Abstract
The Tm-1 gene of tomato confers resistance to Tomato mosaic virus (ToMV). Tm-1 encodes a protein that binds ToMV replication proteins and inhibits the RNA-dependent RNA replication of ToMV. The replication proteins of resistance-breaking mutants of ToMV do not bind Tm-1, indicating that the binding is important for inhibition. In this study, we analyzed how Tm-1 inhibits ToMV RNA replication in a cell-free system using evacuolated tobacco protoplast extracts. In this system, ToMV RNA replication is catalyzed by replication proteins bound to membranes, and the RNA polymerase activity is unaffected by treatment with 0.5 M NaCl-containing buffer and remains associated with membranes. We show that in the presence of Tm-1, negative-strand RNA synthesis is inhibited; the replication proteins associate with membranes with binding that is sensitive to 0.5 M NaCl; the viral genomic RNA used as a translation template is not protected from nuclease digestion; and host membrane proteins TOM1, TOM2A, and ARL8 are not copurified with the membrane-bound 130K replication protein. Deletion of the polymerase read-through domain or of the 3′ untranslated region (UTR) of the genome did not prevent the formation of complexes between the 130K protein and the host membrane proteins, the 0.5 M NaCl-resistant binding of the replication proteins to membranes, and the protection of the genomic RNA from nucleases. These results indicate that Tm-1 binds ToMV replication proteins to inhibit key events in replication complex formation on membranes that precede negative-strand RNA synthesis.
INTRODUCTION
Tomato mosaic virus (ToMV) is a positive-strand RNA virus in the genus Tobamovirus, family Virgaviridae (1). The genome of a tobamovirus encodes at least four proteins (2). The 130K protein and its read-through product, the 180K protein, are involved in RNA replication and thus are referred to as replication proteins (3). The 130K protein contains the methyltransferase and helicase domains, and the read-through region of the 180K protein contains the polymerase domain. The other two tobamoviral proteins, movement protein and coat protein, are not required for RNA replication (4, 5).
When the genomic RNA of a positive-strand RNA virus enters host cells, the replication proteins are translated from the genomic RNA, recognize the genomic RNA as a template for replication, and form replication complexes on intracellular membranes. Replication of all known eukaryotic positive-strand RNA viruses occurs in replication complexes formed on host membranes (6–10), which contain viral genomic RNAs, replication proteins, and host factors (11). To avoid elicitation of host defense systems triggered by viral double-stranded RNA (12), the activity of eukaryotic positive-strand RNA virus RNA-dependent RNA polymerases (RdRps) must be strictly regulated so that they synthesize negative-strand RNA only in the replication complexes that are sequestered from the cytoplasm. Many host factors, including chaperones and enzymes, and cellular membranes are hijacked by viruses for replication (13–16). However, knowledge of the processes leading to replication complex formation and activation of the viral polymerases is limited.
In tobamovirus-infected cells, the replication proteins exist in both membrane-bound and soluble forms (17). Studies have suggested that the membrane-bound forms participate in RNA replication and the soluble forms play a role in RNA silencing suppression (17, 18). The replication proteins of tobamoviruses are not predicted to have membrane-targeting signals or membrane-spanning regions, and how they bind membranes remains obscure. The host TOM1 protein is a putative seven-pass transmembrane protein required for efficient tobamovirus multiplication (19). TOM1 interacts with the helicase domain of tobamovirus replication proteins, and overexpression of TOM1 in tobacco plants or yeast cells increases the proportion of membrane-bound replication proteins compared with soluble proteins (18, 20), suggesting that TOM1 helps tether the replication proteins on membranes. Solubilized replication proteins from the membranes of ToMV-infected cells copurified TOM1 and two other host membrane proteins, TOM2A and ARL8 (20, 21). Together with genetic data, these host proteins are suggested to be components of the ToMV replication complex (22).
Many plant genes for resistance against viruses have been identified (23–26). However, little is known about how the resistance gene products inhibit virus multiplication because viral molecules become detectable in infected cells only after multiple rounds of replication occur. If an early stage of virus multiplication is inhibited, virus-related molecules cannot be detected. In fact, no ToMV-related molecules are detectable in tomato protoplasts harboring the resistance gene Tm-1 (27). Tm-1 encodes a protein of unknown function that binds the ToMV replication proteins (28). Resistance-breaking mutants of ToMV have amino acid substitutions in the replication proteins and thereby escape inhibitory interaction with Tm-1, although a fitness cost is associated with this process (29–31).
We have developed a cell-free translation and replication system for ToMV RNA using evacuolated tobacco BY-2 protoplast lysate (BYL) (32). Such cell-free viral RNA replication systems enable the biochemical analysis of replication processes and have been used to analyze how the replication complex is formed (33–40). Using this in vitro system, we previously demonstrated that the Tm-1 protein inhibits ToMV RNA replication when added before, but not after, the ToMV replication complex is formed on membranes (28). This result indicates that Tm-1 inhibits the formation of the RNA replication complex but not the replication reactions that occur in the replication complex. In this study, we performed a more detailed analysis of Tm-1 action using an in vitro system.
MATERIALS AND METHODS
Viruses.
TLIle (41) is a ToMV strain that is highly sensitive to Tm-1 (31, 42). Strain LT1 (29) is a Tm-1 resistance-breaking mutant. TL130F was described previously (43). TLIle and LT1 mutations were introduced into TL130F to create TLIle130F and LT1130F, respectively. RNAs synthesized from the MluI-linearized plasmids using an AmpliCap-Max T7 High Yield Message Maker kit (Cellscript, Inc., Madison, WI) were used as templates for in vitro translation and replication. To prepare Δ3′ RNA, PmlI-linearized plasmids were used as templates for transcription.
In vitro translation and replication.
BYL was prepared as described previously (44). ToMV RNA (100 ng) was translated in 36 μl of membrane-depleted BYL (mdBYL)-based translation mixture (33, 44) at 23°C for 1 h. mdBYL from transgenic BY-2 cells expressing Tm-1 (31) or from nontransgenic BY-2 cells (4 μl) was added to the translation mixtures and incubated at 23°C for 20 min where indicated. The mixtures (40 μl) were further mixed with pellets of BYL from centrifugation at 30,000 × g (30,000 × g pellets of BYL) {prepared from 50 μl of BYL from nontransgenic BY-2 and suspended in 10 μl of TR buffer [30 mM HEPES-KOH, pH 7.4, 80 mM KOAc (potassium acetate), 1.8 mM Mg(OAc)2 (magnesium acetate), 2 mM DTT (dithiothreitol), with Complete EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland)]}, and incubated at 15°C for 2 h, followed by centrifugation at 16,000 × g for 20 min to obtain supernatants (S16 fractions) and pellets (P16 pellets). The P16 pellets were suspended in 50 μl of TR buffer (P16 fractions). For the 0.5 M NaCl treatment, the P16 pellets were suspended in TR buffer containing 0.5 M NaCl and incubated on ice for 30 min. RdRp reactions and membrane flotation analysis were performed as described previously (21, 44). For micrococcal nuclease (MNase) resistance assays, RNAs synthesized in the presence of [α-32P]GTP were used as templates for translation in mdBYL. After incubation with BYL membranes, 0.5 μl of 0.1 M CaCl2 and 1 μl of 20 U/μl MNase (TaKaRa Bio Inc., Shiga, Japan) were added to 20 μl of the mixture and incubated at 23°C for 30 min, followed by addition of 1 μl of 0.5 M EGTA. RNA was purified from the mixtures by phenol extraction and ethanol precipitation and analyzed by 8 M urea-2.4% PAGE. Where indicated, puromycin was added to 0.4 mM before translation or Triton X-100 was added to 1% before MNase treatment. For detection of the negative-strand RNA, in vitro translation and replication reactions were performed without radioisotopes, followed by RNase protection assays using a 32P-labeled P2P probe (45). 32P-labeled RNA bands were detected by autoradiography using a BAS-2500 imager (GE Healthcare, Piscataway, NJ).
Antibodies.
Anti-ToMV 130K, anti-TOM1, anti-TOM2A (17), and anti-ARL8 (20) antibodies were described previously. Anti-Tm-1 protein rabbit antiserum was raised against an Escherichia coli-expressed hexahistidine-tagged full-length Tm-1 protein.
Immunoprecipitation.
Solubilization of the replication proteins from the P16 fraction with lysophosphatidylcholine (LPC) and subsequent immunoprecipitation using anti-FLAG antibody were performed as described previously (21).
RESULTS
Tm-1 inhibits ToMV negative-strand RNA synthesis.
A previous study demonstrated that ToMV RNA synthesis is inhibited when ToMV RNA is translated in mdBYL, mixed with Tm-1, and then mixed with BYL membranes (28). However, whether negative-strand RNA was synthesized was unclear. To address this point, the genomic RNAs of the ToMV mutants TLIle (a Tm-1-sensitive mutant) and LT1 (a resistance-breaking mutant) were translated in mdBYL, mixed with mdBYL prepared from Tm-1-expressing BY-2 cells or nontransgenic BY-2 cells and with a membrane-containing 30,000 × g pellet of BYL from nontransgenic BY-2 (P30BYL membranes), and then incubated with ribonucleoside triphosphates (NTPs). Following the reactions, total RNA was extracted and subjected to an RNase protection assay using a 32P-labeled probe to detect the negative-strand RNA. In the absence of Tm-1, negative-strand RNAs of TLIle and LT1 accumulated to similar levels. However, when mdBYL prepared from Tm-1-expressing cells was added to the reaction mixture, negative-strand RNA was detected for LT1 but not for TLIle (Fig. 1). Thus, Tm-1 directly inhibits ToMV negative-strand RNA synthesis or inhibits a step in ToMV RNA replication preceding the negative-strand RNA synthesis.
Fig 1.
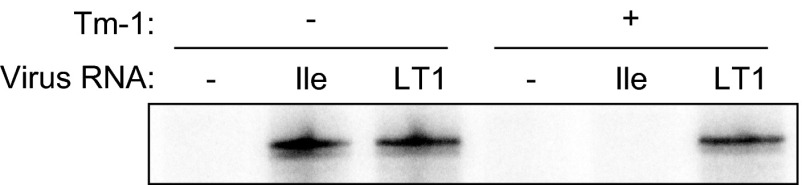
Tm-1 inhibits negative-strand RNA synthesis. In vitro translation and replication of Tm-1-sensitive (Ile) and resistance-breaking (LT1) ToMV strains were performed in the presence (+) or absence (−) of Tm-1. Accumulation of the negative-strand RNA was examined by RNase protection assay using a 32P-labeled probe.
Tm-1 inhibits 0.5 M NaCl-resistant membrane binding of ToMV replication proteins.
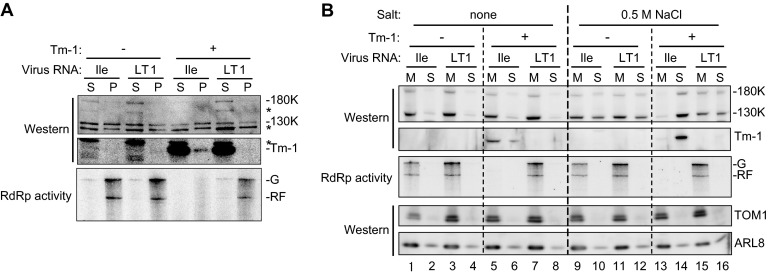
The ability of ToMV replication proteins to bind membranes in the presence of Tm-1 was examined next. TLIle and LT1 RNAs were translated separately in mdBYL, mixed with mdBYL prepared from Tm-1-expressing BY-2 cells or nontransgenic BY-2 cells, and mixed with P30BYL membranes from nontransgenic BY-2. After incubation, the mixtures were centrifuged at 16,000 × g for 20 min to obtain supernatants (S16) and pellets (P16). Membranes were recovered primarily in the P16 fraction. The replication proteins of TLIle and LT1 were detected in both the S16 and P16 fractions irrespective of the presence or absence of Tm-1. RdRp activity was detected only in the absence of Tm-1 for TLIle and fractionated mostly in the P16 fraction (Fig. 2A). This observation is consistent with previous results showing that the RNA polymerase activity of ToMV is associated with membranes (17, 21, 46).
Fig 2.
Effect of Tm-1 addition on the binding of ToMV replication proteins to membranes. (A) Fractionation of ToMV replication proteins by centrifugation. Genomic RNAs of TLIle or LT1 were translated in mdBYL, mixed with mdBYL from Tm-1-expressing (+) or -nonexpressing (−) BY-2 cells, incubated with BYL membranes, and fractionated into soluble (S) and membrane-containing (P) fractions by centrifugation. ToMV replication proteins and the Tm-1 protein were detected by Western blotting. To examine RdRp activity, fractions were incubated with [α-32P]CTP and other ribonucleoside triphosphates. The 32P-labeled RNA products were separated by 8 M urea-2.4% PAGE and visualized by autoradiography. G, genomic RNA; RF, replicative-form RNA. The asterisks represent background signals. (B) Membrane flotation analysis. Membrane-containing pellets prepared as described for panel A were suspended with TR buffer (lanes 1 to 8) or TR buffer containing 0.5 M NaCl (lanes 9 to 16) and subjected to membrane flotation analysis. The membrane (M) fractions and the soluble (S) fractions were collected, and the amounts of the indicated proteins and the activity of ToMV RdRp were examined.
Remarkably, a small fraction of Tm-1 protein was detected in the P16 fraction of the TLIle RNA-translated mixture but not in the P16 fraction of the LT1 RNA-translated mixture. To confirm if the replication proteins and Tm-1 protein recovered in the P16 fraction are actually associated with membranes, the P16 fraction was further analyzed using a membrane flotation centrifugation method. During centrifugation, membranes move in the centrifuge tube from the loading layer at the bottom to the top layer (Fig. 2B, lanes M), while soluble proteins remain in the bottom layer (Fig. 2B, lanes S). The host membrane proteins TOM1 and ARL8 were fractionated mainly into the M fraction (Fig. 2B). In addition to the replication proteins, Tm-1 protein that had been present in the P16 fractions of the TLIle RNA-translated mixture was detected in the M fraction (Fig. 2B, lane 5), indicating that Tm-1 binds membranes. These results suggest that the ability of ToMV replication proteins to bind membranes is not inhibited by Tm-1, and thus, the complex of Tm-1 and the replication proteins is recruited to the membrane surfaces.
Nishikiori et al. reported that a fraction of membrane-bound ToMV replication proteins is dissociated from membranes by treatment with 1 M NaCl-containing buffer, while RdRp activity remains bound to membranes (21). We further found that similar results were obtained when membranes were treated with 0.5 M NaCl. These results indicate that membrane-bound replication proteins exist in at least two forms, those that are not involved in RNA replication but associate with membranes in a 0.5 M NaCl-sensitive manner and those that participate in RNA synthesis and do not dissociate from membranes with NaCl treatment. We then tested whether the TLIle replication proteins bound by Tm-1 dissociate from membranes after treatment with 0.5 M NaCl-containing buffer using a membrane flotation assay. Strikingly, in the presence of Tm-1, TLIle replication proteins, as well as Tm-1 protein, were detected in the S fraction (Fig. 2B, lane 14). The membrane-binding properties of LT1 replication proteins were not affected by Tm-1. Thus, ToMV replication proteins bound by Tm-1 associate with membranes in a 0.5 M NaCl-sensitive manner, in contrast to those involved in RNA replication.
ToMV RNA is not protected from nuclease digestion in the presence of Tm-1.
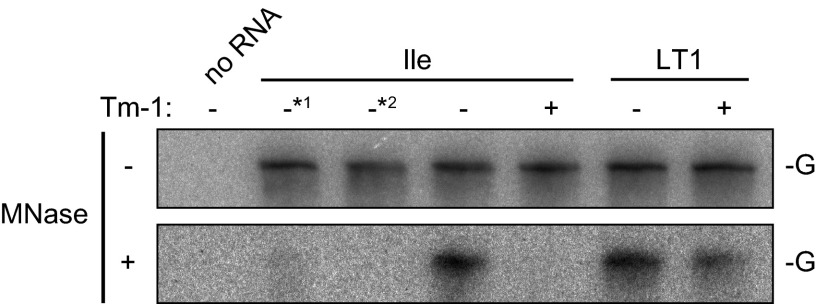
The 1a protein (the counterpart of ToMV 130K protein) of brome mosaic virus (BMV) binds to membranes and recruits and sequesters replication template RNA in a membranous compartment that cytoplasmic macromolecules cannot enter. Accordingly, 1a-recruited template RNA in yeast membrane fractions shows resistance to digestion by nucleases but becomes sensitive when treated with detergents (47). As is the case for BMV in yeast, 32P-labeled ToMV RNA translated in mdBYL and mixed with BYL membranes showed resistance to MNase (Fig. 3). The resistance was not observed when translation in mdBYL was inhibited by puromycin or when Triton X-100 was added prior to MNase treatment (Fig. 3). When Tm-1 was added after translation in mdBYL and before the addition of P30BYL membranes, LT1 RNA showed resistance to the digestion, but Tm-1-sensitive TLIle RNA did not (Fig. 3). Without MNase treatment, the levels of the input RNAs after the reactions with Tm-1 were similar to those after the reactions in the absence of Tm-1 (Fig. 3), suggesting that Tm-1 does not affect the stability of viral RNA.
Fig 3.
Tm-1 inhibits sequestration of ToMV RNA in an isolated membranous compartment. 32P-labeled TLIle and LT1 RNAs were translated in mdBYL. The translation mixtures were mixed with mdBYL from Tm-1-expressing (+) and -nonexpressing (−) BY-2 cells and incubated with membranes. The samples were then divided into two aliquots. One aliquot was treated with MNase, and RNA was extracted and analyzed by PAGE and autoradiography. The other aliquot was directly analyzed for RNA without MNase treatment. For the sample marked *1, puromycin was added before the translation reaction. For the sample marked *2, Triton X-100 was added before MNase treatment. G, genomic RNA.
Negative-strand RNA synthesis is not required for either 0.5 M NaCl-resistant membrane binding of ToMV replication proteins or protection of ToMV RNA from MNase.
Given the above-mentioned results where Tm-1 inhibits negative-strand RNA synthesis, 0.5 M NaCl-resistant membrane association of the replication proteins, and template sequestration, we examined whether these three events take place at once or sequentially by using two ToMV derivatives that are defective in negative-strand RNA synthesis. TLIle130F is a ToMV derivative that has a deletion in the region containing the read-through part of the 180K protein, the 30K protein, and the coat protein, and it encodes the TLIle-type 130K protein tagged by the FLAG peptide at the C terminus (Fig. 4A). TLIleΔ3′ is a ToMV derivative that lacks the 3′-terminal 158-nucleotide sequence with TLIle-type replication proteins (Fig. 4A). For the MNase resistance assay, 32P-labeled full-length TLIle, TLIle130F, and TLIleΔ3′ RNAs were separately translated in mdBYL, incubated with Tm-1-expressing or nontransgenic mdBYL, and then incubated with P30BYL membranes from nontransgenic BY-2, followed by MNase digestion. As was found for full-length TLIle RNA, TLIle130F RNA and TLIleΔ3′ RNA showed resistance to MNase in the absence of Tm-1 but not in the presence of Tm-1 (Fig. 4B). TLIle130F RNA was protected less efficiently than TLIle RNA or TLIleΔ3′ RNA (Fig. 4B), suggesting that the 180K protein or an RNA element lacking in TLIle130F enhances the efficiency of genome sequestration. To examine whether the replication proteins associate with membranes in a 0.5 M NaCl-resistant manner, full-length TLIle, TLIle130F, and TLIleΔ3′ RNAs were translated in mdBYL, incubated with mdBYL from Tm-1-expressing or nontransgenic cells, and then incubated with P30BYL membranes. The 16,000 × g pellet fractions were prepared from the mixtures, suspended in 0.5 M NaCl-containing buffer, and further centrifuged to obtain the 16,000 × g supernatant (S16) and pellet (P16) fractions. In the absence of Tm-1, a large proportion (50 to 79%) of the replication proteins were detected in the P16 fraction (Fig. 4C). The 130K protein expressed from TLIle130F established NaCl-resistant membrane binding less efficiently than that from TLIle or TLIleΔ3′. In the presence of Tm-1, replication proteins synthesized from TLIle130F and TLIleΔ3′ were detected mainly in the S16 fraction (Fig. 4C). These results indicate that neither 0.5 M NaCl-resistant binding of ToMV replication proteins with membranes nor establishment of the resistance of the template RNA to MNase requires the 180K protein or 3′-terminal sequence of ToMV RNA, i.e., negative-strand RNA synthesis, although the 180K protein or an RNA element in the region deleted in TLIle130F may facilitate these processes. Thus, these events probably precede negative-strand RNA synthesis in the ToMV RNA replication cycle.
Fig 4.
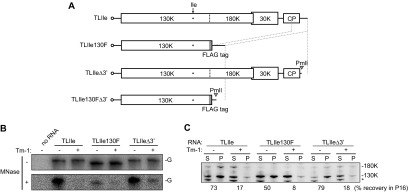
Negative-strand RNA synthesis is not required for the sequestration of ToMV RNA in the membranous compartment or NaCl-resistant membrane binding of the 130K protein. (A) Schematic representation of ToMV RNA derivatives used in Fig. 4 and 5. TLIle130F lacks the read-through region of the 180K polymerase, the 30K protein-coding region, and the 5′ half of the coat protein (CP)-coding region. TLIleΔ3′ is a transcript from a PmlI-linearized plasmid carrying full-length TLIle cDNA and lacks the 3′-terminal 158-nucleotide sequence. TLIle130FΔ3′ is a transcript from a PmlI-linearized plasmid encoding TLIle130F. (B) The read-through region for the 180K protein and the 3′ UTR of ToMV RNA are not required for the nuclease resistance of the genome RNA. 32P-labeled TLIle, TLIle130F, and TLIleΔ3′ RNAs were used as translation templates and analyzed as for Fig. 3. (C) The read-through region for the 180K protein and the 3′ UTR of ToMV RNA are not required for the 130K protein to bind membranes in a 0.5 M NaCl-resistant manner. TLIle, TLIle130F, and TLIleΔ3′ RNAs were translated in mdBYL, incubated with mdBYL from Tm-1-expressing (+) or -nonexpressing (−) BY-2 cells, and then incubated with BYL membranes. After incubation, membrane-containing pellets were prepared by centrifugation at 16,000 × g, suspended in 0.5 M NaCl-containing buffer, and centrifuged again at 16,000 × g to obtain the supernatant (S) and pellet (P) fractions. ToMV replication proteins in each fraction were detected by Western blotting and quantified with a LAS-3000 (Fujifilm, Japan). A typical set of results is shown, and the percentages of the 130K protein recovered in the P16 fractions are indicated. The asterisk represents background signals.
Tm-1 inhibits the association of host membrane proteins with ToMV 130K protein.
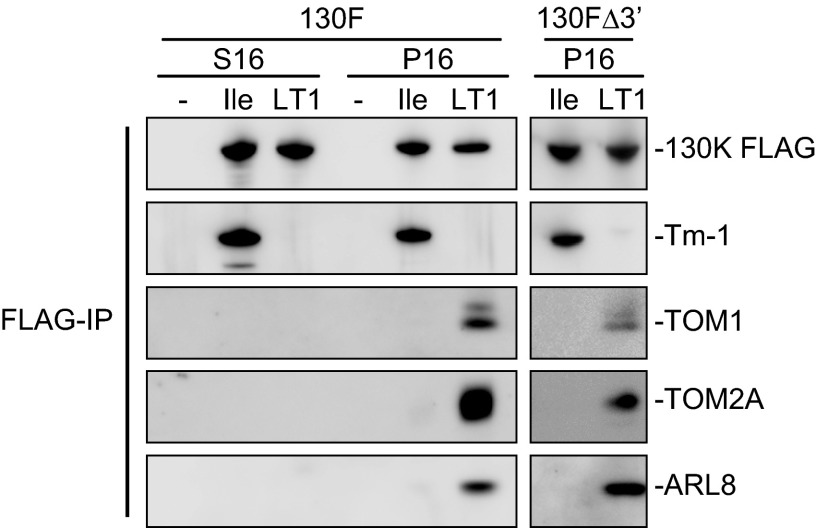
Three host membrane proteins, TOM1, TOM2A, and ARL8, are involved in tobamovirus RNA replication (19, 20, 48). These proteins are copurified with ToMV replication proteins solubilized from the membranes of infected cells (20, 21), suggesting that they are components of the tobamovirus RNA replication complex. In previous studies, epitope-tagged 180K protein was used for copurification experiments. Using ToMV derivatives TLIle130F and TLIle130FΔ3′ and those having LT1-type replication proteins, we examined whether TOM1, TOM2A, and ARL8 are copurified with the 130K protein in the absence of the 180K protein and, if they are, whether Tm-1 prevents copurification. These ToMV-derivative RNAs were translated in mdBYL and subsequently incubated with mdBYL from Tm-1-expressing BY-2 and with BYL membranes. The mixtures were centrifuged to obtain the S16 and P16 fractions, and the P16 fraction was solubilized with LPC. Immunoprecipitates obtained from each fraction with anti-FLAG antibody were analyzed by Western blotting. TOM1, TOM2A, and ARL8 were detected in the immunoprecipitates from the P16 fractions for the LT1130F and LT1130FΔ3′ RNAs (Fig. 5). Thus, the 180K protein and the 3′ untranslated region (UTR) are not required for the association of the 130K protein with these host proteins, indicating that negative-strand RNA synthesis is not required for complex formation of the 130K protein with the host membrane proteins. For TLIle130F and TLIle130FΔ3′ RNAs, these host proteins were not detected in the immunoprecipitates from the P16 fractions, but Tm-1 protein was detected (Fig. 5). TOM1, TOM2A, and ARL8 were not detected in the immunoprecipitates from the S16 fractions, and Tm-1 protein was copurified only with TLIle-type 130K protein (Fig. 5). Thus, a complex of Tm-1 and ToMV replication protein is stable in both soluble and membrane-bound forms, and the latter binds membranes without forming a complex with the host membrane proteins.
Fig 5.
Tm-1 inhibits complex formation of ToMV replication proteins with host membrane proteins. ToMV RNA derivatives encoding the FLAG-tagged 130K protein of either Tm-1-sensitive (Ile) or resistance-breaking (LT1) type with (130F) or without (130Δ3′) the 3′ UTR (Fig. 4A) were translated in mdBYL, incubated with mdBYL from Tm-1-expressing cells, and then incubated with BYL membranes. The mixtures were fractionated into soluble (S16) and membrane (P16) fractions. The 130K protein in the P16 fraction was solubilized with LPC and immunoprecipitated (IP) with anti-FLAG antibody. The precipitates were analyzed by Western blotting to detect the indicated proteins.
DISCUSSION
When ToMV RNA-translated mdBYL is mixed with P30BYL membranes, a fraction of the replication proteins binds membranes; forms a complex with host membrane proteins TOM1, TOM2A, and ARL8; and synthesizes the negative-strand RNA; also, a fraction of ToMV RNA gains resistance to MNase (template sequestration). We found that Tm-1 allows ToMV replication proteins to bind membranes in a 0.5 M NaCl-sensitive manner but inhibits their 0.5 M NaCl-resistant membrane binding, complex formation with the membrane proteins, and template sequestration (called “the three events” here) (Fig. 2, 3, and 5). The three events occurred even with ToMV RNA derivatives incapable of synthesizing negative-strand RNA due to deletions in the 180K protein-coding region or 3′ UTR (Fig. 4 and 5). Tm-1 also inhibited the three events for these ToMV derivatives (Fig. 4 and 5). These results suggest that the three events precede negative-strand RNA synthesis and that the inability of TLIle to synthesize negative-strand RNA in the presence of Tm-1 (Fig. 1) is a consequence of the inhibition of the three events.
Because the sequestered template RNA became sensitive to MNase upon detergent treatment, viral RNA is probably protected in a membranous compartment, as has been seen for other positive-strand RNA viruses, although the structure of the ToMV replication complex has not been visualized by electron microscopy or other techniques. We presume that the template sequestration is coupled with membrane rearrangement. Considering that the three events were not separable by our experiments, the replication proteins' 0.5 M NaCl-resistant membrane binding and complex formation with TOM1 and ARL8 might also be coupled with the membrane rearrangement process. On the other hand, unlike the three events, 0.5 M NaCl-sensitive membrane binding of the replication proteins occurred in the presence of Tm-1, indicating that the replication proteins could bind membranes without forming a complex with the host membrane proteins. Consistently, a previous study demonstrated that TOM1 and ARL8 are not required for the recruitment of the replication proteins to the membranes in the yeast Saccharomyces cerevisiae, while they contribute to the activation of the replication proteins on membranes (20).
We recently determined a crystal structure for the helicase domain of ToMV 130K protein (residues 666 to 1116), which consists of a C-terminal helicase core containing two RecA folds and an N-terminal accessory domain (49). Yeast two-hybrid experiments designed in light of the structure information suggested that ARL8 interacts with the N-terminal accessory domain and TOM1 interacts with both the accessory domain and the helicase core (49). Moreover, the reporter activity in the yeast two-hybrid experiment was much stronger between ARL8 and the accessory domain alone than between ARL8 and the full-length helicase domain polypeptide, suggesting that conformational changes in the 130K protein to expose the ARL8-binding site are associated with the process of complex formation (49). On the other hand, the mutation sites of the resistance-breaking ToMV mutants are located on the surface of the helicase core that is opposite the surface where the accessory domain is located. If Tm-1 binds the region around the mutation site, it is unlikely that Tm-1 competes directly with TOM1 or ARL8 for their binding to the helicase domain, although the binding of the 130K protein with Tm-1 and that with the host membrane proteins were mutually exclusive (Fig. 5). Tm-1 might inhibit the putative conformational change required for ARL8 binding, as well as membrane rearrangement.
Apart from the Tm-1 action, our results showed that negative-strand RNA synthesis is not required for the three events that are likely associated with membrane rearrangement during ToMV replication complex formation. Similarly, BMV 1a protein induces replication complex-like spherules in which replication template RNA is recruited and sequestered, even in the absence of the 2a polymerase (47). In contrast, RNA polymerase activity is required for the formation of replication complex spherules by Flock House virus and Semliki Forest virus (50, 51). In a certain host mutant, negative-strand RNA of tomato bushy stunt virus accumulates to levels similar to that in wild-type cells, but the negative-strand RNA is sensitive to nucleases, unlike in wild-type cells (52). Thus, the requirement for negative-strand RNA synthesis in membrane rearrangement/template sequestration differs from one virus to another.
ACKNOWLEDGMENTS
This study was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, to M.I.
We thank Y. Watanabe for the plasmids carrying TLIle cDNA and members of our laboratory for productive discussions.
Footnotes
Published ahead of print 8 May 2013
REFERENCES
- 1. Adams M, Antoniw J, Kreuze J. 2009. Virgaviridae: a new family of rod-shaped plant viruses. Arch. Virol. 154:1967–1972 [DOI] [PubMed] [Google Scholar]
- 2. Goelet P, Lomonossoff GP, Butler PJ, Akam ME, Gait MJ, Karn J. 1982. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. U. S. A. 79:5818–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ishikawa M, Meshi T, Motoyoshi F, Takamatsu N, Okada Y. 1986. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 14:8291–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takamatsu N, Ishikawa M, Meshi T, Okada Y. 1987. Expression of bacterial chloramphenicol acetyltransferase gene in tobacco plants mediated by TMV-RNA. EMBO J. 6:307–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meshi T, Watanabe Y, Saito T, Sugimoto A, Maeda T, Okada Y. 1987. Function of the 30 kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J. 6:2557–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salonen A, Ahola T, Kääriäinen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256 [DOI] [PubMed] [Google Scholar]
- 8. den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laliberté J-F, Sanfaçon H. 2010. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48:69–91 [DOI] [PubMed] [Google Scholar]
- 11. Mine A, Okuno T. 2012. Composition of plant virus RNA replicase complexes. Curr. Opin. Virol. 2:669–675 [DOI] [PubMed] [Google Scholar]
- 12. Karpala AJ, Doran TJ, Bean AGD. 2005. Immune responses to dsRNA: implications for gene silencing technologies. Immunol. Cell Biol. 83:211–216 [DOI] [PubMed] [Google Scholar]
- 13. Nagy PD, Pogany J. 2012. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 10:137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K. 2011. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411:374–382 [DOI] [PubMed] [Google Scholar]
- 15. Huang Y-W, Hu C-C, Lin N-S, Hsu Y-H. 2012. Unusual roles of host metabolic enzymes and housekeeping proteins in plant virus replication. Curr. Opin. Virol. 2:676–682 [DOI] [PubMed] [Google Scholar]
- 16. Stapleford KA, Miller DJ. 2010. Role of cellular lipids in positive-sense RNA virus replication complex assembly and function. Viruses 2:1055–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hagiwara Y, Komoda K, Yamanaka T, Tamai A, Meshi T, Funada R, Tsuchiya T, Naito S, Ishikawa M. 2003. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 22:344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hagiwara-Komoda Y, Hirai K, Mochizuki A, Nishiguchi M, Meshi T, Ishikawa M. 2008. Overexpression of a host factor TOM1 inhibits tomato mosaic virus propagation and suppression of RNA silencing. Virology 376:132–139 [DOI] [PubMed] [Google Scholar]
- 19. Yamanaka T, Ohta T, Takahashi M, Meshi T, Schmidt R, Dean C, Naito S, Ishikawa M. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. U. S. A. 97:10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishikiori M, Mori M, Dohi K, Okamura H, Katoh E, Naito S, Meshi T, Ishikawa M. 2011. A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication. PLoS Pathog. 7:e1002409. 10.1371/journal.ppat.1002409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishikiori M, Dohi K, Mori M, Meshi T, Naito S, Ishikawa M. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 80:8459–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishibashi K, Miyashita S, Katoh E, Ishikawa M. 2012. Host membrane proteins involved in the replication of tobamovirus RNA. Curr. Opin. Virol. 2:693–698 [DOI] [PubMed] [Google Scholar]
- 23. Kang B-C, Yeam I, Jahn MM. 2005. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43:581–621 [DOI] [PubMed] [Google Scholar]
- 24. Gómez P, Rodríguez-Hernández AM, Moury B, Aranda MA. 2009. Genetic resistance for the sustainable control of plant virus diseases: breeding, mechanisms and durability. Eur. J. Plant Pathol. 125:1–22 [Google Scholar]
- 25. Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW. 2012. Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78:51–65 [Google Scholar]
- 26. Maule AJ, Caranta C, Boulton MI. 2007. Sources of natural resistance to plant viruses: status and prospects. Mol. Plant Pathol. 8:223–231 [DOI] [PubMed] [Google Scholar]
- 27. Watanabe Y, Kishibayashi N, Motoyoshi F, Okada Y. 1987. Characterization of Tm-1 gene action on replication of common isolates and a resistance-breaking isolate of TMV. Virology 161:527–532 [DOI] [PubMed] [Google Scholar]
- 28. Ishibashi K, Masuda K, Naito S, Meshi T, Ishikawa M. 2007. An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:13833–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meshi T, Motoyoshi F, Adachi A, Watanabe Y, Takamatsu N, Okada Y. 1988. Two concomitant base substitutions in the putative replicase genes of tobacco mosaic virus confer the ability to overcome the effects of a tomato resistance gene, Tm-1. EMBO J. 7:1575–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strasser M, Pfitzner AJP. 2007. The double-resistance-breaking Tomato mosaic virus strain ToMV1-2 contains two independent single resistance-breaking domains. Arch. Virol. 152:903–914 [DOI] [PubMed] [Google Scholar]
- 31. Ishibashi K, Mawatari N, Miyashita S, Kishino H, Meshi T, Ishikawa M. 2012. Coevolution and hierarchical interactions of Tomato mosaic virus and the resistance gene Tm-1. PLoS Pathog. 8:e1002975. 10.1371/journal.ppat.1002975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komoda K, Naito S, Ishikawa M. 2004. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. U. S. A. 101:1863–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Komoda K, Mawatari N, Hagiwara-Komoda Y, Naito S, Ishikawa M. 2007. Identification of a ribonucleoprotein intermediate of tomato mosaic virus RNA replication complex formation. J. Virol. 81:2584–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pogany J, Stork J, Li Z, Nagy PD. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 105:19956–19961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barton DJ, Flanegan JB. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mine A, Hyodo K, Tajima Y, Kusumanegara K, Taniguchi T, Kaido M, Mise K, Taniguchi H, Okuno T. 2012. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J. Virol. 86:12091–12104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, Nagy PD. 2010. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175. 10.1371/journal.ppat.1001175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pathak KB, Pogany J, Xu K, White KA, Nagy PD. 2012. Defining the roles of cis-acting RNA elements in tombusvirus replicase assembly in vitro. J. Virol. 86:156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwakawa H-O, Mine A, Hyodo K, An M, Kaido M, Mise K, Okuno T. 2011. Template recognition mechanisms by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J. Virol. 85:497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamamoto H, Watanabe Y, Kamada H, Okada Y. 1997. A single amino acid substitution in the virus-encoded replicase of tomato mosaic tobamovirus alters host specificity. Mol. Plant Microbe Interact. 10:1015–1018 [Google Scholar]
- 42. Ishibashi K, Naito S, Meshi T, Ishikawa M. 2009. An inhibitory interaction between viral and cellular proteins underlies the resistance of tomato to nonadapted tobamoviruses. Proc. Natl. Acad. Sci. U. S. A. 106:8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishikiori M, Meshi T, Ishikawa M. 2012. Guanylylation-competent replication proteins of Tomato mosaic virus are disulfide-linked. Virology 434:118–128 [DOI] [PubMed] [Google Scholar]
- 44. Ishibashi K, Komoda K, Ishikawa M. 2006. In vitro translation and replication of tobamovirus RNA in a cell-free extract of evacuolated tobacco BY-2 protoplasts, p 183–194 Nagata T, Matsuoka K, Inźe D. (ed), Tobacco BY-2 cells: from cellular dynamics to omics, vol 58 Springer, Berlin, Germany [Google Scholar]
- 45. Ishikawa M, Meshi T, Ohno T, Okada Y. 1991. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J. Virol. 65:861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Osman TA, Buck KW. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 48. Tsujimoto Y, Numaga T, Ohshima K, Yano M-A, Ohsawa R, Goto DB, Naito S, Ishikawa M. 2003. Arabidopsis tobamovirus multiplication (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J. 22:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nishikiori M, Sugiyama S, Xiang H, Niiyama M, Ishibashi K, Inoue T, Ishikawa M, Matsumura H, Katoh E. 2012. Crystal structure of the superfamily 1 helicase from tomato mosaic virus. J. Virol. 86:7565–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kopek BG, Settles EW, Friesen PD, Ahlquist P. 2010. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein A, RNA templates, and polymerase activity. J. Virol. 84:12492–12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spuul P, Balistreri G, Hellström K, Golubtsov AV, Jokitalo E, Ahola T. 2011. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J. Virol. 85:4739–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barajas D, Jiang Y, Nagy PD. 2009. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5:e1000705. 10.1371/journal.ppat.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]