Abstract
Ebola virus (EBOV) infections are characterized by deficient T lymphocyte responses, T lymphocyte apoptosis, and lymphopenia in the absence of direct infection of T lymphocytes. In contrast, dendritic cells (DC) are infected but fail to mature appropriately, thereby impairing the T cell response. We investigated the contributions of EBOV proteins in modulating DC maturation by generating recombinant viruses expressing enhanced green fluorescent protein and carrying mutations affecting several potentially immunomodulating domains. They included envelope glycoprotein (GP) domains, as well as innate response antagonist domains (IRADs) previously identified in the VP24 and VP35 proteins. GP expressed by an unrelated vector, but not the wild-type EBOV, was found to strongly induce DC maturation, and infections with recombinant EBOV carrying mutations disabling GP functional domains did not restore DC maturation. In contrast, each of the viruses carrying mutations disabling any IRAD in VP35 induced a dramatic upregulation of DC maturation markers. This was dependent on infection, but not interaction with GP. Disabling of IRADs also resulted in up to a several hundredfold increase in secretion of cytokines and chemokines. Furthermore, these mutations induced formation of homotypic DC clusters, which represent close correlates of their maturation and presumably facilitate transfer of antigen from migratory DC to lymph node DC. Thus, an individual IRAD is insufficient to suppress DC maturation; rather, the suppression of DC maturation and the “immune paralysis” observed during EBOV infections results from a cooperative effect of two or more individual IRADs.
INTRODUCTION
Ebola virus (EBOV), along with Marburg virus, is a member of the family Filoviridae. The virus causes a severe hemorrhagic fever in humans with exceptionally high, up to 90%, mortality and even greater, up to 100%, mortality in nonhuman primates (NHP) (1). After the first recorded outbreak in 1976 (2), outbreaks of EBOV infections have occurred regularly. The latest deadly outbreaks occurred in July 2012 in Uganda and in August to October 2012 in the Democratic Republic of the Congo (DRC), and a new EBOV outbreak started in Uganda in November 2012 (3).
Infections with EBOV lead to “immune paralysis” characterized by deficient T lymphocyte responses, T lymphocyte apoptosis, and lymphopenia, despite the lack of infection of T lymphocytes (4). In contrast, dendritic cells (DC), along with macrophages, are believed to be the primary targets of the virus in both humans and NHP (5–7), yet they do not undergo normal maturation despite the infection (8, 9). Importantly, inactivated preparations of EBOV not only fail to stimulate human DC (8, 9) but actually inhibit activation of DC in response to other stimuli (9). Depending on their maturation state and presentation of antigen, DC promote either T lymphocyte survival or apoptosis (10), the latter of which, if related to interleukin 2 (IL-2) deprivation, can be overcome by a cytokine cocktail (11). Thus, the lack of a functional T lymphocyte response to EBOV may be directly related to the lack of proper DC maturation.
Glycoprotein (GP) is the only envelope protein of EBOV and is a type I transmembrane protein (12). Posttranslational cleavage of the protein results in two major subunits, the N-terminal GP1 and the C-terminal GP2, which are connected by a disulfide bond, and they form trimers (13, 14). GP1 is heavily glycosylated with N- and O-linked carbohydrates and is also C-mannosylated (15, 16). The mucin domain of GP causes rounding and detachment of infected cells, resulting from a strong global downregulation of cell surface proteins and a loss of cell adherence, without causing cell death (17, 18). Another study described GP as the main viral determinant of vascular cell toxicity and injury (19), which was suggested to be mediated by a dynamin-dependent protein-trafficking pathway (20). The GP-induced cytotoxicity was also shown to be mediated by the ERK mitogen-activated protein kinase pathway (21). A single amino acid, aspartic acid, in position 71 of GP1 was suggested to be responsible for cell rounding. Replacement of this amino acid with glutamic acid in the adenovirus type 5-vectored vaccine construct completely abolished cell rounding (22). A 17-amino-acid-long immunosuppressive domain of EBOV is highly similar to the domain in the p15E envelope proteins of nononcogenic retroviruses (23, 24). The biological effects of the immunosuppressive domain of retroviruses have been studied extensively and include disabling the activity of protein kinase C, which is involved in T cell activation, suppression of cell-mediated immunity, and the induction of an imbalance between Th1 and Th2 responses (25). The peptides corresponding to the immunosuppressive domain of EBOV showed strong suppression of activation and strong apoptosis of human CD4+ and CD8+ T cells (24). However, without a direct demonstration of the domain's effects during EBOV infection, its role in pathogenesis remains unknown. The soluble (secreted) GP (sGP) (26, 27), which is an alternative form of the protein produced from the GP gene as a result of alternative transcription, was demonstrated to inhibit neutrophils (28). A recent study demonstrated that sGP competes with anti-GP antibodies and subverts the immune response to induce cross-reactivity with epitopes it shares with membrane-bound GP1 and GP2 (29).
Two EBOV proteins, VP24 and VP35, each antagonize the innate immune response through multiple mechanisms (30). Translocation of phosphorylated STAT1 to the nucleus is necessary for transcriptional activation of multiple genes induced by type I interferon (IFN-I). This translocation is triggered by its interaction with karyopherin α1 (KPNα1) and possibly other NPI-1 proteins (31–33). EBOV VP24 protein interacts with KPNα1, thereby preventing translocation of STAT-I (34), and also interacts with heterogeneous nuclear ribonuclear protein complex C1/C2 (hnRNP C1/C2), partially altering its nuclear transport (35). VP24 also blocks the phosphorylation of p38 (36), which triggers the phosphorylation of transcription factors mediating the IFN response (37). The interaction of VP24 with KPNα1 is completely prevented by a K142A mutation (38).
VP35 blocks the induction and phosphorylation of IFN regulatory factor 3 (IRF-3) and inhibits the IFN production induced by RIG-I signaling (39, 40). Introduction of K319A and R322A mutations in the VP35 protein abrogated its double-stranded RNA (dsRNA) binding activity and severely impaired its ability to suppress IFN-α/β production but did not affect the primary polymerase function of the protein; the resulting virus was attenuated in guinea pigs (41).
The ability of EBOV VP24 and VP35 proteins to antagonize the innate immune response is associated with separate clusters of amino acids located at or around positions 42 and 142 to 146 in VP24 (38) and 239, 240, 305, 309, 312, 319, 322, and 339 in VP35 (38, 42, 43). The mechanisms by which innate immunity is inhibited by these specific amino acids in both VP24 and VP35 partially overlap, and the degree of their dependence on each other is still unclear. Since some of these regions only weakly suppress the IFN response while strongly suppressing other components of innate immunity (38, 42, 43), we will refer to them as the innate response antagonist domains (IRADs), rather than IFN antagonist domains.
Here, we investigated how EBOV infection prevents effective DC maturation. We hypothesized that the deficient adaptive immune response during EBOV infections could result from the effects of one or several of the potentially immunomodulating domains located in GP, or IRADs located in VP24 and VP35. To investigate why EBOV does not cause DC maturation, we used two approaches: (i) a comparison of DC maturation induced by a chimeric virus carrying the envelope GP of EBOV and the rest of the proteins from human parainfluenza virus type 3 (HPIV3) (44) with maturation induced by EBOV and (ii) a comparison of DC maturation induced by a panel of recombinant EBOVs in which the potential immunomodulating domains were disabled by point mutations with that induced by the nonmutated (wild-type [wt]) EBOV. Consistent with previous studies (8, 9), we found that wt EBOV induced no or weak maturation of human DC. However, EBOV GP by itself was found to be a strong inducer of DC maturation, and none of the functional domains in the GP had a significant effect on DC maturation. In contrast, each of the viruses carrying mutations disabling any IRAD induced a dramatic increase in DC maturation and/or an even more dramatic (up to several hundredfold) increase in secretion of chemokines. This was accompanied by the formation of homotypic DC clusters, which represents a close correlate of their maturation and antigen-presenting potential (45). Neither the wt EBOV nor the viruses carrying mutations in GP induced DC clusters. Collectively, these results suggest that the effect of each individual IRAD is insufficient to suppress DC maturation, and thus, the strong suppression of DC maturation by the virus results from the cooperative effect of multiple IRADs.
MATERIALS AND METHODS
Viruses and cells.
To generate the recombinant viruses, we used the EBOV reverse genetics system, which included the “full-length clone,” representing the plasmid carrying the genomic RNA of wt Zaire EBOV under the control of T7 polymerase, previously modified by addition of the transcriptional cassette encoding enhanced green fluorescent protein (EGFP) between the NP and VP35 genes (46), generously provided by J. Towner and S. Nichol (CDC). The system also included the five “support plasmids” encoding the EBOV components of the polymerase NP, VP35, L, and VP30, and also T7 polymerase (47), kindly provided by Y. Kawaoka (University of Wisconsin) and H. Feldmann (NIH). The full-length clone was used to introduce the mutations described in Table 1. To generate the mutations in the GP or VP24 gene, the SalI-SacI fragment of the pEBOV plasmid encompassing a part of EBOV, including the GP gene, was cloned in a pUC19 plasmid digested by the same enzymes. The resulting subclone was subjected to mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), followed by replacement of the corresponding fragment in pEBOV with its mutagenized copy. The mutations in VP35 were introduced in a similar manner, with the exception that the ApaI-NruI fragment of the EBOV plasmid encompassing part of the genome encoding VP35 was cloned in the pcDNA3 plasmid (Invitrogen, Carlsbad, CA) digested by the same enzymes and was subjected to mutagenesis as described above. The recovered recombinant viruses were amplified by two passages in Vero-E6 cells, and the presence of the mutations was confirmed by sequencing the entire viral genomes; no reversions to the wt genotype or occurrence of any other mutations were observed in any part of the genome of any of the viruses. The growth kinetics of the viruses were compared in triplicate Vero-E6 monolayers; cells were infected at a multiplicity of infection (MOI) of 0.1 PFU/cell, incubated for 2 h, washed three times with phosphate-buffered saline, covered with minimal essential medium (MEM) with 2% fetal bovine serum, and incubated for 5 days. Daily aliquots were collected, flash frozen, and used for quantitation of the viruses by plaque titration. Plaques positive for EGFP were counted under a UV microscope. The statistical significance of the differences between the titers for each mutant compared to that for wt EBOV were evaluated by paired-sample t tests. The other viruses used in the study, HPIV3/ΔF-HN/EboGP (44) and HPIV3, were also propagated in Vero-E6 cells, and the concentrations of all the viruses were determined by plaque titration in Vero-E6 cell monolayers under a 0.9% methylcellulose overlay. Plaques of HPIV3/ΔF-HN/EboGP and HPIV3 were counted under bright-field microscopy or after immunostaining with rabbit serum specific for HPIV3, followed by treatment with goat anti-rabbit IgG antibodies conjugated with horseradish peroxidase and staining of the antigen with the 4CN two-component peroxidase substrate system (KPL, Gaithersburg, MD). Work with the EBOV full-length clone was performed in a laboratory approved by the NIH Recombinant DNA Advisory Committee. All work with EBOV was performed in the biosafety level 4 (BSL-4) facility of the Galveston National Laboratory and in the BSL-4 facility of the Robert E. Shope Laboratory (University of Texas Medical Branch [UTMB]). All staff had the appropriate training and U.S. government permissions and registrations for work with the full-length clone and with EBOV.
Table 1.
Recombinant viruses generated for the study
| Virus name | Gene/protein | Mutated amino acids and nucleotides | Description of mutations |
|---|---|---|---|
| EBOV/GP–R587D–K588Da | GP | R587D and K588D; CGT→GAT and AAG→GAC | Disabling the immunosuppressive domain (55) |
| EBOV/GP-I584L | GP | I584L; ATC→CTC | Replacement of the immunosuppressive domain with that of EBOV Reston |
| EBOV/ΔsGP | GP | AAAAAAA→AAGAAGAA | Disabling the expression of sGP and stabilizing the transcriptional editing site by three point mutations (56) |
| EBOV/GP-E71D | GP | E71D; GAA→GAT | Abrogation of cell rounding (22) |
| EBOV/VP24-K142A | VP24 | K142A; AAG→GCG | Disabling of an IRAD (38) |
| EBOV/VP35-R312A | VP35 | R312A; CGT→GCT | Disabling of an IRAD (40, 42, 81, 82) |
| EBOV/VP35-F239A | VP35 | F239A; TTC→GCC | Disabling of an IRAD (57) |
| EBOV/VP35-R322A | VP35 | R322A; CGA→GCA | Disabling of an IRAD (57) |
The virus was not capable of replication.
Analysis of GP by Western blotting.
Infected or mock-infected Vero-E6 cells were harvested at day 1 or 2 postinfection, resuspended in lysis buffer (100 mM Tris-HCl, 0.03% Triton X-100, 5 mM MgCl2, 4% SDS), and heated for 10 min at 95°C. Cell lysates were separated in NuPAGE 4 to 12% Bis-Tris gels, along with Novex Sharp Pre-Stained Protein Standard, and proteins were transferred to nitrocellulose membranes using the iBlot Gel transfer system (Invitrogen). The membranes were incubated with primary rabbit polyclonal antibodies against EBOV strain Mayinga (kindly provided by T. Ksiazek, University of Texas Medical Branch) and secondary goat anti-rabbit IgG antibodies conjugated with horseradish peroxidase (1:2,000; KPL). Protein bands were visualized using the chromogenic 4CN two-component peroxidase substrate system (KPL), scanned, and quantified using ImageJ software, version 1.46 (NIH, Bethesda, MD).
Isolation of monocytes and generation of DC.
Deidentified buffy coats obtained from the blood of healthy adult donors according to a clinical protocol approved by the UTMB Institutional Review Board were provided by the UTMB Blood Bank. CD14+ monocytes were purified by positive selection using anti-CD14 monoclonal-antibody-coated magnetic microbeads according the manufacturer's instructions (QuadroMACS; Miltenyi Biotech, Auburn, CA). The purity of the CD14+ cells was determined to be greater than 98% by flow cytometry using anti-CD14-phycoerythrin (PE) monoclonal antibodies (BD Biosciences, San Jose, CA). The monocytes were placed in 6-well plates at 1.2 × 106 cells per well in Advanced RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Quality Biologicals, Gaithersburg, MD), 2 mM l-glutamine (Invitrogen), 0.05 mM β-mercaptoethanol, 50 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) (R&D Systems, Minneapolis, MN), 16 ng/ml interleukin-4 (IL-4) (R&D Systems), 200 IU/ml penicillin, and 200 μg/ml streptomycin sulfate (Invitrogen). The cells were incubated for 7 days at 37°C, 5% CO2. Following incubation, the cells had phenotypic characteristics of immature DC: CD1a+ CD14low CD38low CD11chigh, as determined by flow cytometry.
Infection of DC.
On day 7 of incubation with IL-4 and GM-CSF, the immature DC were harvested, and their viability was tested by trypan blue staining. Cells were seeded at 6 × 105 per well in 12-well plates; inoculated at an MOI of 2 PFU/cell with the recombinant strains of EBOV listed in Table 1, HPIV3/ΔF-HN/EboGP or HPIV3, or Escherichia coli O55:B5 lipopolysaccharide (LPS) (Sigma, St. Louis, MO) at 1 μg/ml; and incubated at 37°C, 5% CO2. Microscopy of infected DC was performed using an Olympus IX71 microscope available in the BSL-4 facility of the Galveston National Laboratory, and the images were taken using a Hamamatsu ORCA-ER Monochrome digital camera with HCImage Live 4.0 software. To determine viral titers in supernatants, DC were infected as described above, incubated for 3 h, and washed three times, followed by centrifugation at 200 × g for 5 min to remove the unbound virus. Aliquots of supernatants were collected daily, centrifuged as described above to separate the cells, and frozen. Thereafter, viral concentrations were determined by plaque titration in Vero-E6 monolayers covered with medium containing 0.9% methylcellulose; EGFP plaques were counted under a UV microscope on days 3 and 4 postinfection.
Flow cytometry analysis of DC.
DC were analyzed for cell surface expression of multiple markers of maturation by flow cytometry at 40 h postinfection. Most of the DC were collected by pipetting, and the remaining cells, which were attached to the bottoms of the plates, were collected by applying staining buffer (phosphate-buffered saline containing 2% fetal bovine serum and 2 mM EDTA). The cells were pelleted by centrifugation at 200 × g at 4°C for 5 min, the buffer was removed, and the pellet was resuspended in staining buffer. DC were incubated for 20 min on ice in the dark with the monoclonal antibodies anti-CD54-PE (clone HA58), anti-CD80-allophycocyanin (APC)-H7 (clone L307.4), and anti-CD86-peridinin chlorophyll protein (PerCP)-Cy 5.5 (clone FUN-1). In addition, IgG1-PE, IgG1-APC-H7, and IgG2a-PerCp-Cy5.5 were used as the respective isotype control antibodies (all from BD Biosciences, San Jose, CA). Following incubation, the cells were washed three times with the staining buffer and resuspended in 200 μl of the same buffer. The Far Red fluorescent dye (Invitrogen) was used to evaluate cell viability by flow cytometry. Data were acquired using a flow cytometer (FACSCanto II; BD Biosciences) located in the BSL-4 facility of the Galveston National Laboratory or using the LSRII Fortessa (BD Biosciences) at the UTMB Flow Cytometry Core Facility following inactivation of infectivity by formalin treatment according to the approved standard operating procedure (SOP). The data were analyzed using FlowJo 7.6.1 software (Tree Star, Inc., Ashland, OR). The statistical significance of the differences in median fluorescence intensity (MFI) values for each individual mutant compared to that for wt EBOV were evaluated by paired-sample t test. The graphs were generated using Prism 6 software (GraphPad, San Diego, CA).
Cytokine assays.
DC culture supernatants were collected at 40 h postinoculation, inactivated by gamma irradiation according to the approved SOP, and removed from the BSL-4 facility. Concentrations of cytokines were determined using multiplex laser bead technology (Bioplex 200; Bio-Rad, Hercules, CA) by Eve Technologies (Calgary, Canada).
Chemokine-driven migration assay.
Immature DC were stimulated with EBOV, EBOV/VP24-K142A, EBOV/VP35-R312A, or LPS for 48 h, and their ability to migrate in response to a CCL19 concentration gradient was evaluated using polycarbonate Transwell plates with 5-μm-diameter pore size (Corning, Lowell, MA). Cell viability was analyzed by flow cytometry using Far Red fluorescent reactive dye. Live DC were seeded in the upper chamber at 1 × 105 per well in duplicate for each condition, and CCL19 (1 μg/ml; R&D Systems, Minneapolis, MN) was added to the lower chamber. After a 3-h incubation at 37°C, 5% CO2, DC that migrated to the lower chamber were harvested, pelleted, resuspended in 200 μl of fluorescence-activated cell sorter (FACS) buffer, and counted by flow cytometry using FACSCanto II. Data were analyzed with FlowJo software. The average number of DC specifically migrating in response to CCL19 was calculated as follows: average number of stimulated DC that migrated in the presence of CCL19 minus average number of stimulated DC that migrated in the absence of CCL19. The statistical significance of the differences in the numbers of migrating cells for each mutant compared to that for wt EBOV was evaluated by paired-sample t test. The graph was generated using Prism 6 software.
Stimulation of DC with immobilized EBOV.
EBOV and EBOV/VP35-R312A were diluted and adjusted to 1 × 106 PFU/ml and then inactivated at 56°C for 60 min. The lack of infectivity was confirmed by adding the inactivated viruses to Vero-E6 cells at an MOI of 2 PFU/cell. To immobilize the viruses, a 96-well enzyme-linked immunosorbent assay (ELISA) plate (Immulux HB; Dynex, Chantilly, VA) was coated with 150 μl of inactivated virus aliquots adjusted to 2 × 106 PFU/ml or equivalent amounts of live viruses, and the plate was stored overnight at 4°C; equivalent amounts of the viruses were stored overnight at 4°C in tubes. The next day, the wells covered with viruses were washed 6 times with PBS, inactivated viruses or viruses stored at 4°C in tubes were added to separate wells, and 150,000 immature DC were added per well in order to achieve an input MOI of 2 PFU/cell. For each condition, assays were performed in quadruplicate. Cells were collected after 40 h, stained, and analyzed by flow cytometry.
RESULTS
DC are poorly infected by EBOV.
We used recombinant EBOV expressing EGFP (46), referred to here as “wt EBOV,” to characterize EBOV infection of immature human blood monocyte-derived DC. Despite the addition of EGFP as an extra gene, the growth kinetics of the virus were not reduced (46). Immature DC were generated by 7-day-long incubation with IL-4 and GM-CSF. Following infection of immature DC at an MOI of 2 PFU/cell, the infection kinetics were monitored daily by bright-field and UV microscopy (Fig. 1A) and by flow cytometric quantification of EGFP-positive cells (Fig. 1B). Surprisingly, we found that the proportions of EGFP-positive DC at 24 and 48 h postinfection were only 1.3% and 8.3%, respectively, and even on day 5, the number of EGFP-positive cells did not exceed 26.8%. In contrast, Vero-E6 cells were readily infected at the same multiplicity; for example, by day 2, virtually all cells were EGFP positive (Fig. 1A). These data suggest that DC are not readily infected by EBOV.
Fig 1.

DC are not readily infected by EBOV. (A) Bright-field (left and middle) and UV (right) microscopy of Vero-E6 cells (day 2; top) or DC (day 3; bottom) infected or mock infected with wt EBOV. (B) Flow cytometry analysis of expression of EGFP by infected DC on days 1 to 5 after infection with wt EBOV. The experiment with DC was repeated with cells derived from blood from two different donors, which resulted in essentially similar results.
The GP of EBOV, but not EBOV particles, induces the formation of homotypic clusters of DC.
Because GP may affect the immune response through multiple mechanisms, we tested the role of whole GP in suppressing EBOV-induced maturation of DC. To do so, we used three viruses: wt EBOV; HPIV3, which causes only low-level DC maturation (48); and the replication-competent chimeric virus HPIV3/ΔF-HN/EboGP, bearing an envelope consisting solely of EBOV GP and the rest of the proteins of HPIV3 (Fig. 2A). Electron microscopy characterization of HPIV3/ΔF-HN/EboGP particles demonstrated that it is covered by GP spikes identical to those on the surface of EBOV (44). To compare the levels of expression of GP by wt EBOV and by HPIV3/ΔF-HN/EboGP, we infected monolayers of Vero-E6 cells with the two viruses at an MOI of 2 PFU/cell and analyzed the protein by Western blotting and densitometry (Fig. 2B). We found that the expression levels of GP in cells infected by HPIV3/ΔF-HN/EboGP were 84% (day 1) and 87% (day 2) of that in cells infected by wt EBOV. These data suggest that the two viruses express comparable levels of the protein. We infected DC with each of the three viruses at an MOI of 2 PFU/cell. Starting at 40 h postinfection, we observed the formation of large homotypic clusters in DC infected with HPIV3/ΔF-HN/EboGP. In contrast, DC infected with wt EBOV or HPIV3 looked similar to mock-infected DC and showed only occasional small homotypic DC clusters (Fig. 2C). The formation of such clusters is mediated by transforming growth factor β1 (TGF-β1)-induced E-cadherin, in cooperation with several constitutively expressed adhesion molecules (49). This represents a close correlate of their state of maturation (50). The formation of structures similar to such clusters was observed after viral infections in vivo (51) and is believed to facilitate stimulation of T cells by various mechanisms, including the transfer of antigen from migratory DC to lymph node DC (45). The formation of DC clusters by HPIV3/ΔF-HN/EboGP, but not EBOV or HPIV3, indicates that GP alone is insufficient to prevent the formation of clusters during EBOV infections.
Fig 2.
GP of EBOV, not whole EBOV particles, induces the formation of homotypic clusters. (A) Schematic representation of the viral particles used in the study. The EBOV proteins and RNA are depicted in red and pink, respectively, and those of HPIV3 in blue and light blue, respectively; the inserted EGFP gene is depicted in green. (B) Western blot analysis of GP expressed by wt EBOV (lanes 2) or by HPIV3/ΔF-HN/EboGP (lanes 3) on days 1 and 2 postinfection. Lanes 1, protein standards; lanes 4, mock infection. (C) Formation of homotypic clusters of DC infected with HPIV3/ΔF-HN/GP, but not EBOV or HPIV3, on day 2 postinfection.
GP induces effective DC maturation.
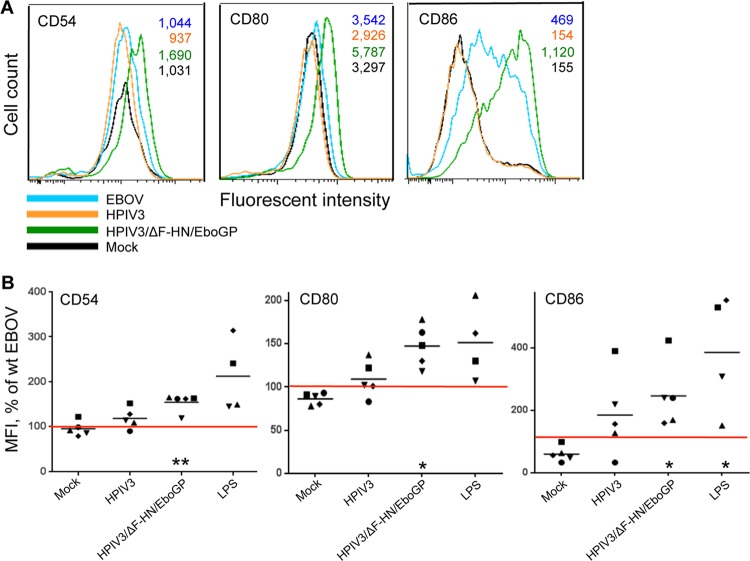
We next employed multiple markers to investigate the effect of GP on DC maturation. They included CD54, CD80 (also known as B7-1), and CD86 (B7-2). CD54 (or ICAM-1) is an intracellular adhesion molecule involved in the interaction of DC with T cells (52). CD80 and CD86 are the ligands for costimulatory receptor CD28 necessary for T cell activation and survival, respectively (53), and for CD152 (or cytotoxic T cell antigen 4 [CTLA-4]), which is an inhibitory receptor (54). DC were infected with wt EBOV, HPIV3, or HPIV3/ΔF-HN/EboGP as described above and analyzed by multicolor flow cytometry at 40 h postinfection. Consistent with the previous results (Fig. 2B), infection with HPIV3/ΔF-HN/EboGP, but not wt EBOV or HPIV3, resulted in a dramatic upregulation of the expression of CD54, CD80, and CD86 (Fig. 3). Specifically, the expression levels of these three molecules were increased by 54%, 47%, and 146% over the level induced by wt EBOV. These data suggest that GP effectively induces DC maturation, which is likely suppressed by some other components of EBOV.
Fig 3.
GP of EBOV induces maturation of DC. (A) Representative flow cytometry data on the expression of maturation markers CD54, CD80, and CD86; the MFI values are shown in the upper right corner. (B) MFI of CD54, CD80, and CD86 normalized to the level of expression induced by wt EBOV in DC from the same donor (100%, indicated by the red horizontal lines). The log10 MFI values ± standard errors (SE) for DC infected with wt EBOV were 3.5 ± 0.2, 3.7 ± 0.1, and 3.0 ± 0.2, respectively, for the three markers of maturation. The values for each donor are indicated by symbols, and the mean values are indicated by horizontal bars. Statistical significance for each treatment compared to wt EBOV: *, P < 0.05; **, P < 0.001.
Generation of EBOV carrying disabling mutations in the putative immunomodulating domains.
The experiments with HPIV3/ΔF-HN/EboGP described above suggest that GP is unlikely to significantly contribute to the suppression of DC maturation. However, the possibility exists that the functional domains present in GP may quantitatively or qualitatively modulate DC maturation. To determine the role of the putative immune-modulating domains of GP, and of the IRADs located in VP24 and VP35, in the formation of homotypic DC clusters, DC maturation, and secretion of cytokines and chemokines, we designed a panel of eight mutant EBOVs, each carrying one individual amino acid substitution in GP, VP24, or VP35 and each expressing EGFP (Fig. 4A). Specifically, we introduced the mutation E71D in GP to disable cell rounding mediated by the protein (22); the mutations R587D and K588D in GP, which disable the protein kinase C phosphotransferase activity of a peptide homologous to the immunosuppressive domain of the retroviral p15E protein (55); and the mutation I584L (24) to convert the immunosuppressive domain of EBOV to that of Reston, which is a filovirus believed to be nonpathogenic for humans, and replaced the GP editing site AAAAAAA with AAGAAGAA, as in our previous study, to transcriptionally stabilize the site and to disable the production of the secreted GP protein (56). We also disabled IRADs in VP24 by introduction of the mutation K142A (38) and in VP35 by individual introductions of the mutation F239A, R312A, or R322A (57, 58). With the exception of VP35 R312A, the effects of the four mutations disabling IRADs were previously tested using plasmid-expressed proteins, and therefore, their roles in EBOV infection remained largely unknown, as did the role of any of the four mutated amino acids in DC maturation. Seven mutated viruses, EBOV/GP-E71D, EBOV/GP-I584L, EBOV/ΔsGP, EBOV/VP24-K142A, EBOV/VP35-R312A, EBOV/VP35-F239A, and EBOV/VP35-R322A, were successfully recovered (Table 1). When the full-length clone with mutations R587D and K588D in the GP gene was used in the virus recovery experiments, isolated EGFP-positive cells were visible under UV microscopy after transfection. However, attempts to propagate the virus were unsuccessful, suggesting that the mutations were incompatible with the ability of EBOV to replicate. Next, we compared the multistep growth kinetics of the mutants in Vero-E6 cells (Fig. 4B), which demonstrated the marginally increased replication of EBOV/GP-I584L and the marginally reduced replication of EBOV/VP35-R312A (Fig. 4B) with no significant difference in any viruses.
Fig 4.
(A) Generation of the recombinant viruses. Recombinant EBOVs were generated that express EGFP and carry amino acid mutations in GP, IRADs located in the VP35 and VP24 proteins, or the RNA-editing site to disable the transcription of sGP and to restore the open reading frame encoding the transcription of the full-length GP1/2 without the requirement for transcriptional editing. The enlarged GP gene is shown in blue, with the functional elements of the protein indicated in yellow, and the EGFP gene is shown in light green. The transcriptional gene start and gene stop signals are shown as green and red bars, respectively, and the introduced mutations are indicated with red arrows. (B) Viral titers in supernatants of Vero cells infected with the indicated viruses (mean ± SE based on triplicate samples). For some values, the error bars cannot be seen due to their small size.
IRADs modulate EBOV infection of DC.
Next, we determined the effects of the mutations on the ability of EBOV to infect DC. DC infected with any of the viruses carrying mutations in GP appeared morphologically identical to those infected with the wt EBOV (data not shown). Surprisingly, UV microscopy demonstrated a greater number of infected cells for EBOV/VP24-K142A than for the wt EBOV. In contrast, for the three VP35 mutants, EGFP-positive cells were either absent or barely visible (Fig. 5A). We next determined the percentages of EGFP-positive cells and the level of EGFP expression by flow cytometry (Fig. 5B and C). These experiments demonstrated that, following infection with the viruses carrying mutations in GP, the percentages and MFIs of EGFP-positive cells were essentially the same as those of cells infected with wt EBOV (data not shown). As expected, after infections with any of the three VP35 mutants, percentages of EGFP-positive DC were strongly reduced compared to that for wt EBOV-infected DC. Consistent with the results of UV microscopy, infection with EBOV/VP24-K142A resulted in an increase in the percentages of EGFP-positive cells. The reason for this phenomenon is unclear. It could be related to suppression of protein translation by the VP24-encoded IRAD, which was disabled by the mutation, or to some other, unknown mechanism.
Fig 5.
Disabling of IRADs alters EBOV infectivity for DC. (A) Bright-field (top) and UV (bottom) microscopy of the same field on day 3 postinfection of DC with the indicated viruses. DC infected with EBOV/VP24-K142A demonstrated greater intensity of EGFP than those infected with wt EBOV, while no EGFP-positive cells were visible among DC infected with the three VP35 mutants. (B) Flow cytometry analysis of EGFP expression on day 2 after infection of DC with the viruses indicated in panel A. Percentages of EGFP+ cells and MFI are shown. The experiment was performed three times. (C) Percentages (left) and MFI (right) of EGFP+ DC on days 1 to 5 postinfection for wt EBOV and the two mutants. The experiment was performed two times. Note that DC from different donors were used in the experiments shown in panels A, B, and C, which had somewhat different levels of susceptibility to the viruses.
A recent study demonstrated abortive replication of influenza virus in mouse DC (59). To determine if the very low level of infection observed for the VP35 mutants was the result of abortive EBOV replication in DC, we quantified infectious wt EBOV in supernatants of infected DC by plaque titration in Vero-E6 cell monolayers. We found that the titers of the tested VP35 mutant were well above the limit of detection and that of the VP24 mutant was comparable to that of the wt EBOV (Fig. 6), indicating effective release of infectious viral particles from the cells. In addition, no differences in the viabilities of DC infected with wt EBOV and mutated viruses were detected (data not shown).
Fig 6.
Viral titers in supernatants of DC infected with the indicated viruses (mean ± SE based on triplicate samples). For some values, the error bars cannot be seen due to their small size.
The formation of homotypic clusters by wt EBOV-infected DC is prevented by the cooperative effect of at least two IRADs.
The viruses described above were used for 40-h-long infections of DC at an MOI of 2 PFU/cell. Consistent with the previous results (Fig. 2C), infection with wt EBOV did not cause the formation of DC clusters. The mutations in the GP gene caused no significant changes in the phenotypes of infected DC compared to wt EBOV (data not shown). Surprisingly, each of the four viruses with mutations in VP24 or VP35 IRADs induced formation of homotypic DC clusters (Fig. 7) similar to that induced by HPIV3/ΔF-HN/EboGP (Fig. 2C). The most dramatic effect was observed for EBOV/VP35-R312A, which induced formation of the largest clusters. These data suggest that none of the VP24 and VP35 IRADs may effectively prevent the formation of homotypic clusters of EBOV-infected DC when acting independently of the other IRADs. Therefore, the formation of clusters by EBOV-infected DC is prevented by the cooperative effect of multiple IRADs.
Fig 7.
Formation of homotypic DC clusters (day 2 postinfection). Disabling any of the four EBOV IRADs by a single point mutation each results in effective formation of homotypic clusters by infected DC.
The effects of individual IRADs are not sufficient to suppress DC maturation.
A multiparameter flow cytometry analysis of infected DC demonstrated, consistently with the experiment shown in Fig. 3, the lack of maturation of cells infected with wt EBOV. In contrast, infections with EBOV/VP35-R312A induced a dramatic upregulation of CD54, CD80, and CD86, by 129%, 48%, and 210% over the wt EBOV level (the data normalized to results with wt EBOV are shown in Fig. 8). Infections with EBOV/VP35-F239A and EBOV/VP35-R322A resulted in comparable increases of all three maturation markers, although the increase of some of them did not reach statistical significance due to large donor-to-donor variations. Infections with EBOV/VP24-K142A resulted in a strong increase in the level of CD86 in DC from three out of six donors. These data indicate that none of the individual IRADs is sufficient to suppress maturation of EBOV-infected DC, and therefore, IRADs located in separate parts of VP35 act cooperatively to suppress DC maturation during EBOV infection.
Fig 8.
Disabling any of the individual EBOV IRADs by a single point mutation each results in effective maturation of infected DC. (A) Expression of maturation markers CD54 and CD86 by DC infected with the indicated viruses; the MFI values are shown in the upper right corner. (B) MFI of CD54, CD80, and CD86 normalized to the level of expression induced by wt EBOV in DC from the same donor (100%, indicated by the red horizontal lines). The log10 MFI values ± SE for DC infected with wt EBOV were 3.5 ± 0.1, 3.5 ± 0.1, and 2.7 ± 0.2 for the three markers of maturation. Values for each donor are indicated by the same symbol in each plot, and the mean values are indicated by horizontal bars. Statistical significance of values for each individual mutant or LPS treatment compared to wt EBOV: *, P < 0.05; **, P < 0.01. Note that most of the DC samples were also included in the experiment shown in Fig. 3.
The mutated EBOVs induce maturation of both infected and uninfected DC.
Even though only a fraction of DC infected with wt EBOV or EBOV/VP24-K142A, and even much smaller fractions of those infected with VP35 mutants, were EGFP positive (Fig. 5A and B), the majority of the DC exposed to any of the mutated viruses appeared to be matured (Fig. 8A and B). To determine how the efficiency of infection affects DC maturation, we compared the levels of expression of the maturation markers in EGFP-positive and EGFP-negative populations of DC infected with each of the three VP35 mutants. We found that infections resulted in upregulation of both EGFP-positive and EGFP-negative cells, with somewhat greater increase of CD54 in EGFP-positive than in EGFP-negative cells and of CD86 in EGFP-negative cells than in EGFP-positive cells, and comparable levels of upregulation of CD80 in the two cell populations (Fig. 9). Overall, no significant difference between EGFP-positive and EGFP-negative DC was observed for any of the three maturation markers. These data demonstrate that effective infection of DC by the mutants, as evidenced by the expression of EGFP, is not absolutely required for the induction of their maturation, which may be caused by a low-level infection or by a transfer of antigen from infected to uninfected cells (see Discussion).
Fig 9.
The mutated EBOVs induce maturation regardless of the level of infection. (A) CD86 expression in EGFP+ (green histograms) and EGFP− (black histograms) DC. (B) Expression of the three maturation markers in EGFP+ and EGFP− DC infected with the four mutants and normalized to the levels of the markers in DC infected with wt EBOV (100%). The log10 MFI values ± SE for EGFP+ DC infected with wt EBOV were 3.1 ± 0.3, 3.4 ± 0.1, and 3.1 ± 0.2 for the three markers of maturation, and the respective values for EGFP− DC were 3.5 ± 0.2, 3.7 ± 0.1, and 2.9 ± 0.2. Values for each donor are indicated by the same symbol in each plot, and the mean values are indicated by horizontal bars. No statistical difference between EGFP+ and EGFP− DC was observed for any of the three markers of activation for any virus.
Disabling at least one IRAD is sufficient to induce secretion of chemokines and cytokines.
We next quantified 65 cytokines and chemokines in the supernatants from DC infected with the mutant or wt EBOV 40 h postinfection by a bead-based multiplex assay. We first tested supernatants of DC isolated from subject 1, which were infected with HPIV3/ΔF-HN/EboGP, EBOV/VP24-K142A, EBOV/VP35-R312A, EBOV/VP35-F239A, or EBOV/VP35-R322A. To determine reproducibility and donor-to-donor variations, the analysis was repeated with cells from subject 2, which were infected with HPIV3/ΔF-HN/EboGP, EBOV/VP24-K142A, and EBOV/VP35-R312A (infections with EBOV/VP35-F239A or EBOV/VP35-R322A were not repeated, since all three VP35 mutants generally demonstrated upregulation of the same set of cytokines and chemokines). Analysis of supernatants of infected DC from subject 2 demonstrated a profile of upregulated cytokines and chemokines similar to that of cells from subject 1. We found that infection with each of the four mutants, as well as HPIV3/ΔF-HN/EboGP, resulted in a dramatic, up to several hundredfold, upregulation of multiple cytokines and chemokines involved in the induction of immune responses (the data normalized to results with wt EBOV are shown in Fig. 10). For example, the level of CXCL10 induced by EBOV/VP35-R312A infection of DC from two donors was increased 243- and 100-fold over that induced by wt EBOV. Importantly, we detected a strong increase in the levels of chemokines in DC infected with EBOV/VP24-K142A compared to wt EBOV. Specifically, the levels of CCL2 in both donors were increased 4- and 10-fold, those of CCL7 9- and 40-fold, those of CCL5 7- and 14-fold, and those of CCL8 17- and 9-fold (Fig. 10).
Fig 10.
Disabling any of the individual EBOV IRADs by a single point mutation each results in effective secretion of chemokines and cytokines by infected DC. The levels of CXCL chemokines (row 1 from top), CCL chemokines (row 2), and cytokines, including IFNs (rows 3 and 4), in supernatants of DC infected with the VP24 and VP35 EBOV mutants, HPIV3 and HPIV3/ΔF-HN/EboGP, normalized to that for DC infected with wt EBOV. The 100% levels in DC infected by wt EBOV are shown by the red lines. Shown are results of two independent experiments, in which DC from subject 1 or DC from subject 2 were used.
The levels of IFN-α and IFN-γ were elevated 12 to 278% in supernatants of DC infected with any of the four mutants, as well as with HPIV3/ΔF-HN/EboGP, over that induced by wt EBOV (Fig. 10). Importantly, cytokines strongly upregulated due to disabling of the individual IRADs included platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF).
Interestingly, the sets of upregulated cytokines and chemokines induced by the VP24 mutant, the VP35 mutants, and HPIV3/ΔF-HN/EboGP were not identical. For example, CCL7 was upregulated by the VP24 mutant, but not the VP35 mutants. In contrast, CXCL10, CCL5, CCL8, and tumor necrosis factor alpha (TNF-α) were upregulated by the three VP35 mutants, but not by the VP24 mutant.
IRADs reduce chemotaxis of EBOV-infected DC.
Since the IRADs appear to block the expression of the maturation markers and the secretion of cytokines and chemokines by the infected DC, we hypothesized that they may also suppress their migration toward a chemokine gradient, which is a key step required for the induction of a T cell response. To test this possibility, we infected DC with wt EBOV, EBOV/VP24-K142A, or EBOV/VP35-R312A or mock infected them as described above for 40 h and placed 100,000 infected cells in the upper chambers of polycarbonate Transwell plates separated from the lower chambers by a membrane with 5-μm-diameter pores and tested their migratory capacity in response to CCL19. DC are polymorphic and typically several micrometers in diameter and therefore are able to slowly migrate through 5-μm pores. The lower chambers were filled with a medium containing the CCL19 chemokine at 1 μg/ml. After a 3-h-long incubation, cells from the lower chamber were collected and quantitated by flow cytometry. Despite the lack of maturation of DC infected with the wt EBOV, we observed some migration at a level exceeding that for the mock-infected DC. However, infection with EBOV/VP24-K142A or EBOV/VP35-R312A increased the number of cells that migrated to the lower chamber over that of DC infected with wt virus in each donor tested (Fig. 11). Specifically, the mean increases for the two viruses were 80% and 382%, although for EBOV/VP24-K142A, the increase was not statistically significant due to the variation between the samples. These data suggest that each of the IRADs located in VP24 and VP35 contributes to the inhibition of migration of infected DC toward a chemokine gradient, thus contributing to the deficient adaptive immune response.
Fig 11.
CCL19-driven migration of infected DC. Shown are the numbers of DC infected with the indicated viruses that migrated to the lower chamber during 3 h. The experiment was performed three times, with DC from each donor shown with the same symbol for each treatment and the mean values indicated by horizontal bars. The 100% level in DC infected by wt EBOV is shown by the dashed line. A statistically significant difference between cells infected with EBOV/VP35-R312A and wt EBOV is indicated by the asterisk (P = 0.002).
EBOV exposure without infection is not sufficient for DC maturation.
Since GP is the only envelope protein of EBOV, and since it was found to effectively induce DC maturation (Fig. 3) despite only a limited level of infection (Fig. 1), we hypothesized that the effect may be caused by direct contact of the virus with immature DC in the absence of actual infection as a necessary step. We therefore immobilized wt EBOV or the mutated EBOV/VP35-R312A, which effectively induces DC maturation (Fig. 8), on a high-adsorption 96-well plate by overnight incubation at 4°C and followed with a wash to remove the unbound virus, as in our previous study with adenovirus 35 (60). The virus bound to the plate could be detected by an ELISA-based assay (data not shown). In addition, viruses in amounts equal to that in the plate were either incubated overnight at 4°C in tubes, which did not result in the reduction of the viral titers (data not shown), or were inactivated by a 60-min-long incubation at 56°C, which resulted in complete loss of infectivity, and were placed in wells of the 96-well plate. Thereafter, DC were added to achieve an MOI of 2 PFU/cell, the plate was incubated for 40 h, and the DC were analyzed by flow cytometry. Incubation of DC in the presence of plate-bound viruses did not result in a significant infection, as only a few EGFP-expressing cells (presumably resulting from a small number of unbound viral particles not removed by the wash) were detected. In contrast, nonbound viruses preincubated at 4°C in a tube caused efficient infection (data not shown). CD86 was upregulated on DC in each of three donors tested after incubation with EBOV/VP35-R312A or with the virus preincubated at 4°C in a tube, but not with the immobilized or heat-inactivated virus (Fig. 12), suggesting that direct contact of DC with the virus without infection is not sufficient to induce their maturation.
Fig 12.
EBOV exposure without infection is not sufficient for DC maturation. The indicated viruses were subjected to 1-h-long incubation at 56°C, overnight incubation at 4°C in tubes, or overnight incubation at 4°C in ELISA plates. The next day, the wells of ELISA plates containing the viruses were washed, and DC were added. Alternatively, DC were added to wells containing the viruses treated in tubes or fresh, untreated viruses. At 40 h, expression of CD86 was measured by flow cytometry. The CD86 MFI values were normalized to the level of expression in mock-infected DC (100%), indicated by the dashed line. Values for each donor are indicated by symbols, and the mean values are indicated by horizontal bars. The P values of the difference between the level of CD86 in DC exposed to EBOV/VP35-R312A preincubated in a plate at 4°C versus the same virus preincubated at 4°C in a tube (*) or untreated virus (**) are 0.02 and 0.007, respectively.
DISCUSSION
This study resulted in several important conclusions about the mechanism by which EBOV suppresses the immune response. First, the lack of DC maturation during EBOV infection results from the cooperative effect of several IRADs located in two different proteins of the virus, VP35 and VP24. Each of the viruses with mutations disabling individual IRADs in either of the two proteins induced a dramatic upregulation of the expression of the classic maturation markers CD54, CD80, and CD86 and/or even more dramatic upregulation of the secretion of cytokines and chemokines compared to that induced by the wt virus. Next, each of the mutated viruses, but not wt EBOV, induced strong formation of homotypic DC clusters, which represents a close correlate of their maturation and presumably facilitates transfer of antigen from migratory DC to lymph node DC (45). In addition, the disabling of an individual IRAD in VP35 or VP24 increased chemokine-driven migration of infected DC. The effective induction of DC maturation by each of the viruses carrying one point mutation each in individual IRADs, but not by wt EBOV, suggests that the effect of each of the IRADs is insufficient to suppress maturation of DC, and thus, the strong suppression of DC maturation by the virus results from the synergistic effect of at least two IRADs (Fig. 13). To our knowledge, no such synergistic effect has been reported for any virus.
Fig 13.
Individual IRADs act synergistically to disable the adaptive immune response during EBOV infections. (A) Summary of the effects of EBOV GP and IRADs located in VP24 and VP35 on DC maturation. ND, not determined; ++++, a very strong effect; +++, a strong effect; ++, a moderate effect; +, a weak effect; −, no effect. (B) Hypothetical model of the effects of individual IRADs located in the VP35 protein on the adaptive immune response. While disabling of any of the IRADs results in effective DC maturation sufficient for the induction of a T cell response (left), the synergistic effect of at least two individual IRADs of wt EBOV effectively blocks DC maturation and the induction of a T cell response (right).
Second, these data demonstrate that GP is a strong inducer of DC maturation and that the induction of DC maturation caused by GP or other components of EBOV particles is suppressed by IRADs located in VP35 and, to a lesser degree, VP24. These data are consistent with the previously published data demonstrating that virus-like particles, which included only the GP, NP, and VP40 proteins, did cause maturation of DC (61) and induced a protective immune response in animal models (62, 63). They are also consistent with a recently published study demonstrating that infection of mouse DC by a recombinant herpes simplex virus expressing EBOV VP35 reduces their maturation (64). The data also explain the high level of protection against EBOV challenge achieved in the NHP model by vectored vaccines carrying GP as a single immunogen (63, 65–67) and the lack of significant protection in response to whole-virion vaccines (reviewed in reference 4), which include all viral proteins, including VP35 and VP24. While the replication-deficient EBOV was protective in rodents (68), it is possible that the immunosuppressive effect of VP35 and VP24 was significantly reduced due to the lack of replication and, therefore, the small amounts of VP35 and VP24 in the cells of vaccinated animals.
Third, these data also help to explain the “immune paralysis” during EBOV infections, which is characterized by massive apoptosis of lymphocytes, lymphopenia, and a deficient immune response (5, 69–72). Even though this study was not designed to investigate the connection between the lack of DC stimulation and the deficient T cell response, the requirement for effective DC maturation and cytokine secretion for stimulation of T cells is well known. As already noted, DC may promote T lymphocyte survival or cause their apoptosis, depending on their maturation state (10).
Fourth, the cytokines strongly upregulated due to the disabling of IRADs included not only those involved in the initiation of the adaptive immune response, but also PDGF and VEGF, two growth factors implicated in angiogenesis following injury of tissues. These two growth factors act together in angiogenesis, including both tissue-generating vessels and microvascular vessels following injury of tissues (73), such as liver tissues (74). Impairment of the vascular system and liver functions is an important characteristic feature of EBOV infections, and therefore, the strong suppression of induction of these two factors by individual IRADs suggests an additional mechanism by which IRADs promote the disease caused by EBOV.
One interesting phenomenon observed in the study is that the levels of DC maturation were comparable in EGFP-positive and EGFP-negative cells. This phenomenon can be explained in two ways. First, maturation could be caused by infection at a very low level, which is insufficient to produce EGFP detectable by flow cytometry but which is nevertheless sufficient to induce DC maturation, a situation similar to that with respiratory paramyxoviruses (48). Second, it is possible that during EBOV infection, EGFP-positive DC transfer viral-peptide–major histocompatibility complex (MHC) complexes to uninfected DC, a phenomenon known as “cross-dressing” (75, 76). Moreover, injection of mice with matured DC induced maturation of some of endogenous splenic DC (77).
An important consideration resulting from this study is related to development of live attenuated or killed vaccines in general. Following the identification of IFN-antagonizing activity of the NS1 protein of influenza A virus in 1998 (78), domains that confer such activities were identified in proteins of many viruses, and the spectra of their activities were found to be broader than IFN-I antagonism alone. The current live attenuated vaccines carry attenuating mutations in sites not related to antagonism of the innate immune response, and killed vaccines are prepared from whole unmodified viral particles. Disabling of IRADs in live attenuated vaccines may attenuate the virus and simultaneously increase their immunogenicity to levels greater than that induced by attenuated vaccines in which attenuation was achieved by introduction of mutations in areas other than IRADs. Use of recombinant viruses with disabled IRADs for the generation of killed vaccines may improve their immunogenicity.
This study will be followed by research in several directions. One obvious direction is investigation of the role of IRADs in stimulation and apoptosis of T cells. Second, it will be important to identify the inhibitory pathways triggered by the IRADs that suppress DC maturation. Identification of these pathways may lead to development of treatments for the disease caused by the virus, which will be based on the blockade of these pathways, which should rescue DC maturation. Third, Marburg virus, which is another virus of the family Filoviridae, is also known to infect DC without inducing their maturation (9). The mechanism by which Marburg virus evades innate immunity is different from that of EBOV and is associated with its VP40 protein (79), in addition to the VP35 protein (80). It will be important to know whether the virus suppresses DC maturation in a manner similar to that described here for EBOV.
ACKNOWLEDGMENTS
The study was supported by a departmental startup grant from the University of Texas Medical Branch (A.B.), the John Sealy Memorial Endowment Fund Pilot Grant “Mechanisms of ‘immune paralysis’ in Ebola infections” (A.B.), and the McLaughlin Fellowship (N.M.L.).
We thank J. Towner and S. Nichol (CDC) for providing the EBOV full-length clone; Y. Kawaoka (University of Wisconsin) and H. Feldmann (NIH) for providing the EBOV NP, VP35, L, VP30, and T7 polymerase plasmids; and T. Garron for excellent technical assistance. We are grateful to P. Halfmann and G. Neumann (University of Wisconsin), J. Towner (CDC), and V. Volchkov (University of Lyon, Lyon, France) for valuable advice.
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Sanchez A, Geisbert TW, Feldmann H. 2007. Filoviridae: Marburg and Ebola viruses, vol 1 Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 2. WHO 1978. Ebola haemmorhagic fever in Zaire, 1976. Bull. WHO 56:271–293 [PMC free article] [PubMed] [Google Scholar]
- 3. CDC 2012. Outbreak postings. http://www.cdc.gov/ncidod/dvrd/spb/outbreaks/index.htm
- 4. Kuhn JH. 2008. Filoviruses. Springer, New York, NY [Google Scholar]
- 5. Geisbert TW, Hensley LE, Gibb TR, Steele KE, Jaax NK, Jahrling PB. 2000. Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab. Invest. 80:171–186 [DOI] [PubMed] [Google Scholar]
- 6. Geisbert TW, Young HA, Jahrling PB, Davis KJ, Larsen T, Kagan E, Hensley LE. 2003. Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am. J. Pathol. 163:2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martinez O, Leung LW, Basler CF. 2012. The role of antigen-presenting cells in filoviral hemorrhagic fever: gaps in current knowledge. Antiviral Res. 93:416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, Pulendran B. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 170:2797–2801 [DOI] [PubMed] [Google Scholar]
- 9. Bosio CM, Aman MJ, Grogan C, Hogan R, Ruthel G, Negley D, Mohamadzadeh M, Bavari S, Schmaljohn A. 2003. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 188:1630–1638 [DOI] [PubMed] [Google Scholar]
- 10. Mailliard RB, Dallal RM, Son YI, Lotze MT. 2000. Dendritic cells promote T-cell survival or death depending upon their maturation state and presentation of antigen. Immunol. Invest. 29:177–185 [DOI] [PubMed] [Google Scholar]
- 11. Kaneko S, Suzuki N, Koizumi H, Yamamoto S, Sakane T. 1997. Rescue by cytokines of apoptotic cell death induced by IL-2 deprivation of human antigen-specific T cell clones. Clin. Exp. Immunol. 109:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanchez A, Kiley MP, Holloway BP, Auperin DD. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29:215–240 [DOI] [PubMed] [Google Scholar]
- 13. Sanchez A, Yang ZY, Xu L, Nabel GJ, Crews T, Peters CJ. 1998. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 72:6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. 1998. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U. S. A. 95:5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feldmann H, Nichol ST, Klenk HD, Peters CJ, Sanchez A. 1994. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 199:469–473 [DOI] [PubMed] [Google Scholar]
- 16. Falzarano D, Krokhin O, Van Domselaar G, Wolf K, Seebach J, Schnittler HJ, Feldmann H. 2007. Ebola sGP—the first viral glycoprotein shown to be C-mannosylated. Virology 368:83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takada A, Watanabe S, Ito H, Okazaki K, Kida H, Kawaoka Y. 2000. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology 278:20–26 [DOI] [PubMed] [Google Scholar]
- 18. Simmons G, Wool-Lewis RJ, Baribaud F, Netter RC, Bates P. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 76:2518–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang ZY, Duckers HJ, Sullivan NJ, Sanchez A, Nabel EG, Nabel GJ. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886–889 [DOI] [PubMed] [Google Scholar]
- 20. Sullivan NJ, Peterson M, Yang ZY, Kong WP, Duckers H, Nabel E, Nabel GJ. 2005. Ebola virus glycoprotein toxicity is mediated by a dynamin-dependent protein-trafficking pathway. J. Virol. 79:547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zampieri CA, Fortin JF, Nolan GP, Nabel GJ. 2007. The ERK mitogen-activated protein kinase pathway contributes to Ebola virus glycoprotein-induced cytotoxicity. J. Virol. 81:1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sullivan NJ, Geisbert TW, Geisbert JB, Shedlock DJ, Xu L, Lamoreaux L, Custers JH, Popernack PM, Yang ZY, Pau MG, Roederer M, Koup RA, Goudsmit J, Jahrling PB, Nabel GJ. 2006. Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified GPs. PLoS Med. 3:e177. 10.1371/journal.pmed.0030177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Volchkov VE, Blinov VM, Netesov SV. 1992. The envelope glycoprotein of Ebola virus contains an immunosuppressive-like domain similar to oncogenic retroviruses. FEBS Lett. 305:181–184 [DOI] [PubMed] [Google Scholar]
- 24. Yaddanapudi K, Palacios G, Towner JS, Chen I, Sariol CA, Nichol ST, Lipkin WI. 2006. Implication of a retrovirus-like glycoprotein peptide in the immunopathogenesis of Ebola and Marburg viruses. FASEB J. 20:2519–2530 [DOI] [PubMed] [Google Scholar]
- 25. Schlecht-Louf G, Renard M, Mangeney M, Letzelter C, Richaud A, Ducos B, Bouallaga I, Heidmann T. 2010. Retroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc. Natl. Acad. Sci. U. S. A. 107:3782–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volchkov VE, Becker S, Volchkova VA, Ternovoj VA, Kotov AN, Netesov SV, Klenk HD. 1995. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology 214:421–430 [DOI] [PubMed] [Google Scholar]
- 27. Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. 1996. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. U. S. A. 93:3602–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang Z, Delgado R, Xu L, Todd RF, Nabel EG, Sanchez A, Nabel GJ. 1998. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 279:1034–1037 [DOI] [PubMed] [Google Scholar]
- 29. Mohan GS, Li W, Ye L, Compans RW, Yang C. 2012. Antigenic subversion: a novel mechanism of host immune evasion by ebola virus. PLoS Pathog. 8:e1003065. 10.1371/journal.ppat.1003065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Basler CF, Amarasinghe GK. 2009. Evasion of interferon responses by Ebola and Marburg viruses. J. Interferon Cytokine Res. 29:511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Najjar I, Fagard R. 2010. STAT1 and pathogens, not a friendly relationship. Biochimie 92:425–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McBride KM, Banninger G, McDonald C, Reich NC. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 21:1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lodige I, Marg A, Wiesner B, Malecova B, Oelgeschlager T, Vinkemeier U. 2005. Nuclear export determines the cytokine sensitivity of STAT transcription factors. J. Biol. Chem. 280:43087–43099 [DOI] [PubMed] [Google Scholar]
- 34. Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 81:13469–13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shabman RS, Gulcicek EE, Stone KL, Basler CF. 2011. The Ebola virus VP24 protein prevents hnRNP C1/C2 binding to karyopherin alpha1 and partially alters its nuclear import. J. Infect. Dis. 204(Suppl. 3):S904–S910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Halfmann P, Neumann G, Kawaoka Y. 2011. The Ebolavirus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase. J. Infect. Dis. 204(Suppl. 3):S953–S956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uddin S, Majchrzak B, Woodson J, Arunkumar P, Alsayed Y, Pine R, Young PR, Fish EN, Platanias LC. 1999. Activation of the p38 mitogen-activated protein kinase by type I interferons. J. Biol. Chem. 274:30127–30131 [DOI] [PubMed] [Google Scholar]
- 38. Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. 2010. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol. 84:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, Garcia-Sastre A, Palese P. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U. S. A. 97:12289–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cardenas WB, Amarasinghe GK, Volchkov VE, Basler CF. 2010. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J. Virol. 84:3004–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hartman AL, Dover JE, Towner JS, Nichol ST. 2006. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol. 80:6430–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, Amarasinghe GK. 2009. Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. U. S. A. 106:411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bukreyev A, Marzi A, Feldmann F, Zhang L, Yang L, Ward JM, Dorward DW, Pickles RJ, Murphy BR, Feldmann H, Collins PL. 2009. Chimeric human parainfluenza virus bearing the Ebola virus glycoprotein as the sole surface protein is immunogenic and highly protective against Ebola virus challenge. Virology 383:348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inaba K, Turley S, Yamaide F, Iyoda T, Mahnke K, Inaba M, Pack M, Subklewe M, Sauter B, Sheff D, Albert M, Bhardwaj N, Mellman I, Steinman RM. 1998. Efficient presentation of phagocytosed cellular fragments on the major histocompatibility complex class II products of dendritic cells. J. Exp. Med. 188:2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. 2005. Generation of EGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology 332:20–27 [DOI] [PubMed] [Google Scholar]
- 47. Neumann G, Feldmann H, Watanabe S, Lukashevich I, Kawaoka Y. 2002. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 76:406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Le Nouen C, Munir S, Losq S, Winter CC, McCarty T, Stephany DA, Holmes KL, Bukreyev A, Rabin RL, Collins PL, Buchholz UJ. 2009. Infection and maturation of monocyte-derived human dendritic cells by human respiratory syncytial virus, human metapneumovirus, and human parainfluenza virus type 3. Virology 385:169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Riedl E, Stockl J, Majdic O, Scheinecker C, Rappersberger K, Knapp W, Strobl H. 2000. Functional involvement of E-cadherin in TGF-beta 1-induced cell cluster formation of in vitro developing human Langerhans-type dendritic cells. J. Immunol. 165:1381–1386 [DOI] [PubMed] [Google Scholar]
- 50. Delemarre FG, Hoogeveen PG, De Haan-Meulman M, Simons PJ, Drexhage HA. 2001. Homotypic cluster formation of dendritic cells, a close correlate of their state of maturation. Defects in the biobreeding diabetes-prone rat. J. Leukoc. Biol. 69:373–380 [PubMed] [Google Scholar]
- 51. McWilliam AS, Marsh AM, Holt PG. 1997. Inflammatory infiltration of the upper airway epithelium during Sendai virus infection: involvement of epithelial dendritic cells. J. Virol. 71:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Segura E, Nicco C, Lombard B, Veron P, Raposo G, Batteux F, Amigorena S, Thery C. 2005. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 106:216–223 [DOI] [PubMed] [Google Scholar]
- 53. Reeves RH, Patch D, Sharpe AH, Borriello F, Freeman GJ, Edelhoff S, Disteche C. 1997. The costimulatory genes Cd80 and Cd86 are linked on mouse chromosome 16 and human chromosome 3. Mamm. Genome 8:581–582 [DOI] [PubMed] [Google Scholar]
- 54. Magistrelli G, Jeannin P, Herbault N, Benoit De Coignac A, Gauchat JF, Bonnefoy JY, Delneste Y. 1999. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur. J. Immunol. 29:3596–3602 [DOI] [PubMed] [Google Scholar]
- 55. Gottlieb RA, Kleinerman ES, O'Brian CA, Tsujimoto S, Cianciolo GJ, Lennarz WJ. 1990. Inhibition of protein kinase C by a peptide conjugate homologous to a domain of the retroviral protein p15E. J. Immunol. 145:2566–2570 [PubMed] [Google Scholar]
- 56. Bukreyev A, Yang L, Zaki SR, Shieh WJ, Rollin PE, Murphy BR, Collins PL, Sanchez A. 2006. A single intranasal inoculation with a paramyxovirus-vectored vaccine protects guinea pigs against a lethal-dose Ebola virus challenge. J. Virol. 80:2267–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, Basler CF, Amarasinghe GK. 2010. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 17:165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF. 2010. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J. Virol. 84:10581–10591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ioannidis LJ, Verity EE, Crawford S, Rockman SP, Brown LE. 2012. Abortive replication of influenza virus in mouse dendritic cells. J. Virol. 86:5922–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Adams WC, Gujer C, McInerney G, Gall JG, Petrovas C, Karlsson Hedestam GB, Koup RA, Lore K. 2011. Adenovirus type-35 vectors block human CD4+ T-cell activation via CD46 ligation. Proc. Natl. Acad. Sci. U. S. A. 108:7499–7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bosio CM, Moore BD, Warfield KL, Ruthel G, Mohamadzadeh M, Aman MJ, Bavari S. 2004. Ebola and Marburg virus-like particles activate human myeloid dendritic cells. Virology 326:280–287 [DOI] [PubMed] [Google Scholar]
- 62. Warfield KL, Bosio CM, Welcher BC, Deal EM, Mohamadzadeh M, Schmaljohn A, Aman MJ, Bavari S. 2003. Ebola virus-like particles protect from lethal Ebola virus infection. Proc. Natl. Acad. Sci. U. S. A. 100:15889–15894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. 2007. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. J. Infect. Dis. 196(Suppl. 2):S430–S437 [DOI] [PubMed] [Google Scholar]
- 64. Jin H, Yan Z, Prabhakar BS, Feng Z, Ma Y, Verpooten D, Ganesh B, He B. 2010. The VP35 protein of Ebola virus impairs dendritic cell maturation induced by virus and lipopolysaccharide. J. Gen. Virol. 91:352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bukreyev A, Rollin PE, Tate MK, Yang L, Zaki SR, Shieh WJ, Murphy BR, Collins PL, Sanchez A. 2007. Successful topical respiratory tract immunization of primates against Ebola virus. J. Virol. 81:6379–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sullivan NJ, Sanchez A, Rollin PE, Yang ZY, Nabel GJ. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605–609 [DOI] [PubMed] [Google Scholar]
- 67. Jones SM, Feldmann H, Stroher U, Geisbert JB, Fernando L, Grolla A, Klenk HD, Sullivan NJ, Volchkov VE, Fritz EA, Daddario KM, Hensley LE, Jahrling PB, Geisbert TW. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 11:786–790 [DOI] [PubMed] [Google Scholar]
- 68. Halfmann P, Ebihara H, Marzi A, Hatta Y, Watanabe S, Suresh M, Neumann G, Feldmann H, Kawaoka Y. 2009. Replication-deficient ebolavirus as a vaccine candidate. J. Virol. 83:3810–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gupta M, Spiropoulou C, Rollin PE. 2007. Ebola virus infection of human PBMCs causes massive death of macrophages, CD4 and CD8 T cell sub-populations in vitro. Virology 364:45–54 [DOI] [PubMed] [Google Scholar]
- 70. Baize S, Leroy EM, Georges-Courbot MC, Capron M, Lansoud-Soukate J, Debre P, Fisher-Hoch SP, McCormick JB, Georges AJ. 1999. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat. Med. 5:423–426 [DOI] [PubMed] [Google Scholar]
- 71. Sanchez A, Lukwiya M, Bausch D, Mahanty S, Sanchez AJ, Wagoner KD, Rollin PE. 2004. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J. Virol. 78:10370–10377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. 2010. Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl. Trop. Dis. 4:e837. 10.1371/journal.pntd.0000837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Luo JZ, Xiong F, Al-Homsi AS, Roy T, Luo LG. 2011. Human BM stem cells initiate angiogenesis in human islets in vitro. Bone Marrow Transplant. 46:1128–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rosmorduc O, Housset C. 2010. Hypoxia: a link between fibrogenesis, angiogenesis, and carcinogenesis in liver disease. Semin. Liver Dis. 30:258–270 [DOI] [PubMed] [Google Scholar]
- 75. Wakim LM, Bevan MJ. 2011. Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature 471:629–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dolan BP, Gibbs KD, Jr, Ostrand-Rosenberg S. 2006. Dendritic cells cross-dressed with peptide MHC class I complexes prime CD8+ T cells. J. Immunol. 177:6018–6024 [DOI] [PubMed] [Google Scholar]
- 77. Yewdall AW, Drutman SB, Jinwala F, Bahjat KS, Bhardwaj N. 2010. CD8+ T cell priming by dendritic cell vaccines requires antigen transfer to endogenous antigen presenting cells. PLoS One 5:e11144. 10.1371/journal.pone.0011144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330 [DOI] [PubMed] [Google Scholar]
- 79. Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krahling V, Basle CF, Muhlberger E. 2010. Marburg virus evades interferon responses by a mechanism distinct from Ebola virus. PLoS Pathog. 6:e1000721. 10.1371/journal.ppat.1000721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, Amarasinghe GK. 2012. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc. Natl. Acad. Sci. U. S. A. 109:20661–20666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hartman AL, Ling L, Nichol ST, Hibberd ML. 2008. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol. 82:5348–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hartman AL, Towner JS, Nichol ST. 2004. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology 328:177–184 [DOI] [PubMed] [Google Scholar]