Abstract
In contrast to orthoretroviruses, processing of foamy viral p71 Gag is limited to a single cleavage site. Nevertheless, Gag maturation is essential for infectivity, but deletion of p3 results in a modest drop in infectivity. Here, we show that Gag processing of p71 to p68 and p3 is essential for full-length cDNA synthesis, while inactivation of Gag cleavage results in cDNAs containing only the RU5 region; cDNAs encompassing the U3 region were almost undetectable.
TEXT
Retroviral Gag and Pol precursor proteins are processed by the viral protease (PR) (1). The Gag and Pol proteins of HIV-1 encode nine functional cleavage sites. Both proteins are processed during maturation (1). Only mature viruses were shown to be infectious. HIV-1 Gag maturation leads to conformational rearrangements of the Gag domains, which are visible by the formation of an electron-dense capsid structure in the maturated virus. Furthermore, it was shown recently that Gag maturation is required for Env clustering on the virus surface (2). Foamy viruses (FV) on the other hand encode single cleavage sites in Gag and in Pol. The prototypic foamy viral p71 Gag protein is processed only at the carboxyl-terminal end to p68 and a p3 peptide. This cleavage was shown to be essential for viral infectivity (3, 4). Eminent structural changes induced by virus maturation were not observed by electron microscopy (EM) (5). In addition, it was shown that processing of FV Pol is not essential for reverse transcriptase (RT) activity (6) but was indispensable for genome integration (7).
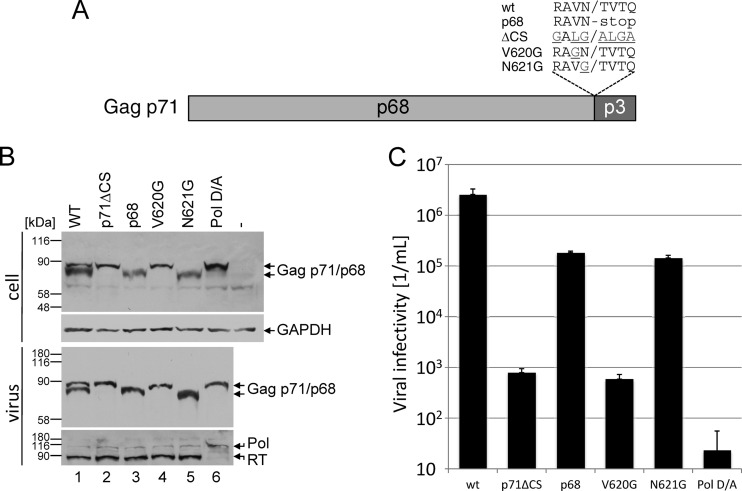
To analyze the influence of prototype FV (PFV) Gag processing on infectivity and to recapitulate experiments that were done before, we inactivated either the Gag cleavage site or expressed only the processed p68 Gag form. First, we exchanged the wild-type Gag cleavage site sequence RAVN-TVTQ with residues GALG-ALGA in the context of the codon-optimized expression plasmid (Fig. 1) (8, 9). This plasmid was named p71ΔCS (Fig. 1). This modified cleavage site is not processed by the viral PR, as shown by Western blotting below. In a second gag mutant, all sequences downstream of p68 were removed and a stop codon was inserted directly downstream of p68, giving rise to the processed p68 form of Gag (Fig. 1). The p71ΔCS and p68 expression constructs were coexpressed in HEK 293T cells (1.8 × 106) with codon-optimized expression constructs for pol-, env-, and a gfp-containing vector genome (10). As an additional control, cells were cotransfected with a previously described protease activity-deficient pol mutant, pPol D/A (5, 9) together with gag, env, and the genome-encoding vector pMD9. Three days after transfection, viral supernatants were collected, filtered, and cleared by size exclusion chromatography using HighTrap Capto Core 700 columns (GE Healthcare, Germany) and phosphate-buffered saline (PBS) (137 mM NaCl, 2.68 mM KCl, 6.46 mM Na2HPO4, 1.15 mM KH2PO4, 0.9 mM CaCl2, 0.5 mM MgCl2). Viruses were collected by ultracentrifugation (201,149 × g, 4°C, 2 h), and Gag processing was visualized by Western blotting as described before (Fig. 1B) (9, 11, 12). All experiments in this study were repeated at least three times. This analysis revealed that wild-type Gag was processed, leading to the characteristic p71-p68 double band, whereas the p71ΔCS mutant remained unprocessed (Fig. 1B, lanes 1 and 2). The recombinant Gag p68 viruses expressed the expected single Gag p68 form (Fig. 1B, lane 3). Viral titers were determined on BHK-21 cells by serial dilutions (Fig. 1C). Viral Gag amounts were quantified, and viral titers were normalized on released Gag. The p71ΔCS recombinant viruses were almost noninfectious (titer reduction more than 3 orders of magnitude), whereas deletion of the complete p3 domain reduced viral titers about 1 order of magnitude. The control transfection with the PR-inactivated Pol led to unprocessed Gag and noninfectious virus as expected. These results indicate that Gag processing is essential for infectivity, while complete deletion of the p3 domain has only limited effects on infectivity.
Fig 1.
Foamy virus infectivity is correlated to Gag maturation. (A) Schema of the Gag p68-p3 cleavage sites used in this study. All plasmids were constructed by site-directed mutagenesis. wt, wild type. (B) Western blotting of purified recombinant with a monoclonal anti-Gag (upper and middle panel) or anti-Pol antibodies (lower panel). Positions of the size markers are indicated. WT, wild type. (C) Viral titers were determined on BHK-21 cells by serial dilutions as described before (8, 9). Error bars indicate the standard deviations of triplicate samples.
To further investigate effects of Gag processing on viral infectivity, we have analyzed the effect of single amino acid exchanges within the Gag cleavage site on Gag processing. We identified that the exchange of amino acid residue N621G led to complete Gag cleavage, whereas the exchange of V620G inhibited processing completely (Fig. 1B). HEK 293T cells were cotransfected with these gag constructs and with expression plasmids for pol and env and pMD9. Viruses were concentrated, and viral titers were determined as described above (Fig. 1C). Gag processing was analyzed by Western blotting (Fig. 1B). In recombinant viruses with Gag N621G, only the processed p68 form of Gag was detected, whereas exchange of V620G led to complete inhibition of cleavage. This mutation resulted in noninfectious virus, while N621G showed a titer reduction similar to the p68 form of Gag (Fig. 1C).
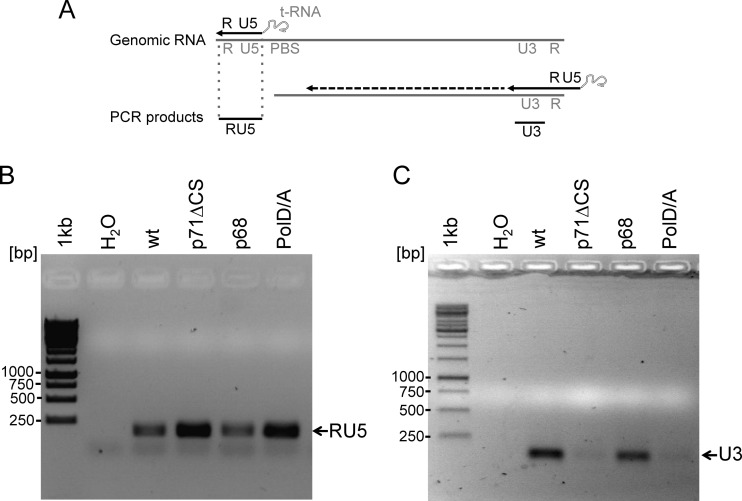
Since structural changes due to Gag maturation were not profound in electron microscopy (5), we sought to analyze effects of Gag cleavage on the RT reaction. The RT reaction occurs late in the foamy viral replication cycle and is almost completed before entry. First we wanted to determine whether the free p3 influences RT activity. Therefore, in vitro RT activity assays using protease-reverse transcriptase (PR-RT) either from prototype or simian FV were performed (13) in the absence of p3 (RT activity, 23.7 ± 1.1 U/μg) or in the presence of equimolar concentrations (12 nM) of PR-RT and p3 (PFV p3 peptide, 5/6-fluorescein-AGDSRAVNTVTQSATSSTDESSSAVTAASGGDQRD) (RT activity, 26.6 ± 1.7 U/μg) or in a 6-fold excess of p3 peptide (RT activity, 25.3 ± 1.4 U/μg). These results indicate that free p3 has no impact on RT activity, leading us to the hypothesis that Gag p71 might inhibit the RT reaction. To investigate this model, recombinant viruses were produced in HEK 293T cells using the Gag p71ΔCS, p68, or pPol D/A expression plasmids in the context of the foamy viral vector system as described above. Viruses were harvested by ultracentrifugation. Viral pellets were dissolved in PBS and treated with RNase-free DNase I to remove contaminating cellular and plasmid DNA. The samples were normalized on relative Gag amounts by quantitative Western blotting (data not shown), and equal amounts were directly used in PCRs using primers in the RU5 and U3 region (Fig. 2). The first PCR detects products of the earliest step of cDNA synthesis, i.e., the strong stop (−) DNA (Fig. 2A), whereas the latter amplifies products after the first template switch (Fig. 2A). The RU5 region-specific PCR led to similar amounts of PCR products, indicating that the initiation of the cDNA synthesis and elongation to the R region were equally efficient for all samples (Fig. 2B). Reverse transcription of the U3 region, after the first strong stop (Fig. 2A), was impaired by Gag p71, since cDNAs encompassing the U3 region of both p71ΔCS and the protease activity-deficient pPol D/A viruses were found to be almost undetectable. The results indicate that Gag processing is essential for the first template switch.
Fig 2.
Foamy virus reverse transcriptase template switch is dependent on Gag processing. (A) Schema of the first steps of the reverse transcription and the position of the PCR products (DNA, black; RNA, gray). (B) Amplification of the RU5 region from viral cDNAs with primers located in R and U5. wt, wild type. (C) PCR with U3-specific primers using concentrated recombinant viruses as the template. The recombinant viruses were analyzed by Western blotting using a monoclonal Gag-specific antibody. The Gag signals of the different mutants were quantified using the AIDA software package (GE Healthcare) and normalized on Gag content, to guarantee equal Gag amounts in the PCRs. Identical amounts of concentrated viruses were used for RU5 and U3 PCR.
In summary, we have shown that Gag processing is essential for FV infectivity, whereas the p3 region itself is not. Nonprocessed p71 seems to inhibit cDNA synthesis at the 1st template switch. We propose that p71 Gag binds to the 3′ end of the viral RNA, thereby blocking the accessibility of the 3′ end, which is required to perform the strand transfer reaction. Foamy virus particles contain a mixture of p68 and p71; thus, complete processing of all Gag molecules is not required for strand transfer. Therefore, blockage of strand transfer appears to be dependent on the concentration of uncleaved p71. We propose that by this mechanism cDNA synthesis is delayed until sufficient amounts of Gag and Pol are cleaved to ensure viral infectivity. This checkpoint is needed, because cDNA synthesis leads to degradation of the protease-activating RNA motif (PARM), which is essential for protease activity in FV (8) and therefore terminates processing of Gag and Pol.
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Coffin JM, Hughes SH, Varmus HE. 1997. Retroviruses Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 2. Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Müller B, Hell SW, Kräusslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528 [DOI] [PubMed] [Google Scholar]
- 3. Enssle J, Fischer N, Moebes A, Mauer B, Smola U, Rethwilm A. 1997. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J. Virol. 71:7312–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baldwin DN, Linial ML. 1999. Proteolytic activity, the carboxy terminus of Gag, and the primer binding site are not required for Pol incorporation into foamy virus particles. J. Virol. 73:6387–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Konvalinka J, Löchelt M, Zentgraf H, Flügel RM, Kräusslich HG. 1995. Active foamy virus proteinase is essential for virus infectivity but not for formation of a Pol polyprotein. J. Virol. 69:7264–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hütter S, Müllers E, Stanke N, Reh J, Lindemann D. 2013. Prototype foamy virus protease activity is essential for intraparticle reverse transcription initiation but not absolutely required for uncoating upon host cell entry. J. Virol. 87:3163–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roy J, Linial ML. 2007. Role of the foamy virus Pol cleavage site in viral replication. J. Virol. 81:4956–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hartl MJ, Bodem J, Jochheim F, Rethwilm A, Rösch P, Wöhrl BM. 2011. Regulation of foamy virus protease activity by viral RNA: a novel and unique mechanism among retroviruses. J. Virol. 85:4462–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spannaus R, Hartl MJ, Wöhrl BM, Rethwilm A, Bodem J. 2012. The prototype foamy virus protease is active independently of the integrase domain. Retrovirology 9:41. 10.1186/1742-4690-9-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stirnnagel K, Lüftenegger D, Stange A, Swiersy A, Müllers E, Reh J, Stanke N, Grosse A, Chiantia S, Keller H, Schwille P, Hanenberg H, Zentgraf H, Lindemann D. 2010. Analysis of prototype foamy virus particle-host cell interaction with autofluorescent retroviral particles. Retrovirology 7:45. 10.1186/1742-4690-7-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Imrich H, Heinkelein M, Herchenröder O, Rethwilm A. 2000. Primate foamy virus Pol proteins are imported into the nucleus. J. Gen. Virol. 81:2941–2947 [DOI] [PubMed] [Google Scholar]
- 12. Heinkelein M, Dressler M, Jarmy G, Rammling M, Imrich H, Thurow J, Lindemann D, Rethwilm A. 2002. Improved primate foamy virus vectors and packaging constructs. J. Virol. 76:3774–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartl MJ, Kretzschmar B, Frohn A, Nowrouzi A, Rethwilm A, Wöhrl BM. 2008. AZT resistance of simian foamy virus reverse transcriptase is based on the excision of AZTMP in the presence of ATP. Nucleic Acids Res. 36:1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]