximal tetanic contraction (1 s, 50 Hz). The force was measured every 5 min. The force output of the wild-type soleus muscle dropped to about 50% of the initial level after 30 min. After completion of the experiments, the length and weight (wet weight) of each muscle was determined and used for normalization to give specific force (in newtons per square centimeter). Results are expressed as means ± standard deviations (SDs). The statistical significance of differences of the means was estimated by one-way analysis of variance (ANOVA) followed by the Newman-Keuls multiple comparison test, performed using GraphPad Prism software version 3.00.
Two-dimensional gel electrophoresis (2-DE) analysis.
Isolated soleus or heart muscle from wild-type or MK2/3−/− mice were ground into a fine powder under liquid nitrogen. Two-dimensional PAGE analysis of the muscle lysates was done by the method of Barjaktarovic et al. (39) with minor modifications. Protein (250 μg) from each sample, dissolved in rehydration buffer {8 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1% DTT, 0.7% Pharmalytes (pH 4 to 7), and 0.001% bromphenol blue{ supplemented with Complete protease inhibitor mixture (Roche Applied Science) and phosphatase inhibitor mixture I (Sigma-Aldrich) was separated on immobilized 24-cm pH gradient polyacrylamide gel strips in the pH range of 4 to 7 for the first, isoelectric focusing (IEF) dimension. After rehydration, the voltage was progressively increased to 10,000 V. IEF was stopped after 65 kV . h was reached. For the second dimension, the equilibrated gel strips were applied to 12% polyacrylamide gels. To detect phosphorylated proteins, gels were first stained with Pro-Q Diamond (Invitrogen) followed by silver staining for total protein detection (40, 41). The gels were comparatively analyzed using the software Delta2D (version 3.4; Decodon, Greifswald, Germany). Spot evaluation, protein digestion, and analysis by electrospray ionization-tandem mass spectrometry (MS) to confirm protein identity were done as described previously (42). For identification of slow and fast myosin light chain (MLC) isoforms in soleus muscle lysates, a narrow pH range (4 to 6.5) in the first dimension was used, and 9 to 20% acrylamide slab gel gradients were used for the second dimension. The resulting two-dimensional (2D) electrophoretograms from two biological replicates were silver stained.
Semiquantitative PCR and quantitative real-time PCR (qPCR).
To assess relative mitochondrial DNA (mtDNA) abundance, total genomic DNA (nuclear and mtDNA) was isolated from whole soleus or heart muscle from WT and MK2/3−/− mice using NucleoSpin tissue kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany). Cytochrome B (Cyt B) was used as a marker for mtDNA, and mouse β-actin was used as a marker for nuclear DNA. Semiquantitative PCR was carried out with GoTaq polymerase (Promega). For each reaction, 250 ng of total DNA was used as the template. Primer pairs (not intron spanning) used for genomic PCR are described below. PCR products were analyzed by 2% agarose gel electrophoresis and visualized with ethidium bromide staining. For qPCR analyzing cDNA levels, total RNA from WT and MK2/3-deficient soleus muscle was purified with a NucleoSpin RNAII kit including on-column rDNase digestion (Macherey-Nagel). Cell lysis was performed with additional proteinase K digestion (10 min, 55°C). mRNA was reverse transcribed with Phusion reverse transcription-PCR (RT-PCR) kit (New England BioLabs GmbH) according to the manufacturer's instructions. qPCR was performed on a Rotor-Gene 2000 real-time PCR thermocycler (Qiagen, Hilden, Germany) using Power SYBR green supermix (Applied Biosystems, Darmstadt, Germany) according to the manufacturer's instructions. Data were quantified using Rotor-Gene Q series software 1.7 (Qiagen). Efficiencies were calculated from the slope of template dilution curves with primers for the genes of interest and the reference gene (18S rRNA) and used for quantification of changes of transcript levels by the ΔΔCT method. The efficiency of all primer sets was between 95% and 105%. Three independent batches of DNA samples were used for qPCR analysis of genomic DNA, and data were presented as means ± SDs and analyzed by the Student t test.
Primer pairs for genomic PCR.
For mouse Cyt B, the forward primer 5′-CAT TTA TTA TCG CGG CCC TA-3′ and reverse primer 5′-TGG GTT GTT TGA TCC TGT TTC-3′ were used. For mouse β-actin, the F primer 5′-TCA ACA CCC CAG CCA TGT A-3′ and R primer 5′-GTG GTA CGA CCA GAG GCA TAC-3′ were used. The primer pairs for qPCR using cDNA follow: for mouse embryonic MyHC (MyHCemb), the F primer 5′-GGA TGG GAA AGT CAC TGT GG-3′ and R primer 5′-GTC CTC TGG CTT AAC CAC CA-3′; for mouse MyHCI/β, the F primer 5′-CGC ATC AAG GAG CTC ACC-3′ and R primer 5′-CTG CAG CCG CAG TAG GTT-3′; for mouse MEF2C, the F primer 5′-CAC CGG AAC GAA TTC CAC TC-3′ and R primer 5′-ATG CGC TTG ACT GAA GGA C-3′; for mouse nuclear respiratory factor 1 (NRF-1), the F primer 5′-AAC CCT GAT GGC ACC GTG TC-3′ and R primer 5′-AGT GAC TGT GGT TGG CAG TTC-3; for mouse Pax7, the F primer 5′-GGC ACA GAG GAC CAA GCT C-3′ and R primer 5′-GCA CGC CGG TTA CTG AAC-3′; for mouse PGC-1α, the F primer 5′-CTG TCA ACG CGT GTC TGT CAG CCC TTG TAA TGT G-3′ and R primer 5′-CTG TCA AGA TCT CCA GCT CCC GAA TGA CGC CAG TCA A-3′; and for 18s rRNA, the F primer 5′-GGA CCA GAG CGA AAG CAT TT-3′ and R primer 5′-TGC CAG AGT CTC GTT CGT TAT-3′.
Northern blot analysis.
Northern blot analysis was performed as described previously (43) with some modifications. Total RNA isolation, RNA separation on 1.2% agarose formaldehyde gels, hybridization with cDNA probes, and autoradiography were done as described previously (43). Total RNA (20 μg) was isolated, fractionated on a formaldehyde agarose gel, and transferred to nitrocellulose. For detection of SERCA2a mRNA, a 272-bp DraI-ClaI fragment from the 3′ untranslated region of rat SERCA2 cDNA (44) was used. To detect Egr-1 mRNA, a 1.6-kb BglII fragment of mouse Egr-1 cDNA was used (45), and to detect Sp1 mRNA, a 2.7-kb EcoRI-HindIII fragment of human Sp1 cDNA (46) was used. The probes were radiolabeled with [α-32P]dCTP by the random priming method as performed previously (43).
Oligonucleotide DNA microarray analysis.
Total RNA from WT and MK2/3-deficient soleus muscle was purified with NucleoSpin RNAII kit (Macherey-Nagel). In total, RNA from muscles from six WT or six MK2/3−/− mice was divided into two pools, each containing muscles from three mice. The microarray-based mRNA expression analysis was performed in the single-color mode as described previously (47) with minor modifications. The whole-mouse genome oligonucleotide microarray V2 (product no. G4846A, design identification [ID] 026655; Agilent Technologies, Böblingen, Germany) used in this study contains 44,397 oligonucleotide probes covering roughly 32,000 murine transcripts. Synthesis of Cy3-labeled cRNA was performed with the one-color Quick Amp labeling kit (catalog no. 5190-0442; Agilent Technologies) according to the manufacturer's recommendations. cRNA fragmentation, hybridization, and washing steps were also carried out exactly as recommended by the one-color microarray-based gene expression analysis protocol, V5.7 (83). Slides were scanned on the Agilent G2565CA microarray scanner system (pixel resolution, 5 μm; bit depth, 20). Data extraction was performed with the Feature Extraction software V10.7.3.1 (Agilent Technologies, Inc.) by using the recommended default extraction protocol file (GE1_107_Sep09.xml). Processed intensity values of the green channel (gProcessedSignal or gPS) were normalized by global linear scaling: all gPS values of one sample were multiplied by an array-specific scaling factor. This scaling factor was calculated by dividing a reference 75th percentile value (set at 1,500 for the whole series) by the 75th percentile value of the particular microarray (“Array I” in the formula shown below). Accordingly, normalized gPS values for all samples (microarray data sets) were calculated by the following formula: normalized gPSArray i = gPSArray i × (1,500/75th percentileArray i). A lower intensity threshold was defined as 1% of the reference 75th percentile value (= 15). All of those normalized gPS values that fell below this intensity border were substituted by the respective surrogate value of 15. Calculation of ratio values of relative gene expression was performed using Excel macros or R-Scripts, and heat map visualization was generated using Excel. A set of transcripts filtered for genes of relevance for muscle fiber type determination and regeneration/repair processes was selected for visualization. Processed signal intensity values of samples WT 2 and MK2/3−/− 1 and 2, respectively, were divided by the respective value of WT sample 1 for each given gene.
Generation of MK2 knockdown cells.
To knock down MK2 expression, the C2C12 muscle cell line was transduced with the SHCLNV Mission short hairpin RNA (shRNA) lentiviral transduction particles (Sigma, Taufkirchen, Germany). Five different lentiviral particles (MK2 shRNAs) (catalogue no. TRCN0000232381 to -5) and the control lentiviral transduction particles (Mission TRC2 Control puro nontarget shRNA) were used. The cells were transduced in 96-well plates at a multiplicity of infection (MOI) of 1 and 2. After transduction, cells were selected with puromycin (2.5 μg/ml). Cell lines containing one of each of the different MK2 shRNAs (clones 1 to 5) as well as the control shRNA (clone 6) were established and analyzed for the best knockdown and also for reduced phosphorylation of MK2 substrate Hsp25. For analysis of substrate phosphorylation, cells were treated with 10 μg/ml anisomycin (Calbiochem, Merck Chemicals, Germany) for 30 min to stimulate the p38α-MK2/3 signaling pathway, as described previously for MEFs (18). The anti-p-Hsp25 (mouse Ser86/human Ser82) antibody was purchased from Invitrogen and anti-murine Hsp25 antibody from Stressgen (Enzo Life Sciences GmbH, Lörrach, Germany).
Preparation of nuclear extracts and electrophoretic mobility shift assays.
Preparation of nuclear extracts (NE) and electrophoretic mobility shift assays (EMSAs) were performed as described previously (48) with some modifications. MK2/3−/− MEFs were transiently transfected with expression plasmid pcDNA3-myc-MK2 and/or pcDNA3-MKK6EE or empty vector. Cells were grown for 48 h followed by NE preparation performed as previously described (48). EMSAs were performed according to the manufacturer's protocol using the Odyssey infrared EMSA kit and IRDye 700 infrared dye-labeled oligonucleotides (Li-Cor Bioscience, Bad Homburg, Germany) using 5% native acrylamide (in 0.5% Tris-borate-EDTA [TBE]) gels. Gels were analyzed with the Odyssey infrared imaging system. Oligonucleotides representing sequences of the human SERCA2 promoter containing overlapping Egr-1/Sp1 consensus binding sites (underlined) (see Fig. 5F) were used as probes: bp −192 to −158 (5′-GCG CGC GGG AGG GGG CGG GGC CTG CGC GGC AGC GT-3′), bp −131 to −103 (5′-GCC GGG AGG AGG GGG CGG GGC CGC GCC GC-3′), or bp −117 to −88 (5′-GCG GGG CCG CGC CGC CCG CGC CGC GCT GGG-3′). For competition experiments, an oligonucleotide probe containing the overlapping Egr-1/Sp1 consensus binding site A at bp −184 to −166 mutated in the nonoverlapping part of the Egr-1 site at bp −184 to −181 as indicated in Fig. 5F was used. Antibody supershift assays were performed by preincubating NE with anti-Egr-1 or anti-Sp1 antisera (Santa Cruz Biotechnology, Inc.).
Isolation of cardiomyocytes.
Isolation of cardiomyocytes was performed as described previously (49). Briefly, the hearts were quickly excised, weighed, mounted on a Langendorff perfusion apparatus, and retrogradely perfused with nominally Ca2+-free solution containing 113 mM NaCl, 4.7 mM KCl, 0.6 mM KH2PO4, 0.6 mM Na2HPO4, 1.2 mM MgSO4, 12 mM NaHCO3, 10 mM KHCO3, 10 mM HEPES, 30 mM taurine, 10 mM 2,3-butanedione monoxime (BDM), 5.5 mM glucose, and 0.032 mM phenol red for 4 min at 37°C (pH 7.4). Then, 7.5 mg/ml liberase TM (Roche), trypsin 0.6%, and 0.125 mM CaCl2 were added to the perfusion solution. Perfusion was continued for 3 min until the heart became flaccid. Ventricular tissue was collected in perfusion buffer supplemented with 5% bovine calf serum and dissociated. Ca2+ reintroduction was performed by stepwise increasing the concentration of Ca2+ from 0.1 to 0.8 mM. For measurements, cells were freshly plated onto superfusion chambers coated with laminin.
Epifluorescence experiments.
Previously described Ca2+ epifluorescence and sarcomere length measurements (49) were performed under superfusion with Tyrode's solution consisting of 140 nM NaCl, 4 mM KCl, 5 mM HEPES, 1 mM MgCl2, 10 mM glucose, and 2 mM CaCl2 (pH 7.4 at 37°C with NaOH). The loading buffers for Ca2+-fluorescent dyes Fluo-3 and Fura-2 loading consisted of normal Tyrode’s (NT) solution (consisting of 140 nM NaCl, 6 mM KCl, 10 mM HEPES, 1 mM MgCl2, 10 mM glucose, and 1 mM CaCl2, pH 7.4, at 37°C with NaOH) with Pluronic F-127 (0.2 mg/ml) in addition to the dye. Myocytes were loaded with either 10 μM Fluo-3 AM or (to measure diastolic Ca2+-levels) 10 μM Fura-2 AM for 15 min. The dye was washed out by superfusion with NT, and 5 min was allowed for complete de-esterification of the dye before starting measurements. All experiments were performed at 37°C. For Fluo-3, excitation was at 480 ± 15 nm, emission was measured at 535 ± 20 nm, and F/F0 was calculated by dividing raw fluorescence F by the baseline fluorescence (F0). In parallel, myocyte contraction was investigated using a sarcomere length detection system (IonOptix). Fura-2 fluorescence ratio was calculated by division of the emission signal obtained by excitation at 340 nm by the emission signal obtained by excitation at 380 nm, with alternation of excitation at 240 Hz by the IonOptix hyperswitch.
Myocytes were field stimulated at 1 Hz until steady state was achieved. To investigate frequency dependency, stimulation frequency was then increased to 4 Hz. SR Ca2+ content was estimated by rapid application of 10 mM caffeine after 1-Hz stimulation. To estimate NCX function, the time constant τ of the caffeine-induced transient was calculated via monoexponential fit (50).
Multiple-sequence alignment.
Nucleotide sequences of the SERCA2 promoter region (common for SERCA2a and -2b isoforms) spanning the Egr-1/Sp1 overlapping site A from several species, bovine (GeneID 540568), dog (GeneID 403878), gibbon (GeneID 100597073), horse (GeneID 791242), human (GeneID 488), mouse (GeneID 11938), pig (GeneID 396875), and rat (GeneID 29693), were aligned with MultAlin 5.4.1.
RESULTS
Abundance and activation of MK2 and MK3 in cross-striated muscles.
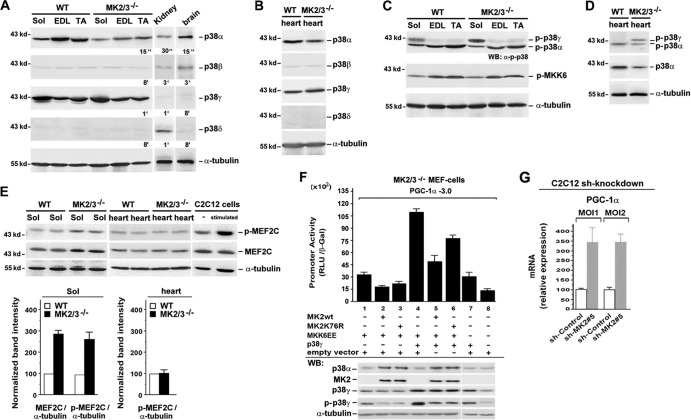
Despite studies investigating the role of p38 MAPK in muscle development and regeneration (13), the precise functions of the downstream protein kinases MK2/3 in cross-striated muscle tissue still have not been determined. We first analyzed protein levels of MK2 and MK3 and determined whether active MK2 and/or MK3 are found in lysates from fast fiber type extensor digitorum longus (EDL) and tibialis anterior (TA), and slow fiber type soleus (Sol) muscles by Western blotting. For detection of lower levels of MK2 and MK3, we performed GST-p38 pulldown experiments. In skeletal and heart muscle lysates, MK2 was detected as two bands (47 and 55 kDa) and MK3 as one band (42 kDa) (18) (Fig. 1A to D). For detection of phosphorylated MK2 and MK3, an antibody that recognizes both pT208-MK2 and pT203-MK3 (18), was used after a GST-p38α pulldown assay (Fig. 1C and D). We demonstrate phosphorylation of MK2 in all muscles and of MK3 in the fast skeletal muscles, reflecting persistent basal activation in the absence of forced contractile activity (Fig. 1C). In heart, detectable phosphorylation was only observed for MK2 (Fig. 1D). Thus, a basal level of activated MK2 is also present in the heart muscle under nonstrenuous conditions.
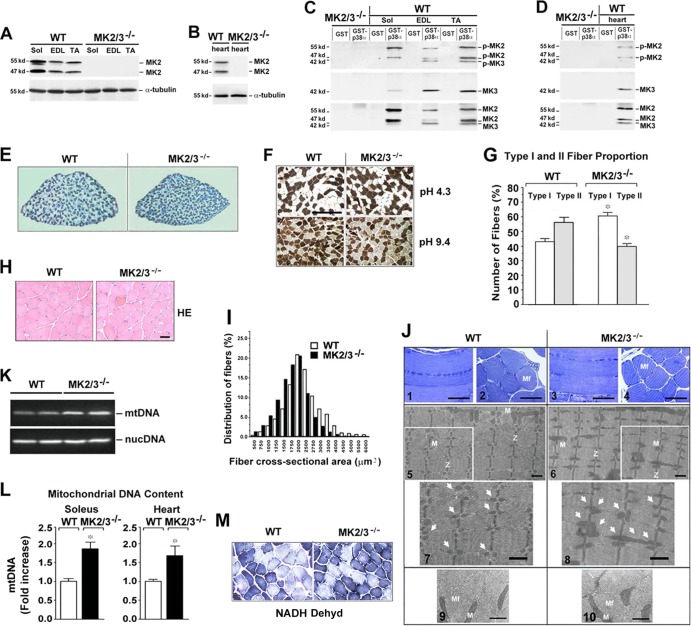
Fig 1.
MK2/3 deficiency increases the number of slow type I fibers and mitochondrial content in soleus muscle. (A to D) Western blot analysis of MK2 and MK3 abundance and phosphorylation in slow type soleus (Sol), fast type extensor digitorum longus (EDL) and tibialis anterior (TA) (A and C) and heart (B and D) muscles. α-Tubulin was used as a loading control. (C and D) GST-p38α pulldown experiments from whole-muscle lysates. To investigate the activity levels, an antibody against p-T222 of human MK2 that recognizes both murine p-MK2 and p-MK3 was used. Blots were stripped and reprobed with anti-MK3 and then with anti-MK2 antibodies. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the blots. (E) Representative metachromatic ATPase staining of cross sections from entire soleus muscles. Slow type I fibers are stained in dark blue, and fast type II fibers in various shades of lighter blue. (F) ATPase fiber type analysis of soleus muscles at different pHs. Type I fibers stained dark at pH 4.3 and light at pH 9.4, and type II fibers exhibited the opposite. Bar, 200 μm. (G) Quantification (shown as a percentage) of type I and II fibers (n = 5) as analyzed in panel F at pH 4.3. Values that were significantly different (P < 0.05) from the values for the same fiber type from WT mice are indicated by an asterisk. (H) Sections of myofibers from WT and MK2/3−/− soleus muscle stained with hematoxylin and eosin (HE). Bar, 15 μm. (I) Frequency histograms of the distribution of cross-sectional areas (CSA) of HE-stained fibers as shown in panel H in whole soleus muscle (distribution of fibers shown as a percentage). (J) Representative light micrographs (panels 1 to 4) and transmission electron micrographs (panels 5 to 10) of longitudinal sections (panels 1, 3, and 5 to 8) and cross sections (panels 2, 4, 9, and 10) from WT and MK2/3-deficient solei. M, mitochondria (also indicated by arrows in panels 7 and 8 [enlargements of sections in panels 5 and 6]); Mf, myofibril; Z, Z-line. Bars, 100 μm (panels 1 to 4), 1 μm (panels 5 to 8), and 0.5 μm (panels 9 and 10). (K and L) Semiquantitative PCR (K) and qPCR (L) analyses of mitochondrial cytochrome B (mtDNA) and nuclear β-actin (nucDNA) DNA levels in soleus and heart muscles. In panel L, fold increase of mitochondrial DNA (mtDNA) relative to nuclear DNA (n = 8) is shown. The ratio of mtDNA to nuclear DNA in WT soleus was set at 1 (mean plus standard error of the mean [error bars] [SEM]. Values that were significantly different (P < 0.01) from those for WT mice are indicated by an asterisk. (M) Staining for NADH dehydrogenase (NADH dehyd) activity in WT and MK2/3−/− soleus.
MK2/3 deficiency alters the fiber type composition of soleus muscle.
In mice, soleus is a slow type muscle composed also of significant amounts of fast type fibers ranging from ∼30 to 60% depending on the strain (51, 52). An altered fiber type pattern was demonstrated by a metachromatic ATPase in situ staining of MK2/3−/− soleus (Fig. 1E) and by comparing staining intensities of ATPase in serial sections incubated at different pHs (Fig. 1F). In wild-type (WT) mice, less than half of the fibers in soleus muscle were of slow type I. In contrast, the number of type I fibers was significantly increased in muscle from DKO mice to about 60% of all fibers, and the number of fast type II fibers was decreased accordingly (Fig. 1F and G). In agreement with increased type I fiber content in muscle from DKO mice, histological analysis by hematoxylin and eosin (HE) staining showed a shift toward a smaller cross-sectional area (CSA) of fibers (Fig. 1H and I). Transmission electron microscopy revealed no structural abnormalities in MK2/3−/− soleus muscle fibers (Fig. 1J), comparable with findings in p38α−/− muscles (11). Furthermore, ultrastructural analysis revealed slightly different mitochondrial patterns in DKO and WT soleus muscles in spite of the genotype-independent heterogeneity in their size (Fig. 1J). Therefore, the relative mitochondrial content in the soleus was estimated by measuring the level of mitochondrial DNA compared with the nuclear DNA levels. Semiquantitative PCR (Fig. 1K) and quantitative real-time PCR (qPCR) (Fig. 1L) demonstrated a relative increase of mitochondrial DNA content in the MK2/3−/− soleus. In accordance, NADH dehydrogenase activity staining indicated increased mitochondrial activity in MK2/3−/− soleus (Fig. 1M), demonstrating an increase in oxidative capacity.
Increased abundance of MyHCI/β and reduced fatigability in MK2/3−/− soleus.
Next, Western blot analysis demonstrated increased MyHCI/β and reduced MyHCII protein levels, and an increased level of the slow isoform of troponin I (TnIs) in MK2/3−/− soleus (Fig. 2A). Also, a slight decrease of MyHCII together with minor MyHCI/β and TnIs abundance was found in EDL and TA muscle from MK2/3−/− mice. High-resolution glycerol gels then demonstrated an increased abundance of MyHCI/β and MyHCIIa, while the levels of MyHCIIb and MyHCIId/x decreased in DKO muscle compared to WT muscle (Fig. 2B). In line with a fast-to-slow shift in MK2/3−/− soleus, the isoform pattern of myosin light chains (MLC) was changed to lower fast alkali/essential (1f and 3f) and regulatory (2f) and to higher slow isoform abundance (1s and 2s) (Fig. 2C). Also, abundance of the cytosolic high-affinity Ca2+-binding protein parvalbumin, highly expressed in fast fibers (53), was diminished in the DKO muscle (Fig. 2C). To evaluate possible functional consequences of the observed changes in fiber type composition and mitochondrial content, we compared fatigue rates of soleus muscles from WT and MK2/3−/− mice. MK2/3−/− soleus fatigued less rapidly than WT soleus (Fig. 2D), in accordance with the shift to type I fibers and the observed increase in oxidative capacity.
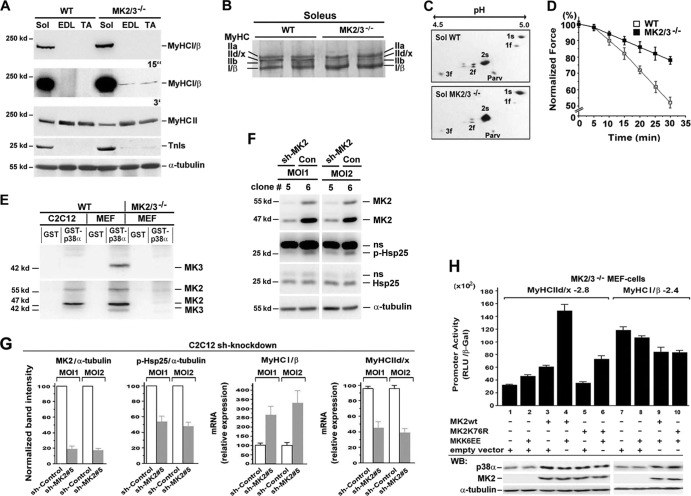
Fig 2.
Increased type I contractile protein level and muscle fatigue resistance in MK2/3−/− soleus. (A) Western blot analysis of extracts from soleus (Sol), EDL, and TA muscles. α-Tubulin was used as a loading control. Exposure times (15 s and 3 min) for enhanced chemiluminescence detection are indicated. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the blots. (B) High-resolution SDS-PAGE of MyHC isoforms in extracts from WT and MK2/3-deficient soleus muscles. (C) 2-DE separation of fast (f) and slow (s) alkali/essential MLC isoforms (isoforms 1 and 3) and regulatory (isoform 2) of soleus (Sol) muscle homogenates. Parv, parvalbumin. (D) Determination of fatigue resistance in WT and MK2/3−/− soleus. Tensions were measured every 5 min and normalized to initial values (n = 15). Specific force (N/cm2) was estimated by length and weight (wet weight) measurements of each muscle. (E) MK2 and MK3 levels in C2C12 muscle cells. WT and MK2/3−/− mouse embryonic fibroblasts (MEFs) were the positive and negative controls, respectively. GST-p38α pulldown assays were analyzed using first anti-MK3 and then anti-MK2 antibodies. (F) Western blot analysis of lysates from MK2 shRNA-transduced (sh-MK2) (clone 5) or control (Con) (clone 6) shRNA-transduced C2C12 cells. Clones were treated with 10 μg/ml anisomycin for 30 min to stimulate the p38α-MK2/3 signaling pathway. α-Tubulin was used as a loading control. MOI1 and MOI2, multiplicity of infection of 1 and 2, respectively; p-Hsp25, phosphorylated Hsp25; ns, nonspecific. (G) Band intensities for MK2 and p-Hsp25 as shown in panel F normalized to those of α-tubulin. Relative gene expression levels of MyHCI/β and MyHCIId/x in control and MK2 knockdown (clone 5) clones as determined by qPCR. The ratios of MK2 or p-Hsp25 to α-tubulin protein and of MyHC to α-tubulin mRNA in cells transduced with control particles were set at 100 (arbitrary units) (mean ± SEM) (n = 4). (H) MK2/3−/− MEFs were transfected with a −2.8 MyHCIId/x or a −2.4 MyHCI/β promoter-luciferase construct and cotransfected as indicated below the bars. Promoter activity is expressed as relative light units per unit of β-galactosidase activity (RLU/β-Gal) (means plus standard deviations [SD] of triplicate data points). Western blot (WB) analysis of p38α, MK2, and α-tubulin protein levels is shown in the bottom blots.
MK2 activity-dependent MyHCIId/x promoter regulation.
To confirm the role of MK2/3 in the regulation of MyHC gene expression, we established a stable shRNA knockdown of MK2 in C2C12 muscle cells (MK3 was not detected [Fig. 2E]) by lentiviral gene transfer. Stable clone5 was identified as the most efficient MK2 knockdown, as measured by the level of phospho-Hsp25 (p-Hsp25), a known MK2 substrate (18) (Fig. 2F and G). In accordance with data from the MK2/3−/− soleus, the specific MK2 knockdown in C2C12 cells resulted in the upregulation of slow MyHCI/β and downregulation of fast MyHCIId/x gene expression (Fig. 2G).
Since fast MyHCIId/x gene expression is partly regulated by p38α/β in myotubes (11, 54), we elucidated whether MyHCIId/x gene expression might be directly affected by the activity of MK2 or whether the effect is just due to the loss of stabilization of p38 by MK2/3 (18) in the DKO mice. Using a −2.8 MyHCIId/x promoter construct in MK2/3-deficient immortalized mouse embryonic fibroblasts (MEFs) (18), we analyzed the influence of reintroduction of MK2 or its catalytically inactive mutant MK2K76R, which restores p38α level (18, 30), on promoter activity. Coexpression of a constitutively active mutant of the direct p38 upstream kinase MKK6 (MAPK kinase 6), MKK6EE, together with WT MK2 strongly induced the promoter activity, indicating a role of MK2 in transcriptional activation (Fig. 2H, lanes 1 and 4). Coexpression of MK2K76R and MKK6EE (Fig. 2H, lane 6) led to similar rescue of the p38α level but only to a somewhat higher MyHCIId/x promoter activity that is far from reaching the level of MK2/MKK6EE. This indicates that catalytic activities of both MK2 and p38α are involved in efficient induction of the MyHCIId/x promoter. In contrast, the activity of a −2.4 MyHCI/β promoter construct was slightly reduced by coexpression of wild-type MK2 (MK2wt) plus MKK6EE in MK2/3−/− MEFs. The catalytic-dead mutant MK2K76R did not change this effect, suggesting a weak inhibitory effect by p38α on the −2.4 MyHCI/β promoter in an MK2 activity-independent manner (Fig. 2H).
Microarray and 2D electrophoretic profiling of MK2/3−/− soleus reveals enhanced expression of the slow myofiber gene program.
To more thoroughly analyze changes that result from the loss of MK2/3 in soleus muscle, we performed a microarray analysis. Numerous genes involved in contraction and energy metabolism were differentially expressed in WT and DKO muscles (Fig. 3A; see Table S1 in the supplemental material). In agreement with data shown in Fig. 2, expression of slow fiber type-specific genes encoding components of the contractile apparatus was increased, whereas fast fiber-specific gene expression was decreased in MK2/3−/− soleus. Analogous changes in the expression of components involved in Ca2+ homeostasis (55) were identified, e.g., reduction of fast type SERCA1 (Atp2a1) and increase of slow type SERCA2a (Atp2a2) gene expression. In addition, increased expression of transcriptional regulators crucial for mitochondrial gene expression and oxidative energy metabolism (24, 25, 56, 57), such as PGC-1α, nuclear respiratory factor 1 (NRF-1), myocyte enhancer factor 2C (MEF2C), as also determined by qPCR (Fig. 3B), and cAMP-responsive element-binding protein 1 (CREB1) may account for increased mitochondrial content (Fig. 1K and L). Accordingly, the altered expression profile in the MK2/3−/− soleus further includes a series of upregulated genes involved in component processes of oxidative energy metabolism, such as fatty acid uptake, biosynthesis/storage, and oxidation, and carbohydrate metabolism (58). Changes also include myoglobin (Mb), another oxidative marker (see Table S1).
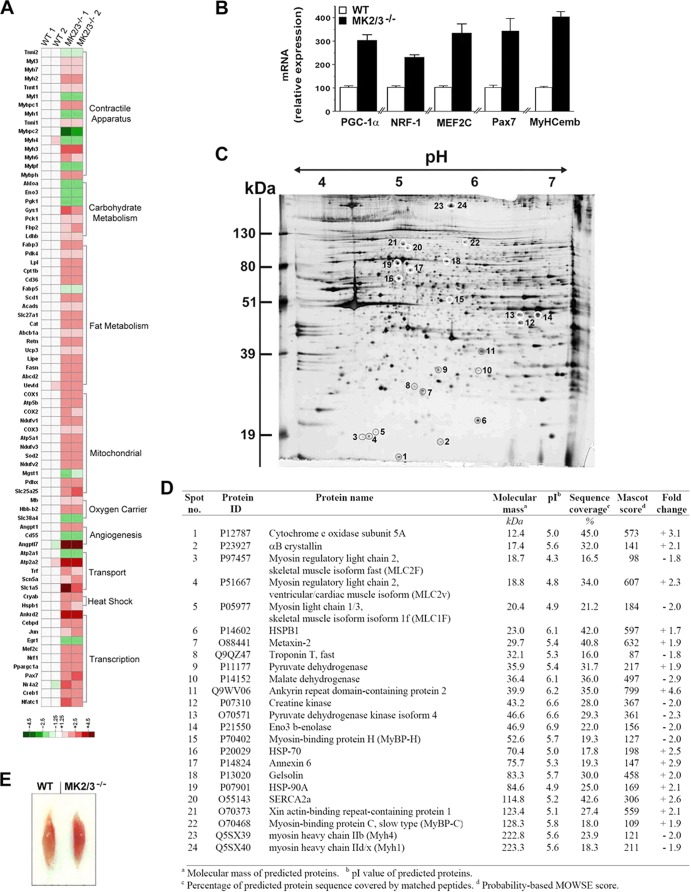
Fig 3.
Increased expression of the type I fiber-specific gene program in MK2/3−/− soleus. (A) Heat map visualization of DNA microarray analysis showing relative expression of genes in WT and MK2/3−/− soleus. A set of transcripts was selected for groups of genes characteristic of fast and slow muscle fiber types (see Table S1 in the supplemental material). Differential mRNA expression was analyzed in four individual samples (two WT samples [WT 1 and 2] and two MK2/3−/− samples [MK2/3−/− 1 and 2]). Each sample was pooled from three mice. Values of WT sample 1 were set to 1 for all genes depicted. The resulting ratios were expressed as relative fold changes and color coded as indicated in the color code key. (B) qPCR analysis of MEF2C, NRF-1, Pax7, PGC-1α, and embryonic MyHC (MyHCemb) mRNA expression, normalized to 18s rRNA levels. The WT values were set at 100 (arbitrary units) (means plus SEM) (n = 4). (C and D) Silver-stained 2-DE reference gel showing primarily proteins characteristic of skeletal muscle fiber types from lysates of MK2/3−/− soleus muscle. Proteins with a marked change in abundance in MK2/3−/− soleus are circled, numbered, and listed in panel D. The Fold change column shows the values compared with the values for the WT soleus of upregulated (+) or downregulated (−) genes. MOWSE, molecular weight search. (E) Representative soleus from WT and MK2/3−/− mice.
In addition, transcripts of embryonic MyHC (Myh3) and α-cardiac MyHC (Myh6) (Fig. 3B; see Table S1 in the supplemental material), both markers of newly forming myofibers in regenerating muscle (59), and transcripts of paired box gene 7 (Pax7) and cadherin 15 (Cdh15) (Fig. 3B; see and Table S1), markers for satellite cell activation (60), are upregulated. These results are in line with increased type I fibers that are thought to have a higher turnover rate during normal physiological maintenance of the muscle (61).
A proteomics approach demonstrated that indeed the abundance of several proteins characteristic for fiber type determination, such as elements of the contractile apparatus and its regulatory machinery, and enzymes of energy metabolism, was changed in the soleus muscle of DKO mice (Fig. 3C and D). In agreement with the observed changes in transcript and protein level of genes of oxidative energy metabolism, MK2/3−/− soleus showed a darker red color compared to WT soleus (Fig. 3E), indicating greater oxidative capacity. Augmented PGC-1α transcript levels (see Table S2 in the supplemental material) and an increase in mitochondrial DNA content (Fig. 1L) indicate similar changes of energy metabolism in cardiac muscle.
The levels of activated p38α and p38γ are differentially affected in MK2/3−/− muscles.
To understand the direct contribution of MK2/3 deficiency to the phenotype of DKO mice, we investigated its effect on p38 in muscle tissue. p38α and p38γ are the main isoforms detected in WT cross-striated muscles (Fig. 4A and B). We previously demonstrated that the absence of MK2/3 leads to a significant reduction of p38α protein content in several tissues and in immune cells due to the failure to form a stabilizing complex (18). In accordance, the total level of p38α protein was reduced in all investigated MK2/3−/− muscles (Fig. 4A and B). However, the p38γ level remained unaffected, suggesting that MK2/3 is dispensable for protein stability of p38γ in skeletal muscles.
Fig 4.
MK2/3 deficiency and shMK2 knockdown alters p38α/γ activation pattern and promotes PGC1-α gene expression. (A to D) Western blot analysis of p38 isoforms and levels of phosphorylated p38α, p38γ, and p-MKK6 in skeletal and heart muscle. p38β and -δ from brain and kidney were used as positive controls. Blots were stripped before incubation with the next antibody. α-Tubulin was used as a loading control. Exposure times for enhanced chemiluminescence detection (indicated) were 15 s, 30 s, 1 min, 3 min, and 8 min (A) and 8 min for p38β and -δ (B). (E) Western blot analysis of phosphorylation of MEF2C in WT and MK2/3−/− soleus and heart muscle. The p38α signaling pathway was stimulated with 10 μg/ml anisomycin for 30 min in C2C12 cells (positive control). α-Tubulin was used as a loading control. For quantification, band intensities were normalized to those of α-tubulin. The levels of expression of proteins from WT soleus were set at 100 (arbitrary units). (F) MK2/3−/− MEFs were transfected with a −3.0 PGC-1α promoter-luciferase construct and cotransfected as indicated below the bars. Promoter activity was expressed as relative light units per unit of β-galactosidase activity (RLU/β-Gal) (means plus SD of triplicate data points). Western blot (WB) analysis with α-tubulin as a loading control is shown in the bottom blots. (G) shRNA-based MK2 knockdown in C2C12 muscle cells. PGC-1α/α-tubulin mRNA levels in cells transduced at a multiplicity of infection of 1 or 2 (MOI1, MOI2) with control particles were set at 100 (arbitrary units) (means plus SEM) (n = 4).
We next analyzed p38 phosphorylation (p-p38) using an antibody that recognizes all p38 isoforms when simultaneously phosphorylated at T181 and Y183 in the activation loop. Two major bands of p-p38 isoforms were detected in all muscles reflecting activated p38α and p38γ (electrophoretic mobility of p38α > p38γ [62]) (Fig. 4C and D). Despite the remarkable reduction of total p38α levels in MK2/3−/− muscles, no striking decline in p-p38α was observed. This finding resembles previous results in neuronal cultures from MK2/3-deficient mice (63). Thus, fiber type changes in MK2/3−/− soleus are most likely direct effects of MK2/3 deficiency and not caused by lower p38α activity levels. Interestingly, p-p38γ was increased in MK2/3−/− soleus (Fig. 4C), while the amount of total p38γ protein remained unchanged. In addition, the levels of p-MKK6, the upstream kinase of p38α and p38γ, were not altered (Fig. 4C), suggesting that the smaller amount of MKK6-competing p38α might account for higher p38γ phosphorylation by MKK6. Likewise, p-p38γ was increased in MK2/3−/− heart (Fig. 4D).
We further investigated the phosphorylation status of MEF2C, a known major substrate of p38α. In MK2/3−/− soleus, the total levels of MEF2C protein were higher (Fig. 4E; also Fig. 3B and see Table S1 in the supplemental material), and an increase of MEF2C phosphorylation demonstrated persistent p38α activity. The levels of total MEF2C and phosphorylated MEF2C in DKO heart were unaltered (see Table S2 in the supplemental material).
Enhanced PGC-1α transcript expression and promoter activation in DKO and MK2 knockdown.
To investigate whether PGC-1α expression correlates with p38γ activity, we performed reporter assays using a −3.0 PGC-1α promoter fragment in DKO MEFs. Overall PGC-1α promoter activity was higher in MKK6EE-transfected cells overexpressing p38γ (Fig. 4F, lanes 4 to 6) than in those exhibiting only endogenous p38γ protein (Fig. 4F, lanes 1 to 3), and Western blot analysis revealed larger amounts of endogenous and exogenous p-p38γ in the absence of WT or mutant MK2. In addition, the highest PGC-1α promoter activity correlates with the strongest signal of phosphorylated p38γ. Furthermore, while the addition of WT MK2 caused a decline in promoter activity, expression of MK2K76R diminished this promoter inhibition without reaching the levels of MK2/3−/− cells. Since coexpression of both WT MK2 and MK2K76R restored p38α protein levels, these data demonstrate an inhibitory effect of catalytic activity of MK2. Taken together, PGC-1α promoter transactivation by increased p38γ signaling correlated with decreased MK2/3 activity could be an important mechanism of PGC-1α regulation.
In addition, increased PGC-1α mRNA levels as detected in MK2/3−/− soleus (Fig. 3A and B) were also detected in C2C12 cells upon MK2 knockdown (Fig. 4G).
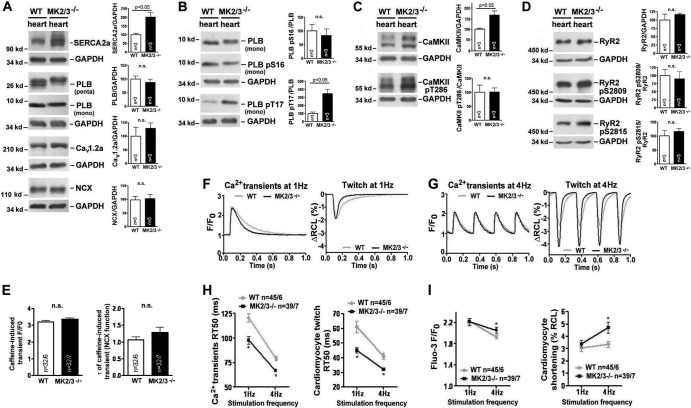
Upregulated SERCA2a abundance and improved contractile force parameters in soleus muscles of MK2/3−/− mice.
In accordance with microarray data (see Table S1 in the supplemental material), expression of both slow type-specific SERCA2a and fast type-specific SERCA1 protein was detected in WT soleus, whereas only SERCA1 was found in EDL and TA (Fig. 5A and B). While MK2/3 deficiency slightly reduced SERCA1 abundance in the soleus, the SERCA2a level was robustly increased, leading to higher total SERCA abundance. A small increase in SERCA2 was also detected in MK2/3−/− EDL and TA (Fig. 5A). In contrast, essential components of Ca2+ homeostasis and excitation-contraction coupling, such as the ryanodine receptor (RyR) and dihydropyridine receptor α1 (DHPRα1), were not altered in MK2/3−/− soleus. The slight increase in the level of slow type-associated PLB (Fig. 5B; see Table S1) was in line with the augmented content of slow fibers. Also, sarcolipin (SLN), another protein regulating SERCA function (64), remained almost unchanged.
Possible functional effects of increased total SERCA2a abundance were determined by force measurements. Specific twitch and tetanus force were higher in MK2/3−/− mice and in MK2−/− mice compared to WT mice (Fig. 5C and D). Notably, this accelerated force is surprising in view of a shift toward more type I fibers. Thus, MK2/3 deficiency leads to the generation of a unique muscle type exhibiting enhanced force and fatigue resistance.
SERCA2 gene expression is regulated via MK2-dependent Egr-1 level and promoter binding.
The strong induction of SERCA2a protein in MK2/3−/− soleus indicates a potential regulatory function of MK2/3 in SERCA2 gene expression. Therefore, we analyzed the activity of SERCA2 promoter reporter constructs in MK2/3-deficient MEFs rescued or not rescued by MKK6EE/MK2 or constitutively active MK2EE alone. A −263 SERCA2 promoter fragment was sufficient for maximal activity in nonrescued cells (Fig. 5E) with more than 2-fold-higher activity than the −2577 construct (all constructs ranging to bp +321), while activity ceased completely in the −68 mutant. In MK2 activity-rescued cells (MKK6EE/MK2), the different SERCA2 promoter constructs were significantly reduced in their activities (Fig. 5E). In addition, constitutively active MK2EE reduced the activity of the −263 construct. Thus, MK2 downregulates the activity of the SERCA2 promoter.
Regulation of SERCA2 promoter activity involves functional Sp1 sites (28), where the transcription factor Egr-1 can act as a repressor when its binding causes the displacement of Sp1 from overlapping Egr-1/Sp1 sites and a reduction of Sp1-dependent transactivation (65–67). Three putative overlapping Egr-1/Sp1 sites were identified in the −263 SERCA2 promoter (Fig. 5F). Besides the increased SERCA2 transcript level, Northern blot analysis revealed reduced Egr-1 mRNA levels in MK2/3−/− soleus muscle, while Sp1 mRNA levels remained constant (Fig. 5G). In addition, Egr-1 overexpression diminished activity of the −263 SERCA2 promoter construct in MK2/3−/− MEFs (Fig. 5H). Activated MK2 was showing similar effects, in contrast to MKK6EE/MK2K76R expression. Together, the data demonstrate a repressive function of MK2 catalytic activity and Egr-1 on the promoter. Mutation of the nonoverlapping parts of the Egr-1 sites in all three Egr-1/Sp1 overlapping regions (A, B, and C sites [Fig. 5F]; mutABC) increased transcriptional activation of the −263 promoter fragment in MK2/3−/− MEFs cotransfected with MKK6EE/MK2 or MK2EE (Fig. 5E), further indicating an MK2-dependent repressive function of Egr-1 binding. Electrophoretic mobility shift assays (EMSAs) using oligonucleotides corresponding to the A, B, and C sites were then performed to determine whether MK2-dependent interference of Egr-1 with Sp1 binding occurred within the SERCA2 promoter. Supershift experiments revealed Sp1 binding to the A and B sites but not to the C site in MK2/3−/− MEFs (Fig. 5I), whereas Egr-1 only weakly bound to the A site. Cotransfection with MKK6EE/MK2 results in increased binding of Egr-1 to the A site, while Sp1 binding was reduced. In accordance, when mutating the three sites alone, the repressive effect of MK2EE on the −263 SERCA2 promoter construct was reduced only with the A-site mutant (Fig. 5E). An increase in the Sp1 to Egr-1 binding ratio on the A site was also found in MK2/3−/− soleus and cardiac muscle compared with WT muscle (Fig. 5J). Nucleotide alignment revealed that the sequence of the bp −186 to −163 human SERCA2a promoter region including the Egr-1/Sp1 overlapping site A is highly conserved (Fig. 5L). Hence, we demonstrate that increased SERCA2 gene transcription is partly due to reduced MK2-dependent Egr-1 expression and repressive function in DKO, identifying SERCA2 as a novel target gene of MK2.
Comparable changes in SERCA2 protein and Egr-1 mRNA as detected in MK2/3−/− soleus were also found in shRNA-based MK2 knockdown C2C12 myotubes (Fig. 5K).
MK2/3 deletion induces SERCA2a abundance in the heart and improves cardiomyocyte function.
SERCA2a abundance was also strongly enhanced in MK2/3−/− heart (Fig. 6A), but Cav1.2a and NCX levels were not significantly changed. Also, rapid caffeine application experiments (50) suggested that NCX function was unaffected by MK2/3 deficiency (Fig. 6E). Interestingly, PLB abundance remained unaltered in MK2/3−/− animals (Fig. 6A), leading to an increased SERCA2a-to-PLB ratio. Since PLB in its nonphosphorylated state inhibits SERCA2a, we further tested the PLB phosphorylation state. While phosphorylation of PLB at the PKA site (S16) was nearly unaltered in MK2/3−/− hearts, we found intense hyperphosphorylation of PLB at the CaMKII site (T17) (Fig. 6B), suggesting enhanced dissociation of PLB from SERCA2a, relieved inhibition, and increased SERCA2a activity. Total CaMKII abundance was significantly higher, while relative CaMKII activation (CaMKII pT286-CaMKII) remained unchanged (Fig. 6C). Moreover, phosphorylation of RyR2 at S2809 (PKA-/CaMKII site) was not changed, and at the CaMKII site, S2815 was slightly, but not significantly, induced (Fig. 6D). Finally, transcript data from microarray analysis (see Table S2 in the supplemental material) were in accordance with the findings on the levels of proteins.
Fig 6.
Augmented SERCA2a level, increased contractility, and accelerated Ca2+ transient decay and twitch relaxation in MK2/3−/− cardiomyocytes. (A to D) Representative Western blots demonstrating the abundance and phosphorylation of indicated Ca2+-handling proteins and CaMKII from WT and MK2/3−/− hearts. GAPDH was used as a loading control. For quantification, the values (abundance relative to GAPDH) for WT were set at 100%, and the MK2/3−/− values were shown as percentages of the WT. Values that were significantly different (P < 0.05) from the WT values are indicated by a bar and the P value. Values were not significantly different (n.s.) from the WT values are also indicated. (E) The Ca2+ content of the SR was estimated on the basis of the amplitude of caffeine-induced Ca2+ transients (left; F/F0 [ratio of fluorescence intensities, i.e., peak fluorescence intensity divided by baseline diastolic fluorescence intensity, in arbitrary units]). The time constant τ of the caffeine-induced Ca2+ transient was calculated to estimate NCX function (right). n.s., not significant. (F and G) Representative original records for MK2/3−/− and WT cardiomyocytes at a stimulation frequency of 1 Hz (F) and 4 Hz (G). (Left) Ca2+ transient amplitude and decay. F/F0 is the ratio of fluorescence intensities, specifically the peak systolic fluorescence intensity divided by baseline diastolic fluorescence intensity (fluorescence intensity in arbitrary units). (Right) Twitch amplitude and relaxation (change in diastolic resting cell length [ΔRCL] as a percentage). (H) Relaxation kinetics of Ca2+ transients (left) and twitches (right) at a stimulation frequency of 1 and 4 Hz. The values are means ± SD (error bars) of time from peak to half relaxation values, the relaxation time (RT50), (in milliseconds). Values that were significantly different (P < 0.05) from the value for the WT at the same stimulation frequency are indicated by an asterisk. (I) Ca2+ transient amplitude (left) and cardiomyocyte shortening (right) at a stimulation frequency of 1 and 4 Hz. Values are the means ± SD of values from Fluo-3 (F/F0) and fractional shortening (percent resting cell length [% RCL]) measurements. Values that were significantly different (P < 0.05) from the value for the WT at the same stimulation frequency are indicated by an asterisk.
SR Ca2+ reuptake by SERCA2a accounts for about 90% of Ca2+ transient decay in murine cardiomyocytes (68). Given the high SERCA2a protein levels and increased PLB phosphorylation in MK2/3−/− heart muscles, we further studied possible functional consequences. We found accelerated decay of the transient increase in intracellular Ca2+ (Ca2+ transient) at 1- and 4-Hz stimulation rates in the DKO cardiomyocytes (Fig. 6F to H), demonstrating enhancement of SERCA2a function. Correspondingly, cardiomyocyte twitch relaxation was also faster in MK2/3-deficient cells than in WT cells (Fig. 6F to H). Furthermore, the amplitude of Ca2+ transients and cardiomyocyte shortening (the shortest systolic length relative to the resting diastolic length, a measure of contractility) were both significantly augmented in the DKO at the more physiological stimulation rate of 4 Hz (Fig. 6I). Thus, these functional changes are in accordance with an increased SERCA2a activity in MK2/3-deficient cardiomyocytes.
When comparing heart weight with total body weight (6.92 ± 0.35 mg/g in DKO mice [n = 5] versus 6.72 ± 0.37 mg/g in WT mice [n = 5]; P = 0.72), we did not find a sign of hypertrophy in MK2/3 DKO mice (see also Table S2 in the supplemental material for unchanged expression of markers of hypertrophy). Furthermore, cardiomyocyte dimensions also did not indicate hypertrophy (cardiomyocyte volume of 36.44 ± 2.29 pl in DKO mice [n = 24] versus 31.44 ± 3.81 pl in WT mice [n = 12]; P = 0.24). The cell length of DKO cardiomyocytes was 122.7 ± 4.1 μm compared to 124.7 μm in the WT cardiomyocytes (P = 0.82), indicating that they were not elongated and suggesting that cardiac dilation had not occurred.
DISCUSSION
In this study, we examined functional roles of MK2/3 in cross-striated muscles using a MK2/3-deficient animal model. We demonstrate that MK2/3 ablation leads to an increase in slow oxidative type I fiber content concomitant with higher fatigue resistance. We further identify SERCA2 and MyHCIId/x as novel MK2/3 target genes. SERCA2 is of highly functional and clinical relevance in cross-striated muscle. We demonstrate a profound increase in the abundance of SERCA2a protein in the absence of MK2/3. Mechanistically, altered SERCA2 promoter activity and SERCA2a protein level can be directly attributed to MK2/3 enzymatic activity via changes in the Egr-1 level and the ratio of Egr-1 to Sp1. Furthermore, faster relaxation and enhanced contractility of MK2/3-deficient cardiomyocytes are in accordance with increased SERCA2a function. Hence, results link MK2/3 to the regulation of calcium dynamics. These findings show the functional relevance of MK2/3 in determining cardiac and skeletal muscle function, generating a novel muscle phenotype exhibiting both reduced fatigability and enhanced force in the DKO (Fig. 7).
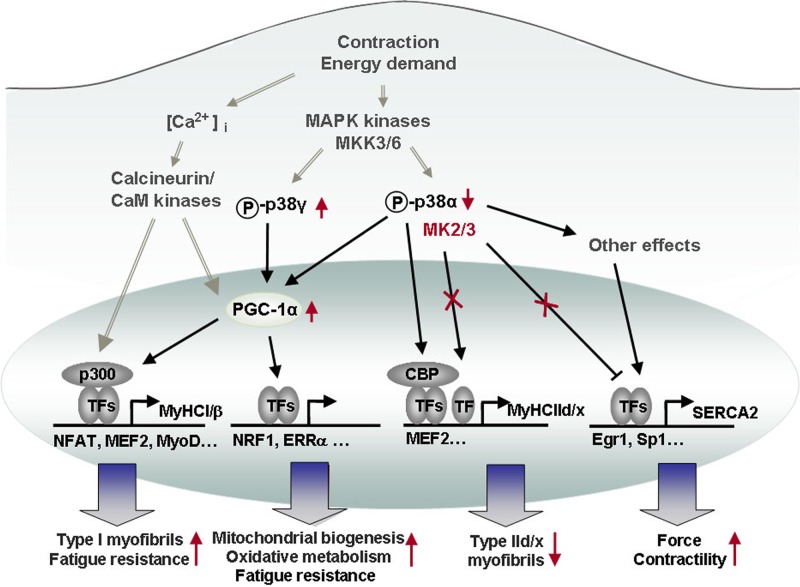
Fig 7.
Known and novel roles of the p38-MK2/3 signaling pathway in cross-striated muscles. In this model, we summarize previously established Ca2+/calcineurin signaling-dependent gene expression (6, 26) and contractile activity-dependent induction of the p38 pathway (16) (gray arrows with two gray lines). We also highlight known effects of p38 signaling on, e.g., the expression of fast MyHCIId/x (26) and PGC-1α (25), as well as reported downstream effects of PGC-1α (57, 82) (black arrows). The latter results were extended in the current study to show that MK2/3 deficiency results in a shift toward slow fiber type I and oxidative energy metabolism. MK2/3 regulates SERCA2 gene transcription via altered Egr-1 binding, and elevated SERCA2a induced by MK2/3 deficiency enhances cross-striated muscle function. Changes arising due to MK2/3 deficiency are denoted in red (↑, upregulation; ↓, downregulation, X, abrogation of signaling). [Ca2+]i, intracellular Ca2+ concentration; TFs, transcription factors; NFAT, nuclear factor of activated T cells; ERRα, estrogen-related receptor alpha.
As previously reported for MK2/3−/− immune cells, the level of p38α protein was also diminished in MK2/3−/− skeletal muscles and heart (30). However, a less robust decline in the basal level of activated p38α compared to total p38α protein indicates a compensatory upregulation of this signaling pathway. Since low-level kinase activities are often sufficient to maintain functional relevance and substrate phosphorylation, we compared the soleus muscle microarray analysis with previously identified p38α and p38γ target genes from small interfering RNA (siRNA)-based p38α and p38γ knockdown C2C12 muscle cells (12). Only a few positively regulated p38α or -γ target genes in C2C12 cells were affected in MK2/3−/− soleus. Many p38α- and p38γ-dependent genes remained unaffected (data not shown). Hence, differential gene expression in the WT versus MK2/3−/− soleus muscle is not merely a consequence of changes in total or activated p38 isoform levels. Nevertheless, increased p-p38γ in MK2/3−/− soleus might at least in part account for metabolic adaptations such as mitochondrial biogenesis and increased oxidative capacity. We demonstrated upregulation of transcripts such as CREB-1, MEF2C, NRF-1, and PGC-1α, in MK2/3-deficient soleus. CREB and MEF2 proteins control PGC-1α transcription (69), and NRF-1 and -2 interact with PGC-1α in the coordinate regulation of nuclear-gene-encoded mitochondrial transcription factors (56). An essential role of p38γ on PGC-1α gene expression has recently been uncovered in a loss-of-function genetic model (70). Robustly enhanced p-p38γ levels and transcriptional activation of the PGC-1α promoter by p38γ in MK2/3−/− cells suggest a mechanism of PGC-1α upregulation by increased p38γ activation. A possible reduction in the known p38α-mediated stabilization of PGC-1α mRNA (71) could be overridden in the DKO mice by the stimulatory effect of p38γ activation. Furthermore, the observed reduction of PGC-1α promoter activity by catalytically active MK2 suggests a complex mechanism of action including different kinases to fine-tune the level of expression of the PGC-1α gene.
Studies of the regulation of the SERCA2a gene provided mechanistic insights in changes resulting from altered p38-MK2/3 signaling. SERCA2a gene expression was previously found to be downregulated by activated p38 signaling in cultured neonatal cardiomyocytes and in vivo (72, 73). In accordance with our recent finding that p38-MK2 signaling plays a role in transcriptional activation of the immediate early gene Egr-1 via an MK2-dependent phosphorylation of serum response factor (SRF) (31), we identified a new target gene regulated by this axis by demonstrating a direct contribution of MK2 catalytic activity in the reduction of SERCA2 gene expression via increased expression and binding of Egr-1 to the SERCA2 promoter.
An accelerated Ca2+ transient decay and faster twitch relaxation were apparent in MK2/3-deficient cardiomyocytes, indicating enhanced SERCA2a activity in the absence of MK2/3, in line with augmented SERCA2a gene expression. The increased SERCA2a-to-PLB ratio permits SERCA2a activity changes, increased Ca2+ transient decay, and augmented contractility (8, 23, 74). In addition, PLB phosphorylation at the CaMKII site (T17) was induced in MK2/3−/− hearts, suggesting a further increase of SERCA2a activity. In accordance with the finding that SERCA2a becomes more relevant at higher stimulation frequencies (75), stimulation with 4 Hz increased Ca2+ transients and augmented contractility in MK2/3-deficient cardiomyocytes. These novel findings identify MK2/3 as a critical factor in modulating Ca2+ handling and cardiac muscle function. The absence of increase in a fetal gene (MyHCβ gene, skeletal alpha-actin gene), atrial natriuretic factor (ANF), or brain natriuretic protein (BNP) expression (see Table S2 in the supplemental material) could indicate that MK2/3 inhibition provides an approach to increase contractility without chronic pathological consequences.
In light of these results, it is tempting to speculate that the increase in total SERCA protein abundance in MK2/3−/− soleus might also contribute to the observed increase in force. In fact, augmented SERCA2a gene expression resulted in an increased SERCA2a-to-SLN ratio in MK2/3−/− soleus. Interestingly, overexpression of SLN in rat soleus reduced maximal Ca2+ transport activity, peak twitch force, and maximal tetanic force (76), indicating that SLN is also a key modulator of SERCA function in skeletal muscle contraction/relaxation in vivo (77). Thus, SLN could regulate SERCA2a function in skeletal muscle in a manner similar to that found in cardiac atria (64). Hence, a central finding of our work is the increase in SERCA2 expression in both heart and skeletal muscle, which is accompanied by unaltered transcript expression and high levels of SERCA2 regulatory proteins, presumably leading to increased SERCA2 activity and the observed changes in muscle performance.
Changes in MK2/3-deficient cross-striated muscles resemble in several ways those occurring during adaptation to increased physiological demands. Particularly high-intensity aerobic exercise training causes increased SERCA2a mRNA and protein levels in cardiomyocytes, while PLB abundance remains unaffected, and also enhances PLB phosphorylation at the CaMKII site but not at the PKA site (8). High-intensity aerobic exercise in skeletal muscle shifts fiber composition toward the composition of type I fibers and increases oxidative energy metabolism (78, 79). Therefore, changes in skeletal and heart muscle of MK2/3-deficient mice resemble the adaptations induced by high-intensity endurance exercise, suggesting a functional relevance of p38-MK2/3 signaling for adaptive processes imposed by training programs.
Our findings of changes in Ca2+ homeostasis due to MK2/3 deficiency might have clinical relevance. Impaired contractility related to disturbed Ca2+ handling is a hallmark of heart failure (80). Enhancing SERCA2a expression/function has been suggested as a novel therapeutic approach for cardiac dysfunction. In fact, virus-mediated SERCA2a gene delivery has been conducted recently in humans with advanced heart failure (81). Hence, our results imply that targeting the MK2/3 pathway provides a mean of enhancing or restoring SERCA2a expression and function, thus improving SR Ca2+ handling and cardiac function.
Thus, extended studies of the mouse model system employed here can help to further understand the mechanisms of skeletal and heart muscle adaptations. This could assist in the development of therapeutics for chronic diseases related to fiber type changes or impaired SERCA2 function in heart that may avoid the broad spectrum of potential side effects associated with p38 inhibition by selectively targeting MK2/3.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Ricci (ETH Zürich, Switzerland) for the gift of anti-p38δ antibody, D. M. Bers (Loyola University, Chicago, IL, USA) for anti-CaMKII and anti-phospho-CaMKII antibodies, F. Wuytack and P. Vangheluwe (Katholieke Universiteit [KU] Leuven, Belgium) for anti-SERCA2a antibody, and R. Davis (University of Massachusetts Medical School, Worcester, MA, USA) for the MKK6EE expression vector. We thank A. Kotlyarov (Medizinische Hochschule Hannover [MHH], Hannover, Germany) for helpful discussions and P. Wetzel (MHH, Hannover, Germany) for initial help with the isolation of muscle fibers. We thank A. Müller-Friedrichsen and B. Piep for expert technical assistance and T. Yakovleva for technical help with the mice.
This work was supported by the Deutsche Forschungsgemeinschaft (SCHE 309/5-1).
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 22 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01692-12.
REFERENCES
- 1. Pette D. 2002. The adaptive potential of skeletal muscle fibers. Can. J. Appl. Physiol. 27:423–448 [DOI] [PubMed] [Google Scholar]
- 2. Williams RS, Neufer PD. 1996. Regulation of gene expression in skeletal muscle by contractile activity, p 1124–1150 In Rowell LB, Shepherd JT. (ed), Handbook of physiology. Section 12. Exercise: regulation and integration of multiple systems. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 3. Booth FW, Baldwin KM. 1996. Muscle plasticity: energy demand and supply processes, p 1075–1123 In Rowell LB, Shepherd JT. (ed), Handbook of physiology. Section 12. Exercise: regulation and integration of multiple systems. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 4. Fluck M. 2006. Functional, structural and molecular plasticity of mammalian skeletal muscle in response to exercise stimuli. J. Exp. Biol. 209:2239–2248 [DOI] [PubMed] [Google Scholar]
- 5. Schiaffino S, Serrano A. 2002. Calcineurin signaling and neural control of skeletal muscle fiber type and size. Trends Pharmacol. Sci. 23:569–575 [DOI] [PubMed] [Google Scholar]
- 6. Bassel-Duby R, Olson EN. 2006. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75:19–37 [DOI] [PubMed] [Google Scholar]
- 7. Clerk A, Cullingford TE, Fuller SJ, Giraldo A, Markou T, Pikkarainen S, Sugden PH. 2007. Signaling pathways mediating cardiac myocyte gene expression in physiological and stress responses. J. Cell. Physiol. 212:311–322 [DOI] [PubMed] [Google Scholar]
- 8. Kemi OJ, Wisloff U. 2010. Mechanisms of exercise-induced improvements in the contractile apparatus of the mammalian myocardium. Acta Physiol. (Oxf.) 199:425–439 [DOI] [PubMed] [Google Scholar]
- 9. Cuenda A, Cohen P. 1999. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. J. Biol. Chem. 274:4341–4346 [DOI] [PubMed] [Google Scholar]
- 10. Marber MS, Rose B, Wang Y. 2011. The p38 mitogen-activated protein kinase pathway–a potential target for intervention in infarction, hypertrophy, and heart failure. J. Mol. Cell. Cardiol. 51:485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perdiguero E, Ruiz-Bonilla V, Gresh L, Hui L, Ballestar E, Sousa-Victor P, Baeza-Raja B, Jardi M, Bosch-Comas A, Esteller M, Caelles C, Serrano AL, Wagner EF, Munoz-Canoves P. 2007. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 26:1245–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H, Xu Q, Xiao F, Jiang Y, Wu Z. 2008. Involvement of the p38 mitogen-activated protein kinase alpha, beta, and gamma isoforms in myogenic differentiation. Mol. Biol. Cell 19:1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puri PL, Wu Z, Zhang P, Wood LD, Bhakta KS, Han J, Feramisco JR, Karin M, Wang JY. 2000. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev. 14:574–584 [PMC free article] [PubMed] [Google Scholar]
- 14. Keren A, Tamir Y, Bengal E. 2006. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 252:224–230 [DOI] [PubMed] [Google Scholar]
- 15. Ruiz-Bonilla V, Perdiguero E, Gresh L, Serrano AL, Zamora M, Sousa-Victor P, Jardi M, Wagner EF, Munoz-Canoves P. 2008. Efficient adult skeletal muscle regeneration in mice deficient in p38beta, p38gamma and p38delta MAP kinases. Cell Cycle 7:2208–2214 [DOI] [PubMed] [Google Scholar]
- 16. Widegren U, Ryder JW, Zierath JR. 2001. Mitogen-activated protein kinase signal transduction in skeletal muscle: effects of exercise and muscle contraction. Acta Physiol. Scand. 172:227–238 [DOI] [PubMed] [Google Scholar]
- 17. Gaestel M. 2006. MAPKAP kinases — MKs — two's company, three's a crowd. Nat. Rev. Mol. Cell Biol. 7:120–130 [DOI] [PubMed] [Google Scholar]
- 18. Ronkina N, Kotlyarov A, Dittrich-Breiholz O, Kracht M, Hitti E, Milarski K, Askew R, Marusic S, Lin LL, Gaestel M, Telliez JB. 2007. The mitogen-activated protein kinase (MAPK)-activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol. Cell. Biol. 27:170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk HD, Gaestel M. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1:94–97 [DOI] [PubMed] [Google Scholar]
- 20. Streicher JM, Ren S, Herschman H, Wang Y. 2010. MAPK-activated protein kinase-2 in cardiac hypertrophy and cyclooxygenase-2 regulation in heart. Circ. Res. 106:1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Periasamy M, Kalyanasundaram A. 2007. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve 35:430–442 [DOI] [PubMed] [Google Scholar]
- 22. Meyer M, Schillinger W, Pieske B, Holubarsch C, Heilmann C, Posival H, Kuwajima G, Mikoshiba K, Just H, Hasenfuss G. 1995. Alterations of sarcoplasmic reticulum proteins in failing human dilated cardiomyopathy. Circulation 92:778–784 [DOI] [PubMed] [Google Scholar]
- 23. Cerra MC, Imbrogno S. 2011. Phospholamban and cardiac function: a comparative perspective in vertebrates. Acta Physiol. (Oxf.) 205:9–25 [DOI] [PubMed] [Google Scholar]
- 24. Arany Z. 2008. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr. Opin. Genet. Dev. 18:426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Puigserver P, Spiegelman BM. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 24:78–90 [DOI] [PubMed] [Google Scholar]
- 26. Meissner JD, Umeda PK, Chang KC, Gros G, Scheibe RJ. 2007. Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway. J. Cell. Physiol. 211:138–148 [DOI] [PubMed] [Google Scholar]
- 27. Meissner JD, Freund R, Krone D, Umeda PK, Chang KC, Gros G, Scheibe RJ. 2011. Extracellular signal-regulated kinase 1/2-mediated phosphorylation of p300 enhances myosin heavy chain I/beta gene expression via acetylation of nuclear factor of activated T cells c1. Nucleic Acids Res. 39:5907–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brady M, Koban MU, Dellow KA, Yacoub M, Boheler KR, Fuller SJ. 2003. Sp1 and Sp3 transcription factors are required for trans-activation of the human SERCA2 promoter in cardiomyocytes. Cardiovasc. Res. 60:347–354 [DOI] [PubMed] [Google Scholar]
- 29. Wankerl M, Boheler KR, Fiszman MY, Schwartz K. 1996. Molecular cloning and analysis of the human cardiac sarco(endo)plasmic reticulum Ca(2+)-ATPase (SERCA2) gene promoter. J. Mol. Cell. Cardiol. 28:2139–2150 [DOI] [PubMed] [Google Scholar]
- 30. Kotlyarov A, Yannoni Y, Fritz S, Laass K, Telliez JB, Pitman D, Lin LL, Gaestel M. 2002. Distinct cellular functions of MK2. Mol. Cell. Biol. 22:4827–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ronkina N, Menon MB, Schwermann J, Arthur JS, Legault H, Telliez JB, Kayyali US, Nebreda AR, Kotlyarov A, Gaestel M. 2011. Stress induced gene expression: a direct role for MAPKAP kinases in transcriptional activation of immediate early genes. Nucleic Acids Res. 39:2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogilvie RW, Feeback DL. 1990. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fiber types I, IIA, IIB and IIC. Stain Technol. 65:231–241 [DOI] [PubMed] [Google Scholar]
- 34. Brooke MH, Kaiser KK. 1970. Three “myosin adenosine triphosphatase” systems: the nature of their pH lability and sulfhydryl dependence. J. Histochem. Cytochem. 18:670–672 [DOI] [PubMed] [Google Scholar]
- 35. Fraysse B, Rouaud T, Millour M, Fontaine-Perus J, Gardahaut MF, Levitsky DO. 2001. Expression of the Na(+)/Ca(2+) exchanger in skeletal muscle. Am. J. Physiol. Cell Physiol. 280:C146–C154 [DOI] [PubMed] [Google Scholar]
- 36. Kubis HP, Gros G. 1997. A rapid electrophoretic method for separating rabbit skeletal muscle myosin heavy chains at high resolution. Electrophoresis 18:64–66 [DOI] [PubMed] [Google Scholar]
- 37. Cannon-Carlson S, Tang J. 1997. Modification of the Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis procedure to eliminate artifacts on reducing and nonreducing gels. Anal. Biochem. 246:146–148 [DOI] [PubMed] [Google Scholar]
- 38. Geers C, Gros G. 1990. Effects of carbonic anhydrase inhibitors on contraction, intracellular pH and energy-rich phosphates of rat skeletal muscle. J. Physiol. 423:279–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barjaktarovic Z, Schutz W, Madlung J, Fladerer C, Nordheim A, Hampp R. 2009. Changes in the effective gravitational field strength affect the state of phosphorylation of stress-related proteins in callus cultures of Arabidopsis thaliana. J. Exp. Bot. 60:779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goodman T, Schulenberg B, Steinberg TH, Patton WF. 2004. Detection of phosphoproteins on electroblot membranes using a small-molecule organic fluorophore. Electrophoresis 25:2533–2538 [DOI] [PubMed] [Google Scholar]
- 41. Schulenberg B, Goodman TN, Aggeler R, Capaldi RA, Patton WF. 2004. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis 25:2526–2532 [DOI] [PubMed] [Google Scholar]
- 42. Lesuffleur T, Porchet N, Aubert JP, Swallow D, Gum JR, Kim YS, Real FX, Zweibaum A. 1993. Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J. Cell Sci. 106(Part 3):771–783 [DOI] [PubMed] [Google Scholar]
- 43. Meissner JD, Kubis HP, Scheibe RJ, Gros G. 2000. Reversible Ca2+-induced fast-to-slow transition in primary skeletal muscle culture cells at the mRNA level. J. Physiol. 523(Part 1):19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu KD, Lee WS, Wey J, Bungard D, Lytton J. 1995. Localization and quantification of endoplasmic reticulum Ca(2+)-ATPase isoform transcripts. Am. J. Physiol. 269:C775–C784 [DOI] [PubMed] [Google Scholar]
- 45. Christy BA, Lau LF, Nathans D. 1988. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc. Natl. Acad. Sci. U. S. A. 85:7857–7861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kadonaga JT, Carner KR, Masiarz FR, Tjian R. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079–1090 [DOI] [PubMed] [Google Scholar]
- 47. Rzeczkowski K, Beuerlein K, Muller H, Dittrich-Breiholz O, Schneider H, Kettner-Buhrow D, Holtmann H, Kracht M. 2011. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J. Cell Biol. 194:581–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Darby TG, Meissner JD, Ruhlmann A, Mueller WH, Scheibe RJ. 1997. Functional interference between retinoic acid or steroid hormone receptors and the oncoprotein Fli-1. Oncogene 15:3067–3082 [DOI] [PubMed] [Google Scholar]
- 49. Backs J, Backs T, Neef S, Kreusser MM, Lehmann LH, Patrick DM, Grueter CE, Qi X, Richardson JA, Hill JA, Katus HA, Bassel-Duby R, Maier LS, Olson EN. 2009. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc. Natl. Acad. Sci. U. S. A. 106:2342–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Neef S, Dybkova N, Sossalla S, Ort KR, Fluschnik N, Neumann K, Seipelt R, Schondube FA, Hasenfuss G, Maier LS. 2010. CaMKII-dependent diastolic SR Ca2+ leak and elevated diastolic Ca2+ levels in right atrial myocardium of patients with atrial fibrillation. Circ. Res. 106:1134–1144 [DOI] [PubMed] [Google Scholar]
- 51. Asmussen G, Schmalbruch I, Soukup T, Pette D. 2003. Contractile properties, fiber types, and myosin isoforms in fast and slow muscles of hyperactive Japanese waltzing mice. Exp. Neurol. 184:758–766 [DOI] [PubMed] [Google Scholar]
- 52. Luedeke JD, McCall RD, Dillaman RM, Kinsey ST. 2004. Properties of slow- and fast-twitch skeletal muscle from mice with an inherited capacity for hypoxic exercise. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 138:373–382 [DOI] [PubMed] [Google Scholar]
- 53. Huber B, Pette D. 1996. Dynamics of parvalbumin expression in low-frequency-stimulated fast-twitch rat muscle. Eur. J. Biochem. 236:814–819 [DOI] [PubMed] [Google Scholar]
- 54. Meissner JD, Chang KC, Kubis HP, Nebreda AR, Gros G, Scheibe RJ. 2007. The p38alpha/beta MAP kinases mediate recruitment of CBP to preserve fast myosin heavy chain IId/x gene activity in myotubes. J. Biol. Chem. 282:7265–7275 [DOI] [PubMed] [Google Scholar]
- 55. Leberer E, Hartner KT, Pette D. 1987. Reversible inhibition of sarcoplasmic reticulum Ca-ATPase by altered neuromuscular activity in rabbit fast-twitch muscle. Eur. J. Biochem. 162:555–561 [DOI] [PubMed] [Google Scholar]
- 56. Gleyzer N, Vercauteren K, Scarpulla RC. 2005. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 25:1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418:797–801 [DOI] [PubMed] [Google Scholar]
- 58. Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, Kang H, Shaw RJ, Evans RM. 2008. AMPK and PPARdelta agonists are exercise mimetics. Cell 134:405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carlson BM. 1968. Regeneration of the completely excised gastrocnemius muscle in the frog and rat from minced muscle fragments. J. Morphol. 125:447–472 [DOI] [PubMed] [Google Scholar]
- 60. Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. 2006. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 119:1824–1832 [DOI] [PubMed] [Google Scholar]
- 61. Gibson MC, Schultz E. 1983. Age-related differences in absolute numbers of skeletal muscle satellite cells. Muscle Nerve 6:574–580 [DOI] [PubMed] [Google Scholar]
- 62. Smith SJ, Fenwick PS, Nicholson AG, Kirschenbaum F, Finney-Hayward TK, Higgins LS, Giembycz MA, Barnes PJ, Donnelly LE. 2006. Inhibitory effect of p38 mitogen-activated protein kinase inhibitors on cytokine release from human macrophages. Br. J. Pharmacol. 149:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thomas T, Timmer M, Cesnulevicius K, Hitti E, Kotlyarov A, Gaestel M. 2008. MAPKAP kinase 2-deficiency prevents neurons from cell death by reducing neuroinflammation–relevance in a mouse model of Parkinson's disease. J. Neurochem. 105:2039–2052 [DOI] [PubMed] [Google Scholar]
- 64. Babu GJ, Bhupathy P, Timofeyev V, Petrashevskaya NN, Reiser PJ, Chiamvimonvat N, Periasamy M. 2007. Ablation of sarcolipin enhances sarcoplasmic reticulum calcium transport and atrial contractility. Proc. Natl. Acad. Sci. U. S. A. 104:17867–17872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Davis W, Jr, Chen ZJ, Ile KE, Tew KD. 2003. Reciprocal regulation of expression of the human adenosine 5′-triphosphate binding cassette, sub-family A, transporter 2 (ABCA2) promoter by the early growth response-1 (EGR-1) and Sp-family transcription factors. Nucleic Acids Res. 31:1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang RP, Fan Y, Ni Z, Mercola D, Adamson ED. 1997. Reciprocal modulation between Sp1 and Egr-1. J. Cell. Biochem. 66:489–499 [PubMed] [Google Scholar]
- 67. Zhang P, Tchou-Wong KM, Costa M. 2007. Egr-1 mediates hypoxia-inducible transcription of the NDRG1 gene through an overlapping Egr-1/Sp1 binding site in the promoter. Cancer Res. 67:9125–9133 [DOI] [PubMed] [Google Scholar]
- 68. Bers DM. 2002. Cardiac excitation-contraction coupling. Nature 415:198–205 [DOI] [PubMed] [Google Scholar]
- 69. Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. 2003. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc. Natl. Acad. Sci. U. S. A. 100:7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pogozelski AR, Geng T, Li P, Yin X, Lira VA, Zhang M, Chi JT, Yan Z. 2009. p38gamma mitogen-activated protein kinase is a key regulator in skeletal muscle metabolic adaptation in mice. PLoS One 4:e7934 doi:10.1371/journal.pone.0007934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol. Cell 8:971–982 [DOI] [PubMed] [Google Scholar]
- 72. Andrews C, Ho PD, Dillmann WH, Glembotski CC, McDonough PM. 2003. The MKK6-p38 MAPK pathway prolongs the cardiac contractile calcium transient, downregulates SERCA2, and activates NF-AT. Cardiovasc. Res. 59:46–56 [DOI] [PubMed] [Google Scholar]
- 73. Liao P, Georgakopoulos D, Kovacs A, Zheng M, Lerner D, Pu H, Saffitz J, Chien K, Xiao RP, Kass DA, Wang Y. 2001. The in vivo role of p38 MAP kinases in cardiac remodeling and restrictive cardiomyopathy. Proc. Natl. Acad. Sci. U. S. A. 98:12283–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E, Park WJ, Hajjar RJ. 2011. SUMO1-dependent modulation of SERCA2a in heart failure. Nature 477:601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bers DM. 2001. Excitation-contraction coupling and cardiac contractile force. Kluwer Academic Press, Dordrecht, The Netherlands [Google Scholar]
- 76. Tupling AR, Asahi M, Maclennan DH. 2002. Sarcolipin overexpression in rat slow twitch muscle inhibits sarcoplasmic reticulum Ca2+ uptake and impairs contractile function. J. Biol. Chem. 277:44740–44746 [DOI] [PubMed] [Google Scholar]
- 77. Tupling AR, Bombardier E, Gupta SC, Hussain D, Vigna C, Bloemberg D, Quadrilatero J, Trivieri MG, Babu GJ, Backx PH, Periasamy M, Maclennan DH, Gramolini AO. 2011. Enhanced Ca2+ transport and muscle relaxation in skeletal muscle from sarcolipin-null mice. Am. J. Physiol. Cell Physiol. 301:841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Linossier MT, Denis C, Dormois D, Geyssant A, Lacour JR. 1993. Ergometric and metabolic adaptation to a 5-s sprint training programme. Eur. J. Appl. Physiol. Occup. Physiol. 67:408–414 [DOI] [PubMed] [Google Scholar]
- 79. Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Deriaz O. 2003. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes 52:2874–2881 [DOI] [PubMed] [Google Scholar]
- 80. Neef S, Maier LS. 2007. Remodeling of excitation-contraction coupling in the heart: inhibition of sarcoplasmic reticulum Ca(2+) leak as a novel therapeutic approach. Curr. Heart Fail. Rep. 4:11–17 [DOI] [PubMed] [Google Scholar]
- 81. Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, Yaroshinsky A, Zsebo KM, Dittrich H, Hajjar RJ. 2011. Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation 124:304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. 2005. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J. Biol. Chem. 280:19587–19593 [DOI] [PubMed] [Google Scholar]
- 83. Agilent Technologies, Inc. 2008. One-color microarray-based gene expression analysis (Quick Amp labeling) protocol: for use with Agilent gene expression oligonucleotide microarrays, version 5.7. Agilent Technologies, Inc., Santa Clara, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.