Abstract
The adenovirus E1A C-terminal region restrains oncogenic transformation through interaction with three distinct cellular protein complexes that include the DYRK1A/1B/HAN11 complex. The E6 proteins of beta-human papillomaviruses (beta-HPVs) also interact with the DYRK1/HAN11 complex. A variant of HPV5 E6 frequently found in epidermodysplasia verruciformis skin lesions interacted less efficiently with DYRK1A/HAN11. The E6 variant and E7 of HPV5 efficiently coimmortalized primary epithelial cells, suggesting that naturally arising variants may contribute potential oncogenic activities of beta-HPV E6 proteins.
TEXT
Functional commonality is a prominent theme by which the oncoproteins of small DNA tumor viruses such as the polyomaviruses, papillomaviruses, and adenoviruses subvert cell growth-regulatory activities. It is well known that the transforming proteins of these viruses target the tumor suppressor protein p53 to inactivate its cell growth-inhibitory activities (reviewed in reference 1). The second example is the targeting of the retinoblastoma protein (pRb) by which the viral proteins such as adenovirus E1A, simian virus 40 (SV40) T antigen, and human papillomavirus (HPV) E7 disable the cell cycle restriction imposed by retinoblastoma family proteins (reviewed in references 2 and 3). Since these viruses infect quiescent target cells, they induce a cellular proliferative state to facilitate their DNA replication by inactivating the cell cycle regulatory checkpoints conferred by antiproliferative molecules such as p53 and pRb. The transforming protein E4-ORF1 of HAdv9 and E6 proteins of high-risk HPVs have been shown to target the epithelial cell tight junction and polarity proteins, such as hDgl-1, Scribble, and PATJ, through conserved PDZ-binding motifs to disrupt epithelial differentiation and promote oncogenesis (4).
Studies on the transforming activities of HAdv5/2 E1A proteins have revealed that the N-terminal region of E1A promotes cell proliferation and inhibits cell differentiation to mediate oncogenic cell transformation (5) by interacting with retinoblastoma family proteins, p300/CBP (6, 7), and p400/TRRAP protein complexes (8–10). In contrast to the N-terminal region, the C-terminal region antagonizes cell proliferation (11) and differentiation activities (12, 13). Deletion of the C-terminal region in HAdv5/2 E1A confers a hypertransforming function to E1A (12, 14–17). The transformed cells that express the E1A C-terminal mutants and the activated Ras oncogene are also highly tumorigenic and metastatic (14, 17). The E1A C-terminal region interacts with three different protein complexes, FOXK1/K2, DYRK1A/1B/HAN11 (referred to here as DYRK1/HAN11), and CtBP1/2 to collectively confer the tumor-restraining activity to the E1A C-terminal region (11).
Of the more than 120 known human papillomaviruses (HPVs), a predominant number belong to the genus beta. Unlike the alpha-HPVs, which are linked to anogenital and head and neck cancers, the beta-HPVs are less extensively studied. Certain beta-HPV species have been linked to the development of nonmelanoma skin cancers (18). More than 40 different beta-HPVs have been identified in the hyperproliferative skin lesions seen in patients suffering from the genetic disorder epidermodysplasia verruciformis (EV) (19–22). About half of these patients develop squamous cell carcinoma (SCC) in sun-exposed areas by the time they reach midlife (22). As the EV lesions progress to SCC, there appears to be a selective prevalence of certain HPV species, such as HPV5 and HPV8 (21). By analogy to alpha-HPVs, it is believed that the E7 and E6 proteins of beta-HPVs may play critical roles in the overall viral biology and in potential oncogenic transformation. The beta-HPV E6 proteins have been reported to suppress UV-induced apoptosis by interacting with the proapoptotic molecule BAK and targeting it for degradation (23, 24). Recent protein-protein interaction studies using affinity purification and mass-spectrometric analyses and yeast two-hybrid interaction approaches identified interactions with cellular proteins such as MAML1, p300, and Ccr4-Not (25–28). These studies have provided novel insights into the functions of beta-HPV E6 proteins.
Earlier, we reported that both HAdv5 E1A and E6 proteins of a subgroup of cutaneous beta-HPVs (HPV14 and 21) contain a Ser/Thr-rich FHA (forkhead-associated domain)-binding motif and interact with the forkhead transcription factors FOXK1 and -K2 to suppress cell proliferation and transformation (11). The E6 protein of HPV20 [prevalent in several SCCs (29)] also contains the FHA-binding motif, but with a single amino acid substitution that prevents interaction with FOXK1/K2. The apparent similarity between HAdv5 E1A and E6 proteins of HPV14/21 prompted us to examine whether the similarity between the E1A C terminus and the E6 proteins of beta-HPVs extends beyond FOXK1/K2 interaction. Here, we report that the E6 proteins of several beta-HPVs interact with the DYRK1/HAN11 complex and certain E6 variants of HPV5 and HPV8 associated with human pathologies are deficient in such interaction.
E6 proteins of beta-HPVs interact with DYRK1A/1B/HAN11 complex.
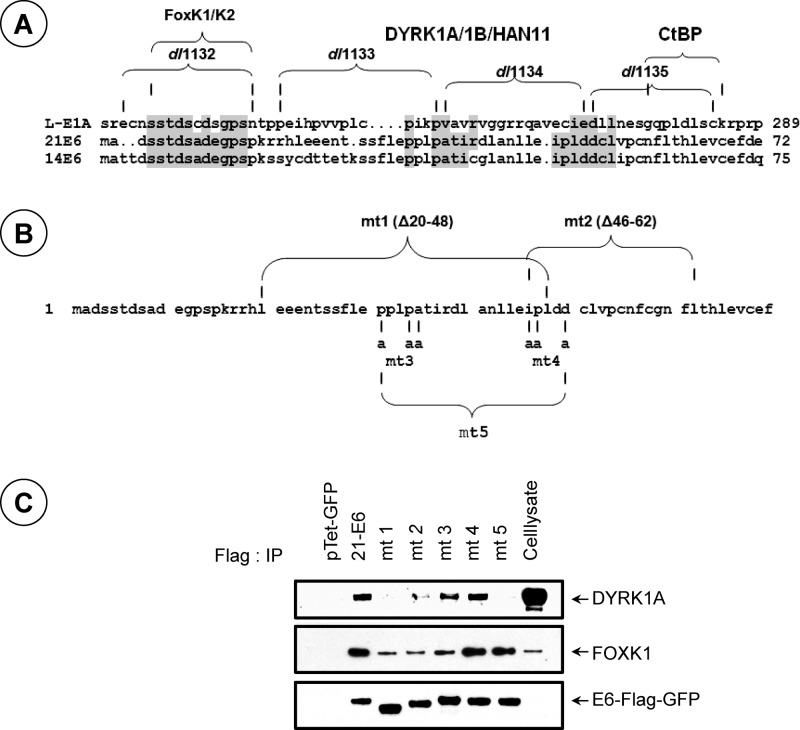
Although no specific sequence motif in HAdv5 E1A that mediates the interaction with DYRK1A/1B has been identified, the CR4 region involved in this interaction is conserved among different HAdv E1A proteins (30). Visual examination of HPV14/21 E6 sequences adjoining the FHA-binding motif revealed a limited sequence similarity (Fig. 1A). We constructed five different mutants in HPV21 E6 (21E6) (Fig. 1B) and carried out coimmunoprecipitation and Western blot analyses to determine their ability to interact with endogenous DYRK1A (Fig. 1C). Immunoprecipitation of the Flag-tagged 21E6-green fluorescent protein (GFP) fusion protein revealed readily detectable interaction of DYRK1A with wild-type (wt) 21E6-GFP. A deletion mutant encompassing the N-terminal residues 20 to 48 (mt 1, Δ20–48) was defective in interaction with DYRK1A. A second deletion mutant, mt 2 (Δ46–62), interacted weakly. Based on these results, we hypothesized that a bipartite sequence motif within the N-terminal region encompassing the sequences deleted within mt 1 and 2 might mediate interaction with DYRK1A. We then made amino acid substitution mutants (mt 3 and mt 4) (Fig. 1B) by converting certain conserved residues within each motif. Immunoprecipitation studies revealed that both mutants interacted with DYRK1A like wt 21E6 (Fig. 1C). In contrast, a mutant (mt 5) that contained a combination of the mutations in mt 3 and mt 4 was deficient in interaction with DYRK1A. It should be noted that mt 5 interacted with FOXK1, showing that the mutation did not alter the overall structure of E6. These results suggest that 21E6 may interact with DYRK1A through two redundant sequence motifs within the N-terminal region.
Fig 1.
Interaction of DYRK1A with HPV21 E6 protein. (A) Homology between the C-terminal region of E1A and the N-terminal regions of 21/14E6. The E1A deletion mutants that are defective in interaction with different cellular protein complexes are indicated. E1A mutants dl1133 and dl1134 are defective in interaction with the DYRK1/HAN11 complex. (B) Mutants of 21E6. (C) Immunoprecipitation analysis of DYRK1A interaction with 21E6. The E6-GFP-Flag fusion proteins of wt 21E6 or E6 mutants (codon optimized) were transiently expressed in 293 cells using the Tet-GFP-inducible vector and immunoprecipitated with the Flag antibody (Ab) as described previously (11). The Western blots were probed with Abs specific to DYRK1A, FOXK1, or the Flag epitope.
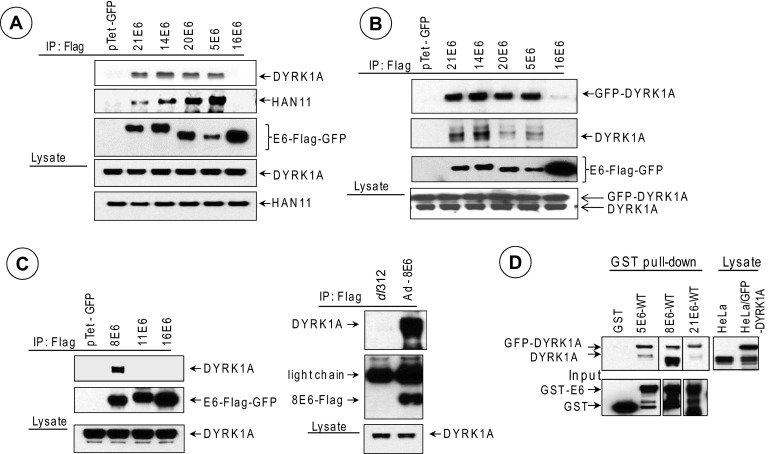
We then tested whether E6 proteins of other beta- and alpha-HPVs interact with DYRK1A. Coimmunoprecipitation/Western blot analysis revealed that the E6-GFP proteins of HPV21, -14, and -20 and HPV5 interacted well with endogenous DYRK1A and HAN11 (Fig. 2A) and endogenous DYRK1B (data not shown). In contrast, 16E6-GFP did not interact with any of the three proteins in spite of relative overexpression of 16E6. We further ascertained the interaction pattern by cotransfection of both E6 and GFP-DYRK1A (Fig. 2B). We also investigated the interaction of DYRK1A with other E6 proteins (Fig. 2C). While DYRK1A interacted with transiently expressed 8E6 (beta-HPV), it did not interact with 11E6 (low-risk alpha-HPV) (Fig. 2C, left). We observed prolific interaction of endogenous DYRK1A with 8E6 expressed from an HAdv5 vector that expresses 8E6 (Fig. 2C, right). We also observed the interaction of DYRK1A and DYRK1A-GFP in a GST pulldown assay with GST-21E6, GST-5E6, and GST-8E6 (Fig. 2D). Thus, the results presented in Fig. 1 and 2 suggest that the E6 proteins of different beta-HPVs (HPV14, -20, -21, -5, and -8) interact with the DYRK1/HAN11 complex, like HAdv5 E1A.
Fig 2.
Interaction of DYRK1/HAN11 complex with different beta-HPV E6 proteins. (A) Interaction with endogenous DYRK1A and HAN11. Flag-tagged E6-GFP fusion proteins were expressed using the Tet-inducible vector and immunoprecipitated with the Flag Ab, and the Western blots were probed with Abs specific to DYRK1A, HAN11, or the Flag epitope. (B) Interaction with GFP-DYRK1A. The different E6-GFP-Flag fusion constructs were cotransfected with a GFP-DYRK1A construct and immunoprecipitated with the Flag Ab, and the Western blots were probed with the DYRK1A Ab or the Flag Ab. Bands corresponding to GFP-DYRK1A and endogenous DYRK1A are indicated. (C) Interaction of DYRK1A with 8E6. The interactions of endogenous DYRK1A with Flag-tagged 8E6-GFP and 11E6-GFP fusion proteins are shown on the left. The interaction of endogenous DYRK1A with Flag-8E6 expressed from an HAdv5 vector in C33A cells is shown on the right. Plasmid vectors that express 5E6-Flag-GFP, 14E6-Flag-GFP, and 20E6-Flag-GFP were described earlier (11). Plasmid vectors that express 8E6, 11E6, and 16E6 as Flag-GFP fusion proteins were constructed by cloning synthetic oligonucleotides containing codon-optimized E6 ORFs in the vector pTet-Flag-GFP. The HAdv5-8E6 recombinant virus was constructed using the transfer vector pLend and the HAdv5 genomic vector pBHGE3 as described previously (38). (D) GST pulldown assay. Bacterially expressed GST, GST-5E6 wt, GST-8E6 wt, and GST-21E6 wt proteins were incubated with the whole-cell lysate from HeLa cells transfected with GFP-DYRK1A and processed as described earlier (14) and analyzed by Western blotting using the DYRK1A Ab. The Western blots of input samples were probed with the GST Ab.
Interaction of DYRK1A with EV and cancer-associated E6 variants.
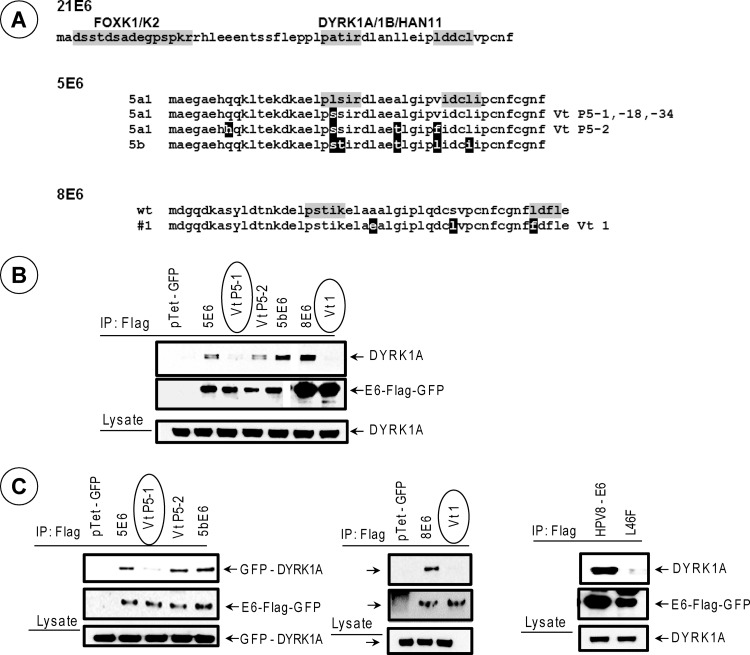
The premalignant and malignant lesions associated with EV contain the DNA sequences of HPV5 and HPV8 DNA (31–33). Sequence analysis of a number of HPV5 (subtype a) isolates from EV lesions were found to contain mutations predominantly in the N-terminal region of E6 (31) that encompasses the DYRK1-interacting region that we have identified here. Although various isolates contained multiple mutations, the E6 open reading frame (ORF) of all EV isolates contained a Ser residue at position 21, while the prototype of 5E6 (subtype a1) contained a Leu residue at that position. We chose to determine the pattern of interaction of DYRK1A with selected number of E6 variant proteins (Vt P5-1, Vt P5-2, and subtype 5b) (Fig. 3). Although GenBank contains only a limited number of HPV8 E6 variant sequences, two of the 8E6 variants found in the data bank also contained multiple mutations in the N-terminal region (Fig. 3A). We also chose to determine the pattern of interaction of DYRK1A with the prototype 8E6 and one of the variants found in a genital carcinoma in situ (34) (designated here as Vt 1). The interaction of selected E6 variants with endogenous DYRK1A (Fig. 3B) or transfected GFP-DYRK1A (Fig. 3C) was determined by immunoprecipitation and Western blot analysis. Among the 5E6 variants examined, variant Vt P5-1 (L21S) interacted with endogenous DYRK1A (Fig. 3B) and GFP-DYRK1A (Fig. 3C) at reduced levels. Similarly, 8E6 Vt 1 was also deficient in interaction with endogenous and GFP-DYRK1A compared to the prototype 8E6. Similar patterns of DYRK1A interactions with 5E6 and 8E6 and the variants were also observed with the GST-E6 pulldown assays (data not shown). In addition to 8E6 Vt 1, a different mutant that was engineered to contain only one of three substitutions (L46→F) also interacted with DYRK1A at a much reduced level in the coimmunoprecipitation studies (Fig. 3C, right). These results suggest that the E6 proteins of some of the EV-associated variants of HPV5 and a cancer-associated variant of 8E6 are deficient in interaction with DYRK1A.
Fig 3.
Interaction of DYRK1A with 5E6 and 8E6 variants. (A) N-terminal amino acid sequences of 5E6 and 8E6 variants. The amino acids that are different from the prototype E6 proteins are highlighted in black. The bipartite motif implicated in DYRK1/HAN11 interaction of 21E6 and the homologous sequences in 5E6 and 8E6 are indicated by grey shading. (B) Interaction of endogenous DYRK1A. The interaction of endogenous DYRK1A with Flag-tagged E6-GFP fusion proteins of 5E6 and 8E6 was determined as for Fig. 1. The variants deficient in DYRK1A interaction are circled. (C) Interaction of GFP-DYRK1A with E6 variants. The interaction of E6 variants with exogenously introduced GFP-DYRK1A was determined as for Fig. 2.
Immortalization of epithelial cells by 5E6.
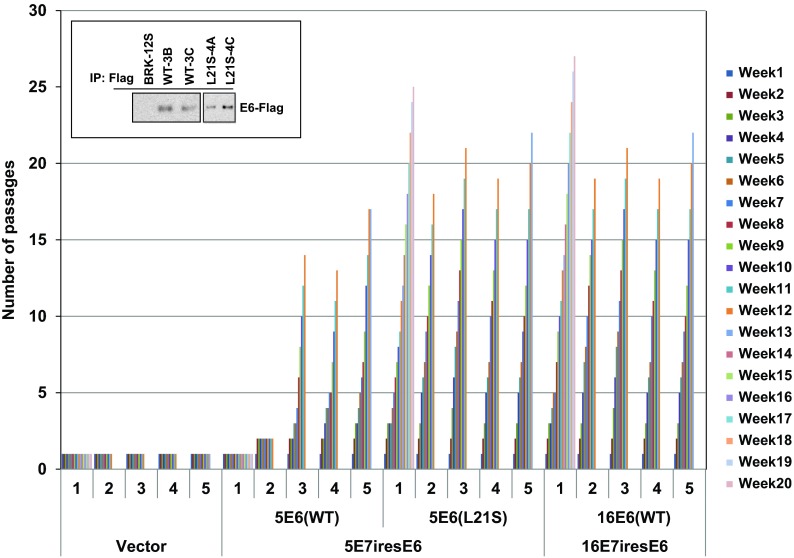
At present there are no well-developed immortalization and cell transformation assays for human beta-HPV E7 and E6 genes. During extensive preliminary studies, we observed that a lentiviral vector that expressed codon-optimized E7 and E6 of HPV5 or HPV16 induced cell proliferation and immortalization of primary baby rat kidney (BRK) cells (Fig. 4), while the lentiviral vectors that expressed nonoptimized E7 and E6 were unable to immortalize BRK cells (results not shown). Also lentiviral vectors that expressed codon-optimized 21E7 and 21E6 or 8E7 and 8E6 were unable to immortalize BRK cells (results not shown). We used the BRK immortalization assay to determine the effect of DYRK1 interaction with HPV5 E6. While BRK cells transduced with the control vector became senescent and did not survive after the first splitting, cells transduced with 5E7-5E6 wt, 5E7-5E6L21S, or 16E7-16E6 proliferated for an additional passage at comparable levels. However, the proliferation of cells expressing 5E6 wt or 5E6L21S differed significantly thereafter. In three of five independent experiments, cells transduced with 5E7-5E6 induced cell proliferation, and cells were passaged beyond 12 to 15 passages. All cells that went beyond passages 12 to 15 were considered immortalized, and they continued to grow beyond these passages. BRK cells transduced with 5E7-5E6L21S induced more efficient cell proliferation in all five independent experiments and resulted in continuously growing immortalized cell lines (Fig. 4). Interestingly, vectors that expressed codon-optimized 16E7 and 16E6 also induced enhanced cell proliferation and immortalized cell lines in all five independent experiments. These results suggest that interaction of DYRK1/HAN11 with 5E6 may retard cell proliferation. This result is reminiscent of HAdv5 E1A mutants that are defective in interaction with DYRK1/HAN11 that also induced cell hyperproliferation and cell transformation (11).
Fig 4.
Immortalization of BRK cells by HPV E7 and E6. Lentiviral constructs that express HA-tagged E7 and Flag-tagged E6 (with an intervening internal ribosome entry site [IRES] sequence for E6 translation) were generated using the vector pCDH–MCS-EF1-Puro (System Biosciences). Lentiviral particles were generated by transfection of 293 T cells with the pCDH-HPV plasmids pVSVG and pCMVΔ8.2ΔVPR (39) using the Lipofectamine reagent. The BRK cells were infected with the virus for 2 to 4 h in the presence of Polybrene (8 μg/ml), and the cells were subjected to puromycin selection (0.25 μg/ml). When the cells reached about 80% confluence, they were split to 1:2, serially. The number of passages was plotted against weeks of propagation. For each construct, columns 1 to 5 show results of five independent immortalization assays. The expression of Flag-E6 proteins in immortalized BRK cell lines that express 5E6 wt or 5E6L21S is shown in the inset. The Flag-E6 proteins were immunoprecipitated with the Flag Ab, and the Western blots were probed with the same Ab.
Here we have demonstrated that the E6 proteins of several beta-HPVs interact with the cellular protein complex DYRK1/HAN11. Previously, we and others showed that the CR4 region of HAdv5/2 E1A interacts with DYRK1 (11, 35) and its cofactor HAN11 (11). Sequence comparison between the E1A CR4 region that is required for DYRK1 interaction and the E6 proteins of HPV21/14 and targeted mutagenesis of HPV21 E6 resulted in our identification of a bipartite sequence element that mediates DYRK1 interaction. In an earlier report, we showed that the E6 proteins of benign HPV21/14 contain a unique N-terminal Ser/Thr-rich motif (FHA-interacting motif) similar to HAdv5 E1A and interact with the transcription factors FOXK1/K2 (11). The FHA-interacting motif is absent in the E6 proteins of other beta-HPV E6 proteins. Like the E6 proteins of HPV21/14, only the E1A proteins of group C HAdvs (nontumorigenic) contain the FHA-interacting motif. In contrast, the E1A CR4 region involved in DYRK1/HAN11 complex is conserved among the E1A proteins of all primate Advs (30). Similarly, our present results indicate that at least five beta-HPV (HPV5, -8, -14, -20, and -21) E6 proteins interact with the DYRK1/HAN11 complex. Thus, the N-terminal regions of the E6 proteins of beta-HPVs appear to be functionally similar to the C-terminal region of E1A. It is unclear why a recent unbiased proteomic study (28) and two hybrid interaction studies (27) did not identify DYRK1/HAN11 interaction with the beta-HPV E6 proteins. A major difference might that we used transient overexpression of different E6 proteins. Further, potential amino acid sequence variations in the N-terminal region of 5E6 and 8E6 used by different groups may also contribute to the difference.
An important outcome of our study is that naturally arising polymorphic variants of 5E6 and 8E6 differed in their ability to interact with the DYRK1/HAN11 complex. While the E6 protein of the prototype strain of HPV5 (strain 5a1) contains a Leu21 residue, the E6 proteins of several EV-associated strains contain a Ser residue at that position. The Ser21 substitution in the E6 protein of 5a1 reduced the level of DYRK1A interaction. Interestingly, the L21S mutation in 5E6 also enhanced the immortalization activity of the 5E7-E6 lentiviral construct. Although the E6 proteins of other variants, such as P5-2 and strain 5b, contain the L21S substitution, they contain additional mutations in the vicinity (Fig. 3A). These E6 proteins interacted with DYRK1A better than the L21S-alone variant (e.g., Vt P5-1), suggesting that other sequences within the N-terminal region of 5E6 influence the level of DYRK1A interaction. In general, 8E6 interacted with DYRK1A more efficiently than other E6 proteins. An 8E6 variant found in a human neoplasm contained three mutations at the N-terminal region (Fig. 3A) and was strongly defective in interaction with DYRK1A (see below).
Among the lentiviral vectors that expressed the E7 and E6 proteins of HPV5, HPV8, HPV21, and HPV16, only vectors that expressed HPV5 or HPV16 proteins were able to extend the life of primary BRK cells, leading to immortalization. The expression of 5E7 and 5E6L21S resulted in efficient proliferation (comparable to that of 16E7-E6) of BRK cells, resulting in immortalization. These results are consistent with the results showing that E1A mutants defective in interaction with DYRK1/HAN11 are hypertransforming (11). Our results raise the possibility that naturally arising variants of 5E6 may contribute to their potential oncogenic activities in humans. Interestingly, variants of HPV16 E6 have been found to be prevalent in invasive cervical carcinomas (36, 37). Analyses of the 5E6 sequences found in the GenBank indicate that most of the mutations in 5E6 EV-associated variants are clustered in the N-terminal region. The mutations in the 8E6 variants were also clustered in the N-terminal region. One of the cancer-associated variants (8E6 Vt 1) that we have analyzed was strongly defective in interaction with the DYRK1/HAN11 complex. Although we were unable to determine the transforming activity of 8E7-E6, the observation that a cancer-associated variant is defective in DYRK1 interaction suggests that potential oncogenic variants of 5E6 (e.g., L21S) and 8E6 (e.g., Vt 1) may be deficient in interaction with the DYRK1/HAN11 complex. As the EV lesions progress to cancer, there appears to be a selective enhancement of HPV5 and HPV8 among different HPVs. It is possible that at least some of the naturally arising variants of E6 (such as 5E6P5-1 and 8E6 Vt 1) may contribute to oncogenic progression of EV lesions.
In summary, our study has discovered a second functional similarity between the C-terminal region of adenovirus E1A and the N-terminal region of the E6 proteins of cutaneous HPVs. By analogy to E1A, the functions encoded by the N-terminal regions of the E6 proteins may have evolved to facilitate viral replication by promoting the level of epithelial differentiation by subverting the functions of cellular protein complexes such as FOXK1/K2 and DYRK1/HAN11. As the cutaneous EV lesions progress to squamous cell carcinoma in sun-exposed areas (22), naturally arising E6 mutants of HPV species such as HPV5 and HPV8 may lose their ability to promote epithelial-cell differentiation in favor of oncogenic transformation. Our results that both E1A and E6 proteins target the DYRK1/HAN11 protein complex may facilitate elucidation of the mechanism by which the viral proteins may alter the functions of DYRK1/HAN11 complex.
ACKNOWLEDGMENTS
This study was supported by research grants CA-84941 and CA-33616.
Footnotes
Published ahead of print 1 May 2013
REFERENCES
- 1. Levine AJ. 2009. The common mechanisms of transformation by the small DNA tumor viruses: The inactivation of tumor suppressor gene products: p53. Virology 384:285–293 [DOI] [PubMed] [Google Scholar]
- 2. DeCaprio JA. 2009. How the Rb tumor suppressor structure and function was revealed by the study of adenovirus and SV40. Virology 384:274–284 [DOI] [PubMed] [Google Scholar]
- 3. McLaughlin-Drubin ME, Munger K. 2009. The human papillomavirus E7 oncoprotein. Virology 384:335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Javier RT. 2008. Cell polarity proteins: common targets for tumorigenic human viruses. Oncogene 27:7031–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chinnadurai G. 2011. Opposing oncogenic activities of small DNA tumor virus transforming proteins. Trends Microbiol. 19:174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arany Z, Newsome D, Oldread E, Livingston DM, Eckner R. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81–84 [DOI] [PubMed] [Google Scholar]
- 7. Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869–884 [DOI] [PubMed] [Google Scholar]
- 8. Deleu L, Shellard S, Alevizopoulos K, Amati B, Land H. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20:8270–8275 [DOI] [PubMed] [Google Scholar]
- 9. Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297–307 [DOI] [PubMed] [Google Scholar]
- 10. Lang SE, Hearing P. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836–2841 [DOI] [PubMed] [Google Scholar]
- 11. Komorek J, Kuppuswamy M, Subramanian T, Vijayalingam S, Lomonosova E, Zhao LJ, Mymryk JS, Schmitt K, Chinnadurai G. 2010. Adenovirus type 5 E1A and E6 proteins of low-risk cutaneous beta-human papillomaviruses suppress cell transformation through interaction with FOXK1/K2 transcription factors. J. Virol. 84:2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fischer RS, Quinlan MP. 1998. The C terminus of E1A regulates tumor progression and epithelial cell differentiation. Virology 249:427–439 [DOI] [PubMed] [Google Scholar]
- 13. Grooteclaes ML, Frisch SM. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823–3828 [DOI] [PubMed] [Google Scholar]
- 14. Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12:469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas JL, Gopalakrishnan S, Quinlan MP. 1991. Modulation of transformation of primary epithelial cells by the second exon of the Ad5 E1A12S gene. Oncogene 6:2093–2103 [PubMed] [Google Scholar]
- 16. Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 92:10467–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Subramanian T, La Regina M, Chinnadurai G. 1989. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene 4:415–420 [PubMed] [Google Scholar]
- 18. Pfister H. 1992. Human papillomaviruses and skin cancer. Semin. Cancer Biol. 3:263–271 [PubMed] [Google Scholar]
- 19. de Villiers EM. 1989. Heterogeneity of the human papillomavirus group. J. Virol. 63:4898–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lutzner MA, Blanchet-Bardon C, Orth G. 1984. Clinical observations, virologic studies, and treatment trials in patients with epidermodysplasia verruciformis, a disease induced by specific human papillomaviruses. J. Investig. Dermatol. 83:18s–25s [DOI] [PubMed] [Google Scholar]
- 21. Orth G. 1986. Epidermodysplasia verruciformis: a model for understanding the oncogenicity of human papillomaviruses. Ciba Found. Symp. 120:157–174 [DOI] [PubMed] [Google Scholar]
- 22. Orth G, Jablonska S, Jarzabek-Chorzelska M, Obalek S, Rzesa G, Favre M, Croissant O. 1979. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 39:1074–1082 [PubMed] [Google Scholar]
- 23. Jackson S, Harwood C, Thomas M, Banks L, Storey A. 2000. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 14:3065–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Underbrink MP, Howie HL, Bedard KM, Koop JI, Galloway DA. 2008. E6 proteins from multiple human betapapillomavirus types degrade Bak and protect keratinocytes from apoptosis after UVB irradiation. J. Virol. 82:10408–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brimer N, Lyons C, Wallberg AE, Vande Pol SB. 2012. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene 31:4639–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Howie HL, Koop JI, Weese J, Robinson K, Wipf G, Kim L, Galloway DA. 2011. Beta-HPV 5 and 8 E6 promote p300 degradation by blocking AKT/p300 association. PLoS Pathog. 7:e1002211. 10.1371/journal.ppat.1002211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. White EA, Kramer RE, Tan MJ, Hayes SD, Harper JW, Howley PM. 2012. Comprehensive analysis of host cellular interactions with human papillomavirus e6 proteins identifies new e6 binding partners and reflects viral diversity. J. Virol. 86:13174–13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fischer SM, Lo HH, Gordon GB, Seibert K, Kelloff G, Lubet RA, Conti CJ. 1999. Chemopreventive activity of celecoxib, a specific cyclooxygenase-2 inhibitor, and indomethacin against ultraviolet light-induced skin carcinogenesis. Mol. Carcinog. 25:231–240 [PubMed] [Google Scholar]
- 30. Pelka P, Ablack JN, Fonseca GJ, Yousef AF, Mymryk JS. 2008. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 82:7252–7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deau MC, Favre M, Orth G. 1991. Genetic heterogeneity among human papillomaviruses (HPV) associated with epidermodysplasia verruciformis: evidence for multiple allelic forms of HPV5 and HPV8 E6 genes. Virology 184:492–503 [DOI] [PubMed] [Google Scholar]
- 32. Feltkamp MC, Broer R, di Summa FM, Struijk L, van der Meijden E, Verlaan BP, Westendorp RG, ter Schegget J, Spaan WJ, Bouwes Bavinck JN. 2003. Seroreactivity to epidermodysplasia verruciformis-related human papillomavirus types is associated with nonmelanoma skin cancer. Cancer Res. 63:2695–2700 [PubMed] [Google Scholar]
- 33. Karagas MR, Nelson HH, Sehr P, Waterboer T, Stukel TA, Andrew A, Green AC, Bavinck JN, Perry A, Spencer S, Rees JR, Mott LA, Pawlita M. 2006. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J. Natl. Cancer Inst. 98:389–395 [DOI] [PubMed] [Google Scholar]
- 34. Wieland U, Jurk S, Weissenborn S, Krieg T, Pfister H, Ritzkowsky A. 2000. Erythroplasia of queyrat: coinfection with cutaneous carcinogenic human papillomavirus type 8 and genital papillomaviruses in a carcinoma in situ. J. Investig. Dermatol. 115:396–401 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Z, Smith MM, Mymryk JS. 2001. Interaction of the E1A oncoprotein with Yak1p, a novel regulator of yeast pseudohyphal differentiation, and related mammalian kinases. Mol. Biol. Cell 12:699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zehbe I, Voglino G, Delius H, Wilander E, Tommasino M. 1998. Risk of cervical cancer and geographical variations of human papillomavirus 16 E6 polymorphisms. Lancet 352:1441–1442 [DOI] [PubMed] [Google Scholar]
- 37. Zehbe I, Wilander E, Delius H, Tommasino M. 1998. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 58:829–833 [PubMed] [Google Scholar]
- 38. Subramanian T, Vijayalingam S, Lomonosova E, Zhao LJ, Chinnadurai G. 2007. Evidence for involvement of BH3-only proapoptotic members in adenovirus-induced apoptosis. J. Virol. 81:10486–10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu Y, Liu C, Zeng L, Lin Z, Dewhurst S, Gartner S, Planelles V. 2003. Efficient gene transfer into human monocyte-derived macrophages using defective lentiviral vectors. Cell. Mol. Biol. (Noisy-le-grand) 49:1151–1156 [PubMed] [Google Scholar]