Abstract
Herpes simplex virus 1 (HSV-1) facilitates virus entry into cells and cell-to-cell spread by mediating fusion of the viral envelope with cellular membranes and fusion of adjacent cellular membranes. Although virus strains isolated from herpetic lesions cause limited cell fusion in cell culture, clinical herpetic lesions typically contain large syncytia, underscoring the importance of cell-to-cell fusion in virus spread in infected tissues. Certain mutations in glycoprotein B (gB), gK, UL20, and other viral genes drastically enhance virus-induced cell fusion in vitro and in vivo. Recent work has suggested that gB is the sole fusogenic glycoprotein, regulated by interactions with the viral glycoproteins gD, gH/gL, and gK, membrane protein UL20, and cellular receptors. Recombinant viruses were constructed to abolish either gM or UL11 expression in the presence of strong syncytial mutations in either gB or gK. Virus-induced cell fusion caused by deletion of the carboxyl-terminal 28 amino acids of gB or the dominant syncytial mutation in gK (Ala to Val at amino acid 40) was drastically reduced in the absence of gM. Similarly, syncytial mutations in either gB or gK did not cause cell fusion in the absence of UL11. Neither the gM nor UL11 gene deletion substantially affected gB, gC, gD, gE, and gH glycoprotein synthesis and expression on infected cell surfaces. Two-way immunoprecipitation experiments revealed that the membrane protein UL20, which is found as a protein complex with gK, interacted with gM while gM did not interact with other viral glycoproteins. Viruses produced in the absence of gM or UL11 entered into cells more slowly than their parental wild-type virus strain. Collectively, these results indicate that gM and UL11 are required for efficient membrane fusion events during virus entry and virus spread.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) encodes at least 11 glycoproteins as well as several membrane-associated proteins which play important roles in viral entry and virus-induced cell fusion. Virus-induced cell fusion is apparent in herpetic lesions and is thought to facilitate virion transmission to adjacent cells without exposure to the host humoral immune system, particularly neutralizing antibody (reviewed in reference 1). Certain mutations in the UL20 gene (2–4), the UL24 gene (5, 6), the UL27 gene encoding glycoprotein B (gB) (7, 8), and the UL53 gene coding for gK (9–14) drastically enhance virus-induced cell fusion (syncytial or syn mutations). Recently, it has been suggested that gB is the sole fusogenic glycoprotein while glycoproteins gD and gH/gL are required to activate gB's fusogenicity in conjunction with specific cellular receptors (15). In this membrane fusion model, binding of gD to its cognate receptors, including nectin-1, herpesvirus entry mediator (HVEM), and other receptors (16–22), is thought to trigger sequential conformational changes in gH/gL and gB, causing fusion of the viral envelope with cellular membranes during virus entry as well as fusion among cellular membranes (23, 24). Extensive membrane fusion can be induced by coexpressing glycoproteins gB, gD, and gH/gL in cell lines (25, 26), suggesting that these glycoproteins are sufficient for membrane fusion. However, virus-induced cell fusion is regulated by a number of other viral proteins, since wild-type viruses cause a limited amount of fusion (27) and a lack of either glycoprotein gK or the membrane protein UL20 severely inhibits membrane fusion (4, 28).
We have shown that HSV-1 gK and UL20 functionally and physically interact and that these interactions are absolutely necessary for their coordinate intracellular transport, cell surface expression, and membrane fusion functions in the HSV-1 life cycle (28, 29). Furthermore, we have shown that a peptide comprised of the amino-terminal 82 amino acids of gK (gKa) expressed in trans complemented gB-mediated cell fusion and may physically interact with gB and gH in infected cells (30). These results suggest that gB-mediated virus-induced cell fusion is regulated via direct interactions with gK and UL20 (30, 31).
Glycoprotein gM is a conserved type III integral membrane protein with multiple transmembrane domains that forms a complex with pUL49.5 (gN) (reviewed in reference 1). Deletion of the gM gene does not abrogate HSV-1 replication but inhibits the ability of the virus to spread (32). gM expression causes relocalization of several membrane proteins from the cell surface to the trans-Golgi network (TGN) (33, 34). Thus, gM may function to retain viral glycoproteins at the TGN or retrieve them from the plasma membrane to the TGN (32). Expression of HSV-1, pseudorabies virus (PRV), and Kaposi's sarcoma-associated herpesvirus (KSHV, or human herpesvirus 8 [HHV-8]) gM and gN in transfected cells inhibited cell fusion caused by simultaneous expression of glycoproteins gB, gD, gH, and gL, suggesting that gM/gN may modulate membrane fusion (34, 35). Also, lack of gM was reported to inhibit virus-induced cell fusion caused by a single amino acid substitution in the carboxyl terminus of gB (A855V; gBsyn) (36, 37).
UL11 is a 96-amino-acid myristoylated and palmitoylated tegument protein anchored into the cytoplasmic side of cell membranes (32, 38). UL11 has been suggested to play a role in recruiting viral proteins to the virion assembly site at the TGN (32). UL11 is known to interact with UL16 and gE through its N-terminal (39–41) and C-terminal (42) domains, respectively. Although absence of UL11 in HSV and PRV revealed only moderate defects in viral replication, the human cytomegalovirus (HCMV, or HHV-5) UL11 homologue is essential for virus replication (32). HSV-1 UL11 was recently shown to form a protein complex with gE, UL16, and UL21 that may be required for efficient virus spread (43).
Recently, we utilized mutant viruses lacking one or more viral genes to show that the deletion of either the gK or UL20 gene produced significantly greater defects in virion envelopment and overall virus replication than deletion of the carboxyl terminus of either gD, UL11, gM, or gE alone or in various combinations (44). Herein, we investigated whether the lack of either gM or UL11 affected the ability of dominant syncytial mutations in either gB or gK to cause extensive virus-induced cell fusion. We found that both gM and UL11 are required for virus-induced cell fusion. Moreover, mutant viruses lacking either gM or UL11 exhibited slower kinetics of entry into Vero cells than the parental virus, suggesting that gM and UL11 are involved in membrane fusion phenomena during both virus-induced cell fusion and virus entry.
MATERIALS AND METHODS
Cells, antibodies, and plasmids.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Rockville, MD). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies-Gibco, Carlsbad, CA), supplemented with 10% fetal calf serum (FCS; Life Technologies-Gibco, Carlsbad, CA) and Primocin antibiotic (InvivoGen, San Diego, CA). Antibodies used include anti-HSV-1 gB, gC, gD, gE, and ICP5 (VP5) monoclonal antibodies (MAbs) (Virusys, Sykesville, MD), Alexa Fluor 488-conjugated goat anti-mouse IgG (Life Technologies-Molecular Probes, Carlsbad, CA), anti-HSV-1 gH MAb (Abcam, Cambridge, MA), anti-FLAG MAb (Sigma, St. Louis, MO), rabbit anti-HSV-1 gM polyclonal antibody (pAb) (a gift from Joel Baines, Cornell University, Ithaca, NY), and rabbit anti-HSV-1 UL11 pAb (a gift from John Wills, Pennsylvania State University, Hershey, PA). For transient-complementation experiments, HSV-1(F) UL10 (gM) and UL11 genes were cloned into pCDNA3.1 plasmid (Life Technologies-Invitrogen, Carlsbad, CA) and named pCgM and pCUL11, respectively.
Construction of HSV-1 mutant viruses.
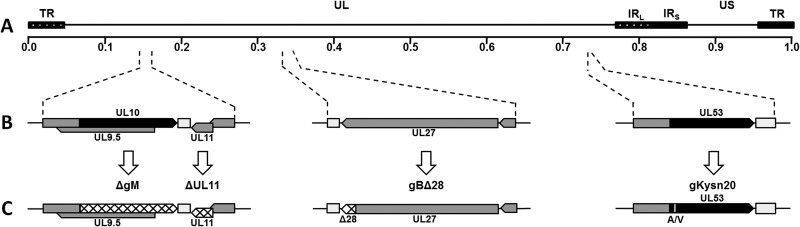
Mutagenesis was accomplished in Escherichia coli using the markerless two-step Red recombination mutagenesis system using synthetic oligonucleotides (45, 46), implemented on the bacterial artificial chromosome (BAC) plasmid pYEbac102 carrying the HSV-1(F) genome (47) (a kind gift from Y. Kawaguchi, University of Tokyo, Tokyo, Japan). Construction of the HSV-1 mutants ΔgM2 (ΔgM) and ΔUL11 was described previously (44). Briefly, the ΔgM recombinant virus was constructed by altering two potential initiation codon sites (from ATG to CTG and from ATG to ATT, respectively) located 57 bp apart at the beginning of the UL10 open reading frame (ORF) (48) (Fig. 1). The ΔUL11 virus was constructed by changing the initiation codon from ATG to CTG. The gBΔ28 recombinant virus was produced by introducing a stop codon causing truncation of gB by 28 amino acids. The gKsyn20 recombinant virus was constructed by introducing a point mutation (Ala to Val) at gK amino acid position 40. The ΔgM and ΔUL11 mutant viruses were used as the backbone for construction of the double mutants gBΔ28/ΔgM, gKsyn20/ΔgM, gBΔ28/ΔUL11, and gKsyn20/ΔUL11 by introducing the designated mutations (Fig. 1).
Fig 1.
Schematic representation of mutant viruses. (A) Prototypic arrangement of the HSV-1(F)-YE102 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) Relative genomic positions and gene arrangements of targeted genes encoding the viral glycoproteins gB (UL27), gM (UL10), and gK (UL53) and membrane protein UL11. (C) Schematic representation of engineered mutations. Mutant viruses containing the ΔgM and ΔUL11 mutations were produced by changing the initiation codons of each gene (hatched regions). Mutant viruses containing gBΔ28 were produced by introduction of a stop codon causing truncation of gB by 28 amino acids (hatched region in gB). Mutant viruses containing gKsyn20 were produced by a point mutation (Ala to Val) at gK amino acid position 40 (white vertical line in gK).
Confirmation of the targeted mutations and recovery of infectious viruses.
HSV-1 BAC DNAs were purified from 50 ml of overnight BAC cultures with the Qiagen large-construct kit (Qiagen, Valencia, CA). Using PCR test primers designed to lie outside the target mutation site(s), all mutated DNA regions were sequenced to verify the presence of the desired mutations in BACs. Viruses were recovered from cells transfected with BAC plasmids as we have described previously (45), and mutations were confirmed by DNA sequencing. To further validate the recombinant viruses, the entire genomes of recovered viruses from BACs were sequenced via Ion Torrent next-generation sequencing (Life Technologies-Invitrogen, Carlsbad, CA). Briefly, total genomic DNA (gDNA) was extracted from the virus-infected Vero cells using the PureLink genomic DNA minikit (Life Technologies-Invitrogen, Carlsbad, CA). High-quality fragment libraries of each virus were prepared from the extracted total gDNAs using the Ion Xpress Plus fragment library kit (Life Technologies-Invitrogen, Carlsbad, CA). The fragment libraries were subsequently applied to Ion 316 chips and were analyzed on the Ion Personal Genome Machine system (Life Technologies-Invitrogen, Carlsbad, CA).
Plaque morphology of mutant viruses on Vero and transiently complementing cells.
Visual analysis of plaque morphology of mutant viruses was performed as we have previously described (28, 45, 49, 50). Confluent monolayers of Vero cells were transfected with 1 μg of gM- or UL11-expressing plasmid (one well of a 12-well tissue culture plate) using Lipofectamine 2000 transfection reagent (Life Technologies-Invitrogen, Carlsbad, CA). At 24 h posttransfection, cells were infected at a multiplicity of infection (MOI) of 0.0001, with each mutant virus lacking gM or UL11. At 48 h postinfection (hpi), the infected cells were fixed with ice-cold methanol. Plaques were visualized after immunohistochemical staining with polyclonal anti-HSV rabbit sera as described earlier (30). More than 50 independent viral plaques were examined to assess relative levels of syncytium formation and select individual plaques as representative plaques for each experiment.
One-step viral growth kinetics.
Analysis of one-step growth kinetics was performed as we have described previously (29, 51). Briefly, nearly confluent Vero cell monolayers in 12-well plates were infected with each virus in triplicate at either a low MOI (0.2) or high MOI (3.0) for 1 h at 4°C. Thereafter, plates were incubated at 37°C and 5% CO2, and virus was allowed to penetrate for 1 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (pH 3.0) for 10 to 15 s, and the plates were incubated at 37°C and 5% CO2. At 12 and 24 hpi, virus stocks were prepared and viral titers were calculated by endpoint titration on Vero cells. Viral plaques were counted after immunohistochemical staining with rabbit anti-HSV-1 pAb as previously described (30).
SDS-PAGE and Western immunoblot assay.
Western immunoblot analysis was carried out essentially as described earlier (30). Briefly, confluent Vero cell monolayers were infected with the indicated virus at an MOI of 3. At 24 hpi, cells were collected by low-speed centrifugation, washed twice with ice-cold phosphate-buffered saline (PBS), and lysed with NP-40 lysis buffer (Life Technologies-Novex, Carlsbad, CA). The collected samples were mixed with SDS-PAGE sample buffer (Bio-Rad, Hercules, CA) at a 1:1 ratio and were electrophoretically separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Tris-HEPES-SDS gradient 4 to 20% gels; Thermo Scientific, Waltham, MA). Following electrophoresis, four identical gels were transferred to a nitrocellulose membrane under a constant current. Membranes were blocked in Tris-buffered saline containing 0.1% Tween 20 (TBST) plus 5% nonfat milk for 1 h at room temperature and were probed with primary MAbs overnight at 4°C. Goat anti-mouse secondary antibody conjugated with horseradish peroxidase (HRP) and enhanced chemiluminescence (ECL) (GE Healthcare, Little Chalfont, United Kingdom) substrate were used for detection purposes.
Quantification of cell-to-cell fusion.
A luciferase-based cell-to-cell fusion assay was performed essentially as described earlier (30). Briefly, a subconfluent monolayer of Vero cells in a 6-well plate was transfected with 2 μg of plasmid containing either the T7 polymerase gene under the cytomegalovirus (CMV) promoter (effector cells) or the T7-dependent luciferase gene (target cells) by using Lipofectamine 2000 transfection reagent. As positive and negative controls, cells were transfected with both of the plasmids simultaneously and with pCAGGS empty vector plasmid, respectively. Twelve hours after each transfection, the two different cell populations, effector and target cells or negative cells and target cells, were detached and reseeded to a new 12-well plate at a 1:1 ratio for experimental groups and negative-control groups, respectively. At 24 h posttransfection, cells were infected with wild-type and mutant viruses at an MOI of 0.2. At 12 and 24 hpi, cells were washed twice with ice-cold PBS and lysed with passive lysis buffer (Promega, Madison, WI). The lysates were clarified by centrifugation at 10,000 × g for 5 min at 4°C and were reacted with luciferase substrate (Promega, Madison, WI). The intensity of luciferase activity was measured by a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) with a 5-s delay and 10-s read.
Analysis of membrane-associated proteins.
Biotinylation of cell surface proteins was used to identify membrane-associated proteins of Vero cells infected with each of the designated viruses. Briefly, surface proteins of Vero cells infected with the designated viruses at an MOI of 3 were biotinylated at 12 hpi and isolated with a Pierce cell surface protein isolation kit (Thermo Scientific, Waltham, MA). The isolated proteins, whole lysates, and flowthrough were analyzed by Western blots with specific antibodies to gB, gC, gD, gH, gM, UL11, and VP5.
Two-way immunoprecipitation experiments.
Immunoprecipitation of proteins was essentially as described previously (52). Vero cell monolayers were infected with HSV-1(F) or HSV-1(F)-YE102-VC1 (having UL20 protein tagged with the 3×FLAG epitope and gK tagged with the V5 epitope) (52). Cellular extracts were prepared at 24 hpi, and the designated proteins were precipitated by protein G magnetic Dynabeads (Life Technologies-Novex, Carlsbad, CA) bound to the corresponding antibodies according to the manufacturer's instructions. The presence of gB, gM, and UL20 was visualized by Western immunoblots using anti-gB, anti-gM, and anti-Flag (UL20) antibodies before or after immunoprecipitation with the same set of antibodies.
Virus penetration kinetics assay.
The kinetics of virus penetration was measured as described previously (29, 52). Briefly, subconfluent monolayers of Vero cells in 12-well tissue culture plates were infected at 4°C for 1 h with approximately 250 PFU of wild-type HSV-1(F), ΔgM mutant, and ΔUL11 mutant viruses. Subsequently, infected cell cultures were incubated at 34°C to allow virus penetration. Immediately thereafter (0 min) and at 30, 60, 120, and 180 min, the virus inoculum was removed and washed once with PBS (pH 7.4), and then the remaining extracellular virus was inactivated by treatment with low-pH PBS (pH 3.0). The cells were washed once with PBS and DMEM without serum sequentially and then overlaid with 1% methylcellulose in DMEM supplemented with 2% FCS. The cells were fixed with ice-cold methanol at 48 hpi, and virus plaques were counted after immunohistochemical staining (30). Mean values and standard deviations of three independent experiments were calculated. To determine the entry kinetics, linear regression slopes during the exponential growth period, from 0 to 120 min, were calculated.
RESULTS
Construction of recombinant viruses.
Deletion of the carboxyl-terminal 28 amino acids of gB (Δ28) or an Ala-to-Val mutation (syn20) in the amino terminus of gK causes extensive virus-induced cell fusion (9, 51, 53). To investigate the role of gM and UL11 in virus-induced cell fusion, we constructed a set of mutant viruses containing these syncytial mutations in the presence or absence of mutations that prevented expression of either the gM or UL11 gene using the HSV-1(F) genome cloned as a BAC (see Materials and Methods). The set of mutant viruses that were constructed included the following: (i) gBΔ28, (ii) gBΔ28/ΔgM, (iii) gBΔ28/ΔUL11, (iv) gKsyn20, (v) gKsyn20/ΔgM, and (vi) gKsyn20/ΔUL11 (Fig. 1 and 2).
Fig 2.
Plaque morphology of mutant viruses in comparison to HSV-1(F)-YE102. Confluent monolayers of Vero cells were infected with wild-type and mutant viruses at an MOI of 0.0001 and immunohistochemically stained at 48 hpi using polyclonal anti-HSV rabbit sera as described in Materials and Methods. (A) Representative viral plaques of HSV-1(F)-YE102 wild-type virus and the syncytial mutant viruses gBΔ28 and gKsyn20 on Vero cells. (B) Representative viral plaques of syncytial mutant viruses lacking gM or UL11 in the absence or presence of transient complementation with either gM- or UL11-expressing plasmid.
Validation of recombinant viruses.
Individual mutant viruses were recovered by transfecting DNA of each constructed mutant BAC into Vero cells, and the entire genomes of the parental HSV-1(F) virus and all mutant viruses were sequenced (see Materials and Methods). Viral genomes matched very well to the published HSV-1(F) genomic sequence (GenBank accession number GU734771.1), with the exception of 37 nucleotide changes resulting in 13 amino acid changes between the published HSV-1(F) and our HSV-1(F) strain, which is derived from pYE102bac (Table 1). Comparison of the HSV-1(F)-pYE102bac with each mutant virus revealed the presence of each engineered mutation, while no other nucleotide changes were observed.
Table 1.
Summary of differences between the published HSV-1(F) and the HSV-1(F) from pYE102baca
| Gene nameb | nt position | aa difference | aa position | Gene description |
|---|---|---|---|---|
| UL1 | 9,604 | gL | ||
| UL6 | 15,564 | Capsid portal protein | ||
| UL8 | 18,753 | Helicase-primase subunit | ||
| 19,678 | Helicase-primase subunit | |||
| UL13 | 26,904 | C→Y | 507 | Tegument serine/threonine protein kinase |
| NC | 34,226 | |||
| 35,711 | ||||
| UL19 | 40,404 | A→T | 15 | Major capsid protein |
| UL21 | 43,555 | Tegument protein | ||
| NC | 47,555 | |||
| UL24 | 47,695 | Nuclear protein | ||
| 47,910 | A→V | 86 | Nuclear protein | |
| 47,911 | A→V | Nuclear protein | ||
| UL27 | 53,638 | gB | ||
| UL34 | 69,681 | S→N | 45 | Nuclear egress membrane protein |
| UL37 | 81,439 | Tegument protein | ||
| 82,015 | Tegument protein | |||
| 82,806 | Tegument protein | |||
| 82,815 | Tegument protein | |||
| UL38 | 84,529 | C→R | 45 | Capsid triplex subunit 1 |
| UL39 | 87,759 | A→V | 424 | Ribonucleotide reductase subunit 1 |
| 88,762 | Ribonucleotide reductase subunit 1 | |||
| UL48 | 103,969 | Transactivating tegument protein VP16 | ||
| 104,340 | T→A | 212 | Transactivating tegument protein VP16 | |
| UL50 | 107,306 | P→T | 134 | Deoxyuridine triphosphatase |
| UL55 | 115,491 | M→I | 35 | Nuclear protein UL55 |
| UL56 | 116,406 | A→T | 137 | Membrane protein UL56 |
| NC | 132,421 | |||
| 134,690 | ||||
| 137,938 | ||||
| US7 | 139,976 | gI |
nt, nucleotide; aa, amino acid.
NC, noncoding region.
Characterization of mutant viruses.
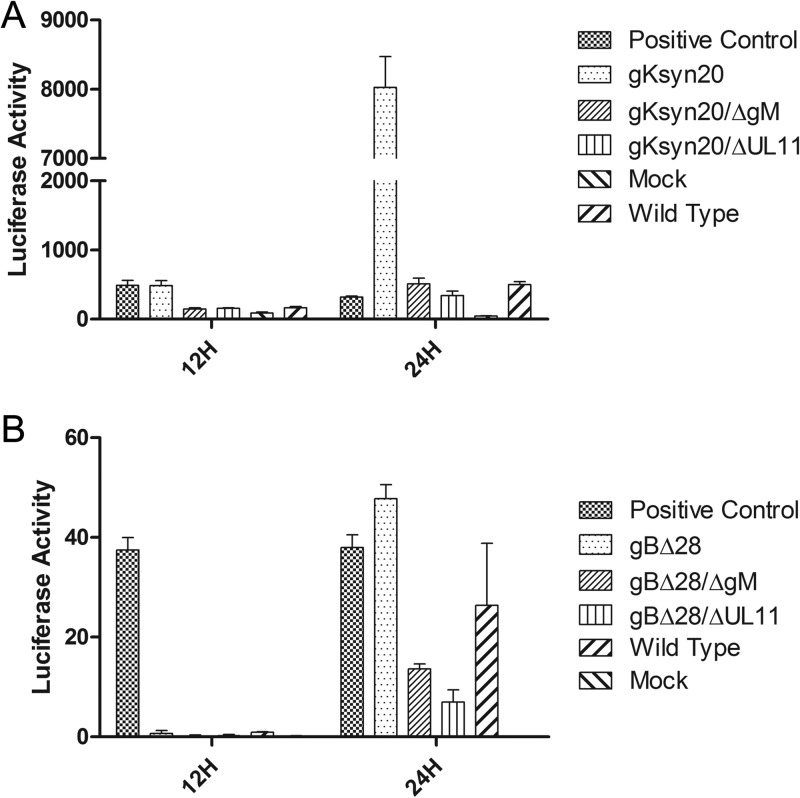
The plaque morphology of each recombinant virus was characterized as described in Materials and Methods. As expected, the recombinant viruses gBΔ28 and gKsyn20 caused extensive cell fusion (Fig. 2A). In contrast, lack of either gM or UL11 expression in the presence of either a gBΔ28 or gKsyn20 mutation caused significantly reduced levels of cell-to-cell fusion and decreased plaque sizes (Fig. 2B). To test whether these plaque changes were due primarily to lack of either gM or UL11 gene expression, complementation assays in which Vero cells were transfected with plasmids expressing either gM or UL11 and subsequently infected with the mutant viruses were performed. These experiments revealed that more than 50% of viral plaques (data not shown) were rescued to either a gBΔ28 or gKsyn20 plaque morphology (Fig. 2B). To quantify the fusogenic capacity of mutant viruses, we utilized a luciferase-based assay to measure cell-to-cell fusion (see Materials and Methods). The gKsyn20 mutation caused much higher virus-induced cell fusion than the gBΔ28 mutation, most likely because this virus spread more rapidly than the gBΔ28 virus. Moreover, the amount of gKsyn20-caused cell fusion was much higher than that obtained by transfection of both luciferase gene and T7 polymerase in the same cell population (positive control). Deletion of either gM or UL11 completely abrogated gKsyn20-mediated cell fusion (Fig. 3A). Similarly, deletion of either gM or UL11 severely inhibited gBΔ28-induced cell fusion, exhibiting cell fusion levels lower than those produced by the wild-type virus (Fig. 3B). To assess the relative effect of each mutation alone or in combination on virus replication, viral titers were obtained at 24 or 48 h after infection of Vero cells at an MOI of either 0.2 or 3.0. All viruses appeared to replicate with efficiencies approximately equal to that of the parental wild-type virus irrespective of the presence of the engineered mutations (Fig. 4).
Fig 3.
Fusion activity of wild-type and mutant viruses. Fusion activity of each virus was quantified by a luciferase-based assay (see Materials and Methods). The extent of fusion was assessed at 12 and 24 hpi for the wild-type virus and all mutant viruses containing the gKsyn20 mutation (A) or the gBΔ28 mutation (B).
Fig 4.
Replication kinetics of the wild-type and mutant viruses. Confluent Vero cell monolayers were infected with each virus in triplicate at either a low MOI (0.2) (A) or a high MOI (3.0) (B), and viral titers were obtained by plaque assay on Vero cells at 24 and 48 hpi. Error bars represent standard deviations.
Effect of mutations on the synthesis and cell surface expression of viral glycoproteins.
The effects of a lack of either gM or UL11 expression on the synthesis of the viral glycoproteins gB, gC, gD, and gM and the membrane-associated protein UL11 were assessed using Western immunoblots of whole-cell lysates. A lack of either gM or UL11 expression was confirmed via the inability of anti-gM or anti-UL11 antibody to detect the presence of either protein in their respective mutant viruses. Overall, neither lack of gM expression nor lack of UL11 expression drastically affected the synthesis of gB, gC, and gD. Furthermore, lack of gM did not affect the synthesis of UL11 and lack of UL11 did not affect gM levels (Fig. 5).
Fig 5.
Characterization of glycoprotein expression by wild-type and mutant viruses. The overall synthesis of each viral protein was assessed by Western immunoblotting. Each lane represents infected cellular extracts from cells infected with either wild-type or mutant virus: 1, wild type; 2, gBΔ28; 3, gBΔ28/ΔgM; 4, gBΔ28/ΔUL11; 5, mock infection; 6, gKsyn20; 7, gKsyn20/ΔgM; 8, gKsyn20/ΔUL11. gB, gC, gD, gM, and UL11 denote antibodies specific for each protein.
Surface glycoprotein expression profiles of Vero cells infected with the designated viruses were analyzed by cell surface biotinylation experiments (Fig. 6). Viral glycoproteins expressed on infected cell surfaces were biotinylated under live conditions. Biotinylated proteins were subsequently isolated by streptavidin immunoprecipitation and analyzed by Western immunoblots (see Materials and Methods). The HSV-1 capsid protein VP5 was included in the immunoblots as a negative control since it is not expected to be expressed in infected cellular plasma membranes. Lack of either gM or UL11 did not affect overall expression of the viral glycoproteins gB, gC, gD, and gH on HSV-1(F)-infected cell surfaces (Fig. 6A). Similar results were obtained for the gBΔ28 virus (Fig. 6B) except in the case of gKsyn20-infected cells, most likely due to rapid loss of fused cells (syncytia) (Fig. 6C).
Fig 6.
Cell surface expression of viral glycoproteins on infected cell surfaces by wild-type and mutant viruses. At 12 hpi, whole lysates were prepared and membrane-associated proteins were isolated (see Materials and Methods). Flowthrough-labeled lanes represent cytoplasmic proteins. Each lane represents wild-type or mutant virus-infected cell extracts. (A) Lane 1, wild type; lane 2, ΔgM; lane 3, ΔUL11. (B) Lane 1, gBΔ28; lane 2, gBΔ28/ΔgM; lane 3, gBΔ28/ΔUL11. (C) Lane 1, gKsyn20; lane 2, gKsyn20/ΔgM; lane 3, gKsyn20/ΔUL11. gB, gC, gD, gH, gM, UL11, and VP5 denote antibodies specific for each protein.
UL20 protein physically interacts with gM in virus-infected cells.
To determine whether gM and UL11 interact directly with gB or the gK/UL20 complex, we performed two-way immunoprecipitation experiments. The results revealed that gM interacted with UL20 but not gB, since gM immunoprecipitates contained UL20 (Fig. 7, lane 2) but not gB (Fig. 7, lane 8) and UL20 immunoprecipitates contained gM (Fig. 7, lane 4). Immunoprecipitations with gB, gD, or gH failed to reveal any interactions with gM (not shown). Similar immunoprecipitations with UL11 failed to reveal any interactions with gB, gC, gD, gH, gM, gK, or UL20 (data not shown).
Fig 7.
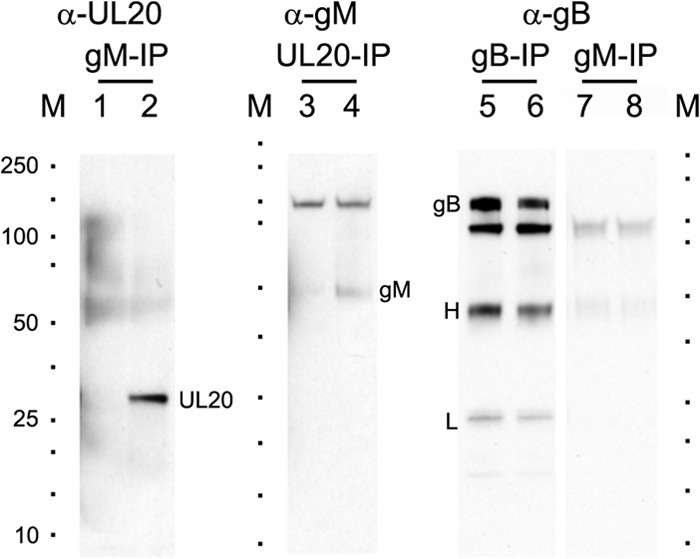
UL20 protein interacts with gM in virus-infected cells. Vero cell monolayers were infected with HSV-1(F) or HSV-1(F)-YE102-VC1 (having UL20 protein tagged with the 3×FLAG epitope and gK tagged with the V5 epitope). Cellular extracts were prepared at 24 hpi, and the presence of gB, gM, or UL20 was detected using Western immunoblots after immunoprecipitation (IP) with designated antibodies. Lanes 1, 3, 5, and 7, antigen from HSV-1(F)-infected cells; lanes 2, 4, 6, and 8, antigen from HSV-1(F)-YE102-VC1-infected cells; lane M demarcates positions of molecular mass markers (10 to 250 kDa; Bio-Rad). Anti-gB (α-gB), anti-gM (α-gM), and anti-FLAG (α-UL20) denote probed antibodies, and gB-IP, gM-IP, and UL20-IP (FLAG-IP) represent antibodies used for immunoprecipitation. gB, H, L, gM, and UL20 denote gB, heavy chain, light chain, gM, and UL20 proteins, respectively.
HSV-1 gM and UL11 are required for wild-type-like virus entry.
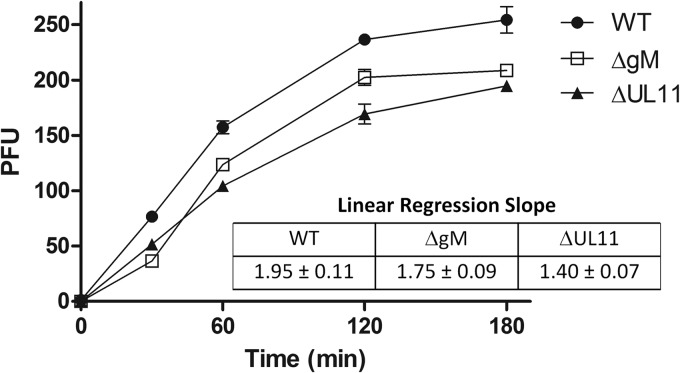
Virus-induced cell fusion and fusion of the viral envelope with cellular membranes during virus entry are thought to occur via similar mechanisms involving viral glycoproteins. Therefore, we tested whether a lack of either gM or UL11 affected the kinetics of virus entry into Vero cells. A lack of either gM or UL11 caused slower entry of both viruses into Vero cells compared to the wild-type virus (Fig. 8). Similarly, lack of either gM or UL11 caused slower virus entry in the presence of either the gBΔ28 or gKsyn20 mutation (data not shown).
Fig 8.
Entry kinetics of HSV-1(F) (WT), ΔgM, and ΔUL11 viruses into Vero cells. Vero cell monolayers were infected with either HSV-1(F), ΔgM, or ΔUL11 virus at 250 PFU per tissue culture well. The kinetics of virus entry at 34°C was measured (see Materials and Methods). Mean values and standard deviations of three independent experiments are shown. The relative efficiency of entry was calculated by obtaining the linear regression slope during the exponential viral growth period (0 to 120 min). The mean value and the standard deviation of each slope are shown.
DISCUSSION
Herpes viral proteins and glycoproteins have been shown to extensively interact within the virion particle and in infected cells. Interactions among the viral glycoproteins gB, gD, gH, gL, and gK and the membrane protein UL20 are known to be involved in regulation of membrane fusion phenomena occurring during virus entry and virus-induced cell fusion. The purpose of this investigation was to assess whether the glycoprotein gM and the membrane-associated protein UL11, both of which are highly conserved in all herpesviruses, are involved in these membrane fusion phenomena. Herein, we show that the absence of either gM or UL11 expression abrogated virus-induced cell fusion caused by strong syncytial mutations in either gB or gK and that gM interacted with the gK/UL20 protein complex. Moreover, virions produced in the absence of gM or UL11 entered more slowly into cells than did their parental wild-type virus. These results suggest that both gM and UL11 directly or indirectly interact and regulate the HSV-1 membrane fusion machinery during virus-induced cell fusion and virus entry.
Glycoprotein gM is a highly hydrophobic glycoprotein predicted to span membranes eight times. It is highly conserved in all alphaherpesviruses, suggesting that it plays an essential role in herpes infections. Previous work indicated that insertional inactivation of the gM gene inhibited cell fusion caused by a syncytial mutation in gB (gBsyn; Ala to Val at gB amino acid 855) (36, 37). This particular syncytial mutation causes significantly smaller amounts of virus-induced cell fusion than the gBΔ28 mutation, with the latter being the strongest gB syncytial mutation causing extensive cell fusion of most cell types in tissue culture (54, 55). Similarly, the gKsyn20 mutation (Ala to Val at gK amino acid 40) causes extensive virus-induced cell fusion in most cell types (30). Our results show that a lack of gM abrogated virus-induced cell fusion caused by either a gKsyn20 or gBΔ28 mutation, in agreement with previous findings (36, 37). Glycoprotein gM has been suggested to function in either retaining viral glycoproteins to TGN membranes or retrieving them from plasma membranes to TGN (32). In contrast to these published findings, deletion of gM did not appear to appreciably impact the overall level of other viral glycoproteins or their relative levels expressed on cell surfaces. Transient expression of HSV-1, PRV, and KSHV gM appeared to inhibit cell fusion caused by simultaneous expression of gB, gD, gH, and gL, suggesting potential interactions between gM and one or more of these viral glycoproteins (34, 35). However, extensive two-way coimmunoprecipitation experiments failed to indicate interactions between gM and either gB or gD, suggesting that gM may indirectly interact with these proteins to regulate cell fusion. Additional protein-protein interaction experiments revealed that gM interacts with the UL20 protein, the interacting partner of gK. We have shown that the gK/UL20 protein complex interacts with gB (31); therefore, it is likely that a lack of gM affects the ability of the gK/UL20 complex to bind and regulate the fusogenic properties of gB. In this regard, gM is a new member of the HSV-1 fusion machinery composed of gB, gH, gD, gL, gK, UL20, and now gM. Recently, it was shown that VP22 forms a protein complex with gM and gE that may affect the functions of these proteins (56); thus, gE may also participate in this glycoprotein network on infected cell surfaces and virion envelopes.
The UL11 protein was of particular interest to these investigations due to its high conservation among all herpesviruses and its ability to interact with the carboxyl terminus of glycoprotein gE and multiple tegument proteins (42). We show that UL11 is expressed on infected cell surfaces at very low levels, if at all, in agreement with the prediction that UL11 is attached to cytoplasmic sides of plasma membranes via its myristoylated and palmitoylated residues and its interaction with the cytoplasmic tail of gE (32, 38–42). Surprisingly, a lack of UL11 severely inhibited both gBΔ28- and gKsyn20-caused virus-induced cell fusion. The gBΔ28 results confirm recent findings that UL11 is required for gBsyn (A855V) but significantly extend these findings, since deletion of the gB carboxyl terminus causes extensive cell fusion in all cell types, unlike the gBsyn (A855V) mutation (55). Moreover, a lack of UL11 inhibited fusion caused by the gKsyn20 mutation, which is a dominant syncytial mutation causing fusion of all cell types tested.
A lack of gE (36, 37) and UL11 (43) was reported to prevent gBsyn-mediated virus-induced cell fusion. Immunofluorescence and flow cytometry experiments failed to detect substantially smaller amounts of gE in cells infected with either gM-null or UL11-null virus (data not shown). This suggests that the observed inhibition of virus-induced cell fusion in the absence of UL11 is not due to a lack of gE cell surface expression. In contrast to our findings, it was recently shown that deletion of UL11 appeared to reduce gE cell surface expression in Vero but not HaCaT cells by fluorescence microscopy (43). It is possible that different results are obtained due to the use of HSV-1 (KOS) (43) versus HSV-1(F) (this study). Additional studies will be needed to resolve whether a lack of UL11 substantially affects gE expression and function on infected cell surfaces.
Virus-induced cell fusion and fusion of the viral envelope with cellular membranes during virus entry are thought to occur via similar mechanisms involving viral glycoprotein gB and cellular receptors (15). We have shown that gK and UL20 are expressed in virion envelopes and are involved in virus entry, since deletion of gK or a small segment of gK that interacts with gB causes virions to enter more slowly than their parental wild-type virus into cells and alter their ability to utilize HSV-1-specific receptors (29, 52, 57). Similarly, virions produced in the absence of either gM or UL11 exhibited slower kinetics of entry into Vero cells than did their parental virus. gM is known to be expressed in virion envelopes (48). Also, UL11 is a structural component of virions (58) and interacts with gE and other tegument proteins in infected cells (43). Recently, it was shown that VP22 bridges a complex between gE and gM (56). Therefore, it is conceivable that UL11 binding to gE can indirectly affect both gE and gM functions, causing both the observed inhibition of virus-induced cell fusion and the slower kinetics of the UL11-null virus in comparison to its parental wild-type virus. Alternatively, it is possible that absence of gM or UL11 affects the incorporation of other viral glycoproteins into virion envelopes, causing the observed inhibition in virus entry. Additional investigations beyond the scope of this work are needed to address this issue in the future.
Collectively, these results suggest that both gM and UL11 are required for virus-induced cell fusion and efficient virus entry, most likely because they directly or indirectly interact with the viral fusion machinery. Glycoprotein gM directly interacts with the gK/UL20 protein complex. We did not detect any UL11 interactions with either gB, gC, gD, gH, gM, or gK/UL20. It is possible that such interactions are weak or of transient nature that cannot be detected via coimmunoprecipitation experiments. UL11 interacts with gE and VP22 interacts with both gE and gM (56), suggesting that gE also participates in indirect interactions with the fusion machinery. Additional experiments are needed to ascertain the intracellular localization of UL11 and its potential functions in virus-induced cell fusion. Overall, it is apparent that multiple protein-protein interactions occur among viral glycoproteins and tegument proteins that regulate membrane fusion phenomena in HSV-1 infections.
ACKNOWLEDGMENTS
This work was supported by NIH NIAID grant AI43000 to K.G.K., and Core Laboratories were supported by NIH NIGMS grant P20GM103458. We acknowledge financial support by the LSU School of Veterinary Medicine.
We acknowledge helpful discussions with other BIOMMED staff.
Footnotes
Published ahead of print 15 May 2013
REFERENCES
- 1. Roizman B, Knipe D, Whitley R. 2007. Herpes simplex viruses, p 2501–2601 Knipe D, Howley P. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Baines JD, Ward PL, Campadelli-Fiume G, Roizman B. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McLean G, Rixon F, Langeland N, Haarr L, Marsden H. 1990. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J. Gen. Virol. 71:2953–2960 [DOI] [PubMed] [Google Scholar]
- 4. Melancon JM, Foster TP, Kousoulas KG. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 78:7329–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacobson JG, Chen SH, Cook WJ, Kramer MF, Coen DM. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161–169 [DOI] [PubMed] [Google Scholar]
- 6. Sanders PG, Wilkie NM, Davison AJ. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277–295 [DOI] [PubMed] [Google Scholar]
- 7. Bzik DJ, Fox BA, DeLuca NA, Person S. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185–190 [DOI] [PubMed] [Google Scholar]
- 8. Pellett PE, Kousoulas KG, Pereira L, Roizman B. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bond VC, Person S. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368–376 [DOI] [PubMed] [Google Scholar]
- 10. Debroy C, Pederson N, Person S. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36–48 [DOI] [PubMed] [Google Scholar]
- 11. Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham FL, Johnson DC. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pogue-Geile KL, Lee GT, Shapira SK, Spear PG. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100–109 [DOI] [PubMed] [Google Scholar]
- 13. Pogue-Geile KL, Spear PG. 1987. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain MP. Virology 157:67–74 [DOI] [PubMed] [Google Scholar]
- 14. Ruyechan WT, Morse LS, Knipe DM, Roizman B. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305–319 [DOI] [PubMed] [Google Scholar]
- 17. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 18. Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436 [DOI] [PubMed] [Google Scholar]
- 19. Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, Arase N, Shiratori I, Tanaka S, Kawaguchi Y, Spear PG, Lanier LL, Arase H. 2008. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13–22 [DOI] [PubMed] [Google Scholar]
- 21. Spear PG, Eisenberg RJ, Cohen GH. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1–8 [DOI] [PubMed] [Google Scholar]
- 22. Spear PG, Longnecker R. 2003. Herpesvirus entry: an update. J. Virol. 77:10179–10185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 25. Muggeridge MI. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017–2027 [DOI] [PubMed] [Google Scholar]
- 26. Turner A, Bruun B, Minson T, Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kousoulas KG, Person S, Holland TC. 1978. Timing of some of the molecular events required for cell fusion induced by herpes simplex virus type 1. J. Virol. 27:505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melancon JM, Luna RE, Foster TP, Kousoulas KG. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79:299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Foster TP, Rybachuk GV, Kousoulas KG. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431–12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chouljenko VN, Iyer AV, Chowdhury S, Chouljenko DV, Kousoulas KG. 2009. The amino terminus of herpes simplex virus type 1 glycoprotein K (gK) modulates gB-mediated virus-induced cell fusion and virion egress. J. Virol. 83:12301–12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chouljenko VN, Iyer AV, Chowdhury S, Kim J, Kousoulas KG. 2010. The herpes simplex virus type 1 UL20 protein and the amino terminus of glycoprotein K (gK) physically interact with gB. J. Virol. 84:8596–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leege T, Fuchs W, Granzow H, Kopp M, Klupp BG, Mettenleiter TC. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J. Virol. 83:896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crump CM, Bruun B, Bell S, Pomeranz LE, Minson T, Browne HM. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517–3527 [DOI] [PubMed] [Google Scholar]
- 34. Klupp BG, Nixdorf R, Mettenleiter TC. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koyano S, Mar EC, Stamey FR, Inoue N. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485–1491 [DOI] [PubMed] [Google Scholar]
- 36. Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75(Part 6):1245–1258 [DOI] [PubMed] [Google Scholar]
- 37. Davis-Poynter N, Bell S, Minson T, Browne H. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 68:7586–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yeh PC, Meckes DG, Jr, Wills JW. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J. Virol. 82:10693–10700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baird NL, Yeh PC, Courtney RJ, Wills JW. 2008. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology 374:315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loomis JS, Bowzard JB, Courtney RJ, Wills JW. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209–12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loomis JS, Courtney RJ, Wills JW. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeh PC, Han J, Chadha P, Meckes DG, Jr, Ward MD, Semmes OJ, Wills JW. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J. Virol. 85:9425–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc. Natl. Acad. Sci. U. S. A. 109:19798–19803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chouljenko DV, Kim IJ, Chouljenko VN, Subramanian R, Walker JD, Kousoulas KG. 2012. Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J. Virol. 86:4262–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee HC, Chouljenko VN, Chouljenko DV, Boudreaux MJ, Kousoulas KG. 2009. The herpes simplex virus type 1 glycoprotein D (gD) cytoplasmic terminus and full-length gE are not essential and do not function in a redundant manner for cytoplasmic virion envelopment and egress. J. Virol. 83:6115–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 47. Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baines JD, Roizman B. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Foster TP, Melancon JM, Olivier TL, Kousoulas KG. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 78:13262–13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fulmer PA, Melancon JM, Baines JD, Kousoulas KG. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 81:3097–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Foster TP, Alvarez X, Kousoulas KG. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jambunathan N, Chowdhury S, Subramanian R, Chouljenko VN, Walker JD, Kousoulas KG. 2011. Site-specific proteolytic cleavage of the amino terminus of herpes simplex virus glycoprotein K on virion particles inhibits virus entry. J. Virol. 85:12910–12918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baghian A, Huang L, Newman S, Jayachandra S, Kousoulas KG. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Foster TP, Melancon JM, Kousoulas KG. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18–29 [DOI] [PubMed] [Google Scholar]
- 55. Silverman JL, Greene NG, King DS, Heldwein EE. 2012. Membrane requirement for folding of the herpes simplex virus 1 gB cytodomain suggests a unique mechanism of fusion regulation. J. Virol. 86:8171–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maringer K, Stylianou J, Elliott G. 2012. A network of protein interactions around the herpes simplex virus tegument protein VP22. J. Virol. 86:12971–12982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chowdhury S, Chouljenko VN, Nadheri M, Kousoulas KG. 2013. The amino terminus of herpes simplex virus 1 glycoprotein K is required for virion entry via the paired immunoglobulin-like type-2 receptor alpha. J. Virol. 87:3305–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]