Abstract
Although infections with “natural” West Nile virus (WNV) and the chimeric W956IC WNV infectious clone virus produce comparable peak virus yields in type I interferon (IFN) response-deficient BHK cells, W956IC infection produces higher levels of “unprotected” viral RNA at early times after infection. Analysis of infections with these two viruses in IFN-competent cells showed that W956IC activated NF-κB, induced higher levels of IFN-β, and produced lower virus yields than WNV strain Eg101. IPS-1 was required for both increased induction of IFN-β and decreased yields of W956IC. In Eg101-infected cells, phospho-STAT1/STAT2 nuclear translocation was blocked at all times analyzed, while some phospho-STAT1/STAT2 nuclear translocation was still detected at 8 h after infection in W956IC-infected mouse embryonic fibroblasts (MEFs), and early viral protein levels were lower in these cells. A set of additional chimeras was made by replacing various W956IC gene regions with the Eg101 equivalents. As reported previously, for three of these chimeras, the low early RNA phenotype of Eg101 was restored in BHK cells. Analysis of infections with two of these chimeric viruses in MEFs detected lower early viral RNA levels, higher early viral protein levels, lower early IFN-β levels, and higher virus yields similar to those seen after Eg101 infection. The data suggest that replicase protein interactions directly or indirectly regulate genome switching between replication and translation at early times in favor of translation to minimize NF-κB activation and IFN induction by decreasing the amount of unprotected viral RNA, to produce sufficient viral protein to block canonical type I IFN signaling, and to efficiently remodel cell membranes for exponential genome amplification.
INTRODUCTION
West Nile virus (WNV) is a positive-sense, single-stranded RNA, enveloped virus of the family Flaviviridae. The WNV genomic RNA is about 11 kb in length, has a capped 5′ end but no poly(A) at the 3′ end, and encodes one open reading frame (ORF). The polyprotein precursor produced is posttranslationally processed by virus and host cell proteases to yield three structural proteins, capsid (C), premembrane (prM), and envelope (E), and seven nonstructural (NS) proteins: glycoprotein NS1, membrane anchor NS2A, membrane anchor and protease cofactor NS2B, protease-helicase NS3, membrane anchors NS4A and NS4B, and methyltransferase–RNA-dependent RNA polymerase (RdRp) NS5 (1).
The majority of known WNV isolates are classified into two lineages (2–4). Both lineages contain low- and high-virulence strains (5–7). Lineage 1 strains cause epidemics in many parts of the world while lineage 2 strains are endemic to Africa. The lineage II virus 956D117B3 (4, 8) is a laboratory-passaged descendant of the WNV prototype strain B956. Archived 956D117B3 RNA and the 1,496 3′ nucleotides (nt) from the lineage 1 strain Eg101 were used to construct the first infectious clone of WNV, SP6WNEg3′/Xba (also referred to as W956IC) (8). W956IC virus and the parental virus 956D117B3 produce similar growth kinetics and peak titers in both Vero cells and C6/36 cells (8). The full-length W956IC cDNA is stable in bacteria and has been used to efficiently rescue virus with engineered mutations (9, 10). Other studies from our lab have shown that WNV Eg101 and other natural WNV strain infections do not activate eukaryotic translation initiation factor 2-alpha kinase 2 (Eif2ak2, also known as PKR) in BHK cells or mouse embryonic fibroblasts (MEFs), but W956IC virus infections do (11). W956IC virus infections also induce stress granule (SG) formation while natural WNV strains do not (12). These unique characteristics of the W956IC virus were attributed to the ability of W956IC virus infections to produce higher levels of “unprotected” viral RNA at early times after infection (12).
Hairpins in viral single-stranded RNAs as well as double-stranded RNA (dsRNA) intermediates formed during viral RNA replication are sensed by cellular dsRNA sensors which then activate the first lines of innate defense. The two cytoplasmic RNA sensors, DEXD/H box helicase retinoic acid-inducible gene l (RIG-I) (13) and melanoma differentiation-associated gene 5 (MDA5), each contain a helicase domain that binds to viral RNA and a caspase recruitment domain (CARD) that recruits the adapter, beta interferon (IFN-β) promoter stimulator 1 (IPS-1) (14). IPS-1 activates the transcription factors (TFs), nuclear factor kappa light-chain enhancer of activated B cells (NF-κB), interferon regulatory factor-3 (IRF-3), and IRF-7, which then induce the expression of various proinflammatory cytokines, apoptosis regulatory proteins, type I IFNs, and interferon-stimulated genes (ISGs) that control virus infections (15, 16). PKR has also been reported to play a role in both NF-κB and type I IFN signaling in response to viral infections (17). Our lab previously reported that although infections in MEFs with WNV Eg101 and other natural WNV strains induce IFN-β production and phosphorylation of signal transducer and activator of transcription 1 and 2 (STAT1 and STAT2, respectively), these TFs were not detected in the nuclei or on the promoters of four IRF-3-independent ISGs in infected MEFs at any of the times after infection analyzed (18).
In the present study, the effects of the efficient early RNA replication of W956IC were investigated in type I IFN-competent MEFs. W956IC infections induced lower yields of virus and higher levels of IFN-β than infections with Eg101. Both PKR and NF-κB were activated in W956IC-infected MEFs. IPS-1 was shown to be required for the production of higher levels of IFN-β, increased levels of ISG expression, and decreased virus yields in W956IC-infected MEFs. In contrast to infections with natural strains of WNV, nuclear translocation of phosphorylated STAT1 and STAT2 was detected up to 8 h after W956IC infection. Analysis of infections with W956IC chimeras that had additional Eg101 gene replacements in IFN-competent MEFs showed that those with lower early viral RNA levels and higher virus yields similar to Eg101 also induced lower early levels of IFN-β and higher early viral protein levels. The data suggest that multiple interactions between viral replicase complex proteins and possibly also between viral nonstructural proteins and cell proteins differentially regulate early viral RNA and protein synthesis levels, most likely by regulating genome switching between replication and translation. Lower intracellular viral RNA levels minimize IFN induction by viral RNA, and higher viral protein levels ensure sufficient viral protein levels to block IFN signaling.
MATERIALS AND METHODS
Cell lines, viruses, and reagents.
Simian virus 40 (SV40)-transformed C3H/He and C57BL/6 MEF lines as well as BHK-21 WI2 cells were maintained as previously described (19). RIG-I+/+ (wild type [WT]), RIG-I−/−, MDA5−/−, and IPS-1−/− MEFs (provided by M. Gale, University of Washington, Seattle, WA) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM glutamine, 1 mM sodium pyruvate, 10 mM HEPES, antibiotic-antimycotic solution (Invitrogen), and 1× nonessential amino acids. p65−/− MEFs and p65-reconstituted p65−/− MEFs (provided by J.-D. Li, Georgia State University, Atlanta, GA) were maintained in DMEM supplemented with 5% fetal bovine serum and 10 μg/ml gentamicin. Human lung carcinoma A549 cells were purchased from ATCC and maintained in F-12K nutrient mixture with (1×) Kaighn modification medium supplemented with 10% fetal calf serum and 1% PenStrep. For the experiments analyzing viral titers, intracellular RNA levels, or cell or viral proteins, cells were seeded the day before infection so that they reached ∼100% confluence by the next day (106 cells/well of a six-well plate for C3H/He and A549 cells and 5 × 105 cells/well for C57BL/6, RIG-I+/+ [WT], RIG-I−/−, MDA5−/−, and IPS-1−/− MEFs). For confocal microscopy experiments, half the number of cells was seeded the day before infection to reach 50% confluence by the next day. Confocal data showed that the majority of the cells (more than 90%) were infected at 24 h after infection at a multiplicity of infection (MOI) of 1 (12).
IPS-1 levels in C3H/He MEFs were reduced by transfection with IPS-1-specific mouse small interfering RNA (siRNA) (On-TargetPlus SMART pool D430028G21RIK; Thermo Scientific Dharmacon) using HiPerFect transfection reagent (Qiagen) according to the manufacturer's protocol. Control cells were transfected with nontargeting siRNA (Santa Cruz Biotechnology, Santa Cruz, CA). After drug selection, cells were cloned using cloning rings, and individual cell lines were amplified.
Stock pools of the WNV Eg101 were prepared by infecting BHK cells at an MOI of 0.1 and harvesting culture fluid 32 h after infection. Clarified culture fluid (∼108 PFU/ml) was aliquoted and stored at −80°C. Pools of WNV W956IC were produced by transfecting BHK monolayers with 1 μg of in vitro-transcribed viral RNA and then harvesting culture fluid 72 h after transfection. Clarified culture fluid (∼107 PFU/ml) was aliquoted and stored at −80°C. A series of chimeric infectious clones containing various WNV Eg101 gene regions inserted into the backbone of W956IC were constructed as described previously (12), and pools of each chimeric virus were made as described above for the W956IC virus. Virus infectivity titers were assessed by plaque assay on BHK monolayers as previously described (20).
Real-time qRT-PCR.
Total cellular RNA was extracted from infected and control cells using TriReagent (Molecular Research Center) according to the manufacturer's protocol. Real-time quantitative reverse transcription-PCR (qRT-PCR) analysis of mouse gene expression was performed with the following Assays-on-Demand 20× primers and TaqMan 6-carboxyfluorescein (FAM)-labeled probe mixes from Applied Biosystems: Mm 00439546_s1 (Ifnb1), Mn00836412 (Oas1a), Mm00516788_m1 (Irf7), and Mm00515191_m1 (Irf1). Intracellular mRNA levels were quantified using an Applied Biosystems 7500 sequence detection system. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an endogenous control and detected using TaqMan mouse GAPDH Control Reagents primers and probe (Applied Biosystems). One-step RT-PCR was performed for each target mRNA and for the endogenous control in a singleplex format using 200 ng of RNA and a TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems). Viral RNAs were detected using previously described TaqMan probes for the NS1 region of the Eg101 and W956IC sequences (9, 12, 19). Mock-infected control samples were collected for each of the time points and analyzed for Ifnb1, Oas1a, Irf1, and Irf7 mRNA expression levels. The expression levels of each of these genes were not found to differ appreciably between the mock samples taken at different times. Therefore, only a single representative mock value was used as the calibrator for estimating the fold change in expression of each gene as described below. Each experiment was repeated at least two times in triplicate. Triplicate threshold cycle (CT) values were analyzed with Microsoft Excel using the comparative CT (ΔΔCT) method of the SDS Applied Biosystems software, which also applied statistical analysis to the data (TINV test in Microsoft Excel). The signals obtained for inducible cellular mRNAs or for viral RNAs were normalized to the GAPDH mRNA signal, and the fold change relative to the uninfected calibrator sample or for the level of viral RNA present at 1 h after infection was calculated and expressed in relative quantification (RQ) units. Error bars represent the standard errors of the means (SE) and indicate the calculated minimum (RQMin) and maximum (RQMax) of the mRNA expression levels based on an RQMin/Max of the 95% confidence level. Differences in expression levels between two samples were considered statistically significant (P value of < 0.05) when the error bars did not overlap.
Northern blot hybridization.
Electrophoresis, transfer, and hybridization of total cellular RNA (5 μg/lane) were performed as described previously (19). The probe used to detect full-length viral RNA corresponded to the 3′-terminal 800 nt of the WNV genome. The sequence of this region was identical in all the virus genomes used in this study.
Confocal microscopy.
MEFs grown to 50 to 70% confluence on 15-mm glass coverslips in wells of a 24-well plate were infected with WNV at an MOI of 5. The cells were fixed by incubation with 4% paraformaldehyde in PBS for 10 min and then permeabilized by ice-cold methanol for 10 min. Coverslips were washed with phosphate-buffered saline (PBS) and then blocked overnight with 5% horse serum (Invitrogen, Carlsbad, CA) in PBS. Coverslips were incubated with rabbit anti-NF-κB p65 antibody (Santa Cruz Biotechnology) diluted 1:200 in the blocking buffer, with anti-dsRNA J2 antibodies (English and Scientific Consulting, Hungary) diluted 1:500 in the blocking buffer, or with rabbit anti-STAT2 antibody (generously provided by C. Schindler, Columbia University, New York, NY) diluted 1:200 for 1 h at room temperature and then washed three times with PBS. Coverslips were then incubated with secondary Alexa Fluor antibodies (Santa Cruz Biotechnology) diluted 1:400 in blocking buffer. In some experiments, 0.5 μg/ml of Hoechst 33342 dye (Invitrogen) was also added. The coverslips were washed with PBS and mounted on glass slides with Prolong Gold Antifade reagent (Invitrogen). Cells were visualized with a 63× oil immersion objective on an LSM 700 laser confocal microscope (Zeiss, Oberkochen, Germany) using LSM 5 (version 4.2) software (Carl Zeiss Inc.). All of the images compared were obtained using the same instrument settings.
Western blotting.
Western blot analysis was carried out as previously described (19). Briefly, the confluent monolayers of cells were either mock infected or infected with Eg101 or W956IC at an MOI of 1. At various times after infection, cell lysates were prepared, and proteins were separated by SDS-PAGE, transferred, and assayed by Western blotting using specific antibodies. Mock-infected samples collected at each time point were also analyzed with each of the antibodies tested. However, since no appreciable change was observed in the intensities of the bands of the mock samples harvested at different times, only one representative mock-infected sample (M) was shown in the figures to save space. The membranes were incubated with a polyclonal primary antibody specific for IRF-3, PKR, actin (Santa Cruz Biotechnology), phospho-IRF-3 (Ser396), phospho-STAT1 (Tyr701), phospho-IκB-alpha (Ser32), phospho-c-Jun (Ser73), STAT1, IκB-alpha (Cell Signaling), WNV NS3, or phospho-PKR (Thr451) (Millipore).
IFN-β protein ELISA.
A commercial capture enzyme-linked immunosorbent assay (ELISA) was used according to the manufacturer's instructions (PBL Biomedical Laboratories) to measure levels of secreted IFN-β protein in cell supernatants.
RESULTS
Analysis of Eg101 and W956IC WNV replication in rodent and human cells.
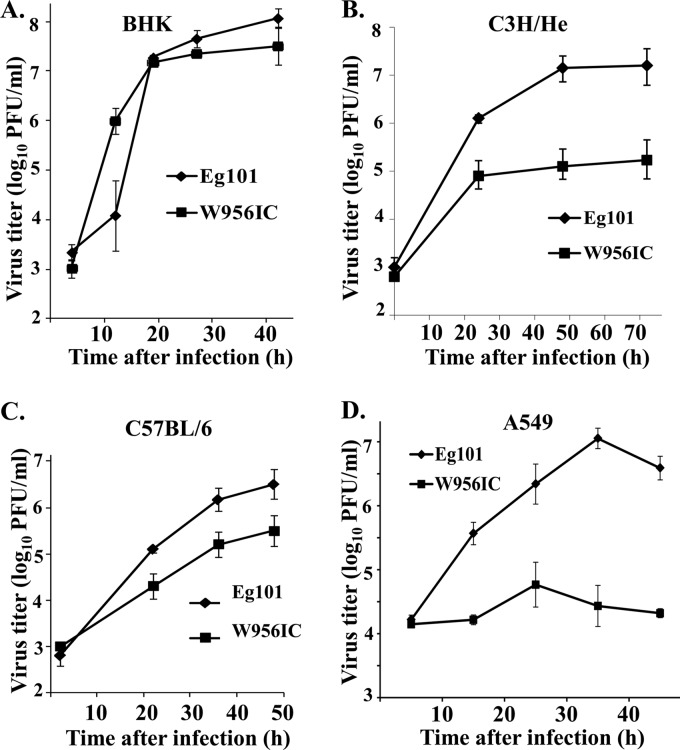
Initial comparisons of the growth characteristics of chimeric W956IC virus with that of parental 956D117B3 virus in both Vero cells and C6/36 cells showed that these two viruses produced similar peak titers (8). Vero cells have a deletion in the IFN-β locus (21, 22), and the mosquito C6/36 cell genome does not encode IFN genes. A previous study from our lab showed that WNV Eg101 and W956IC infections also produced similar peak titers in BHK cells (12). BHK cells do not produce or respond to type I IFN (23, 24). To determine whether W956IC and Eg101 infections produce similar virus yields in cells with a competent type I IFN response, C3H/He and C57BL/6 MEF cell lines were infected at an MOI of 1. W956IC yields at 48 and 72 h from C3H/He MEFs were 100 times lower (Fig. 1B), and W956IC yields from C57BL/6 MEFs were about 10-fold lower (Fig. 1C) than those of Eg101. Consistent with previous data (12), Eg101 and W956IC infections in BHK cells produced similar virus yields at 20 and 28 h after infection, but W956IC-infected BHK cells produced 100 times more virus at 12 h and slightly less virus at 42 h than Eg101 (Fig. 1A). To determine whether the differential response was unique to IFN-competent mouse cells, human A549 cells were infected with either W956IC or Eg101 at an MOI of 1. The results showed that W956IC yields were significantly lower (by 1.5 to 3 logs) than those of Eg101 at 16, 36, and 48 h after infection (Fig. 1D).
Fig 1.
Comparison of WNV Eg101 and W956IC virus production in rodent and human cells. Confluent monolayers of BHK cells (A), C3H/He MEFs (B), C57BL/6 MEFs (C), or A549 cells (D) were infected with Eg101 or W956IC virus at an MOI of 1. Culture fluids were collected at the indicated times after infection, and viral titers were determined by plaque assay on BHK cells. The values shown are averages of duplicate titrations of samples from two independent experiments. Bars represent ± standard deviations.
Comparison of the cellular IFN response to Eg101 and W956IC WNV infections.
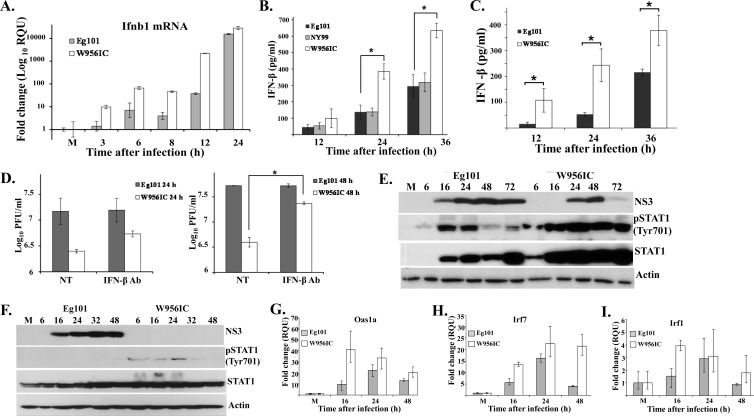
Previous studies of the innate response to WNV in MEFs showed that both an Eg101 infection in C3H/He MEFs (19) and an NY2000 infection in C57BL/6 MEFs (25) efficiently induced IFN-β mRNA expression. The type I IFN response can be activated through the sensing of viral dsRNA by cytoplasmic RNA sensors, and W956IC infections were previously reported to produce higher levels of unprotected intracellular RNA at early times after infection in BHK cells (12). To determine whether Eg101 and W956IC virus infections induced IFN-β mRNA with similar kinetics and to similar levels in MEFs, C3H/He MEFs were infected with Eg101 or W956IC at an MOI of 1, and IFN-β mRNA levels were assessed by real-time qRT-PCR at 3, 6, 8, 12, and 24 h after infection. IFN-β mRNA expression was upregulated 10-fold in W956IC-infected cells but less than 2-fold in Eg101-infected cells at 3 h after infection. At all subsequent times examined, the levels of IFN-β mRNA in W956IC-infected cells were higher than those in Eg101-infected cells (Fig. 2A). The levels of IFN-β protein secreted from C3H/He and C57BL/6 MEFs infected with Eg101 or W956IC were next measured by ELISA. The IFN-β levels produced by W956IC-infected C3H/He (Fig. 2B) and C57BL/6 (Fig. 2C) MEFs were higher than those produced by Eg101-infected MEFs of the same type. Also, the levels of IFN-β induced by a W956IC infection in C3H/He MEFs were higher than those induced in C57BL/6 MEFs, consistent with the lower virus yields produced by the W956IC-infected C3H/He cells (Fig. 1).
Fig 2.
Analysis of induction of IFN-β by WNV Eg101 and W956IC infections. (A) C3H/He MEFs were mock infected or infected with WNV Eg101 or W956IC at an MOI of 1. Replicate cultures were processed to extract total cell RNA or protein at the indicated times after infection. Ifnb1 mRNA levels detected by real-time qRT-PCR were normalized to the levels of GAPDH in the same sample and are shown as the fold change compared to the level of the Ifnb1 mRNA in mock-infected cells. Samples from each qRT-PCR experiment were assayed in triplicate. Error bars represent the standard errors (SE) of the means. Representative data from one of three independent experiments are shown. M, mock infection. The levels of IFN-β protein in culture fluids harvested from WNV-infected C3H/He (B) or C57BL/6 (C) MEFs were determined by ELISA. Data points are averages from two or more independent experiments performed in duplicate. Bars represent ± standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05). (D) C3H/He MEFs were infected with WNV Eg101 or W956IC at an MOI of 5 for 1 h, and then the cultures were incubated with or without neutralizing antibody (Ab) (1,000 neutralizing units/ml) (PBL Interferon Source) against IFN-β for the duration of the infection. Virus titers in culture fluids were determined at 24 and 48 h after infection by plaque assay on BHK cells. The values shown are averages of duplicate titrations from two experiments. Bars represent ± standard deviations. NT, not treated. NS3 and phosphorylated STAT1 (pSTAT1) or total STAT1 in C3H/He (E) or A549 (F) cell lysates were detected by Western blotting with specific antibodies. Actin was used as the loading control. The blots shown are representative of results obtained from three independent experiments. Changes in mRNA expression levels for the ISGs Oas1a (G), Irf7 (H), and Irf1 (I) in infected C3H/He-infected MEFs were assessed by real-time qRT-PCR as described in panel A.
WNV Eg101 was isolated from a human in Egypt in 1951 and has been used extensively to study flavivirus replication and flavivirus-host interactions (1). Since its appearance in New York in 1999, WNV has spread rapidly across the North American continent. The prototype 1999 U.S. isolate, WNV NY99-flamingo382-99 (NY99), and its close relative WNV TX02, isolated in Texas in 2002, have recently become the most frequently used WNV strains for studies of virus replication and the host immune response (26). A phylogenetic analysis of the complete coding sequences of Eg101 and NY99 showed 99.6% amino acid identity (14 amino acid [aa] differences in a total of 3,433) and classified these two strains in the same subclade of lineage 1 (26). To determine whether the amino acid differences between these two WNV strains affected the amount of IFN-β secreted by infected MEFs, IFN-β production by Eg101- and NY99-infected C3H/He MEFs was compared. NY99- and Eg101-infected MEFs produced similar levels of extracellular IFN-β at 12, 24, and 36 h after infection (Fig. 2B). These results indicate that the amino acid differences between Eg101 and NY99 do not affect the efficiency of the induction of a type I IFN response in MEFs.
As a means of analyzing whether IFN-β contributes to the reduction in virus yield in W956IC-infected MEFs, C3H/He MEFs were infected with Eg101 or W956IC at an MOI of 5 (the higher MOI was used to increase the levels of IFN-β as well as virus produced) and then incubated with an IFN-β neutralizing antibody starting immediately after the 1-h virus attachment period, and virus yields were assessed at 24 and 48 h after infection. Virus yields from Eg101-infected MEFs were similar in cultures with or without anti-IFN treatment. However, W956IC virus yields increased at both times analyzed in cultures treated with IFN-β antibody (Fig. 2D). The results indicate that IFN-β plays a role in reducing W956IC but not Eg101 virus production in MEFs.
Phosphorylation of Tyr701 on STAT1, a result of activation of the cellular JAK-STAT signaling pathway by extracellular IFN binding to its receptor, was next assessed by Western blotting in C3H/He MEFs infected with Eg101 or W956IC. Higher levels of STAT1 protein as well as phosphorylation were observed in W956IC-infected cells at every time tested (Fig. 2E), indicating that W956IC virus infections induce a more rapid and robust type I IFN signaling response in MEFs than Eg101 virus infections.
Differences in the ability of flaviviruses to counteract the type I IFN response between human and mouse cells have previously been reported (27–29). Phosphorylation of STAT1 and STAT2 proteins was previously shown to be blocked in human cells infected with natural strains of WNV (30–32), but STAT1 and STAT2 phosphorylation were not inhibited in the mouse cells (Fig. 2E) (18). Phosphorylation of STAT1 on Tyr701 was therefore next compared in A549 human cells infected with Eg101 or W956IC at an MOI of 1. As expected, no STAT1 phosphorylation was observed in the Eg101-infected A549 cell samples (Fig. 2F). Higher levels of STAT1 protein were detected starting at early times in W956IC-infected human cells, indicating that W956IC virus infections induce a more rapid and robust type I IFN signaling response than Eg101 virus infections in human cells. Also, weak phospho-STAT1 bands were observed in W956IC-infected A549 cells at 6, 16, and 24 h after infection (Fig. 2F), indicating that phosphorylation of STAT1 is not completely blocked in W956IC-infected A549 cells.
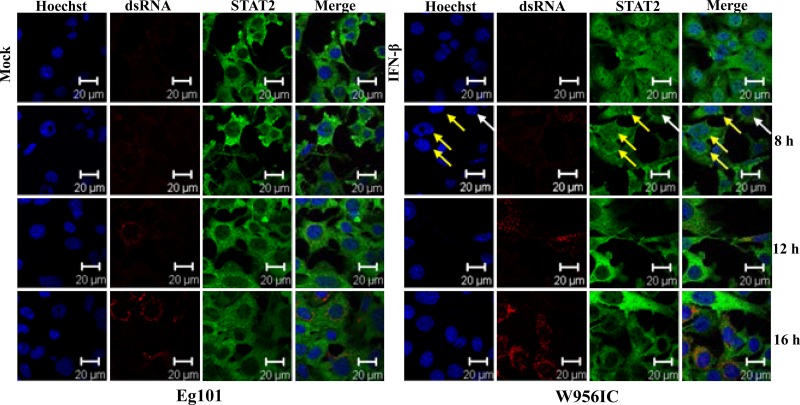
Type I IFN signaling induces the expression of numerous ISGs that mediate the cell antiviral response. However, a previous study from our lab reported that even though STAT1 and STAT2 were phosphorylated in WNV-infected MEFs and even though phospho-STAT1/STAT2 translocation to the nucleus was efficiently blocked, IFN-independent upregulation of the ISGs, Oas1a, Oas1b, Irf7, and Irf1, was still observed (18). To determine whether the higher early IFN induction resulted in increased ISG upregulation, Oas1a, Irf7, and Irf1 mRNA levels were analyzed by real-time qRT-PCR in C3H/He MEFs infected with Eg101 or W956IC at 16, 24, and 48 h after infection. Although the expression of each of these genes increased with time after infection in cells infected with either virus (Fig. 2G to I), the level of upregulation of each of these genes was significantly higher at 16 h after infection in W956IC-infected cells, indicating a more rapid and robust induction of the antiviral state. To determine whether the higher upregulation of these genes at early times after infection with W956IC was dependent on the canonical type I IFN pathway, STAT2 protein nuclear translocation was analyzed by confocal microscopy in C3H/He cells at 8, 12, and 16 h after infection with Eg101 or W956IC. Antibody to dsRNA was used to detect replicating viral RNA. dsRNA was detected in W956IC-infected but not Eg101-infected MEFs at 8 h (Fig. 3). As previously reported, STAT2 was located in the cytoplasm in cells infected with Eg101 at 8, 12, and 16 h after infection (18). In contrast, some STAT2 was observed both in the cytoplasm and nucleus in some cells infected with W956IC at 8 h after infection (Fig. 3, yellow arrows indicate cells with nuclear and cytoplasmic localization of STAT2 while white arrows indicate a cell with predominantly cytoplasmic localization of STAT2) but not at 12 or 16 h after infection. These results indicate that both Eg101 and W956IC infections are able to block STAT translocation to the nucleus but that the blockade was less efficient at early times in W956IC-infected MEFs, allowing some canonical type I IFN signaling to occur, which led to higher ISG expression levels (Fig. 2G to I).
Fig 3.
Analysis of STAT2 nuclear localization. Laser scanning confocal microscopy analyses were performed of STAT2 localization in C3H/He MEFs infected with WNV Eg101 or W956IC at an MOI of 5 for the indicated times or incubated with 1,000 U/ml of IFN-β for 30 min (upper right panel set). Nuclei were stained with Hoechst 33258 dye (blue), STAT2 was detected with anti-STAT2 antibody (green), and infected cells were detected with anti-dsRNA antibody (red). Yellow arrows indicate nuclei that contain STAT2; white arrows indicate a cell with predominantly cytoplasmic STAT2. The results shown are representative of two independent experiments.
Comparison of the extent of activation of transcription factors required for IFN-β gene induction at early times after infection of MEFs infected with Eg101 or W956IC WNV.
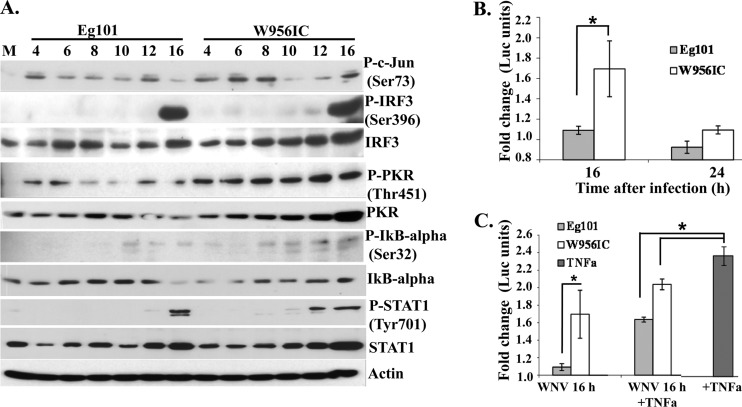
Sensing of viral dsRNA by cytoplasmic RNA sensors, such as RIG-I, MDA5, and PKR, leads to the activation of the transcription factors NF-κB, IRF-3, and ATF2/c-Jun that bind cooperatively to the IFN-β promoter and activate its transcription (16, 33, 34). As previously reported (11), significant PKR activation, as indicated by phosphorylation of Thr451, was detected in W956IC-infected MEFs at all times analyzed (Fig. 4A). Activation of c-Jun, as indicated by phosphorylation of Ser73, was higher in W956IC-infected cells at 6 and 8 h but similar at other times after infection with either Eg101 or W956IC (Fig. 4A). Phospho-IRF-3 (Ser396) bands were not detected until 16 h after infection, and no difference in IRF-3 phosphorylation in response to Eg101 or W956IC infection was observed (Fig. 4A).
Fig 4.
Activation of transcription factors involved in IFN-β gene induction by WNV Eg101 or W956IC infection in MEFs. (A) C3H/He MEFs were infected with WNV Eg101 or W956IC at an MOI of 1. At the indicated times after infection, cell lysates were prepared, and proteins were separated by SDS-PAGE, transferred, and assayed by Western blotting using specific antibodies. Actin was used as the loading control. The blots are representative of results obtained from three independent experiments. P-, phosphorylated. (B) C3H/He MEFs were cotransfected with an NF-κB luciferase reporter vector (pNF-kB-Luc; Stratagene) and a thymidine kinase (TK) promoter-Renilla vector (pTK-RLuc) using Fugene (Roche) transfection reagent. After 24 h, cells were infected with either WNV Eg101 or W956IC at an MOI of 1. Cells were harvested at the indicated times after infection, and luciferase activity was measured using a dual-luciferase assay kit (Promega). Reporter activity was normalized to Renilla luciferase (Luc) activity in the same sample and is shown as fold change compared to the normalized activity in mock-infected cells. The values shown are averages from three independent experiments. Bars represent ± standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05). (C) At 16 h after infection, mouse TNF-α (20 ng/ml) (Biosource) was added to infected and mock-infected cells for 3 h. The data obtained were analyzed as described for panel B.
NF-κB is sequestered in the cytoplasm as an inactive complex that contains the inhibitory subunit IκBα. Phosphorylation of IκBα at Ser32 results in degradation of this protein. Increased phosphorylation of IκBα at Ser32 and smaller amounts of total IκBα were observed in W956IC-infected cell extracts than in Eg101 extracts. As another means of analyzing NF-κB activation, a luciferase reporter driven by a promoter containing tandem repeats of the NF-κB binding site was transiently transfected into C3H/He MEFs, and luciferase activity was analyzed after infection with Eg101 or W956IC at an MOI of 1. At 16 h after infection, the luciferase activity in W956IC-infected cells was significantly higher than that in Eg101-infected cells, while at 24 h, only a slightly higher level was observed in W956IC-infected cells (Fig. 4B). To test whether NF-κB activation is actively blocked by viral products, luciferase activity was analyzed in cells infected with Eg101 or W956IC for 16 h and then treated with tumor necrosis factor alpha (TNF-α; a known NF-κB inducer) for 3 h. Luciferase activity increased slightly in W956IC-infected, TNF-α-treated cells and increased to a greater extent in Eg101-infected, TNF-α-treated cells (Fig. 4C), suggesting that further NF-κB activation could be induced in virus-infected cells. However, the level of activation induced by TNF-α was lower in infected cells than in mock-infected cells, suggesting the possibility that WNV infection may also suppress or not activate a part of the NF-κB pathway activated by TNF-α.
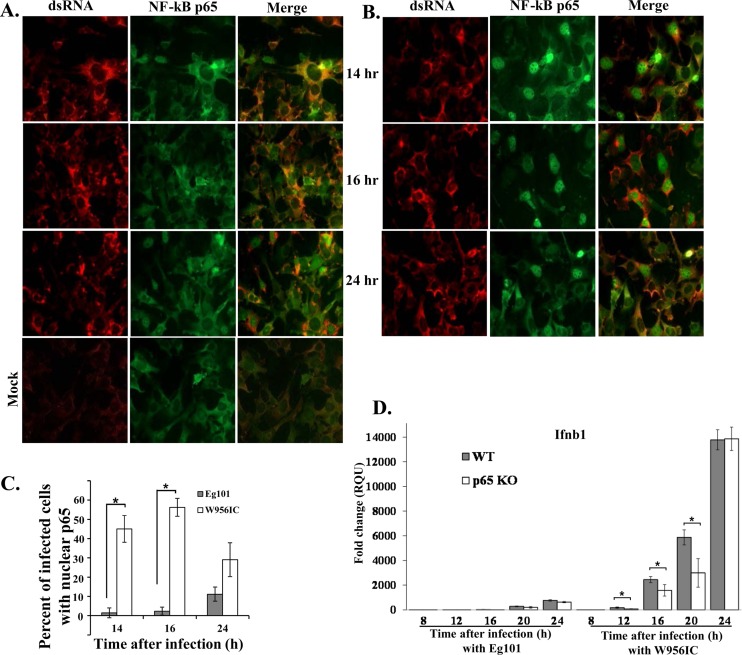
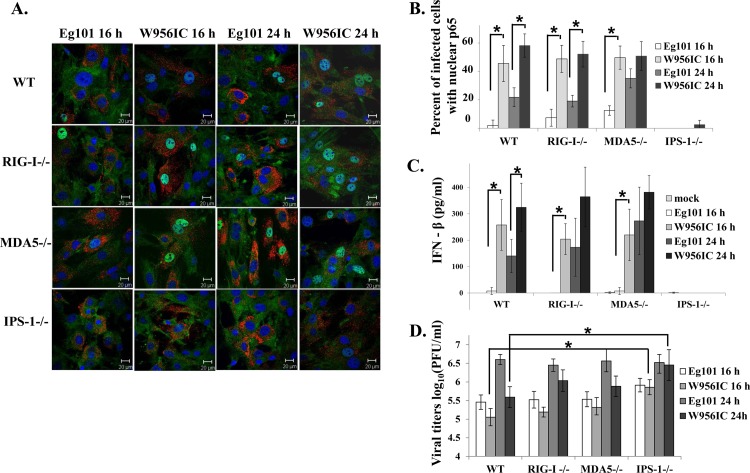
Phosphorylation of IκBα leads to its ubiquitination and subsequent degradation by the proteosome. The loss of IκBα exposes the nuclear localization signal of the RelA/p65 subunit of the NF-κB complex, which results in the translocation of this subunit to the nucleus (35). Nuclear translocation of RelA/p65 was assayed in MEFs infected with Eg101 or W956IC by confocal microscopy. p65 was predominantly cytoplasmic in Eg101-infected cells at 14 and 16 h after infection, and only at 24 h was nuclear p65 detected in some cells (Fig. 5A). In contrast, efficient nuclear localization of p65 was detected in W956IC-infected cells at 14, 16, and 24 h after infection (Fig. 5B and C), confirming that a W956IC infection activates NF-κB more rapidly and efficiently than an Eg101 infection.
Fig 5.
Analysis of NF-κB p65 cellular localization. C3H/He MEFs were infected with Eg101 (A) or W956IC (B) virus (MOI of 5). At the indicated times after infection, cells were stained with anti-dsRNA (red) and anti-NF-κB antibody (green) and analyzed by laser scanning confocal microscopy. (C) Quantification of the percentage of infected cells with nuclear p65 localization was done at the indicated times after infection. At least three fields of cells for each condition were counted, and average values were plotted. Bars represent ± standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05). (D) p65−/− MEFs (p65 KO, where KO is knockout) and p65−/− MEFs reconstituted with p65 (WT) were infected with Eg101 or W956IC at an MOI of 1, and replicate cultures were processed to extract total cell mRNA at the indicated times after infection. Ifnb1 mRNA levels detected by real-time qRT-PCR were normalized to the levels of GAPDH in the same sample and are shown as the fold change compared to the level of the Ifnb1 mRNA in mock-infected cells. Samples from each qRT-PCR experiment were assayed in triplicate. Error bars represent the SE of the mean. Representative data from one of three independent experiments are shown.
Analysis of the role of NF-κB in IFN induction in MEFs infected with Eg101 or W956IC WNV.
To determine whether NF-κB is involved in IFN-β production in W956IC- or Eg101-infected cells, p65−/− and control p65−/− MEF cells reconstituted with p65 were infected with Eg101 or W956IC (MOI of 1), and IFN-β mRNA expression was assessed by real-time qRT-PCR at 8, 12, 16, 20, and 24 h after infection. Low and similar upregulation of IFN-β mRNA expression was observed in Eg101-infected p65−/− and p65-reconstituted MEFs (Fig. 5D), suggesting that NF-κB does not play a major role in mediating IFN-β gene expression in cells infected with natural strains of WNV. However, lower levels of IFN-β mRNA were observed in W956IC-infected, p65−/− MEFs than in the W956IC-infected, p65-reconstituted control MEFs at 12, 16, and 20 h after infection. By 24 h, similar levels of IFN-β mRNA upregulation were observed in both types of cells (Fig. 5D).
Analysis of PKR activation in MEFs infected with Eg101 or W956IC WNV.
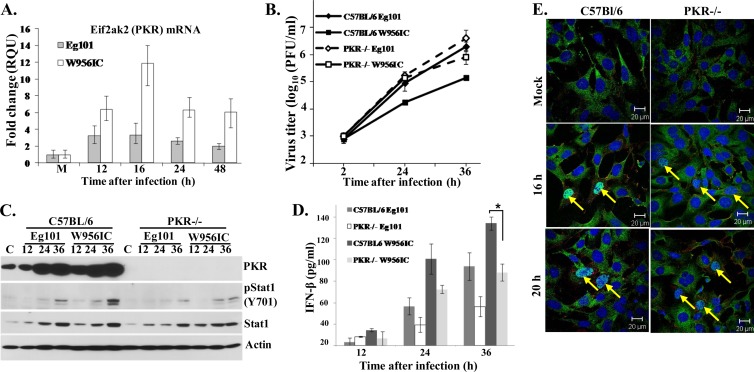
Several previous studies reported that PKR is one of the kinases leading to NF-κB activation during viral infection (reviewed in reference 17), and one of these studies suggested that PKR is involved in NF-κB-mediated type I IFN induction occurring in response to WNV infection (36). The induction of PKR mRNA expression was assessed by real-time qRT-PCR in C3H/He MEFs at 12, 16, 24, or 48 h after infection with Eg101 or W956IC (MOI of 1). The levels of PKR mRNA were higher in W956IC-infected cells than in Eg101-infected cells at all times tested (Fig. 6A). A previous study from our lab (11) as well as data shown in Fig. 4A indicates that the levels of both phospho-PKR and total PKR were significantly higher in C3H/He MEFs infected with W956IC than in those infected with Eg101, consistent with more rapid and efficient upregulation and activation of PKR by a W956IC infection. The effect of the lack of PKR on virus production was analyzed by infecting PKR−/− and control C57BL/6 MEFs with Eg101 or W956IC (MOI of 1) and analyzing virus yields by plaque assay. W956IC virus yields were 7- and 10-fold higher from PKR−/− cells than from control C57BL/6 MEFs at 24 and 36 h, respectively, while Eg101 virus yields were only slightly higher in PKR−/− cells at 36 h after infection (Fig. 6B).
Fig 6.
Induction and activation of PKR in MEFs after infection with WNV Eg101 or W956IC. (A) C3H/He MEFs were infected with WNV Eg101 or W956IC at an MOI of 1. Changes in Eif2ak2 (PKR) mRNA levels were assessed by real-time qRT-PCR at the indicated times after infection. PKR mRNA levels were normalized to the level of GAPDH mRNA in the same sample and are shown as the fold change relative to the levels of PKR mRNA in mock-infected cells. Each sample was assayed in triplicate. Error bars represent the SE of the means. Representative data from one of three independent experiments are shown. M, mock infection. (B) Confluent monolayers of C57BL/6 and PKR−/− MEFs were infected with WNV Eg101 or W956IC at an MOI of 1, culture fluids were collected at the indicated times after infection, and virus titers were determined by plaque assay on BHK cells. The values shown are averages of duplicate titrations of samples from two independent experiments. Bars represent ± standard deviations. (C) At the indicated times (h) after infection, cell lysates were prepared, and cell proteins were separated by SDS-PAGE, transferred, and assayed by Western blotting using specific antibodies. Actin was used as the loading control. The blots shown are representative of results obtained from three independent experiments. (D) The levels of IFN-β protein in supernatants of infected MEFs were determined by ELISA at the indicated times after infection. The values are averages of results from replicate assays done on samples from two independent experiments. Bars represent ± standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05). (E) Laser scanning confocal microscopy of W956IC-infected cells stained with anti-dsRNA (red), anti-NF-κB antibody (green), and Hoechst 33258 dye (blue) at the indicated times after infection.
To assess the extent of type I IFN signaling, phosphorylation at STAT1 Tyr701 was analyzed by Western blotting in control and PKR−/− cell extracts at different times after infection with Eg101 or W956IC. Lower levels of STAT1 phosphorylation were detected in the PKR−/− extracts infected with either type of virus, suggesting that less IFN was produced by these cells than by infected control C57BL/6 cells (Fig. 6C). Analysis of secreted IFN-β by ELISA confirmed that PKR−/− MEFs produced less IFN-β after infection with either Eg101 or W956IC than infected control C57BL/6 MEFs at 24 and 32 h after infection (Fig. 6D). However, the levels of IFN-β were higher in W956IC-infected than in Eg101-infected PKR−/− MEFs. The kinetics of NF-κB nuclear translocation in response to a W956IC infection was next compared by confocal microscopy in C57BL/6 and PKR−/− cells infected with W956IC virus. The numbers of PKR−/− and control cells containing nuclear p65 NF-κB were similar at both 16 and 20 h after infection with W956IC (Fig. 6E). The data suggest that even though W956IC infection upregulates and activates PKR, which does contribute to increased IFN-β production, NF-κB is activated by another pathway in WNV-infected MEFs.
Analysis of NF-κB activation and IFN-β production in RIG-I−/−, MDA5−/−, IPS-1−/−, and wild-type C57BL/6 MEFs infected with Eg101 or W956IC WNV.
The cytoplasmic RNA sensors RIG-I and MDA5 were previously shown to participate in the establishment and maintenance of the innate antiviral response to WNV infection (37, 38). IPS-1 transmits the signal from either of these activated sensors to the TFs needed for type I IFN mRNA transcription and was previously shown to be essential for IFN-β production in WNV-infected MEFs, myeloid dendritic cells (DCs), macrophages, and neurons (25, 39). The data shown in Fig. 4A indicate that among the three TFs that activate IFN-β transcription, NF-κB showed a greater increase in activation as indicated by higher levels of phosphorylation and degradation of IkBα in W956IC-infected MEFs than in Eg101-infected MEFs. Although the mechanisms by which different RNA sensors and TFs control WNV infections are currently under active investigation (40), involvement of RIG-I and MDA5 in activating NF-κB in WNV-infected cells has not previously been reported. To determine whether RIG-I or MDA5 contributes to the observed differential activation of NF-κB, RIG-I+/+ (WT), RIG-I−/−, MDA5−/−, and IPS-1−/− MEFs were infected with Eg101 or W956IC WNV, and the numbers of infected cells containing nuclear p65 were quantified by confocal microscopy. At both 16 and 24 h after infection with W956IC, the number of cells in C57BL/6, RIG-I−/−, and MDA5−/− cultures that contained nuclear NF-κB was significantly higher than that in cultures infected with Eg101 (Fig. 7A and B). In contrast, p65 was predominantly cytoplasmic in IPS-1−/− cells infected with either virus at 16 h after infection, and only a few W956IC-infected IPS-1−/− cells had nuclear p65 (<3%) at 24 h after infection (Fig. 7A and B). The levels of extracellular IFN-β induced in each of the MEF cell lines by Eg101 or W956IC WNV infection were next assessed by ELISA. High levels of IFN-β (300 to 400 pg/ml) were produced by W956IC-infected C57BL/6, RIG-I−/−, and MDA5−/− MEFs at 24 h after infection, while no increase in IFN-β over the level produced by mock-infected cells was detected in infected IPS-1−/− culture fluids (Fig. 7C). The effect of the absence of RIG-I, MDA5, or IPS-1 on virus yields was also assessed. The yields of the Eg101 and W956IC viruses produced by control C57BL/6 MEFs were similar to those produced by MDA5−/− MEFs, while W956IC titers at 24 h were slightly higher in RIG-I−/− MEFs. However, only IPS-1−/− cells produced higher yields of Eg101 at 16 h and of W956IC at both 16 and 24 h (Fig. 7D). The results indicate that IPS-1 is required for efficient activation of NF-κB in response to both Eg101 and W956IC infections and that either RIG-I or MDA5 or an unknown sensor can mediate this activation. These results also confirm the conclusion made from the results of the experiment with neutralizing IFN antibody (Fig. 2D) that the lower yields of W956IC observed are due to the higher early levels of IFN-β induced by W956IC infections.
Fig 7.
Analysis of IPS-1 dependence of NF-κB activation and IFN-β induction in WNV Eg101- and W956IC-infected MEFs. (A) C57BL/6 (WT), RIG-I−/−, MDA5−/−, and IPS-1−/− MEFs were infected with WNV Eg101 or W956IC at an MOI of 1. Laser scanning confocal microscopy of WNV-infected cells stained with anti-dsRNA (red), anti-NF-κB antibody (green), and Hoechst 33258 dye (blue) was performed at the indicated times after infection. (B) Quantification of the percentage of infected cells with nuclear p65 localization was done at the indicated times after infection. At least three fields of cells for each condition were counted, and average values were plotted. Bars represent ± standard deviations. (C) The levels of IFN-β protein in the infected MEF culture fluids were determined by ELISA at the indicated times after infection. The values are averages of results from replicate assays done on samples from two independent experiments. Bars represent ± standard deviations. (D) Viral titers in culture fluids were determined by plaque assay on BHK cells. The values shown are averages of duplicate titrations of samples from two independent experiments. Bars represent ± standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05).
Analysis of the kinetics of viral RNA amplification in cells infected with Eg101 or W956IC WNV.
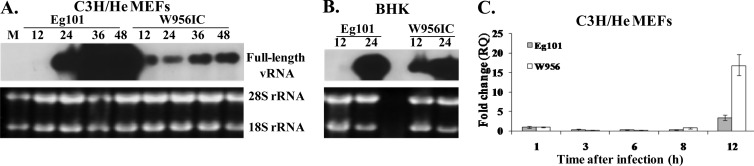
Since RIG-I and MDA5 sense viral dsRNA, the kinetics and efficiency of viral genome RNA amplification were next compared by Northern blotting of total RNA extracted from Eg101- and W956IC-infected, IFN-competent C3H/He and IFN-incompetent BHK cells. Since the sequences of the 3′ 1,496 nt of the W956IC and Eg101 genomes are identical, a DNA probe complementary to the 3′ terminal 800 nt of the Eg101 genome was used to detect intracellular genome RNA of both viruses. Viral RNA was detected at 12 h after W956IC infection in both types of cells but was not detected until 24 h in cells infected with Eg101 (Fig. 8A and B). Previously reported real-time qRT-PCR data (12) also showed higher levels of intracellular W956IC RNA than Eg101 RNA at 12 h after infection in BHK cells. From 24 h after infection with either virus, high levels of intracellular viral RNA were observed in infected BHK cells. In contrast, in C3H/He MEFs, intracellular W956IC RNA levels were significantly lower than Eg101 RNA levels at 24 h after infection and increased only slightly with time after infection (Fig. 8A), consistent with the differential extracellular virus yields produced by these two infections in C3H/He MEFs (Fig. 1B). The viral RNA levels were next quantified by real-time qRT-PCR at earlier times after infection in Eg101- and W956IC-infected C3H/He MEFs. No significant fold change over the amount of viral RNA present at 1 h after infection was detected at 3, 6, or 8 h after infection. However, at 12 h, W956IC RNA increased by ∼15-fold while Eg101 increased by only ∼3-fold (Fig. 8C), confirming the results obtained in the Northern blot analysis. These results indicate that although W956IC viral RNA amplification was more efficient by 12 h after infection in both IFN-deficient and IFN-competent cells, only in the IFN-competent cells was the efficiency of the subsequent exponential amplification of W956IC viral RNA significantly suppressed.
Fig 8.
Detection of viral RNA in C3H/He MEFs and BHK cells infected with Eg101 or W956IC WNV. Northern blot analysis of total cellular RNA from C3H/He (A) and BHK (B) cells infected with WNV Eg101 or W956IC at an MOI of 5 was performed for the indicated periods of time. Total cell RNA (5 μg) was separated on a 1% denaturing agarose gel and transferred to a Hybond-XL membrane. The blot was hybridized to a 32P-labeled DNA probe specific for the 3′ untranslated region of the WNV genomic RNA. (C) Relative quantification of WNV genomic RNA by real-time qRT-PCR. Intracellular viral RNA levels are expressed as the fold change compared to the level of viral RNA at 1 h after infection and normalized to GAPDH mRNA in the same RNA sample. Each sample was assayed in triplicate. Error bars represent the SE of the mean. Representative data from one of three independent experiments are shown.
Comparison of the relative efficiencies of early viral RNA replication and IFN-β induction by additional chimeric WNV viruses.
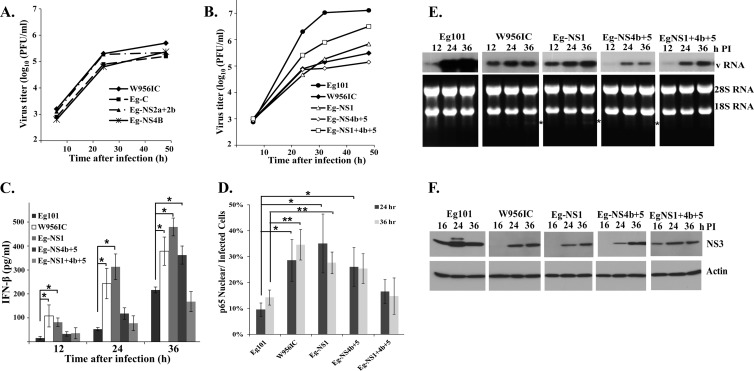
Several flavivirus proteins have previously been reported to counteract the type I IFN response. Also, single nucleotide mutations in the WNV NS2a and NS4b genes were previously reported to affect both the efficiency of viral replication and IFN induction (32, 41–44). A set of chimeric viruses was previously made by replacing one or more of the gene regions in the lineage 2 W956IC backbone with the lineage 1 Eg101 equivalent and used to map the region(s) of the W956IC genome involved in efficient early RNA replication (12). In BHK cells, low early RNA levels similar to those seen in Eg101-infected cells were observed with W956IC chimeras in which the NS1, NS3, and NS4a (NS1+3+4a), NS1+4b+5, or NS4b+5 regions were from Eg101 (12). A subset of these chimeras was used to further analyze the correlation between the level of early intracellular viral RNA and the level of IFN-β induction in IFN-competent C3H/He MEFs. The parental W956IC chimera already contains the C-terminal part of NS5 and the 3′ noncoding region (NCR) from Eg101. Chimeras that also contained the capsid gene (Eg-C), the NS2a and NS2b genes (Eg-NS2a+2b), or the N-terminal part of NS4b (Eg-NS4b) from Eg101 produced growth curves similar to the growth curve of W956IC (Fig. 9A), indicating that none of the amino acid differences between Eg101 and W956IC in these genes accounted for the higher virus yields produced by Eg101 in C3H/He MEFs. The chimera containing the NS4b and the complete NS5 gene from Eg101 (Eg-NS4b+5) produced lower virus yields than the parental W956IC virus (Fig. 9B), and the chimera containing the NS1 gene from Eg101 (Eg-NS1) produced a slightly higher peak titer than W956IC. Only the chimera containing a combination of NS1, NS4b, and complete NS5 gene regions from Eg101 (Eg-NS1+4b+5) produced significantly higher virus titers at all times than W956IC, but these titers were still lower than those produced by Eg101. IFN-β production by cells infected with Eg101, W956IC, or one of the chimeric viruses was next measured by ELISA. As expected, significantly larger amounts of IFN-β were produced by cells infected with W956IC than by cells infected with Eg101 at all times tested (Fig. 8C). The levels of IFN-β produced by the Eg-NS1 chimera were similar to or higher than those produced by W956IC (Fig. 9C). Although the amounts of IFN-β produced by the Eg-NS4b+5 chimera were initially low, like those from Eg101-infected cells (at 12 and 24 h after infection), the levels of IFN-β produced at 36 h after infection were high, similar to those produced by W956IC virus. The Eg-NS1+4b+5 chimera produced much smaller amounts of IFN-β than W956IC, similar to Eg101 (Fig. 9C). Quantification of the number of chimera-infected cells with nuclear p65 by confocal microscopy showed that only the Eg-NS1+4b+5 chimera induced low NF-κB activation similar to that produced by Eg101 (Fig. 9D). The results indicated that lower IFN-β production correlated with lower NF-κB activation.
Fig 9.
Relative replication efficiency and extent of IFN-β induction by additional WNV chimeric viruses. (A and B) C3H/He MEFs were infected with Eg101 or W956IC or another chimera at an MOI of 1, the supernatants were collected at the indicated times after infection, and the viral titers were determined by plaque assay on BHK cells. The values shown are averages of duplicate titrations from two experiments. (C) The levels of IFN-β protein in supernatants from infected C3H/He MEFs were determined by ELISA. The values shown are averages from two or more independent experiments performed in duplicate. Bars represent ± standard deviations. Asterisks indicate values that were statistically significant (*, P < 0.05). (D) Quantification of the percentage of infected cells with nuclear p65 localization was done at the indicated times after infection. At least three fields for each condition were counted, and average values were plotted. Bars represent ± standard deviations. Asterisks indicate values that were statistically significant (*, P < 0.05; **, P < 0.005). (E) Northern blot analysis of total cellular RNA from C3H/He MEFs infected with Eg101, W956IC, or another chimera. Cells were infected at an MOI of 1 for the indicated times. Total cell RNA (5 μg) was separated on a 1% denaturing agarose gel and transferred to a Hybond-XL membrane. The blot was hybridized to a 32P-labeled DNA probe specific for the 3′ untranslated region of the WNV genomic RNA. rRNA detected by ethidium bromide staining was used as a loading control. (F) C3H/He MEFs were infected with WNV Eg101, W956IC, or another chimera at an MOI of 1. At the indicated times after infection, cell lysates were prepared, and proteins were separated, transferred, and assayed by Western blotting using anti-WNV NS3 antibody. Actin was used as the loading control. The blots shown are representative of results obtained from three independent experiments.
To assess whether the amounts of viral RNA produced at early times after chimeric virus infection in C3H/He cells correlated with the levels of IFN-β produced, the levels of intracellular viral RNA were assessed by Northern blotting at 12, 24, and 36 h after infection. At 12 h after infection, viral RNA levels were higher in cells infected with W956IC or Eg-NS1 than in Eg101-infected cells (Fig. 9E). Cells infected with Eg-NS4b+5 or Eg-NS1+4b+5 virus produced low levels of viral RNA at 12 h after infection, similar to Eg101. Also similar to Eg101, these two chimeras produced low levels of IFN at initial times of infection (Fig. 9C). However, even though the levels of intracellular Eg-NS1+4b+5 chimera RNA at later stages of infection were similar to those of W956IC (Fig. 9E), this chimera produced low levels of IFN-β, similar to Eg101, throughout the course of infection (Fig. 9C).
Intracellular viral protein levels were also assessed by Western blotting of NS3 at 16, 24, and 36 h after infection of C3H/He MEFs with W956IC, Eg101, or a chimeric virus. NS3 was detected by 16 h after infection with Eg101 but was not detected until 24 h in W956IC-infected cells (Fig. 9F). Also, NS3 levels were higher in cells infected with Eg101 at all times tested than in cells infected with W956IC. In cells infected with either Eg-NS1 or Eg-NS4b+5 chimeric viruses, NS3 was not detected at 16 h after infection, and viral protein levels were lower than those in Eg101-infected cells at 24 and 36 h (Fig. 9F). However, NS3 was detected at 16 h after infection with the Eg-NS1+4b+5 chimeric virus, similar to what was observed in Eg101-infected cells. This chimera was the only one that produced low levels of IFN-β, similar to Eg101, at all the times analyzed. It was previously reported that cleavage of 28S rRNA by RNase L was an indicator of sustained antiviral activity at late stages of a WNV infection (19). A characteristic rRNA cleavage band (indicated by an asterisk in Fig. 9E) was observed at 36 h after infection in cells infected with the W956IC, Eg-NS1, or Eg-NS4b+5 virus but not in cells infected with the Eg101 or Eg-NS1+4b+5 virus. The detection of this band indicates a high level of upregulation of active Oas proteins, which are ISGs, and production of 2′-5′ oligo(A) by these proteins, leading to high levels of activation of RNase L.
DISCUSSION
A previous study from our lab reported that W956IC infections in IFN-incompetent BHK cells produced higher levels of intracellular viral RNA at early times after infection than natural strains of WNV, but both types of infections produced similar peak yields of extracellular virus (12). Data from the present study showed that in IFN-competent MEFs, W956IC infections produced higher levels of early intracellular viral RNA but also higher levels of IFN-β and lower virus yields than an infection with the natural WNV strain Eg101. The importance of type 1 IFNs in controlling WNV infections has been previously documented (45–47). IFN-β−/− mice are more susceptible to WNV infection, displaying increased mortality, enhanced viral loads, and broader virus tissue tropism (47). Recognition of viral components by cellular sensors results in the coordinated assembly of the IRF-3, ATF-2/c-Jun, and NF-κB TFs on the IFN-β promoter (34). Although multiple studies have characterized the induction of the antiviral state in WNV-infected cells by assessing type I IFN expression, IRF-3 phosphorylation/nuclear translocation, and the expression levels of ISGs (mostly IRF-3-dependent ISGs) (20, 25, 37, 48–50), the involvement of the NF-κB pathway in the establishment of the antiviral state in WNV-infected cells was not previously assessed. Data from the present study showed that at early stages of a WNV infection (before 16 h after infection) when IRF-3 activation was still very low, higher levels of NF-κB activation were detected in MEFs infected with the chimeric W956IC virus than in MEFs infected with the natural WNV strain Eg101 and correlated with significantly higher levels of IFN-β production. Also, the induction of mRNA expression from the IFN-β gene by a W956IC infection at early times after infection was significantly decreased in p65−/− cells, supporting the involvement of this transcription factor in IFN-β gene regulation. However, the levels of IFN-β mRNA were still higher in W956IC-infected p65−/− MEFs than in Eg101-infected MEFs, suggesting that additional TFs are also involved in early IFN-β gene regulation in W956IC-infected cells.
Previous studies on the role of NF-κB in mediating type I IFN induction in response to dsRNA or to various types of RNA virus infections produced contradictory data. Initial studies with Newcastle disease virus (NDV) and vesicular stomatitis virus (VSV) found no role for NF-κB in IFN-β induction (51, 52), while later studies reported a critical role for the p65 subunit in early IFN-β induction and cellular resistance to NDV and VSV replication (53). During the early phase of IFN-β induction in uninfected cells when the levels of activated IRF-3 are low (53), p65 was reported to be important for maintaining autocrine IFN-β signaling, for directly regulating a set of ISGs, and for protecting infected cells from dsRNA/IFN-induced necrotic death (54). It was also previously reported that the production of type 1 IFN was significantly decreased in Japanese encephalitis virus (JEV)-infected cells defective in the NF-κB pathway (55) and that IFN-β production by JEV- and dengue-2 virus-infected cells was reduced when NF-κB activation was blocked by MG132 treatment (56). Although the involvement of PKR in the activation of the NF-κB pathway (17) and in the induction of IFN-β (36) in WNV-infected cells was previously reported, the data from the present study showed that PKR is not essential for the initial induction of IFN-β in response to WNV infection but does play a role in amplifying IFN-β production at later stages of infection. IPS-1 and both of the upstream cytoplasmic RNA sensors RIG-I and MDA5 were previously shown to be involved in mediating the innate antiviral response to a WNV infection (37–39). The data from the present study indicate that although either RIG-I, MDA5, or an unknown sensor can mediate NF-κB activation and IFN-β induction, IPS-1 is required for early viral RNA-mediated signaling leading to NF-κB activation and IFN-β induction in MEFs infected with W956IC. NF-κB activation in cells infected with other flaviviruses, JEV or dengue-2 virus, was previously reported to be dependent on the phosphatidylinositol 3-kinase (PI3K) pathway (56). Although an initial study reported that NF-κB could be activated downstream of IPS-1 in a classical IκB kinase β (IKKβ)-dependent but TRAF6-, TBK1-, and inducible IKK (IKKi)-independent manner (14), a subsequent study demonstrated that activation of NF-κB, Jun N-terminal protein kinase (JNK), and p38, but not IRF-3, was impaired in TRAF6 knockout MEFs in response to VSV infection or poly(I·C) transfection and identified TRAF6 as a critical factor for IFN-α/β induction in response to dsRNA (57). However, TRAF6 was previously shown to modulate only the late phase of an IPS-1-dependent, type I IFN response in WNV-infected MEFs, whereas TRAF3 and TBK1 regulated the early response (25). Since a number of known and yet to be identified IPS-1 downstream molecules are expected to cross talk in mediating the signal to IFN-β and ISGs, further studies are needed to fully elucidate the mechanisms of induction of the antiviral state in WNV-infected cells. The results of a recent study from our lab showed that the initial upregulation of a set of ISGs in MEFs infected with the natural WNV strain Eg101 was only partially dependent on IPS-1 and that upregulation of these ISGs was independent of IFN-β both at early and late times of infection (18). The present study revealed a role for IPS-1 in mediating NF-κB activation and IFN-β induction in cells infected with W956IC virus. Since natural strains of WNV are able to keep early RNA replication at low levels, infections with these viruses do not activate NF-κB to the same extent as a W956IC infection, and infections with natural strains of WNV induce type I IFN gene expression to much lower levels. Previous studies have shown that although flavivirus infections rapidly induce a type I IFN response, various flavivirus proteins block type 1 signaling. Expression of dengue virus nonstructural proteins NS2A, NS4A, and NS4B and the Kunjin virus NS2A, NS2B, NS3, NS4A, and NS4B proteins was shown to inhibit STAT signaling (31, 42, 58). The NS4B of WNV and yellow fever virus (YFV) was reported to block STAT1 activation and ISG induction (31, 32). The NS5 protein of several flaviviruses including West Nile virus has been reported to be a potent suppressor of IFN-mediated JAK-STAT signaling (30, 59, 60). In human cells, the N-terminal residues of NS5 in JEV and dengue virus appear to block Tyk2 phosphorylation and degrade STAT2, respectively (27, 60, 61). Data from several studies have shown that counteraction of the host type I IFN response by flavivirus proteins differs between human and mouse cells. For example, dengue virus is able to counteract human but not mouse STAT2 and STING (27–29). Previous studies reported that an A30P substitution in the Kunjin WNV NS2A protein abrogated its ability to inhibit IFN signaling and attenuated rather than enhanced its virulence in mice (41). Also, infections with TX02 WNV with a D73H mutation in the NS2A protein were reported to result in reduced viremia and a decreased IFN response, while an E249G substitution in either the NY00 or TX02 WNV NS4B protein resulted in lower viral replication in rodent cells (44). Although the majority of the determinants of mouse neuroinvasiveness and neurovirulence previously identified for WNV were located in the E protein (5, 62), a C102S substitution in the NY99 WNV NS4B protein attenuated both neuroinvasiveness and neurovirulence (63). Alignment of the Eg101 and W956IC amino acid sequences showed that none of the previously identified critical residues discussed above differed between the Eg101 and W956IC virus genomes; both genomes had D73 in NS2A, E249 in NS4B, and C102 in NS4B. Therefore, these changes could not account for the observed differences between Eg101 and W956IC in the efficiency of virus replication and IFN induction observed in MEFs.
A sufficient amount of viral protein is required to effectively block IFN signaling initiated during the early phase of the viral replication cycle. The genome of a flavivirus serves as the template for minus-strand RNA synthesis and is the only viral mRNA for viral protein translation. At early times of infection when the number of viral genomes is low, genomes must switch between translation and replication. Natural strains of WNV show a preference for translation over RNA replication at early times after infection. Lower RNA replication and higher translation would have two beneficial effects for the virus in relation to the type I IFN response. Lower levels of early viral RNA would reduce the extent of activation of cytoplasmic RNA sensors leading to IFN production, while higher levels of viral protein would not only enhance the remodeling of endoplasmic reticulum (ER) membranes to form protective replication complex vesicles but also provide more efficient blockage of any canonical IFN signaling that occurred. The data obtained with the additional chimeras in the present study showed that downregulation of early viral RNA replication consistently correlated with enhancement of early viral protein translation and vice versa. Exactly how flaviviruses differentially regulate their early translation and replication functions is not known. One possibility is that interactions between viral nonstructural proteins and possibly also between viral nonstructural proteins and cell proteins regulate genome switching between translation and replication. A previous study from our lab showed that only three of the additional W956IC chimeras made, Eg-NS4b+5, Eg-NS1+4b+5, and Eg-NS1+3+4a, produced low levels of early viral RNA similar to WNV Eg101 (12). In the ORF of the original W956IC chimera, which produces high levels of early RNA, only the 287 C-terminal amino acids of NS5 are from Eg101. The finding that replacement of NS1 and NS4B from Eg101 produced a low early RNA phenotype when the entire NS5 was also from Eg101 but that replacement of NS1, NS3, and NS4A was required to give this phenotype when the NS5 was still a hybrid suggests that multiple interactions between the viral nonstructural proteins are needed to correctly regulate early viral RNA synthesis and template switching. The finding that NS1 is one of the nonstructural proteins involved is consistent with previous reports that NS1 plays a role in viral RNA synthesis and is essential for regulating minus-strand RNA synthesis (64, 65). A previous study reported a genetic interaction between NS1 and NS4A (66). However, a recent study reported genetic and physical evidence supporting an interaction between NS1 and NS4B but not between NS1 and NS4A (67). The authors of the recent study suggested that the interaction between NS1 and NS4B could be indirectly mediated by another viral or host protein.
A sustained negative effect on the extent of amplification of intracellular viral RNA was observed in W956IC virus-infected, IFN-competent cells but not in IFN-incompetent cells. It is not currently known how this is accomplished. One possibility is that this effect could be due to the lower levels of early viral protein resulting in inefficient or nonoptimal remodeling of the ER membrane, which would negatively impact the efficiency of subsequent exponential genome RNA amplification especially if there is an optimal window of opportunity for the generation of the viral replication vesicles in the ER. Alternatively, the effect could be due to the action of one or more of the ISGs that are expressed prior to complete blockage of the canonical IFN signaling pathway.
ACKNOWLEDGMENTS
This work was supported by Public Health Service research grant AI048088 to M.A.B from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by developmental grant U54 AI 057157 (SE-DP-001) to M.A.B from the Southeastern Regional Centers for Excellence for Biodefense and Emerging Infections. J.A.P.-P. and S.C.C. were supported by Molecular Basis of Disease Fellowships from Georgia State University.
Footnotes
Published ahead of print 15 May 2013
REFERENCES
- 1. Brinton MA. 2002. The molecular biology of West Nile virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371–402 [DOI] [PubMed] [Google Scholar]
- 2. Berthet FX, Zeller HG, Drouet MT, Rauzier J, Digoutte JP, Deubel V. 1997. Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J. Gen. Virol. 78:2293–2297 [DOI] [PubMed] [Google Scholar]
- 3. Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R, Bowen M, McKinney N, Morrill WE, Crabtree MB, Kramer LD, Roehrig JT. 2002. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298:96–105 [DOI] [PubMed] [Google Scholar]
- 4. Botha EM, Markotter W, Wolfaardt M, Paweska JT, Swanepoel R, Palacios G, Nel LH, Venter M. 2008. Genetic determinants of virulence in pathogenic lineage 2 West Nile virus strains. Emerg. Infect. Dis. 14:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beasley DW, Li L, Suderman MT, Barrett AD. 2002. Mouse neuroinvasive phenotype of West Nile virus strains varies depending upon virus genotype. Virology 296:17–23 [DOI] [PubMed] [Google Scholar]
- 6. Venter M, Human S, Zaayman D, Gerdes GH, Williams J, Steyl J, Leman PA, Paweska JT, Setzkorn H, Rous G, Murray S, Parker R, Donnellan C, Swanepoel R. 2009. Lineage 2 West Nile virus as cause of fatal neurologic disease in horses, South Africa. Emerg. Infect. Dis. 15:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Venter M, Swanepoel R. 2010. West Nile virus lineage 2 as a cause of zoonotic neurological disease in humans and horses in southern Africa. Vector Borne Zoonotic Dis. 10:659–664 [DOI] [PubMed] [Google Scholar]
- 8. Yamshchikov VF, Wengler G, Perelygin AA, Brinton MA, Compans RW. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294–304 [DOI] [PubMed] [Google Scholar]
- 9. Davis WG, Blackwell JL, Shi PY, Brinton MA. 2007. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 81:10172–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Basu M, Brinton MA. 2011. West Nile virus (WNV) genome RNAs with up to three adjacent mutations that disrupt long distance 5′-3′ cyclization sequence basepairs are viable. Virology 412:220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elbahesh H, Scherbik SV, Brinton MA. 2011. West Nile virus infection does not induce PKR activation in rodent cells. Virology 421:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Courtney SC, Scherbik SV, Stockman BM, Brinton MA. 2012. West Nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J. Virol. 86:3647–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 14. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 15. Baum A, Garcia-Sastre A. 2010. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids 38:1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131–137 [DOI] [PubMed] [Google Scholar]
- 17. Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811 [DOI] [PubMed] [Google Scholar]
- 18. Pulit-Penaloza JA, Scherbik SV, Brinton MA. 2012. Type 1 IFN-independent activation of a subset of interferon stimulated genes in West Nile virus Eg101-infected mouse cells. Virology 425:82–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scherbik SV, Stockman BM, Brinton MA. 2007. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. J. Virol. 81:12005–12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emeny JM, Morgan MJ. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247–252 [DOI] [PubMed] [Google Scholar]
- 22. Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 85:5259–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otsuki K, Maeda J, Yamamoto H, Tsubokura M. 1979. Studies on avian infectious bronchitis virus (IBV). III. Interferon induction by and sensitivity to interferon of IBV. Arch. Virol. 60:249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lam V, Duca KA, Yin J. 2005. Arrested spread of vesicular stomatitis virus infections in vitro depends on interferon-mediated antiviral activity. Biotechnol. Bioeng. 90:793–804 [DOI] [PubMed] [Google Scholar]
- 25. Daffis S, Suthar MS, Szretter KJ, Gale M, Jr, Diamond MS. 2009. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 5:e1000607. 10.1371/journal.ppat.1000607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M., Jr 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 80:9424–9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, Garcia-Sastre A. 2010. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe 8:410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 8:e1002934. 10.1371/journal.ppat.1002934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 84:3503–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo JT, Hayashi J, Seeger C. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 34. Thanos D, Maniatis T. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091–1100 [DOI] [PubMed] [Google Scholar]
- 35. Hayden MS, Ghosh S. 2004. Signaling to NF-κB. Genes Dev. 18:2195–2224 [DOI] [PubMed] [Google Scholar]
- 36. Gilfoy FD, Mason PW. 2007. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 81:11148–11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr 2008. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82:609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, Diamond MS, Gale M., Jr 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 6:e1000757. 10.1371/journal.ppat.1000757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Diamond MS, Gale M., Jr 2012. Cell-intrinsic innate immune control of West Nile virus infection. Trends Immunol. 33:522–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rossi SL, Fayzulin R, Dewsbury N, Bourne N, Mason PW. 2007. Mutations in West Nile virus nonstructural proteins that facilitate replicon persistence in vitro attenuate virus replication in vitro and in vivo. Virology 364:184–195 [DOI] [PubMed] [Google Scholar]
- 44. Puig-Basagoiti F, Tilgner M, Bennett CJ, Zhou Y, Munoz-Jordan JL, Garcia-Sastre A, Bernard KA, Shi PY. 2007. A mouse cell-adapted NS4B mutation attenuates West Nile virus RNA synthesis. Virology 361:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Daffis S, Suthar MS, Gale M, Jr, Diamond MS. 2009. Measure and countermeasure: type I IFN (IFN-alpha/beta) antiviral response against West Nile virus. J. Innate Immun. 1:435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lazear HM, Pinto AK, Vogt MR, Gale M, Jr, Diamond MS. 2011. Beta interferon controls West Nile virus infection and pathogenesis in mice. J. Virol. 85:7186–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Daffis S, Samuel MA, Suthar MS, Keller BC, Gale M, Jr, Diamond MS. 2008. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J. Virol. 82:8465–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fredericksen BL, Smith M, Katze MG, Shi PY, Gale M., Jr 2004. The host response to West Nile Virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737–7747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Daffis S, Samuel MA, Keller BC, Gale M, Jr, Diamond MS. 2007. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 3:e106. 10.1371/journal.ppat.0030106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peters KL, Smith HL, Stark GR, Sen GC. 2002. IRF-3-dependent, NFκB- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 99:6322–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X, Hussain S, Wang EJ, Li MO, Garcia-Sastre A, Beg AA. 2007. Lack of essential role of NF-κB p50, RelA, and cRel subunits in virus-induced type 1 IFN expression. J. Immunol. 178:6770–6776 [DOI] [PubMed] [Google Scholar]
- 53. Wang J, Basagoudanavar SH, Wang X, Hopewell E, Albrecht R, Garcia-Sastre A, Balachandran S, Beg AA. 2010. NF-κB RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J. Immunol. 185:1720–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Basagoudanavar SH, Thapa RJ, Nogusa S, Wang J, Beg AA, Balachandran S. 2011. Distinct roles for the NF-κB RelA subunit during antiviral innate immune responses. J. Virol. 85:2599–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Abraham S, Nagaraj AS, Basak S, Manjunath R. 2010. Japanese encephalitis virus utilizes the canonical pathway to activate NF-κB but it utilizes the type I interferon pathway to induce major histocompatibility complex class I expression in mouse embryonic fibroblasts. J. Virol. 84:5485–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang TH, Liao CL, Lin YL. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation. Microbes Infect. 8:157–171 [DOI] [PubMed] [Google Scholar]
- 57. Yoshida R, Takaesu G, Yoshida H, Okamoto F, Yoshioka T, Choi Y, Akira S, Kawai T, Yoshimura A, Kobayashi T. 2008. TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J. Biol. Chem. 283:36211–36220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79:5414–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chambers TJ, Halevy M, Nestorowicz A, Rice CM, Lustig S. 1998. West Nile virus envelope proteins: nucleotide sequence analysis of strains differing in mouse neuroinvasiveness. J. Gen. Virol. 79:2375–2380 [DOI] [PubMed] [Google Scholar]
- 63. Wicker JA, Whiteman MC, Beasley DW, Davis CT, Zhang S, Schneider BS, Higgs S, Kinney RM, Barrett AD. 2006. A single amino acid substitution in the central portion of the West Nile virus NS4B protein confers a highly attenuated phenotype in mice. Virology 349:245–253 [DOI] [PubMed] [Google Scholar]
- 64. Lindenbach BD, Rice CM. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232–240 [DOI] [PubMed] [Google Scholar]
- 66. Lindenbach BD, Rice CM. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 73:4611–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Youn S, Li T, McCune BT, Edeling MA, Fremont DH, Cristea IM, Diamond MS. 2012. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol. 86:7360–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]