Abstract
In vivo, cytokines noncovalently bind to the extracellular matrix (ECM), to facilitate intimate interactions with cellular receptors and potentiate biological activity. Development of a biomaterial that simulates this type of physiological binding and function is an exciting proposition for designing controlled advanced delivery systems for simulating in vivo conditions in vitro. We have decorated silk protein with sulfonated moieties through diazonium coupling reactions to noncovalently immobilize pro-inflammatory cytokines interleukin-1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α) in such a biomimetic manner. After adsorption of the cytokines to the diazonium-modified silk matrix, constant release of cytokines up to at least 3 days was demonstrated, as an initial step to simulate an osteoarthritic (OA) microenvironment in vitro. Matrix-embedded cytokines induced the formation of multiple elongated processes in chondrocytes in vitro, akin to what is seen in OA cartilage in vivo. Gene expression profiles with this in vitro tissue model of OA showed significant similarities to profiles from explanted OA cartilage tissues collected from patients who underwent total knee replacement surgery. The common markers of OA, including COL, MMP, TIMP, ADAMTS, and metallothioneins, were upregulated at least 35-fold in the in vitro model when compared to the control—non-OA in vitro generated tissue-engineered cartilage. The microarray data were validated by reverse transcriptase–polymerase chain reaction. Mechanistically, protein interaction studies indicated that TNF-α and IL-1β synergistically controlled the equilibrium between MMPs and their inhibitors, TIMPs, resulting in ECM degradation through the MAPK pathway. This study offers a promising initial step toward establishing a relevant in vitro OA disease model, which can be further modified to assess signaling mechanisms, responses to cell or drug treatments and patient-specific features.
Introduction
Osteoarthritis (OA), the most prevalent degenerative joint disease, may be defined as a heterogeneous condition resulting from the interactions of a constellation of factors such as mechanical injury,1 genetic predisposition,2 aging,3,4 gender,5 and body-mass index.6 These factors can lead to defective integrity of articular cartilage and changes in the subchondral bone. Despite the prevalence of arthritis, there is no cure due to the limited understanding regarding the mechanisms of onset and disease progression, as well as the complexity of the disease. Hence establishing a relevant in vitro OA human tissue disease model would have potential as a preclinical investigative platform to assess new clinical strategies and to screen potential drug candidates in patient specific manner. In addition, these tissue constructs can provide improved insight into the mechanisms of disease formation and progression.
Several animal models of OA have been developed either by surgically induced joint instability (e.g., partial or radical meniscectomy, anterior cruciate ligament transection models) or genetically manipulated animal models enabling specific proteins to be overexpressed, mutated or silenced, but these models do not fully recapitulate human pathobiology.7 Further, replicating features of OA in vitro is challenging due to the limited understanding of characteristics of diseased tissue matrix, wide genetic variation among human ethnic populations, subtle phenotypic differences between normal and mutant alleles (OA susceptibility single-nucleotide polymorphisms), and the slow and gradual progression of complex OA pathophysiology in humans. Moreover, the involvement of articular cartilage, subchondral bone, synovium, and synovial fluid makes the establishment of in vitro tissue-engineered model of OA challenging.
OA originates within the cartilage due to wear and tear of matrix, and then the resulting inflammation associated processes propagate to the surrounding of joints. In recent studies inflammation-related features, either due to primary or secondary causes, are highlighted as important characteristics of OA.8 Locally produced tumor necrosis factor alpha (TNF-α) and interleukin-1 beta (IL-1β) are the most well studied inflammatory mediators in early OA pathobiology.9 Macrophages,10 activated synoviocytes,11 or chondrocytes itself,8,11,12 produce inflammatory cytokines TNF-α, IL-1α, IL-8, prostaglandins and tissue-degrading proteases at regions of damaged cartilage. TNF-α can, in turn, trigger the production of several other cytokines (such as IL-8, IL-6, and leukemia inhibitory factor), impair matrix synthesis and remodeling pathways by suppressing synthesis of aggrecan and collagen type 2, eventually leading to accelerated damage of cartilage. After lipopolysaccharide stimulation of whole blood, patients with the highest quartile of TNF-α production reported a six-fold increased risk of OA.13 Intra-articular simultaneous inoculation of IL-1β and TNF-α caused more cartilage destruction compared to the application of only IL-1β.14 IL-1β and TNF-α, synergistically, are known to promote the synthesis of metalloproteinases,15 activate inflammatory enzymes such as nitric oxide synthase, cyclooxygenase 2 and phospholipase A2,16,17 and regulate the apoptotic pathway in chondrocytes, depending upon the expression levels of prostaglandin-E2 and caspase-8.18
In order to generate an in vitro disease model of OA a continuous supply of inflammatory cytokines is considered important. In a first attempt, human articular chondrocytes were cultured on silk protein scaffolds in the presence of either two exogenously added cytokines, or with macrophage-conditioned medium, with a gamut of inflammatory mediators.19 The macrophage-conditioned medium downregulated expression of collagen type 2 and upregulated aggrecan expression. Interestingly, cytokine treatment failed to induce hypertrophy and apoptosis of chondrocytes, which was achieved with the macrophage-conditioned medium. Culturing synoviocytes, chondrocytes and cartilage explants in the presence of IL-1β and TNF-α-containing media caused significant downregulation of lubricin, a superficial zone glycoprotein, which helps to maintain the smoothness of articular cartilage surfaces, whereas exposure to transforming growth factor (TGF-β) significantly upregulated lubricin synthesis.20 Further, continuous exposure of human chondrocytes to reactive oxygen species caused reduction in glycosaminoglycan production and telomere length, leading to premature senescence.21 Human chondrocytes express microRNA-140, which was found to be significantly downregulated in OA cartilage.22 On treating normal chondrocytes with IL-1β in vitro the expression of microRNA-140 was downregulated. Interestingly, transfection of chondrocytes with double-stranded-microRNA-140 downregulated IL-1β-induced disintegrin and metalloproteinase with thrombospondin (ADAMTS)-5 gene expression and recovered IL-1β-induced suppression of aggrecan gene expression.22
The goal of the present study was to further this mimetic strategy; thus, rather than the exogenous addition of inflammatory factors, we utilized matrix-embedded cytokines to simulate the localized tissue microenvironment with more relevance to the in vivo condition. Hence, a prerequisite for developing in vitro OA tissue models is to develop a biomaterial that can mimic at least some characteristics of extracellular matrix (ECM) and offer a system for sustained release of inflammatory mediators, resulting in more subtle but chronic modulation in cellular signaling pathways. The hypothesis for the present study was that biomimetic immobilization of two pro-inflammatory cytokines (IL-1β and TNF-α) to biomaterial scaffolds, from which they can be released at a steady rate, can simulate the microenvironment of cartilage tissue to generate OA-like features. While we recognize the complexity of other factors in OA-related symptoms as mentioned earlier, the central role of inflammation (primary or secondary) in the disease prompted this approach as a useful first step. We used diazonium coupling chemistry to tailor the surface chemistry and hydrophilicity of silk scaffolds to noncovalently adsorb the selected cytokines (Fig. 1) to ensure steady release to chondrocytes and thereby generating an inflammatory microenvironment similar to osteoarthritic tissue. Further, comparative information from transcript profiling from the cartilage of OA disease patients and the in vitro OA-like tissue system offers a route to understand OA pathophysiology.
FIG. 1.
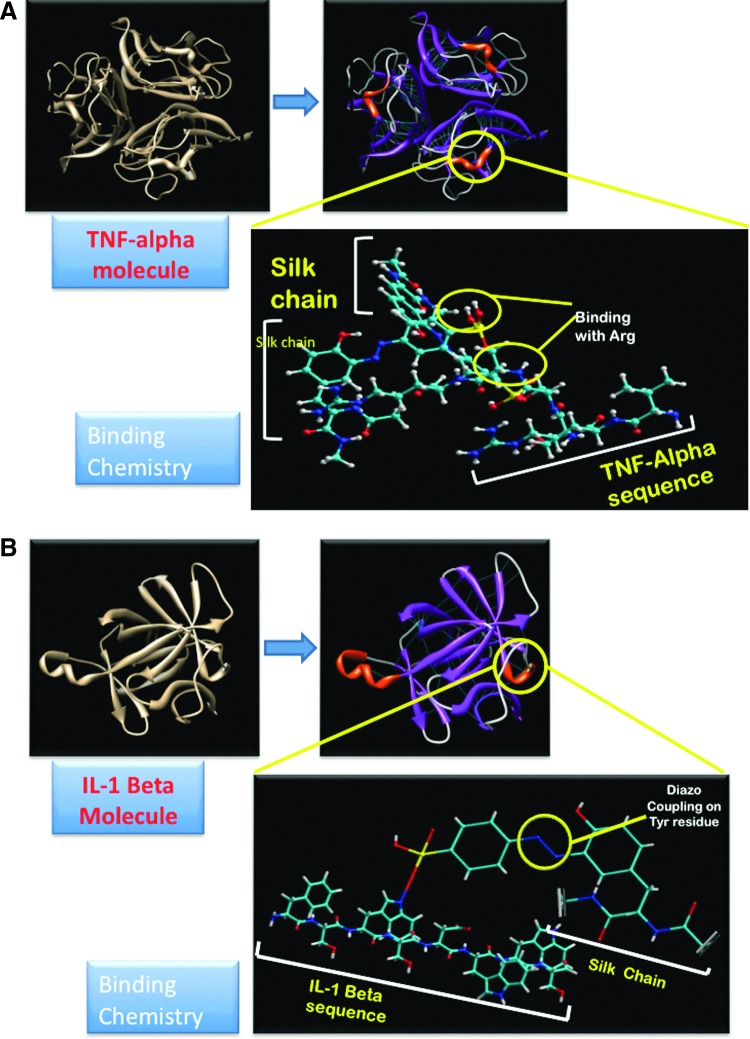
(A) Binding of tumor necrosis factor alpha (TNF-α) to silk polymer chain: the VRSSSR sequence is highlighted in red color. (B) Binding of interleukin-1 beta (IL-1β) to silk polymer chain: the Ser-Asp-Trp-Trp-Ser sequence is highlighted in red color. Color images available online at www.liebertpub.com/tea
Materials and Methods
Scaffold preparation
Preparation of aqueous silk solution
Cocoons from the Bombyx mori silkworm were cut and boiled for 30 min in an aqueous solution of 0.02 M Na2CO3, and rinsed several times with distilled water. The purified silk fibroin fibers were solubilized by dissolving in a 9.3 M LiBr solution at 60°C for 45 min to generate a 20 wt% solution. This solution was then dialyzed (Slide-A-Lyser dialysis cassette, Mol wt cut off value 3.5 KDa; Thermo Scientific) against distilled water for 2 days. This silk solution had a final concentration of ∼9 wt%.
Diazonium coupling with silk fibroin
The diazonium reactions have been described previously.23,24 Three different concentrations of diazonium salt solution were prepared, 10%, 20%, and 30%, which were compared against unmodified silk solution. The corresponding modification of silk was determined by UV-Vis spectrophotometry (SHIMADZU UV-2450). The extent of azo incorporation was tracked at λmax 274 nm for tyrosine. Then, salt-leached porous scaffolds were prepared with this modified fibroin.
ATR-FTIR study
Infrared spectra were measured on an ATR FTIR model Alpha-P, Bruker. All spectra were taken in the spectral range of 4000–500 cm−1 by accumulation of 264 scans and with a resolution of 4 cm−1.
Fluorescence spectra
A Horiba Scientific Fluoromax-4 spectroflurometer was used for studying the binding of IL-1β with the diazotized silk fibroin. The silk fibroin solution was diluted 1000-fold. The cytokine (10 ng/mL) was mixed with diazotized silk solution and allowed to react for 2 h. Samples were excited at 295 nm to produce >95% emission from tryptophan residues and changes in the fluorescent spectra of this reaction mixture were captured at continuous intervals of 5 min until stable (around 120 min). Excitation and emission bandwidths of 200–400 nm were employed, at a scan speed of 240 nm/min.
Adsorption and release kinetics studies
A preweighed silk film (10 mg) was suspended in 5 mL of 10 ng/mL of IL-1β solution and incubated at 37°C while stirring at 20 rpm for 24 h. Supernatants were collected at 5, 30, 60, 180, 360, 720, and 1400 min and residual concentrations of IL-1β were measured by ELISA for human IL-1β (Invitrogen) and adsorption kinetics were plotted. Similarly, release kinetics was studied by immersing films, preadsorbed with IL-1β, into PBS on shaker at 30 rpm. Aliquots were collected after 5, 30, 60, 180, 360, 720, 1400, 2880, and 4320 min and assessed for IL-1β concentration.
Cell culture
Human articular chondrocytes (a gift from Prof. Ivan Martin, University of Basel, Switzerland) were obtained post mortem (within 24 h after death), with ethics approval of University Hospital, Basel, from the knee joint of a 29-year-old patient who had no known clinical history of joint disorders.25 Matrigel (BD Biosciences) was diluted in the ratio of 1:10 and glass cover slips were coated with 500 μL of the diluted Matrigel and gelled in an incubator at 37°C overnight. TNF-α (10 ng/mL) and IL-1β (10 ng/mL) were incubated overnight with the diazotized-silk scaffolds and 2D films. Chondrocytes were cultured on Matrigel-coated cover slips (with or without cytokines), silk films, and cytokine-tethered silk films for 3 days. Films were fixed with 2% glutaraldehyde after 1 and 3 days of culture and processed for SEM imaging. A total of 1.5 million chondrocytes (P2) were seeded on the silk scaffolds and diazonium coupled cytokine-tethered (10 ng/mL of each cytokines) silk scaffolds, and cultured in DMEM supplemented with 4 ng/mL TGF-β1, transferrin, insulin, 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin, and cultured for 3 weeks. Some of the 3D constructs were harvested after 1 week to process for microarray study and other constructs were harvested after 3 weeks to process for histology, gene expression, and biochemical characterization.
Cartilage biopsy collection
Articular cartilage samples were obtained from three patients with OA (2 females and 1 male, ranging from 45 to 71 years), who fulfilled the OA criteria, undergoing total knee replacement surgery at New England Baptist Hospital, Boston, with informed consent. All experiments using OA cartilage biopsy tissue were approved by the Institutional Review Board of Tufts University and New England Baptist Hospital. All patients with OA received analgesic drugs, but no anti-inflammatory drugs 3 weeks before surgery. Three cartilage samples were collected directly from the degraded lesions, in weight-bearing surfaces of the medial femoral condyle part of cartilage of knee joints. In order to study differences in transcript levels from the chondrocytes from the degraded focal lesions and apparently integrated cartilage area from the same patient, one cartilage sample was also collected from the nondegraded part of the non-weight-bearing site of cartilage (henceforth mentioned as “intact cartilage biopsy”) from a 45-year-old male OA patient for microarray study. All samples were immediately processed for RNA extraction for microarray analysis.
DNA microarray
Ten micrograms of RNA from each sample was reverse transcribed and biotin labeled using commercial kits according to the suppliers' instructions (TotalPrep RNA Amplification Kit, Ambion). It was then transcribed and the Biotinylated cRNA was fragmented by treatment at 94°C for 35 min in 40 mM TRIS-acetate, pH 8.1, 100 mM potassium acetate, and 30 mM magnesium acetate and hybridized to human Bead array, HumanRef-6 chips (Illumina), with more than 43,000 transcript probes per sample, which generated whole genome transcription profiles simultaneously for six samples. Two separate hybridizations were performed for each sample. The arrays were scanned using the Illumina Bead station 500GX System and pixel levels were analyzed using commercial software (Beadstudio, Illumina, Inc.). For each of the two hybridizations, two separate data readings were performed. Raw gene expression data were normalized and analyzed with GeneChip Operating Software (genome studio v2010.2).
Functional classification was performed with annotations from the Gene Ontology Annotation Database (www.ebi.ac.uk/GOA). To support the system generated gene list, dendogram and cluster analysis was performed to obtain the correlation among the three groups (tissue-engineered cartilage, tissue-engineered OA disease model, biopsies from OA patients) that were considered for the experiment. Hierarchical cluster analysis was performed with log2-transformed signals normalized by genes and Manhattan correlation as distance measure using Genespring 12.0 software (Agilent).
Analysis of protein interaction networks
To study the interactions occurring between the proteins coded by the differentially regulated genes expressed by the cells from the in vivo and in vitro samples, and databases of known and predicted protein interactions, STRING 9.0 (Search Tool for the Retrieval of Interacting Genes/Proteins) software was used (http://string-db.org/).26 These interactions were studied to gain primary ideas about how the proteins being expressed in the two experimental settings (tissue-engineered OA-like model, biopsies from OA patients) were interacting and if the two systems shared similarities, which could then be analyzed further by pathway analysis.
Gene ontological classification
The gene ontological classification of the gene lists was done using ToppGene suite gene ontology tool.27 The ToppFun (ToppFun: transcriptome, ontology, phenotype, proteome, and pharmacome annotations based gene list functional enrichment analysis) module of the suite was used for the classification of the gene lists. ToppFun detects functional enrichment of a gene list based on Transcriptome, Proteome, Regulome, Ontologies (GO, Pathway), Phenotype (human disease and phenotype), Pharmacome (Drug-Gene associations), literature co-citation, and other features.
Histology
Cartilage biopsies and tissue-engineered constructs were rinsed with PBS, fixed in 4% formalin, washed in PBS, embedded in paraffin, sectioned (7 μm), and stained either with hematoxylin and eosin (H&E) for studying cell morphology or with Safranin-O for presence of sulfated glycosaminoglycans (GAG).
Real-time PCR
Cartilage tissue biopsies from osteoarthritic patients undergoing total knee replacement were collected in RNAlater (Invitrogen). About 30 mg of the degraded and non degraded tissue was used for total RNA isolation by using Trizol (Invitrogen) and RNeasy mini kit (Qiagen). It was then purified by treating with DNase I to eliminate genomic DNA contamination. RNA concentrations and the purity were determined using Agilent 2100 Bioanalyzer (Ambion). Extracted RNA was reverse-transcribed into cDNA using first-strand cDNA Synthesis Kit (Fermentas). Quantitative real-time reverse transcriptase–polymerase chain reaction (RT-PCR) was conducted using SYBRgreen chemistry and QuantiTect primer assay (Qiagen) and Rotorgene Q thermocycler (Qiagen). The reactions were carried out in triplicates for each sample in 25 μL total volume containing 1 μL cDNA and 2.5 μL primer. The primers used included those for GAPDH (QT00079247), ADAMTS 5 (QT00011088), Aggrecan (QT00001365), MMP1 (QT00014581), MMP2 (QT00088396), COMP1 (QT00001050), Collagen 1 (QT00037793), Collagen 2 (QT00049518), and Collagen 10 (QT00096348). The analysis was carried out with the Rotor gene Q software and the relative expression levels were calculated using the 2−(ΔΔc(t)) method with GAPDH as a control.
Biochemical estimation
Constructs were digested with proteinase K solution. GAG content was measured spectrophotometrically after reacting it with dimethylmethylene blue, using chondroitin sulfate as a standard.28 The normalization of GAG content was done against amount of DNA, which was measured using a CyQUANT cell proliferation assay kit (Molecular Probes), with calf thymus DNA as a standard.
Statistical analysis
Raw data without normalization were analyzed by GeneSpring software 12.0 (Agilent). Mean values were obtained for gene readouts after normalization for the duplicates of the samples. Fold ratios were derived by comparing normalized data between two sets, the first set included the comparison of biopsies from the degraded region against tissue-engineered constructs with cytokines (the in vitro OA-like model) and the second set consisted of comparison of biopsies from nondegraded regions to tissue-engineered constructs (the in vitro healthy equivalent tissue model). Gene lists were created with normalized intensities that contained genes with p<0.001 and greater than a fivefold change by log transformation of average difference values. Expression data were median centered and statistical comparisons were done by ANOVA, using the Benjamini and Hochberg correction for false-positive reductions.
Statistical significance for RT-PCR was calculated by independent t tests using SPSS software (SPSS, Inc., an IBM company) and was considered significant at p≤0.05.
Results
UV-visible spectroscopy analysis
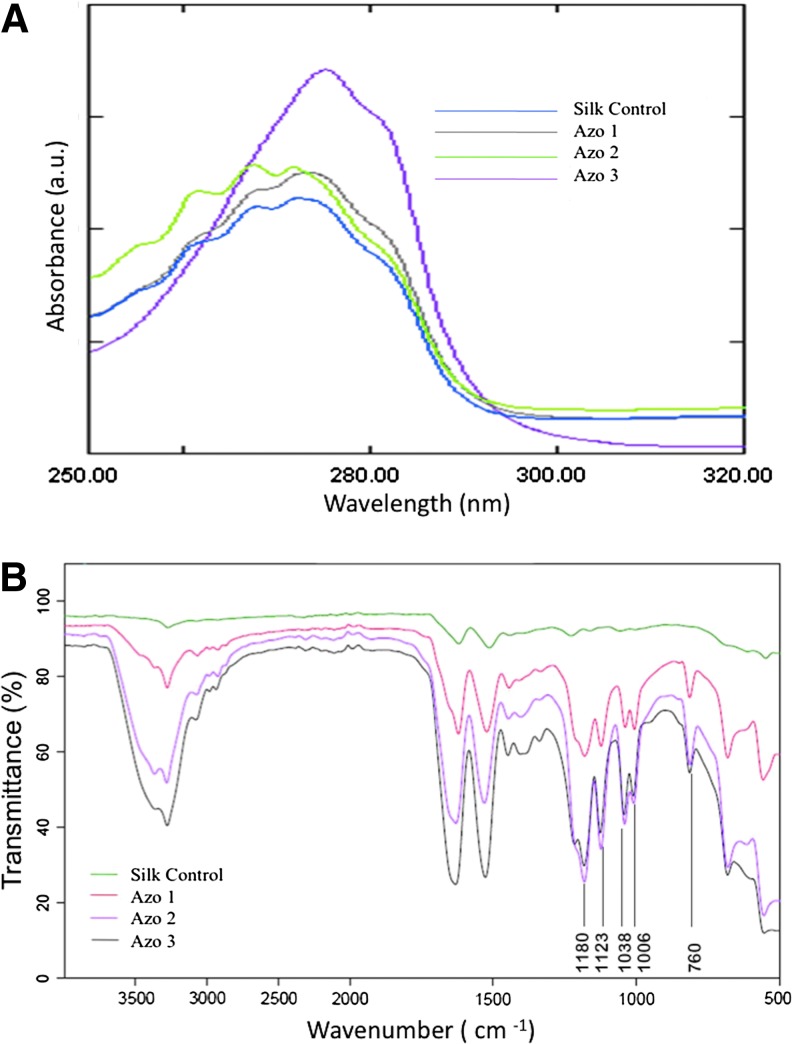
Enhanced diazonium reactions increased the number of modified tyrosines based on the increase of peak intensity at 274 nm (Fig. 2A and Table 1). A maximum of 20% of the total tyrosine residues present were modified with an azo concentration of 30% (w/v).
FIG. 2.
(A) Comparison of the UV-Vis Spectra of unmodified silk and sulfonic acid azosilks modified with increasing equivalents of diazonium salt. (B) ATR-FTIR Spectra showing increase in modification of tyrosine residues with increase in azo concentration. Color images available online at www.liebertpub.com/tea
Table 1.
Estimated Percentage of Tyrosine Groups Modified with Varied Amount of Diazonium Salt
| Sample | Equivalents added | Absorbance | Azo conc. (%) | Silk conc. (%) | % Amino acids modified |
|---|---|---|---|---|---|
| Silk control | 0 | 0.187 | 0 | 4 | 0 |
| AZO 1 | 1 | 0.294 | 10 | 4 | 8 |
| AZO 2 | 2 | 0.365 | 20 | 4 | 14 |
| AZO 3 | 3 | 0.467 | 30 | 4 | 20 |
ATR FTIR analysis
FTIR analysis was performed to further validate the incorporation of sulfate groups to fibroin through the diazo bond. The intensity of amide peaks at 1621 (amide I) and 1522 cm−1 (amide II) increased with the increase in the concentration of the diazonium salt (Fig. 2B). The peaks at 1180, 1123, 1038, and 1006 cm−1 are due to the C-N bond formed as a result of the diazonium reaction, which were absent in the control silk solution. There was a corresponding increase in the intensity of the peaks at 1180, 1123, 1038, and 1006 cm−1 with the increase in the diazo concentration. The diazonium salt incorporation was further confirmed by the S-O (sulfate) stretching vibrations at 760 cm−1 for which there was a constant increase in peak intensity with the increasing concentration of the diazonium salt, while the peak was absent in control silk sample.
Fluorescence emission spectra
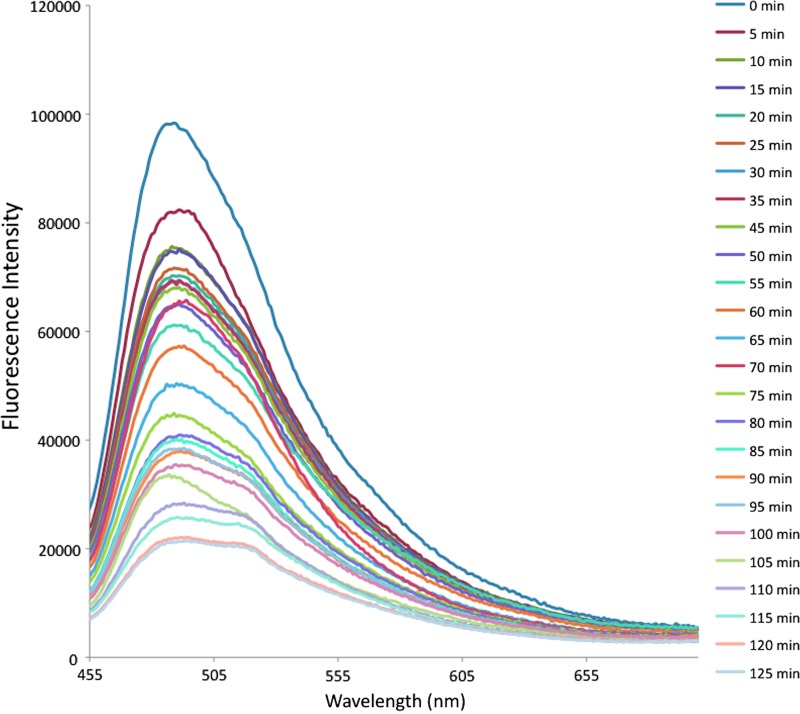
The binding kinetics of IL-1β with diazotized silk fibroin was studied exploiting the intrinsic fluorescence property of tryptophan residues present in the conserved domain having the sequence Phe-Ser-Trp-Ser-Asp-Trp-Trp-Ser of the cytokine IL-1β, which is responsible for the binding of the molecule on to the sulfate moiety.29 There was a continuous quenching in the fluorescence intensity of the tryptophan residues at 470 nm with the increase of time of reaction, which reached a constant after 120 min as a result of binding with the sulfate functionality of diazo modified silk (Fig. 3).
FIG. 3.
Binding kinetics of IL-1β on diazotized silk film. Color images available online at www.liebertpub.com/tea
Adsorption and release kinetics studies
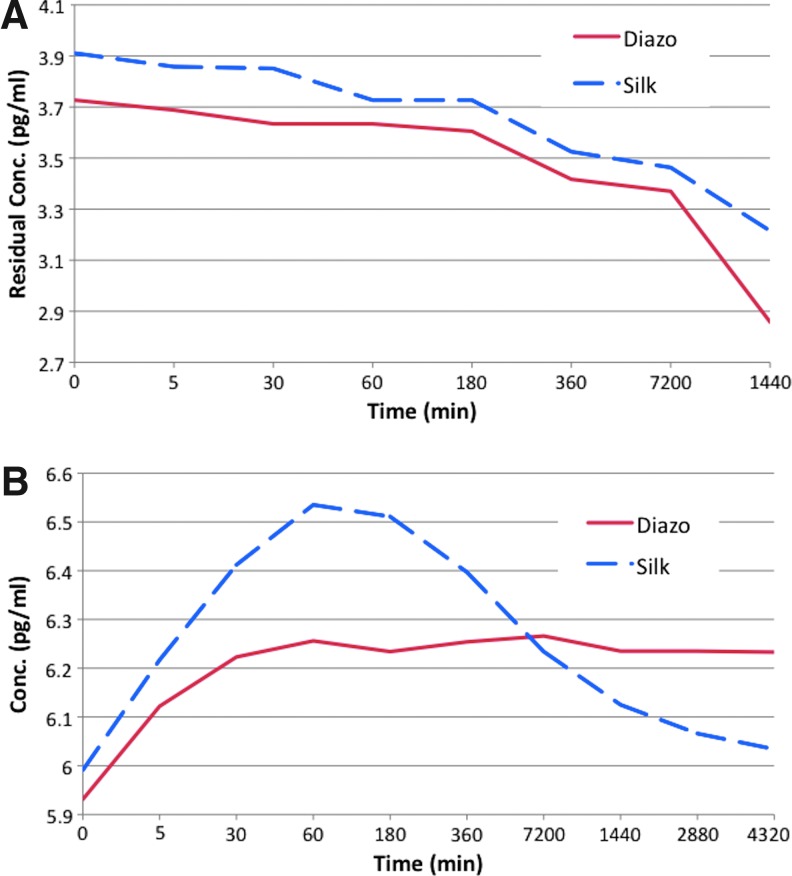
Adsorption studies showed a uniform trend of adsorption of the cytokine by both the silk and diazonium-coupled silk films. The adsorption studies were conducted up to 1 day and showed that the diazonium-coupled silk films adsorbed a slightly higher amount, with statistically significant differences (p<0.05, n=3, SD=0.094). The release experiments showed a significant difference between the diazonium-coupled and unmodified silk films. The silk films showed a burst release after 1 hr followed by subsequent fast release of cytokine from the films (Fig. 4). In contrast, the diazotized silk films demonstrated slow and steady release of the adsorbed cytokine over 3 days. These results indicate that diazotization of silk results in controlled release of the cytokines, a feature hypothesized to help in mimicking the microenvironment in OA-like tissues.
FIG. 4.
(A) Adsorption and (B) release kinetics of cytokines from the silk film as determined by ELISA. Color images available online at www.liebertpub.com/tea
Cell culture and morphological study
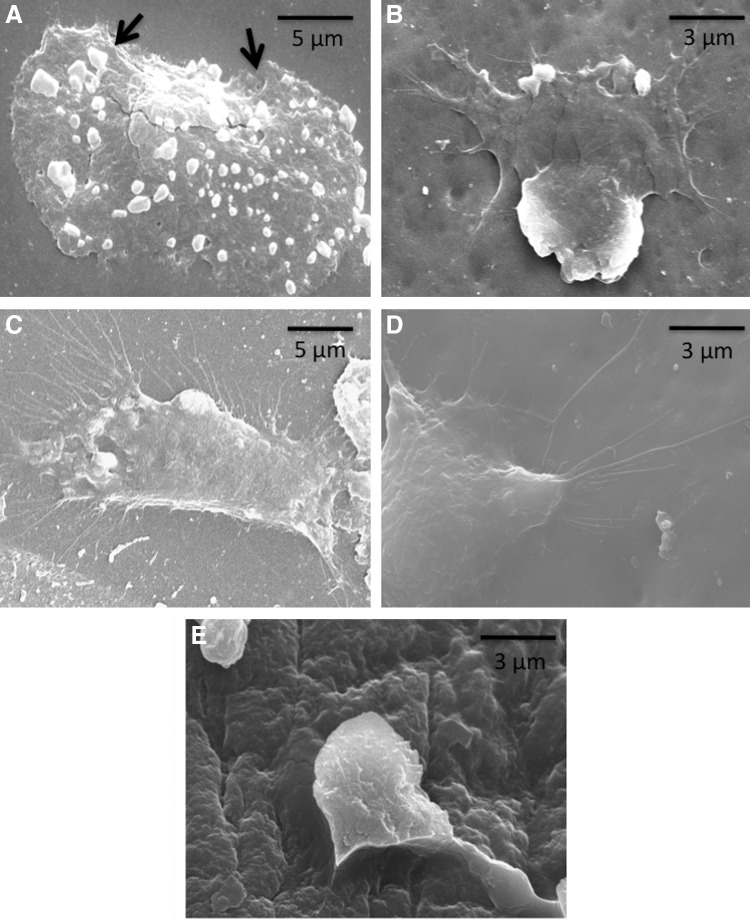
To investigate the role of matrix-tethered cytokines on chondrocyte function, we first documented changes in cell morphology on silk films and Matrigel, with or without anchoring cytokines to the matrix. Chondrocytes grown on silk films showed good attachment and spreading, with smaller cytoplasmic projections of 1–2 μm from the periphery of the cells (Fig. 5A). In contrast, cells on cytokine-immobilized silk films exhibited extensive development of cellular extensions resembling filopodia, and lamellipodia within 8 h (Fig. 5B). The length of these cellular processes was as long as 15 μm after 1 day (Fig. 5C). As additional controls, in order to replicate ECM surface chemistry, cells were cultured over Matrigel layers with or without cytokines. Cells expressed more sprouting cytoplasmic extensions, with anchorage points embedded to the matrix on cytokine-sequestered Matrigel (Fig. 5D), compared to only Matrigel (Fig. 5E).
FIG. 5.
Morphology of chondrocytes on (A) silk film and (B) cytokine immobilized diazotized silk film within 8 h, (C) extensive expression of cellular extensions on cytokine immobilized silk film after 1 day, (D) similar extensions noticed on cytokine-sequestered Matrigel layer, (E) chondrocytes on Matrigel-coated coverslip without cytokines after 1 day (arrows indicate smaller cytoplasmic projections from the periphery of the cells on silk films, compared to cytokine immobilized silk films and Matrigel).
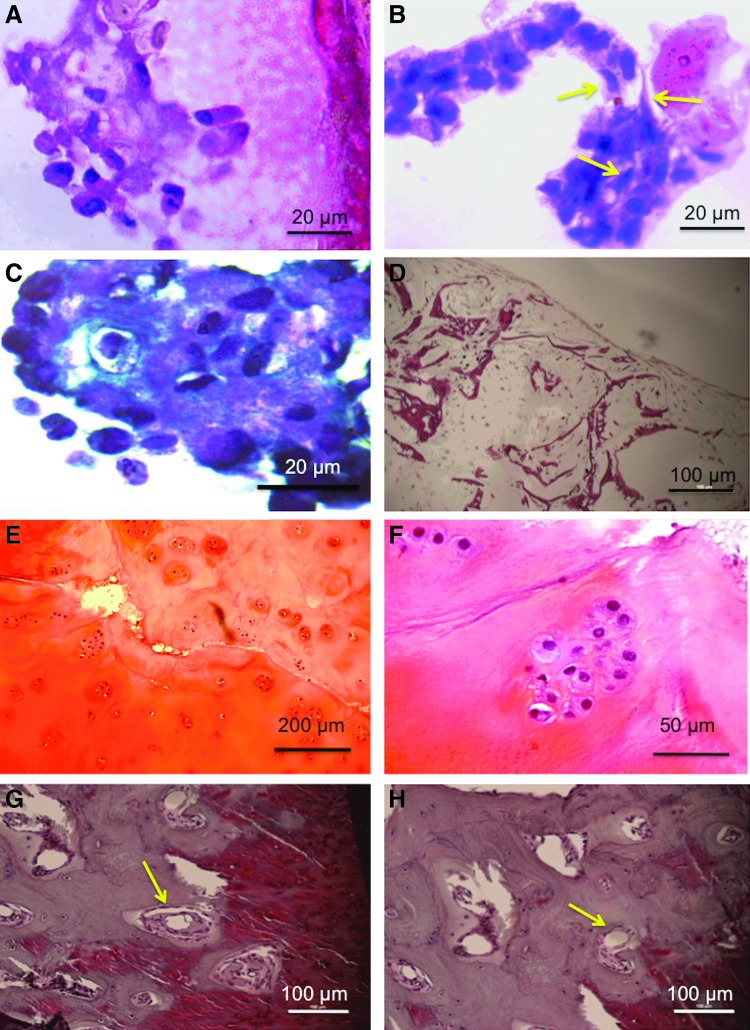
Histological analysis-features of OA
H&E staining of chondrocytes cultured on silk scaffolds showed round morphologies (Fig. 6A), as well as abundant production of ECM. After 4 weeks of culture 84%±5% cells were rounded. In contrast, chondrocytes cultured on cytokine-immobilized silk matrices displayed mixed populations of round and extended cell morphologies, consistent with a fibroblastic phenotype (Fig. 6B); 56%±9% of cells were rounded (p<0.001, n=3). After 21 days of static culture, more intense safranin-O staining was detected in tissue-engineered constructs on the silk scaffolds compared to the in vitro OA tissue models (Fig. 6C, D). GAG/DNA in the in vitro TE cartilage model was 6.5±0.2. In contrast, safranin-O-stained sections of in vitro disease model demonstrated evidence of nominal and nonuniform safranin-O staining, indicating less GAG production and/or retention compared to the tissue-engineered cartilage constructs. GAG/DNA for the in vitro disease model (with cytokines) was 3.1±0.5, validating the histology results.
FIG. 6.
(A) Hematoxylin and eosin (H&E)–stained section of tissue-engineered cartilage showing rounded cell morphology. (B) H&E-stained section of in vitro osteoarthritic (OA) model showing mixed population of rounded and elongated cells (marked by arrows). (C) Safranin-O staining of tissue-engineered cartilage showing the presence of glycosaminoglycans (GAG). (D) Safranin-O staining showing the absence of GAG. (E) H&E-stained cartilage biopsy tissue showing histological features of osteoarthritis such as fibrillation and erosion of GAG deposition, (F) clustered chondrocytes, (G) tide mark and subchondral bone cyst (marked by arrow), and (H) signs of angiogenic invasion in cartilage (marked by arrow). Color images available online at www.liebertpub.com/tea
Histological analysis of OA cartilage biopsies was performed in order to validate the microarray results of the OA cartilage biopsies. In all biopsies from degraded lesions the thickness of cartilage layers appeared to be reduced compared to the normal tissue. Nonuniform depletion of matrix proteoglycans (Fig. 6E), clefts, or fissures was evident, beginning in the superficial layers and progressing inward as the cartilage ECM eroded. Clustering of cells, especially in the middle layers of the cartilage, probably indicates colonies of regenerating chondrocytes (Fig. 6F). Tidemarks that divide the uncalcified cartilage from the deeper calcified cartilage were prominent. Formation of new blood vessels was also visualized (arrow mark) (Fig. 6G), along with subchondral bony cysts between the junction of bone and articular cartilage. The formation of bony outgrowths or osteophytes was observed around the margins of the cartilage and subchondral bone.
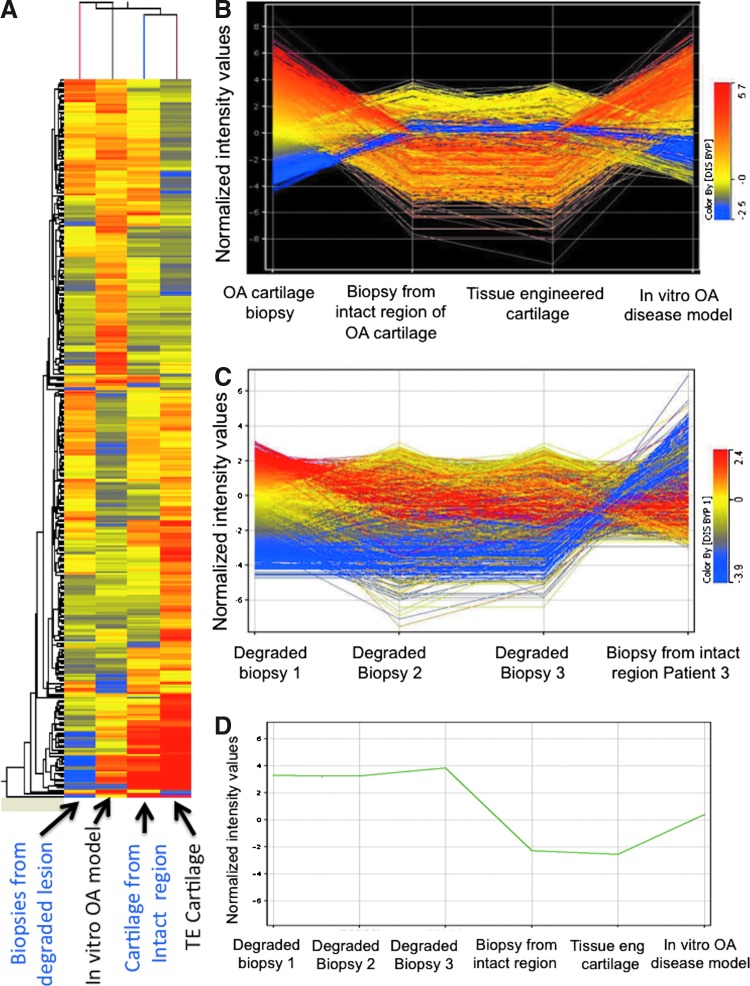
Comparative gene expression profiling
A differential gene expression study was conducted for both the tissue-engineered cartilage with cytokines (in vitro OA-like model) and OA biopsies (in vivo diseased condition), with the tissue-engineered cartilage construct (without cytokine) as the reference (Fig. 7). Over 43,000 transcripts were expressed in all samples. In comparison to the tissue-engineered cartilage, 144 and 168 genes were differentially expressed in the in vitro tissue model and diseased biopsy samples, respectively. Of these, 116 genes were upregulated more than fivefold, while 28 genes were downregulated more than fivefold in the case of the in vitro disease model, compared to tissue-engineered construct. Similarly, in the case of OA biopsy samples 133 genes were upregulated and 35 genes were downregulated significantly, compared to the tissue-engineered constructs.
FIG. 7.
(A) Modulation of gene expression in biopsies from degraded lesion, in vitro OA model, cartilage from intact region, and tissue-engineered cartilage. (B) Extent of the up- or downregulation of each gene in tissue-engineered cartilage, degraded lesion of OA biopsies, intact portion of OA biopsies, and in vitro OA model. (C) Variation among biopsies. (D) Relative expression of vimentin. Color images available online at www.liebertpub.com/tea
Differential gene expression profiles of in vitro disease model and OA cartilage biopsies were compared (Fig. 7). The in vitro diseased tissue model showed strong similarity in gene expression patterns with the OA cartilage biopsies, in that the major categories of genes that were upregulated were those related to cytokines, chemokines, angiogenesis, growth factors, ECM remodeling, ossification, and apoptosis, with higher degrees of expression intensity of OA signaling transcripts in the case of diseased biopsies as compared to the in vitro disease model. In particular, significant upregulation was found in a number of ECM components, including COMP (72.9-fold), COLA1 (6.8-fold), COL8A2 (6.94-fold), COL2A1 (9.76-fold), and Asporin (5.66-fold), as well as matrix degrading components, for example, ADAM28 (5.2-fold), ADAM32 (10.19-fold), and MMP3 (14.67-fold), in OA cartilage biopsies, as compared to the in vitro OA diseased model (Table 2A and 2B). Interestingly, transcripts related to inhibitors of MMPs, such as TIMP1 (53.01-fold) and TIMP2 (5.46-fold) were also upregulated in OA biopsies, in comparison to the in vitro diseased model construct.
Table 2a.
Modulation of Gene Expression in Osteoarthritic Cartilage Biopsy Compared to In Vitro Osteoarthritic Model (More Than Fivefold Upregulated Genes)
| Symbol | Entrez gene ID | Fold change | Definition |
|---|---|---|---|
| HTRA1 | 5654 | 77.95 | Homo sapiens HtrA serine peptidase 1 (HTRA1), mRNA. |
| COMP | 1311 | 72.97 | Homo sapiens cartilage oligomeric matrix protein (COMP), mRNA. |
| MT2A | 4502 | 65.29 | Homo sapiens metallothionein 2A (MT2A), mRNA. |
| CILP | 8483 | 60.07 | Homo sapiens cartilage intermediate layer protein, nucleotide pyrophosphohydrolase (CILP), mRNA. |
| TIMP1 | 7076 | 53.01 | Homo sapiens TIMP metallopeptidase inhibitor 1 (TIMP1), mRNA. |
| ANGPTL4 | 51129 | 38.97 | Homo sapiens angiopoietin-like 4 (ANGPTL4), transcript variant 1, mRNA. |
| PRG4 | 10216 | 33.60 | Homo sapiens proteoglycan 4 (PRG4), mRNA. |
| CDK2AP1 | 8099 | 27.21 | Homo sapiens CDK2-associated protein 1 (CDK2AP1), mRNA. |
| MFGE8 | 4240 | 26.30 | Homo sapiens milk fat globule-EGF factor 8 protein (MFGE8), mRNA. |
| SERPINE2 | 5270 | 17.40 | Homo sapiens serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (SERPINE2), mRNA. |
| MT1G | 4495 | 16.88 | Homo sapiens metallothionein 1G (MT1G), mRNA. |
| ACTB | 60 | 16.10 | Homo sapiens actin, beta (ACTB), mRNA. |
| MMP3 | 4314 | 14.67 | Homo sapiens matrix metallopeptidase 3 (stromelysin 1, progelatinase) (MMP3), mRNA. |
| CD68 | 968 | 13.40 | Homo sapiens CD68 antigen (CD68), mRNA. |
| CAMLG | 819 | 13.36 | Homo sapiens calcium modulating ligand (CAMLG), mRNA. |
| RIS1 | 25907 | 11.90 | Homo sapiens Ras-induced senescence 1 (RIS1), mRNA. |
| SAA2 | 6289 | 11.87 | Homo sapiens serum amyloid A2 (SAA2), mRNA. |
| PRDX1 | 5052 | 11.58 | Homo sapiens peroxiredoxin 1 (PRDX1), transcript variant 2, mRNA. |
| BCL2L13 | 23786 | 11.33 | Homo sapiens BCL2-like 13 (apoptosis facilitator) (BCL2L13), nuclear gene encoding mitochondrial protein, mRNA. |
| G3BP1 | 10146 | 11.30 | Homo sapiens GTPase activating protein (SH3 domain) binding protein 1 (G3BP1), transcript variant 1, mRNA. |
| MT1F | 4494 | 10.77 | Homo sapiens metallothionein 1F (MT1F), mRNA. |
| COL2A1 | 1280 | 9.76 | Homo sapiens collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) (COL2A1), transcript variant 1, mRNA. |
| ITM2B | 9445 | 9.65 | Homo sapiens integral membrane protein 2B (ITM2B), mRNA. |
| PSMC1 | 5700 | 9.58 | Homo sapiens proteasome (prosome, macropain) 26S subunit, ATPase, 1 (PSMC1), mRNA. |
| GSPT1 | 2935 | 9.25 | Homo sapiens G1 to S phase transition 1 (GSPT1), mRNA. |
| NFATC1 | 4772 | 9.01 | Homo sapiens nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (NFATC1), transcript variant 3, mRNA. |
| CCPG1 | 9236 | 8.96 | Homo sapiens cell cycle progression 1 (CCPG1), transcript variant 1, mRNA. |
| SS18L2 | 51188 | 8.93 | Homo sapiens synovial sarcoma translocation gene on chromosome 18-like 2 (SS18L2), mRNA. |
| CYP26C1 | 340665 | 8.86 | Homo sapiens cytochrome P450, family 26, subfamily C, polypeptide 1 (CYP26C1), mRNA. |
| ICA1 | 3382 | 8.80 | Homo sapiens islet cell autoantigen 1, 69 kDa (ICA1), transcript variant 3, mRNA. |
| RCAN1 | 1827 | 8.73 | Homo sapiens regulator of calcineurin 1 (RCAN1), transcript variant 2, mRNA. |
| HOXB8 | 3218 | 8.57 | Homo sapiens homeo box B8 (HOXB8), mRNA. |
| NCOA4 | 8031 | 8.47 | Homo sapiens nuclear receptor coactivator 4 (NCOA4), mRNA. |
| PAGE2B | 389860 | 8.43 | Homo sapiens P antigen family, member 2B (PAGE2B), mRNA. |
| CD44 | 960 | 8.17 | Homo sapiens CD44 molecule (CD44), transcript variant 3, mRNA. |
| CARD14 | 79092 | 7.77 | Homo sapiens caspase recruitment domain family, member 14 (CARD14), transcript variant 2, mRNA. |
| ITGB1 | 3688 | 7.73 | Homo sapiens integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) (ITGB1), transcript variant 1A, mRNA. |
| CTSD | 1509 | 7.71 | Homo sapiens cathepsin D (CTSD), mRNA. |
| CEP27 | 55142 | 7.69 | Homo sapiens centrosomal protein 27 kDa (CEP27), mRNA. |
| SLC6A9 | 6536 | 7.68 | Homo sapiens solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (SLC6A9), transcript variant 1, mRNA. |
| CALM2 | 805 | 7.63 | Homo sapiens calmodulin 2 (phosphorylase kinase, delta) (CALM2), mRNA. |
| PRKRIR | 5612 | 7.53 | Homo sapiens protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) (PRKRIR), mRNA. |
| LPAR1 | 1902 | 7.43 | Homo sapiens lysophosphatidic acid receptor 1 (LPAR1), transcript variant 1, mRNA. |
| NFKBIA | 4792 | 7.36 | Homo sapiens nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (NFKBIA), mRNA. |
| G1P3 | 2537 | 7.14 | Homo sapiens interferon, alpha-inducible protein (clone IFI-6-16) (G1P3), transcript variant 1, mRNA. |
| COL8A2 | 1296 | 6.94 | Homo sapiens collagen, type VIII, alpha 2 (COL8A2), mRNA. |
| MRPL17 | 63875 | 6.93 | Homo sapiens mitochondrial ribosomal protein L17 (MRPL17), nuclear gene encoding mitochondrial protein, mRNA. |
| MATK | 4145 | 6.91 | Homo sapiens megakaryocyte-associated tyrosine kinase (MATK), transcript variant 2, mRNA. |
| COL4A1 | 1282 | 6.80 | Homo sapiens collagen, type IV, alpha 1 (COL4A1), mRNA. |
| EEF2 | 1938 | 6.71 | Homo sapiens eukaryotic translation elongation factor 2 (EEF2), mRNA. |
| PRCP | 5547 | 6.70 | Homo sapiens prolylcarboxypeptidase (angiotensinase C) (PRCP), transcript variant 2, mRNA. |
| MAP3K5 | 4217 | 6.64 | Homo sapiens mitogen-activated protein kinase kinase kinase 5 (MAP3K5), mRNA. |
| TNFRSF8 | 943 | 6.63 | Homo sapiens tumor necrosis factor receptor superfamily, member 8 (TNFRSF8), transcript variant 1, mRNA. |
| SCARA3 | 51435 | 6.59 | Homo sapiens scavenger receptor class A, member 3 (SCARA3), transcript variant 2, mRNA. |
| GABARAPL1 | 23710 | 6.58 | Homo sapiens GABA(A) receptor-associated protein like 1 (GABARAPL1), mRNA. |
| ITGA1 | 3672 | 6.54 | Homo sapiens integrin, alpha 1 (ITGA1), mRNA. |
| BNIP3 | 664 | 6.51 | Homo sapiens BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), nuclear gene encoding mitochondrial protein, mRNA. |
| PSMB3 | 5691 | 6.35 | Homo sapiens proteasome (prosome, macropain) subunit, beta type, 3 (PSMB3), mRNA. |
| PLOD1 | 5351 | 6.26 | Homo sapiens procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 (PLOD1), mRNA. |
| PLD3 | 23646 | 6.26 | Homo sapiens phospholipase D family, member 3 (PLD3), transcript variant 2, mRNA. |
| ATF2 | 1386 | 6.19 | Homo sapiens activating transcription factor 2 (ATF2), mRNA. |
| CCNF | 899 | 6.11 | Homo sapiens cyclin F (CCNF), mRNA. |
| MRPS6 | 64968 | 6.09 | Homo sapiens mitochondrial ribosomal protein S6 (MRPS6), nuclear gene encoding mitochondrial protein, mRNA. |
| PPP1R14B | 26472 | 6.03 | Homo sapiens protein phosphatase 1, regulatory (inhibitor) subunit 14B (PPP1R14B), mRNA. |
| NR2E1 | 7101 | 5.99 | Homo sapiens nuclear receptor subfamily 2, group E, member 1 (NR2E1), mRNA. |
| LASS6 | 253782 | 5.95 | Homo sapiens LAG1 homolog, ceramide synthase 6 (LASS6), mRNA. |
| CD81 | 975 | 5.93 | Homo sapiens CD81 molecule (CD81), mRNA. |
| ERRFI1 | 54206 | 5.92 | Homo sapiens ERBB receptor feedback inhibitor 1 (ERRFI1), mRNA. |
| GLTSCR2 | 29997 | 5.92 | Homo sapiens glioma tumor suppressor candidate region gene 2 (GLTSCR2), mRNA. |
| ANXA7 | 310 | 5.90 | Homo sapiens annexin A7 (ANXA7), transcript variant 2, mRNA. |
| S100A6 | 6277 | 5.87 | Homo sapiens S100 calcium binding protein A6 (S100A6), mRNA. |
| RRAGA | 10670 | 5.86 | Homo sapiens Ras-related GTP binding A (RRAGA), mRNA. |
| DNAJB12 | 54788 | 5.80 | PREDICTED: Homo sapiens DnaJ (Hsp40) homolog, subfamily B, member 12, transcript variant 2 (DNAJB12), mRNA. |
| CD300A | 11314 | 5.77 | Homo sapiens CD300a molecule (CD300A), mRNA. |
| PRAME | 23532 | 5.75 | Homo sapiens preferentially expressed antigen in melanoma (PRAME), transcript variant 5, mRNA. |
| MMEL1 | 79258 | 5.73 | Homo sapiens membrane metallo-endopeptidase-like 1 (MMEL1), mRNA. |
| PXMP4 | 11264 | 5.73 | Homo sapiens peroxisomal membrane protein 4, 24 kDa (PXMP4), transcript variant 1, mRNA. |
| RHOA | 387 | 5.69 | Homo sapiens ras homolog gene family, member A (RHOA), mRNA. |
| ASPN | 54829 | 5.66 | Homo sapiens asporin (ASPN), mRNA. |
| IFI44 | 10561 | 5.64 | Homo sapiens interferon-induced protein 44 (IFI44), mRNA. |
| FTH1 | 2495 | 5.61 | Homo sapiens ferritin, heavy polypeptide 1 (FTH1), mRNA. |
| UBXD2 | 23190 | 5.60 | Homo sapiens UBX domain containing 2 (UBXD2), mRNA. |
| ADD3 | 120 | 5.59 | Homo sapiens adducin 3 (gamma) (ADD3), transcript variant 1, mRNA. |
| EPHA3 | 2042 | 5.59 | Homo sapiens EPH receptor A3 (EPHA3), transcript variant 2, mRNA. |
| SCRG1 | 11341 | 5.59 | Homo sapiens scrapie responsive protein 1 (SCRG1), mRNA. |
| CRTAC1 | 55118 | 5.58 | Homo sapiens cartilage acidic protein 1 (CRTAC1), mRNA. |
| IGFBP7 | 3490 | 5.56 | Homo sapiens insulin-like growth factor binding protein 7 (IGFBP7), mRNA. |
| SCARB2 | 950 | 5.49 | Homo sapiens scavenger receptor class B, member 2 (SCARB2), mRNA. |
| LYST | 1130 | 5.48 | Homo sapiens lysosomal trafficking regulator (LYST), mRNA. |
| TIMP2 | 7077 | 5.46 | Homo sapiens TIMP metallopeptidase inhibitor 2 (TIMP2), mRNA. |
| MYH9 | 4627 | 5.45 | Homo sapiens myosin, heavy chain 9, nonmuscle (MYH9), mRNA. |
| KCNAB3 | 9196 | 5.38 | Homo sapiens potassium voltage-gated channel, shaker-related subfamily, beta member 3 (KCNAB3), mRNA. |
| MARCKSL1 | 65108 | 5.31 | Homo sapiens MARCKS-like 1 (MARCKSL1), mRNA. |
| MYOM1 | 8736 | 5.30 | Homo sapiens myomesin 1 (skelemin) 185 kDa (MYOM1), mRNA. |
| NPAL1 | 152519 | 5.30 | Homo sapiens NIPA-like domain containing 1 (NPAL1), mRNA. |
| EXOC2 | 55770 | 5.29 | Homo sapiens exocyst complex component 2 (EXOC2), mRNA. |
| AK3 | 50808 | 5.28 | Homo sapiens adenylate kinase 3 (AK3), nuclear gene encoding mitochondrial protein, mRNA. |
| RELT | 84957 | 5.24 | Homo sapiens RELT tumor necrosis factor receptor (RELT), transcript variant 1, mRNA. |
| LY96 | 23643 | 5.20 | Homo sapiens lymphocyte antigen 96 (LY96), mRNA. |
| SEPT9 | 10801 | 5.20 | Homo sapiens septin 9 (SEPT9), mRNA. |
| ADAM28 | 10863 | 5.20 | Homo sapiens ADAM metallopeptidase domain 28 (ADAM28), transcript variant 1, mRNA. |
| CCL20 | 6364 | 5.15 | Homo sapiens chemokine (C-C motif) ligand 20 (CCL20), mRNA. |
| VPS52 | 6293 | 5.11 | Homo sapiens vacuolar protein sorting 52 homolog (S. cerevisiae) (VPS52), mRNA. |
| CCNI | 10983 | 5.10 | Homo sapiens cyclin I (CCNI), mRNA. |
| PCOLCE2 | 26577 | 5.08 | Homo sapiens procollagen C-endopeptidase enhancer 2 (PCOLCE2), mRNA. |
| TAPBP | 6892 | 5.08 | Homo sapiens TAP binding protein (tapasin) (TAPBP), transcript variant 3, mRNA. |
| BPIL1 | 80341 | 5.07 | Homo sapiens bactericidal/permeability-increasing protein-like 1 (BPIL1), mRNA. |
| PRRC1 | 133619 | 5.07 | Homo sapiens proline-rich coiled-coil 1 (PRRC1), mRNA. |
| TPH1 | 7166 | 5.07 | Homo sapiens tryptophan hydroxylase 1 (TPH1), mRNA. |
| PROS1 | 5627 | 5.04 | Homo sapiens protein S (alpha) (PROS1), mRNA. |
| CLIC3 | 9022 | 5.03 | Homo sapiens chloride intracellular channel 3 (CLIC3), mRNA. |
| CADM1 | 23705 | 5.02 | Homo sapiens cell adhesion molecule 1 (CADM1), mRNA. |
Table 2b.
Modulation of Gene Expression in Osteoarthritic Cartilage Biopsy Compared to In Vitro Osteoarthritic Model (More Than Fivefold Downregulated Genes)
| Symbol | Entrez gene ID | Fold change | Definition |
|---|---|---|---|
| ADAM32 | 203102 | 10.19 | Homo sapiens ADAM metallopeptidase domain 32 (ADAM32), mRNA. |
| BTNL3 | 10917 | 9.91 | Homo sapiens butyrophilin-like 3 (BTNL3), mRNA. |
| OR7C2 | 26658 | 9.76 | Homo sapiens olfactory receptor, family 7, subfamily C, member 2 (OR7C2), mRNA. |
| CACNG1 | 786 | 9.31 | Homo sapiens calcium channel, voltage-dependent, gamma subunit 1 (CACNG1), mRNA. |
| MANBAL | 63905 | 9.08 | Homo sapiens mannosidase, beta A, lysosomal-like (MANBAL), transcript variant 1, mRNA. |
| GJC1 | 125111 | 8.78 | Homo sapiens gap junction protein, chi 1, 31.9 kDa (GJC1), mRNA. |
| EDA | 1896 | 8.63 | Homo sapiens ectodysplasin A (EDA), transcript variant 4, mRNA. |
| CUEDC1 | 404093 | 8.44 | Homo sapiens CUE domain containing 1 (CUEDC1), mRNA. |
| UBTD1 | 80019 | 8.28 | Homo sapiens ubiquitin domain containing 1 (UBTD1), mRNA. |
| RGS9BP | 388531 | 8.20 | Homo sapiens RGS9 anchor protein (RGS9BP), mRNA. |
| PLXNB1 | 5364 | 8.17 | Homo sapiens plexin B1 (PLXNB1), mRNA. |
| PIGR | 5284 | 8.16 | Homo sapiens polymeric immunoglobulin receptor (PIGR), mRNA. |
| HNRNPG-T | 27288 | 8.11 | Homo sapiens testes-specific heterogenous nuclear ribonucleoprotein G-T (HNRNPG-T), mRNA. |
| ST7L | 54879 | 8.11 | Homo sapiens suppression of tumorigenicity 7 like (ST7L), transcript variant 4, mRNA. |
| PDE1A | 5136 | 7.67 | Homo sapiens phosphodiesterase 1A, calmodulin-dependent (PDE1A), transcript variant 2, mRNA. |
| MAX | 4149 | 7.42 | Homo sapiens MYC associated factor X (MAX), transcript variant 5, mRNA. |
| FGF7 | 2252 | 7.40 | Homo sapiens fibroblast growth factor 7 (keratinocyte growth factor) (FGF7), mRNA. |
| BTBD11 | 121551 | 7.34 | Homo sapiens BTB (POZ) domain containing 11 (BTBD11), transcript variant b, mRNA. |
| CXCR5 | 643 | 7.30 | Homo sapiens chemokine (C-X-C motif) receptor 5 (CXCR5), transcript variant 2, mRNA. |
| TFF1 | 7031 | 7.14 | Homo sapiens trefoil factor 1 (TFF1), mRNA. |
| LTA | 4049 | 7.01 | Homo sapiens lymphotoxin alpha (TNF superfamily, member 1) (LTA), mRNA. |
| ANKRD34B | 340120 | 6.82 | Homo sapiens ankyrin repeat domain 34B (ANKRD34B), mRNA. |
| SUMO1 | 7341 | 6.66 | Homo sapiens SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) (SUMO1), transcript variant 1, mRNA. |
| ECSIT | 51295 | 6.50 | Homo sapiens ECSIT homolog (Drosophila) (ECSIT), mRNA. |
| CDC2L2 | 985 | 6.32 | Homo sapiens cell division cycle 2-like 2 (PITSLRE proteins) (CDC2L2), transcript variant 3, mRNA. |
| RAB40AL | 282808 | 6.25 | Homo sapiens RAB40A, member RAS oncogene family-like (RAB40AL), mRNA. |
| DNMT2 | 1787 | 6.24 | Homo sapiens DNA (cytosine-5-)-methyltransferase 2 (DNMT2), transcript variant e, mRNA. |
| MON2 | 23041 | 6.20 | Homo sapiens MON2 homolog (yeast) (MON2), mRNA. |
| SMPX | 23676 | 6.17 | Homo sapiens small muscle protein, X-linked (SMPX), mRNA. |
| FETUB | 26998 | 6.16 | Homo sapiens fetuin B (FETUB), mRNA. |
| MURC | 347273 | 6.15 | Homo sapiens muscle-restricted coiled-coil protein (MURC), mRNA. |
| K5B | 196374 | 6.02 | Homo sapiens keratin 5b (K5B), mRNA. |
| IRF4 | 3662 | 6.02 | Homo sapiens interferon regulatory factor 4 (IRF4), mRNA. |
| HOXD3 | 3232 | 6.01 | Homo sapiens homeobox D3 (HOXD3), mRNA. |
| CA8 | 767 | 6.00 | Homo sapiens carbonic anhydrase VIII (CA8), mRNA. |
| AK2 | 204 | 5.99 | Homo sapiens adenylate kinase 2, transcript variant AK2B, mRNA. |
| AKAP11 | 11215 | 5.94 | Homo sapiens A kinase (PRKA) anchor protein 11, transcript variant 2, mRNA. |
| PARVB | 29780 | 5.91 | Homo sapiens parvin, beta (PARVB), transcript variant 2, mRNA. |
| SEMA6D | 80031 | 5.87 | Homo sapiens sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphoring) 6D, transcript variant 4, mRNA. |
| APRIN | 23047 | 5.84 | Homo sapiens androgen-induced proliferation inhibitor (APRIN), transcript variant 2, mRNA. |
| VIM | 7431 | 5.83 | Homo sapiens vimentin (VIM), mRNA. |
| ALS2 | 57679 | 5.75 | Homo sapiens amyotrophic lateral sclerosis 2 (juvenile) (ALS2), mRNA. |
| TAF4 | 6874 | 5.72 | Homo sapiens TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135 kDa (TAF4), mRNA. |
| DHRS2 | 10202 | 5.71 | Homo sapiens dehydrogenase/reductase (SDR family) member 2 (DHRS2), transcript variant 1, mRNA. |
| MEF2C | 4208 | 5.71 | Homo sapiens myocyte enhancer factor 2C (MEF2C), mRNA. |
| TRH | 7200 | 5.66 | Homo sapiens thyrotropin-releasing hormone (TRH), mRNA. |
| NTF3 | 4908 | 5.66 | Homo sapiens neurotrophin 3 (NTF3), mRNA. |
| AKAP7 | 9465 | 5.66 | Homo sapiens A kinase (PRKA) anchor protein 7 (AKAP7), transcript variant gamma, mRNA. |
| TTTY5 | 83863 | 5.53 | Homo sapiens testis-specific transcript, Y-linked 5 (TTTY5), noncoding RNA. |
| CAMKK1 | 84254 | 5.51 | Homo sapiens calcium/calmodulin-dependent protein kinase kinase 1, alpha (CAMKK1), transcript variant 3, mRNA. |
| MSH4 | 4438 | 5.51 | Homo sapiens mutS homolog 4 (E. coli) (MSH4), mRNA. |
| OCLM | 10896 | 5.45 | Homo sapiens oculomedin (OCLM), mRNA. |
| APLN | 8862 | 5.45 | Homo sapiens apelin (APLN), mRNA. |
| BTBD11 | 121551 | 5.44 | Homo sapiens BTB (POZ) domain containing 11 (BTBD11), transcript variant 1, mRNA. |
| ACSL5 | 51703 | 5.43 | Homo sapiens acyl-CoA synthetase long-chain family member 5 (ACSL5), transcript variant 1, mRNA. |
| MTUS1 | 57509 | 5.40 | Homo sapiens mitochondrial tumor suppressor 1 (MTUS1), nuclear gene encoding mitochondrial protein, transcript variant 5, mRNA. |
| XCL1 | 6375 | 5.38 | Homo sapiens chemokine (C motif) ligand 1 (CXCL1), mRNA. |
| TCHP | 84260 | 5.37 | Homo sapiens trichoplein, keratin filament binding (TCHP), mRNA. |
| TDRD4 | 54513 | 5.36 | Homo sapiens tudor domain containing 4 (TDRD4), mRNA. |
| KRTAP19-1 | 337882 | 5.33 | Homo sapiens keratin associated protein 19-1 (KRTAP19-1), mRNA. |
| SELS | 55829 | 5.27 | Homo sapiens selenoprotein S (SELS), transcript variant 1, mRNA. |
| TEAD2 | 8463 | 5.26 | Homo sapiens TEA domain family member 2 (TEAD2), mRNA. |
| FLJ40142 | 400073 | 5.26 | Homo sapiens FLJ40142 protein (FLJ40142), mRNA. |
| VEGFB | 7423 | 5.25 | Homo sapiens vascular endothelial growth factor B (VEGFB), mRNA. |
| NLK | 51701 | 5.24 | Homo sapiens nemo-like kinase (NLK), mRNA. |
| TMLHE | 55217 | 5.22 | Homo sapiens trimethyllysine hydroxylase, epsilon (TMLHE), mRNA. |
| ISYNA1 | 51477 | 5.19 | Homo sapiens inositol-3-phosphate synthase 1 (ISYNA1), mRNA. |
| CASP9 | 842 | 5.17 | Homo sapiens caspase 9, apoptosis-related cysteine peptidase (CASP9), transcript variant alpha, mRNA. |
| CFHR3 | 10878 | 5.17 | Homo sapiens complement factor H-related 3 (CFHR3), mRNA. |
| Sept2 | 4735 | 5.15 | Homo sapiens septin 2 (SEPT2), transcript variant 3, mRNA. |
| TRIP4 | 9325 | 5.01 | Homo sapiens thyroid hormone receptor interactor 4 (TRIP4), mRNA. |
One major group of genes that was highly expressed in the in vitro disease and in vivo conditions were the cytokines, chemokines, and their receptors. These genes probably regulate OA pathophysiology as triggers and signaling molecules for modulating cellular functions. The receptors that were highly expressed included ITGB1 (7.73-fold upregulated), RELT (5.24-fold upregulated), EPHA3 (5.59-fold upregulated), LPAR1 (7.43-fold upregulated), PIGR (8.16-fold downregulated), SCARA3 (6.59-fold upregulated), TNFRSF8 (6.63-fold upregulated), SCARB2 (5.49-fold upregulated), CXCR5 (7.3-fold downregulated), GABARA (6.58-fold upregulated), NCOA4 (8.47-fold upregulated), and TRIP4 (5.01-fold downregulated), while the chemokines that were highly expressed included CXCL1 (5.38-fold downregulated), CCL20 (5.15-fold upregulated), and CXCR5 (7.3-fold downregulated) in the OA diseased biopsies with respect to the in vitro tissue diseased model. These genes were not expressed significantly in the intact region of OA cartilage from patients and tissue-engineered cartilage (Supplementary Data S1, S2; Supplementary data are available online at www.liebertpub.com/tea).
A pairwise comparison revealed that genes associated with angiogenesis, such as ANGPTL4, were 38.97-fold upregulated in the OA biopsies as compared to the in vitro disease tissue model, while expression of other angiogenic transcripts (Angio-associated migratory cell protein (AAMP), angiopoietin 1 (ANGPT1), angiotensin II receptor, type 2 (AGTR2), and vascular endothelial growth factor (VEGF)-C) were similarly expressed (within 1.5-fold change), suggesting that angiogenesis is a feature of OA pathogenesis, both in vitro and in vivo.
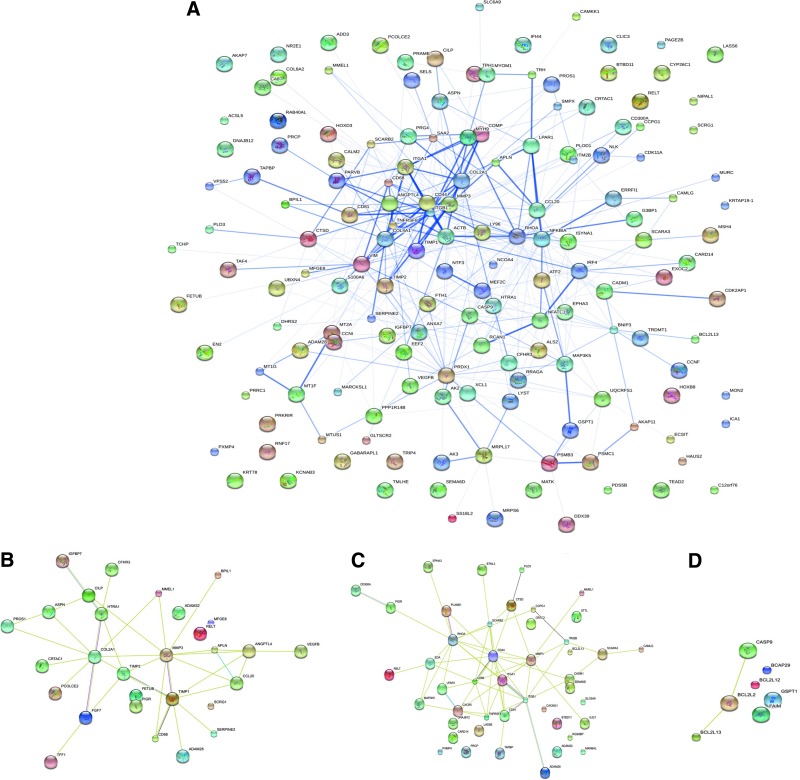
Analysis of the protein–protein interaction network
The protein–protein interaction network analysis showed that genes exhibited a high degree of connection in case of the in vitro OA model (minimum connections on a hub were found to be 4 with a maximum of 15 connections), when compared to the OA biopsy (minimum connections on a hub were found to be 2 with a maximum of 9 connections), indicating greater degree of complexity of the protein interaction network identified among the differentially expressed genes of the in vitro OA model. The protein network obtained for the diseased condition showed three subnetworks (i.e., matrix production, matrix degradation, and apoptosis) that were highly interconnected within themselves and with other networks, contributing to cartilage destruction (Fig. 8A). The protein–protein interaction studies revealed possible roles of these two proinflammatory cytokines in the pathogenesis of OA, as evidenced by the complex network of MMP and the related ADAMTS genes, which were in turn connected to TNF and IL receptor networks. These findings highlight the presence of a stimulating action of these inflammatory cytokines on the action of the metalloproteinases. MMP3 had four major interactants, Col 2A1, COMP, TIMP1, and TIMP2, causing an imbalance which could disrupt cartilage homeostasis (Fig. 8B, C, D). The close connection of CD44 with integrin α1 and integrin β1 provided chondrocytes the ability to sense changes in the extracellular microenvironment. Vimentin was present in a centre of a network, as connected with integrin α1, β-actin, and tumor necrosis factor receptor superfamily member 8.
FIG. 8.
Protein interaction network analysis (confidence view) for (A) upregulated genes in diseased condition, (B) matrix-related proteins, (C) receptors binding, and (D) apoptotic process. Color images available online at www.liebertpub.com/tea
In contrast, the networks presented in tissue-engineered cartilage (without cytokines) showed an absence of the apoptotic subnetwork, but the other two subnetworks of matrix production and matrix degeneration were present but with lower levels of complexity and interconnections (Supplementary Fig. S1).
Gene ontological classification of the gene lists
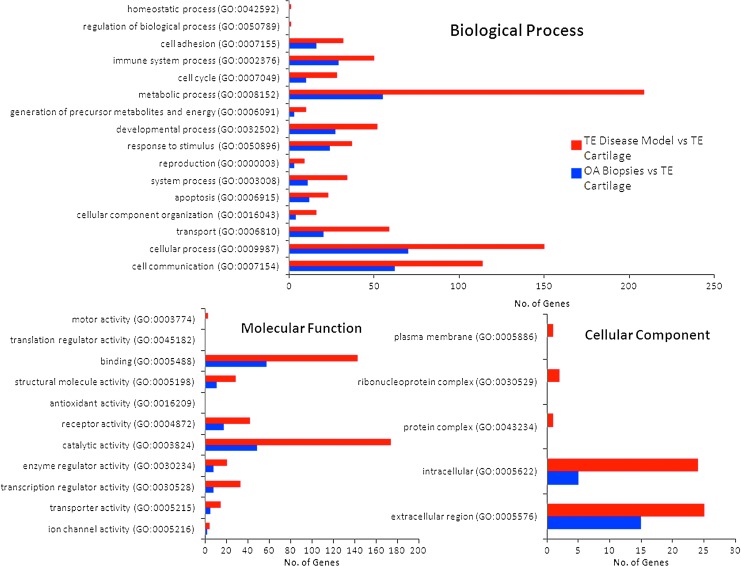
Gene ontology analysis was performed to directly compare differentially expressed genes in OA biopsies and in the in vitro disease model compared to tissue-engineered cartilage, and categorize differentially expressed genes into the three main gene ontology classifications: biological process, molecular function, and cellular components (Fig. 9). Gene ontological classification and analysis demonstrated that the in vitro model of OA expressed the major marker genes of OA, including matrix degeneration and production, apoptosis, chemokines, cytokine and their receptors, angiogenesis, and osteogenic differentiation.
FIG. 9.
Comparative gene ontology of OA biopsies versus tissue-engineered in vitro disease model system, compared to tissue-engineered cartilage. Color images available online at www.liebertpub.com/tea
The in vitro cartilage model showed upregulation of genes in the gene ontological categories for immune response (50 genes), response to external stimulus (37 genes), apoptosis (23 genes), and ECM (25 genes). In contrast, input genes in OA biopsies compared to tissue-engineered cartilage were located in the following major gene ontological categories that signify their biological functions: receptor binding (58 genes), apoptosis (12 genes), cell communication (62 genes), immune system process (29 genes), and ECM (17 genes). These biological functions fall in the pathophysiological cascade of OA, showing the validity of the in vitro disease model. Pathway analysis showed that 8 transcripts encoding the inflammation mediated by chemokine and cytokine signaling pathway were upregulated in OA biopsy, as compared to 12 transcripts in the in vitro disease model. OA biopsy and in vitro disease model expressed 2 and 6 transcripts of interleukin signaling pathway, and 6 and 6 transcripts from Apoptosis signaling pathway, respectively. Interestingly, 8 transcripts related to Wnt signaling pathway were significantly expressed in the in vitro disease model. The Wnt pathway is known to regulate MMPs and aggrecanases. The significant expression of these OA triggering pathways clearly highlighted the validity of the developed in vitro disease model.
Reverse transcriptase–polymerase chain reaction
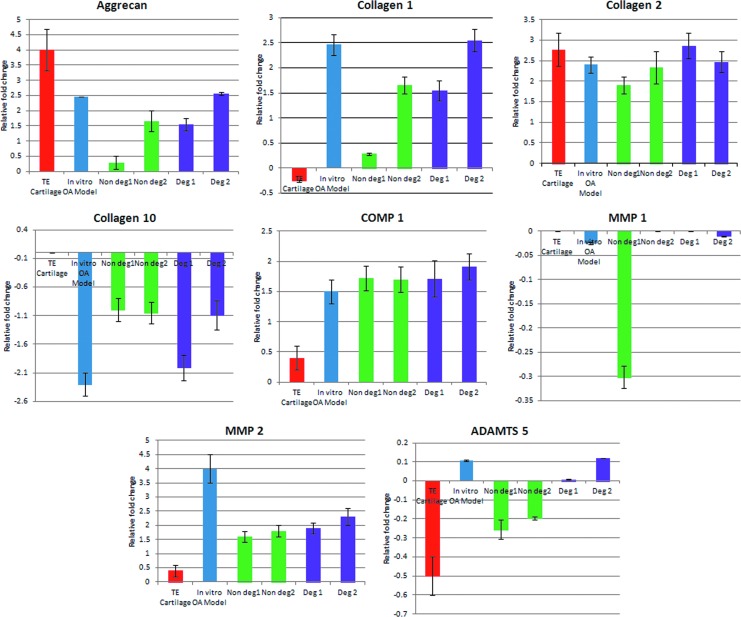
Since the genes implicated in matrix synthesis and matrix degradation are mostly affected by the OA disease condition, eight genes were selected covering these biological processes: collagen 1, collagen 2, collagen 10, aggrecan, COMP1, MMP 1, MMP 2, and ADAMTS5 and were further analyzed by quantitative RT-PCR (Fig. 10). Expression of ADAMTS5 was upregulated in the degraded area of cartilage tissues as well as in vitro disease model, whereas it was downregulated in the nondegraded area of OA cartilage tissues. COMP 1, MMP 2, aggrecan, collagen 1, and collagen 2 were upregulated in all degraded tissues, nondegraded tissues, as well as the in vitro disease constructs, but the level of expression was similar to that of the degraded tissues. MMP 1 and Collagen 10 were downregulated in all degraded tissues, nondegraded tissues, as well as the tissue-engineered constructs, but the level of expression was similar to that of the degraded tissues. Overall, except for the expression of ADAMTS5, the RT-PCR and microarray data were in agreement, corroborating the validation/accuracy of the microarray-based transcriptional analysis. Differences in gene expression levels between tissue-engineered constructs and intact cartilage biopsies were found to be statistically significant (p≤0.05), while that between in vitro OA-like model and degraded biopsies were found to be statistically significant (p<0.05).
FIG. 10.
Relative expression of ADAMTS5, aggrecan, COMP1, collagen 1, collagen 2, collagen 10, MMP1, and MMP2 using tissue-engineered cartilage, engineered in vitro OA-like model, biopsies from nondegraded region (intact cartilage biopsy), and degraded regions from two patients. Color images available online at www.liebertpub.com/tea
Discussion
The current generation of therapeutic modalities is moving toward patient-specific clinical strategies. To further exploit this approach, there is a need to establish patient-specific and tissue-specific disease tissue model systems with which to explore drug response profiles. This goal is being realized with progress in many areas of complex 3D tissue systems in vitro.30
Key features of OA pathogenesis include (a) modulation of metabolic activation of chondrocytes toward dedifferentiated or immature state, (b) hypertrophy and increased apoptosis of chondrocytes,31,32 (c) oxidative stress and inflammatory microenvironment,8,13,33,34 (d) changes in ECM composition with damaged collagen network, depleted GAG, reduced keratan sulfate content, and increased water content, (e) elevated MMPs35 and ADAMTS motif activity,36 (f) growth of osteophytes and thickening of subchondral layer, and (g) neuro/angiogenic invasion.33
In early stages of OA the balance between degradation and synthesis of the pericellular matrix of chondrocytes is disturbed.37 Animal models of secondary (posttraumatic) OA showed enhanced synthesis of collagen 2 and aggrecan,38 as well as abnormal ECM components. Transgenic mice expressing mutant aggrecan, lacking a key aggrecanase cleavage site, demonstrated that erosion of ECM is the key event in the biomechanical failure of the OA joint, irrespective of whether the initiating cause was driven by inflammation (antigen-induced arthritis) or biomechanics (meniscectomy-induced).39 Local immobilization of inflammatory mediators could trigger a cascade of matrix degrading signaling by direct contact with cellular receptors,40 based on immunohistochemical analysis of whole joints of OA-prone guinea pigs. The gradual enhancement and temporal expression and tissue localization of IL-1β occurred with progression of spontaneous OA onset. All these findings corroborate the concept of local inflammation in OA cartilage, rather than systemic inflammation in early stages of degenerative joint disease. Tiku et al. reported that chondrocyte-derived lipid peroxidation end products mediate the oxidation of neighboring collagen fibrils in OA cartilage, leaving them susceptible to degradation.41 El-Bikai et al. supported this concept by showing that ICAM-1, integrin α1β1, MMP13, extracellular signal-regulated kinases 1 and 2, and COX-2 were strongly upregulated in chondrocytes cultured over 4-hydroxynonenal anchored collagen 2 fibrils.42 These findings indicate that the intimate interaction between receptors of chondrocytes with inflammatory mediators anchored to the underlying matrix or modulated matrix composition initiates cascades of signaling pathways triggering early features of OA pathogenesis.
GAG components of the ECM of native cartilage can specifically bind to the heparin binding domains of growth factors and cytokines and immobilize them in the ECM, so that the cellular receptors can intimately bind to these factors and potentiate their biological activity. For example, the formation of a ternary complex among fibroblast growth factors (FGF), their high-affinity receptors, and the proteoglycan heparan sulfate results in activation of FGF.43 This phenomenon has been replicated in numerous studies to develop growth factor delivery systems or immobilization of diffusible or soluble factors in a scaffold, for example, controlled delivery of bone morphogenetic protein by heparin-loaded hydrogels,44 delivery of FGF from chondroitin sulfate-chitosan scaffolds,45 and silk.23,24 The affinity of IL-1β and TNF-α for heparin sulfate46,47 provided the inspiration in the present work for control release of these cytokines, instead of exogenous addition through the media. Immobilization of cytokines to the scaffold may enhance function by facilitating high concentrations of cytokine accumulated on scaffold surfaces or by promoting conformation-dependent interactions of cytokines and their receptors on cell membranes to facilitate signal transduction cascades. Covalent crosslinking of bioactive factors with biomaterials may mask the active sites required for cytokine binding to cell surface receptors and inhibit the internalization of growth factor–receptor complexes by cells. Diazonium coupling of silk scaffold had been reported to enhance cell attachment and proliferation on silk scaffolds23,24 due to enhancement of hydrophilicity of the silk fibroin surface; thus, we utilized this approach in the current study.
The N-terminus of the trimeric TNF-α molecule comprises of two basic arginines inside the short amino-acid sequence Val-Arg-Ser-Ser-Ser-Arg (VRSSSR) (Fig. 1), which is essential for binding of TNF-α to heparin sulfate.48 The same domain could be utilized for the binding of TNF-α to the diazotized silk to simulate in vivo conditions. The binding of IL-1β to heparin sulfate takes place at a conserved domain having the sequence Phe-Ser-Trp-Ser-Asp-Trp-Trp-Ser.29 Both the first and second tryptophan residues in this sequence are essential for tight binding of IL-1β to heparan sulfate.
Wenk et al. reported the highest metabolic activity of mesenchymal stem cells cultured on matrices consisting of 20 sulfonic acid groups per silk fibroin molecule.24 Hence, the optimized concentration of diazonium modified silk sample was used for establishing the in vitro OA model. In the present study the fluorescence of tryptophan molecules present in the binding site of IL-1β was exploited to quantitatively understand the mechanism of attachment of IL-1β to diazo-coupled silk at the sulfonic acid groups.
Matrigel consists of components of ECM such as laminin, collagen 4, proteoglycans (heparan sulfate, glycosaminoglycans), and entactin, which can sequester cytokines. Interestingly, chondrocytes cultured over cytokine-immobilized silk matrices as well as cytokine-enriched Matrigel displayed the presence of multiple elongated processes from cell surfaces, extending up to 15 μm into the surrounding matrix, confirming the feasibility of using diazonium silk protein as an ECM analog. The presence of such extensive elongated fibrous structures had been reported in arthritic human tibia plateaus.49 Holloway et al. reported the presence of extensive elongated fibrous structures in osteoarthritic (femoral head) chondrocytes, extending up to 30 μm, which were found to be networks of intermediate filament vimentin.50 Localization of the chondrocytes expressing these elongated vimentin-rich processes corresponded to the prominent histological lesions and macroscopic signs of OA pathophysiology. However, there is still no clear consensus whether these vimentin-enriched extensions directly contribute to cartilage stiffness, causing localized matrix fracture/degradation through the release of catabolic agents from the cell processes,51 as seen in growth-plate cartilage. Alternatively, these extensions may be expressed only as a direct stress response to localized inflammatory signals. Calamia et al. also reported vimentin was the most abundant protein expressed after cytokine treatment to chondrocytes.52 Interestingly in microarray results, vimentin was 5.8-fold upregulated in the in vitro OA model compared to the OA cartilage biopsies, whereas it was 59.98-fold upregulated in the in vitro OA model compared to tissue-engineered cartilage (Fig. 8D). Lambrecht et al. studied protein expression patterns of articular chondrocytes obtained from intact cartilage from healthy donor, from the visually intact zones and degenerated zones of OA cartilage, and found maximum distorted vimentin organization in degenerated zones of OA chondrocytes compared to other samples.53
Safranin-O staining of in vitro OA model simulated reduced GAG accumulation like OA biopsies. Compared to tissue-engineered cartilage, less accumulation of GAG in the in vitro OA model may indicate that either the chondrocytes had produced less GAG in the presence of the matrix-embedded cytokines, or the sulfated proteoglycans were more depleted.
Microarray results provided further confidence for the successful establishment of an in vitro disease model of OA, with the expression of similar transcripts related with matrix remodeling and matrix degradation, in accordance with the OA diseased cartilage biopsies, compared to the tissue-engineered cartilage. Several transcripts encoding collagen were upregulated related to matrix remodeling of the osteoarthritic cartilage, such as COL4α1, COL8α2, and COL2α1, encoding the alpha chain of one of the nonfibrillar collagens. A COL2α1 transcription factor related to chondrodysplasia-associated OA,54 which is known to be associated with early onset of OA, was upregulated 25.24-fold in the diseased biopsies as well as 12.5-fold in the in vitro disease model as compared to the tissue-engineered cartilage.
Cartilage oligomeric matrix protein (COMP) and cartilage intermediate layer protein (CILP) are known to be strongly upregulated in early OA.55,56 Activation of chondrocytes and synovial cells by proinflammatory cytokines are reported to promote expression of COMP.56 An increase in COMP expression was statistically correlated with the extent of cartilage loss using magnetic resonance images of symptomatic knee OA patients.57 In microarray results, COMP was 72-fold upregulated in OA biopsies compared to the in vitro OA model, whereas RT-PCR results showed similar expression of the COMP transcript in the in vitro OA model and cartilage from nondegraded area of OA cartilage.
Asporin is one of the polymorphisms thought to be strongly associated with OA susceptibility. Asporin was fivefold upregulated in OA biopsies compared to the in vitro disease model. Asporin acts as a negative regulator of matrix homeostasis by inhibiting TGF-β-induced chondrogenesis,58 eventually downregulating sox-9, aggrecanase and collagen 2 expression.
Hypertrophy of chondrocytes is considered as one of the hallmarks of OA, but expression of collagen 10 mRNA was downregulated in all the samples. Some studies have reported contradictory findings concerning hypertrophy markers in OA. Brew et al. demonstrated that expression of collagen 10 was not enhanced during disease progress of OA.59 Bluteau et al. reported an initial downregulation of Collagen 10α1 in early stage and upregulation in later stage in rabbit anterior cruciate ligament rupture model.60 Aigner et al. identified Collagen 10α1 only in the deep zone of OA cartilage using in situ hybridization, whereas collagen 10 expression was detected by immunohistochemistry in the superficial cartilage, but corresponding Collagen 10α1 mRNA could not be observed. This might be due to different half-life of the mRNA and protein.61
In OA, collagenolysis is mediated by collagensase 1 (MMP1), collagenase 2 (MMP8), and collagenase 3 (MMP13), while aggrecanolysis is predominantly mediated by aggrecanase 1 (ADAMTS4) and aggrecanase 2 (ADAMTS5). Transcripts encoding MMP2, MMP3, ADAM28, and ADAMTS32 were upregulated in cartilage biopsies, compared to the in vitro OA-like tissue model. Surprisingly, expression of ADAMTS5 was downregulated from OA cartilage biopsies. In contrast, RT-PCR studies demonstrated that ADAMTS5 was upregulated in the degraded tissues as well as in the in vitro disease model, whereas it was downregulated in the nondegraded part of OA cartilage tissues. Expression of MMP13, which is involved in early cartilage degeneration, and MMP2, which is only significantly detectable in late-stage osteoarthritic cartilage samples,62 was similarly expressed in microarray results of OA biopsies and the in vitro disease model and validated by RT-PCR results.
Surprisingly, TIMPs, which are responsible for the inhibition of MMPs, were highly upregulated in both systems in the microarray results. This finding suggests homeostasis between the action and inhibition of the metalloproteinase genes in early stages of OA, and an imbalance of this dynamic equilibrium probably results in the progression of OA. Bluteau et al. identified the immediate upregulation of TIMP 1 after rupturing of ligament in their rabbit OA model, but the expression level was downregulated after 1 month.60
A group of genes related to oxidative stress was found to be highly upregulated in the OA biopsies, compared to the in vitro OA model. In the present study metallothionin genes, S100A6, MT1F, MT1A, TRIM44, MT2A, and MT1G, were highly upregulated in the osteoarthritic cartilage. These transcripts encode cytosolic protein products, which are mainly involved in the management of oxidative stress in the articular cartilage.
The interconnecting protein interaction studies established that the in vitro tissue model had similarities with the in vivo osteoarthritic condition, providing some confirmation that this is a reasonable starting point as a disease model of OA. For example, the presence of two strong metalloproteinase and cytokine subnetworks that are further interlinked give these two networks their basic similarity. CD44, the major receptor for hyaluronan binding, helps to retain proteoglycan in the ECM and initiate a reparative response when tissue homeostasis is disturbed. Protein networks had shown close connection of CD44 with integrin α1 and integrin β1, which could be considered as part of an attempt by the chondrocyte to repair the damaged matrix. As compared to normal cartilage, increased immunostaining of integrin α1β1 has been reported in osteoarthritic cartilage in a monkey cartilage OA model.63
However, significant dissimilarity in the genes expressed and the protein interaction networks were also highlighted in this study, likely in part due to the differences in ECM as well as differences in cell types present. These are issues that can be added in future iterations of the tissue model system to assess improvements on the similarities reported here as a foundation for a relevant OA tissue model system.
The gene ontological classification further confirmed that in the in vitro disease model using matrix-anchored cytokines resulted in the gene expression profile toward simulating OA-related biological function categories. The genes related to receptor binding were upregulated significantly, showing the effect of cytokine uptake by the chondrocytes that finally leads to the development of OA. Genes related to the ontological category:collagen binding were also highly upregulated in the disease model, showing OA-like gene expression related to matrix restoration. Further genes falling under the ontological category:regulation of apoptosis were highly upregulated, clearly showing the effect of these two proinflammatory cytokines in the apoptosis and subsequent degeneration of cartilage tissue. Other genes related to normal physiological processes like signal transduction via phosphorylation events, intracellular protein kinase cascades, cell proliferation, tissue development, chemotaxis, cell development, regulation of cell communication, and ECM were also significantly differentially expressed.
While the control tissue-engineered cartilage showed the upregulation of genes in the gene ontological categories related to normal physiology of the cartilage tissue, such as G-protein-coupled receptor binding, collagen metabolic process, multicellular organismal metabolic process, and ECM.
The main strength of this study is spatial sequestration of cytokines at the cell–matrix interface to help in forming multimeric signaling complexes with cytokine receptors localized at the focal adhesion sites of chondrocytes and direct the signal transduction to inflammatory cellular response.64 This localization plays pivotal role in OA pathogenesis, because in the absence of such focal adhesion complexes, activation of focal adhesion kinase, tyrosine phosphorylation, as well as propagation of IL-1-induced Ca2+ fluxes do not take place.65 Ultimately, this Ca2+ release is crucial for IL-1 signaling, affecting the downstream pathways involved in matrix destruction.66 Taken together, diazonium-coupled silk matrix could anchor cytokines in a biomimetic manner. This in vitro culture system had been used to study the effect of a localized osteoarthritic inflammatory microenvironment in modulating the cellular morphology, reduced proteoglycan accumulation, and gene expression pattern in vitro and was compared to cartilage biopsies from OA patients. Despite moderated gene expression patterns in comparison to cartilage biopsies, several OA marker transcripts depicting earlier regulatory events were strongly upregulated in the in vitro disease model, compared to the OA cartilage biopsies. Although the present study suggests a path forward to establish in vitro disease models to identify disease origins, such as genetic pathogenesis, there remains a lot to add to the tissue model to more fully recapitulate the in vivo state, including involvement of other cell types, mechanical injury, modulation of ECM composition, effect of matrix degradation products, and aging mechanisms, among others. Other in vitro models are being developed,67 which would offer new strategies to find specific drug targets for the prevention of OA. Hence, as an alternative to organ repair by tissue engineering and regenerative medicine, a paradigm shift would be to utilize tissue-engineered systems to understand and optimize disease tissue constructs to introduce selective or specific features to assess various signaling mechanisms and their response to selective cell or drug treatments.
Conclusions
Putative roles for matrix-bound cytokines in the pathology of OA disease were identified. The data support the hypothesis that diazonium-coupled silk scaffolds simulated local inflammatory microenvironments consisting of the synergistic effects of IL-1β and TNFα, in a biomimetic manner, and are capable of inducing changes in chondrocyte morphology and phenotype that provide a basis for OA-like features. The similarities in these in vitro tissue models to OA patient transcript profiles provide a solid starting point to further improve the system reported here for future therapeutic screening and related assessments. We see this tissue system as a first step toward more complex in vitro tissue designs to emulate the OA disease state, wherein additional inputs can now be added to build in more complexity and relevance.
Supplementary Material
Acknowledgments
This study was supported by a grant from the Department of Biotechnology, Government of India, and a travel grant to S.G. from Indo-US Science and Technology Forum. We also thank the NIH Tissue Engineering Resource Center for help with this study (EB002520).
Disclosure Statement
No competing financial interests exist.
References
- 1.Anderson D.D. Chubinskaya S. Guilak F. Martin J.A. Oegema T.R. Olson S.A., et al. Post-traumatic osteoarthritis: Improved understanding and opportunities for early intervention. J Orthop Res. 2011;29:802. doi: 10.1002/jor.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y. Xu L. Olsen B.R. Lessons from genetic forms of osteoarthritis for the pathogenesis of the disease. Osteoarthritis Cartilage. 2007;15:1101. doi: 10.1016/j.joca.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin J.A. Buckwalter J.A. Telomere erosion and senescence in human articular cartilage chondrocytes. J Gerontol A Biol Sci Med Sci. 2001;56:B172. doi: 10.1093/gerona/56.4.b172. [DOI] [PubMed] [Google Scholar]
- 4.Aigner T. Rose J. Martin J. Buckwalter J. Aging theories of primary osteoarthritis: from epidemiology to molecular biology. Rejuvenation Res. 2004;7:134. doi: 10.1089/1549168041552964. [DOI] [PubMed] [Google Scholar]
- 5.Valdes A.M. Loughlin J. Oene M.V. Chapman K. Surdulescu G.L. Doherty M., et al. Sex and ethnic differences in the association of ASPN, CALM1, COL2A1, COMP, and FRZB with genetic susceptibility to osteoarthritis of the knee. Arthritis Rheum. 2007;56:137. doi: 10.1002/art.22301. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi H. Nakajima M. Ozaki K. Tanaka T. Kamatani N. Ikegawa S. Prediction model for knee osteoarthritis based on genetic and clinical information. Arthritis Res Ther. 2010;12:R187. doi: 10.1186/ar3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritzker K. P. H. Osteoarthritis: joint instability and OA: do animal models provide insights? Nat Rev Rheumatol. 2011;7:444. doi: 10.1038/nrrheum.2011.104. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier J.P. Martel-Pelletier J. Abramson S.B. Osteoarthritis, an inflammatory disease: potential implication for the selection of new therapeutic targets. Arthritis Rheum. 2001;44:1237. doi: 10.1002/1529-0131(200106)44:6<1237::AID-ART214>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Smith M.D. Triantafillou S. Parker A. Youssef P.P. Coleman M. Synovial membrane inflammation and cytokine production in patients with early osteoarthritis. J Rheumatol. 1997;24:365. [PubMed] [Google Scholar]
- 10.Bondeson J. Wainwright S.D. Lauder S. Amos N. Hughes C.E. The role of synovial macrophages and macrophage-produced cytokines in driving aggrecanases, matrix metalloproteinases, and other destructive and inflammatory responses in osteoarthritis. Arthritis Res Ther. 2006;8:R187. doi: 10.1186/ar2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Towle C.A. Hung H.H. Bonassar L.J. Treadwell B.V. Mangham D.G. Detection of interleukin-1 in the cartilage of patients with osteoarthritis: a possible autocrine/paracrine role in pathogenesis. Osteoarthritis Cartilage. 1997;5:293. doi: 10.1016/s1063-4584(97)80008-8. [DOI] [PubMed] [Google Scholar]
- 12.Ollivierre F. Gubler U. Towle C.A. Laurencin C. Treadwell B.V. Expression of IL-1 genes in human and bovine chondrocytes: a mechanism for autocrine control of cartilage matrix degradation. Biochem Biophys Res Commun. 1986;141:904. doi: 10.1016/s0006-291x(86)80128-0. [DOI] [PubMed] [Google Scholar]
- 13.Riyazi N. Slagboom E. de Craen A.J. Meulenbelt I. Houwing-Duistermaat J.J. Kroon H.M., et al. Association of the risk of osteoarthritis with high innate production of interleukin-1 and low innate production of interleukin-10 ex vivo, upon lipopolysaccharide stimulation. Arthritis Rheum. 2005;52:1443. doi: 10.1002/art.21014. [DOI] [PubMed] [Google Scholar]
- 14.Richardson D.W. Dodge G.R. Effects of interleukin-1beta and tumor necrosis factor-alpha on expression of matrix-related genes by cultured equine articular chondrocytes. Am J Vet Res. 2000;61:624. doi: 10.2460/ajvr.2000.61.624. [DOI] [PubMed] [Google Scholar]
- 15.Goldring M.B. Osteoarthritis and cartilage: the role of cytokines. Curr Rheumatol Rep. 2000;2:459. doi: 10.1007/s11926-000-0021-y. [DOI] [PubMed] [Google Scholar]
- 16.Lotz M. Blanco F.J. von Kempis J. Dudler J. Maier R. Villiger P.M., et al. Cytokine regulation of chondrocyte functions. J Rheumatol Suppl. 1995;43:104. [PubMed] [Google Scholar]
- 17.Fernandes J.C. Martel-Pelletier J. Pelletier J.P. The role of cytokines in osteoarthritis pathophysiology. Biorheology. 2002;39:237. [PubMed] [Google Scholar]
- 18.López-Armada M.J. Caramés B. Lires-Deán M. Cillero-Pastor B. Ruiz-Romero C. Galdo F., et al. Cytokines, tumor necrosis factor-μ and interleukin-1 beta, differentially regulate apoptosis in osteoarthritis cultured human chondrocytes. Osteoarthritis Cartilage. 2006;14:660. doi: 10.1016/j.joca.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Sun L. Wang X. Kaplan D.L. A 3D cartilage - Inflammatory cell culture system for the modeling of human osteoarthritis. Biomaterials. 2011;32:5581. doi: 10.1016/j.biomaterials.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones A.R. Flannery C.R. Bioregulation of lubricin expression by growth factors and cytokines. Eur Cell Mater. 2007;13:40. doi: 10.22203/ecm.v013a04. [DOI] [PubMed] [Google Scholar]
- 21.Yudoh K. Nguyen T. Nakamura H. Hongo-Masuko K. Kato T. Nishioka K. Potential involvement of oxidative stress in cartilage senescence and development of osteoarthritis: oxidative stress induces chondrocyte telomere instability and downregulation of chondrocyte function. Arthritis Res Ther. 2005;7:R380. doi: 10.1186/ar1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyaki S. Nakasa T. Otsuki S. Grogan S.P. Higashiyama R. Inoue A., et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy A.R. St John P. Kaplan D.L. Modification of silk fibroin using diazonium coupling chemistry and the effects on hMSC proliferation and differentiation. Biomaterials. 2008;29:2829. doi: 10.1016/j.biomaterials.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wenk E. Murphy A.R. Kaplan D.L. Meinel L. Merkle H.P. Uebersax L. The use of sulfonated silk fibroin derivatives to control binding, delivery and potency of FGF-2 in tissue regeneration. Biomaterials. 2010;31:1403. doi: 10.1016/j.biomaterials.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Barbero A. Grogan S. Schäfer D. Heberer M. Mainil-Varlet P. Martin I. Age related changes in human articular chondrocyte yield, proliferation and post-expansion chondrogenic capacity. Osteoarthritis Cartilage. 2004;12:476. doi: 10.1016/j.joca.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Snel B. Lehmann G. Bork P. Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighborhood of a gene. Nucleic Acids Res. 2000;28:3442. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D.W. Sherman B.T. Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh S. Parker S.T. Wang X. Kaplan D.L. Lewis J.A. Direct-write assembly of microperiodic silk fibroin scaffolds for tissue engineering applications. Adv Func Mater. 2008;18:1883. [Google Scholar]
- 29.Barthelmes K. Reynolds A.M. Peisach E. Jonker H.R.A. Denunzio N.J. Allen K.N., et al. Engineering encodable lanthanide-binding tags into loop regions of proteins. J Am Chem Soc. 2011;133:808. doi: 10.1021/ja104983t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutmacher D.W. Loessner D. Rizzi S. Kaplan D.L. Mooney D.J. Clements J.A. Can tissue engineering concepts advance tumor biology research? Trends Biotechnol. 2010;28:125. doi: 10.1016/j.tibtech.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Jeffrey J.E. Aspden R.M. Cyclooxygenase inhibition lowers prostaglandin E2 release from articular cartilage and reduces apoptosis but not proteoglycan degradation following an impact load in vitro. Arthritis Res Ther. 2007;9:R129. doi: 10.1186/ar2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malemud C.J. Gillespie H.J. The role of apoptosis in arthritis. Curr Rheum Rev. 2005;1:131. [Google Scholar]
- 33.Haywood L. McWilliams D.F. Pearson C.I. Gill S.E. Ganesan A. Wilson D., et al. Inflammation and angiogenesis in osteoarthritis. Arthritis Rheum. 2003;48:2173. doi: 10.1002/art.11094. [DOI] [PubMed] [Google Scholar]
- 34.Sakkas L.I. Platsoucas C.D. The role of T cells in the pathogenesis of osteoarthritis. Arthritis Rheum. 2007;56:409. doi: 10.1002/art.22369. [DOI] [PubMed] [Google Scholar]
- 35.Fosang A.J. Little C.B. Drug insight: aggrecanases as therapeutic targets for osteoarthritis. Nat Clin Pract Rheumatol. 2008;4:420. doi: 10.1038/ncprheum0841. [DOI] [PubMed] [Google Scholar]
- 36.Bondeson J. Wainwright S. Hughes C. Caterson B. The regulation of ADAMTS4 and ADAMTS5 aggrecanases in osteo-arthritis: a review. Clin Exp Rheumatol. 2008;26:139. [PubMed] [Google Scholar]
- 37.Peters H.C. Otto T.J. Enders J.T. Jin W. Moed B.R. Zhang Z. The protective role of the pericellular matrix in chondrocyte apoptosis. Tissue Eng Part A. 2011;17:2017. doi: 10.1089/ten.TEA.2010.0601. [DOI] [PubMed] [Google Scholar]
- 38.Sandy J.D. Adams M.E. Billingham M.E.J. Plaas A. Muir H. In vivo and in vitro stimulation of chondrocyte biosynthetic activity in early experimental osteoarthritis. Arthritis Rheum. 1984;27:388. doi: 10.1002/art.1780270405. [DOI] [PubMed] [Google Scholar]
- 39.Little C.B. Christopher B. Meeker C.T. Golub S.B. Lawlor K.E. Farmer P.J., et al. Blocking aggrecanase cleavage in the aggrecan interglobular domain abrogates cartilage erosion and promotes cartilage repair. J Clin Invest. 2007;117:1627. doi: 10.1172/JCI30765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santangelo K.S. Pieczarka E.M. Nuovo G.J. Weisbrode S.E. Bertone A.L. Temporal expression and tissue distribution of interleukin-1b in two strains of guinea pigs with varying propensity for spontaneous knee osteoarthritis. Osteoarthritis Cartilage. 2011;19:439. doi: 10.1016/j.joca.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiku M.L. Shah R. Allison G.T. Evidence linking chondrocyte lipid peroxidation to cartilage matrix protein degradation. Possible role in cartilage aging and the pathogenesis of osteoarthritis. J Biol Chem. 2000;275:20069. doi: 10.1074/jbc.M907604199. [DOI] [PubMed] [Google Scholar]
- 42.El-Bikai R. Welman M. Margaron Y. Côté J.F. Macqueen L. Buschmann M.D., et al. Perturbation of adhesion molecule-mediated chondrocyte-matrix interactions by 4-hydroxynonenal binding: implication in osteoarthritis pathogenesis. Arthritis Res Ther. 2010;12:R201. doi: 10.1186/ar3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yayon A. Klagsbrun M. Esko J.D. Leder P. Ornitz D.M. Cell Surface, Heparin-like Molecules Are Required for Binding of Basic Fibroblast Growth Factor to Its High Affinity Receptor. Cell. 1991;64:841. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 44.Pratt A.B. Weber F.E. Schmoekel H.G. Mu R. Hubbell J.A. Synthetic extracellular matrices for in situ tissue engineering. Biotechnology. 2004;86:27. doi: 10.1002/bit.10897. [DOI] [PubMed] [Google Scholar]
- 45.F Mi F.L. Shyu S.S. Peng C.K. Wu Y.B. Sung H.W. Wang P.S., et al. Fabrication of chondroitin sulfate–chitosan composite artificial extracellular matrix for stabilization of fibroblast growth factor. J Biomed Mater Res Part A. 2006;76:1. doi: 10.1002/jbm.a.30298. [DOI] [PubMed] [Google Scholar]
- 46.Parish C.R. The role of heparan sulphate in inflammation. Nat Rev Immunol. 2006;6:633. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 47.Kenig M. Gaberc-Porekar V. Fonda I. Menart V. Identification of the heparin-binding domain of TNF-alpha and its use for efficient TNF-alpha purification by heparin–Sepharose affinity chromatography. J Chromatogr B. 2008;867:119. doi: 10.1016/j.jchromb.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 48.Beg O.U. Uddin M. Siddiqi A.R. Analysis of heparin-binding sites in human lipoprotein lipase using synthetic peptides. J Protein Chem. 1998;17:807. doi: 10.1023/a:1020730418999. [DOI] [PubMed] [Google Scholar]
- 49.Bush P.G. Hall A.C. The volume and morphology of chondrocytes within non-degenerate and degenerate human articular cartilage. Osteoarthritis Cartilage. 2003;11:242. doi: 10.1016/s1063-4584(02)00369-2. [DOI] [PubMed] [Google Scholar]
- 50.Holloway I. Kayser M. Lee D.A. Bader D.L. Bentley G. Knight M.M. Increased presence of cells with multiple elongated processes in osteoarthritic femoral head cartilage. OsteoArthritis Cartilage. 2004;12:17. doi: 10.1016/j.joca.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Haudenschild D.R. Chen J. Pang N. Steklov N. Grogan S.P. Lotz M.K., et al. Vimentin contributes to changes in chondrocyte stiffness in osteoarthritiss. J Orthop Res. 2011;29:20. doi: 10.1002/jor.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calamia V. Rocha B. Mateos J. Fernández-Puente P. Ruiz-Romero C. Blanco F.J. Metabolic labeling of chondrocytes for the quantitative analysis of the interleukin-1-beta-mediated modulation of their intracellular and extracellular proteomes. J Proteome Res. 2011;10:3701. doi: 10.1021/pr200331k. [DOI] [PubMed] [Google Scholar]
- 53.Lambrecht S. Verbruggen G. Verdonk P.C. Elewaut D. Deforce D. Differential proteome analysis of normal and osteoarthritic chondrocytes reveals distortion of vimentin network in osteoarthritis. Osteoarthritis Cartilage. 2008;16:163. doi: 10.1016/j.joca.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Ala-Kokko L. Baldwin C.T. Moskowitz R.W. Prockop D.J. Single base mutation in the type II procollagen gene (COL2A1) as a cause of primary osteoarthritis associated with a mild chondrodysplasia. Proc Natl Acad Sci USA. 1990;87:6565. doi: 10.1073/pnas.87.17.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lorenzo P. Bayliss M.T. Heinegard D. Altered patterns and synthesis of extracellular matrix macromolecules in early osteoarthritis. Matrix Biol. 2004;23:381. doi: 10.1016/j.matbio.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Recklies A.D. Baillargeon L. White C. Regulation of cartilage oligomeric matrix protein synthesis in human synovial cells and articular chondrocytes. Arthritis Rheum. 1998;41:997. doi: 10.1002/1529-0131(199806)41:6<997::AID-ART6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 57.Hunter D.J. Li J. LaValley M. Bauer D.C. Nevitt M. DeGroot J., et al. Cartilage markers and their association with cartilage loss on magnetic resonance imaging in knee osteoarthritis: the Boston Osteoarthritis Knee Study. Arthritis Res Ther. 2007;9:R108. doi: 10.1186/ar2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kizawa H. Kou I. Iida A. Sudo A. Miyamoto Y. Fukuda A., et al. An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat Genet. 2005;37:138. doi: 10.1038/ng1496. [DOI] [PubMed] [Google Scholar]
- 59.Brew C.J. Clegg P.D. Boot-Handford R.P. Andrew J.G. Hardingham T. Gene expression in human chondrocytes in late osteoarthritis is changed in both fibrillated and intact cartilage without evidence of generalised chondrocyte hypertrophy. Ann Rheum Dis. 2008;69:234. doi: 10.1136/ard.2008.097139. [DOI] [PubMed] [Google Scholar]
- 60.Bluteau G. Gouttenoire J. Conrozier T. Mathieu P. Vignon E. Richard M., et al. Differential gene expression analysis in a rabbit model of osteoarthritis induced by anterior cruciate ligament (ACL) section. Biorheology. 2002;39:24. [PubMed] [Google Scholar]
- 61.Aigner T. Reichenberger E. Bertling W. Kirsch T. Stoss H. von der Mark K. Type X collagen expression in osteoarthritic and rheumatoid articular cartilage. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;63:205. doi: 10.1007/BF02899263. [DOI] [PubMed] [Google Scholar]
- 62.Aigner T. Zien A. Gehrsitz A. Gebhard P.M. McKenna L. Anabolic and catabolic gene expression pattern analysis in normal versus osteoarthritic cartilage using complementary DNA-array technology. Arthritis Rheum. 2001;44:2777. doi: 10.1002/1529-0131(200112)44:12<2777::aid-art465>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 63.Loeser R.F. Carlson C.S. McGee M.P. Expression of beta 1 integrins by cultured articular chondrocytes and in osteoarthritic cartilage. Exp Cell Res. 1995;217:248. doi: 10.1006/excr.1995.1084. [DOI] [PubMed] [Google Scholar]
- 64.McCulloch C.A. Downey G.P. El-Gabalawy H. Signalling platforms that modulate the inflammatory response: new targets for drug development. Nat Rev Drug Discov. 2006;5:864. doi: 10.1038/nrd2109. [DOI] [PubMed] [Google Scholar]
- 65.Arora P.D. Ma J. Min W. Cruz T. McCulloch C.A. Interleukin-1-induced calcium flux in human fibroblasts is mediated through focal adhesions. J Biol Chem. 1995;270:6042. doi: 10.1074/jbc.270.11.6042. [DOI] [PubMed] [Google Scholar]
- 66.Lo Y.Y. Luo L. McCulloch C.A. Cruz T.F. Requirements of focal adhesions and calcium fluxes for interleukin-1-induced ERK kinase activation and c-fos expression in fibroblasts. J Biol Chem. 1998;273:7059. doi: 10.1074/jbc.273.12.7059. [DOI] [PubMed] [Google Scholar]
- 67.Musumeci G. Loreto C. Carnazza M.L. Strehin I. Elisseeff J. OA cartilage derived chondrocytes encapsulated in poly(ethylene glycol) diacrylate (PEGDA) for the evaluation of cartilage restoration, apoptosis in an in vitro model. Histol Histopathol. 2011;26:1265. doi: 10.14670/HH-26.1265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.