Abstract
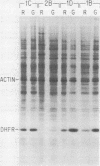
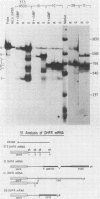
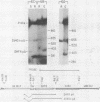
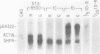
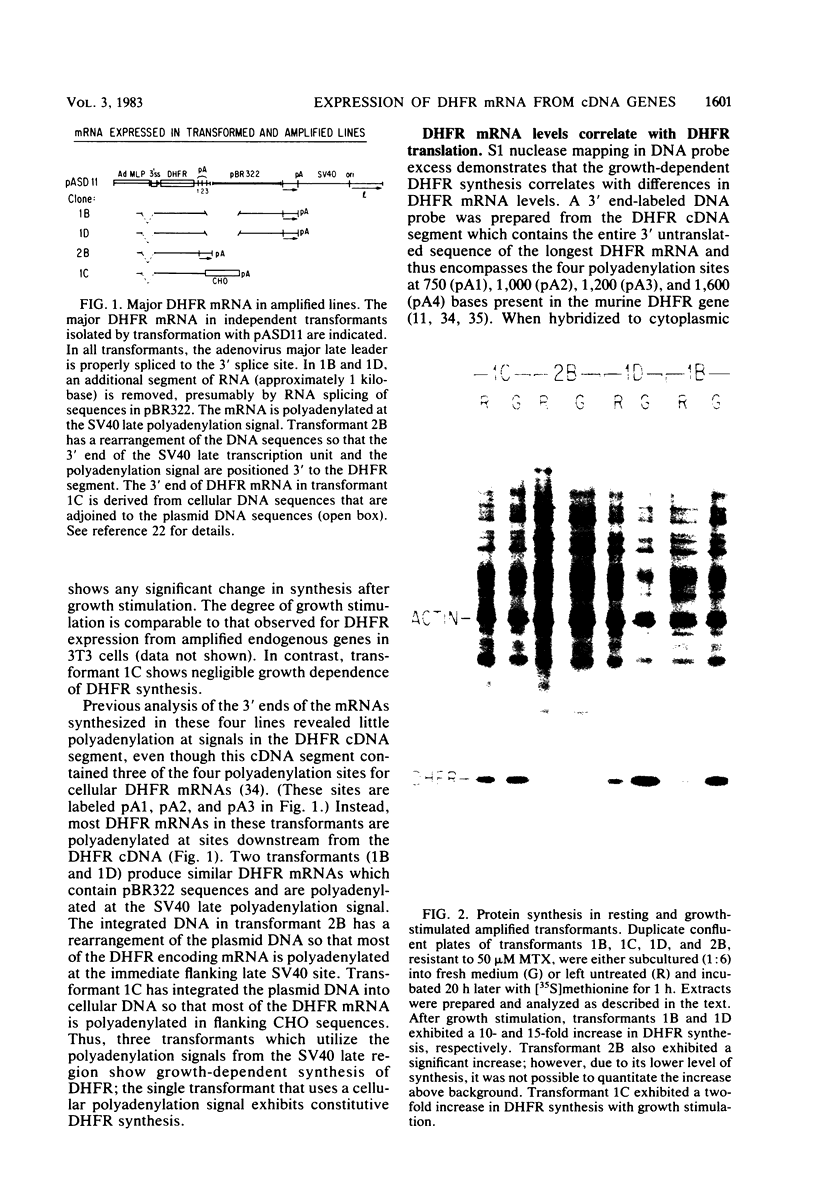
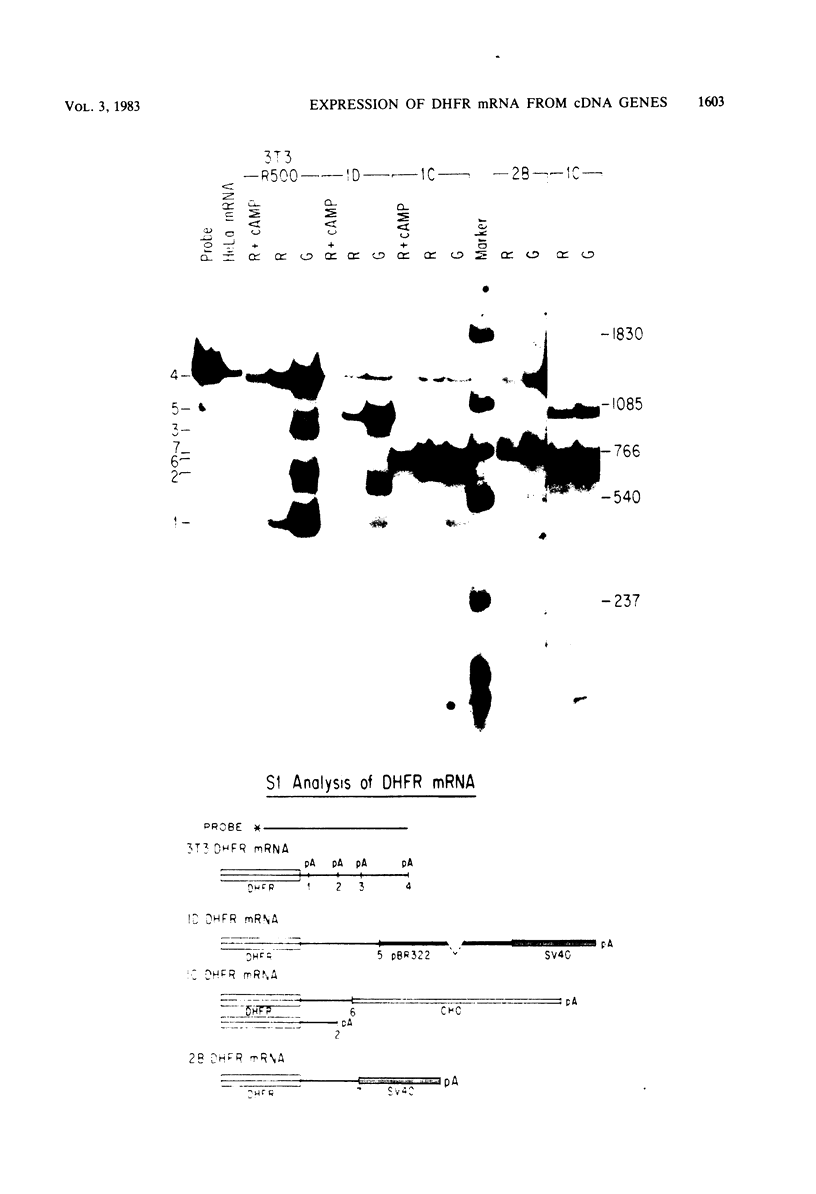
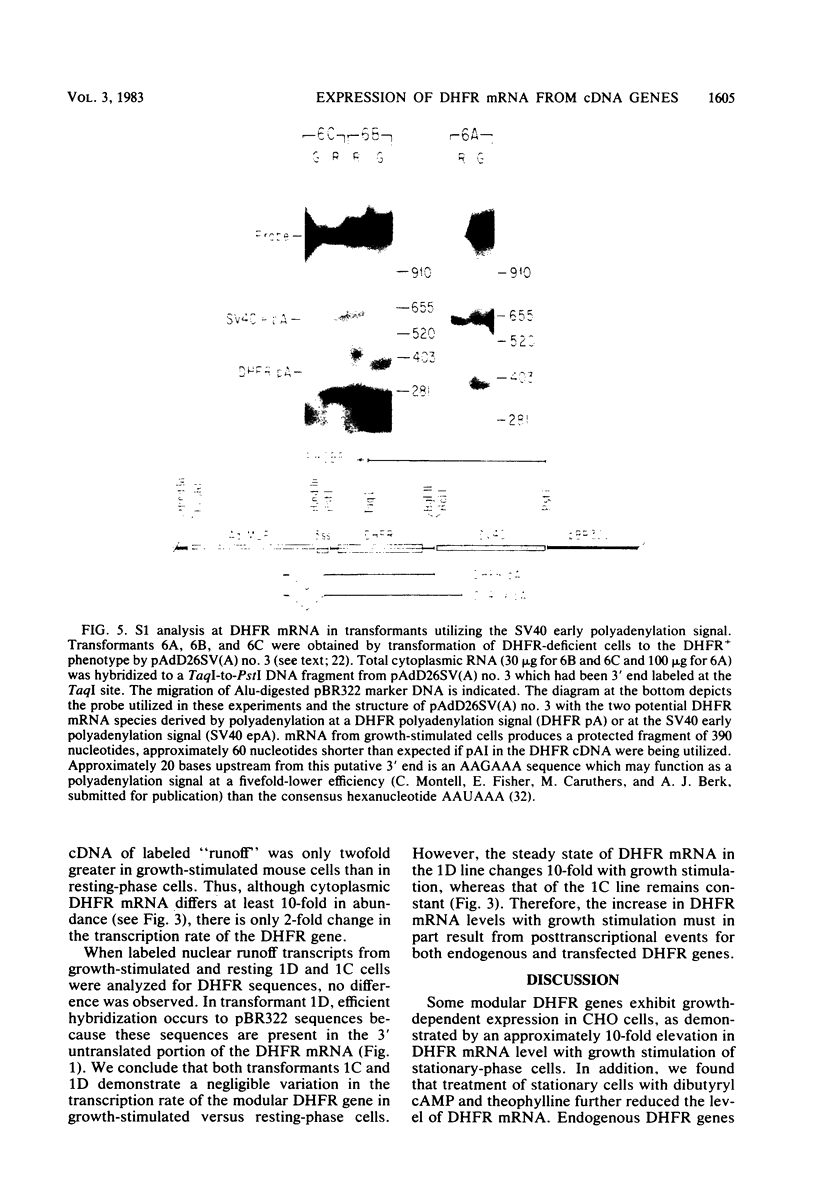
Dihydrofolate reductase (DHFR) synthesis is regulated in a growth-dependent fashion. Dividing cells synthesize DHFR at a 10-fold-higher rate than do stationary cells. To study this growth-dependent synthesis. DHFR genes have been constructed from a DHFR cDNA segment, the adenovirus major late promoter, and fragments of simian virus 40 (SV40) which provide signals for polyadenylation. These genes have been introduced into Chinese hamster ovary cells. The DHFR mRNAs produced in different transformants are identical at their 5' ends, but differ in sequences in their 3' ends as different sites are utilized for polyadenylation. Three transformants that utilize either DHFR polyadenylation signals or the SV40 late polyadenylation signal exhibit growth-dependent DHFR synthesis. The level of DHFR mRNA in growing cells is approximately 10 times that in stationary cells for these transformants. This growth-dependent DHFR mRNA production probably results from posttranscriptional events. In contrast, three transformants that utilize the SV40 early polyadenylation signal and another transformant that utilizes a cellular polyadenylation signal do not exhibit growth-dependent DHFR synthesis. In these three cell lines, the fraction of mRNAs polyadenylated at different sites in a tandem array shifts between growing and stationary cells. These results suggest that the metabolic state of the cell is important in determining either the efficiency of polyadenylation at various sites or the stability of mRNA polyadenylated at various sites.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981 Jul 30;292(5822):420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Alt F. W., Kellems R. E., Schimke R. T. Synthesis and degradation of folate reductase in sensitive and methotrexate-resistant lines of S-180 cells. J Biol Chem. 1976 May 25;251(10):3063–3074. [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birkenmeier E. H., Chiu N., Radonovich M. F., May E., Salzman N. P. Regulation of simian virus 40 early and late gene transcription without viral DNA replication. J Virol. 1979 Mar;29(3):983–989. doi: 10.1128/jvi.29.3.983-989.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brown P. C., Beverley S. M., Schimke R. T. Relationship of amplified dihydrofolate reductase genes to double minute chromosomes in unstably resistant mouse fibroblast cell lines. Mol Cell Biol. 1981 Dec;1(12):1077–1083. doi: 10.1128/mcb.1.12.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Nunberg J. H., Kaufman R. J., Erlich H. A., Schimke R. T., Cohen S. N. Phenotypic expression in E. coli of a DNA sequence coding for mouse dihydrofolate reductase. Nature. 1978 Oct 19;275(5681):617–624. doi: 10.1038/275617a0. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Lopata M. A., MacDonald R. J., Cowan N. J., Rutter W. J., Kirschner M. W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980 May;20(1):95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Ferdinand F. J., Brown M., Khoury G. Characterization of early simian virus 40 transcriptional complexes: late transcription in the absence of detectable DNA replication. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5443–5447. doi: 10.1073/pnas.74.12.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Frearson P. M., Kit S., Dubbs D. R. Induction of dihydrofolate reductase activity by SV40 and polyoma virus. Cancer Res. 1966 Aug;26(8):1653–1660. [PubMed] [Google Scholar]
- Groudine M., Peretz M., Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981 Mar;1(3):281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudewicz T. M., Morhenn V. B., Kellems R. E. The effect of polyoma virus, serum factors, and dibutyryl cyclic AMP on dihydrofolate reductase synthesis, and the entry of quiescent cells into S phase. J Cell Physiol. 1981 Jul;108(1):1–8. doi: 10.1002/jcp.1041080102. [DOI] [PubMed] [Google Scholar]
- Hendrickson S. L., Wu J. S., Johnson L. F. Cell cycle regulation of dihydrofolate reductase mRNA metabolism in mouse fibroblasts. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5140–5144. doi: 10.1073/pnas.77.9.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J., Sharp P. A. Amplification and expression of sequences cotransfected with a modular dihydrofolate reductase complementary dna gene. J Mol Biol. 1982 Aug 25;159(4):601–621. doi: 10.1016/0022-2836(82)90103-6. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J., Sharp P. A. Construction of a modular dihydrofolate reductase cDNA gene: analysis of signals utilized for efficient expression. Mol Cell Biol. 1982 Nov;2(11):1304–1319. doi: 10.1128/mcb.2.11.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J., Sharp P. A., Latt S. A. Evolution of chromosomal regions containing transfected and amplified dihydrofolate reductase sequences. Mol Cell Biol. 1983 Apr;3(4):699–711. doi: 10.1128/mcb.3.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellems R. E., Morhenn V. B., Pfendt E. A., Alt F. W., Schimke R. T. Polyoma virus and cyclic AMP-mediated control of dihydrofolate reductase mRNA abundance in methotrexate-resistant mouse fibroblasts. J Biol Chem. 1979 Jan 25;254(2):309–318. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Leys E. J., Kellems R. E. Control of dihydrofolate reductase messenger ribonucleic acid production. Mol Cell Biol. 1981 Nov;1(11):961–971. doi: 10.1128/mcb.1.11.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani B. D., Slate D. L., Schimke R. T. S phase-specific synthesis of dihydrofolate reductase in Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4985–4989. doi: 10.1073/pnas.78.8.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. S., Palmiter R. D. Transcriptional regulation of the ovalbumin and conalbumin genes by steroid hormones in chick oviduct. J Biol Chem. 1979 Sep 25;254(18):9050–9058. [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. Sequence at the 3' end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974 Nov 29;252(5482):359–362. doi: 10.1038/252359a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Schimke R. T. Nucleotide sequence surrounding multiple polyadenylation sites in the mouse dihydrofolate reductase gene. J Biol Chem. 1982 May 10;257(9):5143–5147. [PubMed] [Google Scholar]
- Shaw A. R., Ziff E. B. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3' coterminal mRNAs and five late families. Cell. 1980 Dec;22(3):905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. S., Johnson L. F. Regulation of dihydrofolate reductase gene transcription in methotrexate-resistant mouse fibroblasts. J Cell Physiol. 1982 Feb;110(2):183–189. doi: 10.1002/jcp.1041100212. [DOI] [PubMed] [Google Scholar]