Abstract
The sphingolipid backbone ceramide (Cer) is a major component of lipid lamellae in the stratum corneum of epidermis and has a pivotal role in epidermal barrier formation. Unlike Cers in other tissues, Cers in epidermis contain extremely long fatty acids (FAs). Decreases in epidermal Cer levels, as well as changes in their FA chain lengths, cause several cutaneous disorders. However, the molecular mechanisms that produce such extremely long Cers and determine their chain lengths are poorly understood. We generated mice deficient in the Elovl1 gene, which encodes the FA elongase responsible for producing C20 to C28 FAs. Elovl1 knockout mice died shortly after birth due to epidermal barrier defects. The lipid lamellae in the stratum corneum were largely diminished in these mice. In the epidermis of the Elovl1-null mice, the levels of Cers with ≥C26 FAs were decreased, while those of Cers with ≤C24 FAs were increased. In contrast, the levels of C24 sphingomyelin were reduced, accompanied by an increase in C20 sphingomyelin levels. Two ceramide synthases, CerS2 and CerS3, expressed in an epidermal layer-specific manner, regulate Elovl1 to produce acyl coenzyme As with different chain lengths. Elovl1 is a key determinant of epidermal Cer chain length and is essential for permeability barrier formation.
INTRODUCTION
An epidermal barrier is essential for terrestrial animals to survive. The mammalian epidermis is composed of four cell layers: the stratum basale (SB), stratum spinosum (SS), stratum granulosum (SG), and stratum corneum (SC) (1, 2). Keratinocytes propagate in the SB and then migrate to the SS, SG, and SC, differentiating along the way (1). In the SG, cells produce lamellar bodies in which the precursors of the lipid lamellae found in the SC are stored until being released into the extracellular space at the interface of the SG and SC. In the outermost layer, the SC, denucleated corneocytes are surrounded by extracellular lipid lamellae, which are mainly composed of ceramides (Cers), cholesterols, and free fatty acids (FAs). Lipid lamellae play essential roles in the epidermal barrier, and Cers are especially important among the components of the lipid lamellae (2, 3).
The sphingolipid backbone Cer is composed of a long-chain base attached to a FA via an amide bond (4). Although in most tissues the chain lengths of the FAs in Cers are 16 to 24 (C16 to C24), epidermal Cers uniquely contain extremely long FAs, up to C36 (2–4). FAs of >C20 are called very-long-chain FAs (VLCFAs), and those of >C24 are referred to as ultra-long-chain FAs (ULCFAs). The epidermis also specifically contains ω-O-acyl Cers (see Fig. S1 in the supplemental material), in which the ω-carbon (the terminus opposite to the carboxyl group) of the FA, carrying C30 to C36, is hydroxylated and esterified. The ester-forming FA is mainly linoleic acid (2, 3). These ULCFA-containing, nonacylated, and ω-O-acylated Cers largely contribute to the permeability barrier of the lipid lamellae, due to their high hydrophobicity. Indeed, a deficiency in the synthesis of ULCFA-containing Cers results in the severe cutaneous disorder ichthyosis in both mice and humans (5–11). Furthermore, decreased levels of Cers and changes in Cer subclasses have been observed in the epidermis of persons with atopic dermatitis or psoriasis (12–16). Recently, shifts of Cers toward shorter chain lengths have also been reported in patients with atopic eczema (17).
In addition to the Cer backbone, sphingolipids also carry a polar head group (phosphocholine for sphingomyelin [SM] or one or more sugars for glycosphingolipids) (4). Sphingolipids are constituents of eukaryotic plasma membranes in most tissues. The simplest glycosphingolipid, glucosylceramide (GlcCer), and SM are both packed into lamellar bodies in the SG for storage. After being released into the extracellular space, GlcCer and SM are hydrolyzed into Cers and used to form lipid lamellae (1, 3).
FAs are elongated by the addition of two carbons through a four-step process (condensation, reduction, dehydration, and reduction), which occurs in the endoplasmic reticulum (9). The rate-limiting step of the FA elongation cycle is the first condensation step, which is catalyzed by elongases. Mammals have seven elongases (ELOVL1 to -7), which differ in their substrate specificities toward acyl coenzyme A's (acyl-CoAs) (9, 18). In vitro assays have revealed that ELOVL1 is active toward saturated and monounsaturated C18 acyl-CoAs (C18-CoAs) to C26-CoAs (18). ELOVL4 exhibits activity toward acyl-CoAs with ≥C24; ELOVL4 is highly expressed in the epidermis and is essential for epidermal barrier formation. Elovl4 mutant mice die soon after birth due to epidermal barrier defects (6, 10, 11). In these mice, production of ULCFAs with ≥C28 is decreased, and ω-O-acyl Cers are nearly abolished. Similar neonatal lethality was observed in knockout mice lacking the CerS3 gene, which encodes a Cer synthase responsible for ULCFA-containing Cers, both nonacylated and ω-O-acylated (8).
To examine the molecular mechanism that regulates the diversified chain lengths of epidermal Cers and connects VLCFA production to ULCFA production, we generated Elovl1 knockout mice. Elovl1-null mice displayed severe epidermal barrier defects and died shortly after birth. In these mice, formation of the lipid lamellae was largely impaired. Mass spectrometry (MS) revealed that the levels of both nonacylated Cers with ≥C26 ULCFAs and ω-O-acyl Cers were greatly reduced in epidermal tissues, indicating that Elovl1 is especially important for the conversion of C24 acyl-CoA to C26 acyl-CoA in late differentiated keratinocytes of the epidermis. In addition, MS analyses of SM have suggested that Elovl1 is mainly involved in the production of C22 and C24 VLCFAs in early differentiated keratinocytes of epidermis and in other tissues.
The ability of Elovl1 to synthesize C24 or C26 acyl-CoAs correlates well with the differential expression of two ceramide synthases, CerS2 and CerS3. CerS2 mRNA is expressed in undifferentiated and early differentiated keratinocytes, while CerS3 mRNA is expressed in early and late differentiated keratinocytes. Interestingly, further studies in HEK 293T cells revealed that heterologous coexpression of ELOVL1 with CerS3 (but not with CerS2) promotes the synthesis of C26 Cer. Thus, Elovl1 is a key determinant for epidermal Cer chain length and is regulated differently in both a differentiation- and tissue-specific manner.
MATERIALS AND METHODS
Generation of Elovl1−/−mice.
The bacterial artificial chromosome (BAC) clone containing the Elovl1 gene (bMQ-173D12) prepared from chromosomal DNAs of Mus musculus AB2.2 (129S7/SvEvBrd-Hprtb-m2) embryonic stem (ES) cells (19) was purchased from the BACPAC Resources Center (Oakland, CA). The Elovl1 targeting vector was constructed by the BAC recombineering method (20). In the targeting vector, the first loxP sequence was inserted into intron 1 of the Elovl1 gene, and the second loxP sequence and subsequent Pgk-Neo (neomycin resistance gene under control of the Pgk promoter) cassette, flanked by FRT sequences, were inserted downstream of exon 8 (Fig. 1A). The targeting vector also contained the Tk-DTA (diphtheria toxin A under control of the thymidine kinase promoter) cassette at the 3′ end of the cloned Elovl1 chromosomal region for negative selection.
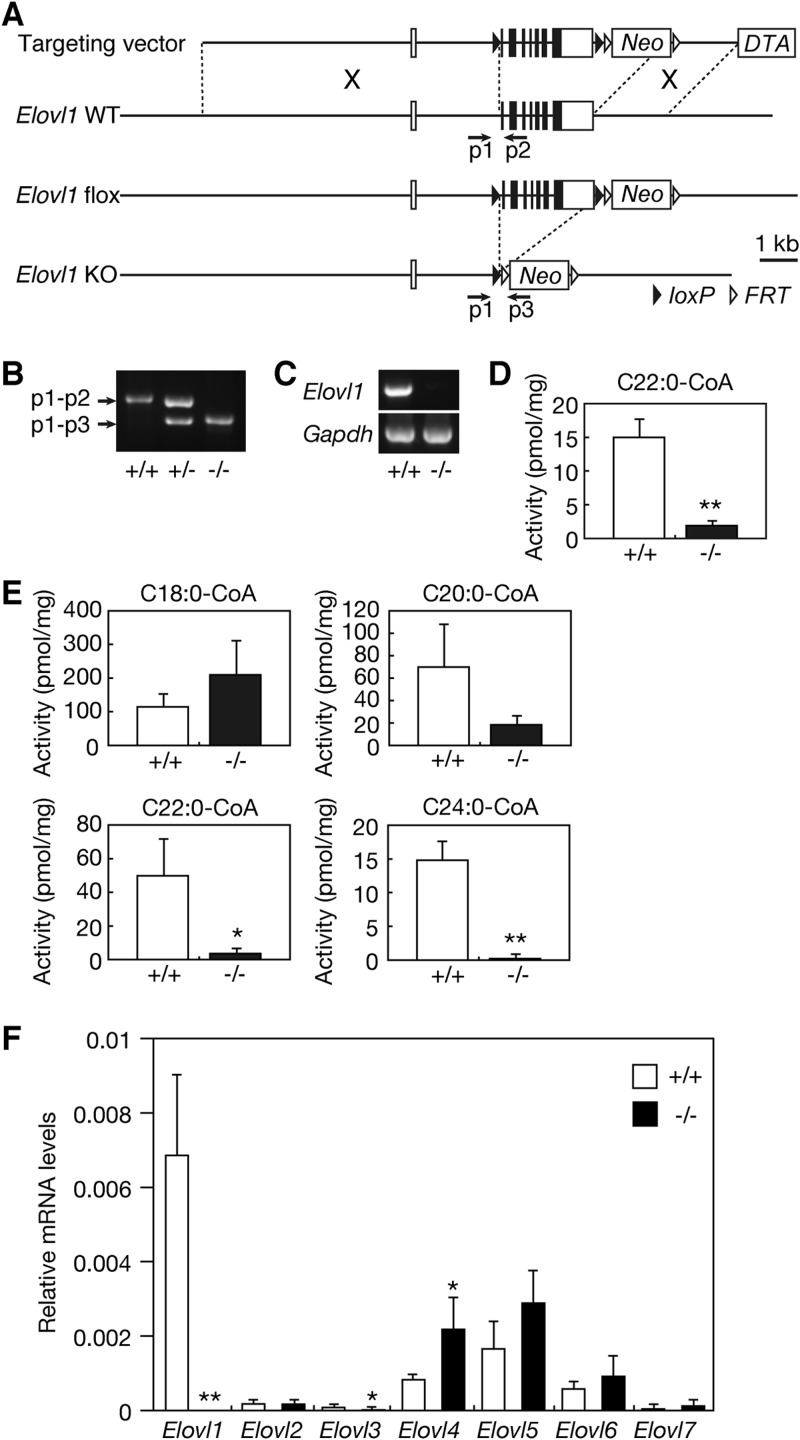
Fig 1.
Generation of Elovl1 knockout mice. (A) Schematic representation of the Elovl1 gene-targeting construct. In the targeting construct, exons 2 to 8 (2.4 kbp) of the Elovl1 gene are flanked by two loxP sequences, leaving an 8-kbp 5′-homologous region and a 2-kbp 3′-homologous region. Neo and DTA represent the positive selection marker, the neomycin resistance gene, and the negative selection marker, the diphtheria toxin A gene, respectively. Homologous recombination between the Elovl1 gene and the targeting construct yielded the Elovl1 flox allele. Crossing Elovl1+/flox mice with CAG-Cre mice produced Elovl1+/− mice. The positions of the primers (p1, p2, and p3) used for genomic PCR are depicted as arrows. KO, knockout. (B) Genomic DNAs prepared from the tails of Elovl1+/+, Elovl1+/−, and Elovl1−/− mice were subjected to PCR using primers p1, p2, and p3. The amplified fragments were separated by agarose gel electrophoresis and stained with ethidium bromide. (C) Total RNAs were prepared from the skin of Elovl1+/+ and Elovl1−/− mice and subjected to RT-PCR using primers specific for the Elovl1 and Gapdh genes. The amplified fragments were separated by agarose gel electrophoresis, followed by staining with ethidium bromide. (D) Total membrane proteins (50 μg) were prepared from the skin of Elovl1+/+ and Elovl1−/− mice and subjected to an in vitro FA elongase assay using 50 μM C22:0-CoA and 0.075 μCi [14C]malonyl-CoA (55 mCi/mmol; Moravek Biochemicals, Brea, CA) as the substrates. After a 30-min incubation at 37°C, lipids were subjected to methanolysis, extraction, separation by reverse-phase TLC, and detection using a BAS-2500 bioimaging analyzer (Fuji Photo Film, Tokyo, Japan). Values were calculated from the amounts of FA methyl ester products and represent the means ± standard deviations (SD) of three independent experiments. A statistically significant difference is indicated (**, P < 0.01; Student's t test). (E) Total membrane proteins (20 μg) were prepared from primary cultures of Elovl1+/+ and Elovl1−/− keratinocytes and subjected to an in vitro FA elongase assay using the indicated acyl-CoA (10 μM) and 0.075 μCi [14C]malonyl-CoA as the substrates. After a 60-min incubation at 37°C, lipids were saponified, acidified, extracted, and separated by normal-phase TLC, followed by detection using a BAS-2500 bioimaging analyzer. Values were calculated from the amounts of FA products and represent the means ± SD of three independent experiments. Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01; Student's t test). (F) Total RNAs were prepared from the skin of Elovl1+/+ and Elovl1−/− mice. SYBR green-based real-time quantitative PCR was performed using primers specific for Elovl1, Elovl2, Elovl3, Elovl4, Elovl5, Elovl6, and Elovl7, and for Gapdh as an internal control. The expression level of each Elovl mRNA was calculated using a standard curve and normalized to that of Gapdh. Values presented are the amount of each Elovl mRNA relative to that of Gapdh and represent the means ± SD from eight independent reactions. Statistically significant differences compared to results for the Elovl1+/+ mice are indicated (*, P < 0.05; **, P < 0.01; Student's t test).
The linearized targeting vector was transfected into E14 ES cells, and G418-resistant clones were selected. Genomic DNAs were prepared from the clones and were subjected to PCR, using primers E1-1 (5′-CTCGATAGCTTGGCTGGACGTAAACTCCTC-3′) and E1-2 (5′-GCCTGTCTGAGTGCTGTGGATGTGCATTCC-3′) to examine each occurrence of homologous recombination at the 3′ region of the Elovl1 gene. Positive clones were then subjected to genomic PCR to investigate homologous recombination at the 5′ region of the Elovl1 gene by using primers E1-3 (5′-GCGCGGCCGCGAGGAGAGTCTTGGAAAAGG-3′) and E1-4 (5′-GCGGTCGACAACCCTAGATCCTGGGATACC-3′). Proper recombination was further confirmed by Southern blotting.
Three positive ES clones were injected into C57BL/6J blastocysts to produce chimeric mice. The resulting male chimera mice were crossed with female C57BL/6J mice, and we obtained Elovl1+/flox mice from two original ES clones. One of these 2 mouse lineages was used for further experiments. F1 offspring with the Elovl1+/flox genotype were crossed with CAG-Cre transgenic mice (21), generating Elovl1+/− mice. The Elovl1+/− mice were maintained by repeated back-crossing with C57BL/6J mice. We used Elovl1−/− mice generated by intercrossing the Elovl1+/− mice, at least in 1 to 3 generations of the back-crossing. Genotyping was performed by PCR using genomic DNAs and the following primers: for discrimination of wild-type and flox alleles, primers p1 (5′-GCGCGGCCGCGAGGAGAGTCTTGGAAAAGG-3′) and p2 (5′-GCGGTCGACAACCCTAGATCCTGGGATACC-3′); for discrimination of wild-type and knockout alleles, primers p1 and p3 (5′-TCGCCTTCTTGACGAGTTCTTCTGAGG-3′).
All mice were housed under standard conditions (temperature, 23 ± 1°C; 12-h light/dark cycle; food and water available ad libitum). Time-pregnant females were obtained by overnight breeding with males. Noon following breeding was considered embryonic day 0.5 (E0.5). Newborn pups were obtained by Cesarean delivery at E18.5 and were maintained in a humidified (60 to 80%), thermostat-controlled chamber (30°C). All animal experiments were approved by the institutional animal care and use committee of Hokkaido University.
RT-PCR.
Total RNAs were isolated from the skin of E18.5 mice by using the NucleoSpin RNA II kit (Machery-Nagel, Dueren, Germany). Reverse transcription-PCR (RT-PCR) was performed using the SuperScript one-step RT-PCR with Platinum Taq (Invitrogen, Carlsbad, CA) and the following primers: for Elovl1, primers E1-5 (5′-CATGCTTTCCAAGGTCATTGAGCTG-3′) and E1-6 (5′-TCTCAGTTGGCCTTGACCTTGGTGG-3′); for Gapdh, primers G-1 (5′-TGGCATTGTGGAAGGGCTCATGACC-3′) and G-2 (5′-TTACTCCTTGGAGGCCATGTAGGCC-3′).
Real-time quantitative PCR (QPCR) was performed by using the one-step SYBR PrimeScript RT-PCR kit II (TaKaRa Bio, Shiga, Japan) on an Mx3005 real-time QPCR system (Agilent Technologies, La Jolla, CA), according to the manufacturer's instructions. Primers E1-5 and E1-6 were used for Elovl1, and primers G-1 and G-2 were used for Gapdh. Other primers used were as follows: for Elovl2, E2-1 (5′-GTTCCTGGACACGATTTTCTTTGTTC-3′) and E2-2 (5′-TTATTGAGCCTTCTTGTCCGTCATGC-3′); for Elovl3, E3-1 (5′-GCTTTGCCATCTACACGGATGACGC-3′) and E3-2 (5′-TCATTGGCTCTTGGATGCAACTTTGC-3′); for Elovl4, E4-1 (5′-GAGGAAGAAAAACAACCAAGTCTCC-3′) and E4-2 (5′-AATTTACTCTCCTTTTGGCTTCCCG-3′); for Elovl5, E5-1 (5′-AAGAACAACCACCAGATCACCGTGC-3′) and E5-2 (5′-TCAATCCTTTCGCTGCTTCCTGGGC-3′); for Elovl6, E6-1 (5′-GAGTTTTTACAATGGACCTGTCAGC-3′) and E6-2 (5′-CTACTCAGCCTTCGTGGCTTTCTTC-3′); for Elovl7, E7-1 (5′-CTGGCTTTATTACTTCTCCAAATTC-3′) and E7-2 (5′-GTATTTTAGTGGCGCTTGCTTTTGC-3′). The transcript level of each gene was normalized with that of Gapdh. Each reaction mixture was incubated at 42°C for 5 min and 95°C for 10 s, followed by 45 cycles at 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s.
Lipid analyses.
Skin was treated with 0.25% trypsin (T4549; Sigma, St. Louis, MO) for 16 h at 4°C, and the epidermis was separated from the dermis by manipulation under a stereomicroscope. The obtained epidermis (40 to 50 mg) was suspended in 2 ml chloroform-methanol-water (30:60:8, vol/vol/vol) and homogenized for 15 min at 50°C. After centrifugation, the supernatant was recovered. The pellet was further subjected to lipid extraction twice, using 2 ml chloroform-methanol-water (10:10:1, vol/vol/vol) and then 2 ml chloroform-methanol (2:1, vol/vol). The supernatant of each extraction step was pooled, and the total was subjected to phase separation by adding 4.8 ml water. After centrifugation, the resulting organic phase was recovered, dried, and suspended in chloroform-methanol (2:1, vol/vol). The lipids were resolved by thin-layer chromatography (TLC) on silica gel 60 high-performance TLC plates (Merck, Whitehouse Station, NJ) as described elsewhere (11).
Lipids for MS analyses were extracted as follows. Chopped tissues (20 mg) mixed with an internal standard, C17:0 SM (500 pmol; Avanti Polar Lipid, Alabaster, AL), were suspended in 500 μl chloroform-methanol (1:2, vol/vol). Lipids were extracted by homogenizing the tissues with zirconia beads in a Micro Smash MS-100R cell disrupter (Tomy, Tokyo, Japan) at 4,500 rpm for 1 min at 4°C. After centrifugation at 2,000 × g for 3 min at 4°C, the supernatant was recovered and divided into two aliquots (each 200 μl). One aliquot was subjected to alkaline treatment with 8.1 μl 4 M KOH in methanol with a 1-h incubation at 37°C, then neutralized with 6.5 μl 5 M HCl. The sample underwent phase separation with the addition of 67 μl chloroform and 134 μl water, and the organic phase was recovered and dried. The lipids obtained were suspended in chloroform-methanol (1:2, vol/vol) containing 0.1% ammonium formate.
MS analyses were performed using a 4000 QTRAP tandem MS (MS/MS) system (AB Sciex, Framingham, MA). Lipid samples were injected into the mass spectrometer by using the nanoflow ion source TriVersa NanoMate (ion spray voltage, 1.6 kV; gas pressure, 0.3 lb/in2; flow rate, 167 nl/min; Advion BioSystems, Ithaca, NY). Ions were detected in the positive ion mode. Parameters used for MS were a scan range of m/z 500 to 1,200, declustering potential of 100 V, and collision energy of 40 to 60 V. Cers and SMs were identified by precursor ion scans (Cer, m/z 264.4; SM, m/z 184.1) and analyzed with Analyst software (version 1.6; AB Sciex). Phosphatidylcholine (PC) and phosphatidylethanolamine (PE) were identified by precursor ion scan (m/z 184.1) and neutral loss scan (m/z 141.0), respectively, and were analyzed by the automated search engine Lipid Search (Mitsui Knowledge Industry, Tokyo, Japan).
In vitro FA elongation assays.
In vitro FA elongation assays were performed as described previously (18).
Skin permeability assays.
Skin permeability assays with toluidine blue were performed essentially as described elsewhere (22). Newborn pups obtained by Cesarean delivery at E18.5 were incubated in methanol for 5 min and rinsed in phosphate-buffered saline (PBS), followed by incubation with 0.1% toluidine blue for 1 h or 24 h. After staining, pups were rinsed with PBS and photographed using a digital camera.
Transepidermal water loss (TEWL) was measured on the dorsal skin of live pups by the ventilated chamber method, using an evaporimeter AS-VT100RS (Asahi Biomed, Yokohama, Japan). Measurements were performed in duplicate for each pup.
H/E staining.
Skins sampled from the backs of E18.5 embryos were fixed with 10% neutral buffered formalin (pH 7.2) for 48 h at room temperature. Fixed skins were dehydrated in methanol followed by xylene cleaning, then infiltrated in paraffin by using an automated tissue processor (Tissue-Tek VIP-6; Sakura, Torrance, CA). Infiltrated tissues were embedded into paraffin wax by a blocking machine (TEC-P-DC; Sakura). Paraffin-embedded skins were cut into 4-μm sections by using a microtome REM-710 (Yamato Kohki, Asaka, Japan). Sections were deparaffinized by treatment with xylene and ethanol, hydrated, stained with hematoxylin and then eosin (H/E), and dehydrated in ethanol and xylene by using an automatic staining system (Tissue-Tek DRS 2000; Sakura). Images were captured using a light microscope (AX70; Olympus, Tokyo, Japan) equipped with a digital color camera (DXM1200; Nikon, Tokyo, Japan).
Transmission electron microscopy.
Skin samples taken from the backs of mouse fetuses at E18.5 were fixed in 5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for >72 h at 4°C. Samples were then postfixed for 60 min in 1% OsO4 in 0.05 M sodium cacodylate buffer (pH 7.4) and for 30 min in 0.5% RuO4 (Electron Microscopy Sciences, Hatfield, PA), each after overnight immersion in 0.1 M sodium cacodylate buffer (pH 7.4). Fixed samples were dehydrated in graded ethanol and propylene oxide and embedded in Epon 812 resin (Taab Laboratories, Berkshire, United Kingdom), followed by an incubation at 70°C for >4 days. The samples were sectioned into ultrathin sections with a thickness of 70 nm, then stained with uranyl acetate and lead citrate. The thin sections were examined with a Hitachi (Tokyo, Japan) H-7100 transmission electron microscope.
Scanning electron microscopy.
Anterior limbs and tails of mouse fetuses at E18.5 were sampled and fixed in 5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for >72 h at 4°C. The samples were soaked in isoamyl acetate, dehydrated in a graded ethanol series, dried in a Hitachi HCP-2 critical point dryer, and coated with platinum-palladium by using a Hitachi E-1030 ion coater. Samples were observed with a Hitachi S-4500 scanning electron microscope.
In situ hybridization.
To construct RNA probes for in situ hybridization, Keratin 14, Involucrin, Elovl1, Elovl4, CerS2, and CerS3 cDNAs were amplified by PCR using the following primers: for Keratin 14, primers K14-1 (5′-TGAACCGCGAGGTGGCCACCAACAG-3′) and K14-2 (5′-TTAGTTCTTGGTGCGCAGGACCTGC-3′); for Involucrin, primers Inv-1 (5′-CCCTGTGAAGGATCTGCCTG-3′) and Inv-2 (5′-GGTTCCTGACACTCCTGGTG-3′); for Elovl1, primers E1-5 and E1-6; for Elovl4, primers E4-1 and E4-2; for CerS2, primers C2-1 (5′-TGATCAATGCTCCAGACCTTGTATGACTACTTCTGG-3′) and C2-2 (5′-GAAGCTCATCAGACTAATAGCAGCC-3′); for CerS3, primers C3-1 (5′-TGGTTCTGGTCGGAGAGATACTGGC-3′) and C3-2 (5′-GAAGCTCATCAGACTAATAGCAGCC-3′). The amplified cDNA fragments were cloned into the pGEM-T Easy vector (Promega, Madison, WI), and each antisense RNA was transcribed from the SP6 promoter while it was labeled with digoxigenin (DIG) by using a DIG RNA labeling mix (Roche Applied Science, Indianapolis, IN) and SP6 RNA polymerase (Roche Applied Science).
In situ hybridization was performed as essentially described elsewhere (23). Briefly, skin isolated from an E18.5 mouse was fixed with 4% paraformaldehyde and hybridized with a digoxigenin-labeled Keratin 14, Involucrin, Elovl1, Elovl4, CerS2, or CerS3 RNA probe. After washes, the hybridized probe was detected using an alkaline phosphatase-conjugated antidigoxigenin antibody (Fab fragment; Roche Applied Science), followed by signal development for 6 to 24 h in a nitroblue tetrazolium–5-bromo-4-chloro-3-indolyl phosphate solution. The sample was postfixed overnight with 3.7% formaldehyde in sodium phosphate buffer (pH 7.4), equilibrated in 30% sucrose, and frozen in Tissue-Tek OCT compound (Sakura Finetek, Alphen aan den Rijn, Netherlands). Sections were cut into 20 or 25 μm by using a cryostat (CM3050S; Leica Biosystems, Wetzlar, Germany) and covered with a glass coverslip by using CC/Mount (Diagnostic Biosystems, Pleasanton, CA). Images were captured using a DM5000B light microscope (Leica Biosystems) equipped with a DFC295 digital color camera (Leica Biosystems).
Plasmids.
The pCE-puro 3×FLAG-1 vector and pCE-puro 3×FLAG-ELOVL1 plasmid have been described previously (18). p3×FLAG-CERS2 and p3×FLAG-CERS3 plasmids, which express 3×FLAG-tagged human CERS2 and CERS3, respectively, under the control of the CMV promoter, were kind gifts of Yukiko Mizutani (Nagoya University) and Yasuyuki Igarashi (Hokkaido University).
Cell culture and transfection.
HEK 293T cells were cultured and transfected as described previously (18). Primary cultures of keratinocytes from E18.5 mouse embryos were prepared using CnT-57 medium (CELLnTEC Advanced Cell Systems AG, Bern, Switzerland) as described elsewhere (24).
[3H]sphingosine labeling assay.
[3H]sphingosine labeling assays were performed as described previously (18).
RESULTS
Generation of Elovl1 knockout mice.
For Elovl1 knockout mice, exons 2 to 8 of the Elovl1 gene, which encodes the entire region of the coding sequence, were first flanked by two loxP sequences (Elovl1flox/+) in ES cells by using gene targeting (Fig. 1A). From the targeted ES cells, Elovl1flox/+ mice were generated. These mice were then crossed with CAG-Cre mice (which express the recombinase Cre widely, including throughout the germ line, under the CAG promoter), generating heterozygous knockout mice (Elovl1+/−) carrying the heterozygous mutation throughout the body (Fig. 1A). Homozygous Elovl1 knockout mice (Elovl1−/−) were obtained by intercrossing Elovl1+/− mice. Disruption of the Elovl1 gene was confirmed by genomic PCR (Fig. 1B) and RT-PCR (Fig. 1C). FA elongation activity toward C22-CoA was greatly reduced (∼13%) in the skin of the Elovl1 knockout mice compared to wild-type mice (Fig. 1D). A similar reduction in FA elongation activity was also observed in brain and liver (see Fig. S2 in the supplemental material).
We also examined FA elongation activities toward various saturated acyl-CoAs by using membrane fractions prepared from primary cultures of keratinocytes. FA elongation activities toward C18:0-CoA and C20:0-CoA were not significantly different between wild-type and Elovl1 knockout keratinocytes, although the Elovl1 knockout keratinocytes exhibited tendencies of increased activity toward C18:0-CoA and decreased activity toward C20:0-CoA compared with wild-type keratinocytes (Fig. 1E). However, FA elongation activities toward C22:0-CoA and C24:0-CoA were significantly decreased (∼7% and <1% for C22:0-CoA and C24:0-CoA, respectively) in the keratinocytes of the Elovl1 knockout mice compared to those of the wild-type mice (Fig. 1E). We also perfomed FA elongation assays using C26:0-CoA, but the signal was too low to detect and quantify. In general, the signals in FA elongation assays decrease as the chain length of acyl-CoA increases. Taken together, these results are consistent with those obtained using membrane fractions of cells overproducing ELOVL1 (18), and they demonstrate that Elovl1 is important for the synthesis of saturated C22- to C26-CoAs.
We next examined the effects of the Elovl1 deletion on the expression of other elongase mRNAs, using quantitative real-time PCR. In wild-type epidermis, the expression of Elovl1 mRNA was the highest among the elongases, followed by that of Elovl5, Elovl4, and Elovl6, respectively (Fig. 1F). The mRNA expression of Elovl2, Elovl3, or Elovl7 in epidermis was quite low. Deletion of the Elovl1 gene caused a significant, ∼2-fold increase in Elovl4 mRNA and a reduction in Elovl3 mRNA (Fig. 1F). The expression levels of other Elovl mRNAs were nearly unaffected.
Elovl1 knockout mice exhibit morphological abnormalities in epidermis and small body size.
Of 29 living pups subjected to genotyping at postnatal day 1 to 28 days (P1 to P28), 9 were Elovl1+/+ and 20 were Elovl1+/−, but no Elovl1−/− mice were found. In contrast, we found living newborn (P0) pups, indicating that Elovl1 knockout mice died within 1 day after birth. Abnormalities in skin morphology and body size were readily observable in the newborn knockout mice (Fig. 2A). The skin of the Elovl1 knockout mice was unusually unwrinkled, shiny, and erythematous (Fig. 2A). Scanning electron microscopy also revealed great differences in skin surface morphology between the wild-type and Elovl1 knockout mice. The skin surface of the tail edge of the wild-type mice was wrinkled, and many projections were observed in a magnified view (Fig. 2B). However, the skin surface of the Elovl1 knockout mice was rather smooth, and few projections were observed, suggesting impaired desquamation (8). Such differences were also observed in the skin surfaces of the anterior limbs (see Fig. S3 in the supplemental material).
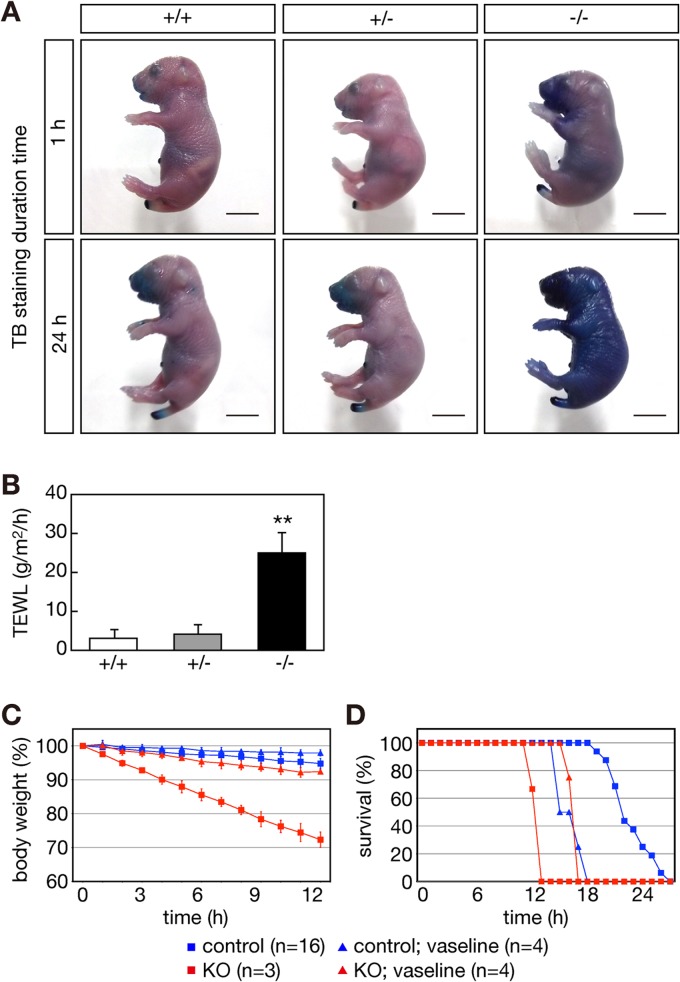
Fig 2.
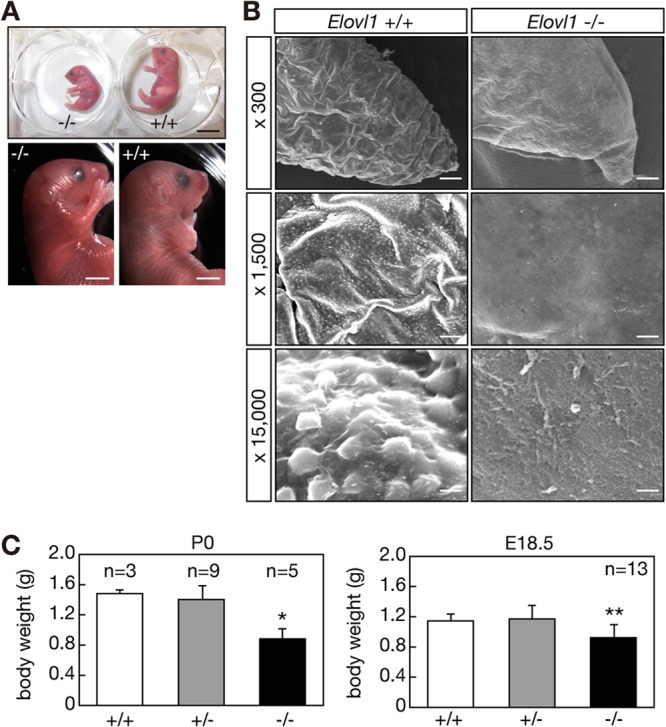
Abnormal skin morphology and reduced body weights in Elovl1 knockout mice. (A) Elovl1+/+ and Elovl1−/− mice at P0 are shown. Bar, 10 mm (upper panel) and 3 mm (lower panels). (B) Tail edges of Elovl1+/+ and Elovl1−/− mice at E18.5 were subjected to electron microscopy. Magnification factors are indicated on the left. Bar, 40 μm (top panels), 8 μm (middle panels), or 0.75 μm (bottom panels). (C) Body weights of Elovl1+/+ and Elovl1−/− mice at P0 and E18.5 were measured and are expressed as the means ± standard deviations of the indicated numbers of mice. Statistically significant differences compared to results for the Elovl1+/+ mice are indicated (*, P < 0.05; **, P < 0.01; Student's t test).
The body weights of the Elovl1−/− mice at P0 were reduced (∼60%) compared to those of the Elovl1+/+ and Elovl1+/− mice (Fig. 2C). The decreased body weights were less prominent before birth. At E18.5, the body weights of the Elovl1−/− mice were ∼80% that of the weights of the Elovl1+/+ and Elovl1+/− mice (Fig. 2C).
Neonatal lethality in Elovl1 knockout mice due to skin barrier defects.
The morphological changes in skin and decreased body weight observed for the Elovl1 knockout mice suggested skin barrier defects and excessive water loss. To test this, we performed a dye exclusion assay. The Elovl1+/+ and Elovl1+/− mice shed external toluidine blue and were unstained (Fig. 3A). However, the Elovl1−/− mice were dyed with toluidine blue after 24 h of incubation. In addition, the TEWL was higher by ∼7-fold in the Elovl1−/− mice compared to that in the Elovl1+/+ or Elovl1+/− mice (Fig. 3B). These results indicated that the skin barrier function of the Elovl1 knockout mice was largely impaired.
Fig 3.
Impaired skin barrier function and neonatal lethality in Elovl1 knockout mice. (A) Elovl1+/+, Elovl1+/−, and Elovl1−/− mice at E18.5 were stained with 0.1% toluidine blue (TB) for 1 h or 24 h and photographed. Bar, 5 mm. (B) TEWL was measured for the Elovl1+/+, Elovl1+/−, and Elovl1−/− mice 30 min after Cesarean delivery at E18.5. Values represent the means ± standard deviations (SD) from seven independent reactions. Statistically significant differences compared to results for the Elovl1+/+ mice are indicated (**, P < 0.01; Student's t test). (C and D) Control (Elovl1+/+ and Elovl1+/−) and Elovl1 knockout (KO; Elovl1−/−) mice at E18.5 with or without the petrolatum coating were housed at 30°C under humid conditions (60 to 80% humidity). (C) Body weights of the mice over time are presented relative to that at 0 h and represent the means ± SD of the indicated numbers of mice. (D) Survival rates of the mice for which data are shown in panel C.
The more prominent decrease in body weight of the Elovl1 knockout mice at P0 compared to that at E18.5 suggested that the body weight was further decreased after birth. Indeed, the Elovl1 knockout mice born by Cesarean delivery at E18.5 lost weight in a time-dependent manner (Fig. 3C). The weight loss reached ∼30% at 12 h. In contrast, little weight loss was observed for wild-type mice. The Elovl1 knockout mice died around 12 h after Cesarean delivery (Fig. 3D). Under the same conditions, in which maternal care and feeding were absent, wild-type mice died 18 to 27 h after Cesarean delivery, probably due to nutrient shortage. When petrolatum was spread over the body of the Elovl1 knockout mice, the body weight loss was suppressed (Fig. 3C), and, accordingly, the lifetime was prolonged to ∼18 h after birth (Fig. 3D). Conversely, petrolatum coating reduced the lifetime of wild-type mice so that wild-type and Elovl1 knockout mice died at similar time points after birth. Attempts to shed the coating might increase energy consumption, resulting in the early death of wild-type mice. In summary, these results indicate that neonatal lethality in Elovl1 knockout mice is attributable to an excessive water loss caused by skin barrier deficiency.
Defects in the formation of lamellar bodies and lipid lamellae in Elovl1 knockout mice.
To examine the cause of the skin barrier defects observed in the Elovl1 knockout mice, we next performed histological analyses. When wild-type epidermis was stained with H/E, several SC layers were observed to be segregated by open spaces, which represented empty lots of lipid lamellae (Fig. 4A). During the staining process, lipid lamellae are removed by dehydration in alcohol, resulting in such open spaces. However, the SC of the Elovl1 knockout mice was compacted, so few open spaces were detected (Fig. 4B), indicating that lipid lamella formation was impaired. On the other hand, structures in the SB, SS, and SG were indistinguishable between the wild-type and knockout mice by this staining.
Fig 4.
Impaired formation of lipid lamellae and lamellar bodies in Elovl1 knockout mice. (A and B) Paraffin sections (4 μm) of skin prepared from Elovl1+/+ (A) and Elovl1−/− (B) mice at E18.5 were subjected to H/E staining. Bright-field images were photographed under a DM5000B light microscope. De, dermis. Bar, 20 μm. (C to H) Sections of skin from E18.5 Elovl1+/+ (C, E, and G) and Elovl1−/− (D, F, and H) mice were subjected to electron microscopy. (C and D) The structures marked by asterisks indicate lipid lamellae present in the lower SC. (E to H) Red arrows indicate lamellar bodies at the interface of the SG and SC. Windows in panels E and F are magnified by 4.5-fold in panels G and H, respectively. Bar, 1 μm (C and D), 500 nm (E and F), or 100 nm (G and H). K, keratohyalin granule.
We also performed electron microscopy on skin samples. Although large lipid lamellar structures were observed in wild-type SC (Fig. 4C), such structures were decreased and significantly smaller in tissue from the Elovl1 knockout mice (Fig. 4D). However, the number of SC layers seemed not to be altered, and ∼10 layers existed in both the wild-type and knockout mice. In wild-type SG, many lamellar bodies with well-organized lipid sheets were present, especially near the boundary to SC (Fig. 4E and G). In contrast, in the SG of the Elovl1 knockout mice, the number and size of lamellar bodies were much smaller than those in wild-type tissues (Fig. 4F), and the lipid structures in the lamellar bodies were not layered but were rather punctate (Fig. 4H).
In situ hybridization demonstrated that expression patterns of Keratin 14, whose levels are high in SB but reduced in SS, were indistinguishable between the wild-type and Elovl1 knockout mice (see Fig. S4 in the supplemental material). The expression patterns of Involucrin mRNA, whose levels are the highest in late differentiated keratinocytes in SG, were similarly indistinguishable (see Fig. S4). These results suggest that keratinocyte differentiation in the Elovl1 knockout mice was not largely affected. High expression of Elovl1 mRNA was detected in the SS and SG of the wild-type mice (see Fig. S4). The expression of Elovl4 mRNA, whose levels in the epidermis were increased in Elovl1 knockout mice (Fig. 1F), was detected in similar patterns in SS and SG in both the wild-type and Elovl1 knockout epidermis (see Fig. S4).
Reduced levels of nonacylated Cers with ≥C26 ULCFAs and ω-O-acyl Cers in Elovl1 knockout mice.
Since ULCFA-containing, nonacylated, and ω-O-acylated Cers are important for skin barrier formation (2, 3), we next examined epidermal Cer composition in the wild-type and Elovl1 knockout mice. First, epidermal lipids were separated by TLC and subjected to cupric acetate-phosphoric acid staining (Fig. 5A). Although the total amounts of cholesterol, FA, PE, PC, and SM in the Elovl1 knockout mice were similar to those in wild-type mice, the amounts of Cer (ω-O-acyl Cers and other Cer species) and GlcCer were reduced (Fig. 5A).
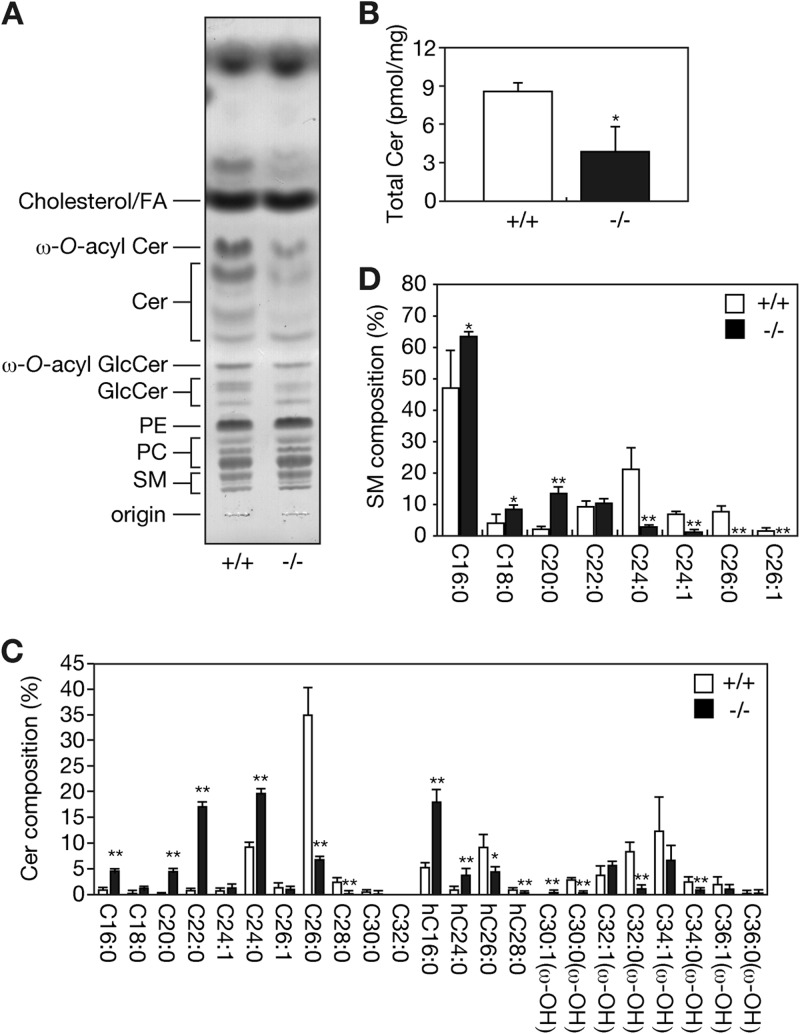
Fig 5.
Reduced levels of Cers containing ≥C26 ULCFAs in Elovl1 knockout mice. (A) Lipids were extracted from the epidermis (25 mg) of Elovl1+/+ and Elovl1−/− mice at E18.5, separated by TLC, and stained with a cupric acetate-phosphoric acid solution. (B to D) Lipids were extracted from the epidermis of Elovl1+/+ and Elovl1−/− mice at E18.5 and treated under alkaline conditions. Cer and SM compositions were determined by electrospray ionization MS using a 4000 QTRAP MS/MS system equipped with the nanoflow ion source TriVersa NanoMate. Cers (B and C) and SMs (D) were identified by precursor ion scans (Cer, m/z 264.4; SM, m/z 184.1) and analyzed by using Analyst software. (B) Values presented are the total amounts of Cers in 1 mg of skin and represent the means ± standard deviations (SD) from three independent reactions. (C) Values presented are the amounts of each Cer species relative to that of total Cer and represent the means ± SD from three independent reactions. (D) Values presented are the amounts of each SM species relative to that of total SM and represent the means ± SD from three independent reactions. Statistically significant differences compared to results for the Elovl1+/+ mice are indicated (*, P < 0.05; **, P < 0.01; Student's t test).
We next examined the Cer species present in more detail by using MS. Total Cer levels in Elovl1 knockout mice were reduced to ∼45% of the wild type (Fig. 5B). Epidermal Cers contained ULCFAs (C26 to C36), and of these, Cers with longer ULCFAs (C30 to C36) were mostly ω-O-acylated (Fig. 5C; see also Fig. S5 in the supplemental material). Note that in Fig. 5C Cer species described as having ω-OH correspond to ω-O-acyl Cers in epidermis, since these compounds received alkaline hydrolysis before MS analysis (see also Fig. S5 in the supplemental material for MS results without alkaline treatment). The amount of ω-O-acyl Cers carrying C30:0, C32:0, and C34:0 was significantly reduced in the Elovl1 knockout mice compared to the amount in the wild-type mice (Fig. 5C). Nonacylated Cer levels with C28:0, C26:0, and α-hydroxy-C26:0 (hC26:0) were also decreased in the knockout mice. Of these, a reduction in nonhydroxylated C26:0 Cer was the most prominent change. Conversely, the amounts of Cers with C16 to C24 FAs were increased or unchanged in the Elovl1 knockout mice. The boundary of increase and decrease in Cer levels in the Elovl1 knockout mice was C24/C26, so the most important elongation step of Elovl1 for epidermal Cer synthesis appears to be the C24-CoA-to-C26-CoA conversion.
We next examined the lipid profiles of SM and the glycerophospholipids PC and PE. SMs in the epidermis of wild-type mice contained up to C26 FAs, and C16:0 SM was the major species present (Fig. 5D). The amounts of SMs with C24:0, C24:1, C26:0, or C26:1 were reduced in the Elovl1 knockout mice, whereas the amounts of shorter SMs were increased. Slight changes in the levels of PC and PE species were also observed in the epidermis of the Elovl1 knockout mice. In general, the levels of PCs and PEs with C42 (perhaps C18 plus C24) and C44 (perhaps C20 plus C24) FAs were decreased, whereas those of PCs and PEs with C36 (perhaps C18 plus C18) and C38 (perhaps C16 plus C22 and/or C18 plus C20) FAs were increased (see Fig. S6 in the supplemental material).
We also examined levels of Cers and SMs in liver and brain. These tissues contained Cers/SMs with C16 to C24 FAs (see Fig. S7 in the supplemental material). The amounts of C22 to C24 Cers/SMs were decreased in the Elovl1 knockout mice, whereas the levels of C16 to C20 Cers/SMs were increased (see Fig. S7). Thus, Elovl1 was responsible for the production of C22- and C24-CoAs in liver and brain, similar to findings for SMs in epidermis but completely different from those of Cers in epidermis. These results suggest that FA elongation by Elovl1 is regulated differently among various tissues or cells.
Differential regulation of Elovl1 by CerS2 and CerS3 to produce acyl-CoAs with different chain lengths.
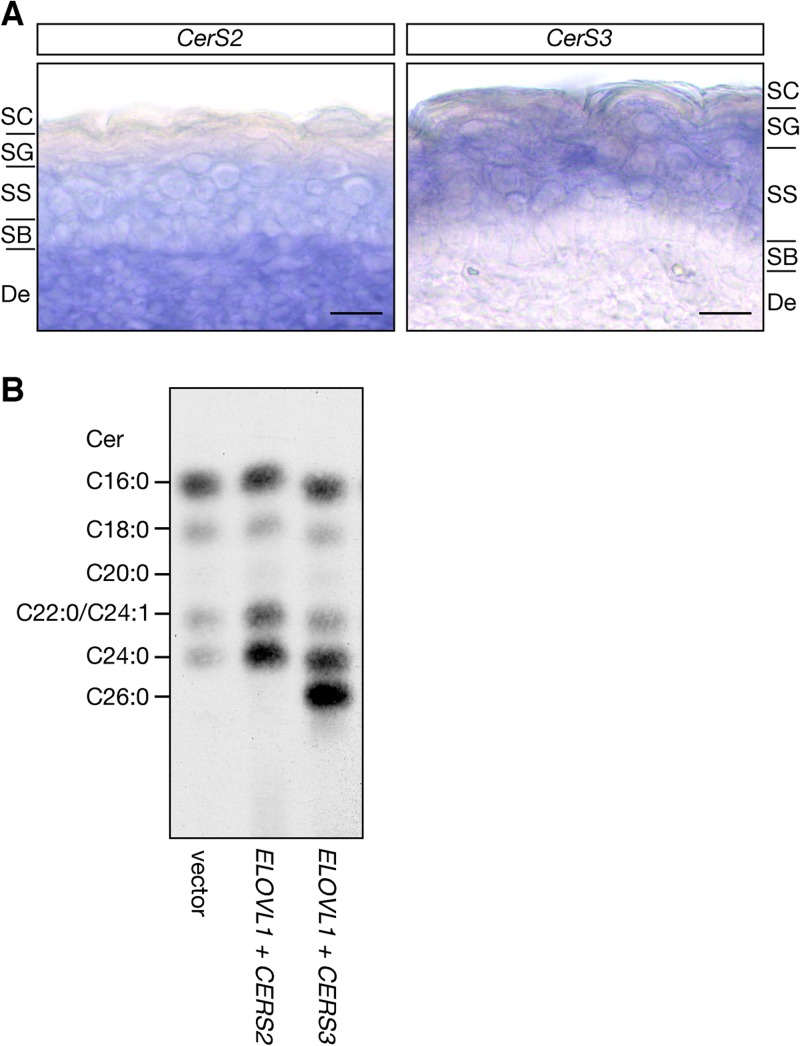
We investigated the mechanisms by which Elovl1 produces acyl-CoAs with different chain lengths in the epidermis. We previously demonstrated that ELOVL1 activity is regulated by the ceramide synthase CERS2, so the production of C24-CoAs is linked to C24 Cer synthesis (18). As demonstrated by the analysis of the CerS3 knockout mice, CerS3 plays an essential role in the production of Cers with ≥C26 ULCFAs and ω-O-acyl Cers (8). We therefore hypothesized that the chain length of acyl-CoAs synthesized by Elovl1 may vary depending on the CerS isozymes with which Elovl1 cooperates. To test this hypothesis, we first examined the expression patterns of CerS2 and CerS3 mRNAs in skin by in situ hybridization. CerS2 mRNA expression was detected in the SB and SS and in the dermis at levels higher than in the SB and SS, but it was nearly undetectable in the SG (Fig. 6A). In contrast, CerS3 mRNA was detected in the SS and SG but not in the SB (Fig. 6A). Thus, the expression of CerS2 and CerS3 is regulated in a layer-specific manner.
Fig 6.
Elovl1 synthesizes C26-CoA in cooperation with CerS3 in the SG. (A) Skin samples isolated from wild-type mice at E18.5 were fixed with 4% paraformaldehyde, hybridized with a digoxigenin-labeled CerS2 or CerS3 RNA probe, and stained with alkaline phosphatase-conjugated antidigoxigenin antibody (Fab fragment) and nitroblue tetrazolium–5-bromo-4-chloro-3-indolyl phosphate solution. Frozen sections (20 or 25 μm) were subjected to microscopic observation under a DM5000B light microscope and photographed. Bar, 50 μm. De, dermis. (B) HEK 293T cells transfected with control vectors or plasmids carrying ELOVL1 and CERS2 or CERS3 were labeled with 2 μCi of [3H]sphingosine for 2 h at 37°C. Lipids were extracted, separated by reverse-phase TLC, and detected by autoradiography. Positions of Cer species are indicated on the left.
We next examined the effect of CerS2 or CerS3 on the chain length of acyl-CoAs synthesized by Elovl1. HEK 293T cells transiently expressing CERS2 or CERS3 together with ELOVL1 were labeled with [3H]sphingosine, and then lipids were extracted and separated by reverse-phase TLC. Control cells transfected with the empty vectors synthesized predominantly C16:0 Cer and, to a lesser extent, C18:0, C22:0/C24:1, and C24:0 Cers (Fig. 6B). As expected considering the involvement of ELOVL1 and CERS2 in the synthesis of C24 Cers, the coexpression of ELOVL1 and CERS2 increased the levels of C22:0/C24:1 and C24:0 Cers. In contrast, the coexpression of ELOVL1 and CERS3 resulted in the synthesis of C26:0 Cer as the major Cer, followed by an increase in C24:0 Cer. These results suggest that isozyme switching from Cers2 to CerS3 during keratinocyte differentiation drives Elovl1 to synthesize C26-CoAs, which are further elongated by Elovl4 to generate Cers with ≥C26 ULCFAs and ω-O-acyl Cers.
DISCUSSION
Cer is a major component of epidermal lipid lamellae (∼50%), and it plays a pivotal role in barrier formation (2, 3). Epidermal Cers are quite unique in that they contain FAs with a wide variety of chain lengths, ranging from C16 to C36. In addition, Cers containing saturated or monounsaturated ULCFAs are observed only in epidermis, while those containing polyunsaturated ULCFAs exist in testis and spermatozoon (9, 25). Cers found in epidermis with ≥C30 FAs are specifically ω-hydroxylated and esterified, mainly with linoleic acid. These ULCFA-containing, nonacylated, and ω-O-acylated Cers are much more hydrophobic than common Cers, so they can function as a barrier to external soluble materials and pathogens or to water loss from inside body.
In the present study, we generated Elovl1 knockout mice and demonstrated that these mice exhibit neonatal lethality due to water loss caused by impaired epidermal barrier formation (Fig. 3). The lamellar bodies in the SG and the lipid lamellae in the SC of these mice were poorly developed and organized (Fig. 4). Major changes in lipid composition were found in the Cers of these mice (Fig. 5). The total amount of Cer in the Elovl1 knockout mice was decreased to ∼45% of the total found in wild-type mice (Fig. 5B). In addition, levels of ULCFA-containing Cers (C26 to C36), with and without acylation, were significantly decreased in the knockout mice (Fig. 5C). The decreases in ULCFA-containing Cers may primarily contribute to the impaired formation of lamellar bodies/lipid lamellae and decreased barrier function. However, slight changes in PC and PE compositions were also observed in the epidermis of the Elovl1 knockout mice (see Fig. S6 in the supplemental material). The amounts of longer PCs and PEs, which probably contain C24 FAs, were decreased. Since PCs and PEs can function as precursors of free FAs, also lipid components of lipid lamellae in SC, it is possible that changes in the PC and PE lipid compositions are also partly responsible for the permeability barrier disruption.
Our previous in vitro FA elongation assays demonstrated that, of 11 acyl-CoAs tested, ELOVL1 was active toward C18:0-, C20:0-, C22:0-, C24:0-, and C26:0-CoAs (18). Of the seven human elongases (ELOVL1 to -7), ELOVL1 was the most potent elongase toward C22:0-CoA and C24:0-CoA. On the other hand, ELOVL3 and ELOVL4 exhibited weak activities toward C22:0-CoA and C24:0-CoA, respectively. The activities of ELOVL1 and ELOVL4 toward C26:0-CoA were comparable. In the Elovl1 knockout mice, Cers with ≥C26 FAs exhibited reduced levels, but they still existed. Therefore, in the absence of Elovl1, Elovl3 and Elovl4 may compensate in the elongation of C22-CoA to C26-CoA. In Elovl4 mutant mice, only small amounts of nonacylated Cers with ULCFAs (≥C28) or ω-O-acyl Cers were present; instead, nonacylated Cer with C26 accumulated (6, 11). These results suggest that Elovl1 and Elovl4 are mainly responsible for the production of ≤C26-CoA and ≥C28-CoA, respectively. Thus, Elovl1 functions in the epidermis to connect the synthesis of long-chain FAs/VLCFAs to the synthesis of ULCFAs.
C24 Cers accumulated in the epidermis of Elovl1 knockout mice, while levels of Cers with ≥C26 FAs decreased (Fig. 5C). However, in the same tissues, the levels of C24 SMs were rather reduced and ≤C20 SMs accumulated (Fig. 5D). So, why did the Elovl1 knockout affect the chain lengths of Cers and SMs differently in the same tissue? The answer may be differences in cell types. The chain lengths of Cers reflect the FA elongation activity present in the late differentiated keratinocytes found in the SG, whereas those of SMs may reflect the FA elongation activity of undifferentiated or early differentiated keratinocytes found in the SB and SS. Keratinocytes propagated in the SB are differentiated to layer-specific cells and exhibit changes in protein expression patterns (1). Since SM is a component of the plasma membrane of mammalian cells, most of the SM in the epidermis may represent SM existing in the major cell population of the epidermis, i.e., undifferentiated or early differentiated keratinocytes. In contrast, most of the Cers in the epidermis represent those in the lipid lamellae of the SC. In the SC, unusually high levels of Cers form the permeability barrier. In contrast, Cer levels are not so high in other cells, including undifferentiated or early differentiated keratinocytes, since Cer is a metabolic intermediate of sphingolipids (4). GlcCers and SMs are stored in the lamellar bodies in the SG and are extruded into the interface of the SG and SC, followed by hydrolysis to Cers to form lipid lamellae (2, 3). Thus, most of the Cer chain lengths in the epidermis reflect the FA elongation activity present in the SG.
In most tissues CerS3, the Cer synthase for ULCFA-containing Cers, is not expressed, whereas CerS2, the Cer synthase for VLCFA-containing Cers such as C22 and C24 Cers, is ubiquitously expressed (26–28). In epidermis, CerS3 is expressed in the upper SS layer and SG layer in a differentiation-dependent manner (8, 29). Although Elovl1 can elongate C18/C20 acyl-CoAs to C28-CoA (18), we speculate that elongation by Elovl1 is normally terminated at C22 or C24 acyl-CoAs to provide them to CerS2. Indeed, we previously revealed that CERS2 increases ELOVL1 activity, probably through formation of a complex (18). For this reason, Elovl1 in undifferentiated and early differentiated epidermis (as well as other general tissues, such as liver and brain) produces C22-/C24-CoAs. However, when present (as in the SG), CerS3 may surpass CerS2 in the regulation of Elovl1, resulting in the elongation of C18/C20 acyl-CoAs further to C26-/C28-CoA. Two lines of evidence demonstrated in this study support this notion (Fig. 6). CerS2 mRNA was expressed in undifferentiated and early differentiated keratinocytes in the SB and SS, respectively, and then was shut down in late differentiated keratinocytes in the SG (Fig. 6A). In a complementary manner, the expression of CerS3 mRNA was apparent in early differentiated keratinocytes in the SS and continued into late differentiated keratinocytes in the SG (Fig. 6A). Moreover, while ELOVL1 produced up to C24-CoAs in cooperation with CERS2, in cooperation with CERS3 it can catalyze one more elongation cycle and produce C26-CoAs (Fig. 6B). Thus, ELOVL1 produces acyl-CoAs with different chain lengths in a CERS isozyme-dependent manner. This may be one of the mechanisms that enable tissues to increase a repertoire of Cers from a limited number of ELOVL and CERS isozymes. It would be interesting to test whether other combinations of ELOVL and CERS isozymes exhibit similar cooperativity.
Recessive mutations in the human ELOVL4 gene cause ichthyosis, along with neuronal disorders (5). In addition, a shift in Cer chain lengths to shorter lengths has been associated with atopic eczema (17). Thus, not only the production of Cers with ULCFAs but also the maintenance of proper chain lengths in Cers are important for skin pathology. Although ELOVL1 mutations have not been discovered in congenital ichthyosis patients, the Elovl1 knockout mice produced in this study exhibited ichthyosis-like cutaneous symptoms. It is possible that ELOVL1 mutations will be found in skin disorders, such as ichthyosis, atopic eczema, and psoriasis. Thus, ELOVL1 is a potential therapeutic target for skin disorders.
It has also been suggested that the modulation of ELOVL1 activity could provide treatment for X-linked adrenoleukodystrophy (X-ALD). X-ALD is caused by mutations in the ABCD1 gene and is characterized by the accumulation of VLCFAs, especially C24:0 and C26:0, in plasma and tissues (30). Knockdown of ELOVL1 in X-ALD fibroblasts reduces the elongation of C22:0-CoA to C26:0-CoA and lowers cellular C26:0 levels (30). Such findings are consistent with results presented here. It is unknown how the accumulation of VLCFAs leads to demyelination in the nervous system and adrenal insufficiency, however, so further studies will be necessary to understand the pathophysiological role of VLCFAs. Elovl1 knockout mice will be useful for this purpose.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to E. A. Sweeney for scientific editing of the manuscript. We thank Kosuke Yusa and Junji Takeda (Osaka University) for kindly providing plasmids for BAC recombineering. We thank Yukiko Mizutani (Nagoya University) and Yasuyuki Igarashi (Hokkaido University) for kindly providing the p3×FLAG-CERS2 and p3×FLAG-CERS3 plasmids. The CAG-Cre mouse was generated by Masaru Okabe (Osaka University) and provided by the Riken BRC through the National Bio-Resource Project of the MEXT, Japan.
This work was supported by a Grant-in-Aid for Scientific Research (B) (23370057) to A.K. and a Grant-in-Aid for Scientific Research (C) (24590073) to T.S. from the Japan Society for the Promotion of Science.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 20 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00192-13.
REFERENCES
- 1.Proksch E, Brandner JM, Jensen JM. 2008. The skin: an indispensable barrier. Exp. Dermatol. 17:1063–1072 [DOI] [PubMed] [Google Scholar]
- 2.Mizutani Y, Mitsutake S, Tsuji K, Kihara A, Igarashi Y. 2009. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie 91:784–790 [DOI] [PubMed] [Google Scholar]
- 3.Uchida Y, Holleran WM. 2008. Omega-O-acylceramide, a lipid essential for mammalian survival. J. Dermatol. Sci. 51:77–87 [DOI] [PubMed] [Google Scholar]
- 4.Kihara A, Mitsutake S, Mizutani Y, Igarashi Y. 2007. Metabolism and biological functions of two phosphorylated sphingolipids, sphingosine 1-phosphate and ceramide 1-phosphate. Prog. Lipid Res. 46:126–144 [DOI] [PubMed] [Google Scholar]
- 5.Aldahmesh MA, Mohamed JY, Alkuraya HS, Verma IC, Puri RD, Alaiya AA, Rizzo WB, Alkuraya FS. 2011. Recessive mutations in ELOVL4 cause ichthyosis, intellectual disability, and spastic quadriplegia. Am. J. Hum. Genet. 89:745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron DJ, Tong Z, Yang Z, Kaminoh J, Kamiyah S, Chen H, Zeng J, Chen Y, Luo L, Zhang K. 2007. Essential role of Elovl4 in very long chain fatty acid synthesis, skin permeability barrier function, and neonatal survival. Int. J. Biol. Sci. 3:111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckl KM, Tidhar R, Thiele H, Oji V, Hausser I, Brodesser S, Preil ML, Onal-Akan A, Stock F, Muller D, Becker K, Casper R, Nurnberg G, Altmuller J, Nurnberg P, Traupe H, Futerman AH, Hennies HC. Impaired epidermal ceramide synthesis causes autosomal recessive congenital ichthyosis and reveals the importance of ceramide acyl chain length. J. Invest. Dermatol., in press [DOI] [PubMed] [Google Scholar]
- 8.Jennemann R, Rabionet M, Gorgas K, Epstein S, Dalpke A, Rothermel U, Bayerle A, van der Hoeven F, Imgrund S, Kirsch J, Nickel W, Willecke K, Riezman H, Grone HJ, Sandhoff R. 2011. Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum. Mol. Genet. 21:586–608 [DOI] [PubMed] [Google Scholar]
- 9.Kihara A. 2012. Very long-chain fatty acids: elongation, physiology and related disorders. J. Biochem. 152:387–395 [DOI] [PubMed] [Google Scholar]
- 10.Li W, Sandhoff R, Kono M, Zerfas P, Hoffmann V, Ding BC, Proia RL, Deng CX. 2007. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int. J. Biol. Sci. 3:120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasireddy V, Uchida Y, Salem N, Jr, Kim SY, Mandal MN, Reddy GB, Bodepudi R, Alderson NL, Brown JC, Hama H, Dlugosz A, Elias PM, Holleran WM, Ayyagari R. 2007. Loss of functional ELOVL4 depletes very long-chain fatty acids (≥C28) and the unique ω-O-acylceramides in skin leading to neonatal death. Hum. Mol. Genet. 16:471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imokawa G, Abe A, Jin K, Higaki Y, Kawashima M, Hidano A. 1991. Decreased level of ceramides in stratum corneum of atopic dermatitis: an etiologic factor in atopic dry skin? J. Invest. Dermatol. 96:523–526 [DOI] [PubMed] [Google Scholar]
- 13.Motta S, Monti M, Sesana S, Caputo R, Carelli S, Ghidoni R. 1993. Ceramide composition of the psoriatic scale. Biochim. Biophys. Acta 1182:147–151 [DOI] [PubMed] [Google Scholar]
- 14.Motta S, Monti M, Sesana S, Mellesi L, Ghidoni R, Caputo R. 1994. Abnormality of water barrier function in psoriasis. Role of ceramide fractions. Arch. Dermatol. 130:452–456 [PubMed] [Google Scholar]
- 15.Di Nardo A, Wertz P, Giannetti A, Seidenari S. 1998. Ceramide and cholesterol composition of the skin of patients with atopic dermatitis. Acta Derm. Venereol. 78:27–30 [DOI] [PubMed] [Google Scholar]
- 16.Bleck O, Abeck D, Ring J, Hoppe U, Vietzke JP, Wolber R, Brandt O, Schreiner V. 1999. Two ceramide subfractions detectable in Cer(AS) position by HPTLC in skin surface lipids of non-lesional skin of atopic eczema. J. Invest. Dermatol. 113:894–900 [DOI] [PubMed] [Google Scholar]
- 17.Janssens M, van Smeden J, Gooris GS, Bras W, Portale G, Caspers PJ, Vreeken RJ, Hankemeier T, Kezic S, Wolterbeek R, Lavrijsen AP, Bouwstra JA. 2012. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients. J. Lipid Res. 53:2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, Igarashi Y, Sassa T, Kihara A. 2010. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. U. S. A. 107:18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, Humphray S, Plumb B, Liu P, Rogers J, Bradley A. 2005. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86:753–758 [DOI] [PubMed] [Google Scholar]
- 20.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65 [DOI] [PubMed] [Google Scholar]
- 21.Matsumura H, Hasuwa H, Inoue N, Ikawa M, Okabe M. 2004. Lineage-specific cell disruption in living mice by Cre-mediated expression of diphtheria toxin A chain. Biochem. Biophys. Res. Commun. 321:275–279 [DOI] [PubMed] [Google Scholar]
- 22.Hardman MJ, Sisi P, Banbury DN, Byrne C. 1998. Patterned acquisition of skin barrier function during development. Development 125:1541–1552 [DOI] [PubMed] [Google Scholar]
- 23.Hauptmann G, Gerster T. 1994. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10:266. [DOI] [PubMed] [Google Scholar]
- 24.Caldelari R, Müller EJ. 2010. Short- and long-term cultivation of embryonic and neonatal murine keratinocytes. Methods Mol. Biol. 633:125–138 [DOI] [PubMed] [Google Scholar]
- 25.Sandhoff R. 2010. Very long chain sphingolipids: tissue expression, function and synthesis. FEBS Lett. 584:1907–1913 [DOI] [PubMed] [Google Scholar]
- 26.Mizutani Y, Kihara A, Igarashi Y. 2005. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 390:263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizutani Y, Kihara A, Igarashi Y. 2006. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem. J. 398:531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH, Jr, Futerman AH. 2008. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283:5677–5684 [DOI] [PubMed] [Google Scholar]
- 29.Mizutani Y, Kihara A, Chiba H, Tojo H, Igarashi Y. 2008. 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J. Lipid Res. 49:2356–2364 [DOI] [PubMed] [Google Scholar]
- 30.Ofman R, Dijkstra IM, van Roermund CW, Burger N, Turkenburg M, van Cruchten A, van Engen CE, Wanders RJ, Kemp S. 2010. The role of ELOVL1 in very long-chain fatty acid homeostasis and X-linked adrenoleukodystrophy. EMBO Mol. Med. 2:90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.