Abstract
Recent studies indicate that expansion of NKG2C-positive natural killer (NK) cells is associated with human cytomegalovirus (HCMV); however, their activity in response to HCMV-infected cells remains unclear. We show that NKG2Chi CD57hi NK cells gated on CD3neg CD56dim cells can be phenotypically identified as HCMV-induced NK cells that can be activated by HCMV-infected cells. Using HCMV-infected autologous macrophages as targets, we were able to show that these NKG2Chi CD57hi NK cells are highly responsive to HCMV-infected macrophages only in the presence of HCMV-specific antibodies, whereas they are functionally poor effectors of natural cytotoxicity. We further demonstrate that NKG2Chi CD57hi NK cells are intrinsically responsive to signaling through CD16 cross-linking. Our findings show that the activity of pathogen-induced innate immune cells can be enhanced by adaptive humoral immunity. Understanding the activity of NKG2Chi CD57hi NK cells against HCMV-infected cells will be of relevance for the further development of adoptive immunotherapy.
INTRODUCTION
Human cytomegalovirus (HCMV) causes severe disease in immunocompromised patients. While the antiviral roles of T cells have been extensively studied and monitored in patients, human studies proving the specific relevance of NK cells against HCMV infection are still very limited. Nevertheless, NK cells are supposed to be important for protection against CMV infections in humans (1). A case report indicated that NK cell deficiency was associated with active HCMV infection (2). Another case report showed that NK cells could control HCMV infection in the absence of T cell help in a Tneg Bneg NKpos SCID patient (3). In transplant recipients, NK cell activity was shown to increase during both primary and recurrent HCMV infection, indicating that NK cells may contribute to recovery (4, 5). In vitro studies have shown that HCMV expresses multiple gene products and a microRNA to modulate the NK cell response, and the mechanisms by which these gene products act have been reviewed (6).
Although NK cells are prototypic innate immune cells, studies on mice show that NK cells also share characteristics of adaptive immune cells (7–9). During murine CMV infection, Ly49H+ NK cells proliferated preferentially, a characteristic of the adaptive immune response. These cells were shown to protect newborn mice from disease (9). In humans, studies showed that HCMV infection selectively expanded NKG2C-positive NK cells in healthy individuals (10, 11). Even in coinfections of HCMV with HIV (12, 13), hantavirus (14), and hepatitis B and hepatitis C viruses (15), the expansion of NKG2C-positive NK cells was exclusively dependent on the HCMV infection. Similar results were also obtained in studies using cells from patients with chronic lymphocytic leukemia (16) and after transplantation (11, 17, 18).
In solid-organ transplant (SOT) recipients with active HCMV infection, the percentage of CD57+ NKG2Chi NK cells increased shortly after the detection of HCMV viremia (11). Clinical studies performed after hematopoietic stem cell transplantation (HCT) and umbilical cord blood (UCB) transplantation confirmed an expansion of NKG2C+ NK cells during the acute phase of HCMV reactivation (17, 18). In humans, CD56dim and CD57 are expressed preferentially by subsets of NK cells with a mature phenotype which may define a subpopulation of highly differentiated NK cells (19, 20). CD57-positive NK cells exhibit a higher cytotoxic capacity, higher sensitivity to stimulation via CD16, and decreased responsiveness to cytokines (20). Thus, we hypothesized that NKG2Chi CD57hi NK cells may possess unique functional properties in HCMV infection.
Myeloid cells are an important site of HCMV latency and reactivation (21). Macrophages can act as antigen-presenting cells upon HCMV infection and can secret cytokines that lead to T and NK cell activation (22, 23). Furthermore, they can be obtained from peripheral blood mononuclear cells (PBMCs) to perform experiments in vitro. The HCMV strain TB40/E exhibits a broad cell tropism, including endothelial- and monocyte-derived cells (24, 25). In addition, TB40/E is available as a bacterial artificial chromosome (BAC) and is suitable for genetic manipulation (26).
NK cells are considered the main effectors of antibody-dependent cell-mediated responses, but evidence for a role of this process in immune defense against HCMV is sparse (27). Our new assay, using autologous macrophages and the HCMV strain TB40/E, enables us to investigate the role of the NKG2Chi CD57hi NK cell response to HCMV-infected macrophages and to measure the additional effect of HCMV antibodies.
MATERIALS AND METHODS
Ethics statement.
All buffy coats were purchased from the Transfusion Center of the Ulm University Hospital (Institut für Klinische Transfusionsmedizin und Immungenetik Ulm GmbH, Ulm, Germany) and were randomly obtained from anonymized healthy blood donors. All blood donors gave written informed consent to approve and authorize the use of their blood for medical, pharmaceutical, and research purposes.
Cells.
Human erythroleukemia cell line K562 (DMSZ), Burkitt lymphoma cell line Raji, and Fc receptor (FcR)-positive mouse mastocytoma cell line P815 (all from American Type Culture Collection) and monocytes, macrophages, PBMCs, and purified NK cells were cultured in RPMI 1640 medium (Gibco/Invitrogen) supplemented with 10% fetal bovine serum (FBS). Primary human foreskin fibroblasts (HFFs) were cultured in minimum essential medium (MEM) (GIBCO/Invitrogen) supplemented with 10% FBS. Macrophage colony-stimulating factor (M-CSF) (R&D Systems)-stimulated monocyte-derived macrophages were obtained from human buffy coats as previously described (22). PBMCs were frozen in 90% FBS containing 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen. In order to preserve the cytotoxic activity of NK cells, the cryoprotectant was added and removed slowly, as previously described (28). NK cells were enriched by negative selection from thawed PBMCs using a human NK cell enrichment kit (Miltenyi).
Preparation of viral stocks and macrophage infections.
HCMV strain TB40/E was propagated in HFFs. For preparation of virus stocks, infectious supernatants from HFF cultures were harvested at 5 to 7 days postinfection. Cellular debris was removed by centrifugation at 2,800 × g for 10 min, and virus particles were precipitated from the supernatants by ultracentrifugation (70,000 × g for 70 min at 10°C). Then, the pellet was resuspended in RPMI–10% FBS medium. Viral stocks were frozen at −80°C and thawed before use. The infectious titer of HCMV preparations was determined as the 50% tissue culture infective dose (TCID50) using HFFs on 96-well plates. Macrophages were infected using a multiplicity of infection (MOI) of 5 PFU/macrophage for 24 h before further experiments.
Immunofluorescence.
To determine the infection rates, macrophages were fixed at 24 h postinfection with 80% acetone and incubated with HCMV immediate early antigen (IEA) antibodies (Argene-Biosoft), followed by staining with Alexa Fluor 555 (AF555)-conjugated goat anti-mouse immunoglobulins (Molecular Probes/Invitrogen). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). The number of IEA and DAPI signals was determined in three frames per well with an automated counting feature of the Zeiss AxioVision microscope. The infection rate was calculated as the ratio of IEA-positive nuclei to total DAPI-positive nuclei.
To determine HCMV antibody binding, infected macrophages were fixed 72 h postinfection with precooled methanol for 10 min at 4°C, incubated for 20 min at 4°C with FcR blocking reagent (Miltenyi), and incubated for 120 min at 4°C with the dilutions of anti-HCMV Ig or human serum/plasma controls indicated in Fig. 3. During the last 30 min of incubation, rabbit serum (1:10; Sigma) was added to block unspecific binding of secondary antibodies. After washing, cells were incubated for 1 h at 4°C with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-human IgG (Dako). Infected and noninfected cells were stained as described above. Microphotographs were generated with a Zeiss Observer.
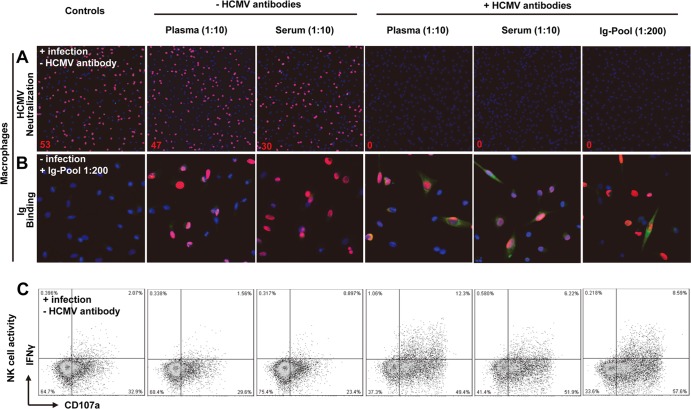
Fig 3.
Establishment of HCMV antibody-dependent NK cell-mediated responses to infected autologous macrophages. (A) Two hundred-microliter cell-free suspensions of TB40/E with or without HCMV antibodies were used in neutralization experiments on macrophages. Virus suspensions and indicated Ig preparations (autologous HCMV-antibody-positive plasma/serum with 1:10 dilution or Ig pool with 1:200 dilution) were incubated for 30 min and then inoculated onto the cells. The infection rates were assessed after 24 h of incubation. The presence of HCMV IEA (red fluorescence) indicates infected macrophages, and cell nuclei are stained in blue (DAPI). The infection rates are indicated in red numbers. Original magnification, ×40. (B) Macrophages were used for staining 3 days postinfection. Methanol-fixed macrophages were incubated with the indicated Ig preparations, followed by FITC–anti-human IgG staining. The presence of HCMV IEA (red fluorescence) indicates infected macrophages, human IgG binding on macrophages is shown by green fluorescence, and nuclei are stained in blue (DAPI). Original magnification, ×100. (C) Thawed PBMCs (1 × 106) were cocultured with TB40/E-infected macrophages (1 × 105) for 48 h. Then, surface expression of CD107a and IFN-γ production of NK cells was assessed after incubation with the indicated HCMV-specific antibodies (seronegative plasma and serum used as controls) and anti-CD107a for an additional 5 h. The results of one representative experiment from an HCMV-seropositive donor without NKG2Chi CD57hi NK cells are shown.
Serum/plasma preparation and neutralization assay.
Pooled immunoglobulins for intravenous use Gamunex (Talecris Biotherapeutics) was purchased commercially and used at a 1:200 dilution. Aliquots of autologous plasma were collected from buffy coats for subsequent cell isolation, and HCMV-IgG-negative and -positive individual serum/plasma samples were collected from whole blood by centrifugation at 4,000 rpm for 20 min. All sera/plasma were heated to 56°C for 30 min and stored at −20°C. HCMV IgG serology was determined with an enzyme-linked fluorescence assay (VIDAS CMV IgG; bioMérieux). For neutralization studies, medium containing TB40/E (MOI of 5) was incubated with the Ig preparations (using seronegative plasma/serum [1:10] as the control) indicated below for 30 min at 37°C, and then the indicated mixtures were added to the macrophages.
Flow cytometry.
The following monoclonal antibodies (MAbs) were used: peridinin chlorophyll protein (PerCP)–Cy5.5–anti-CD3 antibody (UCHT1), allophycocyanin (APC)- or phycoerythrin (PE)–Cy7–anti-CD56 antibody (B159), PE- or FITC–anti-CD107a antibody (H4A3), PE–anti-NKp46 antibody (9E2/NKp46), PE–anti-NKG2D antibody (1D11), FITC–anti-leukocyte Ig-like receptor 1 (LIR-1) antibody (GHI/75), PE–anti-CD57 antibody (NK-1), PE–anti-CD158b antibody (CH-L), FITC–anti-CD69 antibody (FN50), FITC–anti-CD94 antibody (HP-3D9), Pacific Blue–anti-CD16 antibody (3G8) (all from BD), Alexa Fluor 488 (AF488)- or PE–anti-NKG2C antibody (134591; R&D Systems), PE–anti-NKG2A antibody (131411; R&D Systems), PE–anti-interleukin-12Rβ2 (IL-12Rβ2) antibody (305719; R&D Systems), APC–anti-CD57 antibody (HCD57; BioLegend), FITC–anti-2B4 antibody (C1.7; BioLegend), APC-Vio770- or PE–anti-gamma interferon (IFN-γ) antibody (45-15, Miltenyi), PE–Vio770–anti-CD56 antibody (AF12-7H3, Miltenyi), Aqua Live/Dead fixable dead cell dye (Invitrogen), and anti-DNAX accessory molecule 1 (DNAM-1) antibody (clone 4; gift from Stipan Jonjić, Department of Histology and Embryology, Medical Faculty, University of Rijeka, Croatia). The chimeric human/mouse anti-CD20 MAb rituximab (Roche; obtained from the Pharmacy Department at the Ulm University Hospital) was used with an optimum concentration of 5 μg/ml. The antibodies used for redirect degranulation assay were purified mouse anti-human NKG2C antibody (134591; R&D Systems), purified mouse anti-human CD16 antibody (3G8; BioLegend), and an isotypic control (MOPC-21; BioLegend). The optimal concentrations for the MAb coating (NKG2C, 1 μg/ml, and CD16, 100 μg/ml) were based on maximum staining by flow cytometry (29). Eight-color staining was modified from a published NK cell panel (30). Cells were fixed and analyzed using a FACSCalibur or FACSCanto II (BD Biosciences).
NK cell degranulation and IFN-γ production assay.
After coculture of NK and target cells, monensin (GolgiStop, 2 μM; BD), brefeldin A (5 mg/ml; Sigma), and anti-CD107a MAb (20 μl/ml) were added for 5 h. Then, CD107a surface expression and IFN-γ production were analyzed by flow cytometry. The K562 cell line was used as a positive control for degranulation. To investigate the HCMV antibody-dependent NK cell-mediated response, HCMV antibodies were added together with anti-CD107a MAb. Raji cells were used as a positive control for the rituximab-dependent NK cell-mediated response.
Redirect degranulation assay.
For redirect degranulation assays, P815 cells were coated with purified anti-NKG2C antibody, anti-CD16 antibody, and the corresponding isotypic control antibodies at 37°C for 60 min. Effectors and antibody-coated P815 cells were cocultured for 4 h at an effector/target ratio of 10:1 in 96-well round-bottom plates in the presence of anti-CD107a MAb.
Statistical analysis.
The Mann-Whitney U test was performed for the data shown in Figure 1A. The nonparametric Wilcoxon signed rank sum test was used to compare the subset with NKG2C-negative NK cells paired from the same donor. Results were considered significant at a two-sided P value of 0.05.
Fig 1.
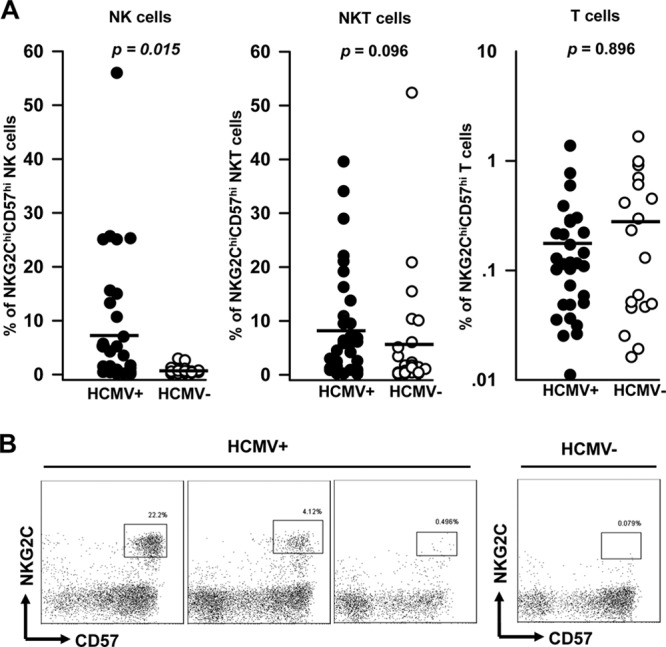
NKG2Chi CD57hi NK cells are associated with HCMV seropositivity. (A) Freshly isolated PBMCs from HCMV-seropositive (n = 35) and -seronegative (n = 24) healthy donors were stained with PerCP–Cy5.5–anti-CD3 (UCHT1), APC–anti-CD56 (B159), PE–anti-CD57 (NK-1), and AF488-NKG2C (134591) antibodies. After gating on lymphocytes, the percentages of NKG2Chi CD57hi NK cells gated on CD3− CD56dim (left), NKG2Chi CD57hi NKT cells gated on CD3+ CD56+ (middle), and NKG2Chi CD57hi T cells gated on CD3+ CD56− (right) are shown. Statistical significance is indicated at the top (Mann-Whitney U test). The horizontal lines represent the mean for each group. (B) Results of representative four-color staining to identify NKG2Chi CD57hi NK cells from 3 HCMV-seropositive donors and 1 seronegative donor are shown.
RESULTS
Identification of the NKG2Chi CD57hi NK cell subset in HCMV-seropositive healthy donors.
The identification of a CD57+ NKG2Chi NK cell subset has been reported after gating on CD3neg CD56dim CD16+ cells (11). To avoid underestimation of the role of NKG2Chi CD57hi NK cells in functional studies due to CD16 downregulation (31) as a consequence of the gating strategy, we applied a four-color staining strategy. We gated NKG2Chi CD57hi NK cells within CD3neg CD56dim populations from freshly isolated PBMCs. As shown in Figure 1A, NKG2Chi CD57hi NK cells can be detected exclusively in many of the HCMV-seropositive healthy donors. In contrast, the occurrence of NKG2Chi CD57hi NKT cells and NKG2Chi CD57hi T cells is not dependent on the HCMV serostatus (Fig. 1A) and no correlation between the percentages of NKG2Chi CD57hi NK cells and T cells was found in the seropositive donors. As shown in Figure 1B, HCMV-seropositive donors possessed substantially different amounts of NKG2Chi CD57hi NK cells. Concerning our further functional analyses, we defined donors with at least 3% NKG2Chi CD57hi NK cells within the CD3neg CD56dim population as NKG2Chi CD57hi NK cell positive. By using this cutoff, 15 of 35 (42%) HCMV-seropositive donors were NKG2Chi CD57hi NK cell positive, but none of the 24 HCMV-seronegative donors were positive for this subset (Fig. 1A). In all NKG2Chi CD57hi NK cell-positive donors, NKG2Chi NK cells expressed a high level of CD57 (clone NK-1). Therefore, we define the subset as CD57hi rather than CD57+.
NKG2Chi CD57hi NK cells express distinct inhibitory and activating receptors and show low responsiveness to HCMV-infected autologous macrophages.
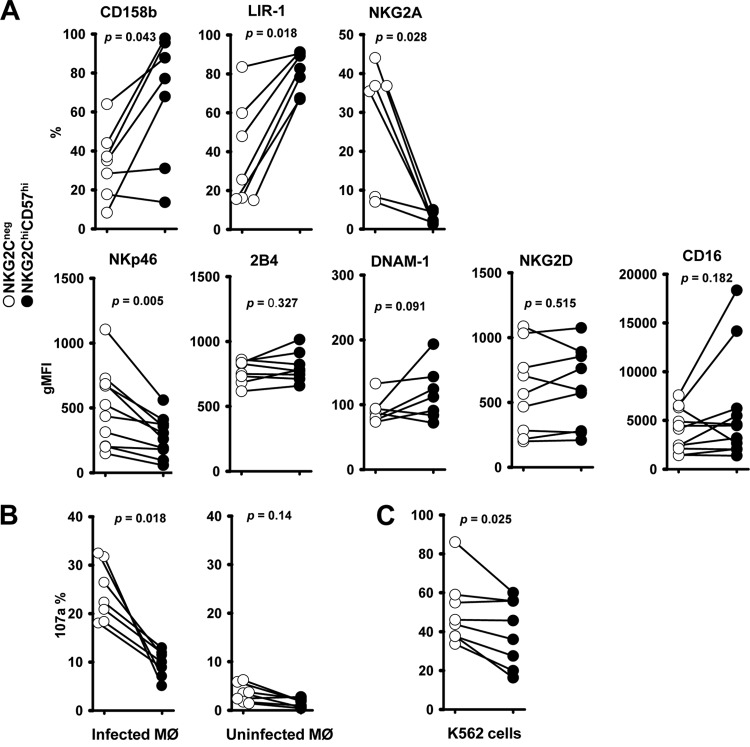
We compared the expression levels of inhibitory and activating receptors on NKG2Chi CD57hi and NKG2C-negative NK cells. As shown in Figure 2A, where the paired results from the same donor are connected by a line, NKG2Chi CD57hi NK cells (Fig. 2A, black circles) expressed significantly different amounts of the inhibitory CD158b, LIR-1, and NKG2A receptors than NKG2C-negative NK cells (Fig. 2A, white circles). CD158b is the most frequently expressed inhibitory killer cell Ig-like receptor (KIR) in NKG2C-positive NK cells (15, 17). The inhibitory leukocyte Ig-like receptor 1 (LIR-1) shows a high binding affinity for the HCMV glycoprotein UL18, which has been shown to act as a major histocompatibility complex (MHC) class I homolog (32). The inhibitory receptor NKG2A interacts specifically with the nonclassical MHC class I molecule HLA-E (33). For the first time, we could also show that, compared to NKG2C-negative NK cells, NKG2Chi CD57hi NK cells expressed lower levels of the activating receptor NKp46 (Fig. 2A, bottom), which has recently been described to be important for the NK cell response to HCMV-infected macrophages (23). In agreement with previous data (10, 11, 15), the expression levels of the activating receptors 2B4, DNAM-1, NKG2D, and CD16 were not significantly different (Fig. 2A, bottom).
Fig 2.
Expression of inhibitory and activating receptors on NKG2Chi CD57hi NK cells (A) and their degranulation to HCMV-infected autologous macrophages (B) and K562 cells (C). (A) Expression of CD158b, LIR-1, NKG2A, NKp46, 2B4, DNAM-1, NKG2D, and CD16 on NKG2Chi CD57hi NK cells and NKG2C-negative NK cells from NKG2Chi CD57hi NK cell-positive donors. The paired circles from one donor are connected by a line. Percentages (CD158b, LIR-1, and NKG2A) or geometric mean fluorescence intensities (gMFI) (NKp46, 2B4, DNAM-1, NKG2D, and CD16) are shown. (B) Thawed PBMCs (1 × 106) were cocultured with TB40/E-infected macrophages (1 × 105) or uninfected macrophages for 48 h. Then, surface expression of CD107a on NK cells was assessed 5 h after the addition of anti-CD107a. (C) Thawed PBMCs (1 × 106) were cultured for 48 h and afterwards cocultured with K562 cells (1 × 105) for 5 h in the presence of anti-CD107a, and then surface CD107a expression was assessed. (A, B, and C) The percentages of positive cells were determined on NKG2Chi CD57hi NK cells and NKG2C-negative NK cells. The nonparametric Wilcoxon signed rank sum test was used to compare NKG2Chi CD57hi NK cells with NKG2C-negative NK cells matched from the same donor.
In order to assess the functional capacity of these cells, we compared NKG2Chi CD57hi and NKG2C-negative NK populations with respect to their degranulation induced either by TB40/E-infected autologous macrophages (Fig. 2B) or MHC class I-negative K562 cells (Fig. 2C). We found that NKG2Chi CD57hi NK cells were less responsive than NKG2C-negative NK cells to both targets (Fig. 2B and C). NKG2C-positive NK cells also showed less degranulation than NKG2C-negative NK cells in response to HCMV-infected dendritic cells (34). However, a recent study suggested that NKG2C-positive NK cells may be involved in the resolution of CMV DNAemia episodes (35). The obvious question was why NKG2Chi CD57hi NK cells from HCMV-seropositive donors did not exhibit an enhanced antiviral effect.
NK cell-mediated response to HCMV-infected autologous macrophages is antibody dependent.
We hypothesize that the low degranulation capacity shown by NKG2Chi CD57hi NK cells in vitro was due to the lack of an important component normally present in the blood of donors and that HCMV antibodies might be required for an efficient NKG2Chi CD57hi NK cell response against HCMV-infected cells. Since there was no accepted standardized assay to measure HCMV-specific antibody-dependent NK cell-mediated responses, we established an autologous assay system using three essential components: HCMV antibodies binding to infected cells, target cells expressing HCMV antigens, and autologous NK cells. First, we demonstrated that plasma or serum from HCMV-positive donors can efficiently neutralize HCMV infection of macrophages. As shown in Figure 3A, comparable percentages of macrophages expressed immediate early HCMV antigens (Fig. 3A, red fluorescence) when infected with the virus alone or in the presence of plasma or serum obtained from HCMV-seronegative individuals. When macrophages were infected in the presence of plasma or serum obtained from HCMV-seropositive donors, the viral infectivity was completely abolished. We also tested a commercially available HCMV antibody preparation produced from a large pool of human plasma, because it contained a broad spectrum of HCMV antibodies (36). Second, although the fluorescence results on infected macrophages were relatively faint at a 1:10 dilution, some of the infected macrophages (Fig. 3B, red fluorescence) showed clear binding of human anti-HCMV IgG (Fig. 3B, green fluorescence). This indicates that the HCMV-infected macrophages did express HCMV antigens which could be recognized by HCMV antibodies. Third, we measured IFN-γ production and CD107a expression as indicators of the biological activity of the autologous NK cells. All three HCMV antibody preparations can efficiently enhance NK cell production of IFN-γ, as well as degranulation, in response to infected macrophages (Fig. 3C, last three graphs), while this effect is not induced by plasma or serum obtained from HCMV-seronegative donors (Fig. 3C, second and third graphs). HCMV antibodies could not elicit any response with uninfected macrophages (data not shown).
NKG2Chi CD57hi NK cells are highly responsive to infected autologous macrophages through HCMV antibody-dependent stimulation.
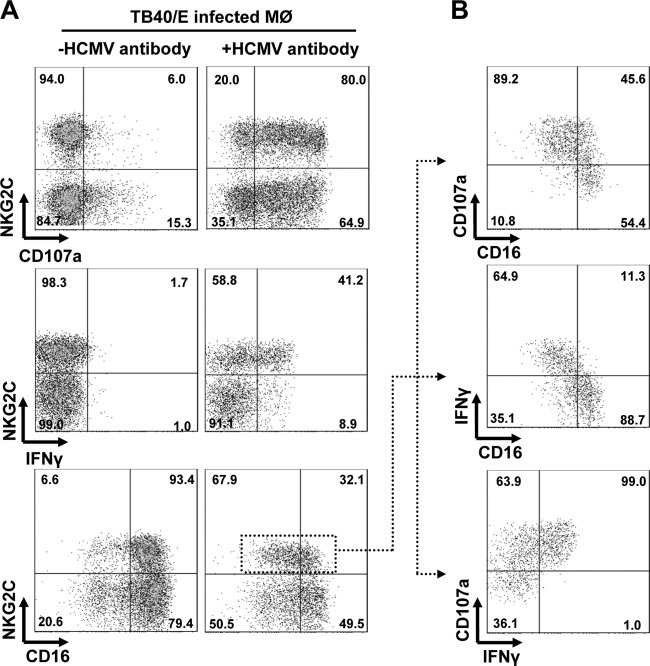
We next examined whether the NKG2Chi CD57hi NK cell subset response is also modulated by HCMV antibodies. We found that NKG2Chi CD57hi NK cells exhibit enhanced degranulation and IFN-γ production in response to HCMV-infected autologous macrophages in the presence of HCMV antibodies (Fig. 4A, right). This enhanced effect was observed in all donors tested. To examine whether this response was also dependent on the spectrum and amount of HCMV antibodies, we compared the effect of the autologous plasma with that of commercial pooled human Ig in our assay. No differences in the response of NKG2Chi CD57hi NK cells to HCMV-infected macrophages could be found (data not shown). The concentration of HCMV antibodies needed in our functional assay was even lower than the physiological concentration in humans. It is known that antibody-dependent NK cell-mediated responses occur in parallel with the CD16 downregulation of NK cells (31). This loss of CD16 is thought to prevent a continuous stimulation of NK cells and activation-induced cell death (37, 38). In the HCMV antibody-mediated NK cell response, CD16 was clearly downregulated (Fig. 4A). Similar downregulation was also observed in the rituximab-mediated NK cell response to Raji cells, which we included as a control. Our data also indicate that CD16-downregulated NKG2Chi CD57hi NK cells show enhanced degranulation and IFN-γ production (Fig. 4B, top and middle). Furthermore, the CD107a expression correlates with the IFN-γ production of NKG2Chi CD57hi NK cells (Fig. 4B, bottom). We also studied purified NK cells and found that NKG2Chi CD57hi NK cells were again highly responsive to infected macrophages in the presence of HCMV antibodies (data not shown). To study NKG2Chi CD57hi NK cell responses to superantigen stimulation, we applied staphylococcal enterotoxin B (SEB), which can induce lymphokine release (39). The SEB-stimulated responses were only observed in NKG2C-negative NK cells, and SEB stimulation did not downregulate CD16 expression (data not shown). This is consistent with the published data showing that NKG2C-positive NK cells respond differently from NKG2C-negative NK cells after cytokine stimulation (15).
Fig 4.
NKG2Chi CD57hi NK cells are highly responsive to HCMV antibody-dependent stimulation. (A) PBMCs (1 × 106) were cocultured with TB40/E-infected macrophages (1 × 105) for 48 h. Then, surface expression of CD107a, CD16, and IFN-γ production of NK cells was assessed after 5 h in the absence (left) or presence (right) of autologous plasma (1:10 dilution) and anti-CD107a. The indicated percentages of positive cells were determined as percentage of NKG2Chi CD57hi NK cells and NKG2C-negative NK cells. (B) CD16 and CD107a expression and IFN-γ production of the NKG2Chi CD57hi NK cell subset. First, NKG2Chi CD57hi NK cells were gated. Then, the percentages of positive cells were determined as percentage of CD16-positive and CD16-negative NK cells (top and middle) or as percentage of IFN-γ-positive and IFN-γ-negative NK cells (bottom). (A, B) The results of one representative experiment of four from NKG2Chi CD57hi NK cell-positive donors are shown.
NKG2C receptor contributes to NKG2Chi CD57hi NK cell activation, and co-cross-linking of NKG2C enhances CD16-mediated response.
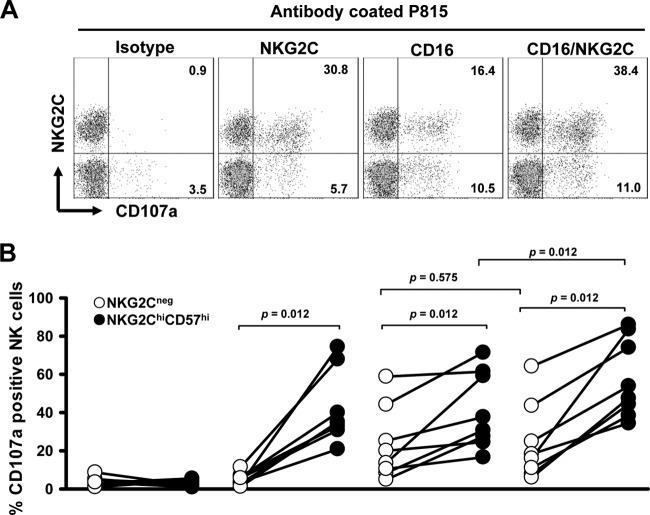
To elucidate whether a greater responsiveness to signaling through the CD16 was an intrinsic property of the NK cell subset and whether the NKG2C receptor augmented CD16-mediated stimulation, we monitored the induction of CD107a expression on NK cells after coculture with CD16 and/or NKG2C antibody-coated P815 cells. It has been shown before that antibody-coated cells induce the NK cell response better than CD16 antibody-coated plastic plates (40). The NKG2C receptor contributed to CD57+ NKG2Chi NK cell activation when stimulated on NKG2C antibody-coated plates (11). In Figure 5, we show the representative results for one donor (Fig. 5A) and the cumulated results from all experiments (Fig. 5B). In our assay, NKG2C cross-linking also induced degranulation of NKG2Chi CD57hi NK cells, while it did not activate NKG2C-negative NK cells (Fig. 5). NKG2Chi CD57hi NK cells were more responsive to CD16-coated P815 cells (Fig. 5), and we could show that both NK cell subpopulations express the same amount of CD16 (Fig. 2A). This indicates that NKG2Chi CD57hi NK cells are intrinsically more responsive to signaling through CD16. Furthermore, we show that co-cross-linking of NKG2C resulted in enhanced degranulation of NKG2Chi CD57hi NK cells in response to CD16-coated cells. Such an enhanced effect was not observed in NKG2C-negative NK cells. These data suggest that NKG2C can initiate NKG2Chi CD57hi NK cell activation and may also contribute to antibody-dependent stimulation.
Fig 5.
NKG2C induces degranulation of NKG2Chi CD57hi NK cells and augments degranulation through CD16 stimulation. (A) Representative staining of redirected degranulation against antibody-coated P815 cells. Thawed PBMCs (1 × 106) were cocultured with P815 cells (1 × 105) which had been precoated with the isotype control MAb (IgG1), anti-NKG2C MAb, or/and anti-CD16 MAb for 4 h. The percentages of CD107a-positive cells were determined on NKG2Chi CD57hi NK cells and NKG2C-negative NK cells. (B) Percentages of CD107a-positive cells on NKG2Chi CD57hi NK cells and NKG2C-negative NK cells from different donors are shown. The paired circles from one donor are connected by a line.
DISCUSSION
Increasing evidence indicates that NK cells play an important role in the control of HCMV and that HCMV infection shapes the NK cell receptor repertoire (41). Our data add important new information to this field, in particular by demonstrating that NKG2Chi CD57hi NK cells exhibit high responsiveness to HCMV-infected autologous macrophages only in the presence of antiviral antibodies.
We demonstrate that, despite a high interdonor variation concerning the relative frequencies, NKG2Chi CD57hi NK cells exclusively appear in HCMV-seropositive donors. The reason for the variability in the occurrence of this NK cell subset in HCMV-seropositive donors is not yet clear but might be related to the status of HCMV latent infection, e.g., the time of primary infection or whether and when the individual donor experienced an HCMV reactivation. This could be assumed in analogy to the fact that, in a study including SOT recipients with reactivated active HCMV infection, a significant number of CD57+ NKG2Chi NK cells appeared within 1 to 2 weeks after the detection of HCMV viremia in all patients (11). Another possibility might be that the NKG2Chi CD57hi NK cell expansion is associated with a particular setup of killer cell immunoglobulin-like receptors (KIR). A cohort study showed that self-specific KIR NK cell expansion was most pronounced in the NKG2C-positive NK cells. Most of the expanded subsets expressed at least one HLA-C-binding inhibitory KIR (41). Furthermore, the NKG2C gene copy number may also contribute to the percentage of viable NKG2Chi CD57hi NK cells. Recently, a study in children reported that a homozygous NKG2C+/+ genotype was associated with increased absolute numbers of NKG2C-positive NK cells (42).
It is known that NK cell activity is regulated by activating and inhibitory receptors. It has been shown in antibody-blocking experiments that the activating receptors NKp46, 2B4, and DNAM-1 contribute to the NK cell response to HCMV-infected macrophages (23). The receptor NKp46 is an important mediator of NK cell cytotoxicity. It has been reported to interact with hemagglutinins derived from influenza and parainfluenza viruses (43). 2B4 (also called CD244) expressed on mature human NK cells is clearly activating or coactivating (44). It is a member of the SLAM family of membrane receptors. The ligand for 2B4 is CD48, a cell surface glycoprotein expressed broadly on hematopoietic cells. DNAX accessory molecule 1 (DNAM-1) receptor (also called CD226) is a member of the Ig superfamily. CD112 (also known as polio virus receptor [PVR]) and CD155 (also called nectin-2) have been identified as ligands for DNAM-1. It regulates both NK cell migration and cellular activation (45). We could show that NKG2Chi CD57hi NK cells expressed lower levels of the activating receptor NKp46 but equal levels of 2B4 and DNAM-1 compared to the levels in NKG2C-negative cells. A lower level of expression of NKp46 may contribute to the reduced responsiveness of NKG2Chi CD57hi NK cells against infected macrophages in the absence of HCMV antibodies. We also show that NKG2Chi CD57hi NK cells express more inhibitory CD158b. CD158b (predominately KIR2D/L3) is the most frequently expressed KIR in NKG2C-positive NK cells (15, 17). It is also possible that CD158b might downregulate the response of the NKG2Chi CD57hi NK subset. Previous data support the idea that self-inhibitory KIRs could damp the NKG2C-mediated activation (15). The exact contributions of NKP46 and CD158b need further investigation.
The response of NKG2C-positive NK cells could be different in healthy subjects than in patients suffering from active HCMV infection. We and others could show that, in the absence of antibodies, NKG2C-positive NK cells are less responsive to HCMV-infected cells and K562 cells (15, 34). Another study showed that NKG2C-positive NK cells produced increasing amounts of IFN-γ after exposure to K562 cells when the NK cells were obtained from transplant recipients after HCMV reactivation (17). However, the underlying mechanism for this upregulation remains unknown. To study whether acute infection contributes to the responsiveness of NKG2C-positive NK cells, the assay presented here will be helpful.
We demonstrate in functional studies that infected autologous macrophages can be utilized to quantitatively determine HCMV antibody-dependent NK cell-mediated responses. In our assay, NKG2Chi CD57hi NK cells are highly responsive to HCMV antibody-dependent stimulation. It had been shown that CD57-positive NK cells do respond better than CD57-negative NK cells to CD16 stimulation (20); however, in our assay, we did not find NKG2Cneg CD57pos NK cells to be highly responsive in response to HCMV antibody stimulation. This discrepancy might be explained by different experimental setups or by the fact that the previous study neither took into account the HCMV serostatus of their donors nor included the analysis of NKG2C-positive NK cells. We also found that CD16 was clearly downregulated on activated NK cells after HCMV antibody stimulation. This demonstrates that including CD16 to identify the NKG2Chi CD57hi NK cell subset would indeed lead to an underestimation of the role of these cells in antibody-dependent assays. Moreover, NKG2Chi CD57hi NK cells are intrinsically more responsive to signaling through CD16. Still, the exact mechanism for the enhanced responses to CD16 stimulation is unknown. During the preparation of the manuscript, a study was published in February that is mainly in line with our findings (46). Zhang et al. describe a molecular signature for HCMV-induced NK cells. Their study shows that FcRγ-deficient NK cells, which appear in HCMV-positive donors and also mostly express the activating receptor NKG2C, show an enhanced response to both herpes simplex virus 1 (HSV-1)- and HCMV-infected allogeneic fibroblasts in the presence of virus-specific antibodies. The cross-reactivity with HSV-infected fibroblasts through HSV-specific antibody stimulation may be explained by our finding that the HCMV-induced NK cells are intrinsically more responsive to CD16 stimulation. The published FcRγ deficiency may provide the clue for the finding that HCMV-induced NK cells respond more robustly through CD16 stimulation. We could show that NKG2Chi CD57hi NK cells are also more responsive to NKp46 cross-linking (data not shown). Both CD16 and NKp46 share the same signaling adaptors, FcRγ and CD3ζ. FcRγ deficiency may enhance the signaling when CD16 and NKp46 have to exclusively use CD3ζ, which contains three immunoreceptor tyrosine-based activation motifs (ITAM) (47). A recent study showed that 38% of 151 HCMV-positive donors exhibited expanded HCMV-induced NK cells, among whom 8 donors had expanded NKG2C-negative NK cells (41). These NKG2C-negative cells expressed activating KIRs but shared the same functional profiles as NKG2C-positive NK cells. It would be interesting to investigate whether these HCMV-induced NKG2C-negative NK cells are FcRγ deficient. These outliers may help to refine the phenotype of HCMV-induced NK cells. Altogether, these phenotypic results have to be related to a functional characterization of the cells. Using autologous HCMV-infected macrophages, we demonstrate that HCMV-induced NK cells are functionally poor effectors of natural cytotoxicity but become strong effectors in the presence of HCMV-specific antibodies. In our experience, the coculture of NK cells with infected fibroblasts for 2 days provides less-reproducible results than using infected autologous macrophages. NKG2C, self-inhibitory KIRs, and FcRγ are most likely to relate to their expansion and function (10, 11, 41, 46). The most intriguing question is how these receptors and adaptors are modulated by HCMV. Understanding this mechanism may reveal fundamental properties of human NK cells.
The role of HCMV antigens for NK cell expansion and function has not been identified. HCMV encodes the MHC class I homolog glycoprotein UL18, which can bind directly to NKG2C (48). The glycoprotein UL40 can upregulate the cell surface expression of HLA-E (49), which in turn is recognized by NKG2C (33). Furthermore, HCMV US genes have been characterized for their capacity to damp the surface expression of MHC class I in infected cells (6). We have preliminary results using HCMV US 2, 3, 6, and 11 and UL18 and UL40 minus mutants in our experimental settings and have not found a contribution of these genes so far.
Although our in vitro assay mimics the physiological role of NK cells during HCMV infection, it has several limitations. First, due to limited resources, we were not able to include other types of autologous cells that are also important for HCMV infection (e.g., endothelial cells). Second, we randomly obtained buffy coats from anonymized healthy blood donors, and therefore, we could not perform kinetic studies on the function of this NK cell subset.
NKG2C-positive NK cell populations are stable in healthy adult donors (41, 46), and they expand and contract dynamically in active CMV infection (11, 17). NK cells undergo apoptosis after long-term IL-12 exposure in vitro (50), and CD57-positive NK cells express less IL-12Rβ2 chain mRNA than CD57-negative NK cells (20), which suggests that NKG2Chi CD57hi NK cells might also be less responsive to IL-12. We also found that NKG2Chi CD57hi NK cells survived preferentially after long-term IL-12 exposure with HCMV-infected macrophages (data not shown). This resistance to apoptosis may be an important feature of NKG2C-positive NK cells during active HCMV infection.
Our study demonstrates that it is extremely critical to take into account the donor HCMV serostatus in further human NK cell research, especially for studies related to NKG2C, CD57, and ADCC. It also suggests that large cohorts should be analyzed in future studies to clarify the occurrence of the NK cell subsets in different populations and their potential clinical relevance. Therefore, it would be necessary to analyze (i) the genotypic and phenotypic profiles of NKG2C, CD16, KIR receptors, and FcRγ adaptor, (ii) the longitudinal functional profiles of NKG2Chi CD57hi NK cells, and (iii) the correlation of the in vitro functional studies with the outcomes of patients with HCMV infections.
ACKNOWLEDGMENTS
We thank Ingrid Bennett for secretarial work, Marlies Just for technical help, and members of our institute for continuous discussion.
This work was supported by grants from the DFG Program International Graduate School in Molecular Medicine, Ulm University, and the Carl Zeiss Stiftung Infection Biology of Human Macrophages (no. 0563-2.8/287/1 D.3482).
Z.W. designed and performed research, analyzed data, and wrote the paper. C.S., G.F., J.R., and R.S. provided reagents and contributed to the writing of the paper. C.B. and L.W. performed research and analyzed data. T.M. designed research, analyzed data, and wrote the paper.
The authors declare no financial or commercial conflict of interest.
Footnotes
Published ahead of print 1 May 2013
REFERENCES
- 1. Lee SH, Miyagi T, Biron CA. 2007. Keeping NK cells in highly regulated antiviral warfare. Trends Immunol. 28:252–259 [DOI] [PubMed] [Google Scholar]
- 2. Biron CA, Byron KS, Sullivan JL. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320:1731–1735 [DOI] [PubMed] [Google Scholar]
- 3. Kuijpers TW, Baars PA, Dantin C, van den Burg M, van Lier RA, Roosnek E. 2008. Human NK cells can control CMV infection in the absence of T cells. Blood 112:914–915 [DOI] [PubMed] [Google Scholar]
- 4. Hertenstein B, Hampl W, Bunjes D, Wiesneth M, Duncker C, Koszinowski U, Heimpel H, Arnold R, Mertens T. 1995. In vivo/ex vivo T cell depletion for GVHD prophylaxis influences onset and course of active cytomegalovirus infection and disease after BMT. Bone Marrow Transplant. 15:387–393 [PubMed] [Google Scholar]
- 5. Venema H, van den Berg AP, van Zanten C, van Son WJ, van der Giessen M, The TH. 1994. Natural killer cell responses in renal transplant patients with cytomegalovirus infection. J. Med. Virol. 42:188–192 [DOI] [PubMed] [Google Scholar]
- 6. Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod'homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, Wang EC, Griffin CA, Davison AJ. 2008. Modulation of natural killer cells by human cytomegalovirus. J. Clin. Virol. 41:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2:951–956 [DOI] [PubMed] [Google Scholar]
- 8. O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. 2006. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 7:507–516 [DOI] [PubMed] [Google Scholar]
- 9. Sun JC, Beilke JN, Lanier LL. 2009. Adaptive immune features of natural killer cells. Nature 457:557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104:3664–3671 [DOI] [PubMed] [Google Scholar]
- 11. Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, Nixon DF, Lanier LL. 2011. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A. 108:14725–14732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guma M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, Lopez-Botet M. 2006. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J. Infect. Dis. 194:38–41 [DOI] [PubMed] [Google Scholar]
- 13. Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E, Moretta A, Mavilio D. 2010. Chronic HIV-1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co-infection. AIDS 24:27–34 [DOI] [PubMed] [Google Scholar]
- 14. Bjorkstrom NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, Michaelsson J, Malmberg KJ, Klingstrom J, Ahlm C, Ljunggren HG. 2011. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J. Exp. Med. 208:13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A, Hervier B, Theodorou I, Martinot M, Debre P, Bjorkstrom NK, Malmberg KJ, Marcellin P, Vieillard V. 2012. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur. J. Immunol. 42:447–457 [DOI] [PubMed] [Google Scholar]
- 16. Petersen L, Roug AS, Skovbo A, Thysen AH, Eskelund CW, Hokland ME. 2009. The CD94/NKG2C-expressing NK cell subset is augmented in chronic lymphocytic leukemia patients with positive human cytomegalovirus serostatus. Viral Immunol. 22:333–337 [DOI] [PubMed] [Google Scholar]
- 17. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Verges S, Lanier LL, Weisdorf D, Miller JS. 2012. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119:2665–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Della Chiesa M, Falco M, Podesta M, Locatelli F, Moretta L, Frassoni F, Moretta A. 2012. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood 119:399–410 [DOI] [PubMed] [Google Scholar]
- 19. Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Bjorklund AT, Flodstrom-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, Malmberg KJ. 2010. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116:3853–3864 [DOI] [PubMed] [Google Scholar]
- 20. Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, Lanier LL. 2010. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood 116:3865–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sinclair J, Sissons P. 2006. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 87:1763–1779 [DOI] [PubMed] [Google Scholar]
- 22. Bayer C, Varani S, Wang L, Walther P, Zhou S, Straschewski S, Bachem M, Soderberg-Naucler C, Mertens T, Frascaroli G. 2013. Human cytomegalovirus infection of m1 and m2 macrophages triggers inflammation and autologous T-cell proliferation. J. Virol. 87:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Romo N, Magri G, Muntasell A, Heredia G, Baia D, Angulo A, Guma M, Lopez-Botet M. 2011. Natural killer cell-mediated response to human cytomegalovirus-infected macrophages is modulated by their functional polarization. J. Leukoc. Biol. 90:717–726 [DOI] [PubMed] [Google Scholar]
- 24. Riegler S, Hebart H, Einsele H, Brossart P, Jahn G, Sinzger C. 2000. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J. Gen. Virol. 81:393–399 [DOI] [PubMed] [Google Scholar]
- 25. Sinzger C, Eberhardt K, Cavignac Y, Weinstock C, Kessler T, Jahn G, Davignon JL. 2006. Macrophage cultures are susceptible to lytic productive infection by endothelial-cell-propagated human cytomegalovirus strains and present viral IE1 protein to CD4+ T cells despite late downregulation of MHC class II molecules. J. Gen. Virol. 87:1853–1862 [DOI] [PubMed] [Google Scholar]
- 26. Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359–368 [DOI] [PubMed] [Google Scholar]
- 27. Couzi L, Pitard V, Sicard X, Garrigue I, Hawchar O, Merville P, Moreau JF, Dechanet-Merville J. 2012. Antibody-dependent anti-cytomegalovirus activity of human gammadelta T cells expressing CD16 (FcgammaRIIIa). Blood 119:1418–1427 [DOI] [PubMed] [Google Scholar]
- 28. Duske H, Sputtek A, Binder T, Kroger N, Schrepfer S, Eiermann T. 2011. Assessment of physiologic natural killer cell cytotoxicity in vitro. Hum. Immunol. 72:1007–1012 [DOI] [PubMed] [Google Scholar]
- 29. Goodridge JP, Witt CS, Christiansen FT, Warren HS. 2003. KIR2DL4 (CD158d) genotype influences expression and function in NK cells. J. Immunol. 171:1768–1774 [DOI] [PubMed] [Google Scholar]
- 30. Eller MA, Currier JR. 2012. OMIP-007: phenotypic analysis of human natural killer cells. Cytometry A 81:447–449 [DOI] [PubMed] [Google Scholar]
- 31. Bowles JA, Weiner GJ. 2005. CD16 polymorphisms and NK activation induced by monoclonal antibody-coated target cells. J. Immunol. Methods 304:88–99 [DOI] [PubMed] [Google Scholar]
- 32. Chapman TL, Heikeman AP, Bjorkman PJ. 1999. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 11:603–613 [DOI] [PubMed] [Google Scholar]
- 33. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391:795–799 [DOI] [PubMed] [Google Scholar]
- 34. Magri G, Muntasell A, Romo N, Saez-Borderias A, Pende D, Geraghty DE, Hengel H, Angulo A, Moretta A, Lopez-Botet M. 2011. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood 117:848–856 [DOI] [PubMed] [Google Scholar]
- 35. Munoz-Cobo B, Solano C, Benet I, Costa E, Remigia MJ, de la Camara R, Nieto J, Lopez J, Amat P, Garcia-Noblejas A, Bravo D, Clari MA, Navarro D. 2012. Functional profile of cytomegalovirus (CMV)-specific CD8(+) T cells and kinetics of NKG2C(+) NK Cells associated with the resolution of CMV DNAemia in allogeneic stem cell transplant recipients. J. Med. Virol. 84:259–267 [DOI] [PubMed] [Google Scholar]
- 36. Fouts AE, Chan P, Stephan JP, Vandlen R, Feierbach B. 2012. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin. J. Virol. 86:7444–7447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grzywacz B, Kataria N, Verneris MR. 2007. CD56(dim)CD16(+) NK cells downregulate CD16 following target cell induced activation of matrix metalloproteinases. Leukemia 21:356–359, author reply, 359 [DOI] [PubMed] [Google Scholar]
- 38. Liu Q, Sun Y, Rihn S, Nolting A, Tsoukas PN, Jost S, Cohen K, Walker B, Alter G. 2009. Matrix metalloprotease inhibitors restore impaired NK cell-mediated antibody-dependent cellular cytotoxicity in human immunodeficiency virus type 1 infection. J. Virol. 83:8705–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. D'Orazio JA, Burke GW, Stein-Streilein J. 1995. Staphylococcal enterotoxin B activates purified NK cells to secrete IFN-gamma but requires T lymphocytes to augment NK cytotoxicity. J. Immunol. 154:1014–1023 [PubMed] [Google Scholar]
- 40. Parsons MS, Zipperlen K, Gallant M, Grant M. 2010. Killer cell immunoglobulin-like receptor 3DL1 licenses CD16-mediated effector functions of natural killer cells. J. Leukoc. Biol. 88:905–912 [DOI] [PubMed] [Google Scholar]
- 41. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, Sverremark-Ekstrom E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaelsson J, Ljunggren HG, Malmberg KJ. 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noyola DE, Fortuny C, Muntasell A, Noguera-Julian A, Munoz-Almagro C, Alarcon A, Juncosa T, Moraru M, Vilches C, Lopez-Botet M. 2012. Influence of congenital human cytomegalovirus infection and the NKG2C genotype on NK-cell subset distribution in children. Eur. J. Immunol. 42:3256–3266 [DOI] [PubMed] [Google Scholar]
- 43. Koch J, Steinle A, Watzl C, Mandelboim O. 2013. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 34:182–191 [DOI] [PubMed] [Google Scholar]
- 44. Lanier LL. 2008. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lanier LL. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274 [DOI] [PubMed] [Google Scholar]
- 46. Zhang T, Scott JM, Hwang I, Kim S. 2013. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J. Immunol. 190:1402–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lanier LL. 2003. Natural killer cell receptor signaling. Curr. Opin. Immunol. 15:308–314 [DOI] [PubMed] [Google Scholar]
- 48. Kaiser BK, Pizarro JC, Kerns J, Strong RK. 2008. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc. Natl. Acad. Sci. U. S. A. 105:6696–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GW. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031. [DOI] [PubMed] [Google Scholar]
- 50. Huang Y, Lei Y, Zhang H, Zhang M, Dayton A. 2011. Interleukin-12 treatment down-regulates STAT4 and induces apoptosis with increasing ROS production in human natural killer cells. J. Leukoc. Biol. 90:87–97 [DOI] [PubMed] [Google Scholar]