Abstract
Human immunodeficiency virus type 1 (HIV-1) variants show considerable geographical separation across the world, but there is limited information from Central America. We provide the first detailed investigation of the genetic diversity and molecular epidemiology of HIV-1 in six Central American countries. Phylogenetic analysis was performed on 625 HIV-1 pol gene sequences collected between 2002 and 2010 in Honduras, El Salvador, Nicaragua, Costa Rica, Panama, and Belize. Published sequences from neighboring countries (n = 57) and the rest of the world (n = 740) were included as controls. Maximum likelihood methods were used to explore phylogenetic relationships. Bayesian coalescence-based methods were used to time HIV-1 introductions. Nearly all (98.9%) Central American sequences were of subtype B. Phylogenetic analysis revealed that 437 (70%) sequences clustered within five significantly supported monophyletic clades formed essentially by Central American sequences. One clade contained 386 (62%) sequences from all six countries; the other four clades were smaller and more country specific, suggesting discrete subepidemics. The existence of one large well-supported Central American clade provides evidence that a single introduction of HIV-1 subtype B in Central America accounts for most current cases. An introduction during the early phase of the HIV-1 pandemic may explain its epidemiological success. Moreover, the smaller clades suggest a subsequent regional spread related to specific transmission networks within each country.
INTRODUCTION
Central America has a significant human immunodeficiency virus type 1 (HIV-1) epidemic, with an estimated adult prevalence that ranges from 0.2% to 2.3% (1). Belize has the highest prevalence (2.3%) and Nicaragua and Costa Rica the lowest (0.2% and 0.3%, respectively) (2). WHO estimates that approximately 200,000 people in Central America live with HIV, with Guatemala, Honduras, and El Salvador contributing the largest number of cases (3–5) (see Table S1 in the supplemental material). The first cases of HIV-1 in Central America were reported among men who have sex with men (MSM) in the middle 1980s, but since then the virus has primarily spread heterosexually in the general population (6). Today, heterosexual transmission accounts for 70% of the cases, even though HIV-1 prevalence is still higher among MSM, female sex workers (FSWs), prisoners, and some ethnic groups, such as the Kuna population in Panama and the Garifuna population in Honduras (3, 7–12).
The distributions of HIV-1 group M subtypes and circulating recombinant forms (CRFs) differ considerably across the world. HIV-1 group M subtype B dominates the epidemics in North America and Western and Central Europe, whereas subtype B and BF recombinants predominate in South America (13). The information on the molecular epidemiology of HIV-1 in Central America is still limited, but available data suggest that HIV-1 group M subtype B predominates (14–17).
As a consequence of rapid evolution, HIV-1 gene genealogies (phylogenetic trees) contain important information concerning the patterns, processes, and dynamics of viral spread at the population level (18–20). In the present study, we employed phylogenetic methods to obtain, for the first time, detailed information about HIV-1 origin and epidemiological history in Central America. We found five monophyletic HIV-1 group M subtype B clades, suggesting multiple independent introductions into Central America, possibly dating back to the 1960s to 1970s and followed by early population subdivision in several country-specific subepidemics.
MATERIALS AND METHODS
Study population.
The study included plasma, serum, or dried-blot-spot samples from 632 individuals who were diagnosed as HIV-1 positive in Belize, Honduras, El Salvador, Nicaragua, Costa Rica, and Panama during 2002 to 2010. For the purpose of this study, we generated 302 new sequences and included 330 published sequences recently obtained from Honduras (16, 21), Belize (22), and Panama (23). The study included samples from the general population in Honduras (n = 305), Costa Rica (n = 38), Nicaragua (n = 52), Panama (n = 37), and Belize (n = 21) and from MSM and FSWs from El Salvador (n = 78 and n = 38, respectively) and Honduras (n = 36 and 27, respectively).
Ethical approvals were obtained from the Regional Medical Ethics Board in each Central American country, as well as in Sweden, and by the Associate Director for Science at Division of Global HIV/AIDS, CDC. All study participants signed an informed consent form and completed a study questionnaire.
pol gene sequencing.
Sequence analysis of the protease (PR) and the first part of the reverse transcriptase (RT) of the HIV-1 pol gene (nucleotides 2268 to 3257 of reference strain HXB2, PR nucleotides 16 to 297, and RT nucleotides 1 to 708) was performed on the plasma or serum samples by using a published protocol (21, 22, 24). Dried-blood-spot samples were sequenced using another published genotyping assay developed by the CDC (25, 26).
HIV-1 subtype classification and analysis of drug resistance mutations.
Subtype was determined by uploading sequences individually into the REGA HIV-1 automated subtyping tool v2.0 (27). Subtyping was confirmed with phylogenetic analysis using Mega v5 and the neighbor-joining method with GTR+G (i.e., generalized time-reversible model plus gamma distribution) estimated distances and 1,000 bootstrap replicates (28). Drug resistance mutations were defined on the basis of the standardized list of mutations for treated (29) and untreated (30) patients.
Data sets for molecular epidemiology of HIV-1 group M subtype B.
Detailed phylogenetic analyses were performed on 625 study subjects infected with HIV-1 group M subtype B. Non-subtype B sequences were too few for meaningful analyses. Subtype B control sequences were obtained in the following way. (i) Related sequences to each study sequence were retrieved by BLAST searches in the Los Alamos HIV (LANL) database (www.hiv.lanl.gov), which generated 137 sequences after removal of duplicates. (ii) All subtype B sequences spanning our PR-plus-RT sequence fragment with known sampling year and country were downloaded from the LANL and GenBank (www.ncbi.nlm.nih.gov) (n = 15,084). Sequences that had already been retained from the BLAST searches were omitted, and from the remaining sequences, we first selected most of the available strains from other Central American neighboring countries (Bahamas, Cuba, Dominican Republic, Haiti, Jamaica, Mexico, and Trinidad and Tobago) (n = 57). Then we made three random selections of 740 sequences from the rest of the world in order to have approximately a 1:1 ratio with our Central America data set. Consequently, we analyzed three independent data sets in which our Central American sequences, sequences from the BLAST search, and sequences from neighboring countries were kept constant, but the random sequences from the rest of the world varied. We also analyzed a fourth data set in which all available pol sequences from Haiti (n = 21) were included. Finally, in order to avoid the ascertainment bias due to the overrepresentation of North American sequences in the HIV databases and to better investigate the relationship between the Central American epidemic and the epidemic in Haiti, which has been reported as the oldest HIV-1 epidemic in the Americas (31), we created five additional data sets, each one including all available pol sequences from Haiti (n = 21) and a random subsample of sequences from North America (42 strains), Central America (21 strains), South America (42 strains), and Europe/Oceania (42 strains). For each data set, two different outgroups were tested: HIV-1 group M subtype D sequences (K03554 and EF633445) or subtype C (U46016) and H (AF005496) sequences.
Sequence alignment and phylogenetic analysis.
Multiple alignments were generated using MUSCLE (32), either using all positions or excluding codons associated with drug resistance, according to the 2010 International AIDS Society (IAS) list of nucleoside reverse transcriptase inhibitor (NRTI), nonnucleoside reverse transcriptase inhibitor (NNRTI), and protease inhibitor (PI) mutations, with the exception of minor PI mutations (29).
Phylogenetic signal was tested by means of Xia's test (33) and likelihood mapping analysis (34). Model selection was executed by using PhyML implemented in TOPALi (35). The TVM+I+G model (i.e., transversion model plus proportion of invariable sites plus gamma distribution) was the model with the lowest Bayesian information criterion (BIC) (73322), followed by the GTR+I+G model (i.e., generalized time-reversible model plus proportion of invariable sites plus gamma distribution) (73344), while all other models had BICs of >20 points compared to GTR+I+G. Maximum likelihood (ML) phylogenetic analysis was performed on the alignment. The parallel version of the FastTree software (36) with the GTR+I+G model (a posteriori 20-parameter gamma optimization) was used (since the TVM+I+G model is not implemented in the FastTree software). The tree topology search was carried out using a mix of nearest-neighbor interchanges and subtree/prune/regraft moves. The reliability of each tree split was calculated by a Shimodaira-Hasegawa (S-H) test (37). For the smaller data sets, we also inferred Bayesian trees with MrBayes, using the TVM+I+G model. For each data set, eight Markov chain Monte Carlo (MCMC) runs were carried out in parallel for 107 generations with sampling every 10,000 generations. Convergence among independent runs was assessed by checking that the standard deviation of split frequencies was below 0.01 at the end of the runs (which was the case for each run). Maximum clade credibility trees were obtained from posterior distribution of trees with TreeAnnotator v.1.6.1 included in the BEAST software package (38).
Epidemiological/transmission clusters were inferred using the phyloPart program (39), with a patristic distance threshold that would maximize the number of clusters and would not be higher than the 25th percentile of the overall patristic distance distribution (see Table S2 in the supplemental material). Only subtrees of the phylogenetic tree with a P value of ≤0.1 as obtained by the Shimodaira-Hasegawa test were considered eligible for phyloPart clustering. Furthermore, a cluster was defined as a monophyletic clade only if it included at least 10 sequences.
Molecular clock analysis.
To obtain a Bayesian estimate of HIV-1 group M subtype B origin in Central America, a data set was assembled that included all sequences from the five Central American monophyletic clades, as well as several HIV-1 non-B, group M strains used as controls. The evolutionary rate (number of nucleotide substitutions per site per year) and the time of the most recent common ancestor (TMRCA) were inferred by the Markov chain Monte Carlo (MCMC) approach implemented in the BEAST software package version 1.6.1 (38). The analysis was performed assuming either a strict or a relaxed molecular clock, with different coalescent priors (constant population size, and Bayesian skyline plot) and a uniform prior for the root height with mean and standard error chosen according to previously published estimates for group M origin (the year 1921 ± 13 years) (40). An MCMC chain was run for 100,000,000 generations with sampling every 10,000th generation. Proper mixing of the MCMC was assessed by calculating the effective sampling size (ESS) for each parameter (38). All ESS values were >200, indicating sufficient mixing of the Markov chain. The results were visualized in Tracer v.1.5.
Different clock models were compared by calculating the Bayes factor (BF), which is the ratio of the marginal likelihoods of the two models (41). We calculated approximate marginal likelihoods for each model via importance sampling (1,000 bootstraps) using the harmonic mean of the sampled likelihoods (with the posterior as the importance distribution). The difference (in loge space) of marginal likelihood between any two models is the loge of the Bayes factor, loge(BF). Evidence against the null model (i.e., the one with lower marginal likelihood) is indicated by 6 > [2 · loge(BF)] > 2 (positive), 10 > [2 · ·loge(BF)] >6 (strong), and [2 · loge(BF)] > 10 (very strong). BF calculations were performed with Tracer v.1.5.
RESULTS
HIV-1 subtype classification.
Almost all sequences (625 of 632 [98.9%]) were classified as subtype B (Table 1). Five (0.8%) sequences were classified as subtype C, and two (0.3%) sequences had previously been identified as unique recombinant forms (URFs), URF_AD and URF_AK (21). Table 1 lists the baseline characteristics of the 632 patients. The median age of the participants was 29 years (range, 0 month to 71 years); the proportions of female and male were almost equal, 49% and 51%, respectively. The predominant transmission route was heterosexual (n = 443 [70.6%]): 346 of these patients were classified as belonging to the general population, whereas 65 patients were FSWs. The remaining patients reported the following routes of transmission: homosexual (18.1%), from mother to child (10.6%), and by blood products (0.6%). Two-thirds of the study participants (66%) reported that they never had been exposed to antiretroviral therapy (ART). Interestingly, 32 patients were Garifunas, a population descending from Caribe, Arawak, and West African people living in relatively isolated communities, about which few HIV epidemiological data are available to date.
Table 1.
Characteristics of the Central America HIV-1 study subjects
| Characteristica | Result for: |
||||||
|---|---|---|---|---|---|---|---|
| Belize | El Salvador | Honduras | Nicaragua | Costa Rica | Panama | Total | |
| Sequences, no. (%) | 21 (3.3) | 116 (18.4) | 368 (58.2) | 52 (8.2) | 38 (6) | 37 (5.9) | 632 (100) |
| HIV-1 subtypes, no. (%) | |||||||
| B | 17 (81) | 116 (100) | 366 (99.5) | 51 (98.1) | 38 (100) | 37 (100) | 625 (98.9) |
| Non-B | 4 (19) | 0 | 2 (0.5) | 1 (1.9) | 0 | 0 | 7 (1.1) |
| Collection date (yr) | 2004–2010 | 2008 | 2002–2007 | 2009 | 2009 | 2004–2009 | 2002–2010 |
| Gender, no. (%) | |||||||
| Male | 12 (57.1) | 78 (67.2) | 184 (50) | 0 | 31 (81.6) | 14 (37.8) | 319 (50.5) |
| Female | 9 (42.9) | 38 (32.8) | 184 (50) | 52 (100) | 7 (18.4) | 22 (59.5) | 312 (49.4) |
| Not known | 0 | 0 | 0 | 0 | 0 | 1 (2.7) | 1 (0.1) |
| Age, median (range) | 4 mo (0–48 mo) | 25 yr (18–52 yr) | 32 yr (2–68 yr) | 34 yr (19–57 yr) | 32 yr (20–71 yr) | 20 yr (1 mo to 43 yr) | 29 yr (0 mo to 71 yr) |
| Transmission route, no. (%) | |||||||
| Heterosexual | 0 | 38 (32.8) | 290 (78.8) | 52 (100) | 38 (100) | 28 (75.7) | 446 (70.6) |
| Homosexual | 0 | 78 (67.2) | 36 (9.8) | 0 | 0 | 0 | 114 (18.1) |
| Mother to child | 21 (100) | 0 | 38 (10.3) | 0 | 0 | 8 (21.6) | 67 (10.6) |
| Blood products | 0 | 0 | 4 (1.1) | 0 | 0 | 0 | 4 (0.6) |
| Not known | 0 | 0 | 0 | 0 | 0 | 1 (2.7) | 1 (0.2) |
| ART exposure, no. (%) | |||||||
| No | 4 (19) | 116 (100) | 213 (57.9) | 10 (19.2) | 38 (100) | 36 (97.3) | 417 (66) |
| Yes | 14 (66.7) | 0 | 155 (42.1) | 42 (80.8) | 0 | 0 | 211 (33.4) |
| Not known | 3 (14.3) | 0 | 0 | 0 | 0 | 1 (2.7) | 4 (0.6) |
| Resistance mutations, no. (%) | |||||||
| No resistance | 10 (58.8) | 106 (91.4) | 254 (69.4) | 35 (68.6) | 25 (65.8) | 34 (91.9) | 464 (74.2) |
| Any resistance | 7 (41.2) | 10 (8.6) | 112 (30.6) | 16 (31.4) | 13 (34.2) | 3 (8.1) | 161 (25.8) |
| Resistant to any PI | 0 | 1 (0.7) | 47 (12.8) | 2 (3.9) | 0 | 0 | 50 (8) |
| Resistant to any NRTI | 1 (5.9) | 4 (3.4) | 99 (27) | 11 (21.6) | 12 (31.6) | 3 (8.1) | 130 (20.8) |
| Resistant to any NNRTI | 7 (41.2) | 9 (7.8) | 97 (26.5) | 15 (29.4) | 2 (5.3) | 1 (2.7) | 131 (21) |
| Sequences per clade, no. (%) | |||||||
| Clade I | 5 (23.8) | 54 (46.5) | 321 (87.2) | 3 (5.8) | 2 (5.3) | 1 (2.7) | 386 (61.1) |
| Clade II | 5 (23.8) | 0 | 5 (1.4) | 0 | 0 | 0 | 10 (1.6) |
| Clade III | 0 | 1 (0.9) | 14 (3.8) | 0 | 0 | 0 | 15 (2.4) |
| Clade IV | 0 | 13 (11.2) | 1 (0.3) | 0 | 0 | 0 | 14 (2.2) |
| Clade V | 0 | 0 | 0 | 11 (21.2) | 0 | 0 | 11 (1.7) |
| Outside clades I–V | 11 (52.4) | 48 (41.4) | 27 (7.3) | 38 (73) | 36 (94.7) | 36 (97.3) | 196 (31) |
PI, protease inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor.
Antiretroviral drug resistance-associated mutations.
Mutations associated with antiretroviral drug resistance were detected in 161 (26%) of the sequences: 130 (62%) among 211 individuals with previous ART and 31 (7.5%) among 417 treatment-naive individuals. In the ART group, the prevalence of resistance mutations to any NRTIs was 53%, that to any NNRTIs was 49%, that to any PIs was 23%, that to the combination of at least one NRTI and at least one NNRTI was 42%, and that to at least one drug from each of the three drug classes (NRTIs plus NNRTIs plus PIs) was 14%. The most commonly observed mutations were M184V (46%), T215Y (26%), M41L (23%), K103N (23%), D67N (18%), and K70R (16%).
We found that 31 of 417 (7.5%; 95% confidence interval [CI], 5.1 to 10.4%) of the sequences from the treatment-naive patients had at least one mutation associated with transmitted drug resistance (TDR) (30). The prevalence of resistance mutations to any NRTI was 5.1%, that to any NNRTI was 3.6%, and that to any PI was 0.2%. The most commonly observed mutations were M41L (3.4%), K103N (2.4%), M184V (1.4%), P255H (1%), and T215Y (0.7%).
Five HIV-1 subtype B monophyletic clades in Central America.
Detailed phylogenetic analyses were carried out on the 625 subtype B sequences. There was no evidence of substitution saturation in the full or the reduced (i.e., after exclusion of positions associated with drug resistance) alignment, as demonstrated by the Xia's saturation test (P < 0.0001). Likelihood mapping analysis showed that the exclusion of drug resistance mutations improved the phylogenetic signal (52.2% versus 64.7% of the quartets localized in the center of the triangle). Therefore, all subsequent phylogenetic analyses were done on the reduced alignment.
To place the sequences from the six Central American countries in the broader context of the HIV-1 group M subtype B pandemic, we inferred eight different ML phylogenetic trees. Each tree was rooted either with HIV-1 group M subtype D sequences or with subtype C and H sequences and included the 625 subtype B sequences from the six study countries and different subsets of subtype B control sequences obtained by a combination of BLAST searches and selection of sequences from neighboring countries, as well as random sampling of worldwide sequences (see Materials and Methods). As expected, each tree showed in general a star-like topology with little resolution, but all trees resulted in identical clustering for the Central American sequences (not shown). The ML tree, including all 21 available pol sequences from Haiti and rooted with HIV-1 group M subtype D sequences, is shown in Fig. 1. By using each of our 625 Central American sequences in a BLAST search, 100 of the 137 (73%) sequences found were from the United States, while very few sequences were found from other American countries. (The origins of all control sequences are given in Table S3 in the supplemental material.) However, it is difficult to draw firm (or any) conclusions from this finding because of the ascertainment bias due to the overrepresentation of U.S. sequences in the databases.
Fig 1.
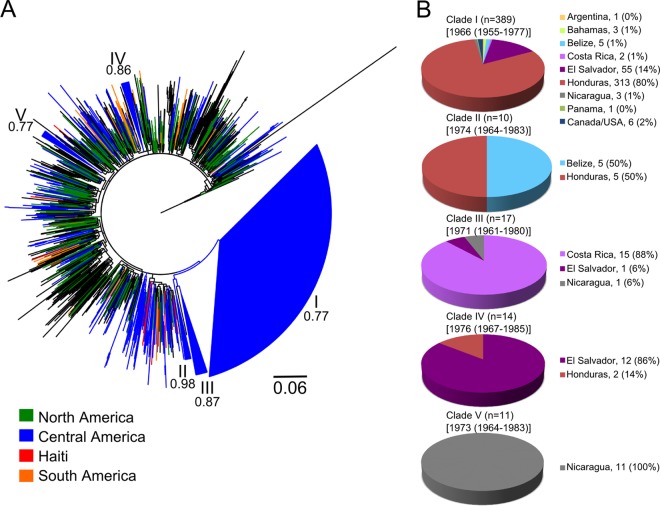
Maximum likelihood phylogenetic analysis of HIV-1 group M subtype B pol sequences. The data set included 625 HIV-1B strains from six Central American countries, including Honduras (366 strains), El Salvador (116 strains), Nicaragua (52 strains), Costa Rica (38 strains), Panama (37 strains), and Belize (17 strains), and 807 non-Central America control sequences downloaded from GenBank and the Los Alamos HIV database, including 67 sequences from Central America neighboring countries and 740 sequences from the rest of the world (including 21 Haitian sequences). The tree was rooted by using subtype D sequences as the outgroup. The tree showing the tip labels with the full name of each strain is given in Fig. S1 in the supplemental material. (A) The maximum likelihood tree shows five significantly supported monophyletic clades (numbers inside the monophyletic clades correspond to approximate likelihood ratio test S-H-like values) formed essentially by Central American sequences. Branches are scaled in nucleotide substitutions per site according to the bar at the bottom of the figure and are colored to indicate the regional origin of each American strain according to the legend in the figure, while black branches represent strains from the rest of the world (Europe, Asia, and Africa). (B) Distribution of the Central American sequences in each of the clades.
The phyloPart analyses of the ML tree identified five statistically supported monophyletic clades formed almost exclusively by Central American sequences (Fig. 1; see Fig. S1 in the supplemental material). These five clades were robust to the selection of worldwide control sequences or the sequences used as the outgroup, as they were present also in the other ML trees (not shown). Clade I was very large, contained the majority of our Central American subtype B sequences (n = 389 [62%]) (Fig. 1), and was represented in all six Central American study countries. Despite our efforts to find closely related database sequences using BLAST searches, clade I was almost exclusively Central American and included only six non-Central American sequences (four from the United States and two from Canada). There were four additional smaller sequence clusters that also fulfilled our criteria for statistically supported clades (clades II to V). Clade II included 10 strains (5 from Belize and 5 from Honduras), clade III included 17 Central American sequences (15 from Costa Rica, 1 from Nicaragua, and 1 from El Salvador), clade IV included 14 sequences (12 from El Salvador and 2 from Honduras), and clade V included 11 Nicaraguan sequences (Fig. 1). In the ML tree rooted with subtype D sequences (Fig. 1), clade III shared a most recent common ancestor with a sequence from Japan (see Fig. S1 in the supplemental material) and, in turn, with clades II and I. However, the monophyletic origin of these clades, indicating that clades II and III are just subclades of the major Central American clade I, should be taken with caution since support for this clustering pattern was extremely low (P < 0.5, S-H test) and the clustering was not present in the tree rooted with subtype C and H sequences (data not shown).
Multivariate testing showed that there were significantly higher odds (P < 0.05) for strains from MSM and mother-to-child transmission cases to be part of a cluster compared to strains from heterosexual subjects. However, the distribution of risk behavior within the five Central American clades showed no significant differences compared to the complete data set. The remaining Central American sequences fell outside the five monophyletic clades and were either intermixed in the trees or belonged to additional clades that did not meet our criteria for significant support (see Materials and Methods).
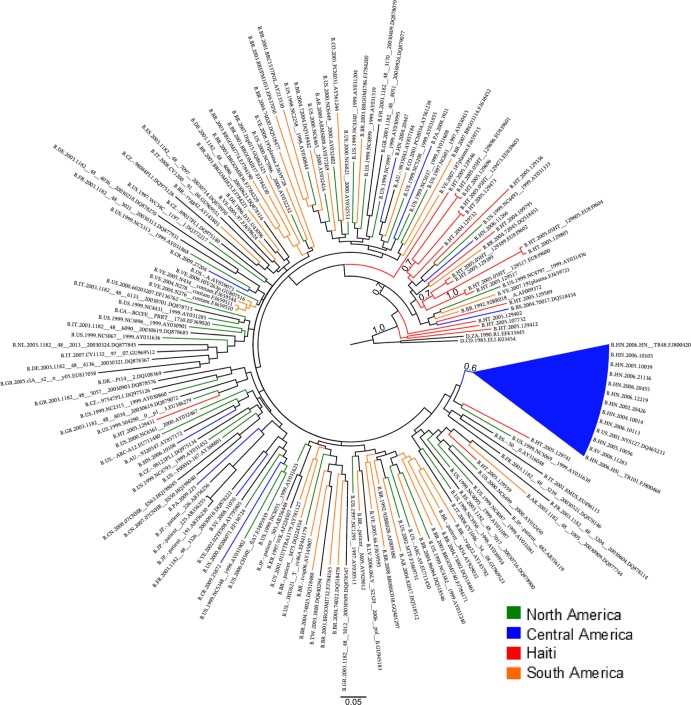
The fact that a single well-supported clade included the 62% of the Central American sequences suggests that a single introduction was responsible for a majority of the current HIV-1 cases in the region. Moreover, the existence of other four smaller clades where strains belonged to either a single country (clade V), two countries (clades II and IV), or three countries (clade III) may represent later and/or independent introductions of subtype B into Central America. There was no well-supported pattern clustering the Central American clades with control sequences from a specific region or country. In order to examine the positioning of our Central American sequences relative to Haitian sequences and the root of the tree, we needed to take into account the strong ascertainment bias in the pol alignment that included 650 Central American sequences but only 21 (available) Haitian sequences, even though both epidemics are roughly the same size (200,000 individuals). To provide a proper comparison, phylogenies were inferred from five additional data sets, each one including the Haitian sequences and a random subsample of sequences from North America (42 strains), Central America (21 strains), South America (42 strains), and Europe/Oceania (42 strains). In each tree rooted with HIV-1 subtype D sequences, the Haitian strains were basal with high posterior probability in the Bayesian analysis (Fig. 2; see Fig. S2 in the supplemental material). This is in agreement with a scenario by Gilbert et al. of a Haitian origin of HIV/AIDS in the Americas (31). Interestingly, the result could not be reproduced when more divergent sequences (subtypes C and H) were used as outgroups (data not shown). The large Central American clade I usually clustered in proximity to U.S. and Haitian strains (see Fig. S2), although the support was low (P < 0.5) in each tree. Taken together, our data indicate that the five HIV-1B Central American clades belong to the U.S.-pandemic subtype B clade.
Fig 2.
Phylogenetic analysis of HIV-1 group M subtype B pol sequences using a subsample of the reference sequences. Shown is the Bayesian maximum clade credibility tree inferred from a data set including the 21 available Haitian sequences and a random subsample of sequences from North America (42 strains), Central America (21 strains), South America (42 strains), and Europe/Oceania (42 strains). The numbers along the monophyletic branches correspond to approximate likelihood ratio test S-H-like values. Branch lengths in nucleotide substitutions per site were scaled according to the bar at the bottom of each tree. Trees from four additional data sets with randomly selected sequences are given in Fig. S2 in the supplemental material.
TMRCA and evolutionary rate of HIV-1 subtype B in Central America.
Different molecular clock models (strict and relaxed) were tested to infer the timing of the introductions of the five HIV-1 subtype B clades in Central America. The Bayes factor (BF) using either a constant population size or Bayesian skyline plot (BSP) coalescent prior favored the relaxed over the strict clock model (see Table S4 in the supplemental material). Moreover, under the relaxed molecular clock, the coefficient of variation 95% highest posterior density (HPD) intervals did not include the value 0 using either a constant (95% HPD, 0.2818 to 0.4628) or BSP (95% HPD, 0.2133 to 0.3339) coalescent prior, in agreement with a better fit of the relaxed over the strict clock model. The BSP coalescent prior also performed significantly better than the constant population size prior in the case of either a strict (BF = 13.3) or relaxed (BF = 18.1) molecular clock. Therefore, the relaxed clock model with a BSP prior was used to investigate the evolutionary rate and the TMRCA of the five Central American HIV1-B clades. Several studies, on the basis of different methods, estimated HIV-1 group M TMRCA in 1921 to 1931 using env sequences (40, 42) and in 1902 to 1939 (median, 1920) using pol sequences (43), while the origin of subtype B has been placed around the early 1960s (44). These estimates were used as internal controls to verify the robustness of the Bayesian inference. The median evolutionary rate estimated for the five clades ranged from 8.0 × 10−4 to 1.1 × 10−3 nucleotide substitutions/site/year (Table 2). The TMRCA median estimate for Central American clade I was 1966, while estimates for clades II to IV ranged from 1971 to 1976 (Table 2 and Fig. 1). However, the 95% highest posterior density (HPD) intervals were rather large, possibly due to the strong star-like signal in the pol alignments (see Table S5 in the supplemental material), and placed the origin of the five Central American clades between the mid-1950s and the mid-1980s (Table 2).
Table 2.
Bayesian estimates of the mean TMRCA for the HIV-1 subtype B sequences
| Sequence type | TMRCA (yr)a | 95% HPD (yr)b | Evolutionary ratec |
|---|---|---|---|
| Clades | |||
| I | 1966 | 1955–1977 | 9.0 × 10−4 (4.0 × 10−4-1.4 × 10−3) |
| II | 1974 | 1964–1983 | 1.0 × 10−3 (5.0 × 10−4-1.6 × 10−3) |
| III | 1971 | 1961–1980 | 1.1 × 10−3 (6.0 × 10−4-1.8 × 10−3) |
| IV | 1976 | 1967–1985 | 8.0 × 10−4 (4.0 × 10−4-1.4 × 10−3) |
| V | 1973 | 1964–1983 | 1.1 × 10−3 (6.0 × 10−4-1.7 × 10−3) |
| Global B | 1957 | 1944–1968 | 1.1 × 10−3 (6.0 × 10−4-1.7 × 10−3) |
| Group M | 1921 | 1908–1933 | 9.0 × 10−4 (4.0 × 10−4-1.4 × 10−3) |
Median estimate of TMRCA.
Lower and upper boundaries in 95% highest posterior density (HPD) intervals.
Median evolutionary rate in no. of nucleotide substitutions/site/year. The 95% HPD intervals are given in parentheses.
DISCUSSION
The present study is the first in-depth description of HIV-1 molecular epidemiology in Central America based on a large number of samples collected in six of the seven Central American countries. The findings show that subtype B is predominant in the region, which is congruent with reports from neighboring countries such as Mexico, Colombia, and Venezuela (45–47), as well as smaller reports from Central America (14–17). Interestingly, however, while HIV-1 group M subtype B strains worldwide usually intermix in a star-like phylogeny, indicative of the panmictic structure of the epidemic, strains from Central America appeared to be highly compartmentalized. The phylogenetic analysis showed that the majority of the current subtype B cases in Central America appeared to originate from a single early introduction (95% HPD 1955 to 1977). Furthermore, four additional statistically supported monophyletic clades consisting of at least 10 Central American sequences appeared to have evolved independently, suggesting the existence of discrete subepidemics within different countries, which originated from separate introductions. The fact that the largest clade included 62% of the strains may suggest that of the possible multiple subtype B introductions in the region, one was very successful and gave rise to a regional epidemic, particularly affecting Honduras and El Salvador. The origin of the major Central American clade occurred relatively soon (median estimate, 1966) after the emergence of the most recent common ancestor of HIV-1 group M subtype B (median estimate, 1957), which may explain in part the early compartmentalization of the epidemic. It is important to note, however, that our median estimate of HIV-1 group M subtype B TMRCA (1957), based on pol sequences, is 1 decade earlier than current estimates (1966) based on env (31). This may be due, at least in part, to the higher star-like signal (phylogenetic noise) in the pol gene, resulting in an unrealistically deep divergence date, although the 95% HPD intervals (1944 to 1968) of our estimates are still overlapping with the ones reported in the literature. The existence of independent monophyletic clades also implies the presence of separate transmission networks for subtype B spread within the Central American region. Therefore, our data may provide valuable information for targeted intervention and prevention.
The five Central American clades appeared to belong to the U.S./pandemic clade of HIV-1 group M subtype B (31), but there was no clear pattern clustering subtype B strains from Central America with specific sequences from the rest of the world. Interestingly, our analyses did show that some strains from Haiti were closely related to strains belonging to the major Central American clade, although statistical support was not significant, suggesting a possible common ancestor between these two epidemics. In addition, the Bayesian trees showed Haitian sequences basal to the HIV-1 group M subtype B phylogeny, in agreement with the results of Gilbert et al. (31). It is also important to note that, in spite of the observed phylogenetic noise in the pol data sets analyzed in the present work, most of the Central American sequences still clustered within a highly supported monophyletic clade, which strengthens the main finding that a single introduction is responsible for the majority of the current cases in the region.
The observation that Central American clades and strains may have originated in the U.S. or Haitian strains is expected and in agreement with known early epidemiological data, as well as other phylogenetic analyses (31), geographical proximity, known tourism, and legal and illegal immigration patterns. However, given the low resolution in the phylogenetic trees and the unbalanced sampling from other American countries, no firm conclusions can be drawn and the route of the earliest HIV-1 transmissions in Central America remains to be investigated.
We found evidence of TDR in 7.5% of the 417 treatment-naive patients from the six Central American countries, which is a moderate level according to WHO criteria (48). This is the first comprehensive report of TDR in Central America. The results have to be interpreted in light of the history of antiretroviral therapy in the countries, which initially often involved uncontrolled monotherapy and dual therapy with antiretroviral drugs that were sold illegally or sent from patients' relatives in the United States, whereas in more recent years, almost all therapy is provided as combination therapy through national treatment programs.
Some limitations of our study should be mentioned. First, we cannot exclude the possibility that the sampled population is not fully representative of all HIV-1 cases in the six Central American countries included in the study because the samples represent only a fraction of all HIV infections in the region. Furthermore, sampling was unbalanced between countries. It is possible that a more balanced sampling would show additional clades and/or that clades II to V are more widely spread across Central America than suggested by our analyses. Consequently, while our study is the most comprehensive investigation of HIV-1 spread in Central America to date, the full picture will require additional sampling from several other countries, including Guatemala, which was not sampled at all. It is also possible that the coalescence times for clades II to V would be more similar to those of clade I (i.e., earlier TMRCAs) had they been more thoroughly sampled. Another limitation was the use of partial, rather than full, genome sequences, which could underestimate the proportion of recombinant strains and possibly may limit the possibilities to resolve completely the deepest portions of the trees. However, our preliminary results suggest that the subtypes and clustering of Honduran strains are similar in analyses based on env V3 sequences.
In conclusion, this is the first comprehensive study of how and when HIV-1 has entered and spread in Central America, which is a region with a substantial HIV/AIDS burden. HIV-1 group M subtype B predominates, although sporadic non-B strains were identified. Phylogenetic and molecular clock analysis showed one major well-supported monophyletic cluster compatible with a single early subtype B introduction accounting for most current cases, as well as a subsequent expansion into regional subepidemics, which deserves further investigation in order to understand the ecological factors driving subtype B's successful emergence and dissemination in Central America.
Supplementary Material
ACKNOWLEDGMENTS
We thank in particular all study participants. Special thanks goes to Michael Worobey for providing the data sets analyzed in the paper by Gilbert et al. and two anonymous reviewers for helpful comments.
This study was funded by the Swedish International Development Cooperation Agency (Sida) on behalf of a bilateral collaboration with the National Autonomous University of Honduras, the Network for Research and Training in Tropical Diseases in Central America (NeTropica) under project no. 06-R-2010, and the Swedish Medical Research Council. The research leading to these results also received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under Chain grant agreement no. 223131 and EuroCoord grant agreement no. 260694.
Footnotes
Published ahead of print 24 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01602-12.
REFERENCES
- 1. USAID February 2011. HIV/AIDS health profiles for the Central America Region. USAID, Washington, DC: www.usaid.gov [Google Scholar]
- 2. USAID April 2011. HIV/AIDS health profiles for the Latin America and the Caribbean Region. USAID, Washington, DC: www.usaid.gov [Google Scholar]
- 3. Ministerio de Salud de El Salvador, Programa Nacional de ITS/VIH/SIDA November 2009. Situación del VIH/SIDA en El Salvador. Ministerio de Salud de El Salvador, San Salvador, El Salvador [Google Scholar]
- 4. Ministerio de Salud de Guatemala, Programa Nacional de ITS/VIH/SIDA June 2009. Reporte epidemiológico del VIH/SIDA en Guatemala. Ministerio de Salud de Guatemala, Guatemala City, Guatemala [Google Scholar]
- 5. Ministerio de Salud de Honduras, Programa Nacional de ITS/VIH/SIDA June 2011. Reporte epidemiológico del VIH/SIDA en Honduras. Ministerio de Salud de Honduras, Tegucigalpa, Honduras [Google Scholar]
- 6. Bastos FI, Caceres C, Galvao J, Veras MA, Castilho EA. 2008. AIDS in Latin America: assessing the current status of the epidemic and the ongoing response. Int. J. Epidemiol. 37:729–737 [DOI] [PubMed] [Google Scholar]
- 7. Ministerio de Salud de El Salvador 2010. Universidad del Valle de Guatemala, USAID, CDC, PASMO. Encuesta Centroamericana de vigilancia del comportamiento sexual y prevalencia de VIH/ITS en poblaciones vulnerables (ECVC El Salvador). Ministerio de Salud de El Salvador, San Salvador, El Salvador [Google Scholar]
- 8. Paz-Bailey G, Jacobson JO, Guardado ME, Hernandez FM, Nieto AI, Estrada M, Creswell J. 2011. How many men who have sex with men and female sex workers live in El Salvador? Using respondent-driven sampling and capture-recapture to estimate population sizes. Sex. Transm. Infect. 87:279–282 [DOI] [PubMed] [Google Scholar]
- 9. Paz-Bailey G, Morales-Miranda S, Jacobson JO, Gupta SK, Sabin K, Mendoza S, Paredes M, Alvarez B, Monterroso E. 2009. High rates of STD and sexual risk behaviors among Garifunas in Honduras. J. Acquir. Immune Defic. Syndr. 51(Suppl 1):S26–S34 [DOI] [PubMed] [Google Scholar]
- 10. Sabido M, Giardina F, Hernandez G, Fernandez VH, Monzon JE, Ortiz R, Montoliu A, Casabona J. 2009. The UALE Project: decline in the incidence of HIV and sexually transmitted infections and increase in the use of condoms among sex workers in Guatemala. J. Acquir. Immune Defic. Syndr. 51(Suppl 1):S35–S41 [DOI] [PubMed] [Google Scholar]
- 11. Soto RJ, Ghee AE, Nunez CA, Mayorga R, Tapia KA, Astete SG, Hughes JP, Buffardi AL, Holte SE, Holmes KK. 2007. Sentinel surveillance of sexually transmitted infections/HIV and risk behaviors in vulnerable populations in 5 Central American countries. J. Acquir. Immune Defic. Syndr. 46:101–111 [PubMed] [Google Scholar]
- 12. UNAIDS 2010. Global Report: UNAIDS report on the global epidemic. UNAIDS Secretariat, Geneva, Switzerland [Google Scholar]
- 13. Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahumada-Ruiz S, Casado C, Toala-Gonzalez I, Flores-Figueroa D, Rodriguez-French A, Lopez-Galindez C. 2008. High divergence within the major HIV type 1 subtype B epidemic in Panama. AIDS Res. Hum. Retroviruses 24:1461–1466 [DOI] [PubMed] [Google Scholar]
- 15. Lloyd B, O'Connell RJ, Michael NL, Aviles R, Palou E, Hernandez R, Cooley J, Jagodzinski LL. 2008. Prevalence of resistance mutations in HIV-1-infected Hondurans at the beginning of the National Antiretroviral Therapy Program. AIDS Res. Hum. Retroviruses 24:529–535 [DOI] [PubMed] [Google Scholar]
- 16. Murillo W, Paz-Bailey G, Morales S, Monterroso E, Paredes M, Dobbs T, Parekh BS, Albert J, Rivera IL. 2010. Transmitted drug resistance and type of infection in newly diagnosed HIV-1 individuals in Honduras. J. Clin. Virol. 49:239–244 [DOI] [PubMed] [Google Scholar]
- 17. Taylor-Castillo L, Leon-Bratti MP, Solano-Chinchilla A, Herrera-Martinez G, Boza-Cordero R, Leon B, Luftig RB, Visona K. 2010. Variability in HIV-1 partial genomic sequences in Costa Rican patients: analysis with different bioinformatics tools. Rev. Panam. Salud Publica. 27:23–31 [DOI] [PubMed] [Google Scholar]
- 18. Gray RR, Tatem AJ, Lamers S, Hou W, Laeyendecker O, Serwadda D, Sewankambo N, Gray RH, Wawer M, Quinn TC, Goodenow MM, Salemi M. 2009. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS 23:F9–F17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Markov PV, Pepin J, Frost E, Deslandes S, Labbe AC, Pybus OG. 2009. Phylogeography and molecular epidemiology of hepatitis C virus genotype 2 in Africa. J. Gen. Virol. 90:2086–2096 [DOI] [PubMed] [Google Scholar]
- 20. Pybus OG, Rambaut A. 2009. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 10:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murillo W, de Rivera IL, Parham L, Jovel E, Palou E, Karlsson AC, Albert J. 2010. Prevalence of drug resistance and importance of viral load measurements in Honduran HIV-infected patients failing antiretroviral treatment. HIV Med. 11:95–103 [DOI] [PubMed] [Google Scholar]
- 22. Parham L, de Rivera IL, Murillo W, Naver L, Largaespada N, Albert J, Karlsson AC. 2011. Short communication: high prevalence of drug resistance in HIV type 1-infected children born in Honduras and Belize 2001 to 2004. AIDS Res. Hum. Retroviruses 27:1055–1059 [DOI] [PubMed] [Google Scholar]
- 23. Ahumada-Ruiz S, Flores-Figueroa D, Toala-Gonzalez I, Thomson MM. 2009. Analysis of HIV-1 pol sequences from Panama: identification of phylogenetic clusters within subtype B and detection of antiretroviral drug resistance mutations. Infect. Genet. Evol. 9:933–940 [DOI] [PubMed] [Google Scholar]
- 24. Lindstrom A, Albert J. 2003. A simple and sensitive ‘in-house’ method for determining genotypic drug resistance in HIV-1. J. Virol. Methods 107:45–51 [DOI] [PubMed] [Google Scholar]
- 25. Yang C, McNulty A, Diallo K, Zhang J, Titanji B, Kassim S, Wadonda-Kabondo N, Aberle-Grasse J, Kibuka T, Ndumbe PM, Vedapuri S, Zhou Z, Chilima B, Nkengasong JN. 2010. Development and application of a broadly sensitive dried-blood-spot-based genotyping assay for global surveillance of HIV-1 drug resistance. J. Clin. Microbiol. 48:3158–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou Z, Wagar N, DeVos JR, Rottinghaus E, Diallo K, Nguyen DB, Bassey O, Ugbena R, Wadonda-Kabondo N, McConnell MS, Zulu I, Chilima B, Nkengasong J, Yang C. 2011. Optimization of a low cost and broadly sensitive genotyping assay for HIV-1 drug resistance surveillance and monitoring in resource-limited settings. PLoS One 6:e28184. 10.1371/journal.pone.0028184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, Seebregts C, Snoeck J, van Rensburg EJ, Wensing AM, van de Vijver DA, Boucher CA, Camacho R, Vandamme AM. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797–3800 [DOI] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. 2010. Update of the drug resistance mutations in HIV-1: December 2010. Top. HIV Med. 18:156–163 [PubMed] [Google Scholar]
- 30. Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks CF. 2008. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir. Ther. 13(Suppl 2):25–36 [PubMed] [Google Scholar]
- 31. Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M. 2007. The emergence of HIV/AIDS in the Americas and beyond. Proc. Natl. Acad. Sci. U. S. A. 104:18566–18570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia X, Xie Z, Salemi M, Chen L, Wang Y. 2003. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 26:1–7 [DOI] [PubMed] [Google Scholar]
- 34. Strimmer K, von Haeseler A. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. U. S. A. 94:6815–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25:126–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114–1116 [Google Scholar]
- 38. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prosperi MC, Ciccozzi M, Fanti I, Saladini F, Pecorari M, Borghi V, Di Giambenedetto S, Bruzzone B, Capetti A, Vivarelli A, Rusconi S, Re MC, Gismondo MR, Sighinolfi L, Gray RR, Salemi M, Zazzi M, De Luca A. 2011. A novel methodology for large-scale phylogeny partition. Nat. Commun. 2:321. 10.1038/ncomms1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suchard MA, Weiss RE, Sinsheimer JS. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013 [DOI] [PubMed] [Google Scholar]
- 42. Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789–1796 [DOI] [PubMed] [Google Scholar]
- 43. Salemi M, Strimmer K, Hall WW, Duffy M, Delaporte E, Mboup S, Peeters M, Vandamme AM. 2001. Dating the common ancestor of SIVcpz and HIV-1 group M and the origin of HIV-1 subtypes using a new method to uncover clock-like molecular evolution. FASEB J. 15:276–278 [DOI] [PubMed] [Google Scholar]
- 44. Deng X, Liu H, Shao Y, Rayner S, Yang R. 2008. The epidemic origin and molecular properties of B′: a founder strain of the HIV-1 transmission in Asia. AIDS 22:1851–1858 [DOI] [PubMed] [Google Scholar]
- 45. Delgado E, Leon-Ponte M, Villahermosa ML, Cuevas MT, Deibis L, Echeverria G, Thomson MM, Perez-Alvarez L, Osmanov S, Najera R. 2001. Analysis of HIV type 1 protease and reverse transcriptase sequences from Venezuela for drug resistance-associated mutations and subtype classification: a UNAIDS study. AIDS Res. Hum. Retroviruses 17:753–758 [DOI] [PubMed] [Google Scholar]
- 46. Eyzaguirre L, Bautista CT, Ayala C, Acosta J, Negrete M, Sateren WB, Montano SM, Sanchez JL, Carr JK. 2006. First case of HIV type 1 subtype F among men who have sex with men in Colombia. AIDS Res. Hum. Retroviruses 22:808–811 [DOI] [PubMed] [Google Scholar]
- 47. Rivera-Morales LG, Novitsky VA, Trujillo JR, Lavalle-Montalvo C, Cano-Dominguez C, Ramos-Jimenez J, Jimenez-Rios E, Flores-Flores L, Lopez-Guillen P, Gilbert P, Vannberg F, Tamez-Guerra R, Rodriguez-Padilla C, Essex M. 2001. The molecular epidemiology of HIV type 1 of men in Mexico. AIDS Res. Hum. Retroviruses 17:87–92 [DOI] [PubMed] [Google Scholar]
- 48. Myatt M, Bennett DE. 2008. A novel sequential sampling technique for the surveillance of transmitted HIV drug resistance by cross-sectional survey for use in low resource settings. Antivir. Ther. 13(Suppl 2):37–48 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.