Abstract
The formation, maintenance, and repair of epithelial barriers are of critical importance for whole-body homeostasis. However, the molecular events involved in epithelial tissue maturation are not fully established. To this end, we investigated the molecular processes involved in renal epithelial proximal-tubule monolayer maturation utilizing transcriptomic, metabolomic, and functional parameters. We uncovered profound dynamic alterations in transcriptional regulation, energy metabolism, and nutrient utilization over the maturation process. Proliferating cells exhibited high glycolytic rates and high transcript levels for fatty acid synthesis genes (FASN), whereas matured cells had low glycolytic rates, increased oxidative capacity, and preferentially expressed genes for beta oxidation. There were dynamic alterations in the expression and localization of several adherens (CDH1, -4, and -16) and tight junction (TJP3 and CLDN2 and -10) proteins. Genes involved in differentiated proximal-tubule function, cilium biogenesis (BBS1), and transport (ATP1A1 and ATP1B1) exhibited increased expression during epithelial maturation. Using TransAM transcription factor activity assays, we could demonstrate that p53 and FOXO1 were highly active in matured cells, whereas HIF1A and c-MYC were highly active in proliferating cells. The data presented here will be invaluable in the further delineation of the complex dynamic cellular processes involved in epithelial cell regulation.
INTRODUCTION
The primary function of epithelial cells is to form a selective barrier between two adjacent compartments. However, epithelial cells are not merely barriers but possess many other functions qualifying them as one of the most diverse group of cells in the human body. Epithelial cells can be considered the body's regulators, functioning in concert to allow precise homeostatic regulation. The loss of regulated epithelial function gives rise to many disease states, including chronic inflammation, fibrosis (leading to end-stage organ failure), and cancer. Indeed, the vast majority of all diagnosed cancers are of epithelial origin (termed carcinomas). In recent years, the use of epithelial cell cultures has allowed detailed studies of many cellular and molecular aspects of epithelial cell physiology and pathophysiology.
In the kidney alone, more than 12 morphologically distinct types of epithelial cells have been described (1). The proximal tubule is the main site of reabsorption in the nephron and is considered a “leaky” epithelium due to the expression of pore-forming claudins 2 and 10 in the tight junctions (2). The high transport rates of the proximal tubule are driven primarily by the Na+/K+ ATPase, creating a high energy demand that is predominantly fulfilled by oxidative metabolism (3). In addition, this part of the nephron is one of the areas most sensitive to chemical and ischemic injury (4–6).
The formation, maintenance, and repair of epithelial barriers are of critical importance for the maintenance of whole-body homeostasis and thus are very tightly regulated processes. Generally, the processes involved in epithelial proliferation are the opposite of those required for cell differentiation (7). Both states involve the sensing of the external environment, providing the cell with critical information pertaining to orientation, neighboring cells, and tissue density. These signals are relayed to the nucleus to activate transcription and alter processes involved in energy metabolism (8), differentiated function (7), proliferation (9), and motility (10). Tight and adherens junctions exert central control in the regulation of cell-to-cell contact and tissue density (11, 12). A low number of intercellular interactions favors epithelial cell proliferation, whereas a high density of intercellular contacts halts proliferation and promotes differentiation, allowing the establishment of specific tissue functions.
The transition of epithelial cells from proliferating, highly energy-consuming cells to a mature transporting epithelium is complex, involving the simultaneous activation and inactivation of several distinct pathways. In order to characterize these processes at a molecular level, we used a simple cell culture system where renal epithelial cells were seeded at low density in culture dishes. Primary human proximal-tubule cells and a telomerase-immortalized human proximal-tubule cell line (RPTEC/TERT1) were studied over a maturation process of 16 days. Transcriptomics and metabolomics were used to investigate the processes involved in epithelial monolayer maturation.
MATERIALS AND METHODS
All chemicals were purchased from Sigma-Aldrich unless otherwise stated.
Cell culture.
Human primary proximal-tubule cells were prepared as previously described (13). Both primary and RPTEC/TERT1 (14) cells were cultured in hormonally defined medium that consisted of a 1:1 mixture of Dulbecco's modified Eagle's medium (DMEM; catalog no. 11966; Invitrogen) and Ham's F-12 nutrient mix (catalog no. 21765; Invitrogen) supplemented with 2 mM Glutamax (Invitrogen), 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml sodium selenite, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 ng/ml epithelial growth factor, and 36 ng/ml hydrocortisone. Routinely, cells were cultured in 10-cm dishes (Sarstedt) at 37°C in a 5% CO2 humidified atmosphere and subcultured by trypsinization. Medium was replaced three times per week, and the volumes were 2 ml for 6 wells, 10 ml for 10-cm dishes, and 1 ml for coverslips. RPTEC/TERT1 cells were used at passages 76 to 100. Primary cells were used at passage 3.
For studying epithelial monolayer formation, cells were freshly seeded to a density of 30% surface area and followed over the following 16-day interval. Medium was replaced every day.
Transcriptomic and array analysis.
Cells were cultured on 6-well plates, and RNA was isolated by cell lysis in RLT buffer (Qiagen) containing 1% (vol/vol) β-mercaptoethanol and further processed with a Qiagen RNeasy Protect cell minikit. For the synthesis of biotinylated cRNA, a MessageAmp II aRNA amplification kit (Ambion Inc.) with several modifications (15) was applied using the Theonyx automatic pipetting robot (Aviso GmbH). To optimize the cleanup, the Agencourt RNAClean (Beckman Coulter) method was used instead of a column-based method. RNA quality was controlled using a 28S/18S ratio with an Agilent Bioanalyzer 2100, and the RNA quantity was assessed with a NanoDrop (Thermo Fisher Scientific) ND-1000 photometer before processing. Amplified biotin-labeled cRNA (750 ng) was hybridized in a hybridization chamber for 20 h at 58°C on Illumina human HT12 v3 BeadChips, which assay 48,802 probes per sample. Afterwards, the chips were washed and stained with 1 μg/ml streptavidin-conjugated Cy3 (Amersham Biosciences). Chips were then dried by centrifugation, and fluorescence was detected by confocal laser scanning with an Illumina bead array reader (Illumina) at 532 nm and a 0.8-μm resolution. Using Illumina BeadStudio software, the data from all beads with the same probe were condensed to one value and further to ensure array quality based on different control bead parameters. Quantile normalization was applied to remove nonbiological variance between arrays (16).
Selection of significantly deregulated genes and cluster analysis of the unsupervised samples were performed using BRB ArrayTools (version 4) (17). Hierarchical clustering of samples using centered correlation and average linkage was carried out, and samples demonstrating a correlation lower than 0.5 to their biological replicates were considered outliers and excluded from further analysis (three primary-cell samples). A class comparison was conducted between all time points within each cell model with a cutoff of 1.5-fold change and a significance level for P of <0.001 for the univariate test. The log2 fold change value of the differentially expressed probes (DEP) is represented here as the ratio of the value at the later time point to that at the earlier time point. DEPs from the RPTEC/TERT1 and primary-cell data sets were intersected according to direction of change. Thus, only DEPs that changed in the same direction in both cell models were used for further analysis. The combined list (1,630 DEPs) was then subjected to a time course analysis in BRB ArrayTools with a threshold false discovery rate of 0.01. The expression values of the resulting 1390 DEPs were normalized by “mean centering” (expression values of each probe were divided by the mean expression of that probe over all time points within each cell model). Heat maps were generated using TreeView (http://rana.lbl.gov/EisenSoftware.htm). Where multiple probes existed for the same gene, the probe with the highest variance across the RPTEC/TERT1 time course was chosen. Finally, a list of 1,238 differently expressed genes (DEG) reflecting gene alterations during the time course of monolayer formation was generated.
Quantitative real-time PCR (qPCR).
At 1, 7, and 16 days after seeding, RNA was harvested from RPTEC/TERT1 cells on 10-cm dishes as described above. cDNA was synthesized from 500 ng of total RNA using a Dynamo cDNA synthesis kit (Biozyme). qPCRs were performed using 5× HOT FIREPol EvaGreen qPCR Mix Plus (Medibena) on a Rotor-Gene Q (Qiagen) according to the manufacturer's protocol. Three biological samples were analyzed with 4 technical replicates each. A standard curve was generated using a dilution series of a reference RPTEC/TERT1 sample. Primer pairs used for amplification of the target genes are given in Table 1.
Table 1.
qPCR primer sequences
| Gene symbol | Forward primer | Reverse primer |
|---|---|---|
| ATP1A1 | GGGAGCTGCTCTGTGCTTTT | CTTATCACGTCCAACCCCCT |
| ATP1B1 | GCTTATCACGTCCAACCCCCT | GGCACATCGCCACAATCTTC |
| BBOX1 | ACCTTCAAGATGAATCCAGGTGA | GACCACATCCCAGTCAGCAT |
| BBS1 | GTCCCCGTCTTCCTAGAGGT | GGGGTGCTTGGAGTCTCTTC |
| CDKN1A | GGCGGCAGACCAGCATGACAGATT | GCAGGGGGCGGCCAGGGTAT |
| CDC25A | TCGTCTGAAGAAGCTCTGAGG | CCCTCTCCAAATGTCACACA |
| FASN | GATGACCGTCGCTGGAAGG | AATCTGGGTTGATGCCTCCG |
Transcription factor (TF) prediction analysis.
The complete list of 1,390 DEPs and their mean-centered expression values were uploaded into Ingenuity Pathway Analysis (IPA) version 9.0 (1,353 mapped genes; in cases of multiple probes for one gene, the probe with the maximum absolute log2 ratio was used, and for prediction of TF activation, direct and indirect relationships were considered where confidence at an experimental level was available), which includes a TF analysis feature. TF analysis computes an overlap P value (Fisher's exact test) assessing whether there is a statistically significant overlap between the genes in the data set and the genes that are regulated by a TF. The activation state of the TF is predicted by a second parameter, the z score, that reflects the expected causal effects between a TF and its targets based on the expression direction of the genes in the data set regulated by the given TF. A z score greater than 2 predicts significant activation and a score lower than −2 points to an inhibition of the given TF. For the refined selection of TFs, only TFs with an overlap P value of ≤0.001, a z score of ≥2 or ≤−2 at least at one time point in both cell models and more than 10 altered target molecules were considered.
Transcription factor activity assays.
Nuclear extracts from RPTEC/TERT1 cells cultured in 10-cm dishes at 1 day (subconfluent) and 16 days (matured) after seeding were washed and scraped into ice-cold hypotonic buffer [10 mM HEPES-NaOH, pH 7.9, containing 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), protease inhibitor cocktail (catalog no. P8340; Sigma), phosphatase inhibitor cocktail (catalog no. P0044; Sigma), and 2 mM activated Na3VO4]. The cell suspensions were incubated for 20 min on ice, and then 10% (vol/vol; final concentration, 0.58%) Igepal CA-630 was added to lyse the cells. Samples were centrifuged at 21,000 × g for 1 min, and the resulting pellets were resuspended in high-salt buffer [20 mM HEPES-NaOH, pH 7.9, containing 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% (vol/vol) glycerol, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail, phosphatase inhibitor cocktail and 2 mM activated Na3VO4]. Samples were incubated for 30 min on ice with periodic vortexing and were subsequently centrifuged at 21,000 × g for 10 min at 4°C. The resulting supernatants were removed as the nuclear extracts. The protein content of nuclear extracts of RPTEC/TERT1 cells was measured using the bicinchoninic acid (BCA) method according to the manufacturer's protocol (Pierce, Thermo Scientific). For the assays of TP53, FOXO1, and c-MYC, 20 μg nuclear extract per well was used, and for HIF1A, 30 μg nuclear extract per well was used. Levels of transcriptionally active TP53, HIF1A, FOXO1, and c-MYC were determined by using TransAM transcription factor enzyme-linked immunosorbent assays (ELISAs) (Active Motif) according to the manufacturer's protocol.
Cell cycle analysis.
At indicated time points, RPTEC/TERT1 cells cultured on 6-well plates, were washed twice in phosphate-buffered saline (PBS) and harvested using 1 ml phenol red-free 5% (wt/vol) trypsin–2% (wt/vol) EDTA solution per well after prolonged incubation at 37°C. Cell pellets were washed three times with ice-cold PBS and fixed in 100% ice-cold methanol. The fixed cells were stored at 4°C until analysis. On the day of analysis, cells were washed thrice with ice-cold PBS and stained with propidium iodide (PI)-RNase staining buffer (5 μg/ml PI, 200 μg/ml RNase) (Becton, Dickinson Biosciences). Cell cycle distribution was assessed using a Becton, Dickinson FACScan (Becton, Dickinson Biosciences) and BD Cell Quest Pro 4.0.2 software (25 000 positive hits, custom inclusion criteria). The data were then analyzed using BD ModFit.
Glycolysis and glycogen measurements.
Lactate was measured in the supernatant medium with a colorimetric assay, as previously described (18). Glucose was measured in the supernatant medium from the same samples using a colorimetric hexokinase-based assay, as previously described (19). Glycogen was measured in methanol extracts by boiling the insoluble fractions in 0.03 N HCl, followed by addition of 10 mM sodium acetate (pH 4.9), incubation of samples with or without amyloglucosidase (24 U/ml, from Aspergillus niger; catalog no. 10115; Sigma), and subsequent determination of glucose. Glycogen content was calculated as the difference between glucose levels with and without amyloglucosidase conversion.
1H nuclear magnetic resonance (NMR) spectroscopy and metabolic profile data processing.
For 1H NMR spectroscopy, 1 ml of cell culture medium (supernatant) was centrifuged for 5 min at 150 × g to remove dead cells and subsequently stored at −80°C to await NMR analysis. 1H NMR spectroscopy and data processing for metabolomic investigation were performed as described by Ellis et al. (20). Briefly, to 550 μl supernatant, 50 μl of D2O [containing 0.2% (wt/vol) TSP (trimethylsilyl propionate] was added, vortexed, and then centrifuged (13,000 × g, 10 min) to remove insoluble material before the mixture was transferred to a clean NMR tube. All reagents were checked prior to sample preparation by obtaining a one-dimensional 1H NMR spectrum to ensure that they contained no contaminants that may interfere with the downstream spectroscopic analysis. CPMG (Carr-Purcell-Meiboom-Gill) spectra were acquired on a 600-MHz Bruker spectrometer for each sample to suppress broad protein peaks. NMR spectroscopic data were imported and manipulated in Matlab (Mathworks) using in-house software written and compiled by T. M. D. Ebbels, H. C. Keun, J. T. Pearce, and R. Cavill. Using this software, the 1H NMR spectra were automatically phased, baseline corrected, and referenced and normalized to the TSP resonance at δ 0.
Immunofluorescence analysis.
RPTEC/TERT1 cells were cultured on 18-mm glass coverslips in 24-well plates and fixed in 100% ice-cold methanol for 10 min at −20°C at three states: subconfluent, newly confluent, and as a matured monolayer. Fixed cells were incubated at room temperature in blocking buffer (5% [wt/vol] bovine serum albumin [BSA; Calbiochem, Merck], 1% [vol/vol] Triton X-100 in PBS) for 30 min followed by incubation with primary antibody for 1 h. All antibodies were diluted in antibody diluent in a 1% BSA, 0.2% Triton X-100 PBS solution. Mouse anti-E-cadherin (no. 610182; BD Transduction Laboratories) was used at 0.31 μg/ml, rabbit anti-cadherin 4 (HPA015613; Sigma) at 0.44 μg/ml, rabbit anti-cadherin 16 (Abcam, ab80320) at 10 μg/ml, mouse anti-claudin 2 (no. 32-5600; Invitrogen) at 1.6 μg/ml, rabbit anti-claudin 3 (SAB4500435; Sigma) at 2 μg/ml, rabbit anti-claudin 10 (ab52234; Abcam) at 3.3 μg/ml and rabbit anti-ZO3 (no. 36-4000; Zymed, Invitrogen) at 1.25 μg/ml. Cells were incubated with 2.5 μg/ml of the appropriate Alexa 488-conjugated secondary antibody (no. A11059 and A21206; Invitrogen) for 35 min. Repeated washing with PBS was performed between the incubation steps. Cells were mounted in 3 mg/ml p-phenylene-diamine glycerol on a microscope slide and sealed with nail varnish. Fluorescence images were obtained using a Zeiss Axiophot fluorescence microscope mounted with a 63× oil immersion objective (1.4 numerical aperture; Zeiss). Images were captured with a cooled charge-coupled device (CCD) camera (Spot Diagnostics) using Metavue image processing software (Molecular Devices).
Western blot analysis.
RPTEC/TERT1 cells were lysed in RIPA buffer including protease inhibitor cocktail (catalog no. 8340; Sigma) and 10 μl/ml phosphatase inhibitor (catalog no. P0044; Sigma) at various time points. Samples were incubated for 30 min on ice and then centrifuged at 8,000 × g for 5 min to separate RIPA buffer-soluble and -insoluble fractions. Protein concentrations of the soluble fraction samples were determined by the BCA method according to the manufacturer's protocol (Pierce). The insoluble fraction was resuspended directly in 90 μl Laemmli buffer. Soluble (35 to 40 μg total protein in Laemmli buffer) and insoluble (15 μl of the resuspended pellet) samples were run on 4 to 12% bis-Tris minigels (Invitrogen). Gels for blots of proteins of <40 kDa were run in NuPage MES (morpholineethanesulfonic acid) SDS running buffer and those of proteins of >40 kDa in NuPage MOPS (morpholinepropanesulfonic acid) SDS running buffer (Invitrogen) at 200 V for 35 min and 50 min, respectively. Proteins were transferred onto methanol-activated low-fluorescence Immobilon-P membranes (Millipore, Merck) in 2× NuPage transfer buffer (Invitrogen) containing 20% methanol (for proteins of <40 kDa) or 10% methanol (for proteins of 40 to 110 kDa) for 30 or 60 min, respectively (maximum, 20 V) using a semidry transfer system (Invitrogen). Membranes were blocked in 5% (wt/vol) BSA (Calbiochem)–Tris-buffered saline-Tween (TBST) for 1 h and probed with primary antibodies (2 h at room temperature [RT] or overnight at 4°C). All antibodies were diluted in TBST, except for anti-cadherin 4, anti-cadherin 16, and anti-ZO3, which were diluted in 5% BSA-TBST. Mouse anti-E-cadherin (no. 610182; BD Transduction) was used at 0.1 μg/ml, rabbit anti-cadherin 4 (HPA015613; Sigma) at 0.44 μg/ml, rabbit anti-cadherin 16 (ab80320; Abcam) at 1 μg/ml, mouse anti-claudin 2 (no. 32-5600; Invitrogen) at 0.8 μg/ml, rabbit anti-claudin 3 (SAB4500435; Sigma) at 2 μg/ml, rabbit anti-claudin 10 (ab52234; Abcam) at 2 μg/ml, and rabbit anti-ZO3 (no. 36-4000; Zymed, Invitrogen) at 2.5 μg/ml. Mouse anti-beta actin (no. A2228; Sigma) at 0.1 μg/ml was used as an internal control. Blots were incubated with the appropriate secondary antibody conjugated to Cy3 or Cy5 (1:5,000; no. PA45011V and PA443009V; GE Healthcare) for 1 h at RT. Bands were detected using an Image Quant Las 4000 imager (GE Healthcare) and visualized and quantified with Image Quant TL image analysis software (GE Healthcare). For graphical representation, target values were normalized to the respective beta-actin band and expressed as the increase (fold) over maximum intensity.
Oxygen consumption analysis.
Oxygen was measured using oxygen sensor spots (PreSens) mounted on the inside of the upper lid of 10-cm dishes, connected on the outside to the accompanying fiber optic AD system Fibox 3 (PreSens). One hour before initiation of measurement, medium was renewed and equilibrated to 21% oxygen and 37°C in a 5% CO2 humidified atmosphere. Dishes were sealed with PetriSeal adhesive tape, and oxygen in the headspace was measured over a 15-min period at 3, 6, and 24 h after sealing. The measurement cycle was repeated every second day from day 1 to day 18 after seeding. Oxygen consumption was determined using an exponential decline fitting for greatest correlation and then integrated for the area over the curve (AOC). To adjust for cell number, 10 or more randomly spaced microscope images per dish were taken with a Zeiss Axiovert 100 microscope at a magnification of 320× before and after each measurement cycle, and cells were counted in an area of 0.08 mm2 using the Image J cell counting tool (National Institutes of Health). Average total cell number of a 10-cm dish was then calculated at the respective time points.
In addition, oxygen consumption measurements were performed in the presence of a 10 μM concentration of the oxidative phosphorylation uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) in order to elucidate the maximum oxygen consumption capacity. Measurements were conducted as described above for a maximum of 8 h. FCCP treatment was conducted in three states: subconfluent (day 1), newly confluent (day 6), and matured (days 25 to 30).
Stereology.
Total RPTEC/TERT1 cell volume was estimated by a stereology approach using light and electron microscopy, as previously described in detail (21). Briefly, newly confluent and matured RPTEC/TERT1 cells were washed with PBS, fixed in 1% (vol/vol) glutaraldehyde, and then postfixed in 1% (wt/vol) OsO4. Monolayers were dehydrated and embedded in Polybed (Polysciences) for transmission electron microscopy (TEM). Relative volume values (VV) of nuclei, mitochondria, and other subcellular structures (e.g., agglomerates of glycogen) were estimated at a magnification of ×5,000 by point counting (22) utilizing a square lattice and the relationship Vv = Pi/Pr × d, where Pi is the number of lattice cross points overlying the structure of interest i, Pr is the total number of lattice cross points over the reference structure r, and d is the lattice square length. Absolute values can be obtained by referring the relative values to the mean average cell volume of RPTEC/TERT1 cells. The latter value was obtained from the number of cells per unit area of culture dish and the mean thickness of the monolayer assessed from electron micrographs of strictly perpendicularly cut cell monolayers.
Statistics.
Statistical tests were applied to data in GraphPad Prism 5.02 and are specified in the legends. The error bars in the figures represent standard deviation (SD), and the n values are given in the legends.
RESULTS
Cells transition from mesenchymal to cobblestone epithelial morphology during the maturation process.
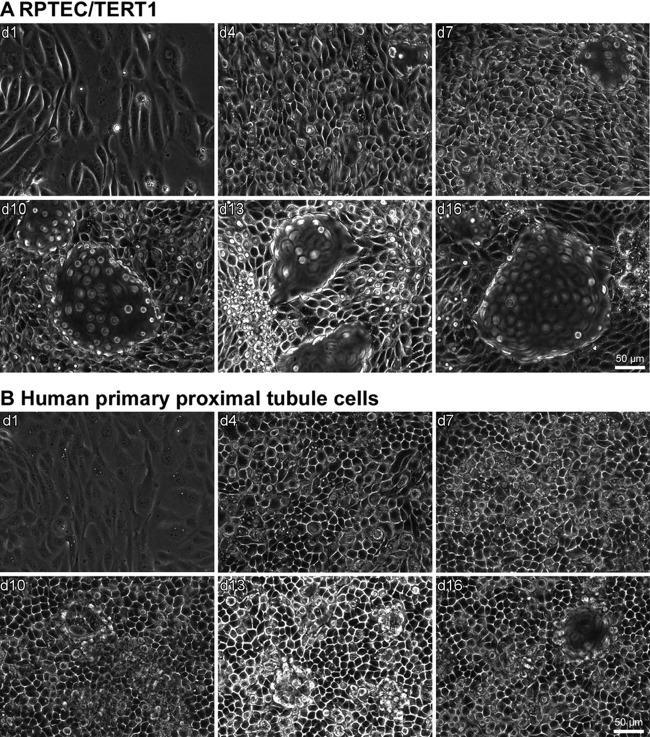
Phase-contrast images at different times of monolayer maturation showed that both primary and RPTEC/TERT1 cells matured from spindle-shaped subconfluent cells to cobblestone-shaped epithelial cells (Fig. 1). In addition, both cell types developed extensive dome formation at the later time points that are indicative of unidirectional transport of water and solutes and thus of a matured epithelium.
Fig 1.
Morphology of RPTEC/TERT1 (A) and primary cells (B) over the experimental time course. Phase-contrast images were captured at the indicated time points after seeding. Domes are visible as structures that are out of focus. Micrographs were obtained with a Zeiss Axiovert 100 microscope at a magnification of ×320.
The number of transcriptomic alterations decreased over the time course of the experiment.
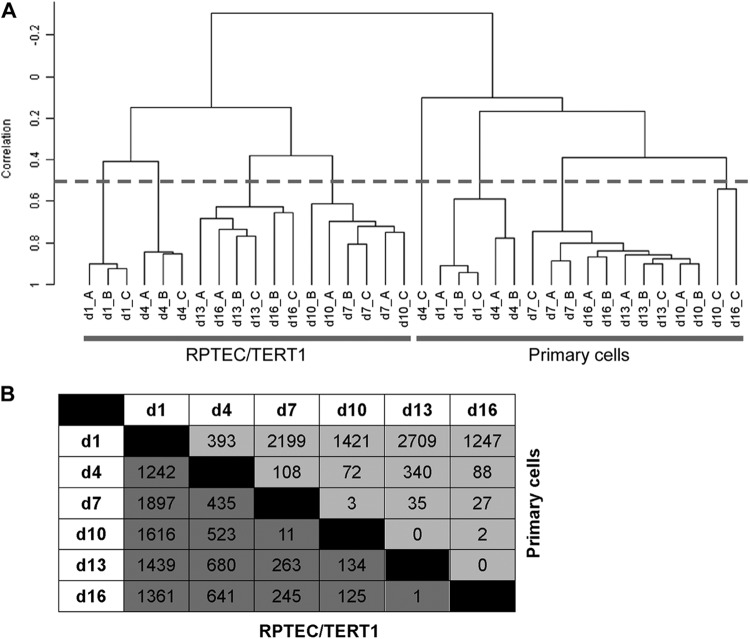
At 3-day intervals over the 16-day monolayer maturation process, RNA was harvested for whole-genome microarray analysis. Hierarchical clustering of the unsupervised samples revealed a high correlation (≥0.5) among the time-matched replicates (Fig. 2A). RPTEC/TERT1 and primary cells clustered separately from each other. In both cell types, two time-dependent clusters appeared (correlation ≥ 0.4), grouping early time points (days 1 and 4) away from later time points (days 7 to 16) (Fig. 2A). Class comparisons (1.5-fold change; P < 0.001) conducted between two time points uncovered a decreasing number of differentially expressed probes (DEPs) over the time of monolayer maturation, indicating a stabilization of the transcriptome as the monolayer matured (Fig. 2B).
Fig 2.
Overview of gene alterations over monolayer formation. (A) Hierarchical clustering. Dendrogram of the hierarchical clustering (centered correlation and average linkage) of the unsupervised samples. The dashed line indicates a correlation of 0.5. Samples that had a correlation of <0.5 with their biological replicates were excluded from further analysis (n = 3). (B) Matrix showing the number of delta differentially expressed probes (DEP) (1.5-fold change; P < 0.001) between two time points for each cell model. Dark and light gray boxes indicate the numbers of DEPs for RPTEC/TERT1 and primary cells, respectively.
Adherens junction proteins mediating cell adhesion increased during epithelial maturation, whereas those that support cell migration decreased.
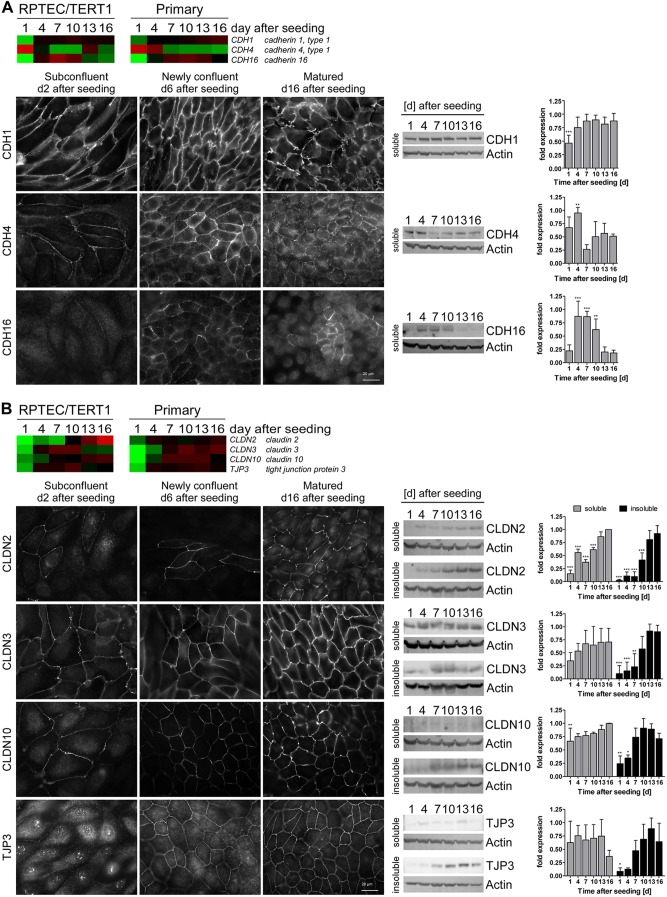
Since adherens junction (AJ) proteins are key players in mediating cell-cell contacts (23), we investigated the protein expression and localization of cadherins that were altered in the transcriptomic data. mRNA levels for cadherin 1 (encoded by CDH1; also known as E-cadherin) increased, whereas levels of the cadherin 4 gene (CDH4) decreased over time, exhibiting the highest expression on days 1 and 4 (Fig. 3A). Cadherin 16 (CDH16) mRNA and protein levels were highest at intermediate time points. CDH1 exhibited increased junctional targeting and abundance during maturation, whereas the opposite was true for CDH4 (Fig. 3A). CDH16, on the other hand, exhibited maximal junctional expression at confluence and became more diffuse after monolayer maturation (Fig. 3A). In the matured epithelium, CDH16 was junctionally expressed in a very small subset of the population which corresponded to cells at the apex of some domes (Fig. 3A).
Fig 3.
Adherens (A) and tight (B) junction protein expression during monolayer formation and maturation. (Top) Heat maps of adherens junction (AJ) and tight junction (TJ) protein genes. Average mean-centered mRNA expression values (log2) per time point and cell type were used to generate the heat maps. Cell colors reflect the value and direction of the expression value relative to the mean expression over all time points (red indicates higher expression and green indicates lower expression compared to the mean) (n = 3). (Bottom) Immunofluorescence staining and Western blots including band intensity quantification (RIPA buffer-soluble and -insoluble fractions; minimum n = 4) of AJ and TJ proteins in RPTEC/TERT1 cells at different time points after seeding. Band intensities were normalized first to actin and then to the maximum intensity per replicate. Values are means plus SD; statistical significances of changes in Western blot band intensities at day 16 were analyzed using one-way analysis of variance (ANOVA) with the Bonferroni multiple comparison test post hoc. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Tissue-specific tight junction proteins exhibited enhanced expression in the matured monolayer.
The tight junction (TJ) complex is one of the main regulators of cell density and contact inhibition (24) and also plays a role in the maintenance and regulation of the epithelial barrier. The transcript levels of genes for the TJ proteins claudin (CLDN-2, -3, and -10) and tight junction protein 3 (TJP3; also known as ZO3) increased over the time course (Fig. 3B). A similar expression pattern was observed at protein level (Fig. 3B). Additionally, CLDN2, -3, and -10 exhibited increased junctional expression over the maturation period. TJP3 exhibited predominant nuclear expression in proliferating cells but was exclusively junctionally expressed after confluence was reached (Fig. 3B). Additionally, TJP3 increased in protein abundance over the maturation period.
Transcriptome alterations associated with arrest in G0/G1 cell cycle phase.
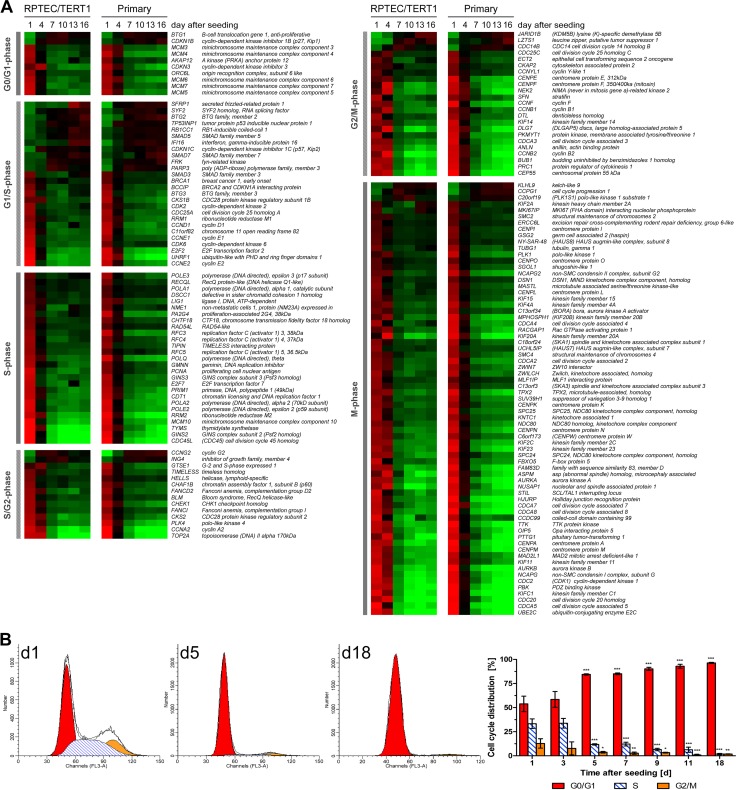
As expected, a large number of the differentially expressed genes are associated with cell cycle (Fig. 4A). For the most part, the expression levels of these genes were higher at early time points (day 1 and 4) and lower at the later time points (day 7 to 16). Many of the G1/S-phase genes exhibited the opposite expression pattern, however. This was observed for B-cell translocation gene 1, anti-proliferative (BTG1), cyclin-dependent kinase inhibitor 1B (CDKN1B, encoding a protein also known as p27), cyclin G2 (CCNG2), inhibitor of growth family, member 4 (ING4), CDC14 cell division cycle 14 homolog B (CDC14B), lysine (K)-specific demethylase 5B (JARID1B), leucine zipper, putative tumor suppressor 1 (LZTS1), polo-like kinase 1 substrate 1 (C20ORF19, encoding a protein also known as PLK1S1), cell cycle progression 1 (CCPG1), and kelch-like 9 (KLHL9).
Fig 4.
Cell cycle alterations over monolayer formation and maturation. (A) Heat map of cell cycle genes identified in the DEG list. Genes were assigned to G0/G1, G1/S, S, S/G2, G2/M, or M phase of the cell cycle and then ranked within a cell cycle phase according to the difference between day 1 and 16 values in the RPTEC/TERT1 data set. Average mean-centered mRNA expression values (log2) per time point and cell type were used to generate the heat map. Cell colors reflect the expression value and direction relative to the mean expression over all time points (red indicates higher expression and green indicates lower expression compared to the mean) (n = 3). (B) Cell cycle distribution of RPTEC/TERT1 cells at different time points after seeding. Values are means ± SD; statistical significance of changes compared to day 1 was analyzed using two-way ANOVA with the Bonferroni multiple comparison test post hoc. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (n = 3 to 6).
The gene expression pattern of these cell cycle-associated genes points to a highly proliferating status until day 4, followed by arrest in the G0/G1 phase. This was confirmed by cell cycle analysis using flow cytometry (Fig. 4B), which indicated that at a subconfluent state (day 1), 53.8% (±8.0%) of the cells were in G0/G1 phase, 33.3% (±5.0%) in S phase and 12.8% (±4.9%) in G2/M phase. Cell cycle distribution was similar on day 3 and shifted at day 5, with most cells being in G0/G1 phase (84.5% ± 0.7%), whereas the fractions in S phase (11.8% ± 0.7%) and G2/M phase (3.7% ± 0.8%) were decreased. This trend continued until day 18, where 96.4% (±0.6%) of the cells were in G0/G1 phase, 1.8% (±0.3%) were in S phase, and 1.8% (±0.7%) were in G2/M phase.
During the maturation process, cells switch from a highly glycolytic and oxygen-consuming metabolism to predominately oxidative metabolism and beta-oxidation of fatty acids.
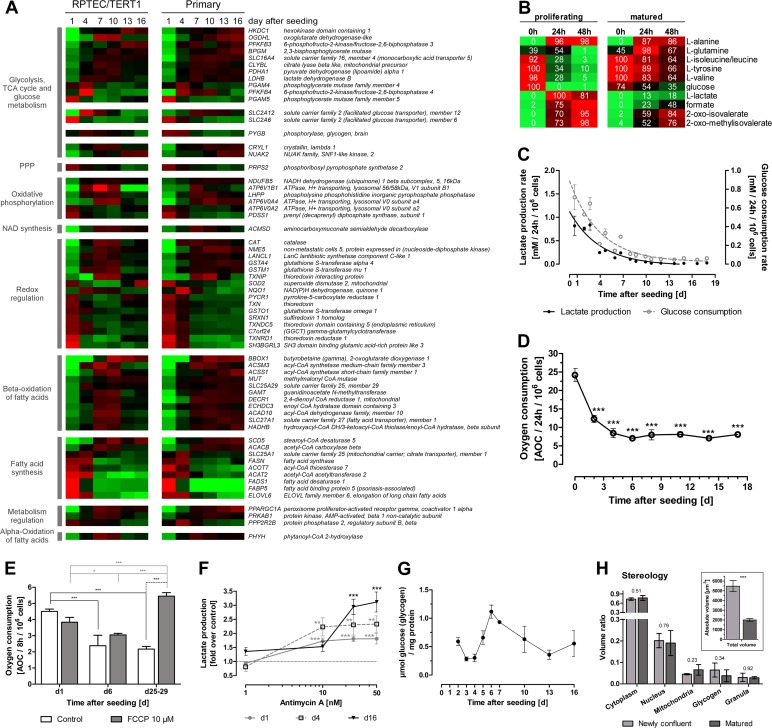
A number of genes involved in energy metabolism (glycolysis, tricarboxylic acid [TCA] cycle, oxidative phosphorylation, redox regulation, and fatty acid metabolism) exhibited altered expression during monolayer maturation (Fig. 5A). Two main expression patterns could be distinguished: a peak expression at early time points (day 1 to 4) and a peak expression at intermediate time points (day 7 to 10). Early highly expressed genes comprised those involved in fatty acid synthesis, including solute carrier family 25, member 1 (SLC25A1) and fatty acid synthase (FASN), as well as genes implicated in redox regulation, such as thioredoxin (TXN), thioredoxin reductase (TXNRD1), sulfiredoxin 1 homolog (SRXN1), glutathione S-transferase omega 1 (GSTO1), and gamma-glutamylcyclotransferase (GGCT). Two genes encoding members of the less well characterized class 3 glucose transporters, SLC2A6 and SCL2A12 (also known as GLUT6 and 12), exhibited the highest expression levels on day 1 and 4, respectively. Genes involved in redox regulation displayed the highest expression at intermediate time points and included catalase (CAT), glutathione S-transferase alpha 4 (GSTA4), mu 1 (GSTM1), thioredoxin interacting protein (TXNIP), and NAD(P)H dehydrogenase, quinone 1 (NQO1). Genes involved in glycolysis and TCA cycle also demonstrated peak expression at intermediate time points and comprised lactate dehydrogenase B (LDHB), pyruvate dehydrogenase (lipoamide) alpha 1 (PDHA1), and oxoglutarate dehydrogenase-like (OGDHL) as well as genes implicated in the beta-oxidation of fatty acids, such as butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (BBOX1), acyl-coenzyme A synthetase medium-chain family member 3 (ACSM3), short-chain family member 1 (ACSS1), and solute carrier family 25 and member 29 (SLC25A29). Additionally, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB) isoforms, responsible for the synthesis and degradation of fructose-2,6-bisphosphate, displayed an altered expression profile: PFKFB3 was decreased in proliferating cells, whereas PFKFB4 was increased.
Fig 5.
Alterations in energy metabolism and mitochondrial function over monolayer formation and maturation. (A) Heat map of energy metabolism-associated genes. Genes are grouped according to their function in different metabolic pathways and then ranked within the group to the difference between day 1 and 16 values in the RPTEC/TERT1 data set. Average mean-centered mRNA expression values (log2) per time point and cell type are represented in red (higher expression compared to the mean) and green (lower expression compared to the mean) (n = 3). (B) Metabolic alterations measured by NMR in RPTEC/TERT1 supernatant. Values are minima and maxima, in percent, normalized per metabolite. Colors represent abundances of the respective metabolites, where red indicates high abundance and green indicates low abundance (n = 3). (C) Lactate production and glucose consumption rates over RPTEC/TERT1 maturation time. n = 3. (D) Oxygen consumption rate over RPTEC/TERT1 maturation time. Statistical significance was analyzed using one-way ANOVA with the Bonferroni multiple comparison test post hoc (n = 3). (E) Oxygen consumption of RPTEC/TERT1 cells after treatment with oxidative uncoupler FCCP for 8 h. Two-way ANOVA with the Bonferroni multiple comparison test post hoc was used to analyze statistical significance (n = 3). (F) Lactate production of RPTEC/TERT1 cells after treatment with the electron transfer chain complex III inhibitor antimycin A for 8 h. Statistical significance was analyzed using one-way ANOVA with the Bonferroni multiple comparison test post hoc to time-matched control (n = 3). (G) Glycogen storage at different time points after seeding (n = 3). (H) Relative volume of RPTEC/TERT1 cell fractions, as determined by stereology. Insert depicts the calculated absolute volume of newly confluent and matured cells. Statistical significance was analyzed by applying a two-tailed unpaired t test. P values are indicated above the bars (n = 3). (B to H) Experiments were performed with RPTEC/TERT1 cells. (C to H) Values are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. PPP, pentose phosphate pathway.
NMR profiling of supernatant medium in proliferating and matured RPTEC/TERT1 cells demonstrated a decreased utilization of glutamine, tyrosine, valine, isoleucine, and glucose in the matured epithelium (Fig. 5B). The glutamine and alanine profiles are of particular interest, since the utilization of both spiked at 24 h, most likely due to catabolism of the medium supplement l-alanyl-l-glutamine (Glutamax, 2 mM). Membrane-bound aminopeptidases hydrolyze the dipeptide, releasing alanine and glutamine into the culture medium. Alanine did not seem to be further used by the cells when they were either proliferating or matured, but glutamine was consumed faster in proliferating cells. Additionally, there was a decreased rate of production of oxo-isovalerate, oxo-methylisovalerate, formate, and lactate in the matured epithelium. In order to compare the kinetics of glycolysis between these two states, we measured lactate and glucose in the supernatant medium over the maturation process with biochemical assays. Glucose utilization and lactate production steadily decreased over the time course and reached a steady state at day 8 (Fig. 5C).
We originally hypothesized that this glycolytic switch would be paralleled by a switch to oxidative metabolism. However, oxygen consumption measurements revealed that proliferating cells consumed significantly more oxygen than the matured epithelium (Fig. 5D). To examine further the respiration states at different time points after seeding, we applied the uncoupler FCCP, which disrupts the proton gradient across the mitochondrial inner membrane. This causes an increase in the intracellular concentration of ADP stimulating the transfer of electrons to oxygen molecules (25). FCCP treatment can thus be used to measure the capacity of the electron transfer system. Proliferating cells and newly confluent cells showed no altered respiration with FCCP compared to their time-matched control (Fig. 5E), whereas FCCP treatment of matured monolayers resulted in a significantly higher rate of oxygen consumption than was seen in time-matched control and proliferating cells (Fig. 5E). Inhibition of complex III of the electron transfer chain by antimycin A caused a small increase in lactate production in proliferating cells, whereas nonproliferating cells reverted to a highly glycolytic metabolism (Fig. 5F). Stereological analysis of matured cells demonstrated a 2.5-fold decrease in cell volume (Fig. 5H, inset) paralleled by a slight, although not significant, increase in mitochondrial volume (Fig. 5H). In line with this trend is the observation that peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PPARGC1A), a gene involved in mitochondrial biogenesis, exhibited increased expression in matured cells (Fig. 5A). A trend pointing to a reduction in glycogen stores was also observed in matured cells by stereological analysis (Fig. 5H). Biochemical measurement of glycogen revealed the highest glycogen storage after the most intense aerobic glycolysis phase, which declined afterwards to basal levels (Fig. 5G). During this peak, the gene encoding the glycogen-degrading enzyme glycogen phosphorylase, brain (PYGB), was highly expressed (Fig. 5A).
Taken together, these data suggest that proliferating cells run both oxidative and glycolytic metabolism at full capacity (Fig. 5C to E), while matured cells exhibit lower energy requirements but have a higher capacity for oxidative metabolism. Furthermore, disruption of oxidative phosphorylation with antimycin A reactivates glycolysis in matured monolayers (Fig. 5F).
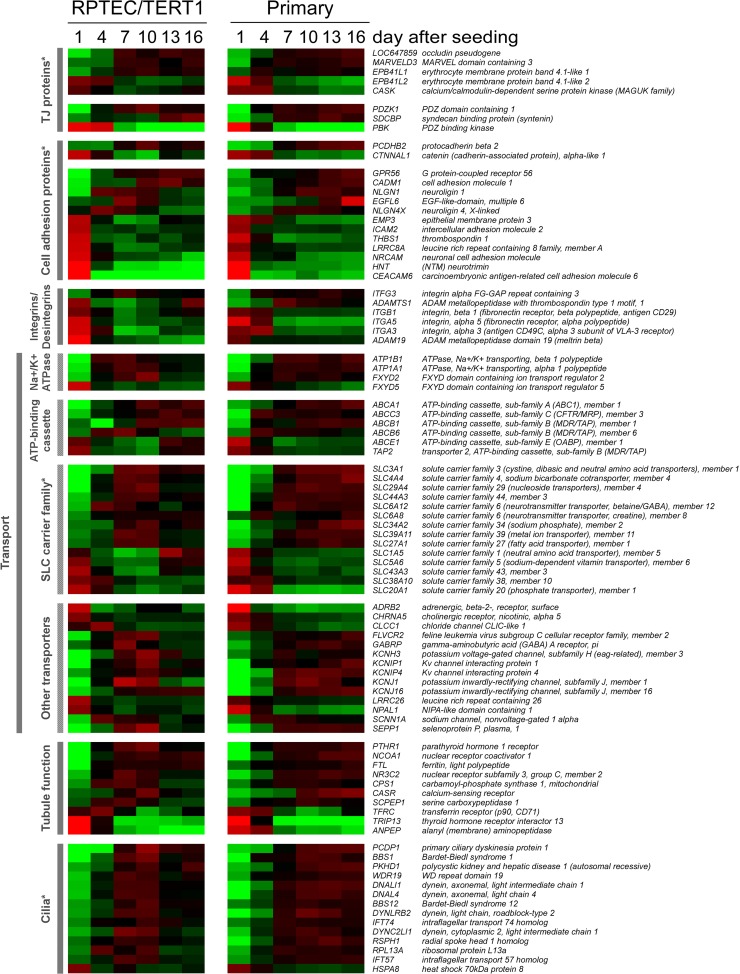
Genes associated with transport, proximal-tubule differentiation, and ciliogenesis were increased in the matured epithelium.
The proximal tubule is the main site in the nephron for solute and nutrient reabsorption. Interestingly, the transcript levels of genes encoding three subunits of the Na+/K+ ATPase (ATP1A1, ATP1B1, and FXYD2), which is the driving force for transport processes in the proximal tubule, increased over time, whereas subunit FXYD5, associated with decreased cell adhesiveness (26), showed the opposite expression profile (Fig. 6). Several members of the ATP-binding cassette transporter family and solute carrier family exhibited altered expression over the time course. Expression of several genes involved in important proximal-tubular functions, including serine carboxypeptidase 1 (SCPEP1) (27) and parathyroid hormone 1 receptor (PTHR1) (14), were increased during the maturation process, whereas expression of a few also decreased, for example, alanyl (membrane) aminopeptidase (ANPEP) (Fig. 6). We also identified an augmentation in the expression of several genes implicated in cilium biogenesis and maintenance, including dynein, cytoplasmic 2, light intermediate chain 1 (DYNC2LI1), Bardet-Biedl syndrome 1 and 12 (BBS1, BBS12), intraflagellar transport 57 homolog (IFT57), and primary ciliary dyskinesia protein 1 (PCDP1) (Fig. 6).
Fig 6.
Functional grouping of genes exhibiting time-dependent expression during epithelial monolayer maturation. Genes were ranked within the allocated group according to the difference between day 1 and 16 values in the RPTEC/TERT1 data set. Cell colors reflect the value and direction of the expression value relative to the mean over all time points (red indicates higher expression and green indicates lower expression compared to the mean) (n = 3). An asterisk indicates that the group contains members that are shown in previous heat maps.
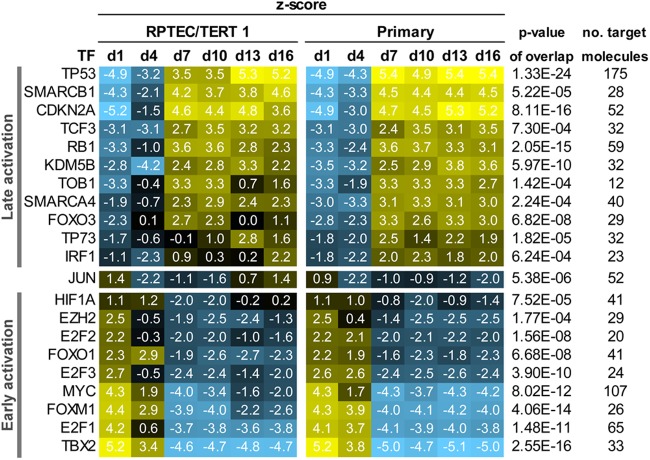
Identification of transcription factor master regulators of proliferation and maturation.
The list of differentially expressed probes over the time course was subjected to transcription factor (TF) analysis in IPA. The refined analysis yielded 9 TFs with predicted early activation, 12 activated at later time points, and 1 (JUN) that was activated at both times (Fig. 7). Among the early active TFs were TBX2, FOXM1, MYC, HIF1A, and E2F1-3, and among the later active TFs, TP53, CDKN2A, RB1, KDM5B, and FOXO3 were identified.
Fig 7.
Predicted transcription factors (TFs) involved in the maturation process. Colors reflect the regulation z score and direction of regulation: yellow indicates activation and blue indicates inhibition of the given TF. All TFs in this table had a P value of <0.001 for the overlap and a z score of >2 or <−2 at at least one time point in both cell models.
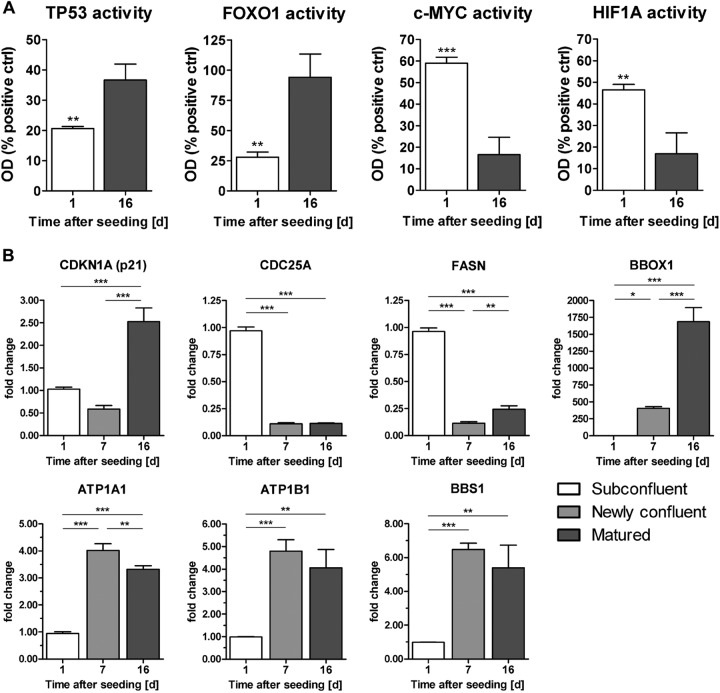
From four TFs of the list of predicted TFs activity-ELISAs were performed. These assays confirmed that TP53 had the highest activity in matured cells, whereas HIF1A and c-MYC exhibited the highest activity in proliferating cells (Fig. 8A). In contrast to the prediction of its activation state, FOXO1 was highly active in matured cells.
Fig 8.
(A) Activation levels of TFs TP53, FOXO1, c-MYC, and HIF1A in RPTEC/TERT1 nuclear extracts at days 1 and 16 after seeding. Values are means plus SD (n = 3). Statistical significance was analyzed by applying a two-tailed unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) qPCR of selected genes in RPTEC/TERT1 cells at subconfluent (day 1 after seeding), newly confluent (day 7 after seeding), and matured (day 16 after seeding) time points. Values are changes in copy number (fold) relative to biological replicate 1 on day 1. Values are means and SD (n = 3). Statistical significance was analyzed using one-way ANOVA with the Bonferroni multiple comparison test post hoc. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Transcript levels of some selected genes were quantified in independent samples by qPCR (Fig. 8B). We selected a panel of genes involved in cell cycle regulation [CDKN1A (p53 driven) and CDC25A (c-MYC driven)], fatty acid metabolism (FASN and BBOX1), transport (ATP1A1 and ATP1B1), and ciliogenesis (BBS1). The gene cyclin-dependent kinase inhibitor 1A (CDKN1A; the protein is also known as p21), which mediates the p53-dependent cell cycle G1 phase arrest, was significantly more highly expressed only in the matured monolayer (day 16 after seeding). The gene for cell division cycle 25A (CDC25A), which promotes the transition from G1 to the S phase of the cell cycle and is a target of c-MYC, exhibited an expression profile contrary to that of CDKN1A and was highly expressed in the subconfluent and newly confluent (day 1 and day 7 after seeding, respectively) monolayers, which corresponded to the microarray data. The gene for fatty acid synthase (FASN) exhibited reduced expression in the newly confluent monolayer, whereas the transcript levels of the genes for butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (BBOX1), a gene involved in l-carnitine synthesis, dramatically increased in the newly confluent monolayer and further increased in the matured monolayer; the same expression trend was observed in the microarray data. Expression of genes for the alpha and beta subunits (ATP1A1 and ATP1B1, respectively) of the Na+/K+ ATPase increased in the newly confluent monolayer, and high expression was sustained in the matured monolayer, consistent with the microarray data. The gene for the protein Bardet-Biedl syndrome 1 (BBS1) is implicated in cilium function, and it exhibited augmented transcript levels beginning in the newly confluent monolayer and continued to be highly expressed in the matured monolayer, as in the microarray data. The qPCR data for the selected genes corresponded well to the microarray expression data.
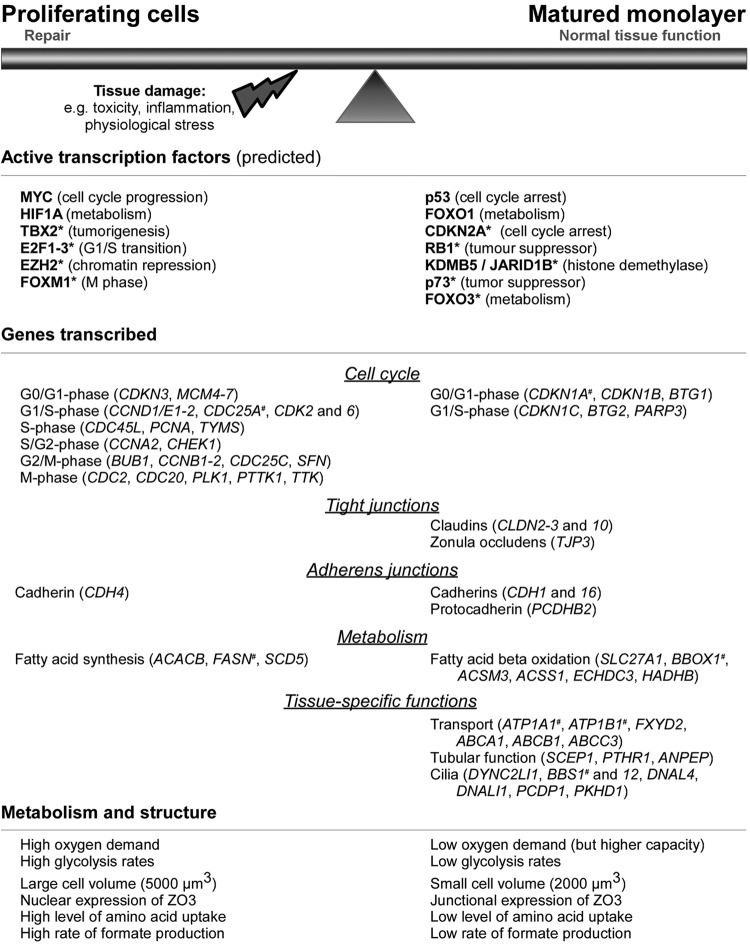
Figure 9 summarizes the main results.
Fig 9.
Summary of key mechanisms and players in epithelial monolayer maturation. *, predicted transcription factor; #, expression confirmed using qPCR.
DISCUSSION
Both primary human renal proximal-tubule cells and the telomerase-expressing human proximal-tubule cell line RPTEC/TERT1 exhibited similar growth rates and similar timelines of transition from mesenchymal-type morphology to confluent monolayers and ultimately dome-forming monolayers. Cadherins are integral members of the adherens junctions that function as intercellular adhesion molecules (28, 29). Here we observed an increase of CDH1 (also known as E-cadherin) and CDH16 mRNA with a decrease in CDH4 mRNA over the maturation time. CDH1 protein was expressed in high abundance at the cell-to-cell junctions throughout the maturation process. CDH1 is pivotal for tissue formation and forms homo-trans-interactions with neighboring cells and homo-cis-interactions within its own cell plasma membrane. The combination of trans and cis interactions promotes the formation of a lattice-type adherens belt (30). CDH4 exhibited sparse junctional localization at the early time points, but junctional expression increased in the intermediate phase. Total protein expression was high at days 1 and 4 and decreased thereafter. These findings support the role of CDH4 as a facilitator of cell motility (31). CDH16 is a nonclassical cadherin lacking the histidine-alanine-valine adhesion recognition sequence and is thought to play a role in maintenance of terminally differentiated tubules rather than initiation of renal morphogenesis (32). Here, CDH16 was absent in proliferating cells, but junctional expression increased dramatically at confluence. In the matured epithelium, there was a decrease in CDH16 total protein, and expression was limited to the apices of cells in dome-forming regions. It has previously been shown that cells at the apices of domes are often mitotic (33), and thus it is plausible that CDH16 plays a role in maintenance of the epithelial barrier in dividing cells of the matured epithelium.
The expression of the claudin family of intercellular TJ proteins determines the paracellular selectivity of different epithelial barriers (34). Here, the expression of genes for several claudin subtypes increased during epithelial maturation, including CLDN2 and CLDN10. These subtypes are highly expressed in the human proximal tubule in vivo (35), and their pore-forming properties are thought to be the reason for the relative leakiness of this region compared to more distal segments (2, 36–38). We recently demonstrated that oxidative stress decreases the expression of CLDN2 and CLDN10 (39), which would support the notion that cell injury can cause a reversal of the differentiation process (40). Other members of the tight junction (TJ) complex, the zonula occludens (ZO) protein family, act as sensors of epithelial compactness and regulate proliferation through their interaction with and sequestration of transcription modifiers (7, 12, 41–43). In our transcriptomics data, genes involved in the cell cycle were highly enriched at the early time points, and cell cycle analysis showed that the majority of cells arrest in G0/G1 as soon as 5 days after seeding. A small proportion of cell turnover remains thereafter. We observed an induction of TJP3 (also known as ZO3) mRNA expression over time and a strong increase in protein levels. Additionally, we could show that TJP3 is expressed in the nucleus of proliferating cells, whereas cell-to-cell junction localization was observed exclusively in the matured epithelium. Thus, it is likely that TJP3 supports proliferation in subconfluent cells but promotes cell cycle inhibition with increasing cell density, as has been previously described for TJP1 (also known as ZO1) and TJP2 (also known as ZO2) (44).
Proliferating cells require larger amounts of energy than nonproliferating cells for biomass synthesis (nucleotides, proteins, and membranes) (45). In our model, proliferating cells exhibited an increased utilization of glucose and amino acids, including glutamine, with a concomitant increased production of lactate and formate. Matured monolayers also exhibited lower oxygen consumption and lactate production rates, further indicating a decreased energy requirement. However, we demonstrated that their oxidative capacity was higher using the uncoupling agent FCCP, and stereology analysis revealed a slightly increased mitochondrial volume. In addition, transcript levels for peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PPARGC1A), encoding a crucial factor for mitochondrial biogenesis (46) and fatty acid beta oxidation (47, 48), were increased after confluence was reached. We also observed an increase in the mRNA for the catalytic α subunit of pyruvate dehydrogenase (PDHA1). In mitochondria, pyruvate dehydrogenase (PDH) converts pyruvate to acetyl coenzyme A (acetyl-CoA), which is used as a precursor for fatty acid synthesis or is fed into the TCA cycle (49). In addition, genes involved in fatty acid synthesis were highly expressed in proliferating cells, whereas genes implicated in beta-oxidation were predominantly expressed in matured cells. We have previously observed that exposure of differentiated renal and hepatic cells to toxins increases lactate production (18), which is likely due to impairment of mitochondrial oxidative respiration. Thus, to establish whether low glycolysis rates in matured cells are truly dependent on mitochondrial respiration, we measured glycolysis rates after exposure to the cytochrome c reductase inhibitor antimycin A. Matured cells reverted back to glycolytic metabolism, thus indicating that (i) their energy requirement is primarily fulfilled by oxidative metabolism and (ii) they can switch to glycolysis when oxidative metabolism does not meet their energy requirements.
The Na+/K+ ATPase is the driving force of numerous secondary active transport processes generating an electrical and chemical gradient across the cell membrane (50). In the proximal tubule, approximately 77% of the energy supplied by oxidative metabolism is invested in the Na+/K+ ATPase (3). This pump is located on the basolateral membrane in most epithelial cells and consists of a catalytic α subunit, a β subunit, and tissue-specific FXYD proteins (51). FXYD2 is expressed in human fetal liver, pancreas, and kidney (52), as well as in rat, pig, and dog kidneys, but was not detected in several established renal cell lines (NRK-52E, LLC-PK, and MDCK) (53). Here, the mRNA levels of the α1 (ATP1A1) and β1 (ATP1B1) subunits as well as FXYD2 were increased at confluence and maintained at high levels. In contrast, FXYD5 was highly expressed in proliferating cells (day 1) and decreased on subsequent days. FXYD5 overexpression has previously been shown to decrease CDH1 and alpha catenin levels (26). Thus, our data suggest that the expression of tissue-specific subunits of this pump is also promoted when the cells reach confluence.
Several genes involved in the cilium biogenesis and cilium function exhibited increased expression over the maturation process, including PCDP1, BBS1, and BBS12. Renal epithelial cells contain a nonmotile primary cilium (14), which is thought to be involved in the maintenance of tubular integrity and luminal fluid sensing. Mounting evidence suggests a role for the deregulation of specific cilia proteins in renal cystic disease, including polycystic kidney disease and Bardet-Biedl syndrome (BBS) (54). BBS arises due to mutations in one of the several BBS-encoding genes or chaperones and has multiple nonrenal manifestations, although renal dysfunction is the major cause of morbidity and mortality (55). We recently demonstrated that carcinogen exposure can induce the loss of primary cilia and that BBS1 is one of the genes heavily attenuated by potassium bromate and ochratoxin A exposure (56).
Utilizing the temporal alterations in the transcriptome, we performed a transcription factor activation prediction using IPA. The analysis revealed p53 (TP53) as the most active transcription factor in the matured epithelium, which was confirmed by quantifying p53 TF activity. This highlights the central role of p53 in epithelial maturation by promoting cell cycle arrest, oxidative phosphorylation, and suppression of glycolysis, as has been shown within different cellular contexts (57, 58). Other transcription factors that were predicted to be activated in the matured epithelium included CDKN2A (p16) and retinoblastoma 1 (RB1). CDKN2A disrupts cyclin D complexes (59), preventing cyclin D-mediated retinoblastoma protein (RB) phosphorylation and causing G1 cell cycle arrest (60). In addition, hypophosphorylated RB1 binds and inhibits E2F transcription factors (61, 62). E2Fs have a central role in mediating G1/S transition (63), and E2F1, -2, and -3 were predicted to be active during proliferation. The highest predicted activity for transcription factors during proliferation was TBX2 (T-box 2). This transcription factor is overexpressed in several cancers and promotes proliferation and metastasis by repressing p14ARF (an alternating reading frame product of CDKN2A), CDKN1A (also known as p21), N-myc downregulated gene 1 (NDRG1) and CDH1 (64). Additionally, c-MYC and HIF1A exhibited higher transcriptional activity rates in proliferating cells. Both transcription factors induce the transcription of pyruvate dehydrogenase kinase 1 (PDK1) (65, 66), whose product phosphorylates and inactivates PDH, thereby promoting glycolysis. Here, PDHA1 mRNA was expressed at very low levels in highly proliferating cells. FOXM1 (forkhead box M1) and FOXO1 (forkhead box O1) were also predicted to be active in proliferating cells. FOXM1 stimulates proliferation by regulating the genes involved in G1/S, S, G2/M, and M cell cycle phase progression. It also trans-activates MYC (67).
In contrast to its predicted activation state (early activation), FOXO1 exhibited higher activity in the matured epithelium. The FOXO family of TFs are tumor suppressors and have multiple roles in cell cycle inhibition, apoptosis, cellular stress response, and regulation of energy metabolism (68). The gene for insulin-like growth factor binding protein 1 (IGFBP1), a target of FOXO1 (69, 70), is highly expressed during proliferation in the microarray data set, consistent with the predicted activation. On the other hand, some regulated FOXO1 targets involved in cell cycle inhibition, like the CDKN1B protein (also known as p27) (71) and the CDKN1C protein (also known as p57) (72), or in the regulation of energy metabolism, like the PPARGC1A protein (73), are expressed at low levels in the subconfluent monolayer. Interestingly, Nowak et al. (74) reported that expression of FOXO1 and FOXO3 was induced by E2F1, another predicted early activated TF. The FOXO TFs have overlapping targets and activities, and their specificity depends on cell type, cellular context, and environmental stimuli (75). FOXO1 is ubiquitously expressed but is highest in insulin-responsive tissues, whereas FOXO3, which was predicted in this study to be in the group of late-activated TFs, is primarily expressed in kidney, brain, heart, and ovaries. Constitutively expression of FOXO1 in mouse liver resulted in the suppression of de novo lipogenesis, upregulation of genes involved in gluconeogenesis and protein and amino acid catabolism, whereas genes implicated in glycolysis, pentose phosphate shunt, and fatty acid and sterol synthesis were downregulated (76).
In summary, this study provides an extensive unbiased characterization of the transcriptional, metabolic, and functional alterations during epithelial monolayer maturation of proximal tubular epithelial cells (summarized in Fig. 9). We show that proliferating cells have a higher energy demand with both increased glycolysis and oxygen consumption and exhibit an immature cell-to-cell junction complex with nuclear TJP3 expression and high activity of HIF1A and c-MYC. The matured monolayers have lower energy demands, express TJP3, claudin 2, and claudin 10 at the intercellular junctions, and exhibit high activities of p53 and FOXO1. Given the main role for these processes in physiological function and disease states such as cancer and fibrosis, these data will be of great benefit in the further delineation of these complex cellular pathways. Finally, these data will serve as a reference for the development of differentiation strategies of inducible pluripotent stem cells to a proximal-tubule phenotype.
ACKNOWLEDGMENTS
This work was primarily supported by the European Union's Seventh Framework Programme (FP7/2007-2013), project Predict-IV, grant agreement 202222. Additional funding from the Sixth Framework Project carcinoGENOMICS (PL037712) is also acknowledged.
Footnotes
Published ahead of print 22 April 2013
REFERENCES
- 1. Jennings P, Koppelstaetter C, Lechner J, Pfaller W. 2008. Renal culture models: contribution to the understanding of nephrotoxic mechanisms, p. 223–250 In Broe ME, Porter GA. (ed), Clinical nephrotoxins: renal injury from drugs and chemicals, 3rd ed Springer, New York, NY [Google Scholar]
- 2. Muto S, Hata M, Taniguchi J, Tsuruoka S, Moriwaki K, Saitou M, Furuse K, Sasaki H, Fujimura A, Imai M, Kusano E, Tsukita S, Furuse M. 2010. Claudin-2-deficient mice are defective in the leaky and cation-selective paracellular permeability properties of renal proximal tubules. Proc. Natl. Acad. Sci. U. S. A. 107: 8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gstraunthaler G, Pfaller W, Kotanko P. 1985. Interrelation between oxygen consumption and Na-K-ATPase activity in rat renal proximal tubule suspension. Ren. Physiol. 8: 38–44 [DOI] [PubMed] [Google Scholar]
- 4. Pfaller W, Gstraunthaler G. 1998. Nephrotoxicity testing in vitro–what we know and what we need to know. Environ. Health Perspect 106(Suppl. 2): 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jennings P. 2009. Epithelial cell cultures as tools to study human disease and the adverse effects of pharmaceuticals and chemicals. Habilitation thesis Medizinische Universität Innsbruck, Innsbruck, Austria [Google Scholar]
- 6. Wilmes A, Crean D, Aydin S, Pfaller W, Jennings P, Leonard MO. 2011. Identification and dissection of the Nrf2 mediated oxidative stress pathway in human renal proximal tubule toxicity. Toxicol. In Vitro 25: 613–622 [DOI] [PubMed] [Google Scholar]
- 7. Lima WR, Parreira KS, Devuyst O, Caplanusi A, N′Kuli F, Marien B, Van Der Smissen P, Alves PM, Verroust P, Christensen EI, Terzi F, Matter K, Balda MS, Pierreux CE, Courtoy PJ. 2010. ZONAB promotes proliferation and represses differentiation of proximal tubule epithelial cells. J. Am. Soc. Nephrol. 21: 478–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moncada S, Higgs EA, Colombo SL. 2012. Fulfilling the metabolic requirements for cell proliferation. Biochem. J. 446: 1–7 [DOI] [PubMed] [Google Scholar]
- 9. Ullrich SJ, Anderson CW, Mercer WE, Appella E. 1992. The p53 tumor suppressor protein, a modulator of cell proliferation. J. Biol. Chem. 267: 15259–15262 [PubMed] [Google Scholar]
- 10. Syed V, Mak P, Du C, Balaji KC. 2008. Beta-catenin mediates alteration in cell proliferation, motility and invasion of prostate cancer cells by differential expression of E-cadherin and protein kinase D1. J. Cell. Biochem. 104: 82–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyoshi J, Takai Y. 2005. Molecular perspective on tight-junction assembly and epithelial polarity. Adv. Drug Deliv. Rev. 57: 815–855 [DOI] [PubMed] [Google Scholar]
- 12. Balda MS, Garrett MD, Matter K. 2003. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160: 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jennings P, Koppelstaetter C, Aydin S, Abberger T, Wolf AM, Mayer G, Pfaller W. 2007. Cyclosporine A induces senescence in renal tubular epithelial cells. Am. J. Physiol. Renal Physiol. 293: F831–F838 [DOI] [PubMed] [Google Scholar]
- 14. Wieser M, Stadler G, Jennings P, Streubel B, Pfaller W, Ambros P, Riedl C, Katinger H, Grillari J, Grillari-Voglauer R. 2008. hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Renal Physiol. 295: F1365–1375 [DOI] [PubMed] [Google Scholar]
- 15. Zidek N, Hellmann J, Kramer PJ, Hewitt PG. 2007. Acute hepatotoxicity: a predictive model based on focused Illumina microarrays. Toxicol. Sci. 99: 289–302 [DOI] [PubMed] [Google Scholar]
- 16. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- 17. Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. 2007. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 3: 11–17 [PMC free article] [PubMed] [Google Scholar]
- 18. Limonciel A, Aschauer L, Wilmes A, Prajczer S, Leonard MO, Pfaller W, Jennings P. 2011. Lactate is an ideal non-invasive marker for evaluating temporal alterations in cell stress and toxicity in repeat dose testing regimes. Toxicol. In Vitro 25: 1855–1862 [DOI] [PubMed] [Google Scholar]
- 19. Jennings P, Aydin S, Bennett J, McBride R, Weiland C, Tuite N, Gruber LN, Perco P, Gaora PO, Ellinger-Ziegelbauer H, Ahr HJ, Kooten CV, Daha MR, Prieto P, Ryan MP, Pfaller W, McMorrow T. 2009. Inter-laboratory comparison of human renal proximal tubule (HK-2) transcriptome alterations due to cyclosporine A exposure and medium exhaustion. Toxicol. In Vitro 23: 486–499 [DOI] [PubMed] [Google Scholar]
- 20. Ellis JK, Athersuch TJ, Cavill R, Radford R, Slattery C, Jennings P, McMorrow T, Ryan MP, Ebbels TM, Keun HC. 2011. Metabolic response to low-level toxicant exposure in a novel renal tubule epithelial cell system. Mol. Biosyst. 7: 247–257 [DOI] [PubMed] [Google Scholar]
- 21. Pfaller W, Gstraunthaler G, Loidl P. 1990. Morphology of the differentiation and maturation of LLC-PK1 epithelia. J. Cell. Physiol. 142: 247–254 [DOI] [PubMed] [Google Scholar]
- 22. Weibel ER. 1979. Morphometry of the human lung: the state of the art after two decades. Bull. Eur. Physiopathol. Respir. 15: 999–1013 [PubMed] [Google Scholar]
- 23. Giepmans BN, van Ijzendoorn SC. 2009. Epithelial cell-cell junctions and plasma membrane domains. Biochim. Biophys. Acta 1788: 820–831 [DOI] [PubMed] [Google Scholar]
- 24. Balda MS, Matter K. 2009. Tight junctions and the regulation of gene expression. Biochim. Biophys. Acta 1788: 761–767 [DOI] [PubMed] [Google Scholar]
- 25. Terada H. 1990. Uncouplers of oxidative phosphorylation. Environ. Health Perspect. 87: 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ino Y, Gotoh M, Sakamoto M, Tsukagoshi K, Hirohashi S. 2002. Dysadherin, a cancer-associated cell membrane glycoprotein, down-regulates E-cadherin and promotes metastasis. Proc. Natl. Acad. Sci. U. S. A. 99: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee TH, Streb JW, Georger MA, Miano JM. 2006. Tissue expression of the novel serine carboxypeptidase Scpep1. J. Histochem. Cytochem. 54: 701–711 [DOI] [PubMed] [Google Scholar]
- 28. Baum B, Georgiou M. 2011. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J. Cell Biol. 192: 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yonemura S. 2011. Cadherin-actin interactions at adherens junctions. Curr. Opin. Cell Biol. 23: 515–522 [DOI] [PubMed] [Google Scholar]
- 30. Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, Troyanovsky RB, Ben-Shaul A, Frank J, Troyanovsky SM, Shapiro L, Honig B. 2011. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 19: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson E, Theisen CS, Johnson KR, Wheelock MJ. 2004. R-cadherin influences cell motility via Rho family GTPases. J. Biol. Chem. 279: 31041–31049 [DOI] [PubMed] [Google Scholar]
- 32. Thomson RB, Igarashi P, Biemesderfer D, Kim R, Abu-Alfa A, Soleimani M, Aronson PS. 1995. Isolation and cDNA cloning of Ksp-cadherin, a novel kidney-specific member of the cadherin multigene family. J. Biol. Chem. 270: 17594–17601 [DOI] [PubMed] [Google Scholar]
- 33. Mullin JM, Fluk L, Tchao R. 1985. Mitosis in domes of renal epithelial (LLC-PK[1]) cell cultures. Mol. Physiol. 8: 317–328 [Google Scholar]
- 34. Li J, Ananthapanyasut W, Yu AS. 2011. Claudins in renal physiology and disease. Pediatr. Nephrol. 26: 2133–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kirk A, Campbell S, Bass P, Mason J, Collins J. 2010. Differential expression of claudin tight junction proteins in the human cortical nephron. Nephrol. Dial. Transplant. 25: 2107–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Furuse M, Furuse K, Sasaki H, Tsukita S. 2001. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J. Cell Biol. 153: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenthal R, Milatz S, Krug SM, Oelrich B, Schulzke JD, Amasheh S, Gunzel D, Fromm M. 2010. Claudin-2, a component of the tight junction, forms a paracellular water channel. J. Cell Sci. 123: 1913–1921 [DOI] [PubMed] [Google Scholar]
- 38. Van Itallie CM, Rogan S, Yu A, Vidal LS, Holmes J, Anderson JM. 2006. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am. J. Physiol. Renal Physiol. 291: F1288–F1299 [DOI] [PubMed] [Google Scholar]
- 39. Limonciel A, Wilmes A, Aschauer L, Radford R, Bloch KM, McMorrow T, Pfaller W, van Delft JH, Slattery C, Ryan MP, Lock EA, Jennings P. 2012. Oxidative stress induced by potassium bromate exposure results in altered tight junction protein expression in renal proximal tubule cells. Arch. Toxicol. 86: 1741–1751 [DOI] [PubMed] [Google Scholar]
- 40. Jennings P, Limonciel A, Felice L, Leonard MO. 2012. An overview of transcriptional regulation in response to toxicological insult. Arch. Toxicol. 87: 49–72 [DOI] [PubMed] [Google Scholar]
- 41. Capaldo CT, Koch S, Kwon M, Laur O, Parkos CA, Nusrat A. 2011. Tight function zonula occludens-3 regulates cyclin D1-dependent cell proliferation. Mol. Biol. Cell 22: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, Balda MS. 2006. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol. Cell. Biol. 26: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Traweger A, Lehner C, Farkas A, Krizbai IA, Tempfer H, Klement E, Guenther B, Bauer HC, Bauer H. 2008. Nuclear Zonula occludens-2 alters gene expression and junctional stability in epithelial and endothelial cells. Differentiation 76: 99–106 [DOI] [PubMed] [Google Scholar]
- 44. Bauer H, Zweimueller-Mayer J, Steinbacher P, Lametschwandtner A, Bauer HC. 2010. The dual role of zonula occludens (ZO) proteins. J. Biomed. Biotechnol. 2010: 402593. 10.1155/2010/402593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mulukutla BC, Khan S, Lange A, Hu WS. 2010. Glucose metabolism in mammalian cell culture: new insights for tweaking vintage pathways. Trends Biotechnol. 28: 476–484 [DOI] [PubMed] [Google Scholar]
- 46. Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. 2000. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 106: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burgess SC, Leone TC, Wende AR, Croce MA, Chen Z, Sherry AD, Malloy CR, Finck BN. 2006. Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha)-deficient mice. J. Biol. Chem. 281: 19000–19008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vega RB, Huss JM, Kelly DP. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 20: 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sugden MC, Holness MJ. 2003. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am. J. Physiol. Endocrinol. Metab. 284: E855–862 [DOI] [PubMed] [Google Scholar]
- 50. Lingrel JB, Kuntzweiler T. 1994. Na+,K+-ATPase. J. Biol. Chem. 269: 19659–19662 [PubMed] [Google Scholar]
- 51. Geering K. 2008. Functional roles of Na,K-ATPase subunits. Curr. Opin. Nephrol. Hypertens. 17: 526–532 [DOI] [PubMed] [Google Scholar]
- 52. Kim JW, Lee Y, Lee IA, Kang HB, Choe YK, Choe IS. 1997. Cloning and expression of human cDNA encoding Na+,K+-ATPase gamma-subunit. Biochim. Biophys. Acta 1350: 133–135 [DOI] [PubMed] [Google Scholar]
- 53. Therien AG, Goldshleger R, Karlish SJ, Blostein R. 1997. Tissue-specific distribution and modulatory role of the gamma subunit of the Na,K-ATPase. J. Biol. Chem. 272: 32628–32634 [DOI] [PubMed] [Google Scholar]
- 54. Yoder BK. 2007. Role of primary cilia in the pathogenesis of polycystic kidney disease. J. Am. Soc. Nephrol. 18: 1381–1388 [DOI] [PubMed] [Google Scholar]
- 55. Forsythe E, Beales PL. 2013. Bardet-Biedl syndrome. Eur. J. Hum. Genet. 21: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Radford R, Slattery C, Jennings P, Blaque O, Pfaller W, Gmuender H, Van Delft J, Ryan MP, McMorrow T. 2012. Carcinogens induce loss of the primary cilium in human renal proximal tubular epithelial cells independently of effects on the cell cycle. Am. J. Physiol. Renal Physiol. 302: F905–F916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Puzio-Kuter AM. 2011. The role of p53 in metabolic regulation. Genes Cancer 2: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang PY, Zhuang J, Hwang PM. 2012. p53: exercise capacity and metabolism. Curr. Opin. Oncol. 24: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. 1998. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395: 237–243 [DOI] [PubMed] [Google Scholar]
- 60. Shapiro GI, Edwards CD, Rollins BJ. 2000. The physiology of p16(INK4A)-mediated G1 proliferative arrest. Cell Biochem. Biophys. 33: 189–197 [DOI] [PubMed] [Google Scholar]
- 61. Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. 1991. The E2F transcription factor is a cellular target for the RB protein. Cell 65: 1053–1061 [DOI] [PubMed] [Google Scholar]
- 62. Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. 1993. The retinoblastoma protein binds to a family of E2F transcription factors. Mol. Cell. Biol. 13: 7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. DeGregori J, Johnson DG. 2006. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6: 739–748 [DOI] [PubMed] [Google Scholar]
- 64. Lu J, Li XP, Dong Q, Kung HF, He ML. 2010. TBX2 and TBX3: the special value for anticancer drug targets. Biochim. Biophys. Acta 1806: 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. 2007. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 27: 7381–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim JW, Tchernyshyov I, Semenza GL, Dang CV. 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3: 177–185 [DOI] [PubMed] [Google Scholar]
- 67. Wierstra I, Alves J. 2007. FOXM1, a typical proliferation-associated transcription factor. Biol. Chem. 388: 1257–1274 [DOI] [PubMed] [Google Scholar]
- 68. van der Vos KE, Coffer PJ. 2011. The extending network of FOXO transcriptional target genes. Antioxid. Redox Signal. 14: 579–592 [DOI] [PubMed] [Google Scholar]
- 69. Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. 1999. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 274: 17184–17192 [DOI] [PubMed] [Google Scholar]
- 70. Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O'Brien R, Granner DK. 2000. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 275: 30169–30175 [DOI] [PubMed] [Google Scholar]
- 71. Medema RH, Kops GJ, Bos JL, Burgering BM. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404: 782–787 [DOI] [PubMed] [Google Scholar]
- 72. Takano M, Lu Z, Goto T, Fusi L, Higham J, Francis J, Withey A, Hardt J, Cloke B, Stavropoulou AV, Ishihara O, Lam EW, Unterman TG, Brosens JJ, Kim JJ. 2007. Transcriptional cross talk between the forkhead transcription factor forkhead box O1A and the progesterone receptor coordinates cell cycle regulation and differentiation in human endometrial stromal cells. Mol. Endocrinol. 21: 2334–2349 [DOI] [PubMed] [Google Scholar]
- 73. Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. 2003. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52: 642–649 [DOI] [PubMed] [Google Scholar]
- 74. Nowak K, Killmer K, Gessner C, Lutz W. 2007. E2F-1 regulates expression of FOXO1 and FOXO3a. Biochim. Biophys. Acta 1769: 244–252 [DOI] [PubMed] [Google Scholar]
- 75. Calnan DR, Brunet A. 2008. The FoxO code. Oncogene 27: 2276–2288 [DOI] [PubMed] [Google Scholar]
- 76. Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG. 2006. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 281: 10105–10117 [DOI] [PubMed] [Google Scholar]