Abstract
The L polymerase of bunyaviruses replicates and transcribes the viral genome. While replication products are faithful copies of the uncapped genomic RNA, transcription products contain capped 5′ extensions which had been cleaved from host cell mRNAs. For La Crosse virus (LACV; genus Orthobunyavirus), the nuclease responsible for host cell mRNA cleavage is located at the N terminus of the L protein, with an active site of five conserved amino acids (H34, D52, D79, D92, and K94) surrounding two Mn2+ ions (J. Reguera, F. Weber, and S. Cusack, PLoS Pathog. 6:e1001101, 2010). Here, we present reverse genetics systems and L mutants enabling us to study bunyaviral genome replication in the absence of transcription. Transcription was evaluated with an enhanced minigenome system consisting of the viral polymerase L, nucleocapsid protein N, a negative-sense minigenome, and—to alleviate antiviral host responses—a dominant-negative mutant (PKRΔE7) of the antiviral kinase protein kinase R (PKR). The transcriptional activity was strongly reduced by mutation of any of the five key amino acids, and the H34K, D79A, D92A, and K94A LACV L mutants were almost entirely silent in transcription. The replication activity of the L mutants was measured by packaging of progeny minigenomes into virus-like particles (VLPs). All mutant L proteins except K94A retained full replication activity. To test the broader applicability of our results, we introduced the homolog of mutation D79A (D111A) into the L sequence of Rift Valley fever virus (RVFV; genus Phlebovirus). As for LACV D79A, the RVFV D111A was incapable of transcription but fully active in replication. Thus, we generated mutants of LACV and RVFV L polymerases that are specifically deficient in transcription. Genome replication by bunyavirus polymerases can now be studied in the absence of transcription using convenient reverse genetics systems.
INTRODUCTION
The family Bunyaviridae is one of the most ubiquitous and widespread virus groups in the world, causing significant harm in humans, domestic animals, and plants (1). Bunyaviruses infecting animals are classified into four genera which all contain important human pathogens, e.g., La Crosse virus (LACV; genus Orthobunyavirus), Rift Valley fever virus (RVFV; genus Phlebovirus), Crimean-Congo hemorrhagic fever virus (genus Nairovirus), or Hantaan virus (HTNV; genus Hantavirus).
Bunyaviruses are enveloped, replicate in the cytoplasm, and bud into the Golgi apparatus (1, 2). They have a genome consisting of three single-stranded RNA (ssRNA) segments of negative or ambisense polarity. The genome segments encode four common structural proteins: the viral polymerase on the large (L) segment, two envelope glycoproteins (Gn and Gc) on the medium (M) segment, and the viral nucleocapsid protein (N) on the smallest (S) segment. Untranslated regions (UTRs) on both the 3′ and the 5′ ends serve as promoters for transcription and replication and mediate the packaging of the genome segments into viral particles (3–5). Both the viral genomic RNA (vRNA) and the antigenomic copy RNA (cRNA) are encapsidated by N protein, and only these ribonucleoprotein particles (RNPs) are functional templates for the viral polymerase L. The mRNA synthesis of bunyaviruses is initiated with 12- to 18-nucleotide (nt)-long, capped oligonucleotides that are cleaved from host cell mRNAs by an endonuclease activity of the L protein (“cap-snatching”). Moreover, the mRNAs are truncated at the 3′ end relative to the genome template (2, 6, 7). Viral RNA replication, in contrast, occurs primer independently by faithful copying of the vRNA or cRNA templates. Thus, during transcription the L polymerase produces 3′-truncated mRNAs that are capped and contain nontemplated nucleotides at their 5′ ends, whereas replication results in full-length positive- and negative-sense copies of the genomic RNA.
In general, the infection cycle of bunyaviruses consists of the stages attachment, entry, primary mRNA transcription, genome replication, secondary mRNA transcription, RNP encapsidation, particle assembly, and exit. To study these individual steps, reverse genetics systems have been established which are based on expression of recombinant proteins and RNAs in mammalian cells (reviewed in reference 8). The RNA synthesis activity of the viral polymerase can be measured by using a so-called “minigenome” (or “minireplicon”) system, in which L and N expressed from cDNA plasmids encapsidate, replicate, and transcribe a negative-stranded reporter gene containing viral UTRs. If plasmid constructs for the viral glycoproteins (GPs) are cotransfected, the artificial RNPs can be packaged into virus-like particles (VLPs) that are released into the supernatant (9–12). These VLPs can also infect new cells and express the encoded reporter gene. They are therefore also called transcription and replication-competent VLPs (trVLPs), to distinguish them from noninfectious, merely physical VLPs (8). trVLPs are, however, not expressing any viral gene. Therefore, reporter expression in naive trVLP-infected cells is solely performed by the copackaged L polymerases, thus reflecting viral primary transcription (9). In most cases, reporter expression in the trVLP-infected cells needs to be supported by L and N expressed in trans, allowing replication and secondary transcription of the VLP minigenome. Minigenome systems have been set up for several (but not all) bunyaviruses and other negative-stranded RNA viruses, whereas trVLP systems exist for only a few of them (8). Possibly, the efficiency of the standard protocol is not high enough for the less active virus polymerases. For bunyaviruses, the reporter-based reverse genetics systems that exist so far are suitable for studying viral primary transcription, assembly, and overall RNA synthesis (i.e., the sum of transcription and replication activities). They have been useful to characterize the role of the envelope proteins for virus assembly and morphogenesis (11, 13, 14), to identify key amino acids involved in RNP packaging (15), and to show that primary transcription is a target of the antiviral interferon responses (9, 16). For RNA replication, the central activity of the RNA polymerase to amplify the viral genome, no comparable approach exists. Here, we present experimental systems and L polymerase mutants that enable us to better dissect and study the infection cycle of bunyaviruses. First, we optimized the performance of reverse genetics systems by suppressing the antiviral kinase protein kinase R (PKR). Second, we identified endonuclease-negative mutants of LACV L polymerase that are still active in replication. Minigenome and trVLP systems based on these mutants allow us to study genome replication in the absence of transcription. Moreover, a key mutation introduced into the L polymerase gene of RVFV led to a similar phenotype, confirming the conserved function of the affected amino acid for bunyavirus mRNA transcription. Thus, the systems we present open up a way to run select parts of the bunyavirus replication cycle and to make genome replication accessible to specific manipulation.
MATERIALS AND METHODS
Chemicals, cells, and viruses.
Geneticin G418 (Biochrom AG) was dissolved in H2O to 100 mg/ml and used at a concentration of 1 mg/ml cell culture medium. BHK-21, CV-1, Huh-7, and 293T cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). BSR-T7/5 cells (17) were additionally provided with G418.
Plasmid constructs.
The expression construct pI.18-HA-PKR-ΔE7 expresses the N-terminal double-stranded RNA (dsRNA)-binding domain of PKR, thus mimicking the natural dominant-negative splice variant PKR-ΔE7 (18). It was generated by PCR amplification of nt 1 to 522 of the human PKR cDNA, using a 5′ primer with a hemagglutinin (HA) epitope tag sequence and Phusion Hot Start II DNA polymerase (NEB). The PCR product was cloned into the BamHI/XhoI site of the expression vector pI.18 and verified by DNA sequencing. Detailed cloning strategies and primer sequences are available upon request.
The expression and reporter constructs for the minigenome and VLP systems of LACV, namely, pTM-LACV_L, pTM-LACV_N, pI.18-LACV_M (encoding the GPs), pHH21-LACV-vMRen, and pTM1-FF-Luc, have been described previously (12, 19, 20). The expression constructs contain the appropriate coding sequences under the control of a T7 promoter and the encephalomyocarditis virus internal ribosome entry site (pTM1 backbone) or under the control of an RNA polymerase II promoter (pI.18 backbone). The minigenome reporter plasmid (pHH21 backbone) contains a human RNA polymerase I promoter and terminator to express a LACV Renilla luciferase (Ren-Luc) minigenome in an antisense orientation flanked by the untranslated regions of the LACV M vRNA. The T7-driven expression constructs for the LACV L mutants (pTM-LACV_L H34A, pTM-LACV_L H34K, pTM-LACV_L E48A, pTM-LACV_L D52A, pTM-LACV_L D79A, pTM-LACV_L D92A, pTM-LACV_L K94A, and pTM-LACV_L K108A) were described previously (21).
The expression and reporter constructs for the minigenome and VLP systems of RVFV, namely, pI.18-RVFV_L, pI.18-RVFV_N, and pI.18-RVFV_M (expressing the L, N, and GP coding sequences, respectively), as well as the reporter plasmids pHH21-RVFV-vMRen (RNA polymerase I-driven Ren-Luc reporter construct for RVFV) and pGL3-control, have been described previously (9, 22). Mutant plasmid pI.18-RVFV_L D111A was generated by PCR mutagenesis and verified by DNA sequencing. Primers and experimental details are available upon request.
The T7 polymerase expression construct pCAGGS-T7 was kindly provided by Ramon Flick (UTMB Texas).
Basic LACV minigenome assay.
Subconfluent monolayers of CV-1 cells seeded in 6-well dishes were transfected with 250 ng each of plasmid pHH21-LACV-vMRen, pTM-LACV_L, pTM-LACV_N, 500 ng pCAGGS-T7, and 100 ng pTM1-FF-Luc using Nanofectin transfection reagent (PAA) in 200 μl 150 mM NaCl. In addition, 250 ng of pI.18-HA-PKRΔE7 was added to the plasmid mix. At 72 h posttransfection, cells were lysed in 200 μl passive lysis buffer (Promega) and measured as described by the manufacturer.
Production of LACV and RVFV VLPs.
For production of LACV trVLPs, Huh-7 donor cells were seeded and transfected as for a minigenome assay (using pI.18-HA-PKRΔE7 to inhibit PKR), but additionally transfected with 500 ng pI.18-LACV_M. At 48 h postinfection, cells were lysed to determine reporter activities, and cell supernatants were harvested. The cell supernatants (2 ml) were treated with 50 U of Benzonase for 2.5 h at 37°C to remove residual plasmid DNA. An aliquot of 250 μl supernatant was then used to infect BHK-21 cells, which were pretransfected with 250 ng each of pTM-LACV_L and pTM-LACV_N and 500 ng pCAGGS-T7 (indicator cells). After an incubation period of 24 h, cells were lysed and luciferase activities were determined.
RVFV trVLPs were produced by a method similar to that described previously (9), except that CV-1 cells were used as donor cells and the incubation time of donor cells was extended to 72 h.
Primer extension analysis.
Primer extension was performed essentially as described by Robb et al. (23). Briefly, 293T cells in 6-well plates were transfected with plasmids of the LACV minigenome system, and total RNA was isolated 72 h posttransfection using TRIzol (life technologies). About 14 μg RNA was used per assay. The RNA was first treated with RNase-free DNase, and after inactivation, reverse transcription was started using the Superscript III kit (Invitrogen) and specific radiolabeled primers. Positive-sense Ren-Luc transcripts were detected with primer 5′-CTT TGT TCT GGA TCA TAA ACT TTC GA-3′ (yielding a 93-nt single-stranded DNA [ssDNA] product), negative-sense transcripts were detected with 5′-CAA ATC GTT CGT TGA GCG AGT TCT CA-3′ (yielding a 179 nt ssDNA product), and 5S rRNA used as loading control was detected with 5′-ACC CTG CTT AGC TTC CGA GA-3′ (yielding a 100-nt-long ssDNA product). Products were resolved by 8% denaturing PAGE and visualized by autoradiography.
Real-time RT-PCR.
Total cellular RNA was isolated with the RNeasy minikit (Qiagen) and eluted in 30 μl of double-distilled water (ddH2O). An aliquot of 400 ng RNA was then used as a template for cDNA synthesis, using the QuantiTect reverse transcription kit (Qiagen) and specific reverse primers (see below). mRNA levels of human β-actin were detected with primers 5′-AAA GAC CTG TAC GCC AAC ACA GTG CTG TCT GG-3′ (forward) and 5′-CGT CAT ACT CCT GCT TGC TGA TCC ACA TCT GC-3′ (reverse), and levels of positive-sense Ren-Luc RNA were detected with primers 5′-AAC GCG GCC TCT TCT TAT TT-3′ (forward) and 5′-ACC AGA TTT GCC TGA TTT GC-3′ (reverse), using the QuantiTect SYBR green reverse transcription-PCR (RT-PCR) kit (Qiagen) and a StepOne cycler (Applied Biosystems). The values obtained for Ren-Luc RNA were normalized against β-actin mRNA levels using the threshold cycle (ΔΔCT) method (24).
Database resources and software.
The structure illustrations were drawn with PyMol. Bunyavirus L sequences were derived from GenBank entries AAO33758.1 (LACV), AEC14282.1 (RVFV), and ABJ74147.1 (HTNV). Primary sequence alignments were performed using ClustalW (25) and visualized using TeXshade (26), both implemented in the Biology WorkBench (http://workbench.sdsc.edu) (27). Structural alignments were performed by Least Square Superpose (28), using the CCP4 package, and visualized with PyMol.
The structure for the LACV endonuclease domain may be found in the Protein Data Bank (PDB) under entry no. 2xi5.
RESULTS
Inhibition of the antiviral kinase PKR enhances activity of the LACV minigenome system.
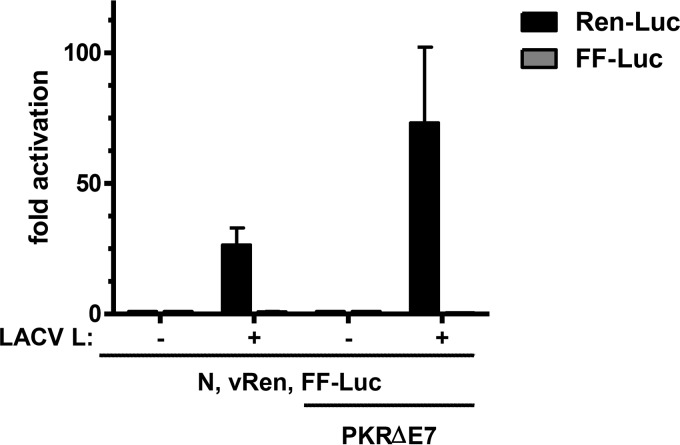
It is well established that bunyaviruses are sensitive to the antiviral kinase PKR (29–32), which is constitutively expressed (33). This raised the possibility that the performance of standard bunyaviral reverse genetics systems is suboptimal due to PKR present in the transfected cells. We therefore generated a plasmid construct for a dominant-negative mutant of PKR, the splice variant PKR-ΔE7 (18), and investigated its influence on the LACV minigenome system. Cells were transfected with the PKR-ΔE7 construct along with plasmids encoding LACV L, LACV N, a Renilla luciferase (Ren-Luc) minigenome, and a firefly luciferase (FF-Luc) control construct, and assayed for reporter activities. Strikingly, coexpression of the dominant-negative PKR mutant largely enhanced activity of the LACV minigenome system (Fig. 1). Whereas normal reporter activity was in the range of 25-fold over background (set by omitting the L polymerase construct in the plasmid mix), coexpression of PKR-ΔE7 amplified it to 70-fold over background. Similar results were obtained with the LACV VLP system and with the minigenome and VLP systems for RVFV (data not shown). We concluded that inhibition of PKR has a general enhancing effect on bunyaviral reverse genetics systems. The PKR-ΔE7 mutant was therefore included in all subsequent experiments.
Fig 1.
Inhibition of PKR improves performance of the LACV minigenome system. CV-1 cells were transfected with T7-driven expression constructs for LACV N, LACV L, and an RNA polymerase I-driven minigenome construct, termed vRen. The vRen minigenome encompasses the Renilla luciferase (Ren-Luc) gene in negative sense, flanked by 5′ and 3′ UTRs of the LACV M segment. In addition to these constructs, the basic transfection mix contained an expression construct for the T7 polymerase and a T7-driven reporter construct for the firefly luciferase (FF-Luc). Endogenous PKR was inhibited by coexpression of a dominant-negative mutant (PKRΔE7). At 72 h posttransfection, Ren-Luc and FF-Luc activities were determined, and luciferase counts were normalized to the negative controls in which the L plasmid was omitted. Mean values and standard deviations from three independent experiments are shown.
Transcription and replication activity of LACV L polymerase mutants.
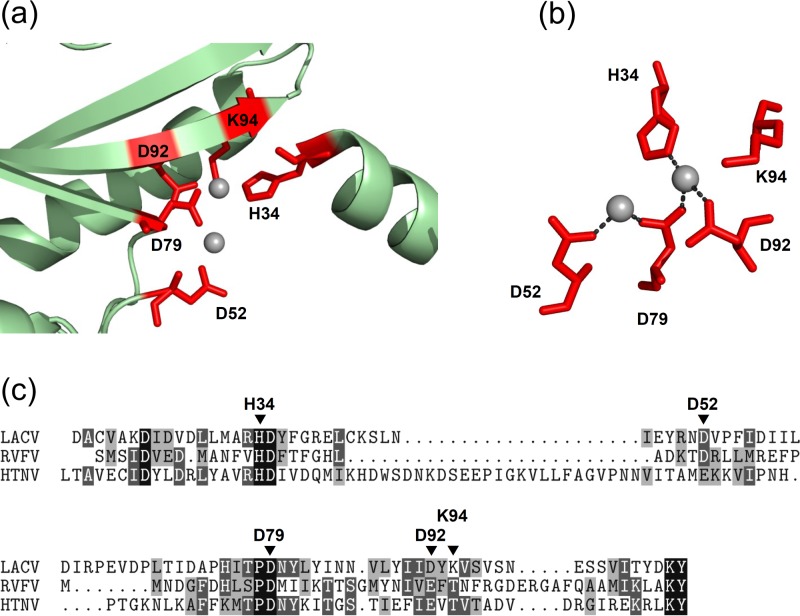
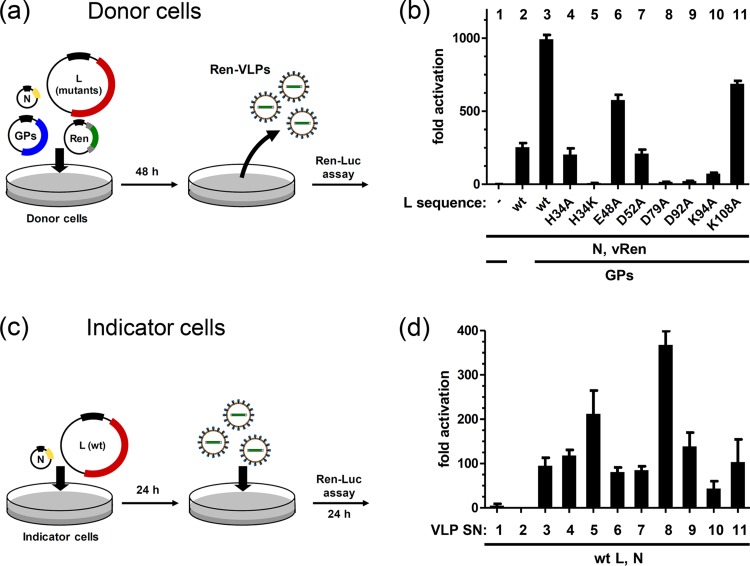
We have recently solved the atomic structure of the LACV domain responsible for cleavage of the donor RNA required for cap-snatching (21). This domain is located at the N terminus (amino acids [aa] 1 to 183) of the L protein and encodes a Mn2+-dependent nuclease of the PD-(D/E)xK superfamily. The active site consists of alpha helices and an anti-parallel beta sheet with two cations in the center (Fig. 2a). Five amino acids (H34, D52, D79, D92, and K94) surround two Mn2+ ions. H34, D79, and D92 hereby form metal binding site 1, D52 and D79 form metal binding site 2, and K94 (which is not directly binding the metal ions) contributes catalytic activity (Fig. 2b). The LACV endonuclease domain displays a strong sequence similarity to polymerases of phleboviruses and hantaviruses, which includes the key residues of the active site (Fig. 2c). Previously, we had introduced point mutations into the full-length LACV L sequence and demonstrated that exchange of any of the 5 key residues against alanine results in a loss of transcriptional activity in a standard minigenome assay (21). It remained unclear, however, whether the ability of the mutant polymerases to replicate a template RNA was also destroyed or whether it was preserved. To solve this question, we devised a VLP-based transcomplementation assay that enables us to measure the replication activity of L mutants. trVLPs were produced in so-called donor cells by coexpressing the minigenome system (using wild-type [wt] or mutant L sequences) with a construct for the viral envelope GPs (Fig. 3a). Since both 5′ and 3′ UTRs are required for packaging of minigenome RNA into particles, trVLPs provide a convenient selection system for full-length replication products. The trVLPs were then used to infect fresh cells (so-called “indicator cells”) expressing wt L and N to transcribe and replicate the VLP-expressed minigenome (Fig. 3c). Specific reporter activity in the indicator cells can only be produced if the recombinant N and L of the donor cells had encapsidated and replicated the template RNA, followed by packaging into VLPs. Thus, reporter activity in the transcomplemented indicator cells can serve as a measure for replication of the minigenome RNA in the donor cells. Figure 3b shows the reporter activities in the VLP donor cells. The minigenome system alone already showed significant activity over the background (compare columns 1 and 2), as expected. Interestingly, coexpression of the viral envelope GPs resulted in a massive increase in activity, most likely due to spread of trVLPs in the cell culture (column 3). When in this setup the wt L was exchanged against point mutants in the endonuclease domain, a clear impact was observed. The L mutants of the active site, namely, H34A, H34K, D52A, D79A, D92A, and K94A, displayed a strongly reduced or even completely absent transcription activity, whereas mutants with mutations outside the active site (E48A and K108A) were less affected. The residual activity of some of the active site mutants is different from what we observed previously using a standard minigenome system (21). The experimental setup employed here, however, displays a much elevated sensitivity due to (i) inclusion of the dominant-negative PKRΔE7 mutant, (ii) an increase in expression time from 24 h (21) to 48 h, and (iii) the coexpression of the envelope GPs, which apparently further boosts the performance. Thus, a basic minigenome system alone may not be sensitive enough to detect subtle differences. However, even under conditions of largely increased sensitivity, the D79A, D92A, and K94A mutants were almost silent. This indicates a nonredundant role of these amino acids for transcriptional activity. Moreover, mutation of the H34 residue to lysine (H34K mutant) had a similarly strong impact, whereas its mutation to alanine (H34A mutant) allowed residual transcription activity. Most likely, the strong positive charge of the introduced lysine residue disturbs the binding of the Mn2+ ion at metal binding site 1 and may also sterically hinder ion recruitment. The small and neutral alanine, in contrast, disturbs metal binding neither by charge nor by size (but neither does it contribute to it) and apparently allows residual endonuclease activity.
Fig 2.
The endonuclease domain of LACV. (a) Representation of the LACV endonuclease domain (21). Side chains of residues important for forming the active center are shown as red sticks and labeled. Manganese ions are shown as gray spheres. Note that part of alpha helix α2 was left out to improve visibility of the active center. (b) Amino acid side chains of the endonuclease active site with secondary structure elements removed and shown from a different angle than in panel a to illustrate that K94 is not involved in ion binding. Metal coordination is illustrated with dashed lines. (c) Alignment of the L amino acid sequences of LACV, RVFV, and HTNV encompassing the endonuclease active site. Key residues of the LACV sequence are marked.
Fig 3.
VLP-based assay for determining the amino acids essential for transcription and replication. Shown are schematic outlines of experiments and data obtained using different L mutants. (a) Procedure used to generate LACV trVLPs. Donor cells were transfected with constructs for the minigenome system as described in the legend to Fig. 1 as well as the LACV envelope glycoproteins (GPs). A set of L mutants was used in parallel to wt L. As negative controls, L (column 1) or GPs (column 2) were omitted. After 48 h, cellular lysates and supernatants containing VLPs were harvested. (b) Minigenome system reporter activities were measured in cell lysates. (c) Outline of the VLP detection assay. Indicator cells expressing wt L and N were superinfected with trVLP supernatants. (d) Ren-Luc minigenome reporter activities of indicator cells as determined after 24 h of VLP infection. Activities of the cotransfected FF-Luc indicated comparable transfection efficiencies (data not shown). Mean values and standard deviations from three independent experiments are shown. SN, supernatant.
The minigenome system activities measured in the donor cells are dependent on the production of capped Ren-Luc reporter mRNAs (21). To evaluate the ability of the L endonuclease mutants to replicate (i.e., to produce uncapped full-length copies of the minigenome), we transferred the trVLP-containing supernatants onto indicator cells expressing N and wt L and measured reporter activity. Strikingly, the transcriptional activity of all trVLPs could be rescued by wt L in trans, irrespective of the particular L variant used to produce their RNPs (Fig. 3d). In fact, all endonuclease mutants except for K94A could be transcomplemented to similar reporter levels as wt L, indicating full replication activity in the VLP-producing donor cells. Of note, the transcriptionally silent H34K and, in particular, D79A mutants displayed an even higher VLP activity than wt L did. Apparently, loss of metal binding (H34K and D79A) can lead to a complete absence of transcription but an increase in replication. Loss of the catalytic residue K94, in contrast, impairs both transcription and replication.
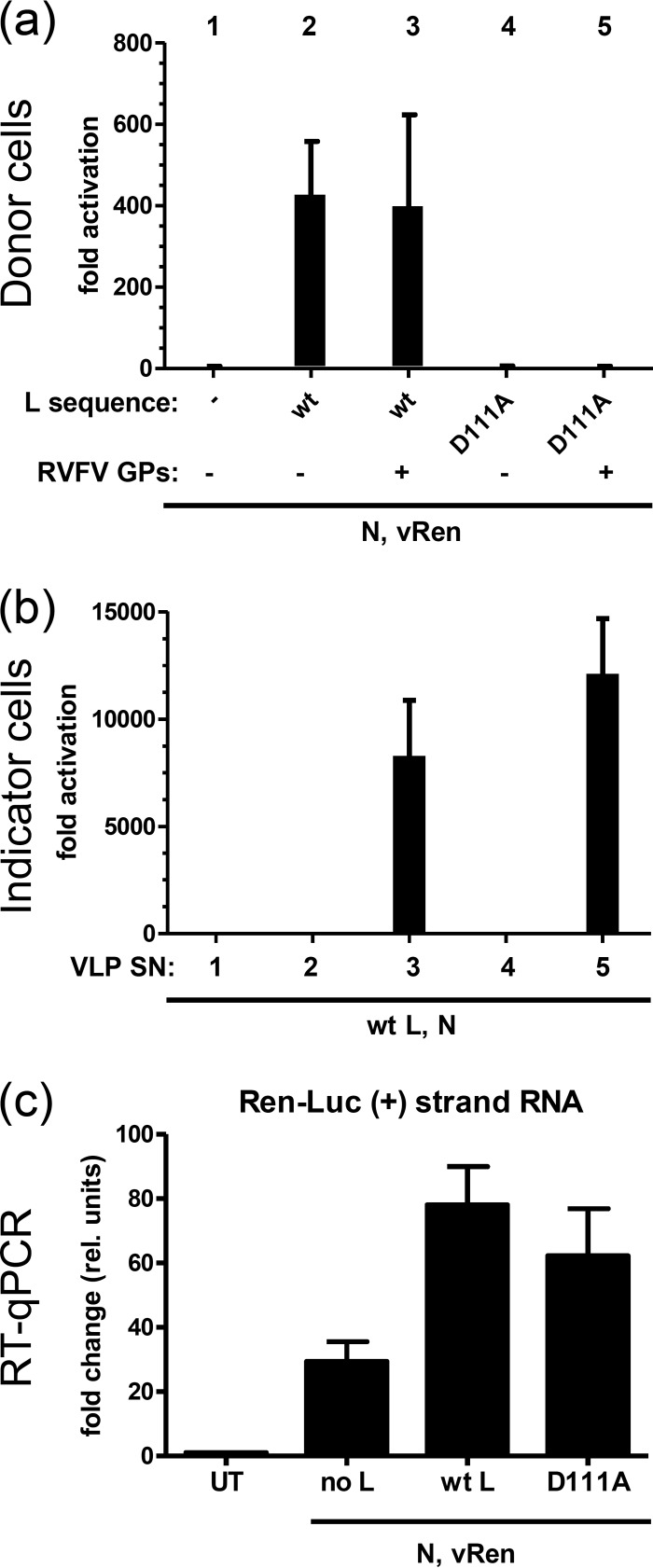
To bolster our confidence in the VLP data, we assayed the replication activity of the mutant with the most striking phenotype (D79A) by two other, independent methods. In the LACV minigenome system, the Ren-Luc minigenome plasmid is transcribed by the cellular RNA polymerase I to a negative-sense RNA, whereas the positive-stranded mRNA and cRNA should only be produced by coexpressed LACV L polymerase. First, we analyzed the RNA products of wt L and the D79A mutant by primer extension, an established method to study viral RNA polymerase activity (23). Cells were transfected with the components of the minigenome system, and total RNA was isolated 72 h later. Positive-strand and negative-strand minigenome RNAs were reverse transcribed using specific, radiolabeled Ren-Luc primers (see Materials and Methods). The labeled ssDNA products were separated by gel electrophoresis and visualized by autoradiography. As shown in Fig. 4a, both wt L and the D79A mutant generated comparable signals of positive-strand and negative-strand minigenome RNAs, whereas omission of an L plasmid resulted in only background amounts. Thus, the D79A mutant, which does not produce translatable mRNA (Fig. 3b), still produces positive-strand cRNA and negative-strand vRNA. To compare the two polymerases in a more quantitative manner, a second assay was performed. Total RNAs of cells harboring the 72-h LACV minigenome system were tested by strand-specific real-time RT-PCR. With this sensitive method, the polymerase-negative control shows production of some background positive-sense Ren-Luc RNA, but coexpression of wt LACV L (along with N) substantially increased the amounts of positive-sense Ren-Luc RNA (Fig. 4b). This indicates the specific production of mRNA and cRNA. Importantly, replacement of wt L with the transcription-negative D79A mutant also led to substantial amounts of positive-sense Ren RNA, indicating production of cRNA. Neither primer extension analysis nor strand-specific RT-PCR can demonstrate full-length RNA replication products. Nonetheless, the data from both of these assays are in agreement with the results of the VLP transcomplementation assay, allowing the conclusion that the transcriptionally inactive D79A mutant remains capable of replication.
Fig 4.
Production of positive-sense RNA by LACV polymerase variants. Comparison of wt L and the D79A mutant. Cells were transfected with constructs for the minigenome system (Fig. 1). Untransfected cells (UT) were used as a reference, and plasmid mixes without L (no L) were used as a negative control. At 72 h after transfection, total RNA was isolated and levels of positive-sense minigenome RNA were determined. (a) Primer extension analysis. RNA was reverse transcribed using radiolabeled primers specific for positive-sense (+) Ren-Luc transcripts or negative-sense (−) Ren-Luc transcripts and for cellular 5S rRNA transcripts. (b) Strand-specific real-time RT-PCR. Note that the amount of the negative-sense vRen minigenome plasmid was lowered to 5 ng per well to the reduce background signal. rel., relative. Mean values and standard deviations from three independent experiments are shown.
A replication-only mutant of RVFV L.
Given that the sequence of the endonuclease domain is highly conserved (21, 34), we wondered whether the results obtained with LACV could be applied to another bunyavirus. RVFV is an obvious candidate due to its medical and economic importance and the availability of reverse genetics tools (reviewed in reference 35). Moreover, for Toscana phlebovirus, an endonuclease activity of the N terminus was demonstrated (34). Thus, we introduced a D-to-A mutation into the RVFV L polymerase at amino acid position 111, the equivalent of D79 of LACV L (Fig. 2c). The mutation was tested the same way as for LACV. First, we investigated the activity of the RVFV L mutant in a minigenome system and found that the D111A mutation completely abrogated transcription by RVFV L (Fig. 5a). However, when a construct for RVFV envelope GPs was coexpressed, the VLP assay shows that D111A produced trVLPs, which can be transcomplemented by wt L (Fig. 5b). In agreement with this, strand-specific real-time RT-PCR revealed a comparable upregulation of plus-sense Ren-Luc RNA for both wt L and the D111A mutant (Fig. 5c). Since the D111A mutant is entirely unable to produce translatable RNA (i.e., mRNA), we conclude that the RT-PCR signal is derived from cRNA. Thus, residue D111 apparently plays a key role in transcription by RVFV L polymerase.
Fig 5.
A transcription-deficient L mutant of RVFV. (a) trVLP production. Donor cells were transfected with VLP plasmid mixes for RVFV, using wt L or the D111A mutant of RVFV L. Omission of L constructs and envelope GPs served as controls. Supernatants containing VLPs were harvested after 72 h of transfection, and reporter activities of the cell lysates were measured. (b) Reporter activities of VLP-infected indicator cells expressing RVFV N and wt L after 24 h of incubation. Only values for the Ren-Luc minigenome reporter are shown. Reporter activities of the cotransfected FF-Luc plasmid indicated comparable transfection efficiencies (data not shown). Mean values and standard deviations from three independent experiments are shown. (c) Quantification of positive-sense RVFV minigenome RNA. Cells were transfected with plasmids of the RVFV minigenome system (with 10 ng vRen plasmid), and 72 h later, RNA levels were determined by strand-specific real-time quantitative RT-PCR (RT-qPCR) as indicated for Fig. 4. rel., relative. Mean values and standard deviations from three independent experiments are shown.
Taken together, we have generated L polymerase mutants for the orthobunyavirus LACV and the phlebovirus RVFV with a selectively inactivated transcription activity and present reverse genetics systems that allow us to measure genome replication in a convenient manner.
DISCUSSION
The L polymerase is the largest structural protein of the bunyaviruses and responsible for transcribing and replicating the viral genome. In common with other segmented negative-sense RNA viruses like arenaviruses and orthomyxoviruses, bunyaviruses initiate mRNA transcription with capped primers that were cleaved from host cell RNAs. Recent structural and biochemical analyses revealed that the N terminus of LACV L protein harbors the endonuclease activity essential for cleaving the capped host cell RNA primers (21). Homologous domains were described for arenaviruses (34) and orthomyxoviruses (36, 37). For LACV, five highly conserved amino acids were shown to be central for the endonuclease activity. Four amino acids (H34, D52, D79, and D92) are forming complexes with two central Mn2+ ions acting as cofactors, and amino acid K94 most likely is responsible for binding the H2O molecules necessary for phosphodiester bond cleavage. In addition, K108 presumably binds the substrate RNA. Mutation of the metal-binding residues H34, D52, D79, and D92 as well as of the catalytic K94 completely abrogated the in vitro endonuclease activity of the isolated N terminus, whereas mutation of K108 allowed for some residual activity (21). Accordingly, all endonuclease-negative mutants were shown to be inactive in the standard LACV minigenome system. However, when we employed the new, optimized minigenome setup, we noted that the H34A and D52A mutants retained some clearly detectable transcription activity. H34 is contributing to metal binding site 1, whereas D52 is the only amino acid exclusively committed to metal binding site 2. Possibly, the lower Mn2+ binding ability of these mutants is tolerated to some extent. In addition, metal binding site 2 may play a minor role compared to metal binding site 1 since it has a much lower ion-binding affinity than metal binding site 1 and was not saturable under in vitro conditions (21).
It was previously unclear whether the endonuclease-negative mutants of LACV L maintained the ability to replicate their template RNA (21). We took advantage of the LACV trVLP system to test the presence of RNA replication products in a convenient manner. The minigenome contained within the infecting trVLPs is thereby amplified in indicator cells by wt L and N in trans. Since the packaging of minigenome (and genome) RNAs into particles is known to be dependent on both the 5′ and the 3′ UTRs (3–5), the trVLP system ensures that only full-length copies of the vRNA were picked up and transferred to the indicator cells. Use of the trVLP transcomplementation system as a proxy for genome replication needs to be taken with some caution, as VLP formation also depends on RNP encapsidation, RNP packaging, and particle assembly. Nonetheless, with all other experimental parameters left constant, the employment of different L endonuclease mutants for trVLP formation showed that most transcription-negative L mutants must have maintained a replication activity that seems at least equal to that of wt L. The only exception was K94A, which had apparently lower replication ability. On the other hand, the replication activity of the two transcription-negative H34K and D79A mutants seems to even exceed that of wt L. Thus, the data confirm that both metal binding and the catalytic residue are important for transcriptional activity. Interestingly, loss of metal binding (H34K and D79A mutants) results in a complete absence of transcription but seems to increase replication. Loss of the catalytic residue K94, in contrast, impairs both transcription and replication. It is possible that the Mn2+ ions are important for initial recruitment of the host mRNA substrate to the endonuclease site, whereas the catalytic K94 acts at a later step. An mRNA substrate bound to the endonuclease site but not further processed (the K94A mutant) may block proper transcription but also inhibit switching to the replication mode. Mutants lacking the metal binding site may be unable to recruit a host mRNA substrate and therefore be free to switch to the replication mode. Although this model is highly speculative, the systems and mutants described here could be instrumental for testing it.
The ability of the D79A L mutant (which had the strongest replication-only phenotype in the VLP system) to produce positive-strand RNA was independently confirmed by primer extension and by real-time RT-PCR assay. Moreover, we mutagenized the D79A-equivalent position in the L polymerase of RVFV, D111, and observed a similar phenotype. The D111A mutant entirely lost its transcriptional activity but maintained its ability to replicate its template. Together with the fact that the Toscana phlebovirus N terminus has endonuclease activity (34), this confirms that the structure of endonuclease domain is highly conserved between the different bunyavirus genera (21, 34).
As mentioned before, the related arenaviruses have a homologous endonuclease domain at the N terminus of the L protein, as revealed by the atomic structure for the lymphocytic choriomeningitis virus (LCMV) (34). The LCMV endonuclease structure did not contain the central Mn ions, but a structural alignment with the LACV domain enables the identity of five putative active site amino acids to be hypothesized (Fig. 6a). LCMV residue E51 thereby locates in the equivalent position of LACV residue H34, and LCMV D66, D89, E102, and K115 are equivalent to LACV D52, D79, D92, and K94, respectively. However, despite the overall similarity of the endonuclease domains, there are some differences. Structurally, several LACV or LCMV amino acid equivalents are holding slightly different relative positions (Fig. 6a). Functionally, not all mutations of those key amino acids have comparable effects on transcription or replication. Based on the literature on LCMV (34) and on the data presented here for LACV, it appears that the LACV/LCMV equivalents D79/D89 and D92/E102 display largely similar phenotypes (Fig. 6b). In both cases, transcription is entirely abrogated, whereas replication is preserved. In contrast, mutations of the putative equivalents H34 and E51 show differences between LACV and LCMV. While both endonuclease mutants have lost transcriptional activity to some extent, the LACV H34A mutant maintains wt replication activity, while the corresponding LCMV E51A mutant displays reduced replication activity. For the putative equivalents D52/D66 and K94/K115A, a functional comparison is not possible due to a lack of data for LCMV. Thus, despite clear similarities, neither the structures nor the mutational phenotypes seem completely comparable between bunyaviruses and arenaviruses.
Fig 6.
Structural and functional comparison of bunyaviral and arenaviral endonucleases. (a) Structural alignment of the endonuclease domains of LACV (21) and LCMV (34). Individual amino acids of LACV and LCMV are shown as red and blue sticks, respectively. (b) Functional comparison of the impacts individual mutations have on Ren-Luc reporter activity (representing transcription) and on replication activity.
Transcription and replication of the bunyavirus genome are regulated not only by the polymerase, but also at several other levels. Previous investigations using Bunyamwera virus have unveiled a role of sequence elements in the UTRs (38, 39) and suggest a contribution of N protein to the transcription-replication switch (40, 41). Together with our results, these studies indicate that genome replication of orthobunyaviruses and phleboviruses can occur independently of genome transcription.
An interesting side result of our studies was that inhibition of the double-stranded RNA(dsRNA)-dependent protein kinase, PKR, robustly enhanced the performance of the LACV (and RVFV [data not shown]) minigenome systems. PKR can be activated by viral dsRNA or single-stranded RNA (ssRNA) containing a 5′-triphosphate group and a short stem-loop structure (42, 43). It phosphorylates several cellular targets, among them NF-κB and the α subunit of the eukaryotic translation initiation factor eIF2, thereby leading to upregulation of innate immune genes and to a strong translational arrest of cellular and viral mRNA. Possibly, the positive-stranded background transcripts, which are synthesized from the minigenome plasmid in a promoter-independent fashion (Figs. 4 and 5), are forming dsRNA hybrids with the intended negative-strand RNAs transcribed from the RNA polymerase I promoter. Moreover, transcription and replication by the minigenome system are likely to trigger PKR activation in a similar manner to bunyavirus infection itself (30–32). Thus, the specific inhibition of PKR may aid in the establishment of reverse genetics systems of bunyaviruses in general and other RNA viruses as well.
In summary, we have generated L polymerase mutants for the medically and economically important bunyaviruses LACV and RVFV that have lost the ability to transcribe mRNAs while maintaining genome replication activity. Similar mutants had recently been proven instrumental for establishing a new model of the influenza virus polymerase function (44). A reverse genetics system for RVFV was used to demonstrate that the antiviral host cell protein MxA inhibits viral primary transcription (9). Whether MxA also affects viral genome replication, however, remains unclear. In general, viral RNA synthesis products are known to activate pattern recognition receptors (RIG-I [retinoic acid-inducible gene I], MDA5 [melanoma differentiation-associated antigen 5], and TLR3 [Toll-like receptor 3]) and antiviral effectors (PKR, OAS [2′-5′-oligoadenylate synthetase], and ADAR [adenosine deaminase acting on RNA]) of the host cell. Although these events are central for the innate immune response (45), the exact nature of those viral RNA products is still not entirely solved (46–49). The mutants and protocols described here will facilitate future studies addressing these questions on bunyavirus polymerase function and host cell responses.
ACKNOWLEDGMENTS
Work in the authors' laboratories is supported by grants We2616/5-2 and SFB 1021 from the Deutsche Forschungsgemeinschaft and grant 01 KI 0705 from the Bundesministerium für Bildung und Forschung (to F.W.), the EMBL, the Agence Nationale de la Recherche (ANR) (to S.C.), and an EMBO long-term fellowship (to J.R.). S.C. and J.R. were supported by a French Agence Nationale de Recherche grant (ANR-GUI-AAP-04).
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Elliott RM, Weber F. 2009. Bunyaviruses and the type I interferon system. Viruses 1:1003–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walter CT, Barr JN. 2011. Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 92:2467–2484 [DOI] [PubMed] [Google Scholar]
- 3. Flick K, Katz A, Overby A, Feldmann H, Pettersson RF, Flick R. 2004. Functional analysis of the noncoding regions of the Uukuniemi virus (Bunyaviridae) RNA segments. J. Virol. 78:11726–11738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kohl A, Lowen AC, Leonard VH, Elliott RM. 2006. Genetic elements regulating packaging of the Bunyamwera orthobunyavirus genome. J. Gen. Virol. 87:177–187 [DOI] [PubMed] [Google Scholar]
- 5. Terasaki K, Murakami S, Lokugamage KG, Makino S. 2011. Mechanism of tripartite RNA genome packaging in Rift Valley fever virus. Proc. Natl. Acad. Sci. U. S. A. 108:804–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Albarino CG, Bird BH, Nichol ST. 2007. A shared transcription termination signal on negative and ambisense RNA genome segments of Rift Valley fever, sandfly fever Sicilian, and Toscana viruses. J. Virol. 81:5246–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ikegami T, Won S, Peters CJ, Makino S. 2007. Characterization of Rift Valley fever virus transcriptional terminations. J. Virol. 81:8421–8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoenen T, Groseth A, de Kok-Mercado F, Kuhn JH, Wahl-Jensen V. 2011. Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 91:195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Habjan M, Penski N, Wagner V, Spiegel M, Overby AK, Kochs G, Huiskonen JT, Weber F. 2009. Efficient production of Rift Valley fever virus-like particles: the antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology 385:400–408 [DOI] [PubMed] [Google Scholar]
- 10. Overby AK, Popov V, Neve EP, Pettersson RF. 2006. Generation and analysis of infectious virus-like particles of Uukuniemi virus (Bunyaviridae): a useful system for studying bunyaviral packaging and budding. J. Virol. 80:10428–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi X, Kohl A, Leonard VH, Li P, McLees A, Elliott RM. 2006. Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. J. Virol. 80:8089–8099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soldan SS, Hollidge BS, Wagner V, Weber F, Gonzalez-Scarano F. 2010. La Crosse virus (LACV) Gc fusion peptide mutants have impaired growth and fusion phenotypes, but remain neurotoxic. Virology 404:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Overby AK, Popov VL, Pettersson RF, Neve EP. 2007. The cytoplasmic tails of Uukuniemi virus (Bunyaviridae) GN and GC glycoproteins are important for intracellular targeting and the budding of virus-like particles. J. Virol. 81:11381–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi X, Kohl A, Li P, Elliott RM. 2007. Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis. J. Virol. 81:10151–10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Overby AK, Pettersson RF, Neve EP. 2007. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J. Virol. 81:3198–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li S, Koromilas AE. 2001. Dominant negative function by an alternatively spliced form of the interferon-inducible protein kinase PKR. J. Biol. Chem. 276:13881–13890 [DOI] [PubMed] [Google Scholar]
- 19. Blakqori G, Kochs G, Haller O, Weber F. 2003. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J. Gen. Virol. 84:1207–1214 [DOI] [PubMed] [Google Scholar]
- 20. Weber F, Dunn EF, Bridgen A, Elliott RM. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67–74 [DOI] [PubMed] [Google Scholar]
- 21. Reguera J, Weber F, Cusack S. 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 6:e1001101. 10.1371/journal.ppat.1001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Habjan M, Penski N, Spiegel M, Weber F. 2008. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J. Gen. Virol. 89:2157–2166 [DOI] [PubMed] [Google Scholar]
- 23. Robb NC, Smith M, Vreede FT, Fodor E. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90:1398–1407 [DOI] [PubMed] [Google Scholar]
- 24. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beitz E. 2000. TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 16:135–139 [DOI] [PubMed] [Google Scholar]
- 27. Subramaniam S. 1998. The Biology Workbench—a seamless database and analysis environment for the biologist. Proteins 32:1–2 [PubMed] [Google Scholar]
- 28. Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256–2268 [DOI] [PubMed] [Google Scholar]
- 29. Carlton-Smith C, Elliott RM. 2012. Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera orthobunyavirus replication. J. Virol. 86:11548–11557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287. 10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Streitenfeld H, Boyd A, Fazakerley JK, Bridgen A, Elliott RM, Weber F. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 77:5507–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morin B, Coutard B, Lelke M, Ferron F, Kerber R, Jamal S, Frangeul A, Baronti C, Charrel R, de Lamballerie X, Vonrhein C, Lescar J, Bricogne G, Gunther S, Canard B. 2010. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 6:e1001038. 10.1371/journal.ppat.1001038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boshra H, Lorenzo G, Busquets N, Brun A. 2011. Rift Valley fever: recent insights into pathogenesis and prevention. J. Virol. 85:6098–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918 [DOI] [PubMed] [Google Scholar]
- 37. Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913 [DOI] [PubMed] [Google Scholar]
- 38. Barr JN, Rodgers JW, Wertz GW. 2005. The Bunyamwera virus mRNA transcription signal resides within both the 3′ and the 5′ terminal regions and allows ambisense transcription from a model RNA segment. J. Virol. 79:12602–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barr JN, Wertz GW. 2005. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J. Virol. 79:3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eifan SA, Elliott RM. 2009. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J. Virol. 83:11307–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walter CT, Bento DF, Alonso AG, Barr JN. 2011. Amino acid changes within the Bunyamwera virus nucleocapsid protein differentially affect the mRNA transcription and RNA replication activities of assembled ribonucleoprotein templates. J. Gen. Virol. 92:80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811 [DOI] [PubMed] [Google Scholar]
- 43. Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 2007. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 318:1455–1458 [DOI] [PubMed] [Google Scholar]
- 44. Jorba N, Coloma R, Ortin J. 2009. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5:e1000462. 10.1371/journal.ppat.1000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 46. Baum A, Garcia-Sastre A. 2011. Differential recognition of viral RNA by RIG-I. Virulence 2:166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bowzard JB, Davis WG, Jeisy-Scott V, Ranjan P, Gangappa S, Fujita T, Sambhara S. 2011. PAMPer and tRIGer: ligand-induced activation of RIG-I. Trends Biochem. Sci. 36:314–319 [DOI] [PubMed] [Google Scholar]
- 48. Jensen S, Thomsen AR. 2012. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 86:2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243:91–98 [DOI] [PubMed] [Google Scholar]