Abstract
Hepatitis C virus (HCV) RNA forms an unusual interaction with human microRNA-122 (miR-122) that promotes viral RNA accumulation in cultured human liver cells and in the livers of infected chimpanzees. GB virus B (GBV-B) is a hepatotropic virus and close relative of HCV. Thus, GBV-B has been used as a surrogate system to study HCV amplification in cultured cells and in infected tamarins. It was discovered that the 5′-terminal sequences of GBV-B RNA, like HCV RNA, forms an Argonaute 2-mediated complex with two miR-122 molecules that are essential for accumulation of GBV-B subgenomic replicon RNA. However, sequences in miR-122 that anneal to each viral RNA genome were different, suggesting distinct overall structural features in HCV:miR-122 and GBV-B:miR-122 complexes. Surprisingly, a deletion that removed both miR-122 binding sites from the subgenomic GBV-B RNAs rendered viral RNA amplification independent from miR-122 and Argonaute 2. This finding suggests that structural features at the end of the viral genome dictate whether miR-122 is required to aid in maintaining viral RNA abundance.
INTRODUCTION
Viruses are obligate intracellular parasites and as such interact extensively with pathways in the infected host. For example, DNA viruses such as polyomaviruses, adenoviruses, and herpesviruses express viral microRNAs (miRNAs) and use the miRNA regulatory pathway to regulate both host and viral RNAs (1–3). Roles for viral miRNAs include modulation of the immune system and regulation of virus latency (3). Although RNA viruses do not encode miRNAs, they interact extensively with cellular miRNAs (2). Knowledge of virus-miRNA interactions may represent novel antiviral targets.
In a unique interaction between a miRNA and an RNA virus, the liver-specific miRNA, miR-122, promotes accumulation of hepatitis C virus (HCV) genomic RNA in infected cells (4). miR-122 slightly stimulates HCV mRNA translation (5) but mostly stabilizes the HCV RNA genome (6–8). This is an unusual role for a miRNA since they typically repress translation and destabilize target mRNAs. The precise mechanism by which miR-122 augments HCV RNA accumulation has not been confirmed, but it requires the host miRNA pathway protein Argonaute 2 (Ago2) and direct binding between miR-122 at two adjacent binding sites located near the 5′ terminus of the HCV genome (8–11). Recent evidence suggests that miR-122 may have an additional role, independent from HCV genome stabilization, proposed to affect viral replication (6). However, data suggests that miR-122 is not essential for replication, at least for HCV subgenomic replicon RNA (12), and does not alter the rate, or the elongation phase, of RNA synthesis by the viral RNA-dependent RNA polymerase (13, 14); thus, a role for miR-122 during viral RNA replication remains to be confirmed.
The mechanism of action of miR-122 requires a unique binding interaction between miR-122 and the HCV genome (7). Unlike translational repression by miR-122, which requires annealing of the miRNA “seed” binding to the target mRNA, augmentation of HCV RNA accumulation requires annealing of the seed sequence and nucleotides 3′ of the seed region with the 5′ terminus of the HCV genome. This interaction creates a 3′ overhang that likely masks the noncapped 5′ terminus of the HCV genome from host RNA degradation enzymes (7).
GB virus B (GBV-B) is a hepatotropic virus and a close relative of HCV. GBV-B was first isolated from tamarins injected with serum from a human who had contracted hepatitis (15). However, the origin of the virus is unknown since it cannot be resolved whether a virus present in the human serum, or endogenous to the tamarins, initiated the infection (16). GBV-B replicates efficiently in tamarins and causes a self-limiting hepatic infection lasting approximately 4 months. However, GBV-B infections in marmosets generate lower virus titers and occasionally induce chronic liver infections (15, 17). Since HCV can only infect humans and chimpanzees, GBV-B has been suggested as a possible surrogate model for chronic HCV infection.
Based on the liver tropism and close relationship to HCV, we hypothesized that GBV-B may also rely on an interaction with miR-122. Examination of the GBV-B 5′ untranslated region (5′ UTR) sequence revealed two potential sequence elements that could base pair with the seed sequences in miR-122 (Fig. 1A). By using a GBV-B subgenomic replicon carrying a luciferase reporter gene, we have confirmed that, like HCV, miR-122 augments GBV-B replication levels through direct interactions with two putative miR-122 seed sequence sites, and with the 5′ terminus of the genome. However, De Tomassi et al. isolated a GBV-B mutant that lacks both miR-122 binding sites and can replicate as efficiently as wild-type replicon RNA (18). We have demonstrated that the mutant genome grows independently of miR-122 and the miRNA machinery, because mutant replicon RNA abundance is completely insensitive to miR-122 supplementation, miR-122 sequestration, and Ago2 depletion. These data suggest that GBV-B uses miR-122 to support efficient viral RNA accumulation in a manner similar to that of HCV but indicates that miR-122-independent RNA accumulation is possible when the miR-122 sites are removed. These results indicate that GBV-B is a possible surrogate model to assess HCV antiviral strategies that target miR-122 and that GBV-B can be used to study the mechanism of miR-122 augmentation of viral RNA accumulation.
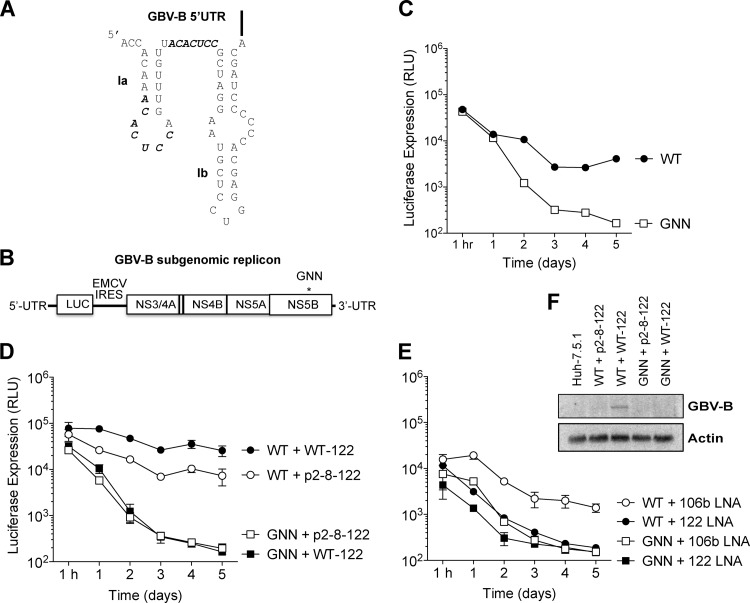
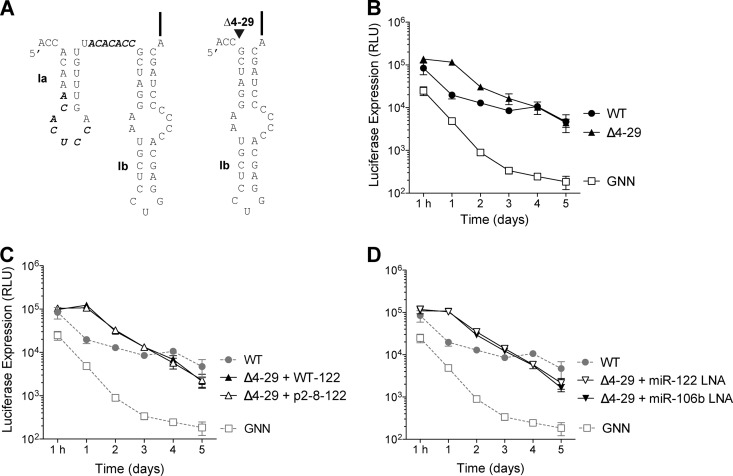
Fig 1.
GBV-B is sensitive to miR-122 levels. (A) Predicted structure of the GBV-B 5′ UTR, including stem-loops Ia and Ib (nucleotides 1 to 61). miR-122 seed matches are indicated with bold letters. (B) Diagram of the GBV-B luc-RepB dicistronic subgenomic replicon RNA used in the present study. The replicon contains the GBV-B 5′ UTR (nucleotides 1 to 459), which includes the GBV-B IRES, followed by the firefly luciferase reporter. The EMCV IRES drives translation of the nonstructural proteins NS3-NS5B, and the replicon is flanked by the GBV-B 3′ UTR. An asterisk indicates a mutation in the active-site residues of the GBV-B polymerase (NS5B) present in the nonreplicating negative control replicon RNA (luc-RepB GNN). (C) The wild-type (WT) GBV-B replicon, but not the GNN GBV-B replicon replicates in Huh-7.5.1 cells as indicated by luciferase expression. (D) Exogenous expression of WT-122, but not negative control p2-8-122, increases WT GBV-B replicon levels. The luciferase levels from GNN GBV-B replicons in combination with the exogenous expression of WT-122 and p2-8-122 are also indicated. (E) Sequestration of miR-122, but not miR-106b/93, using antisense locked nucleic acids (LNAs), results in loss of GBV-B replicon RNA. Sequestration of miR-122 and mir-106b/93 LNA in combination with GNN GBV-B replicons are also indicated. The data show the average of at least three experiments and are representative of at least six independent experiments. Error bars represent the standard deviation of the mean and are frequently invisible due to low variability between experiments. (F) Northern blot analysis of GBV-B electroporated Huh-7.5.1 cells. GBV-B subgenomic replicon RNA accumulation was measured by Northern blotting 5 days after electroporation in Huh-7.5.1 cells. Wild-type (WT) and GNN GBV-B subgenomic replicon RNA co-electroporated with either WT-122 or p2-8-122 are indicated. WT GBV-B subgenomic replicon RNA was only detectable upon exogenous addition of WT-122.
MATERIALS AND METHODS
Cell culture.
Huh-7.5.1 cells were kindly provided by Francis V. Chisari. Huh-7.5.1 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 1% nonessential amino acids, and 200 μM l-glutamate (Gibco).
Plasmid and mutagenesis.
The GBV-B neo-RepB and GBV-B neo-RepB Δ4-29 replicon constructs were obtained from Cinzia Traboni and were described previously (18). To create the GBV-B luc-RepB replicon plasmid, neo was removed and replaced with luciferase by digestion with AscI and PmeI. The luciferase sequence was transferred from pHCVrep AB12 (19) after the internal SapI site was removed, but the luciferase coding sequence was retained, by using a QuikChange site-directed mutagenesis protocol (Stratagene, La Jolla, CA) and the mutagenic primers luc mut SapI s (5′-ATT TGA AGA AGA ACT GTT TTT A-3′) and luc mut SapI as (5′-TCG TAA AAA CAG TTC TTC TTC A-3′). A GBV-B nonreplicative mutant replicon, luc-RepB GNN, was created by mutating the GDD active site of the viral polymerase, NS5B, to GNN by using the primers GNN mut s (5′-CTT ATT TGC GGC AAT AAT TGC ACC GTA ATT-3′) and GNN mut as (5′-AAT TAC GGT GCA ATT ATT GCC GCA AAT AAG-3′).
To create GBV-B site 1 (A12) and GBV-B site 2 (A27) luc-repB plasmids, the forward primers S1:p4 (5′-ACG GAT CCG GTA ATA CGA CTC ACT ATA GAC CAC AAA CAC ACC AGT TTG-3′) and S2:p4 (5′-ACG GAT CCG GTA ATA CGA CTC ACT ATA GAC CAC AAA CAC TCC AGT TTG TTA CAC ACC GCT AGG-3′) were used, along with the reverse primer AscI-REV (5′-ATG GCG CGC CCT GTT TGA GTA GAA ATA-3′). PCRs were performed using Taq DNA polymerase under the following conditions: (i) 95°C for 3 min; (ii) 95°C for 1 min, 59°C for 1 min, and 72°C min for 30 cycles; (iii) 72°C for 7 min; and, finally, (iv) 4°C for an indefinite period of time. Products were digested and inserted into the parent vector at BamHI and AscI restriction sites. G2G3 site 1 (A12) and GBV-B G2G3 site 2 (A27) GBV-B luc-repB plasmids were created from the site 1 (A12) and site 2 (A27) GBV-B plasmids using the forward primer G2G3 (5′-ACG GAT CCG GTA ATA CGA CTC ACT CAC TAT CAC TAT AGA GGA CAA ACA C-3′) with the AscI-REV primer and inserted into the parent vector as described above.
miRNAs, small interfering RNA (siRNAs), and locked nucleic acids (LNAs).
All miRNAs were synthesized by the Stanford Protein and Nucleic Acid facility. The sequences of the miRNAs were as follows: wild-type miR-122 (WT-122), 5′-UGG AGU GUG ACA AUG GUG UUU GU-3′; negative control, p2-8-122, 5′-UAA UCA CAG ACA AUG GUG UUU GU-3′; p4-122, 5′-UGG UGU GUG ACA AUG GUG UUU GU-3′; and p4,15,16-122, 5′-UGG UGU GUG ACA AUC CUG UUU GU-3′. Guide strands p2-8-122 and p4,15,16-122 were annealed to the corresponding mutant passenger (*) strands (p2-8–122*, 5′-AAA CGC CAU UAU CUG UGA GGA UA-3′; p4,15,16–122*, 5′-AAA CGG GAU UAU CAC ACU AAA UA-3′) to maintain base-pairing interactions of the duplex. All other guide strands were duplexed with the wild-type passenger strand (WT-122*, 5′-AAA CGC CAU UAU CAC ACU AAA UA-3′).
The antisense LNA oligonucleotide to miR-122 (SPC3649, 5′-CcAttGTcaCaCtCC-3′) was kindly provided by Santaris Pharma a/s. Capital letters in this sequence indicate LNAs, and this oligonucleotide has a complete phosphorothioate backbone. The antisense LNA oligonucleotide to miR-106b/93 5′-TgCaCkGwCaGcAcT-3′ was synthesized by Exiqon. A “K” (keto) indicates an G or T base, and a “W” (weak) indicates and an A or T base; capital letters indicate LNAs. For gene knockdown, the siRNA siAgo2 target sequence 5′-CAG ACU CCC GUG UGU CCU ATT-3′ and the siControl target sequence 5′-GAG AGU CAG UCA GCU AAU CAC TT-3′ (11) were synthesized by Dharmacon.
Replicon RNA transcription and transfection.
The GBV-B replicon RNA was synthesized with T7 MEGAScript kit (Ambion) according to the manufacturer's instructions after linearizing plasmids with SapI. After treatment with Turbo DNase to remove template DNA and lithium chloride precipitation, the RNA was resuspended in sterile RNase-free water and introduced into Huh-7.5.1 cells by electroporation. Briefly, 10 μg of GBV-B RNA was mixed with 4 × 106 Huh-7.5.1 cells suspended in 400 μl of Cytomix (10 mM KH2PO4, 120 mM KCl, 0.15 mM CaCl2, 25 mM HEPES, 2 mM EDTA, 5 mM MgCl2 [pH 7.6]) in 0.4-cm-gap-width electroporation cuvettes (Bio-Rad). Electroporation consisted of a single pulse of current delivered by the Gene Pulser II electroporation device (Bio-Rad), set at 270 V, 950 μF, and ∞ resistance. After 10 min at room temperature, electroporated cells were resuspended in 9 ml of medium, and 1 ml was plated/six-well dish for downstream luciferase assays.
For microRNA or antisense LNA assays, 20 μl of 20 μM microRNA duplexes or 2 μl of 50 μM antisense LNAs were co-electroporated along with GBV-B replicon RNA. At 2 days after electroporation, another 20 μl of 20 μM miRNAs were introduced by transfection using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested at 1 h, and 1 to 5 days posttransfection for downstream analysis.
For siRNA knockdown experiments, 4 μl of 20 μM siRNA duplex was electroporated into cells. The electroporated cells were grown in culture for 3 days to establish gene knockdown, and then re-electroporated with GBV-B RNA and siRNA. Cells were harvested at 1 h and 1 to 5 days after a second electroporation for downstream analysis.
Luciferase assays.
For luciferase assays, each well was lysed with 200 μl of 1× cell culture lysis buffer, and 25 μl of sample was used, along with 50 μl of Firefly luciferase reagent (Promega). Luciferase assays were carried out on a Monolight 2010 luminometer with an integration time of 10 s. The data shown are of three independent experiments carried out simultaneously. However, each experiment was also carried out in at least two other independent replicates at other times. While the overall trends were the same, the absolute luciferase expression differed between experiments done at different times, most likely due to differences in the cell preparation from 1 day to another. Thus, our data are representative of at least six different technical replicates of each experiment.
Total RNA isolation and Northern blot analysis.
Total cellular RNA was isolated by using TRIzol reagent (Life Technologies) and the recommended protocol. For Northern blot analysis, 10 μg of total RNA were separated in 1% agarose gels containing 1× 3-(N-morpholino)propanesulfonic acid (MOPS) buffer and 2.2 M formaldehyde and transferred to Zeta-Probe membranes (Bio-Rad). Membranes were hybridized in UltraHyb hybridization buffer (Clontech) to random-primed 32P-labeled DNA probes (RadPrime DNA labeling system; Invitrogen) complementary to GBV-B (nucleotides 1 to 472) or γ-actin (nucleotides 685 to 1171). Autoradiographs were quantitated by using ImageQuant (GE Healthcare).
Argonaute qRT-PCR.
Small RNA knockdown of Ago2 was confirmed by quantitative reverse transcription-PCR (qRT-PCR) using the Applied Biosystems Ago2 gene expression assay Hs00293044_m1 at 72 h after knockdown, as described previously (11).
RESULTS
GBV-B replicon RNA abundance is modulated by miR-122.
Upon inspection of the GBV-B genome, we observed that the GBV-B 5′ UTR contained two potential seed match sequences to miR-122 (Fig. 1A). To test whether miR-122 affects GBV-B RNA accumulation, we created a GBV-B subgenomic replicon containing the GBV-B 5′ UTR, including the GBV-B internal ribosomal entry site (IRES) and a luciferase reporter, followed by the nonstructural genes (NS3-5B) and the 3′ UTR of GBV-B (Fig. 1B). Although full-length GBV-B can infect and replicate in primary tamarin and marmoset hepatocytes (20–22), it does not replicate in cultured liver cell lines (23). However, cells harboring stable GBV-B subgenomic replicon RNAs expressing a neomycin gene can be selected from transfected Huh-7 cell lines (18, 24). Using GBV-B subgenomic replicon RNA carrying a luciferase gene, we found that Huh-7.5.1 cells (compared to Huh-7 and Huh-7.5 cells) had the greatest capacity to support transient GBV-B replication (data not shown) (25). Indeed, transfected wild-type GBV-B subgenomic replicons accumulated in Huh-7.5.1 cells, whereas a mutated GBV-B subgenomic replicon, where the viral polymerase active site (GDD) contained an inactivating mutation (GNN), expressed from 10- to 24-fold less luciferase (Fig. 1C). In addition, MK0608, an HCV RNA-dependent RNA polymerase inhibitor, decreased luciferase expression from wild-type GBV-B subgenomic replicons to levels similar to that of GNN controls (data not shown). Therefore, transient replication of the GBV-B luciferase-subgenomic replicon system can be used to study the requirements for GBV-B replication in Huh-7.5.1 cells.
To examine whether GBV-B subgenomic replicon abundance is modulated by miR-122, miR-122 was either overexpressed or sequestered in Huh-7.5.1 cells (Fig. 1D and E). Exogenous addition of wild-type miR-122 mimetics (WT-122) resulted in an ∼3.4-fold increase in GBV-B accumulation compared to the exogenous addition of a control microRNA, p2-8–122, in which the entire seed region of miR-122 (nucleotides 2 to 8) is mutated (7) (Fig. 1D). This suggests that miR-122 abundance influences GBV-B accumulation in Huh-7.5.1 cells and that miR-122 is limiting for GBV-B subgenomic replicon RNA in Huh-7.5.1 cells. In contrast, sequestration of miR-122 by an antisense LNA resulted in loss of GBV-B RNA abundance in Huh-7.5.1 cells (Fig. 1E), compared to treatment with a control LNA, that sequesters an unrelated microRNA, miR-106b/93. The loss of GBV-B subgenomic replicon abundance after miR-122 sequestration mirrored subgenomic replicon abundance in replicons containing the inactivating GNN mutations in the RNA-dependent RNA polymerase (Fig. 1E). GBV-B subgenomic replicon RNA was only detectable by Northern blotting under conditions whereby replicon RNA accumulation was boosted by the exogenous addition of WT-122 (Fig. 1F), suggesting that GBV-B replicon RNA accumulation is just below the limit of detection for Northern blot analyses in Huh-7.5.1 cells and that miR-122 augments GBV-B RNA accumulation. These data suggest that GBV-B subgenomic replicon accumulation is dependent on the abundance of miR-122 in Huh-7.5.1 cells.
The GBV-B 5′ UTR contains two functional miR-122 binding sites.
GBV-B contains two adjacent potential miR-122 seed sequences in its 5′ UTR (Fig. 2A). To examine whether the sites are functional miR-122 sites, we introduced mutations in the seed sequences of the potential miR-122 sites, whereby the uridine residues at nucleotides 12 or 27 of GBV-B subgenomic replicons were mutated to adenosines (Fig. 2). A synthetic miR-122 in which the adenosine at nucleotide position 4 (p4) is mutated to a uridine (p4-122) is predicted to interact with the GBV-B site 1 (A12) or site 2 (A27) mutants (p4-122, Fig. 2A to D). The miR-122 molecules bound to site 1 or site 2 can be examined independently if both sites need to be occupied by a miR-122 having complementary seed sequences, and endogenous wild-type miR-122 can function at the wild-type (nonmutated) site in the presence of transfected mutant p4-122 duplexes.
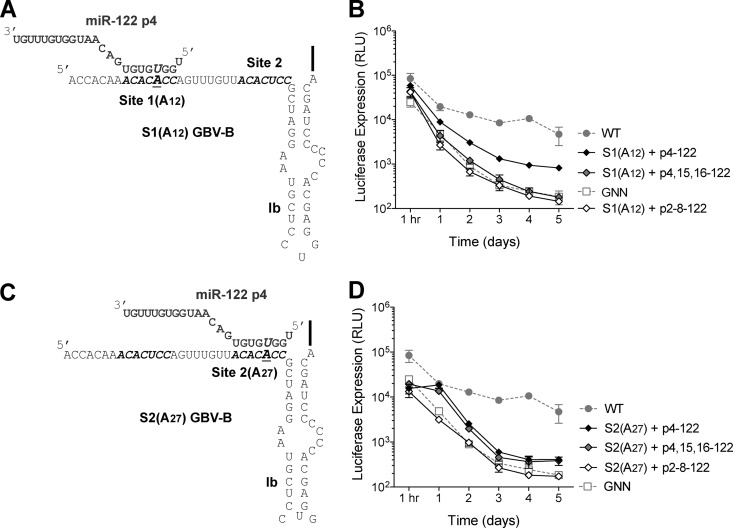
Fig 2.
GBV-B has two functional miR-122 sites. (A) Mutations were made within site 1 (A12) GBV-B 5′ UTR as indicated. miR-122 p4 is indicated in gray and the S1(A12) GBV-B 5′ UTR is indicated in black. miR-122 with a mutation at position 4 (p4) is able to bind to the mutated site 1, while the wild-type site 2 is free to bind to endogenous miR-122 (the binding of wild-type miR-122 is not shown). (B) Addition of compensatory p4-122, but not p4,15,16-122 or p2-8-122, is able to rescue RNA accumulation of GBV-B Site 1 (A12) replicon. (C) Mutations were made within site 2 (A27) GBV-B 5′ UTR as indicated. miR-122 with a mutation at position 4 (p4) is able to bind to the mutated site 2, while the wild-type site 1 is free to bind to endogenous miR-122 (not shown). miR-122 p4 is indicated in gray and the S2(A27) GBV-B 5′ UTR is indicated in black. (D) The addition of compensatory p4-122, p4,15,16-122, but not p2-8-122, is able to rescue RNA accumulation of GBV-B site 2 (A27) replicon. Luciferase expression from WT and GNN GBV-B replicons are also indicated (gray). The data show the average of at least three experiments and are representative of at least six independent experiments. Error bars represent the standard deviation of the mean.
When site 1 (A12) mutant GBV-B replicon RNA was introduced into Huh-7.5.1 cells, it was only able to accumulate in the presence of synthetic miR-122 containing a compensatory mutation (p4-122, Fig. 2B). In the presence of the negative control, p2-8–122, site 1 (A12) GBV-B RNA was unable to accumulate and displayed a similar luciferase profile to GBV-B subgenomic replicons containing inactivating GNN mutations (Fig. 2B). There was consistently a 4.8- to 10.9-fold increase in abundance in p4-122 transfected cells compared to p2-8–122 transfected cells at 4 days after electroporation. Similarly, when site 2 (A27) GBV-B RNA (Fig. 2C) was introduced into Huh-7.5.1 cells, it was only able to accumulate in the presence of miR-122 molecules containing a compensatory mutation (p4-122, Fig. 2D). There was consistently a 2.0- to 2.5-fold increase in abundance in p4-122 transfected cells compared to p2-8–122 transfected cells at 4 days after electroporation. Interestingly, rescue of neither site 1 (A12) nor site 2 (A27) GBV-B RNA was able to reach wild-type levels of GBV-B RNA accumulation. This is similar to what is seen in rescue with HCV miR-122 site mutants (7, 11) and may be due to decreased availability of Argonaute-loaded p4-122 mimetics compared to endogenous wild-type miR-122 or to decreased replication capacity of viral RNA having point mutations. These results suggest that both site 1 and site 2 in the GBV-B 5′ UTR are functional miR-122-binding sites. Both sites are required for GBV-B subgenomic replicon accumulation since mutation of either site independently resulted in a failure to accumulate in Huh-7.5.1 cells in the absence of exogenously introduced compensatory miR-122 molecules (Fig. 2).
Site 1-bound, but not site 2-bound miR-122 interacts with the 5′ terminus of the GBV-B genome.
To analyze the requirement for miR-122 annealing to sites outside of the canonical miRNA seed sequence, we analyzed accumulation of GBV-B replicon RNAs having point mutations to nucleotides near the 5′ terminus of the genome (Fig. 3). The rationale of this experiment was based on the similarity of the GBV-B 5′ terminus to that of HCV and the model for miR-122 binding beyond base pairing of seed sequences in the interaction of HCV with miR-122 (7) (Fig. 4A). The site 1-bound miR-122 molecule interacts with the 5′ terminus of the HCV genome and is thought to confer protection of the 5′ terminus from recognition by exonucleases or cellular sensors of RNA (7). Like in HCV, GBV-B contains cytosine nucleotides at positions 2 and 3 at the 5′ terminus that could potentially form additional interactions with miR-122 at sites 15 and 16. Thus, nucleotides 2 and 3 of the GBV-B genome were changed to guanosine residues (G2G3) in combination with mutations at site 1 (A12) or site 2 (A27) (Fig. 3A and C) to monitor affects of miR-122 molecules binding to site 1 or site 2 individually.
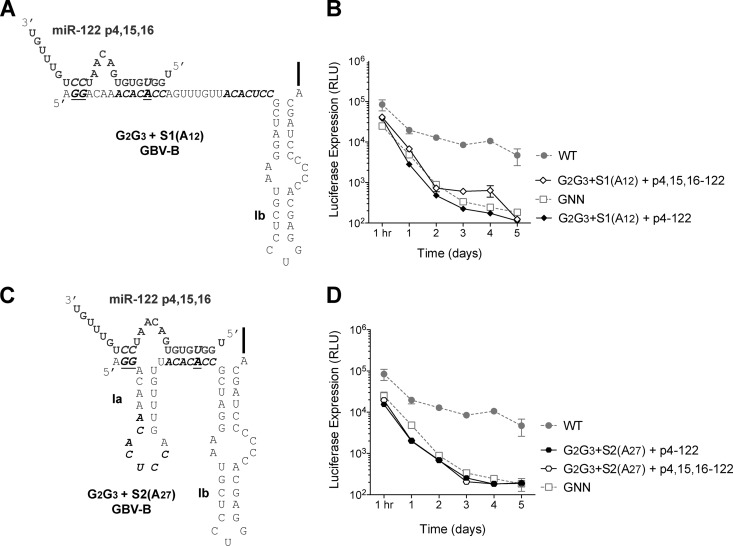
Fig 3.
Site 1-bound miR-122 interacts with the 5′ terminus of GBV-B replicon. (A) Mutations were made at nucleotides 2 and 3 (G2G3) of the GBV-B 5′ UTR in combination with site 1 (A12) as indicated. miR-122 p4,15,16 is indicated in gray and the G2G3 plus the S1(A12) GBV-B 5′ UTR is indicated in black. (B) Addition of p4,15,16-122, but not p4-122, is able to rescue RNA accumulation of the GBV-B G2G3 plus the site 1 (A12) replicon. (C) Mutations were made at nucleotides 2 and 3 (G2G3) of the GBV-B 5′ UTR in combination with site 2 (A27) as indicated. miR-122 p4,15,16 is indicated in gray, and the G2G3 plus the S2(A27) GBV-B 5′ UTR is indicated in black. (D) The addition of neither p4,15,16-122 nor p2-8-122 was able to rescue RNA accumulation of GBV-B G2G3 plus the site 2 (A27) replicon. Luciferase expression from WT and GNN GBV-B replicons is also indicated (gray). The data show the average of at least three experiments and are representative of at least six independent experiments. Error bars represent the standard deviation of the mean.
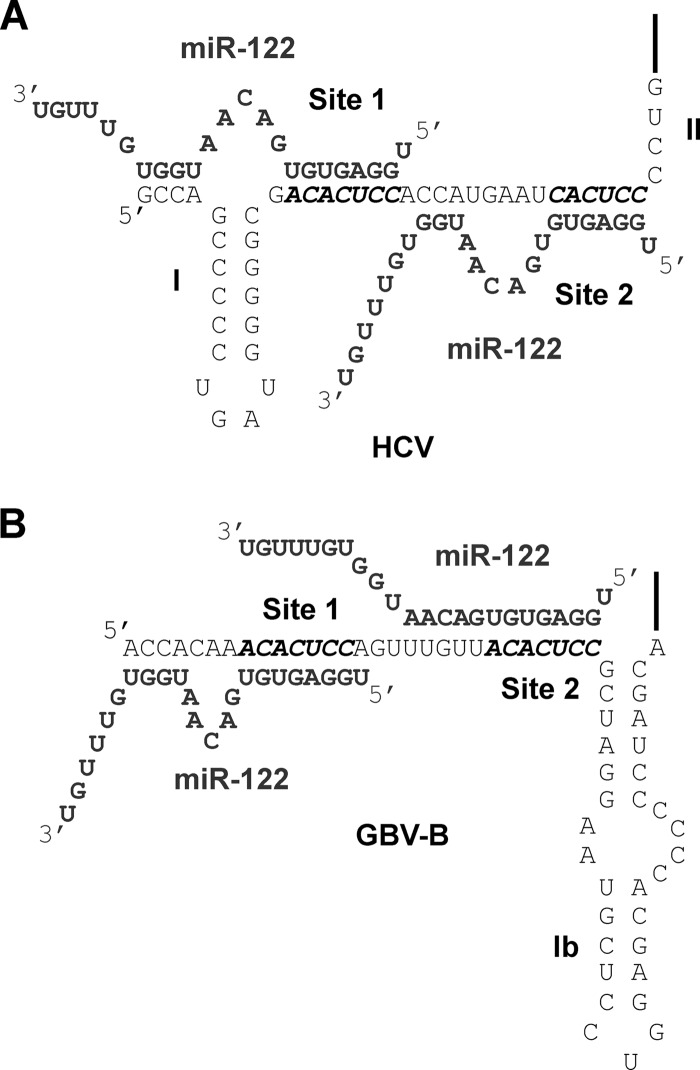
Fig 4.
Model of HCV:miR-122 and GBV-B:miR-122 interactions. (A) The HCV genome is indicated in black and the miR-122 molecules are indicated in gray. (B) The GBV-B 5′ UTR is indicated in black, and the miR-122 molecules are indicated in gray. The miR-122 seed sites 1 and site 2 are also indicated.
Transfected G2G3 site 1 (A12) GBV-B subgenomic replicons failed to accumulate in the presence of p4-122 (Fig. 3B). However, in the presence of miR-122 molecules containing a compensatory mutation at nucleotides 15 and 16 (p4,15,16-122), which could potentially bind to nucleotides 2 and 3 of the 5′ terminus, G2G3 site 1 (A12) GBV-B subgenomic replicons were able to accumulate (Fig. 3B). In particular, there was consistently a 2.6- to 4.8-fold increase in the abundance in p4,15,16-122-transfected cells compared to p4-122-transfected cells at 4 days after electroporation (Fig. 3B). In further support of the importance of miR-122 nucleotides 15 and 16 for site 1-bound miR-122, p4-122 but not p2-8–122 nor p4,15,16-122 was able to rescue site 1 (A12) GBV-B subgenomic replicons (Fig. 2B). Consistent with a model whereby nucleotides 15 and 16 of site 1-bound miR-122 interact with nucleotides 2 and 3 of the GBV-B genome (Fig. 4B). In contrast, neither exogenous introduction of p4-122 or p4,15,16-122 was able to rescue accumulation of G2G3 site 2 (A27) GBV-B subgenomic replicons at any time point (Fig. 3D). Moreover, miR-122 molecules with mutations at nucleotides 15 and 16 (p4,15,16-122) were able to rescue site 2 (A27) GBV-B replicon RNA accumulation (Fig. 2D), further suggesting that nucleotides 15 and 16 of site 2-bound miR-122 are dispensable for GBV-B replicon RNA accumulation. These results suggest a model for GBV-B:miR-122 interactions in which site 1-bound miR-122 interacts with the 5′ terminus of GBV-B (Fig. 4B). Curiously, the same six-nucleotide loop sequences are predicted to form in site 1-bound GBV-B:miR-122 and site 1-bound HCV:miR-122 complexes (Fig. 4).
The GBV-B replicon deletion mutant, Δ4-29, is not modulated by miR-122.
Results, thus far, have demonstrated that GBV-B has two functional miR-122 sites that are required for GBV-B accumulation in Huh-7.5.1 cells. Therefore, we were intrigued by the selection of a deletion mutant identified by De Tomassi et al. (18) that was able to accumulate in Hep3B cells, a cell line that does not contain sufficient levels of miR-122 to support HCV replication (12, 26). The Hep3B-adapted GBV-B mutant contains a deletion that spans nucleotides 4 to 29 of the GBV-B genome, thus removing both miR-122 seed sequences (Fig. 5A). Interestingly, this deletion was demonstrated to have no significant effect on GBV-B translation but allowed for approximately 2- and 6-fold increases in subgenomic replicon RNA accumulation over wild-type replicons in Huh-7 and Hep3B cells, respectively (18). To test the miR-122 dependence on accumulation of the deletion mutant, a luciferase subgenomic replicon was generated that harbored this deletion, termed Δ4-29 GBV-B luc (Fig. 5A). After transfection of Δ4-29 into Huh-7.5.1 cells, RNA accumulated to similar levels as wild-type GBV-B, particularly after 4 days of culture (Fig. 5B). Because Δ4-29 does not contain miR-122 seed sequences, we next tested whether Δ4-29 RNA accumulation was dependent on miR-122 abundance. Interestingly, Δ4-29 RNA accumulation was neither affected by exogenous overexpression of miR-122 (Fig. 5C) or by sequestration of miR-122 using antisense LNAs (Fig. 5D). Thus, Δ4-29 represents a mutant that is able to overcome the requirement for miR-122 in accumulation of GBV-B subgenomic replicons in cell culture.
Fig 5.
The GBV-B Δ4-29 replicon is insensitive to miR-122. (A) Wild-type GBV-B and Δ4-29 GBV-B replicon 5′ UTRs. miR-122 sites are indicated in boldface and italics. (B) The Δ4-29 GBV-B replicon accumulates to wild-type levels in Huh-7.5.1 cells. (C) The Δ4-29 GBV-B replicon is insensitive to exogenous expression of wild-type (WT) miR-122. (D) The GBV-B Δ4-29 replicon is insensitive to sequestration of miR-122 using an antisense miR-122 LNA. WT and GNN GBV-B replicons are also indicted (gray). The data show the average of at least three experiments and are representative of at least six independent experiments. Error bars represent the standard deviations of the mean.
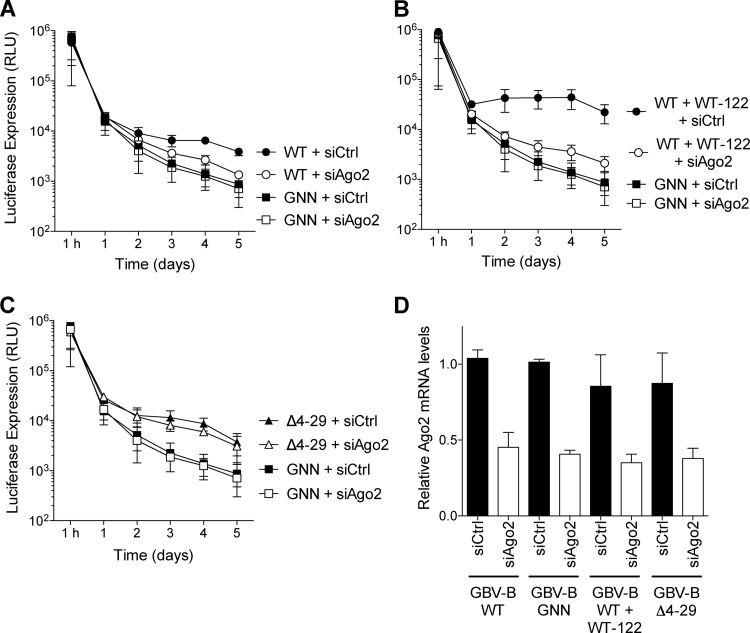
Wild-type GBV-B, but not Δ4-29, RNA abundance is regulated by the Ago2 protein.
miRNAs are typically delivered to their target mRNAs by an Argonaute (Ago) protein (27). A dependence on Ago2 for formation of HCV:miR-122 interactions has been recently demonstrated (11). Therefore, we tested whether GBV-B was also dependent on Ago2. Small RNA interference was used to deplete Ago2 levels in Huh-7.5.1 cells (Fig. 6). Wild-type GBV-B subgenomic replicon RNA abundance was diminished ∼5-fold after depletion of Ago2, compared to control siRNA depletion (siCtrl) (Fig. 6A). Exogenous expression of wild-type miR-122 enhanced GBV-B replicon RNA abundance at least 10-fold in control siRNA-treated cells compared to Ago2-siRNA-treated cells (Fig. 6B). In contrast, the deletion mutant, Δ4-29, was insensitive to Ago2 depletion (Fig. 6C). In all cases, Ago2 mRNA abundances were depleted by >50% in Huh-7.5.1 cells (Fig. 6D). These results suggest that Ago2 is required for wild-type GBV-B subgenomic replicon accumulation in Huh-7.5.1 cells. As demonstrated by the deletion mutant, Δ4-29, the absence of miR-122 sites in the 5′ UTR abolishes the requirement for Ago2, suggesting that Ago2's effects are mediated through miR-122.
Fig 6.
Wild-type GBV-B replicon, but not the deletion mutant Δ4–29, is sensitive to Argonaute 2 (Ago2) levels. (A) Wild-type GBV-B replicon is sensitive to Ago2 knockdown (siAgo2). (B) Addition of exogenous wild-type miR-122 results in a more dramatic sensitivity of the wild-type (WT) GBV-B replicon to Ago2 knockdown. (C) The Δ4-29 GBV-B replicon is insensitive to Ago2 knockdown. (D) Quantitative RT-PCR of Ago2 knockdown compared to control (Ctrl) siRNA at 72 h after knockdown. GNN GBV-B replicon treatment with Ago2 knockdown (siAgo2) and control siRNA knockdown (siCtrl) is also indicated. The data show the average of at least three experiments and are representative of at least six independent experiments. Error bars represent the standard deviations of the mean.
DISCUSSION
GBV-B is a hepatotropic virus classified within the Flaviviridae family, within the Hepacivirus genus, and is closely related to HCV with ca. 28% amino acid identity (28, 29). We hypothesized that GBV-B might also display a dependence on the liver-specific miRNA, miR-122, for viral RNA accumulation.
To test this hypothesis, we developed a luciferase reporter expression assay based on transient GBV-B replication. Several years ago, a stable replication system was established for GBV-B subgenomic replicons expressing a neomycin resistance gene (neo), through G418 drug selection in cultured Huh-7 cells (24). To establish a transient replication assay for GBV-B, we modified the GBV-B subgenomic replicon RNA to express a firefly luciferase reporter gene instead of a neomycin resistance gene (Fig. 1B). By assaying luciferase expression at various time points after electroporation of replicon RNA into Huh-7.5.1 cells, we observed 10- to 24-fold-higher luciferase expression from wild-type GBV-B replicons than from nonreplicative (GNN) replicons thus confirming GBV-B subgenomic RNA replication (Fig. 1C). In addition, treatment of wild-type GBV-B subgenomic replicons with MK0608, an HCV RNA-dependent RNA polymerase inhibitor caused similar decreases in luciferase expression to that of GNN controls (data now shown). The levels of luciferase expression by GBV-B replicons were lower than that observed using similar subgenomic replicons based on the efficiently replicating HCV JFH-1 virus strain (12) but were comparable in level and dynamics to those observed using first-generation HCV replicons based on the HCV Con1 strain (30). In addition, as for HCV, GBV-B replication efficiency (based on luciferase expression) was enhanced in Huh-7.5.1 cells compared to Huh-7 (data not shown). This was not surprising since Huh 7.5.1 cells are known to have defects in the RIG-I signaling pathway (25, 31), an arm of the innate immune response responsible for recognition of viral RNA. This pathway is known to limit replication of several viruses, including GBV-B, and is impeded by major proteases of HCV (32–35) and GBV-B (36).
Using the GBV-B transient replication system, it was found that GBV-B accumulation correlated with miR-122 abundance in the Huh-7.5.1 cells. Exogenous overexpression of miR-122 caused a 3.4-fold increase in GBV-B accumulation and miR-122 sequestration, using an antisense LNA, diminished GBV-B subgenomic replicon accumulation to levels similar to that of nonreplicating control (GNN) subgenomic replicons.
Sequence analysis and experimental evidence argued that GBV-B has two functional miR-122 sites in its 5′ UTR. Upon comparison to the 5′ UTR of HCV (see Fig. 4), we noted that the sequences between the two miR-122 sites had the same spacing, a distance that has been demonstrated to provide miRNAs in close proximity to cooperatively regulate gene expression (37). Our data also indicated that, as in HCV, interaction of miR-122 with site 1 creates a 3′ overhang (7). We noted that the 5′ terminus of the GBV-B genome is similar to that found in HCV, where nucleotides 1 to 4 could functionally pair with nucleotides 13 to 17 of miR-122. Specifically, GBV-B RNA accumulation was only observed when site 1-bound miR-122 was able to interact with the 5′ terminus of GBV-B at nucleotides 2 and 3. This result suggests that, like HCV, site 1-bound miR-122 interacts with the 5′ terminus of GBV-B creating a 3′ overhang that may protect the viral genome from nucleases or cellular sensor of RNA and is likely to play a similar role in GBV-B as in HCV (see models, Fig. 4). This is further supported by the requirement for nucleotides 15 and 16 of miR-122 for rescue of site 1 (A12) GBV-B accumulation (Fig. 2B). In contrast to HCV, site 2-bound miR-122 does not appear to have requirements for nucleotides 15 and 16, because a site 2 (A27) GBV-B mutant was able to accumulate to similar RNA abundance in the presence of p4-122 or p4,15,16-122 (Fig. 2D). Examination of the GBV-B 5′ UTR sequence flanking site 2, indicates that GBV-B lacks nucleotides complementary to nucleotides 13 to 17. However, nucleotide complementarities suggest that site 2-bound miR-122 may have an extended seed sequence, although this has not been tested directly (see model, Fig. 4B). Further characterization will be required to determine which of the other residues of site 1- or site 2-bound miR-122 molecules are important for GBV-B RNA accumulation. These residues may form additional interactions with the GBV-B genome, with cellular RNAs, or may form interactions with host or viral proteins to mediate their effects on viral RNA accumulation.
We have demonstrated that GBV-B has two functional miR-122 sites that are required for GBV-B subgenomic replicon accumulation in Huh-7.5.1 cells. However, our data cannot distinguish whether miR-122 binding to the GBV-B 5′ UTR enhances GBV-B RNA stability, promotes more efficient replication, or perhaps does both. In a recent publication, miR-122 was found to be required to stabilize the HCV 5′ UTR from degradation by host exoribonuclease Xrn-1 (6). However, their data also showed that knockdown of Xrn-1 can increase genome stability but cannot restore genome replication in the absence of miR-122, suggesting that genome stabilization was likely not the sole role for miR-122 in the HCV life cycle. The authors speculated that miR-122 might also play an essential role during the process of HCV replication.
We were therefore intrigued by the selection several years ago of a replication competent GBV-B mutant, Δ4-29 (18), that had a deletion that encompasses both functional miR-122 sites. De Tomassi et al. introduced GBV-B subgenomic replicons into various human liver-derived cell lines and selected clones able to replicate in those lines (18). Upon sequencing of selected clones, they detected a mutant, Δ4-29, which had a deletion from nucleotides 4 to 29 that was adapted for growth in Hep3B cells, a cell line we now know expresses undetectable levels of miR-122, but can support HCV replication if miR-122 is provided exogenously (12, 26). The deletion mutant did not differ from wild-type GBV-B subgenomic replicons in viral translation but was able to accumulate approximately 2- to 6-fold higher RNA levels in Huh-7 and Hep3B cells, respectively (18). To investigate dependence on miR-122, we assembled a luciferase-containing Δ4-29 GBV-B subgenomic replicon RNA and found that it amplified without a requirement for miR-122 and accumulated to similar levels as wild-type replicon RNA in Huh-7.5.1 cells. However, Δ4-29 GBV-B RNA accumulation had different kinetics. Luciferase expression levels were higher than wild-type levels between 1 h and 1 day after electroporation, but during subsequent days were similar to that of the wild type GBV-B (Fig. 5B). We speculate that miR-122-independent replication of Δ4-29 may be due to evolution of a miR-122-independent mechanism to protect the 5′ end from degradation. Protection of the 5′ end may be mediated by structural modifications induced by the deletion, or perhaps by its proximity to RNA-binding proteins that associate with the GBV-B IRES. Alternatively, deletion of Δ4-29 may abolish the association of an inhibitory factor. We believe it unlikely that miR-122-independent replication of Δ4-29 GBV-B is related to the defective RIG-I status of Huh 7.5.1 cells since Δ4-29 GBV-B can replicate as efficiently or better than wild-type GBV-B in Hep3B cells (18) (data not shown), which have been demonstrated to have an intact RIG-I pathway (26). Overall, these results suggest that a deletion of nucleotides 4 to 29 is somehow able to overcome the requirement for miR-122 in accumulation of GBV-B subgenomic RNA.
The ability of GBV-B to overcome the requirement for miR-122 is remarkable because sequestration of miR-122 in HCV-infected chimpanzees using an antisense LNA showed no emergence of resistant virus by deep sequencing (38). Although it would be interesting to analyze genome replication and virion assembly of full-length GBV-B genomes having the Δ4-29 mutation, these experiments were not feasible in the present study since full-length GBV-B replication can only be analyzed in primary tamarin hepatocytes in vitro (where it replicates poorly and does not make viral particles) and in tamarins and marmosets in vivo (20, 21, 23, 39). However, we do not think that our data indicate that HCV could easily evolve mechanisms to overcome the requirement for miR-122. Based on previous predictions, nucleotides in the miR-122 seed match sequences of HCV form a stem structure in the negative-strand intermediate that is important for replication (40). Hence, HCV may not be able to tolerate deletion of the miR-122 binding sites in its 5′ UTR. Interestingly, the deletion of nucleotides 4 to 29 of GBV-B has a minimal effect on the predicted secondary structure of the remaining GBV-B negative strand 3′ UTR (data not shown).
Finally, the requirement for the Ago2 protein in GBV-B subgenomic replicon accumulation was investigated. Similar to the mechanism of miRNA-directed translational suppression, the mechanism of miR-122-mediated augmentation of viral RNA accumulation requires Ago2, which based on the model for miR-122 augmentation of HCV is likely targeted to the GBV-B 5′ UTR (8, 10, 11, 41). Like HCV, wild-type GBV-B RNA accumulation was diminished after Ago2 depletion. In contrast to wild-type GBV-B, Δ4-29 RNA abundance was not affected by Ago2 depletion. This finding suggests that the absence of miR-122 sites in the 5′ UTR abolishes the requirement for Ago2 and that the primary role of Ago2 in supporting GBV-B replication is through miR-122 activity.
Our data also suggest a new model for the organization of the GBV-B 5′ UTR. Previously, predictive models based on consensus sequences of the HCV and GBV-B 5′ UTRs suggested a model whereby the GBV-B 5′ UTR contains two stem-loops referred to as SL1a and 1b, whereas HCV only contains a single SL1 (Fig. 4) (42). Our data suggest that miR-122 binds to site 1 and has additional interactions with the 5′ terminus. This model suggests that SL1a is unlikely to form in the presence of miR-122 and suggests an extended single-stranded 5′ terminus of GBV-B in cell culture. This is not surprising since, unlike the high-affinity SL1 of HCV, SL1a of GBV-B has a much lower affinity since it is composed of primarily A-U base pairs.
Both HCV and GBV-B usurp miR-122 to mediate viral RNA accumulation. Since miR-122 is expressed primarily in the liver, this suggests that it is likely a key factor in regulating cell tropism. HCV replication in the liver provides access to numerous factors such as those of the lipid metabolism pathway (cholesterol biosynthesis and VLDL pathway components), which are known to have key roles during the HCV life cycle (43). Similarly, GBV-B is also likely to require liver specific factors for replication or virion production. In addition, recent identification of related viruses from horses and rodents, nonprimate hepacivirus (NPHV) and rodent hepacivirus (RHV), respectively, also have putative miR-122 binding sites, but whether these viruses exhibit liver tropism remains to be confirmed and analysis of the influence of miR-122 on the life cycles of these viruses awaits the development of in vitro replication systems (44–46). These viruses have all been classified as hepaciviruses within the Flaviviridae family, and it is tempting to speculate that they may have evolved from a common hepatotropic ancestor (47).
In summary, our data demonstrate that GBV-B uses the liver-specific miR-122 to accumulate in human liver-derived cells. Like HCV, GBV-B has two tandem miR-122 sites in its 5′ UTR and site 1-bound miR-122 forms an interaction with the 5′ terminus of the GBV-B genome, which likely plays a role in protection from nucleases or possibly masks detection by cellular sensors of RNA. Similar to HCV, this interaction is likely to be mediated by Ago2. However, GBV-B seems to be able to overcome the requirement for miR-122 by deleting the miR-122 sites. The deletion mutant is able to accumulate to levels similar to those of the wild type and is insensitive to the levels of miR-122 and Ago2. The similarities to HCV–miR-122 interactions suggest that the two viruses likely share a mechanism and hence GBV-B may represent a surrogate system in which to investigate the mechanism of action of miR-122 on viral RNA accumulation. In addition, the GBV-B culture system developed here allows a novel platform to screen for factors required for miR-122-dependent (wild-type) and miR-122-independent (Δ4-29) viral RNA accumulation. Further elucidation of host and viral factors associated with these complexes is a promising strategy to provide novel antiviral targets to limit GBV-B and HCV replication.
ACKNOWLEDGMENTS
We are grateful to Cinzia Traboni and Licia Tomei for providing the GBV-B neo-RepB and GBV-B Δ4-29 neo-RepB replicon constructs and to Francis V. Chisari for kindly providing Huh-7.5.1 cells. We thank Karla Kirkegaard for kindly providing MK0608. We also thank Annette Martin for valuable discussions during the preparation of the manuscript.
S.M.S. is an Amgen Fellow of the Life Sciences Research Foundation. S.M.S. also acknowledges the National CIHR Research Training Program in Hepatitis C (NCRTP-HepC), as well as the American Liver Foundation, for postdoctoral fellowships. P.S. was supported by National Institutes of Health grants AI 069000 and AI 47365. J.A.W. acknowledges funding provided by the Natural Science and Engineering Research Foundation (NSERC-RGPIN-342475) and the Saskatchewan Health Research Foundation (RAPID 1927).
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kincaid RP, Sullivan CS. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 8:e1003018. 10.1371/journal.ppat.1003018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 5. Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. 2013. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 110:1881–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Machlin ES, Sarnow P, Sagan SM. 2011. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. U. S. A. 108:3193–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. 2012. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U. S. A. 109:941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jopling CL, Schutz S, Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roberts AP, Lewis AP, Jopling CL. 2011. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 39:7716–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilson JA, Zhang C, Huys A, Richardson CD. 2011. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J. Virol. 85:2342–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thibault PA, Huys A, Dhillon P, Wilson JA. 2012. MicroRNA-122-dependent and -independent replication of hepatitis C virus in Hep3B human hepatoma cells. Virology 431:179–190 [DOI] [PubMed] [Google Scholar]
- 13. Norman KL, Sarnow P. 2010. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J. Virol. 84:666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Villanueva RA, Jangra RK, Yi M, Pyles R, Bourne N, Lemon SM. 2010. miR-122 does not modulate the elongation phase of hepatitis C virus RNA synthesis in isolated replicase complexes. Antivir. Res. 88:119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J. Gen. Virol. 92:233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akari H, Iwasaki Y, Yoshida T, Iijima S. 2009. Non-human primate surrogate model of hepatitis C virus infection. Microbiol. Immunol. 53:53–57 [DOI] [PubMed] [Google Scholar]
- 17. Iwasaki Y, Mori K, Ishii K, Maki N, Iijima S, Yoshida T, Okabayashi S, Katakai Y, Lee YJ, Saito A, Fukai H, Kimura N, Ageyama N, Yoshizaki S, Suzuki T, Yasutomi Y, Miyamura T, Kannagi M, Akari H. 2011. Long-term persistent GBV-B infection and development of a chronic and progressive hepatitis C-like disease in marmosets. Front. Microbiol. 2:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Tomassi A, Pizzuti M, Traboni C. 2003. Hep3B human hepatoma cells support replication of the wild-type and a 5′-end deletion mutant GB virus B replicon. J. Virol. 77:11875–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. U. S. A. 100:2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beames B, Chavez D, Guerra B, Notvall L, Brasky KM, Lanford RE. 2000. Development of a primary tamarin hepatocyte culture system for GB virus-B: a surrogate model for hepatitis C virus. J. Virol. 74:11764–11772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chavez D, Guerra B, Lanford RE. 2009. Antiviral activity and host gene induction by tamarin and marmoset interferon-alpha and interferon-gamma in the GBV-B primary hepatocyte culture model. Virology 390:186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rijnbrand R, Yang Y, Beales L, Bodola F, Goettge K, Cohen L, Lanford RE, Lemon SM, Martin A. 2005. A chimeric GB virus B with 5′ nontranslated RNA sequence from hepatitis C virus causes hepatitis in tamarins. Hepatology 41:986–994 [DOI] [PubMed] [Google Scholar]
- 23. Buckwold VE, Collins B, Hogan P, Rippeon S, Wei J. 2005. Investigation into the ability of GB virus B to replicate in various immortalized cell lines. Antivir. Res. 66:165–168 [DOI] [PubMed] [Google Scholar]
- 24. De Tomassi A, Pizzuti M, Graziani R, Sbardellati A, Altamura S, Paonessa G, Traboni C. 2002. Cell clones selected from the Huh7 human hepatoma cell line support efficient replication of a subgenomic GB virus B replicon. J. Virol. 76:7736–7746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsuura Y. 2012. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J. Virol. 86:1382–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fabian MR, Sonenberg N. 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19:586–593 [DOI] [PubMed] [Google Scholar]
- 28. Beames B, Chavez D, Lanford RE. 2001. GB virus B as a model for hepatitis C virus. ILAR J. 42:152–160 [DOI] [PubMed] [Google Scholar]
- 29. Muerhoff AS, Leary TP, Simons JN, Pilot-Matias TJ, Dawson GJ, Erker JC, Chalmers ML, Schlauder GG, Desai SM, Mushahwar IK. 1995. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J. Virol. 69:5621–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krieger N, Lohmann V, Bartenschlager R. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U. S. A. 102:2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 102:17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 103:6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 36. Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, Lanford RE, Weinman SA, Lemon SM, Martin A, Li K. 2007. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J. Virol. 81:964–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saetrom P, Heale BS, Snove O, Jr, Aagaard L, Alluin J, Rossi JJ. 2007. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 35:2333–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin A, Bodola F, Sangar DV, Goettge K, Popov V, Rijnbrand R, Lanford RE, Lemon SM. 2003. Chronic hepatitis associated with GB virus B persistence in a tamarin after intrahepatic inoculation of synthetic viral RNA. Proc. Natl. Acad. Sci. U. S. A. 100:9962–9967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Friebe P, Bartenschlager R. 2009. Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly. J. Virol. 83:11989–11995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang C, Huys A, Thibault PA, Wilson JA. 2012. Requirements for human Dicer and TRBP in microRNA-122 regulation of HCV translation and RNA abundance. Virology 433:479–488 [DOI] [PubMed] [Google Scholar]
- 42. Rijnbrand R, Abell G, Lemon SM. 2000. Mutational analysis of the GB virus B internal ribosome entry site. J. Virol. 74:773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alvisi G, Madan V, Bartenschlager R. 2011. Hepatitis C virus and host cell lipids: an intimate connection. RNA Biol. 8:258–269 [DOI] [PubMed] [Google Scholar]
- 44. Bukh J. 2011. Hepatitis C homolog in dogs with respiratory illness. Proc. Natl. Acad. Sci. U. S. A. 108:12563–12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. 2012. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 86:6171–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, Chauhan LV, Duraisamy R, Sanchez Leon M, Jain K, Vandegrift KJ, Calisher CH, Rice CM, Lipkin WI. 2013. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio 4:e00216–13. 10.1128/mBio.00216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simmonds P. 2013. The origin of hepatitis C virus. Curr. Top. Microbiol. Immunol. 369:1–15 [DOI] [PubMed] [Google Scholar]