Abstract
Dengue virus is a major human pathogen that has four serotypes (DENV1 to -4). Here we report the cryoelectron microscopy (cryo-EM) structures of immature and mature DENV1 at 6- and 4.5-Å resolution, respectively. The subnanometer-resolution maps allow accurate placement of all of the surface proteins. Although the immature and mature viruses showed vastly different surface protein organizations, the envelope protein transmembrane (E-TM) regions remain in similar positions. The pivotal role of the E-TM regions leads to the identification of the start and end positions of all surface proteins during maturation.
INTRODUCTION
Dengue virus (DENV), the cause of dengue fever, infects 100 million people worldwide every year. It is a member of the Flaviviridae family (1), with four serotypes: DENV1, -2, -3, and -4. Infection with DENV usually causes a self-limiting fever accompanied by rashes and joint pain in patients but might lead to dengue hemorrhagic fever and dengue shock syndrome, which may be fatal.
DENV consists of an ∼500-Å-diameter protein shell embedded in a host-derived lipid membrane and encapsidates an 11-kb single-stranded positive-sense RNA genome. The dengue genome encodes three structural proteins, the core (or capsid), the premembrane (prM), and the envelope (E) protein, that form the virus particle, as well as seven nonstructural proteins (1) that are involved in replication of the virus genome. The newly synthesized immature DENV has a spiky appearance (2) (Fig. 1A) and is typically noninfectious unless it is complexed with certain antibodies (3). Virus maturation occurs during transportation of the virus particle through the trans-Golgi component network (TGN). The acidic environment of these compartments induces structural rearrangement of the virus surface proteins. During this initial maturation process, the furin protease cleaves prM molecules on the virus into pr and M. After leaving the cell, the cleaved pr dissociates from the virus surface, resulting in smooth, fully mature infectious virus particles (4) (Fig. 1B).
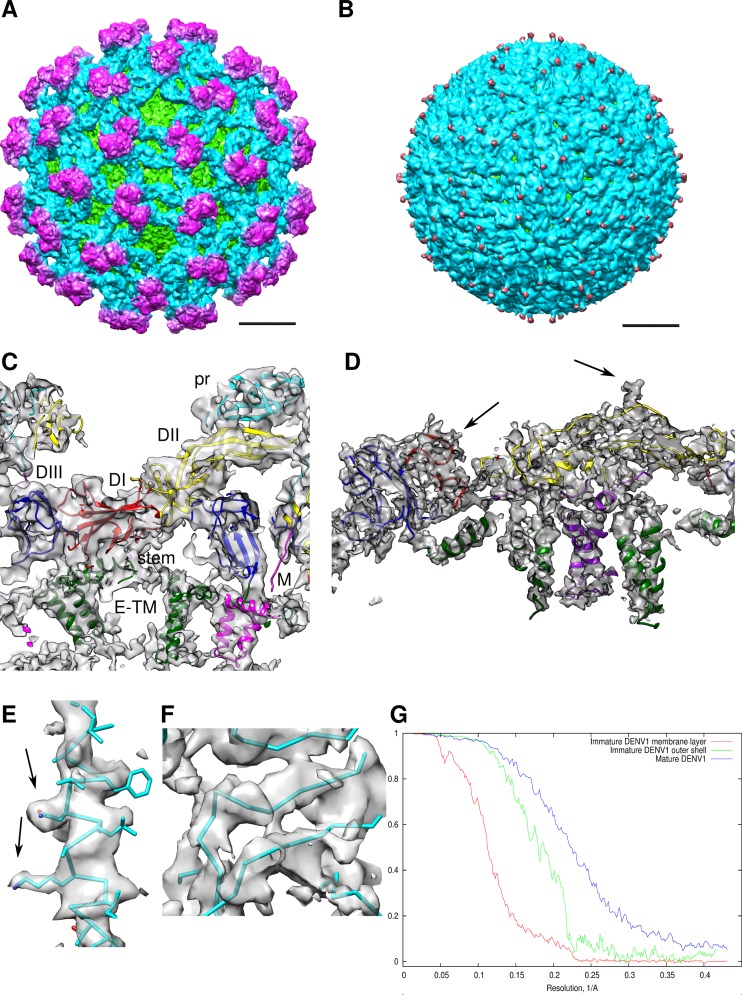
Fig 1.
Cryo-EM structures of the DENV1. (A) Cryo-EM density of immature virus. The scale bar is 100 Å. The density is colored radially: radii between 0 and 160 Å, between 161 and 210 Å, between 211 and 270 Å, and 271 Å and above are colored in yellow, green, light blue, and magenta, respectively. For clarity, the cryo-EM maps in panels A and B were low-pass filtered to an 8-Å resolution. (B) Cryo-EM density of mature DENV1. The density is colored radially: radii between 0 to 160 Å, between 161 and 210 Å, between 211 and 250 Å, and 251Å and above are colored in yellow, green, light blue, and magenta, respectively. (C and D) Zoomed-in view of fitted surface proteins into the cryo-EM density maps (gray) of immature (C) and mature (D) virus. Black arrows in panel D point to density corresponding to glycosylation sites. DI, DII, and DIII of the E ectodomain are colored in red, yellow, and blue, respectively. The pr part of the prM molecule is colored in cyan, and the M protein is colored in purple. The E stem and transmembrane (TM) regions are colored in dark green. (E) E protein stem helix α2 fitted into the 4.5-Å-resolution mature DENV1 virus map. Densities of some side chains are resolved (black arrows). The side chain models were not determined experimentally and are shown to indicate the protein sequence. (F) Close-up view of a part of the E protein DI main chain in the 4.5-Å-resolution map of the mature DENV1 virus. The densities for β-strands are separated. (G) Fourier shell correlation versus resolution plots for the individual lipid bilayer (red) and the E-prM protein shell (green) of the immature DENV1 map and the shell including the lipid bilayer and the outer surface protein of the mature DENV1 (blue).
The E protein is the major structural component of the viral surface. The ectodomain of E protein (5) contains three distinct domains, DI, DII, and DIII (also shown in Fig. 1C), which are connected by flexible links that allow rearrangement of domains during virus assembly, maturation, and infection (5–7). DIII is involved in attachment to host cell receptors, whereas DII is responsible for fusion to the host endosomal membrane during infection (8). The ectodomain is connected to the stem made from amphipathic helices α1 and α2. The stem, in turn, is anchored to the virus lipid membrane by two transmembrane (TM) α-helices, TM1 and TM2.
The crystal structure of an E-prM complex (6) shows that pr has a β-barrel fold and caps the fusion loop of the E protein, consistent with its function in preventing the newly synthesized virus from fusing back into the cell during maturation. The furin cleavage site lies between the pr molecule and the ectodomain of M protein, which exists as a linear polypeptide chain (6). The prM protein also has a stem region with a single amphipathic α-helix followed by two TM α-helices (9) (Fig. 2B). The core protein (10) associates with the viral RNA (11); however, it is not observed in cryoelectron microscopy (cryo-EM) reconstructions of flaviviruses (2, 9), indicating that the core proteins do not form icosahedral structures in the virus particle.
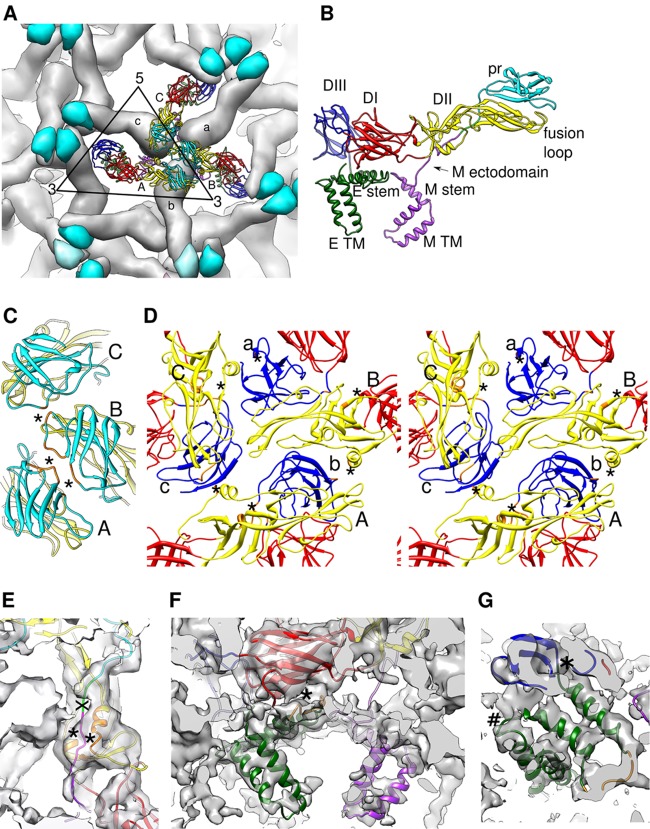
Fig 2.
Organization of the E and prM ectodomains in the immature DENV1 and their interactions. (A) Organization of the E and prM protein trimeric spikes on the surface of the virus. Three molecules in the asymmetric unit are shown as ribbons and are labeled A, B, and C. Symmetry-related molecules are shown as surfaces, and E protein surfaces are colored in gray whereas the pr surfaces are colored in cyan. The symmetry-related molecules are labeled a, b, and c. The black triangle represents one asymmetric unit. (B) Structure of an E protein complexed with prM protein. Color schemes are defined in Fig. 1. (C) Interactions between pr molecules (cyan) at the tip of the spike. Residues are identified as interacting by having a distance of less than 8 Å between Cα chains. (C to F) The interacting residues are colored in orange, and the interacting interface is indicated by an asterisk. (D) Stereodiagram showing the interactions between DII (yellow) of the molecules (labeled A, B, and C) in the asymmetric unit and DIII (blue) from neighboring symmetry-related molecules (labeled a, b, and c). (E) Interactions between M molecule (pink) and the DII (yellow) of the E ectodomain. The furin cleavage site on the prM protein is colored in green and marked with an “X.” (F) Interactions of the E stem region (dark green) with DI of the E protein (red). (G) Fit of the E protein stem α-helices into the 6-Å cryo-EM map. The density that connects the DIII of the E protein to the stem region (indicated by “*”) and the stem to the transmembrane region (indicated by “#”) can be observed.
Cryo-EM structures of immature (2) and mature (9, 12) DENV2 have been described previously. Both are icosahedral structures with three E and M (prM in immature virus) heterodimers per asymmetric unit. The surface of the immature virus (Fig. 1A) contains 60 spikes, each made from three prM-E heterodimers (2, 6). In contrast, the mature virus (Fig. 1B) has a smooth surface assembled from 90 E protein dimers organized in a characteristic “herringbone” pattern (13) with the M protein lying underneath (9). Until very recently, the cryo-EM reconstructions of homologous DENV2 have been calculated to a resolution of 12.5 Å for the immature virus (5) and 9.5 Å for mature virus (9). Although it is possible to define the domain organization of the surface proteins at these resolutions, it is not possible to identify molecular interactions between the components that drive assembly, maturation, or infection. A 7-Å-resolution cryo-EM reconstruction of a complex of mature DENV1 with Fab molecules has also been published recently (14); however, this structure likely does not represent the infectious virus particle, as Fab binding may induce some structural changes (15). A very recent publication on a 3.5-Å cryo-EM reconstruction of mature DENV2 (12) allows a comparison and validation of our findings. Here we report 4.5- and 6-Å structures of the mature and immature dengue serotype 1 virus, respectively. At these resolutions, component proteins can be accurately placed, revealing essential molecular interactions between molecules that guide assembly, drive maturation, and confer stability to the infectious virus. Furthermore, a previously unobserved connection between the TM and stem regions in the immature virus is now clearly visible, allowing us to unambiguously deduce the start and end positions of the E and M proteins during virus maturation.
MATERIALS AND METHODS
Purification of immature and mature DENV.
Production and purification of immature (2) and mature (13) dengue virus had been described previously. Briefly, immature DENV1 (DEN1/SG/07K3640DK1/2008) (16) was grown in C6/36 cells in a 10-cell stack containing minimum essential medium (MEM; Gibco) and 10% (vol/vol) fetal bovine serum (FBS) (Gibco) and infected at a multiplicity of infection (MOI) of 1 at 29°C. At 2 h postinfection, the medium was replaced with MEM supplemented with 2% (vol/vol) FBS and 40 mM NH4Cl and incubated for 2 days. Mature virus was grown under similar conditions except using RPMI 1640 (Gibco) medium containing 25 mM HEPES and 10% (vol/vol) FBS with no NH4Cl used. Cells were infected at an MOI of 0.1, and the virus was harvested 96 h postinfection.
The purification steps were similar for both samples. Briefly, the harvested medium was centrifuged at 6,000 rpm for 30 min, and the supernatant was precipitated with 8% (wt/vol) PEG 8000 (Sigma-Aldrich) by incubation at 4°C overnight. The suspension was centrifuged, and the pellet was resuspended in NTE buffer (12 mM Tris, pH 8.0, 120 mM NaCl, 1 mM EDTA). The virus was purified through a 24% (wt/vol) sucrose cushion and a linear 10 to 30% (wt/vol) potassium tartrate-glycerol gradient. The virus band was collected, buffer exchanged to NTE buffer, and concentrated in an Amicon Ultra centrifugal filter (100 kDa; Millipore). The purity and concentration of DENV were estimated using Coomassie blue-stained SDS-PAGE gel.
Cryo-electron microscopy and image processing.
Copper grids covered with lacey carbon and a layer of thin carbon were used for both samples. A 2.5-μl sample was pipetted onto the grid and then blotted with filter paper for 2 s before being flash frozen in liquid ethane using FEI Vitrobot Mark IV (The Netherlands). Image acquisitions were done by using an FEI Titan Krios electron microscope operating at 300 kV at liquid nitrogen temperature with a nominal magnification of 75,000 and an electron dose of 18 e−/Å2 at a defocus range of 0.7 to 3.7 μm. The images were collected with a Gatan 4K by 4K charge-coupled device (CCD) camera with a pixel size of 1.2 Å. Virus particles were selected manually using the boxer tool from an EMAN (17) software package. The astigmatic defocus parameters for each micrograph were estimated with CTFFIND (18). A total of 13,204 and 14,126 particles were selected for the immature and mature virus reconstructions, respectively.
3D reconstruction, structure validation, and model building.
The three-dimensional (3D) reconstructions for both data sets were performed in a similar way: the orientation assignment for all images was carried out using the MultiPath Simulated Annealing protocol (19), followed by 3D reconstruction using the make3d program from EMAN (17). DENV2 immature (16-Å-resolution) (2) and mature (9.5-Å-resolution) (9) virus maps were used as starting models. For both structures, after ∼25 cycles of iterations, the reconstructions had converged. To produce the immature and mature virus maps, 10,588 and 9,447 images were selected from the data sets, respectively. Resolution was determined by plotting the Fourier shell correlation coefficient between reconstructions generated from two half-data sets with a cutoff value of 0.5 (Fig. 1G).
The crystal structures of the DENV2 soluble fragment of E protein (Protein Data Bank [PDB] code 1TG8) (5) for mature virus and the prM-E ectodomain complex (6) (PDB code 3C6E) for immature virus were used to fit into the cryo-EM maps by using Chimera (20). The fitted structures were then modified to contain a DENV1 amino acid sequence. The missing stem and transmembrane helices were added using Coot (21). In the immature virus map, only part of the M ectodomain density is continuous: the density that connects to the pr molecule and the density near domain II of the E protein. The visible part of the M ectodomain was then connected to the nearest M stem region. Molecular dynamic flexible fitting (MDFF) (22), with symmetry restraints (23) calculated using NAMD (24), was used to produce the molecules in the asymmetric unit of the final virus structures. It involved 3,000 steps of energy minimization, followed by 5,000,000 steps of molecular dynamics run and another 3,000 steps of minimization. The structures obtained have correct polypeptide chain geometry and were observed to fit the electron density well.
Protein structure accession numbers.
The cryo-EM maps of the immature (EMD-2141) and mature (EMD-2142) DENV1 were deposited in the Electron Microscopy Databank. The coordinates of the modeled component proteins for immature (PDB code 4B03) and mature (PDB code 4AZX) virus were deposited in the Protein Data Bank.
RESULTS
The immature DENV1 is approximately 600 Å in diameter and has a highly contoured spiky surface (Fig. 1A). The overall resolution of the cryo-EM map is 6 Å; however, the TM layer density has poorer resolution (8 Å) (Fig. 1G), probably due to slight differences in the symmetry-related positions of the TM regions. Distinct shapes of three pairs of structurally independent E and pr proteins are clearly visible in each spike in the asymmetric unit (Fig. 1C and 2A). The crystal structure of a complex of soluble fragments of DENV2 E and prM (6) (70% protein sequence identity to DENV1) was fitted into the cryo-EM map as a rigid body and served as a base from which the complete structures of the DENV1 E and prM proteins, including the stem and TM regions, were built (Fig. 1C and 2B, F, and G).
The contacts between proteins were identified by measuring the distances between Cα positions, with pairs less than 8 Å apart considered to be interacting. In each spike, the arrangement of the prM-E complexes is stabilized at two sites. The first interaction between pr molecules occurs at the tip of the spike (Fig. 2A and C) and is likely responsible for holding the spike structure together. The second site of interaction is between DII of one E protein and DIII of another E protein from a neighboring spike (Fig. 2D), linking the spikes to make a shell. The small number of contacts between molecules suggests that the virus structure is labile and can be easily disrupted during virus maturation. The N terminus of the M protein (Fig. 2B) contains a highly conserved hydrophobic patch that interacts with two α-helices (residues 209 to 215 and 258 to 265) of the E protein DII (Fig. 2E). The underside of the E protein DI (residues 19 to 25, a conserved hydrophobic sequence) contacts the highly conserved hydrophobic loop between helices α1 and α2 of its stem region (Fig. 2F). There was no obvious density in the cryo-EM map that would correspond to the known capsid protein structure, consistent with the previous results, indicating that the capsid protein is either disordered or differently ordered in different virus particles (5).
At a 4.5-Å resolution, the map of the mature DENV1 (Fig. 1B) shows well-resolved densities for α-helical structures in the stem and TM region (Fig. 1D). Densities of several side chains were readily visible (Fig. 1E), as was the separation of β-strands within the E protein (Fig. 1F). However, similar to 4.5-Å-resolution crystal structures, most side chains are not resolved; hence, protein-protein interactions were identified as having a distance of less than 8 Å between Cα positions. A superposition of Cα chains of the E and M proteins with those from the 3.5-Å DENV2 cryo-EM structure (12) did not show any significant deviations.
The surface of the mature virus contains 180 copies of E protein organized into 30 “rafts” of three dimers lying parallel to each other (Fig. 3A). Each raft is stabilized by intra- and interdimeric contacts between ectodomains of the E protein (Fig. 3B and C, respectively), with DII playing a dominant role. In contrast, the interaction between rafts is highly dependent on DIII (Fig. 3D to F). At the 5-fold vertex, DIIIs from different rafts have extensive contacts with each other (Fig. 3D). Near the 3-fold vertex, the hinge between DIII and DI interacts with DI of a neighboring E protein from another raft (Fig. 3E). The DI-DIII hinge of an E protein from the middle dimer in the raft also contacts DII from the E protein in an adjacent raft (Fig. 3F).
Fig 3.
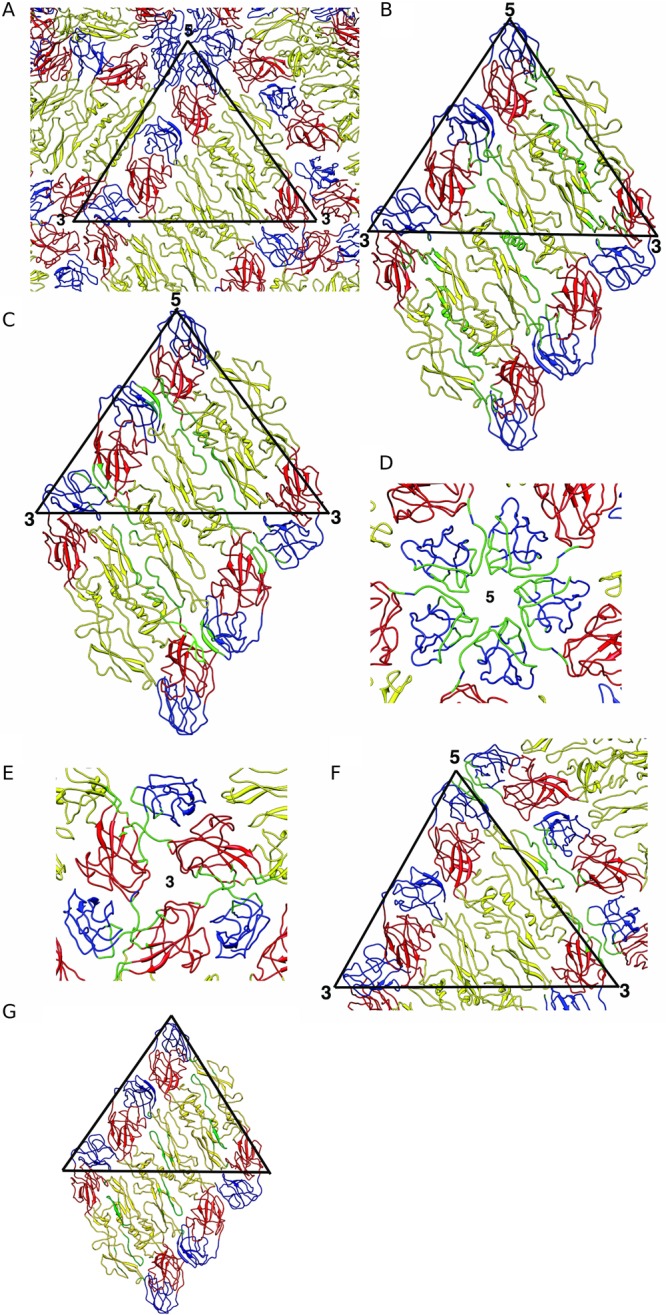
Organization of ectodomain E on the mature DENV1 and their interactions. (A) Organization of ectodomain E on the surface of DENV1. DI, DII, and DIII are colored in red, yellow, and blue, respectively. The black triangle represents an asymmetric unit. (B) Interactions between the molecules within the E ectodomain dimers. A raft containing two asymmetric units is shown here. (B to F) Interacting residues are colored in green. (C) Interdimeric interactions between the E ectodomains in the raft. (D) Interactions between DIIIs (blue) near the 5-fold vertex. (E) Interactions between E ectodomains near the 3-fold vertex. (F) Interactions between two E ectodomain rafts that form the herringbone pattern on the virus surface. (G) Binding site of the pr molecule on the E protein (6), shown in mature virus. Residues bound by the pr molecule are highlighted in green.
Unlike the immature virus, the TM and stem regions of E proteins of the mature virus interact with the M protein dimer (Fig. 4A). The ectodomain of M (the polypeptide chain preceding the stem helix) was thought to be a linear polypeptide lying along the ectodomain of E based on the 12.5-Å-resolution immature DENV2 map (6). However, the M ectodomain was not observed in the 9.5-Å-resolution mature DENV2 map (9). In our map, the resolution is such that the N terminus of M (residues 1 to 27) (Fig. 4B) is observed and can be fully traced, similarly to the 3.5-Å-resolution DENV2 structure (12). It interacts with the stem helix (residues 27 to 39) from the other M molecule in the dimer (Fig. 4C). The C-terminal end of the TM regions also interacts with the same region of the opposite M protein (Fig. 4C). The N-terminal end of the M protein makes contacts with two E protein loops, the loop that connects E stem α2 to the TM region and another loop that connects E α1 and α2 (Fig. 4D). Examination of the interactions between E ectodomain and its stem region and M protein shows that the underside (facing the virus membrane) from DI of E protein interacts with the entire E stem α1, the highly conserved loop, and α2. The underside of E DII has extensive contacts with the M ectodomain (Fig. 4E), similar to those observed in the immature structure (Fig. 2E and F).
Fig 4.
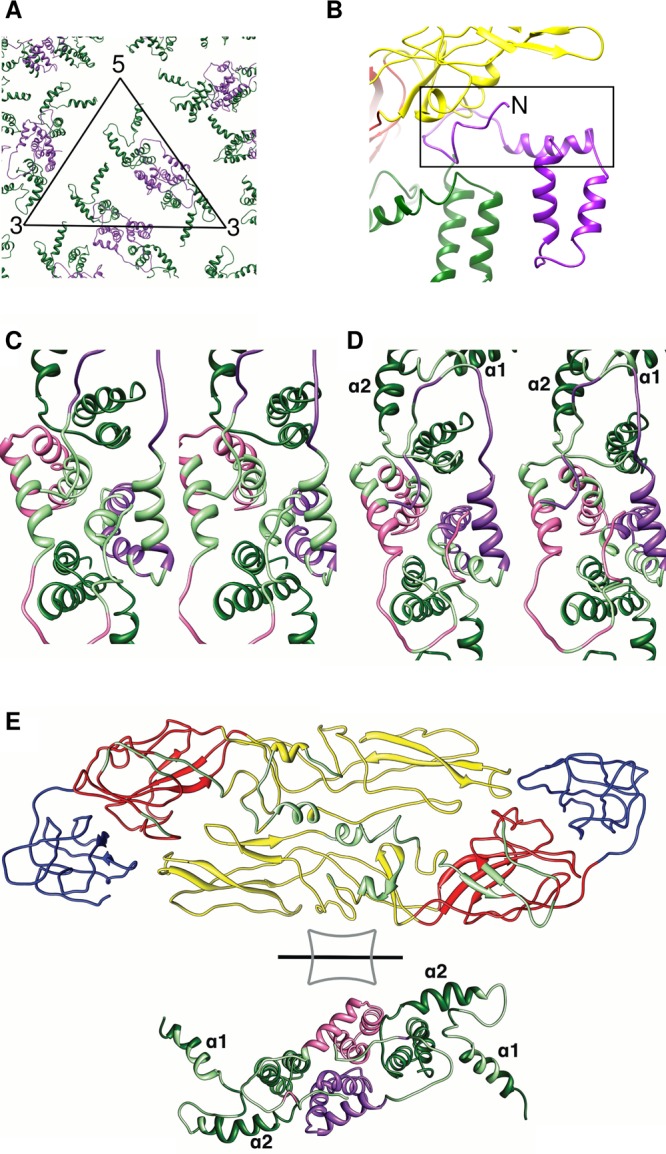
Organization of membrane-associated proteins in the mature DENV1 virus. (A) Organization of the stem and TM regions of E proteins (dark green) and M proteins (purple) in the virus. The black triangle represents an asymmetric unit. The M protein forms homodimeric interactions. (B) M ectodomain (N-terminal residues 1 to 26) and stem region (N-terminal residues 27 to 39) (black rectangle box) in the mature DENV. Domains DI and DII of the interacting E protein are shown in red and yellow, respectively. (C) Stereodiagram of the interactions of the M protein (purple) with the other molecule (pink) in the homodimeric structure. (C to E) Interacting residues are colored in light green. (D) Stereodiagram of the interactions of the M protein with the stem and TM regions of the E protein. (E) Open-book view of the interactions between the E ectodomain and the M protein as well as the stem and TM regions of E protein. View of the E ectodomain from inside the virion while the TM regions are viewed from the outside.
DISCUSSION
Comparison of the amino acid sequences of the E proteins of DENV1 and DENV2 shows that 8% of the residues are nonconserved. Heparin is an attachment factor on cells that likely binds to stretches of positive charges on the DENV surface. Infection of cells with DENV2 was shown to be highly dependent on binding to heparan sulfate, while the other serotypes were less dependent (25). Comparison of the surface charges of the E proteins on the DENV1 and DENV2 (12) cryo-EM structures showed that DENV2 has more positively charged residues (Fig. 5A).
Fig 5.
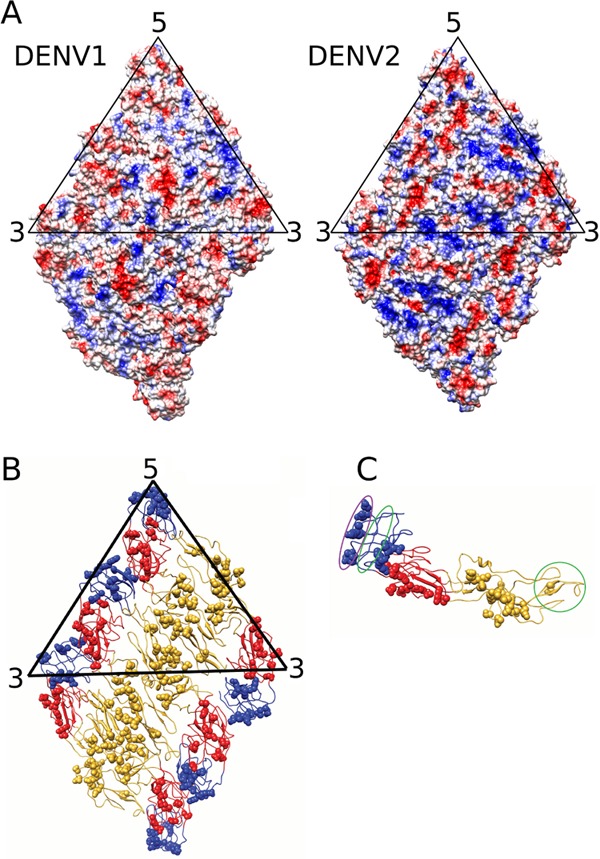
Comparing the surface E proteins between the mature DENV1 and DENV2. (A) Electrostatic charges on the surfaces of DENV1 and DENV2. Surfaces of a raft consisting of two asymmetric units of the E ectodomain are shown. Positive and negative charges are colored in blue and red, respectively. The DENV2 surface contains larger numbers of positively charged residues. (B) Location of nonconserved residues on an E ectodomain raft. The nonconserved residues (spheres) are present on all domains of the E ectodomain protein except at the hinge between DI and DII, the hinge between DI and DIII, and the fusion loop at the tip of DII. (C) Location of nonconserved residues on an E ectodomain, side view. The epitope recognized by highly potent serotype-specific antibodies is located at the lateral side of DIII (purple circle). The epitopes bound by weakly neutralizing antibodies, which are generally cross-reactive to all serotypes, are circled in green.
Most of the nonconserved residues between DENV1 and DENV2 congregate on the surface-exposed regions on the virus (Fig. 5B), suggesting that the variations of residues defining the different serotypes may have been evolved as a means of evading the host immune system. Nonconserved residues are present on all surface-exposed regions of the E protein domains with the exception of the hinge between DII and DIII, the hinge between DI and DII, and the fusion loop at the tip of DII (Fig. 5B). The highly potent DIII antibodies are serotype specific and bind to the lateral ridge of DIII (26), consistent with the location of the nonconserved regions in DIII (Fig. 5C). Weaker antibodies that are generally flavivirus cross-reactive bind to either the conserved regions next to the lateral ridge on DIII (26) or the fusion loop at the tip of DII (27) (Fig. 5C).
When exposed to the low-pH environment of the TGN during maturation, the surface proteins of immature virus undergo extensive rearrangement to adopt a structure similar to that of the mature virus except with regard to the pr molecule attached to each E protein (4). Analysis of the pr binding site on the E proteins (6) in the cryo-EM mature virus structure (Fig. 3G) showed that bound pr molecules would prevent dimeric interactions between E protein monomers (Fig. 3B). This suggests that the packing of surface E protein in the immature virus structure at low pH is likely loose. This loose structure may allow the reversibility of the surface structural change of immature virus structure observed when pH is reversed from a neutral- to a low-pH environment (4). Hence, after the prM molecule is cleaved by furin, the dimeric structure of the E protein becomes stabilized when the virus is released into the neutral-pH environment outside the cell, thus preventing the previously reversible structural change and locking the structure into a stable lattice.
In addition to the rearrangements of the ectodomain of the E and prM proteins, there are also significant movements of the stem and TM regions during DENV maturation (Fig. 6). By aligning the orientation of the TM regions of E protein from the immature and mature virus structures (Fig. 6A), movements of α1 and α2 of the E stem region can be visualized. Helix α1 is rotated by about 180 degrees relative to α2, and α2 is rotated approximately 45 degrees counterclockwise relative to TM1. The rotation of the E stem helices likely plays an important role in facilitating the large structural rearrangements of E ectodomains during maturation. This is because the contacts between the stem α1 and the DI of the E ectodomain remain the same in the immature (Fig. 2F) and mature (Fig. 4E) virus, so the stem α1 probably moves together with the ectodomain as a rigid body.
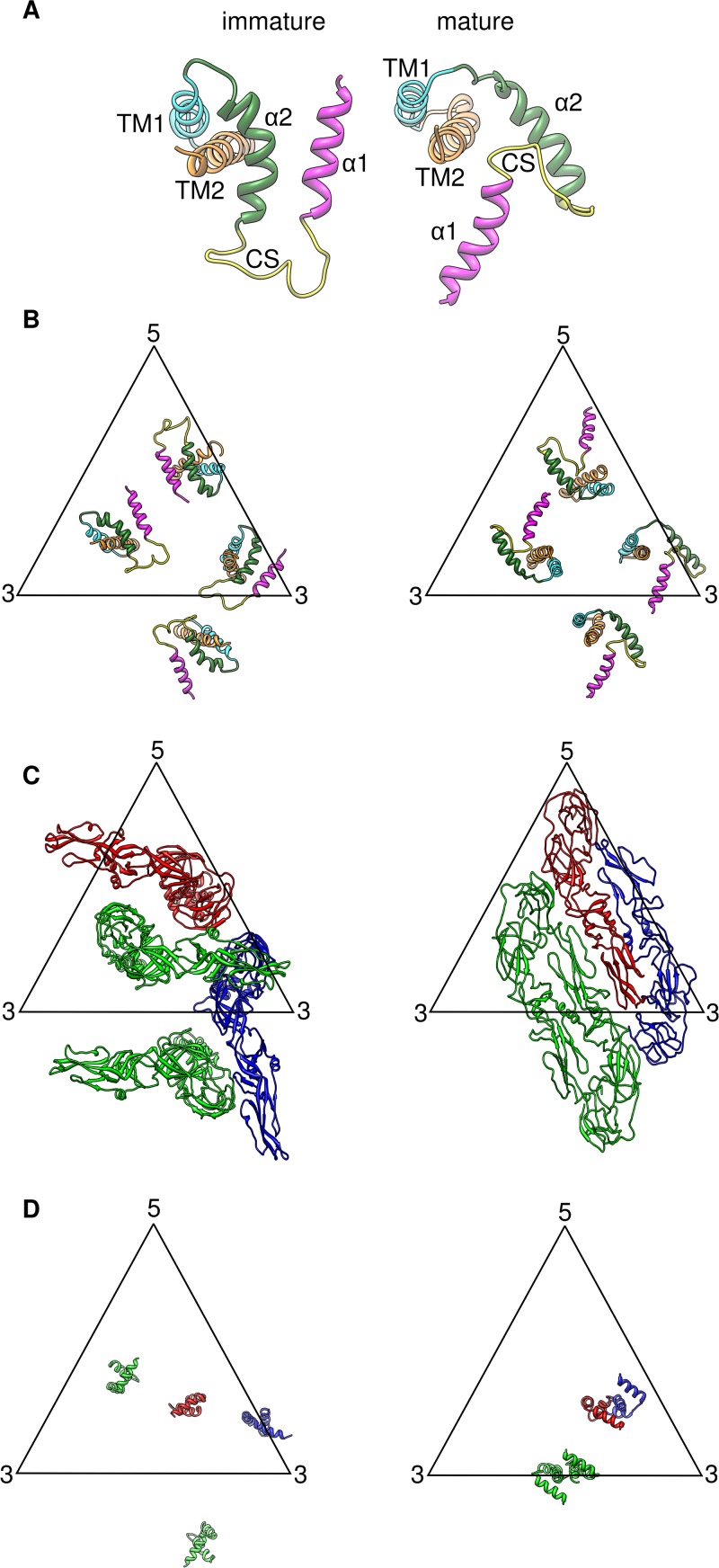
Fig 6.
Movement of surface proteins during DENV maturation. (A) Movement of the E stem region during maturation. The positions of TM1 and TM2 of the E proteins of the immature and mature virus are aligned so as to compare the positions of the stem α1 and α2. Helix α1 is rotated by about 180 degrees relative to α2, and α2 is rotated approximately 45 degrees counterclockwise relative to TM1. (B) Positions of the stem and TM regions of the E protein in immature (left) and mature (right) structures. There are some localized rotations occurring in the stem region and also the TM region of the E proteins during maturation. However, the position of the E transmembrane in the asymmetric unit of the virus did not change significantly between the immature and mature virus. (C) Rearrangement of the E ectodomain molecules in the immature and mature structures. The three independent E protein molecules in the asymmetric unit are colored red, green, and blue. The positions of their TM regions allow us to identify which position the E molecule in the immature virus structure moves into in the mature virus structure—red moves to red, green moves to green, and blue moves to blue. (D) Movements of the M protein. The three independent M protein molecules in the asymmetric unit are colored in red, green, and blue. There is significant movement of the M proteins in the virus membrane.
Although there are significant movements of the stem regions of the E protein, the TM regions stay at approximately the same positions in the asymmetric unit during virus maturation (Fig. 6B). Thus, we are able to identify which position a particular E protein in the immature virus moved to in the mature virus structure (Fig. 6C).
Significant rotations of the E ectodomain are required during maturation, and it is unlikely that such a transformation could happen simultaneously on all immature virus spikes without extensive clashing (Fig. 6C). Thus, it is plausible that maturation is a sequential process in which one set of spike proteins rearranges before the other set. Indeed, such partial maturation has been observed for DENV2 (28). As for the M stem and TM regions, the structure of the molecule in the immature virus is similar to that in the mature virus. Even though there is significant movement of the M proteins during maturation, we can also identify the start and end positions of these M proteins (Fig. 6D). This is possible because the interaction of the M ectodomain (residues 1 to 27) with the α-helix (residues 258 to 265) in the DII of the E protein is similar between the immature (Fig. 2E) and mature (Fig. 4E) virus, and hence, the movement of the M protein should follow that of the E proteins. It is interesting that the TM helices in E protein do not move as much as M (Fig. 6). One possible explanation is that the TM helices of E are longer than those of M (Fig. 2B) and therefore have more contacts with the membrane, making them less mobile.
E protein has been shown to have an increased affinity for the stem region of M protein when exposed to acidic pH (mimicking the TGN) (29). It is thus plausible that the trimer-to-dimer structural transition of the E proteins on the virus surface during maturation is due to the changes in the quaternary organization of the M proteins. However, what stimulates the movement of the M proteins during maturation remains largely unknown.
In conclusion, the subnanometer-resolution structures of the immature and mature DENV1 illustrate the conformational rearrangement of the surface proteins during maturation and show the intermolecular interactions between the viral proteins that stabilize specific structural features necessary for various stages of the viral life cycle.
ACKNOWLEDGMENTS
We thank Eng-Eong Ooi for providing the DENV serotype 1 strain PVP159 (DEN1/SG/07K3640DK1/2008).
This research has been supported by a BMRC grant (R-913-300-019-304) and an NRF fellowship (R-913-301-015-281).
Q.Z. and J.L.T. produced and purified the immature and mature DENV and conducted part of the image processing. T.-S.N. collected the cryo-EM images. V.A.K. and S.-M.L. performed the cryo-EM reconstructions and fitting of molecules. V.A.K. performed the molecular dynamic flexible fitting. S.-M.L., V.A.K., Q.Z., and J.L.T. wrote the manuscript.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 1 May 2013
REFERENCES
- 1. Lindenbach BDR, CM 2001. Flaviviridae: the viruses and their replication, p 991–1042 In Knipe DM, et al. (ed), Fields virology, vol II Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zybert IA, van der Ende-Metselaar H, Wilschut J, Smit JM. 2008. Functional importance of dengue virus maturation: infectious properties of immature virions. J. Gen. Virol. 89:3047–3051 [DOI] [PubMed] [Google Scholar]
- 4. Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837 [DOI] [PubMed] [Google Scholar]
- 5. Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830–1834 [DOI] [PubMed] [Google Scholar]
- 7. Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319 [DOI] [PubMed] [Google Scholar]
- 8. Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291–298 [DOI] [PubMed] [Google Scholar]
- 9. Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dokland T, Walsh M, Mackenzie JM, Khromykh AA, Ee KH, Wang S. 2004. West Nile virus core protein; tetramer structure and ribbon formation. Structure 12:1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khromykh AA, Westaway EG. 1996. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 141:685–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. 2013. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat. Struct. Mol. Biol. 20:105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, Chan SH, Smith KG, Chan AH, Zou G, Ooi EE, Kemeny DM, Tan GK, Ng JK, Ng ML, Alonso S, Fisher D, Shi PY, Hanson BJ, Lok SM, MacAry PA. 2012. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci. Transl. Med. 4:139ra83. 10.1126/scitranslmed.3003888 [DOI] [PubMed] [Google Scholar]
- 15. Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15:312–317 [DOI] [PubMed] [Google Scholar]
- 16. Low J, Ooi E, Tolfvenstam T, Leo Y, Hibberd M, Ng L, Lai Y, Yap G, Li C, Vasudevan S, Ong A. 2006. Early dengue infection and outcome study (EDEN)—study design and preliminary findings. Ann. Acad. Med. Singapore 35:783–789 [PubMed] [Google Scholar]
- 17. Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 18. Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347 [DOI] [PubMed] [Google Scholar]
- 19. Liu X, Jiang W, Jakana J, Chiu W. 2007. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J. Struct. Biol. 160:11–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. 2008. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16:673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan KY, Gumbart J, McGreevy R, Watermeyer JM, Sewell BT, Schulten K. 2011. Symmetry-restrained flexible fitting for symmetric EM maps. Structure 19:1211–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin YL, Lei HY, Lin YS, Yeh TM, Chen SH, Liu HS. 2002. Heparin inhibits dengue-2 virus infection of five human liver cell lines. Antiviral Res. 56:93–96 [DOI] [PubMed] [Google Scholar]
- 26. Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. 2007. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 81:12816–12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J. Virol. 80:12149–12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Plevka P, Battisti AJ, Junjhon J, Winkler DC, Holdaway HA, Keelapang P, Sittisombut N, Kuhn RJ, Steven AC, Rossmann MG. 2011. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 12:602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Q, Hunke C, Yau YH, Seow V, Lee S, Tanner LB, Guan XL, Wenk MR, Fibriansah G, Chew PL, Kukkaro P, Biukovic G, Shi PY, Shochat SG, Gruber G, Lok SM. 2012. The stem region of premembrane protein plays an important role in the virus surface protein rearrangement during dengue maturation. J. Biol. Chem. 287:40525–40534 [DOI] [PMC free article] [PubMed] [Google Scholar]