Abstract
Avian influenza A viruses, such as the highly pathogenic avian H5N1 viruses, sporadically enter the human population but often do not transmit between individuals. In rare cases, however, they establish a new lineage in humans. In addition to well-characterized barriers to cell entry, one major hurdle which avian viruses must overcome is their poor polymerase activity in human cells. There is compelling evidence that these viruses overcome this obstacle by acquiring adaptive mutations in the polymerase subunits PB1, PB2, and PA and the nucleoprotein (NP) as well as in the novel polymerase cofactor nuclear export protein (NEP). Recent findings suggest that synthesis of the viral genome may represent the major defect of avian polymerases in human cells. While the precise mechanisms remain to be unveiled, it appears that a broad spectrum of polymerase adaptive mutations can act collectively to overcome this defect. Thus, identification and monitoring of emerging adaptive mutations that further increase polymerase activity in human cells are critical to estimate the pandemic potential of avian viruses.
INTRODUCTION
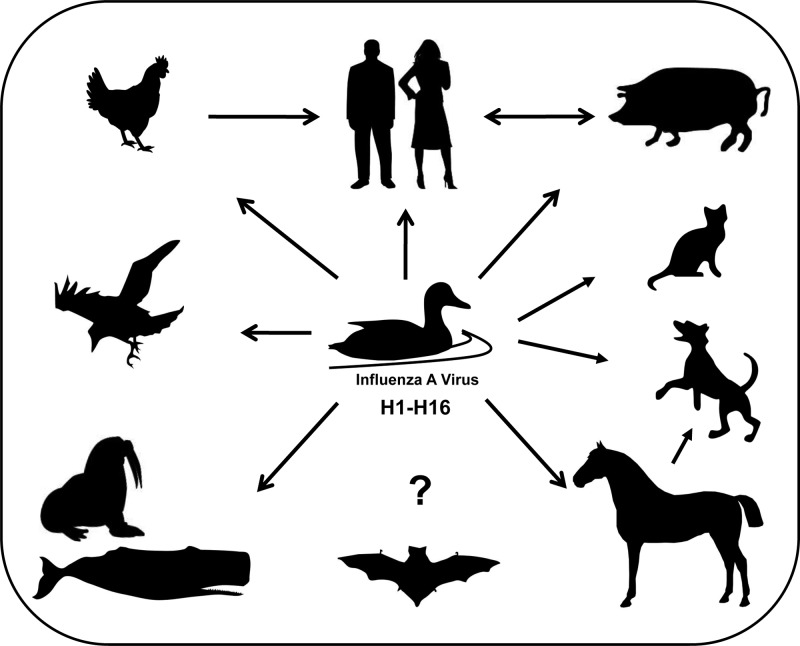
Although the natural reservoirs of influenza A viruses lacking adaptive mutations that increase polymerase activity in mammalian cells (1–53) are wild birds (54), mammals are frequently infected with influenza viruses of avian origin. These zoonotic transmissions can cause severe disease in different mammals, including cats, dogs, horses, pigs, and humans (55–57), due to the lack of preexisting immunity in these species to the new influenza virus strain (Fig. 1). Fortunately, most of these infections are so-called dead-end infections and are not further transmitted within the new species due to several barriers. However, on rare occasions influenza A viruses can indeed break the species barrier and establish an entirely new virus lineage in a mammalian species, as exemplified by the human pandemic of 1918 (58), the Eurasian classical swine influenza virus lineage (59), H3N8 influenza viruses in horses (56), and possibly H17N10-like influenza viruses in bats (60). Although 16 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes have been identified in wild birds, human infection has been documented only for H1, H2, H3, H5, H7, and H9 (61–64) and only H1, H2, and H3 have been stably introduced into the human population (56, 65, 66). In the majority of cases, human pandemics were a result of genetic reassortment events whereby the circulating human virus strains acquired one or more gene segments from avian or swine sources (54). However, since humans in certain regions of the globe are constantly exposed to H5N1 influenza viruses, there exists a serious concern that this subtype might acquire mutations to stably cross the species barrier and start a new pandemic (67, 68).
Fig 1.
Host range of influenza A viruses. Wild water birds represent the natural reservoir of influenza A viruses, from which they can be transmitted to a wide variety of other hosts, including horses, cats, dogs, whales, seals, wild flying birds, chicken, pigs, and humans. Only recently, influenza A virus has also been detected in bats, although the origin is unclear.
REPLICATION OF INFLUENZA A VIRUSES
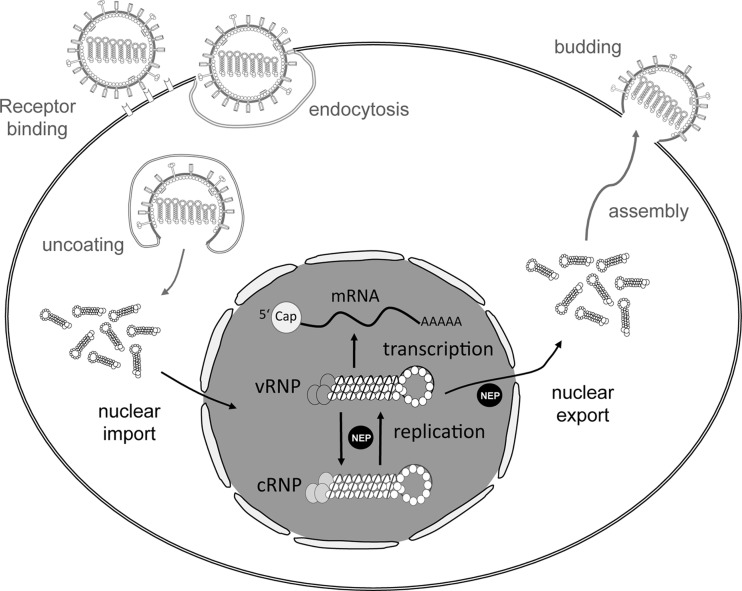
Influenza viruses belong to the family of Orthomyxoviridae, possessing a single-stranded negative-sense RNA genome that is comprised of eight segments (69). The ends of each genome segment are short complementary elements that form the viral promoter and are recognized by the viral RNA-dependent RNA polymerase, which is composed of the three subunits PB2, PB1, and PA (70–74). Together, the viral polymerase, the nucleoprotein NP, and the viral RNA genome form the ribonucleoprotein (RNP) complex, which is responsible for viral mRNA synthesis and genome replication. Upon infection, viral RNPs (vRNPs) are released into the cytoplasm (75) and transported into the nucleus, where the RNPs perform all of their enzymatic functions (76, 77). First, primary transcription is initiated and viral mRNA is synthesized from the viral genome (vRNA) (Fig. 2). vRNA also serves as a template for synthesis of a full-length copy (cRNA), which serves as a template for subsequent synthesis of new viral genomes (78). Viral mRNA synthesis is primed using a cellular capped pre-mRNA which is bound by the PB2 subunit (30, 79) and cleaved by the endonucleolytic domain of the polymerase subunit PA (39, 80). Synthesis of both c- and vRNA is apparently primer independent (81–83). Recent evidence suggests that mRNA is synthesized by viral polymerase complexes which are resident on vRNPs (in “cis”), whereas vRNA is synthesized from the cRNA template using soluble viral polymerase complexes (in “trans”) (84). However, the mechanism of cRNA synthesis and the regulation of the switch between transcription and genome replication remain largely unknown (85). The viral nucleoprotein (NP) is one major candidate to regulate the switch between transcription and replication (86), presumably by direct interaction with the viral polymerase (87), although this role of NP has been questioned more recently (88).
Fig 2.
Illustration of the influenza A virus replication cycle. Influenza A virus particle binds to the cellular receptor and enters the cell by endocytosis. The viral ribonucleoproteins (vRNPs) are released into the cytoplasm upon acidification of the endosome. vRNPs are transported into the nucleus, where transcription and replication occur. Replication is supported by the viral nuclear export protein NEP. For genome replication, negative-sense viral RNA is transcribed into plus-sense cRNA that is complexed by the viral polymerase and NP to form the cRNP and serves as a template for vRNA synthesis. After export from the nucleus, vRNPs are assembled into new viral particles at the plasma membrane and released from the cell.
Interestingly, in addition to the viral nucleoprotein, the nuclear export protein NEP (NS2) has been shown to be involved in the regulation of polymerase activity (11, 89, 90). In contrast to NP, it is not an essential component of the RNP complex, as it is not required for transcriptional activity in a polymerase reconstitution assay (Fig. 3). In this system, the effect of NEP on viral RNA synthesis was shown to be concentration dependent. While very high NEP concentrations completely abrogated polymerase activity (11), recent data demonstrate that small amounts of NEP stimulate the synthesis of vRNA and cRNA and, depending on the experimental system, also mRNA (11, 89). The mechanism by which NEP enhances vRNA and/or cRNA synthesis and regulates viral polymerase activity remains unknown, but it appears to occur independently of its previously described function as a mediator of vRNP nuclear export (11, 89).
Fig 3.
Experimental systems for identification of adaptive mutations of influenza A viruses. Several experimental systems are used to identify adaptive mutations that facilitate host adaptation. These include the following. (A) Bioinformatics analysis of bird- and human-derived influenza virus genome sequences. Such analyses are frequently used to identify unique nucleotide changes that occur only in humans but not in avian species that are selected in the specific host. (B) The polymerase reconstitution assay allows functional analysis of adaptive mutations in the polymerase subunits or NP by reverse genetics. Expression plasmids for the polymerase subunits PB2, PB1, and PA as well as NP and a viral genome analog, harboring a reporter gene instead of the viral protein, are transfected into cells, and reporter gene activity can be measured 24 h later. With this approach, the activities of polymerases from different influenza virus strains can be compared and analyzed in human and avian cells. (C) Different animal models, including mice and ferrets, are used to determine pathogenicity and airborne transmissibility of bird- and human-derived influenza A virus strains. (D) Biochemical analysis can be performed with virion-derived RNPs or recombinant proteins from different expression systems to study polymerase activity in vitro.
The discovery of small viral RNAs (svRNAs) generated during viral replication (91, 92) has furthermore paved the way for a new understanding of influenza virus polymerase regulation. svRNAs are 22 to 27 nucleotides in length, corresponding to the 5′ ends of the genomic viral RNA segments, and are synthesized from the 3′ end of the cRNA by the viral polymerase. Functional characterization indicates that svRNAs directly interact with the PA subunit and could provide a segment-specific guide for the viral polymerase to the cRNA templates, thereby promoting synthesis of new genomic vRNAs (93). This suggests that svRNAs are involved in the switch from mRNA transcription to genome replication (91) and that NEP mediates the generation of these svRNAs by stimulating cRNA synthesis (93).
During a replication cycle, at least 10 different viral proteins are expressed to establish efficient propagation of the virus in the infected host (40, 94–97). Besides the requirement of these proteins for essential functions during the viral life cycle, multiple interactions with host factors, such as DNA-dependent RNA polymerase II (Pol II), are required to snatch cap structures from host mRNAs (98) and to function as part of the cellular splicing machinery (99, 100) as well as to function as factors for nuclear trafficking (101). In the case of transmission of avian influenza viruses to mammalian hosts, introduction of adaptive mutations into viral proteins is essential to ensure optimal functionality and virus-host protein interactions in the cellular environment of the new host.
VIRAL FACTORS IMPORTANT IN HOST ADAPTATION
The hemagglutinin (HA) is one of the key factors which determines the host range of influenza A viruses, since it mediates attachment and entry of the virus into target cells. Adaptation from avian to human hosts has been shown to target three main properties of HA. A switch in receptor specificity from avian (α-2,3-linked) to human (α-2,6-linked) sialic acids as well as stabilization of the stalk region to allow endosomal membrane fusion at the optimal pH is crucial for efficient transmission between mammals (67, 68) and host adaptation (102). Other important factors include appropriate HA glycosylation, the length of the stalk region in the neuraminidase (NA), and specific differences in codon usage (103–106).
As an antagonist of the alpha/beta interferon (IFN-α/β)-mediated host immune response, the NS1 protein plays a critical role during zoonotic transmission (107). The nonessential viral protein PB1-F2 was shown to be able to antagonize the interferon response (108). Additionally, the viral nucleoprotein NP mediates escape from restriction by the interferon-stimulated human MxA protein (50, 109, 110). Polymerase activity of various avian influenza A viruses was shown to be strongly impaired in mammalian cells, restricting replication of avian viruses in mammals (10, 111, 112). Low polymerase activity results not only in fewer copies of vRNA for packaging into new viral particles but also in reduced mRNA synthesis and expression of viral proteins. Perhaps most important for zoonotic viruses, low polymerase activity results in fewer opportunities for the virus to create new variant genomes containing potentially beneficial mutations. For this reason, mutations in the polymerase subunits that increase transcriptional activity are fundamental for avian influenza viruses to adapt to the human host.
THE DEFECT OF AVIAN H5N1 POLYMERASES REMAINS ELUSIVE
The molecular basis underlying the low polymerase activity of avian influenza A viruses in human cells remains a mystery. However, several mechanisms have been postulated to be responsible for this constraint. One popular hypothesis suggests that cellular factors are responsible for the inhibition: either the presence of a negative factor in mammalian cells which leads to inhibition of avian polymerases or the absence of a positive factor in mammalian cells—or the inability of avian viruses to bind such a positive factor—needed for high polymerase activity has been proposed to account for the restriction of avian polymerases in human cells (113, 114). Furthermore, it has long been unclear which transcriptional activity of avian polymerase—synthesis of viral mRNA, cRNA, or vRNA or of all three—is restricted in human cells. Recent work has provided evidence that vRNA synthesis, and thereby vRNP accumulation, is significantly reduced in avian influenza virus-infected cells, possibly due to the generation of defective complementary RNPs (cRNPs), which also results in reduced mRNA synthesis (11). These results suggest that only one enzymatic process may be constrained in human cells; however, further work is required to more precisely characterize this defect in avian polymerases.
ADAPTIVE MUTATIONS IN THE VIRAL POLYMERASE
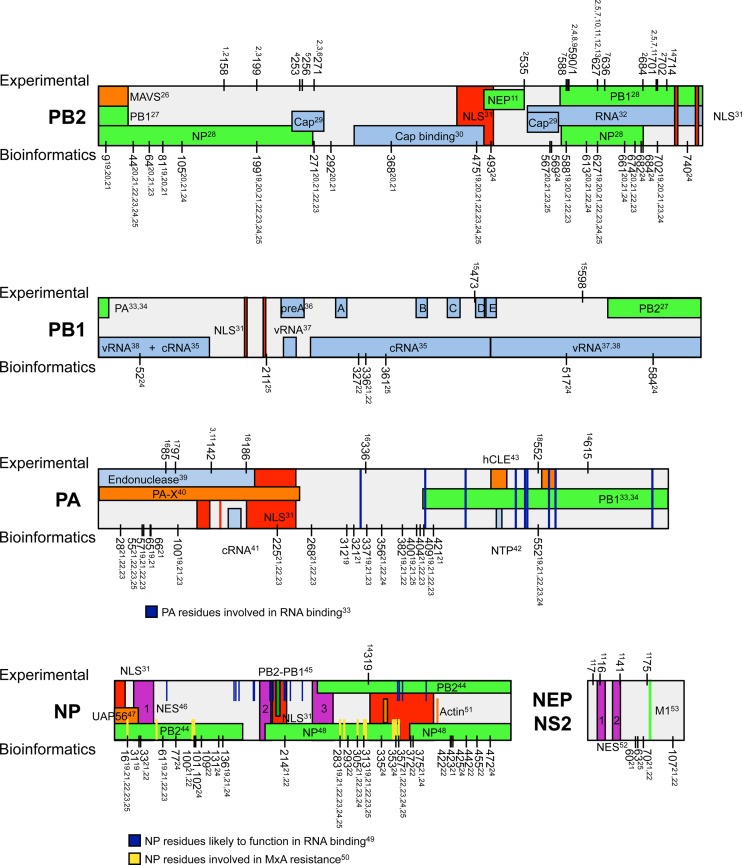
The significance of enhanced polymerase activity during cross-species transmission of avian viruses to humans is highlighted by the occurrence of numerous adaptive mutations in the viral polymerase proteins during natural infections of mammals (including humans), as well as in experimental infection of animal models. Based on different experimental systems (Fig. 3), adaptive mutations conferring enhanced polymerase activity in mammals have been identified in all three subunits of the viral polymerase, NP, and NEP (Fig. 4) (115, 116). Interestingly, most mutations reside in the PB2 subunit, where they localize in two clusters: in the N and C termini of the protein.
Fig 4.
Described mutations increasing polymerase activity in mammalian cells. Published data were analyzed to screen for predicted host-adaptive amino acids (Bioinformatics) and mutations experimentally shown to increase activity of an avian influenza virus polymerase in the context of mammalian cells or to increase pathogenicity in a viral infection (Experimental) (115, 116). Functional domains are indicated in green (interaction with viral proteins), red (involved in nuclear localization), purple (involved in nuclear export), orange (interaction with cellular proteins), yellow (MxA resistance), and blue (RNA binding).
The most often-observed and well-described mutation codes for a lysine instead of a glutamate at position 627 of the PB2 protein (PB2-E627K) and is solely sufficient for replication of several avian influenza viruses in mammals (3, 10, 12, 111). Amino acid 627 is located in the C-terminal region of the PB2 protein which includes the eponymous “627 domain” in addition to the importin alpha-binding domain (117). The change from glutamate to lysine at this position has been proposed to influence host adaptation on multiple levels. It has been shown to increase transmissibility of avian viruses between mammals (118), presumably due to increased polymerase activity at lower temperatures, as found in the human upper respiratory tract (33 to 35°C) compared to the avian intestinal tract (38 to 40°C) (12, 119). In addition, assembly of new vRNPs was suggested to be impaired during replication of avian influenza virus in mammals due to unstable binding between NP and the PB2 subunit, a defect that was rescued by the mutation PB2-E627K (113, 120–122). Furthermore, the emergence of this adaptive mutation in mammalian cells and mice was shown to be dependent on the origin of the viral nucleoprotein (123). Whereas NP of avian origin provoked the appearance of E627K, NP of a human-derived H5N1 virus did not (123). These data indicate a strong correlation between NP and PB2 in host adaptation and the occurrence of adaptive mutations in either protein. However, there is also a deviating interpretation proposing that the reduced binding between PB2 and NP is an indirect observation and is a consequence of the restricted avian influenza virus RNA replication rather than of an alteration in NP-PB2 binding affinity (2).
The interaction of the viral polymerase with host cellular factors also seems to be affected by the species-specific amino acid at PB2 627. Structural investigations of the PB2 627 domain revealed that the change from glutamate to lysine alters the electrostatic surface potential of this domain. Interestingly, a change in shape and surface potential is also seen for the SR polymorphism at positions 590 and 591 in PB2, which is suggested to compensate for the lack of PB2-E627K in the 2009 pandemic H1N1 virus (4, 8, 9, 124). The change in surface potential is hypothesized to influence the interaction with cellular factors, such as α-importins (32, 125–127). Furthermore, in the case of the PB2-binding DEAD box RNA helicase DDX17/p72, the human form of the protein has been shown to stimulate mRNA and vRNA synthesis of human-adapted H5N1 polymerases (PB2-627K) but not their avian precursor, while chicken DDX17 supports growth of avian but not human-adapted H5N1 virus (128). However, it remains to be shown whether the species-specific activity of DDX17 is a determining factor driving human adaptation of avian H5N1 viruses.
Nonetheless, the precise function during species transmission of the PB2 C terminus, which harbors most of the identified adaptive mutations (Fig. 4), remains largely unknown. Strong homology to the cellular RNA-binding clamp-loader complex and increased RNA-binding activity of the recombinant PB2 C-terminal domain harboring the human 627K signature suggest a role in RNA binding (32). However, decreased binding to the viral promoter and a decrease in primed in vitro polymerase activity assays was observed with full-length PB2 627K (129, 130), leaving the role of the 627 domain in RNA binding unresolved.
Several other important polymerase activity-enhancing mutations are found in the PB2 C-terminal domain (2, 7, 9, 14, 18, 118) (Fig. 4). The adaptive mutation PB2-701N is located in the well-defined importin alpha-binding domain and can partially compensate for the lack of PB2-627K in avian viruses (118). The mutation could be shown to influence the interaction with various importin alpha isoforms, which are responsible for the nuclear import of PB2 as well as of other viral proteins (125, 126). Mutation to PB2-701N causes avian viruses to switch dependency from importin alpha 3 to importin alpha 7, which increases viral replication and pathogenicity in mice (126, 131). On the molecular level, recent data suggest that PB2-701N, together with PB2-714R, increases the cap-binding efficiency of the PB2 subunit but decreases primed in vitro polymerase activity, an effect also observed with PB2-627K (132). Adaptive mutations in the N terminus of PB2 include exchanges at positions PB2-158G, -199S, -253N, -256G, and -271A (1–6). However, mechanistic insights into the effects of these mutations on polymerase activity and enhanced replication of avian influenza viruses in mammals are not yet available.
In the PA subunit, adaptive mutations seem to cluster in the endonuclease-containing N terminus (39) (Fig. 4). This region overlaps with the newly identified frameshift product PA-X that modulates host response to infection (40). However, it is not clear whether such N-terminal adaptive mutations impact PA-X function. Interestingly, the N terminus of PA is also associated with viral genome promoter binding (41, 133) and regulation of cRNA/vRNA synthesis (134), although it remains to be shown whether these mutations might affect the RNA-binding feature of PA. While additional mutations were identified at various positions in PA, including 336M, 552S, and 615N (3, 11, 14, 16–18), in PB1 only mutations at positions 473 and 598 were shown to increase polymerase activity of avian polymerases in mammalian cells (15).
Although the mutation PB2-E627K has been shown to efficiently adapt the polymerase of avian viruses to human cells, only 40% of H5N1 influenza viruses isolated to date from humans have acquired this mutation (13/01/14 NCBI database) (135), indicating that other mechanisms of adaptation to humans might have evolved. The lack of PB2-E627K can be partially compensated by mutations at other positions in the polymerase subunits, but none of these mutations has a comparable potency to increase polymerase activity (11, 13), and it is conceivable that mutations in other viral factors are required to overcome species barriers.
ADAPTIVE MUTATIONS IN THE VIRAL NUCLEOPROTEIN
To date, few adaptive mutations in NP have been identified as required for the efficient growth of avian influenza viruses in mammalian hosts (Fig. 4). These include N319K, which enhances viral replication in mammalian cells (14, 136) by affecting interaction of NP with host importin-a isoforms (126, 131). Although several other NP mutations were obtained by adaptation of human influenza A virus strains to mice or guinea pigs (137–140), it remains to be shown whether these mutations are also required for avian strains to efficiently replicate in mammalian cells. Multiple potentially adaptive mutations in NP were identified in silico by sequence comparisons of avian and human influenza A viruses (Fig. 4). Some of these adaptive mutations (G16D, L283P, F313Y, Q357K) are required to escape from restriction by the interferon-stimulated human MxA protein (50).
NEP: A NEW ADAPTIVE FACTOR
Recent work suggests that NEP is required for the adaptation of some avian H5N1 viruses, specifically those lacking the PB2 E627K mutation (11). Single mutations in NEP from human H5N1 isolates (Fig. 4) were found to be sufficient for NEP to stimulate viral RNA synthesis from avian polymerases in human cells to overcome polymerase restriction and, in conclusion, facilitate adaptation to the new host (11).
The adaptive mutations found in H5N1 human isolates are situated in both the N terminus (M16I and Y41C) and the C terminus (E75G) of NEP (Fig. 4). Interestingly, the C terminus of NEP alone is sufficient to regulate viral polymerase activity (89, 93). This suggests that the N terminus may act as a regulatory domain whose tertiary conformation relative to that of the C-terminal domain determines the protein's cofactor activity. Adaptive mutations may thus affect the interaction between the N-terminal and C-terminal domains, thereby increasing the polymerase activity-enhancing property of NEP through conformational changes. However, further investigations are required to substantiate this hypothesis.
MUTATIONS IN THE NONCODING REGIONS (NCRs)
Although naturally occurring adaptive mutations in the noncoding regions of the viral genomes are not known, artificial introduction of cRNA promoter-like bases into the 3′-end vRNA promoter, especially at positions 3 (G ★ A), 5 (U ★ C), and 8 (C ★ U), referred to as the up-promoter, is also able to rescue the restricted avian polymerase activity in the polymerase reconstitution assay in human cells (2, 83, 141–144).
As expected, during viral infection with recombinant viruses bearing the up-promoter mutations in either the PA- or PB1-encoding segment, segment-specific increases in cRNA and mRNA synthesis were observed (145). However, despite elevated PA or PB1 protein expression, these viruses were attenuated (145) and might explain why this strategy to increase polymerase activity is not observed in natural isolates.
It was shown that the up-promoter mutations lead to increased base pairing in the promoter (141). This might positively influence polymerase activity by changing the promoter structure. Alternatively, the nature of the nucleotides might be the important factor for increased RNA replication, as was suggested by an extensive promoter study (142).
Despite extensive studies on the promoter regions (first and last 12 to 13 nucleotides of the viral genome), very little is known about the impact of mutations in the remaining noncoding regions (NCRs) downstream of the polymerase-binding site (146). Besides their involvement in packaging (147, 148), few studies addressed the regulatory role of these NCRs in translation, mRNA transcription, and RNA replication. Intriguingly, these studies indicated a strong impact of the NCRs on genome replication (149–152), but further detailed analyses are needed to demonstrate a possible involvement of the NCRs in adaptation processes.
CONCLUDING REMARKS
A contribution of the polymerase complex, and particularly the PB2 subunit, in host adaptation to mammals has been known since the 1970s (153). Interestingly, a comparable pattern of adaptive mutations in PB2 was found not only in mammals but also in different flightless bird species (Ratitae), including ostrich, emu, and rhea. These include the mutations PB2-591K, -627K, and -701N (154, 155), suggesting related adaptive mechanisms occurring in these species after infection with avian influenza A viruses.
However, the number of adaptive mutations arising in the polymerase subunits also indicates a certain flexibility to overcome the restricted polymerase activity of avian viruses after species transmission and might explain the appearance of multiple compensatory mutations in influenza A virus strains, which have not adopted the human-like PB2-E627K mutation, to successfully replicate in human cells (11, 14, 18).
The ability of each of PB2-627K, NEP, and the artificial up-promoter to almost completely rescue the defect of an avian influenza virus polymerase in mammalian cells raises the hypothesis of a functional linkage of these three strategies, especially considering that adaptive mutations in NEP were observed only in human H5N1 isolates harboring PB2-627E (11). Interestingly, in all three cases, viral genome replication is boosted (2, 11, 89, 145), which corresponds to a potential defect in producing sufficient quantities of vRNA. To date, the exact defect of the avian polymerase in producing insufficient amounts of vRNA has not been known but could involve different steps in viral replication such as insufficient or inappropriate encapsidation of vRNA and/or cRNA, sequence errors introduced into noncoding regions during replication of vRNA, reduced promoter clearance by altered secondary structures, or localization of the RNP to an inappropriate cellular compartment. Furthermore, it remains to be shown whether host-specific factors contribute to this defect.
Increasing evidence indicates that NEP plays a highly versatile role during influenza virus infection, being involved in early and late phases of viral infection. It seems intriguing that a low-molecular-mass (14.8-kDa) protein such as NEP can harbor interaction sites for at least five different viral and cellular proteins (PB1, PB2, M1, CRM1, F1Fo-ATPase). These different interactions must be tightly regulated by as-yet-unknown mechanisms that could involve phosphorylation (156) and sumoylation (157) of NEP.
Clearly, further work is required to elucidate mechanistical insights into the adaptation of the avian influenza virus polymerase in mammals. The fact that the viral polymerase adapts in different bird species, but also that the polymerase PB2 and PA genes of the 2009 pandemic were of avian origin and partially adapted to humans in swine, highlights the necessity of investigating the role of different animal species in the adaptation process of avian influenza A virus to humans. Unraveling the precise function of NEP in viral replication and transcription processes might further help to elucidate the defect of the avian influenza virus polymerase in mammals, which would substantially increase our knowledge about influenza virus host adaptation.
ACKNOWLEDGMENTS
We thank Geoffrey Chase and Ervin Fodor for critically reading the manuscript and Hardin Bolte for generating the figures.
This study was funded by the Bundesministerium für Bildung und Forschung (FluResearchNet) and the Deutsche Forschungsgesellschaft (SCHW 632/11-2).
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Zhou B, Li Y, Halpin R, Hine E, Spiro DJ, Wentworth DE. 2011. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza A viruses in mice. J. Virol. 85:357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cauldwell AV, Moncorge O, Barclay WS. 2012. Unstable polymerase-NP interaction is not responsible for avian influenza polymerase restriction in human cells. J. Virol. 87:1278–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim JH, Hatta M, Watanabe S, Neumann G, Watanabe T, Kawaoka Y. 2010. Role of host-specific amino acids in the pathogenicity of avian H5N1 influenza viruses in mice. J. Gen. Virol. 91:1284–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mok CK, Yen HL, Yu MY, Yuen KM, Sia SF, Chan MC, Qin G, Tu WW, Peiris JS. 2011. Amino acid residues 253 and 591 of the PB2 protein of avian influenza virus A H9N2 contribute to mammalian pathogenesis. J. Virol. 85:9641–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manzoor R, Sakoda Y, Nomura N, Tsuda Y, Ozaki H, Okamatsu M, Kida H. 2009. PB2 protein of a highly pathogenic avian influenza virus strain A/chicken/Yamaguchi/7/2004 (H5N1) determines its replication potential in pigs. J. Virol. 83:1572–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. 2010. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J. Virol. 84:4395–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foeglein A, Loucaides EM, Mura M, Wise HM, Barclay WS, Digard P. 2011. Influence of PB2 host-range determinants on the intranuclear mobility of the influenza A virus polymerase. J. Gen. Virol. 92:1650–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamada S, Hatta M, Staker BL, Watanabe S, Imai M, Shinya K, Sakai-Tagawa Y, Ito M, Ozawa M, Watanabe T, Sakabe S, Li C, Kim JH, Myler PJ, Phan I, Raymond A, Smith E, Stacy R, Nidom CA, Lank SM, Wiseman RW, Bimber BN, O'Connor DH, Neumann G, Stewart LJ, Kawaoka Y. 2010. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 6:e1001034. 10.1371/journal.ppat.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mänz B, Brunotte L, Reuther P, Schwemmle M. 2012. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat. Commun. 3:802. 10.1038/ncomms1804 [DOI] [PubMed] [Google Scholar]
- 12. Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. 2007. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 3:1374–1379. 10.1371/journal.ppat.0030133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2010. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J. Virol. 84:1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U. S. A. 102:18590–18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu C, Hu WB, Xu K, He YX, Wang TY, Chen Z, Li TX, Liu JH, Buchy P, Sun B. 2012. Amino acids 473V and 598P of PB1 from an avian-origin influenza A virus contribute to polymerase activity, especially in mammalian cells. J. Gen. Virol. 93:531–540 [DOI] [PubMed] [Google Scholar]
- 16. Bussey KA, Desmet EA, Mattiacio JL, Hamilton A, Bradel-Tretheway B, Bussey HE, Kim B, Dewhurst S, Takimoto T. 2011. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J. Virol. 85:7020–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song MS, Pascua PN, Lee JH, Baek YH, Lee OJ, Kim CJ, Kim H, Webby RJ, Webster RG, Choi YK. 2009. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model. J. Virol. 83:12325–12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mehle A, Dugan VG, Taubenberger JK, Doudna JA. 2012. Reassortment and mutation of the avian influenza virus polymerase PA subunit overcome species barriers. J. Virol. 86:1750–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naffakh N, Massin P, Escriou N, Crescenzo-Chaigne B, van der Werf S. 2000. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J. Gen. Virol. 81:1283–1291 [DOI] [PubMed] [Google Scholar]
- 20. Miotto O, Heiny A, Tan TW, August JT, Brusic V. 2008. Identification of human-to-human transmissibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinformatics 9(Suppl 1):S18. 10.1186/1471-2105-9-S1-S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miotto O, Heiny AT, Albrecht R, Garcia-Sastre A, Tan TW, August JT, Brusic V. 2010. Complete-proteome mapping of human influenza A adaptive mutations: implications for human transmissibility of zoonotic strains. PLoS One 5:e9025. 10.1371/journal.pone.0009025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen GW, Chang SC, Mok CK, Lo YL, Kung YN, Huang JH, Shih YH, Wang JY, Chiang C, Chen CJ, Shih SR. 2006. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 12:1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finkelstein DB, Mukatira S, Mehta PK, Obenauer JC, Su X, Webster RG, Naeve CW. 2007. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 81:10292–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamuri AU, Dos Reis M, Hay AJ, Goldstein RA. 2009. Identifying changes in selective constraints: host shifts in influenza. PLoS Comput. Biol. 5:e1000564. 10.1371/journal.pcbi.1000564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Allen JE, Gardner SN, Vitalis EA, Slezak TR. 2009. Conserved amino acid markers from past influenza pandemic strains. BMC Microbiol. 9:77. 10.1186/1471-2180-9-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patel D, Schultz LW, Umland TC. 2013. Influenza A polymerase subunit PB2 possesses overlapping binding sites for polymerase subunit PB1 and human MAVS proteins. Virus Res. 172:75–80 [DOI] [PubMed] [Google Scholar]
- 27. Sugiyama K, Obayashi E, Kawaguchi A, Suzuki Y, Tame JR, Nagata K, Park SY. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poole E, Elton D, Medcalf L, Digard P. 2004. Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology 321:120–133 [DOI] [PubMed] [Google Scholar]
- 29. Honda A, Mizumoto K, Ishihama A. 1999. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 4:475–485 [DOI] [PubMed] [Google Scholar]
- 30. Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500–506 [DOI] [PubMed] [Google Scholar]
- 31. Hutchinson EC, Fodor E. 2012. Nuclear import of the influenza A virus transcriptional machinery. Vaccine 30:7353–7358 [DOI] [PubMed] [Google Scholar]
- 32. Kuzuhara T, Kise D, Yoshida H, Horita T, Murazaki Y, Nishimura A, Echigo N, Utsunomiya H, Tsuge H. 2009. Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue. J. Biol. Chem. 284:6855–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, Ge R, Rao Z, Liu Y. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126 [DOI] [PubMed] [Google Scholar]
- 34. Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131 [DOI] [PubMed] [Google Scholar]
- 35. Gonzalez S, Ortin J. 1999. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18:3767–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leahy MB, Dessens JT, Weber F, Kochs G, Nuttall PA. 1997. The fourth genus in the Orthomyxoviridae: sequence analyses of two Thogoto virus polymerase proteins and comparison with influenza viruses. Virus Res. 50:215–224 [DOI] [PubMed] [Google Scholar]
- 37. Li ML, Ramirez BC, Krug RM. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gonzalez S, Ortin J. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913 [DOI] [PubMed] [Google Scholar]
- 40. Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maier HJ, Kashiwagi T, Hara K, Brownlee GG. 2008. Differential role of the influenza A virus polymerase PA subunit for vRNA and cRNA promoter binding. Virology 370:194–204 [DOI] [PubMed] [Google Scholar]
- 42. de la Luna S, Martinez C, Ortin J. 1989. Molecular cloning and sequencing of influenza virus A/Victoria/3/75 polymerase genes: sequence evolution and prediction of possible functional domains. Virus Res. 13:143–155 [DOI] [PubMed] [Google Scholar]
- 43. Huarte M, Sanz-Ezquerro JJ, Roncal F, Ortin J, Nieto A. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 75:8597–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Biswas SK, Boutz PL, Nayak DP. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marklund JK, Ye Q, Dong J, Tao YJ, Krug RM. 2012. Sequence in the influenza A virus nucleoprotein required for viral polymerase binding and RNA synthesis. J. Virol. 86:7292–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu M, Liu X, Cao S, Zhao Z, Zhang K, Xie Q, Chen C, Gao S, Bi Y, Sun L, Ye X, Gao GF, Liu W. 2012. Identification and characterization of three novel nuclear export signals in the influenza A virus nucleoprotein. J. Virol. 86:4970–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Momose F, Basler CF, O'Neill RE, Iwamatsu A, Palese P, Nagata K. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elton D, Medcalf E, Bishop K, Digard P. 1999. Oligomerization of the influenza virus nucleoprotein: identification of positive and negative sequence elements. Virology 260:190–200 [DOI] [PubMed] [Google Scholar]
- 49. Ye Q, Krug RM, Tao YJ. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444:1078–1082 [DOI] [PubMed] [Google Scholar]
- 50. Mänz B, Dornfeld D, Götz V, Zell R, Zimmermann P, Haller O, Kochs G, Schwemmmle M. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 9:e1003279. 10.1371/journal.ppat.1003279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Digard P, Elton D, Bishop K, Medcalf E, Weeds A, Pope B. 1999. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J. Virol. 73:2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang S, Chen J, Chen Q, Wang H, Yao Y, Chen J, Chen Z. 2013. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 87:767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paterson D, Fodor E. 2012. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 8:e1003019. 10.1371/journal.ppat.1003019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuiken T, Rimmelzwaan G, van Riel D, van Amerongen G, Baars M, Fouchier R, Osterhaus A. 2004. Avian H5N1 influenza in cats. Science 306:241. [DOI] [PubMed] [Google Scholar]
- 56. Horimoto T, Kawaoka Y. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3:591–600 [DOI] [PubMed] [Google Scholar]
- 57. Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396 [DOI] [PubMed] [Google Scholar]
- 58. Taubenberger JK, Kash JC. 2011. Insights on influenza pathogenesis from the grave. Virus Res. 162:2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. 1981. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull. World Health Organ. 59:75–78 [PMC free article] [PubMed] [Google Scholar]
- 60. Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. 1999. Human infection with influenza H9N2. Lancet 354:916–917 [DOI] [PubMed] [Google Scholar]
- 62. Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A. 2004. Human illness from avian influenza H7N3, British Columbia. Emerg. Infect. Dis. 10:2196–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. U. S. A. 97:9654–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125 [DOI] [PubMed] [Google Scholar]
- 66. Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Noda T, Sugita Y, Aoyama K, Hirase A, Kawakami E, Miyazawa A, Sagara H, Kawaoka Y. 2012. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 3:639. 10.1038/ncomms1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang TS, Palese P, Krystal M. 1990. Determination of influenza virus proteins required for genome replication. J. Virol. 64:5669–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl. Acad. Sci. U. S. A. 84:8140–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee MT, Bishop K, Medcalf L, Elton D, Digard P, Tiley L. 2002. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hagen M, Chung TD, Butcher JA, Krystal M. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fodor E, Pritlove DC, Brownlee GG. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Holsinger LJ, Nichani D, Pinto LH, Lamb RA. 1994. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 68:1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Whittaker G, Bui M, Helenius A. 1996. Nuclear trafficking of influenza virus ribonuleoproteins in heterokaryons. J. Virol. 70:2743–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Whittaker G, Bui M, Helenius A. 1996. The role of nuclear import and export in influenza virus infection. Trends Cell Biol. 6:67–71 [DOI] [PubMed] [Google Scholar]
- 78. Lamb RA, Choppin PW. 1983. The gene structure and replication of influenza virus. Annu. Rev. Biochem. 52:467–506 [DOI] [PubMed] [Google Scholar]
- 79. Ulmanen I, Broni B, Krug RM. 1983. Influenza virus temperature-sensitive cap (m7GpppNm)-dependent endonuclease. J. Virol. 45:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918 [DOI] [PubMed] [Google Scholar]
- 81. Deng T, Sharps JL, Brownlee GG. 2006. Role of the influenza virus heterotrimeric RNA polymerase complex in the initiation of replication. J. Gen. Virol. 87:3373–3377 [DOI] [PubMed] [Google Scholar]
- 82. Zhang S, Wang J, Wang Q, Toyoda T. 2010. Internal initiation of influenza virus replication of viral RNA and complementary RNA in vitro. J. Biol. Chem. 285:41194–41201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vreede FT, Gifford H, Brownlee GG. 2008. Role of initiating nucleoside triphosphate concentrations in the regulation of influenza virus replication and transcription. J. Virol. 82:6902–6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jorba N, Coloma R, Ortin J. 2009. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5:e1000462. 10.1371/journal.ppat.1000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Resa-Infante P, Jorba N, Coloma R, Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8:207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Portela A, Digard P. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83:723–734 [DOI] [PubMed] [Google Scholar]
- 87. Newcomb LL, Kuo RL, Ye Q, Jiang Y, Tao YJ, Krug RM. 2009. Interaction of the influenza A virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J. Virol. 83:29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Turrell L, Lyall JW, Tiley LS, Fodor E, Vreede FT. 2013. The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat. Commun. 4:1591. 10.1038/ncomms2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Robb NC, Smith M, Vreede FT, Fodor E. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90:1398–1407 [DOI] [PubMed] [Google Scholar]
- 90. Bullido R, Gomez-Puertas P, Saiz MJ, Portela A. 2001. Influenza A virus NEP (NS2 protein) downregulates RNA synthesis of model template RNAs. J. Virol. 75:4912–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, ten Oever BR. 2010. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U. S. A. 107:11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Umbach JL, Yen HL, Poon LL, Cullen BR. 2010. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio 1:e00204–10. 10.1128/mBio.00204-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Perez JT, Zlatev I, Aggarwal S, Subramanian S, Sachidanandam R, Kim B, Manoharan M, Tenoever BR. 2012. A small-RNA enhancer of viral polymerase activity. J. Virol. 86:13475–13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Selman M, Dankar SK, Forbes N, Jia JJ, Brown GE. 2012. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg. Microbes Infect. 1:e42. 10.1038/emi.2012.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. 2013. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 87:2455–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306–1312 [DOI] [PubMed] [Google Scholar]
- 97. Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza a virus m2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 8:e1002998. 10.1371/journal.ppat.1002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Engelhardt OG, Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16:329–345 [DOI] [PubMed] [Google Scholar]
- 99. Lamb RA, Lai CJ. 1982. Spliced and unspliced messenger RNAs synthesized from cloned influenza virus M DNA in an SV40 vector: expression of the influenza virus membrane protein (M1). Virology 123:237–256 [DOI] [PubMed] [Google Scholar]
- 100. Lamb RA, Lai CJ. 1984. Expression of unspliced NS1 mRNA, spliced NS2 mRNA, and a spliced chimera mRNA from cloned influenza virus NS DNA in an SV40 vector. Virology 135:139–147 [DOI] [PubMed] [Google Scholar]
- 101. Kemler I, Whittaker G, Helenius A. 1994. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology 202:1028–1033 [DOI] [PubMed] [Google Scholar]
- 102. Zaraket H, Bridges OA, Russell CJ. 2013. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus replication and pathogenesis in mice. J. Virol. 87:4826–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Matrosovich M, Stech J, Klenk HD. 2009. Influenza receptors, polymerase and host range. Rev. Sci. Tech. 28:203–217 [DOI] [PubMed] [Google Scholar]
- 104. Anhlan D, Grundmann N, Makalowski W, Ludwig S, Scholtissek C. 2011. Origin of the 1918 pandemic H1N1 influenza A virus as studied by codon usage patterns and phylogenetic analysis. RNA 17:64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. 2008. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog. 4:e1000079. 10.1371/journal.ppat.1000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rabadan R, Levine AJ, Robins H. 2006. Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes. J. Virol. 80:11887–11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ayllon J, Russell RJ, Garcia-Sastre A, Hale BG. 2012. Contribution of NS1 effector domain dimerization to influenza A virus replication and virulence. J. Virol. 86:13095–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. 2011. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 7:e1002067. 10.1371/journal.ppat.1002067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Dittmann J, Stertz S, Grimm D, Steel J, Garcia-Sastre A, Haller O, Kochs G. 2008. Influenza A virus strains differ in sensitivity to the antiviral action of Mx-GTPase. J. Virol. 82:3624–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J. Virol. 85:8133–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, Webster RG, Hoffmann E. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Clements ML, Subbarao EK, Fries LF, Karron RA, London WT, Murphy BR. 1992. Use of single-gene reassortant viruses to study the role of avian influenza A virus genes in attenuation of wild-type human influenza A virus for squirrel monkeys and adult human volunteers. J. Clin. Microbiol. 30:655–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mehle A, Doudna JA. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4:111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Moncorge O, Mura M, Barclay WS. 2010. Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J. Virol. 84:9978–9986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 62:403–424 [DOI] [PubMed] [Google Scholar]
- 116. Ping J, Keleta L, Forbes NE, Dankar S, Stecho W, Tyler S, Zhou Y, Babiuk L, Weingartl H, Halpin RA, Boyne A, Bera J, Hostetler J, Fedorova NB, Proudfoot K, Katzel DA, Stockwell TB, Ghedin E, Spiro DJ, Brown EG. 2011. Genomic and protein structural maps of adaptive evolution of human influenza A virus to increased virulence in the mouse. PLoS One 6:e21740. 10.1371/journal.pone.0021740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tarendeau F, Crepin T, Guilligay D, Ruigrok RW, Cusack S, Hart DJ. 2008. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 4:e1000136. 10.1371/journal.ppat.1000136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5:e1000252. 10.1371/journal.ppat.1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Massin P, van der Werf S, Naffakh N. 2001. Residue 627 of PB2 is a determinant of cold sensitivity in RNA replication of avian influenza viruses. J. Virol. 75:5398–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Labadie K, Dos Santos Afonso E, Rameix-Welti MA, van der Werf S, Naffakh N. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271–282 [DOI] [PubMed] [Google Scholar]
- 121. Rameix-Welti MA, Tomoiu A, Dos Santos Afonso E, van der Werf S, Naffakh N. 2009. Avian influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J. Virol. 83:1320–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ng AK, Chan WH, Choi ST, Lam MK, Lau KF, Chan PK, Au SW, Fodor E, Shaw PC. 2012. Influenza polymerase activity correlates with the strength of interaction between nucleoprotein and PB2 through the host-specific residue K/E627. PLoS One 7:e36415. 10.1371/journal.pone.0036415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bogs J, Kalthoff D, Veits J, Pavlova S, Schwemmle M, Manz B, Mettenleiter TC, Stech J. 2011. Reversion of PB2-627E to -627K during replication of an H5N1 clade 2.2 virus in mammalian hosts depends on the origin of the nucleoprotein. J. Virol. 85:10691–10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Liu Q, Qiao C, Marjuki H, Bawa B, Ma J, Guillossou S, Webby RJ, Richt JA, Ma W. 2012. Combination of PB2 271A and SR polymorphism at positions 590/591 is critical for viral replication and virulence of swine influenza virus in cultured cells and in vivo. J. Virol. 86:1233–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14:229–233 [DOI] [PubMed] [Google Scholar]
- 126. Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 4:e11. 10.1371/journal.ppat.0040011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hudjetz B, Gabriel G. 2012. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS Pathog. 8:e1002488. 10.1371/journal.ppat.1002488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, Chase G, Martinez-Sobrido L, Schwemmle M, Garcia-Sastre A. 2011. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2:e00151–11. 10.1128/mBio.00151-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Nakazono Y, Hara K, Kashiwagi T, Hamada N, Watanabe H. 2012. The RNA polymerase PB2 subunit of influenza A/HongKong/156/1997 (H5N1) restricts the replication of reassortant ribonucleoprotein complexes [corrected]. PLoS One 7:e32634. 10.1371/journal.pone.0032634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Aggarwal S, Dewhurst S, Takimoto T, Kim B. 2011. Biochemical impact of the host adaptation-associated PB2 E627K mutation on the temperature-dependent RNA synthesis kinetics of influenza A virus polymerase complex. J. Biol. Chem. 286:34504–34513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gabriel G, Klingel K, Otte A, Thiele S, Hudjetz B, Arman-Kalcek G, Sauter M, Shmidt T, Rother F, Baumgarte S, Keiner B, Hartmann E, Bader M, Brownlee GG, Fodor E, Klenk HD. 2011. Differential use of importin-alpha isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang S, Wang Q, Wang J, Mizumoto K, Toyoda T. 2012. Two mutations in the C-terminal domain of influenza virus RNA polymerase PB2 enhance transcription by enhancing cap-1 RNA binding activity. Biochim. Biophys. Acta 1819:78–83 [DOI] [PubMed] [Google Scholar]
- 133. Hara K, Schmidt FI, Crow M, Brownlee GG. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80:7789–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Huarte M, Falcon A, Nakaya Y, Ortin J, Garcia-Sastre A, Nieto A. 2003. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77:6007–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Gabriel G, Abram M, Keiner B, Wagner R, Klenk HD, Stech J. 2007. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J. Virol. 81:9601–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sakabe S, Ozawa M, Takano R, Iwastuki-Horimoto K, Kawaoka Y. 2011. Mutations in PA, NP, and HA of a pandemic (H1N1) 2009 influenza virus contribute to its adaptation to mice. Virus Res. 158:124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ince WL, Gueye-Mbaye A, Bennink JR, Yewdell JW. 2013. Reassortment complements spontaneous mutation in influenza A virus NP and M1 genes to accelerate adaptation to a new host. J. Virol. 87:4330–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 84:8607–8616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ye J, Sorrell EM, Cai Y, Shao H, Xu K, Pena L, Hickman D, Song H, Angel M, Medina RA, Manicassamy B, Garcia-Sastre A, Perez DR. 2010. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog. 6:e1001145. 10.1371/journal.ppat.1001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Neumann G, Hobom G. 1995. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76(Pt 7):1709–1717 [DOI] [PubMed] [Google Scholar]
- 142. Flick R, Hobom G. 1999. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J. Gen. Virol. 80(Pt 10):2565–2572 [DOI] [PubMed] [Google Scholar]
- 143. Crescenzo-Chaigne B, van der Werf S, Naffakh N. 2002. Differential effect of nucleotide substitutions in the 3′ arm of the influenza A virus vRNA promoter on transcription/replication by avian and human polymerase complexes is related to the nature of PB2 amino acid 627. Virology 303:240–252 [DOI] [PubMed] [Google Scholar]
- 144. Widjaja I, de Vries E, Rottier PJ, de Haan CA. 2012. Competition between influenza A virus genome segments. PLoS One 7:e47529. 10.1371/journal.pone.0047529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Belicha-Villanueva A, Rodriguez-Madoz JR, Maamary J, Baum A, Bernal-Rubio D, Minguito de la Escalera M, Fernandez-Sesma A, Garcia-Sastre A. 2012. Recombinant influenza A viruses with enhanced levels of PB1 and PA viral protein expression. J. Virol. 86:5926–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Gultyaev AP, Fouchier RA, Olsthoorn RC. 2010. Influenza virus RNA structure: unique and common features. Int. Rev. Immunol. 29:533–556 [DOI] [PubMed] [Google Scholar]
- 147. Noda T, Kawaoka Y. 2010. Structure of influenza virus ribonucleoprotein complexes and their packaging into virions. Rev. Med. Virol. 20:380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J. Gen. Virol. 91:313–328 [DOI] [PubMed] [Google Scholar]
- 149. Zheng H, Palese P, Garcia-Sastre A. 1996. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology 217:242–251 [DOI] [PubMed] [Google Scholar]
- 150. Ng SS, Li OT, Cheung TK, Malik Peiris JS, Poon LL. 2008. Heterologous influenza vRNA segments with identical non-coding sequences stimulate viral RNA replication in trans. Virol. J. 5:2. 10.1186/1743-422X-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Suzuki Y, Kobayashi Y. 2013. Evolution of complementary nucleotides in 5′ and 3′ untranslated regions of influenza A virus genomic segments. Infect. Genet. Evol. 13:175–179 [DOI] [PubMed] [Google Scholar]
- 152. Furuse Y, Oshitani H. 2011. Evolution of the influenza A virus untranslated regions. Infect. Genet. Evol. 11:1150–1154 [DOI] [PubMed] [Google Scholar]
- 153. Almond JW. 1977. A single gene determines the host range of influenza virus. Nature 270:617–618 [DOI] [PubMed] [Google Scholar]
- 154. Rigoni M, Shinya K, Toffan A, Milani A, Bettini F, Kawaoka Y, Cattoli G, Capua I. 2007. Pneumo- and neurotropism of avian origin Italian highly pathogenic avian influenza H7N1 isolates in experimentally infected mice. Virology 364:28–35 [DOI] [PubMed] [Google Scholar]
- 155. Shinya K, Makino A, Ozawa M, Kim JH, Sakai-Tagawa Y, Ito M, Le QM, Kawaoka Y. 2009. Ostrich involvement in the selection of H5N1 influenza virus possessing mammalian-type amino acids in the PB2 protein. J. Virol. 83:13015–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Hutchinson EC, Denham EM, Thomas B, Trudgian DC, Hester SS, Ridlova G, York A, Turrell L, Fodor E. 2012. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 8:e1002993. 10.1371/journal.ppat.1002993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Pal S, Santos A, Rosas JM, Ortiz-Guzman J, Rosas-Acosta G. 2011. Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 158:12–27 [DOI] [PubMed] [Google Scholar]