Abstract
The adenovirus L4-22K protein is multifunctional and critical for different aspects of viral infection. Packaging of the viral genome into an empty capsid absolutely requires the L4-22K protein to bind to packaging sequences in cooperation with other viral proteins. Additionally, the L4-22K protein is important for the temporal switch from the early to late phase of infection by regulating both early and late gene expression. To better understand the molecular mechanisms of these key functions of the L4-22K protein, we focused our studies on the role of conserved pairs of cysteine and histidine residues in the C-terminal region of L4-22K. We found that mutation of the cysteine residues affected the production of infectious progeny virus but did not interfere with the ability of the L4-22K protein to regulate viral gene expression. These results demonstrate that these two functions of L4-22K may be uncoupled. Mutation of the histidine residues resulted in a mutant with a similar phenotype as a virus deficient in the L4-22K protein, where both viral genome packaging and viral gene expression patterns were disrupted. Interestingly, both mutant L4-22K proteins bound to adenovirus packaging sequences, indicating that the paired cysteine and histidine residues do not function as a zinc finger DNA binding motif. Our results reveal that the L4-22K protein controls viral gene expression at the posttranscriptional level and regulates the accumulation of the L4-33K protein, another critical viral regulator, at the level of alternative pre-mRNA splicing.
INTRODUCTION
Human adenoviruses (Ad) consist of a nonenveloped icosahedral capsid with a linear double-stranded viral genome of ∼36,000 bp. Many questions remain about the basic biology of Ad infection. In particular, virus assembly for complex eukaryotic viruses is relatively poorly understood. Understanding the molecular mechanisms of the viral life cycle is critical for development of strategies for treatment of Ad infections and for the use of Ad as a gene therapy vector. The Ad5 genome contains five early transcription units (E1A, E1B, E2, E3, and E4), which encode ∼25 proteins that are expressed before viral DNA replication, and a set of delayed mRNAs, which encode proteins IX and IVa2, synthesized at the onset of DNA replication. These early and intermediate transcripts encode proteins with various roles during infection, including transcriptional regulation, viral DNA replication, inhibition of cellular antiviral responses, and inhibition of immune responses (1–4). Following DNA replication, a single major late transcription unit (MLTU) is transcribed and includes five different groups of mRNAs (L1 to L5) that encode capsid structural proteins and proteins that promote virus assembly, direct Ad genome packaging, and serve regulatory functions (2). The Ad major late promoter (MLP) drives transcription from the MLTU regions L1 to L5, producing all late mRNAs by alternative splicing and polyadenylation of a primary transcript. Prior to DNA replication, the MLP is active at low levels, with transcription proceeding only as far as the L3 region and mRNA production restricted to the L1-52/55K and i leader proteins (5). The specific activation of the MLP requires binding of Ad-infected cell-specific transcription factor complexes to the downstream element (DE) of the MLP (6). The Ad L4-22K protein is thought to be the minimal factor required for MLP activation, but additional components, such as IVa2, are required to obtain the maximum activation observed at late times of infection (7). Following DNA replication, the MLP is fully activated, and transcription continues to the L4 and L5 regions. The L4 region encodes four proteins: L4-100K, a translation enhancer protein that targets specific late mRNAs (8, 9); protein pVIII, a structural protein of the viral capsid; L4-33K, a viral splicing factor (10); and L4-22K. The L4-22K protein has been shown to bind to the packaging domain in vitro and in vivo (11) and is required for viral DNA packaging into the empty capsid (12). L4-22K also is associated with other functions pertaining to the regulation of Ad late gene expression (12, 13).
Following late-stage activation of the MLP, L4 poly(A) site usage is prominent (14). It is only after this increase in expression of L4 gene products that the L2, L3, and L5 poly(A) sites are used, generating the complete late viral gene expression profile. The timing of L4 expression fits well with the idea that its products have regulatory functions in late viral gene expression (13, 15). However, the essential role of L4 family proteins at this stage in the viral life cycle creates a paradox, since their expression is achieved only as a consequence of the activation of late-phase expression. To resolve this paradox, Morris et al. (16) identified a novel, intermediate-phase Ad5 promoter, the L4 promoter (L4P), that directs the expression of the L4-22K and L4-33K proteins independent from the MLP. L4P is active in its natural context, and the amount of L4-22K protein expressed via L4P is sufficient to induce the early-to-late transition in MLTU activity. Following polyadenylation, Ad late primary transcripts are spliced so that each mature mRNA contains the untranslated tripartite leader sequence (TPL), composed of leaders 1, 2, and 3. The TPL enhances translation of mRNAs during the late phase of Ad infection and can also increase the efficiency of mRNA export from the nucleus (17, 18). The TPL is spliced to one of many alternative 3′ splice sites, generating the 20 or more cytoplasmic mRNAs that encode the Ad late proteins (5). The individual 3′ acceptor splice and poly(A) sites within the main MLTU body have different efficiencies of usage that change as infection proceeds. The L4-33K protein serves as a viral splicing factor in the L1 unit (10). The L1 unit has a common 5′ donor splice site that can be joined to one of two alternative 3′ acceptor splice sites, forming the L1-52/55K or IIIa mRNAs. During early times of infection, the L1-52/55K acceptor splice site is preferentially used, while the IIIa acceptor splice site is not active until late times of infection. The L4-33K protein is required for the early-to-late shift in L1 alternative splicing patterns (10) and plays a positive role in regulating L1 splice site selection (5).
Although the role of L4-33K in alternative splicing is well understood, the exact role of L4-22K in the regulation of Ad late gene expression is still unclear. Morris and Leppard established that L4-22K has an important role in regulating the pattern of MLTU gene expression, and this role is independent of the effect of L4-33K on late RNA splicing (13). In a separate study, the analysis of the phenotypes of L4-22K mutant viruses confirmed that L4-22K is important for the transition from early to late viral gene expression (12). Both studies presented evidence indicating that L4-22K acts at a posttranscriptional level, affecting late gene mRNA production and/or stability, and that this effect was on specific Ad late RNAs, not on a global scale. Prior to being linked to a regulatory function in viral gene expression, the L4-22K protein was first attributed to a role in packaging of the Ad genome into the empty capsid (11). Ad packaging is dependent on a cis-acting region located at the left end of the genome. This region, termed the packaging domain, consists of seven AT-rich repeats with the consensus sequence 5′-TTTG-N8-CG-3′ (19). There are four different Ad particle forms identified by CsCl equilibrium centrifugation: empty capsids devoid of viral DNA, light assembly intermediates that contain the left end of the viral genome, heavy assembly intermediates that contain the full viral genome and precursor forms of certain capsid proteins, and mature virus particles that contain the full-length genome with proteolytically processed capsid proteins. Efficient packaging is required for the formation of mature virus particles, and a complex of DNA and proteins at the packaging domain plays a key role. The IVa2 and L4-22K proteins bind to the CG and TTTG motifs of the packaging repeats, respectively (11, 20, 21) and play an important role in viral DNA encapsidation. IVa2 and L4-22K mutant viruses produce empty particles containing no viral DNA (12, 22). In vivo studies have shown that the L4-22K and IVa2 proteins are dependent on each other for binding to the packaging domain (12). In addition to IVa2 and L4-22K, the L1-52/55K and IIIa proteins also play roles in the packaging process (23, 24).
The L4-22K protein contains a conserved pair of cysteine residues and a conserved pair of histidine residues near its C terminus. Given the configuration of these amino acid pairs (CXXXC and HXXXH separated by 24 amino acids), we sought to explore their potential to function as a putative zinc finger of the L4-22K DNA binding domain. Two different L4-22K mutant viruses were constructed and analyzed: one with substitution mutations in the cysteine residues and the other with substitution mutations in the histidine residues. Analyses of these mutant viruses showed that the double cysteine mutant uncouples the functions of L4-22K in Ad DNA packaging from the regulation of viral late gene expression. The cysteine residues were not required for binding of the L4-22K protein to the packaging sequences, so the observed defect in mature virus particle production was not due to aberrant DNA binding. We found that the double histidine mutant had a complex phenotype with defects observed in genome packaging and expression of specific mRNA transcripts. We further confirmed that L4-22K acts at the level of late gene mRNA production to drive the early-to-late phase transition during the Ad life cycle and that this function is specific to a unique set of mRNA transcripts. We identified proteins L4-33K and L4-pVIII as primary targets for L4-22K regulation and found that L4-22K is required for efficient splicing of the L4-33K mRNA transcript and subsequent expression of the protein. Thus, the Ad L4-22K protein has two distinct functions to promote specific patterns of viral late gene expression and viral genome encapsidation.
MATERIALS AND METHODS
Cells, viruses, and infections.
N52.E6-Cre cells are derived from the E1-expressing cell line N52.E6 (25), a gift from G. Schiedner and S. Kochanek, Center for Molecular Medicine (ZMMK), University of Ulm, Ulm, Germany. Cre recombinase was not important for these experiments. N52.E6-Cre cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% Fetalclone III serum (HyClone), penicillin, and streptomycin. A549 cells (ATCC) were maintained in Dulbecco's modified Eagle medium supplemented with 10% bovine calf serum (HyClone), penicillin, and streptomycin. The TetC4-22K-ΔC (12) and TetC4-33K (37) cell lines express the Ad5 L4-22K and L4-33K proteins, respectively, under the control of a Tet-inducible promoter. The TetC4-22K-ΔC protein contains a truncation of the C-terminal 23 amino acids of Ad5 L4-22K that are not required for activity. TetC4 cells and the TetC4 cell line derivatives were maintained in Dulbecco's modified Eagle medium supplemented with 8% fetal bovine serum, 200 μg/ml Geneticin, 200 μg/ml hygromycin B, 100 μg/ml blasticidin S, and 0.1 μg/ml doxycycline. Doxycycline was removed from the culture medium 36 to 48 h prior to conducting experiments to induce expression of L4-22K and L4-33K.
The construction of viruses containing the L4-22K point mutations is described below. The L4-22K-C137S/C141S mutant virus was propagated in N52.E6-Cre cells, and the L4-22K-H166Q/H170Q and the L4-Δ22K viruses were propagated in TetC4-22K-ΔC cells. Virus particles were purified by cesium chloride equilibrium centrifugation, as described below. All infections were carried out in N52.E6-Cre, TetC4-22K-ΔC, or TetC4-33K cells with a multiplicity of infection of 100 to 200 particles/cell of each virus stock, unless otherwise noted, as previously described (26).
Plasmids, mutagenesis, and cloning.
Expression plasmids were generated by using pcDNA3 (Invitrogen Life Technologies). Expression plasmids pcDNA3-22K and pcDNA3-33K were previously described (11). Plasmid pcDNA3-22K-C137S/C141S was constructed by introducing double serine substitution mutations into the L4-22K gene by site-directed mutagenesis (QuikChange mutagenesis; Stratagene). The pair of cysteines were mutated to serines, and an NheI restriction site was introduced by using forward primer 5′-GCCATAGTTGCTAGCTTGCAAGACAGTGGGGGCAACATC-3′ and reverse primer 5′GATGTTGCCCCCACTGTCTTGCAAGCTAGCAACTATGGC-3′, corresponding to Ad5 nucleotides (nt) 26591 to 26619. The underlined nucleotides correspond to the mutation sites. Plasmid pcDNA3-22K-H166Q/H170Q was constructed in a similar manner by introducing double glutamine substitution mutations into the L4-22K gene. The pair of histidines were mutated to glutamines and a ScaI restriction site was introduced using forward primer 5′-GTAACATCCTGCAGTACTACCGTCAACTCTACAGCCCAT-3′ and reverse primer 5′-ATGGGCTGTAGAGTTGACGGTAGTACTGCAGGATGTTAC-3′, corresponding to Ad5 nt 26679 to 26717. The underlined nucleotides correspond to the mutation site. Each mutated segment was then recombined into an infectious clone, pTG3602 (27), as described previously (26). Each mutated viral genome was excised from the pTG3602 plasmid by PacI digestion and used to transfect N52.E6-Cre cells (pTG3602-22K-C137S/C141S) or TetC4-22K-ΔC cells (pTG3602-22K-H166Q/H170Q) by using Lipofectamine 2000 reagent (Invitrogen) according to the instructions provided by the manufacturer. Viruses were isolated after 7 days by plaque purification and propagated in the cell lines specified above. Particles for each virus stock were purified by two successive rounds of cesium chloride equilibrium centrifugation as described below. Mutant virus sequences were confirmed by DNA sequencing and restriction enzyme digestion and Southern blot analysis for the presence of the indicated mutations. For simplicity, these mutant viruses are referred to as C137S/C141S and H166Q/H170Q, indicating their amino acid substitution mutations. The construction of Δ22K virus was described previously (12).
Expression plasmid pcDNA3-L4 was generated by PCR cloning of the regions from Ad5 nt 26195 to 27086 into pcDNA3. Note that this region contains the coding regions for both L4-22K (Ad5 nt 26195 to 26785) and L4-33K (26195 to 27086) (Fig. 1A). Plasmid pcDNA3-L4-C137S/C141S was constructed by introducing double serine substitution mutations to the L4-22K gene in the background of the pcDNA3-L4 plasmid. Expression plasmid pcDNA3-L4-22K− was constructed by introducing a stop codon (TAG) in the coding region of 22K by changing Ad5 nt 26533 to 26536 from GTTA to CTAG, in the background of the pcDNA3-L4 plasmid. Similarly, plasmid pcDNA3-L4-33K− was constructed by introducing a stop codon in the coding region of 33K by changing Ad5 nt 26814 to 26816 from ACA to TAG in the background of the pcDNA3-L4 plasmid.
Fig 1.
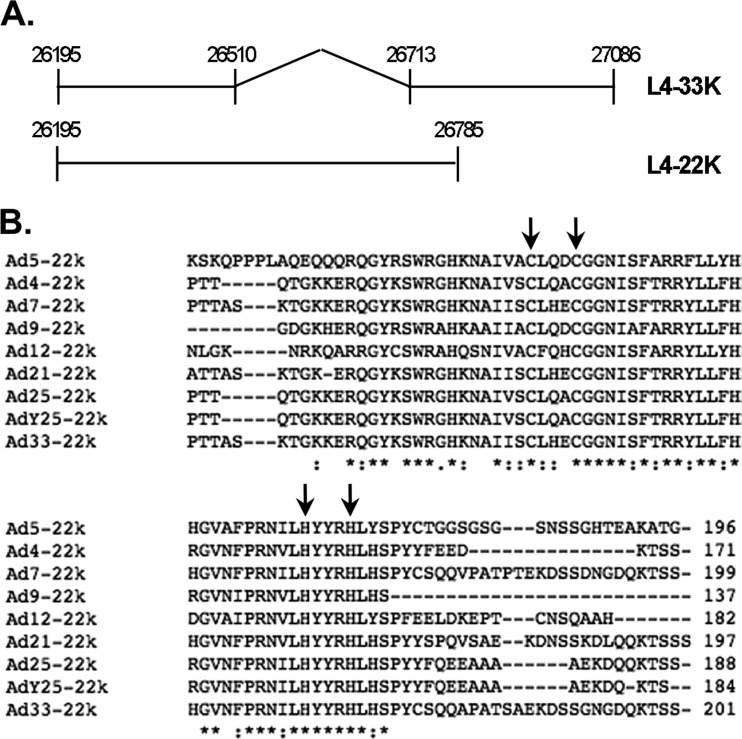
The Ad5 L4-22K protein. (A) Schematic diagram of the coding sequences of the L4-22K and L4-33K proteins. Numbers correspond to Ad5 nucleotides. The intron of L4-33K is indicated by the gap and angled lines. (B) ClustalW protein sequence alignment of the L4-22K carboxy termini of L4-22K of human (Ad5, Ad4, Ad7, Ad9, Ad12), simian (Ad21, Ad25, Ad33), and chimpanzee (AdY25) Ad serotypes. The consensus symbols are as follows: the asterisk indicates positions which have a single, fully conserved residue, while the colon and period indicate conservation between groups of strongly and weakly similar properties, respectively. The cysteine and histidine residues that were mutated in this study are indicated with arrows and correspond to residues C137, C141, H166, and H170. Amino acid numbers are indicated to the right of the alignment.
Replication assay and quantitative PCR.
Infected N52.E6-Cre cells were harvested at 2 and 24 h postinfection (hpi). Cells were washed in phosphate-buffered saline (PBS), pelleted, resuspended in cold isotonic buffer containing NP-40 (140 mM NaCl, 10 mM Tris [pH 7.4], 1.5 mM MgCl2, 0.6% NP-40), and incubated on ice for 10 min. Following another round of centrifugation at 2,000 × g, the pellet containing nuclei was resuspended in PBS and DNA was prepared using a DNeasy blood and tissue kit (Qiagen) following the manufacturer's protocol. The amount of viral DNA was quantified with a DyNAmo HS SYBR green quantitative PCR (qPCR) kit (Finnzymes). Primer pairs surrounding the packaging domain were used to quantify viral DNA, and appropriate primers were used to quantify glyceraldehyde 3-phosphate dehydrogenase (GAPDH) copy number, as described previously (12). The genome replication was calculated as the value of the viral genome copy number normalized by the GAPDH copy number. The value calculated for each virus at each time point was divided by that of Ad5 wild-type (WT) at 2 hpi (input) and is shown as the relative viral genome copy number.
Plaque assays and immunofluorescence analysis.
CsCl-purified virus particles (described below) from Ad5 WT, C137S/C141S, H166Q/H170Q, and Δ22K were used to infect A549 and TetC4-22K-ΔC cells. After 7 to 10 days, the number of plaques were counted, and the ratio of particles (determined as described below) to plaques was calculated as the the number of particles/PFU. Infectivity was directly measured in a fluorescence focus assay by infecting A549 cells grown on coverslips with different dilutions of particles from Ad5 WT, C137S/C141S, and H166Q/H170Q. Eighteen hours postinfection, cells were fixed with methanol and processed for immunofluorescence by using an antibody against the DNA binding protein (DBP; provided by Arnold Levine, Princeton University) and a secondary goat anti-mouse antibody conjugated to fluorescein isothiocyanate (FITC; Invitrogen). For each coverslip, the average number of DBP-positive cells was taken from 10 random fields and quantified as the fluorescent focus units (FFU) per field. The particle/FFU ratio was calculated as the number of particles/total FFU in the well.
Virus growth and cesium chloride equilibrium gradients.
For virus growth curves, N52.E6-Cre cells were infected with Ad5 WT, C137S/C141S, or H166Q/H170Q, harvested at 6, 12, 20, 28, and 36 hpi, and lysed by four cycles of freeze-thawing. A specific volume of each lysate was used to infect A549 cells grown on coverslips and processed for immunofluorescence, as described above. Virus growth was calculated as the log of the fluorescent focus units per volume of lysate used (log FFU/ml). To compare virus growth on three different cell lines, N52.E6-Cre, TetC4-22K-ΔC, and TetC4-33K cells were infected with Ad5 WT, C137S/C141S, or H166Q/H170Q or mock infected. For TetC4-22K-ΔC and TetC4-33K cells, two sets of mock-infected cells were prepared, one with doxycycline removed from the medium to induce expression of 22K and 33K and one with doxycycline maintained in the medium. Cells were harvested 48 hpi; a portion was used for whole-cell extract preparation for Western blot analysis (described below), and the rest was subjected to four cycles of freeze-thawing. Virus lysates were used to infect A549 cells grown on coverslips and processed for immunofluorescence as described above. Virus yield was measured as the FFU/ml.
For cesium chloride equilibrium gradients, cells were infected with Ad5 WT or C137S/C141S. At 72 to 96 hpi, cells and medium were collected and the cells were pelleted, resuspended in 6 ml of TD buffer (25 mM [pH 7.4], 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4), and frozen and thawed for four cycles. The lysates were cleared by centrifugation and added to step gradients of 1.25g/cm3 and 1.40g/cm3 cesium chloride in TD buffer and spun at 175,587 × g at 15°C for 1 h. Visible bands at each step were collected and mixed with 1.34g/cm3 CsCl and spun for 20 h at 174,618 × g at 15°C. The visible band was collected from the equilibrium gradient, and the absorbance was measured at a wavelength of 260 nm and converted to densities by using the following calculation: 1 optical density at 260 nm (OD260) unit = 1 × 1012 particles/ml (28).
Gel mobility shift assays.
Nuclear extracts from N52.E6-cre cells infected with Ad5 WT, C137S/C141S, H166Q/H170Q, or Δ22K or mock infected were prepared 24 hpi, as described previously (11, 19). Six micrograms of nuclear extract was incubated with 0.5 to 1.25 μg of poly(dI-dC), with and without anti-22K antibody, for 15 min at room temperature. Following this incubation, 120,000 to 150,000 cpm of 32P-labeled DNA probe was added to each reaction mixture and incubated for an additional 30 min at room temperature. Packaging sequence probes used in the assays contained A repeats 1 and 2, corresponding to Ad5 nt 236 to 282. The gel mobility shift assays were performed as described previously (22).
Western blot analyses.
N52.E6-Cre, TetC4-22K-ΔC, or TetC4-33K cells were mock or virus infected, and whole-cell extracts were prepared, as described previously (12). Protein concentrations were determined, and equal amounts of protein were loaded on SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membranes (Whatman). The membrane was blocked for 1 h at room temperature in a solution of 1% casein–Tris-buffered saline (TBS) followed by an incubation with a designated antibody in 1% casein–TBS with sodium azide for 1 h at room temperature, or overnight at 4°C. The membrane was washed four times for 5 min in TBS with 0.5% Tween 20 and incubated with 800 IRdye800-conjugated goat anti-rabbit immunoglobulin G (Rockland Immunochemicals, Gilbertsville, PA) for 45 min, followed by four more washes in TBS with 0.5% Tween 20 for 5 min each. Two final washes in TBS were done, and the membrane was scanned using an Odyssey system (LiCOR, Lincoln, NE). For L4-22K detection, the SDS-PAGE gel was transferred to a polyvinylidene difluoride (PVDF; GE Healthcare) membrane, blocked with 3% bovine serum albumin–TBS for 1 h at room temperature, and probed with primary antibody overnight at 4°C. All washes and secondary antibody incubations were performed with 5% nonfat milk–TBS containing 0.05% Tween 20. Proteins were detected using the following antibodies: rabbit polyclonal antibodies to fiber and penton (gifts from Carl Anderson, Brookhaven National Laboratory); rabbit polyclonal antibody to VII (Daniel Engel, University of Virginia); rabbit polyclonal antibody to hexon (GeneTex, Inc., Irvine, CA); rabbit polyclonal antibodies to L4-22K and L4-33K (11); rabbit polyclonal antibody to VIII (William Wold, St. Louis University); rabbit polyclonal antibodies to IVa2 and L1-52/55K (19); rabbit polyclonal antibody to IIIa (24); rabbit polyclonal antibody to V (David Matthews, University of Bristol); rabbit polyclonal antibody to L4-100K (37); rabbit polyclonal antibody to VI (Christopher Wiethoff, Loyola University Chicago); rabbit polyclonal antibody to E1A (Santa Cruz Biotechnology); rabbit polyclonal antibody to γ-tubulin (Sigma). Additional information on Western blot analyses and antibodies was provided previously (12).
Northern blots and reverse transcriptase PCR (RT-PCR).
Infected N52.E6-Cre cells were harvested at 24 h or 48 hpi, and the cytoplasmic fraction was extracted using an RNeasy isolation kit for cytoplasmic RNA from animal cells (Qiagen). Four to 12 μg of total cytoplasmic RNA was separated on a 1% agarose gel containing 2.2 M formaldehyde and transferred to a nylon membrane (GE Healthcare). The filter was then stained in a solution of 0.02% methylene blue and 0.3 M sodium acetate, pH 5.5, to detect rRNAs. The stain was removed and the filter hybridized with 0.5 to 1 μg of a designated DNA probe 32P-labeled by random priming (random primer DNA labeling kit; TaKaRa Bio), followed by a series of washes, as described previously (29). DNA probes were constructed using primers surrounding the coterminal 3′ end of each late gene family: Ad5 L1 (nt 13025 to 13751), Ad5 L2 (nt 16834 to 17452), Ad5 L3 (nt 21573 to 22322), Ad5 L4 (nt 26769 to 27590), and Ad5 L5 (nt 31920 to 32465). The fragments were extended by PCR and resolved through 1% agarose followed by gel extraction (Qiagen).
A 2.5-μg aliquot of RNA was used with oligo(dT) to generate cDNA in a 20-μl reaction mixture, using the SuperScript II reverse transcriptase kit (Invitrogen). Two units of Escherichia coli RNase H was added to remove RNA complementary to the cDNA, and the mixture was incubated at 37°C for 20 min. A 1-μl portion of the RT reaction mixture was added to a PCR mixture in a total 12-μl reaction volume for 20 cycles. Each reaction mixture included a forward primer complementary to Ad5 tripartite leader 3 (nt 9700 to 9719) and a reverse primer within the intron of L4-33K (nt 26682 to 26703). PCR products were resolved on a 1% agarose gel, and the fragments were extracted for sequencing. Equal portions of each cDNA were used in a PCR with primers GAPDH-1 (5′-ACCCAGAAGACTGTGGATGG-3′) and GAPDH-2 (5′-TTCTAGACGGCAGGTCAGGT-3′) to compare levels of GAPDH.
In vivo analysis of alternative splicing.
N52.E6 cells were transfected in 100-mm dishes with specific expression plasmids and using Lipofectamine 2000 reagent (Invitrogen). Twenty micrograms of each expression plasmid was cotransfected with 200 ng of pEGFP-C1 expression plasmid (Clontech), and cells were harvested 48 h after transfection. A portion of cells from each transfected dish was used for whole-cell extract preparation and Western blot analysis, while the rest of the cells harvested were used for total cytoplasmic RNA preparation. For Western blot analysis, protein levels were normalized to levels of green fluorescent protein (GFP), based on quantification using an Odyssey infrared imaging system (Li-COR) to account for any differences in transfection efficiencies. Samples were separated on an SDS-PAGE gel, transferred to a nitrocellulose membrane, and analyzed for protein levels as described above. For primary antibodies, rabbit anti-L4-22K and L4-33K antibodies (11) and rabbit anti-GFP antibodies (Abcam) were used. For mRNA analysis, cytoplasmic RNA was prepared as described above. mRNA levels were normalized to levels of GFP based on quantification using phosphorimaging analysis to account for any differences in transfection efficiencies. Samples were used for Northern blotting, as described above, with a designated DNA probe 32P-labeled by random priming. The DNA probe used to detect levels of L4-22K and L4-33K was constructed by amplification of Ad5 nt 26769 to 27590 by PCR, resolution of the fragment through 1% agarose, and gel extraction. The DNA probe used to detect levels of GFP was constructed by digesting plasmid pEGFP-C1 with NheI and BamHI, followed by gel purification of the digested fragment.
RESULTS
Generation and characterization of L4-22K mutant viruses: viral genome replication and infectivity.
The Ad5 L4-22K and L4-33K proteins are encoded by the L4 region and share an N terminus (Fig. 1A). The L4-33K protein is the product of an additional splicing event, while the L4-22K protein is translated from the unspliced version of this transcript, with each protein having a unique C terminus. Using ClustalW multiple sequence alignment (30), we found that the unique C terminus of L4-22K, amino acids 116 to 175, is highly conserved among human Ad serotypes (Ads 4, 5, 7, 9, and 12) and many nonhuman primate Ads (Ads 21, 25, Y25, and 33) (Fig. 1B). The C-terminal 23 amino acids of Ad5 L4-22K is not conserved and is not essential for its functions (12). The conservation in the C-terminal region of L4-22K suggests a crucial and unique function of this domain. Within this region, there are two conserved cysteine residues (C137and C141) and two conserved histidine residues (H166 and H170) that are clustered within 30 amino acids and have general spacing requirements consistent with features of a zinc finger motif (31, 32). We postulated that these four conserved residues constituted a putative DNA binding motif involved in the DNA-protein interaction of L4-22K with the packaging domain. In order to assess the role of these conserved residues in L4-22K activity, two mutant L4-22K viruses were generated and characterized in this study: C137S/C141S, with conservative, double serine substitution mutations at amino acids 137 and 141, and H166Q/H170Q, with conservative, double glutamine substitution mutations at amino acids 166 and 170. The C137S/C141S and H166Q/H170Q mutant viruses were propagated in N52.E6-Cre and TetC4-22K-ΔC cell lines, respectively. The C137S/C141S mutant virus grew poorly in N52.E6-Cre cells, but these cells were used to eliminate the possibility of recombination with the endogenous L4-22K gene in the TetC4-22K-ΔC cell line to generate a revertant virus. The H166Q/H170Q mutant virus grew poorly in both N52.E6-Cre and TetC4-22K-ΔC cell lines (discussed below), but enough purified virus was produced by the complementing cell line to conduct experiments. The TetC4-22K-ΔC cell line expresses an L4-22K protein with a truncation of the C-terminal 23 amino acids, which are not required for activity (12), greatly reducing the possibility of homologous recombination to generate a revertant virus with the H166Q/H170Q mutant. Mutant virus sequences were confirmed by DNA sequencing and Southern blot analysis for the presence of the indicated mutations; these mutant virus stocks were pure from any Ad5 WT contamination (data not shown).
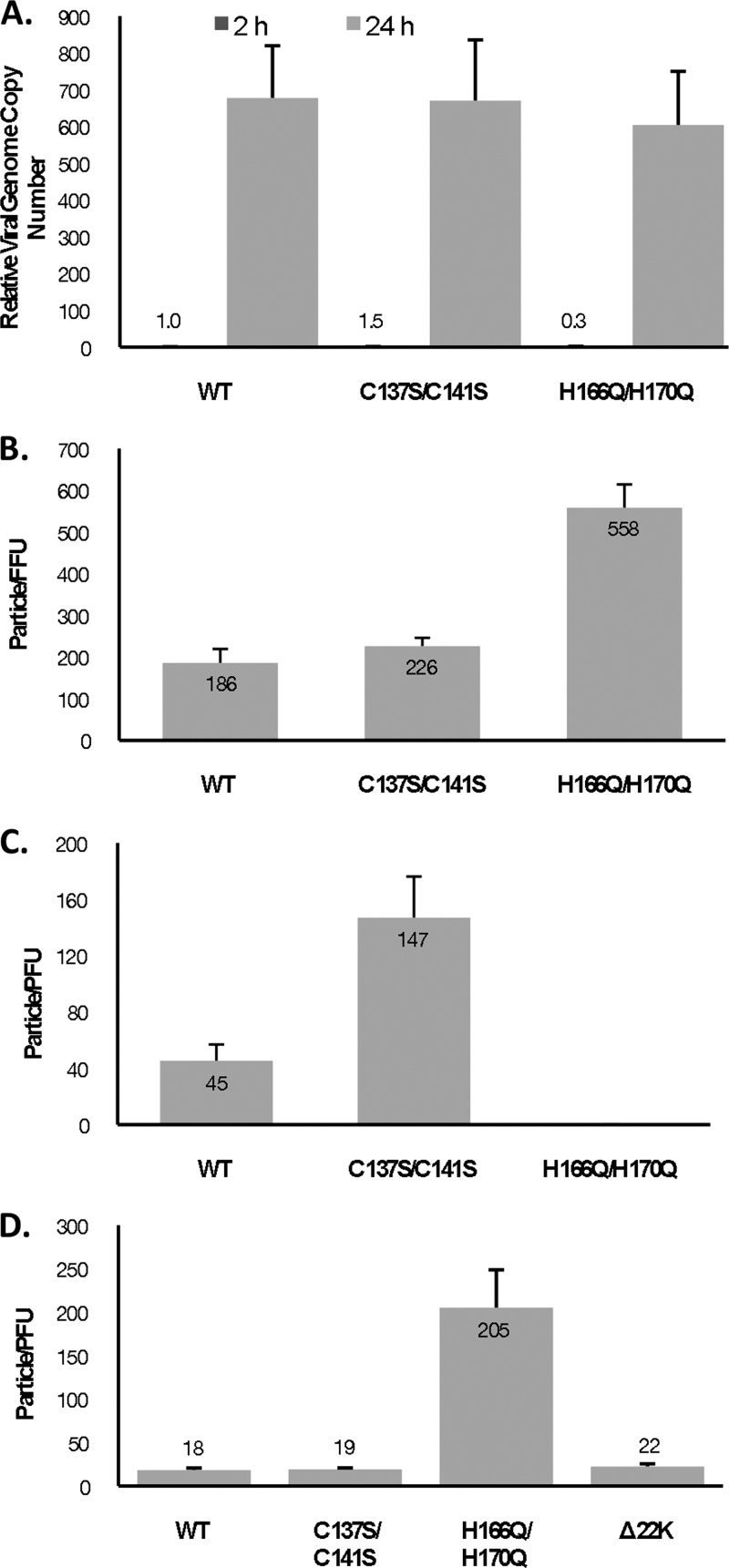
We evaluated viral DNA replication with the C137S/C141S and H166Q/H17 mutant viruses by qPCR and found that both mutant viruses replicated to similar levels as Ad5 WT (Fig. 2A). To test the infectivity of the mutant viruses, virus particles were used in a fluorescent focus assay in A549 cells (Fig. 2B). The particle/FFU ratio for each virus was calculated, and the results showed similar infectivities between Ad5 WT and the C137S/C141S mutant virus, while a marginal 3-fold decrease in infectivity was observed with the H166Q/H170Q mutant. Infectivity was also examined in a plaque assay in A549 cells (Fig. 2C) and the L4-22K-ΔC complementing cell line (Fig. 2D), and the particle/PFU ratio for each virus was calculated. Consistent with the DNA replication and infectivity results, there was only a modest difference in the particle/PFU ratio between Ad5 WT and the C137S/C141S mutant virus in A549 or TetC4-22K-ΔC cells. In contrast, no plaques were observed with the H166Q/H170Q mutant virus in A549 cells, even though DNA replication and infectivity were not significantly affected. To determine if providing WT L4-22K in trans would rescue plaque formation with the H166Q/H170Q mutant, plaque assays were performed in TetC4-22K-ΔC cells (Fig. 2D). We observed that the particle/PFU ratio with the H166Q/H170Q mutant virus was increased ∼10-fold compared to Ad5 WT; the complementing cell line was able to efficiently rescue plaque formation with an L4-22K null mutant, consistent with previous results (12). We concluded that the C137S/C141S and H166Q/H17 mutant viruses had levels of viral genome replication and infectivity comparable to Ad5 WT, while plaque formation was severely affected with the H166Q/H170Q mutant virus. The L4-22K protein in trans only partially rescued plaque formation with the H166Q/H170Q mutant.
Fig 2.
Properties of L4-22K mutant viruses. (A) Viral genome replication in N52.E6-Cre cells infected with Ad5 WT, C137S/C141S, or H166Q/H170Q. Values were normalized to GAPDH and compared to that of Ad5 WT at 2 hpi. (B) The particle/FFU ratios of Ad5 WT and C137S/C141S and H166Q/H170Q mutant viruses were determined in A549 cells. (C and D) The particle/PFU ratios of Ad5 WT and C137S/C141S and H166Q/H170Q mutant viruses were determined in A549 cells (C) and TetC4-22K-ΔC cells (D).
The conserved cysteine residues in the C-terminal region of L4-22K are important for viral genome packaging.
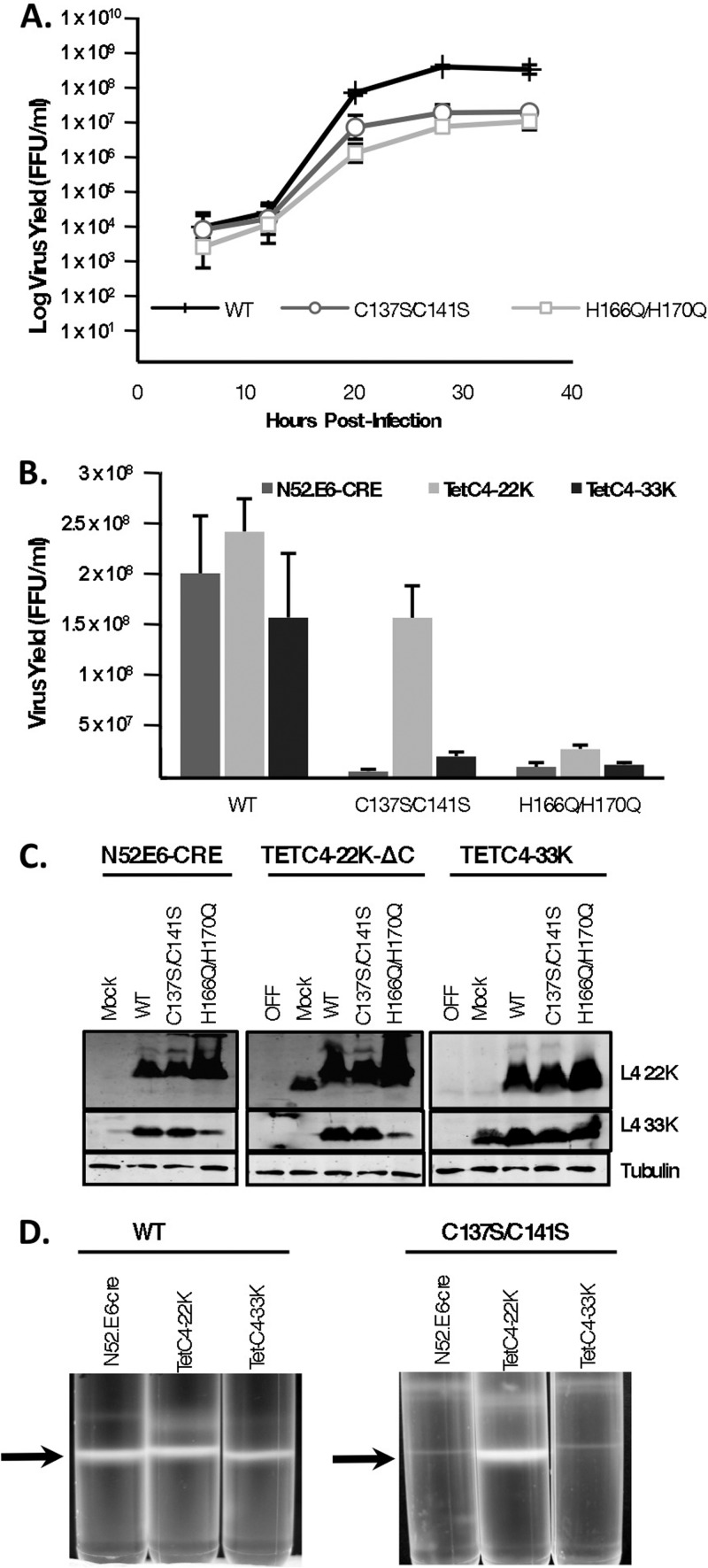
To analyze the effects of the L4-22K point mutations on overall viral growth properties, single-step growth curves were performed. The C137S/C141S and H166Q/H170Q mutants were reduced ∼25- to 40-fold in the production of infectious progeny in N52.E6-Cre cells compared to Ad5 WT (Fig. 3A). Since L4-22K was shown to be required for genome packaging in vivo (11, 12) and L4-33K regulates late gene expression on the posttranscriptional level (5, 10), we wanted to examine the contribution of these two proteins to the observed growth defect phenotype with the mutant viruses. In order to examine if the defect in virus growth exhibited by the two L4-22K mutants could be restored by complementation with WT L4-22K protein, the mutant viruses were grown in TetC4-22K-ΔC cells and their growth was compared to that on N52.E6-Cre cells. Overall virus yield was analyzed using a fluorescent focus assay, and results were calculated as the total FFU/ml (Fig. 3B). Consistent with the single-step growth curves, we observed a 30-fold reduction in overall virus yield with the C137S/C141S mutant virus in N52.E6-Cre cells compared to Ad5 WT, a phenotype that was almost fully rescued by providing L4-22K in trans using the L4-22K complementing cell line. With the H166Q/H170Q mutant, virus yield was modestly increased in TetC4-22K-ΔC cells. Analysis of protein expression showed that with the C137S/C141S mutant virus, both L4-22K and L4-33K proteins were expressed to WT levels in N52.E6-Cre and TetC4-22K-ΔC cells and, therefore, decreased production of these proteins did not contribute to the decrease in infectious virus yield with this mutant (Fig. 3C, left and middle panels). With the H166Q/H170Q mutant virus, L4-22K protein expression was significantly increased in both N52-E6.Cre and TetC4-22K-ΔC cells, while expression of L4-33K was decreased in both cell lines. Note that L4-22K-ΔC protein expressed in the complementing cell line ran slightly faster than WT L4-22K protein due to the C-terminal truncation. We recently showed that mutation of Ad5 L4-33K impairs the viral genome packaging process, but not virion assembly, resulting in the production of only empty capsids (37). To determine if the observed decrease in L4-33K protein levels contributed to the H166Q/H170Q mutant phenotype, we infected TetC4-33K cells with Ad5 WT, C137S/C141S, or H166Q/H170Q viruses and analyzed virus yield (Fig. 3B) and L4-22K and L4-33K protein expression (Fig. 3C, right panel). Supplementation with WT L4-33K in trans modestly increased C137S/C141S mutant virus yield. This result was not surprising, since there was no indication that the L4-33K protein had any contribution to the phenotype of this mutant virus. Interestingly, although growth in the TetC4-33K cells supplemented the levels of L4-33K protein with H166Q/H170Q-infected cells (Fig. 3C, right panel), we did not see any associated increase in virus yield with this mutant (Fig. 3B). The possibility that overproduction of L4-22K protein with the H166Q/H170Q mutant had any effect on production of infectious progeny was also ruled out, since providing L4-33K protein during infection reduced the levels of L4-22K protein expression to those seen with Ad5 WT (Fig. 3C, right panel).
Fig 3.
Viral growth kinetics of L4-22K mutant viruses. (A) N52.E6-Cre cells were infected with Ad5 WT or C137S/C141S or H166Q/H170Q mutant viruses and harvested at the designated time points after infection, and cell lysates were used to infect A549 cells to measure infectious virus yield (FFU/ml) in a fluorescent focus assay. (B) N52.E6-Cre, TetC4-22K-ΔC, or TetC4-33K cells were infected with Ad5 WT or C137S/C141S or H166Q/H170Q mutant viruses and harvested at 48hpi, and cell lysates were used to infect A549 cells to measure infectious virus yield (FFU/ml) in a fluorescent focus assay. (C) A portion of the cell lysate from the experiment shown in panel B was used to prepare whole-cell extracts for Western blot analysis of L4-22K and L4-33K protein levels. Tubulin was used as a loading control. (D) CsCl equilibrium density gradient profiles of virus particles produced from Ad5 WT- or C137S/C141S-infected N52-E6.Cre, TetC4-22K-ΔC, or Tet-C4-33K cells. Arrows indicate mature virus particles.
To determine if the reduction in infectious virus production with the mutant viruses was due to a packaging defect, we analyzed virus particle production in vivo by using CsCl equilibrium density gradient centrifugation with the three cell lines (Fig. 3D). Consistent with the results presented in Fig. 3B, supplementation of L4-22K, but not L4-33K, to C137S/C141S mutant virus infections augmented production of mature virions. Note that mature particle production was consistent among the three cell lines with Ad5 WT infection. We did not observe any significant increase in mature particle production with H166Q/H170Q-infected TetC4-22K-ΔC cells or TetC4-33K cells compared to N52.E6-Cre cells (data not shown). We conclude that the C137 and C141 residues in the C-terminal region of L4-22K play a role in viral genome packaging, and mutation of these residues dramatically decreases the production of infectious virus without affecting the levels of L4-22K or L4-33K protein expression.
The conserved cysteine and histidine residues are not required for L4-22K protein to bind to packaging sequences.
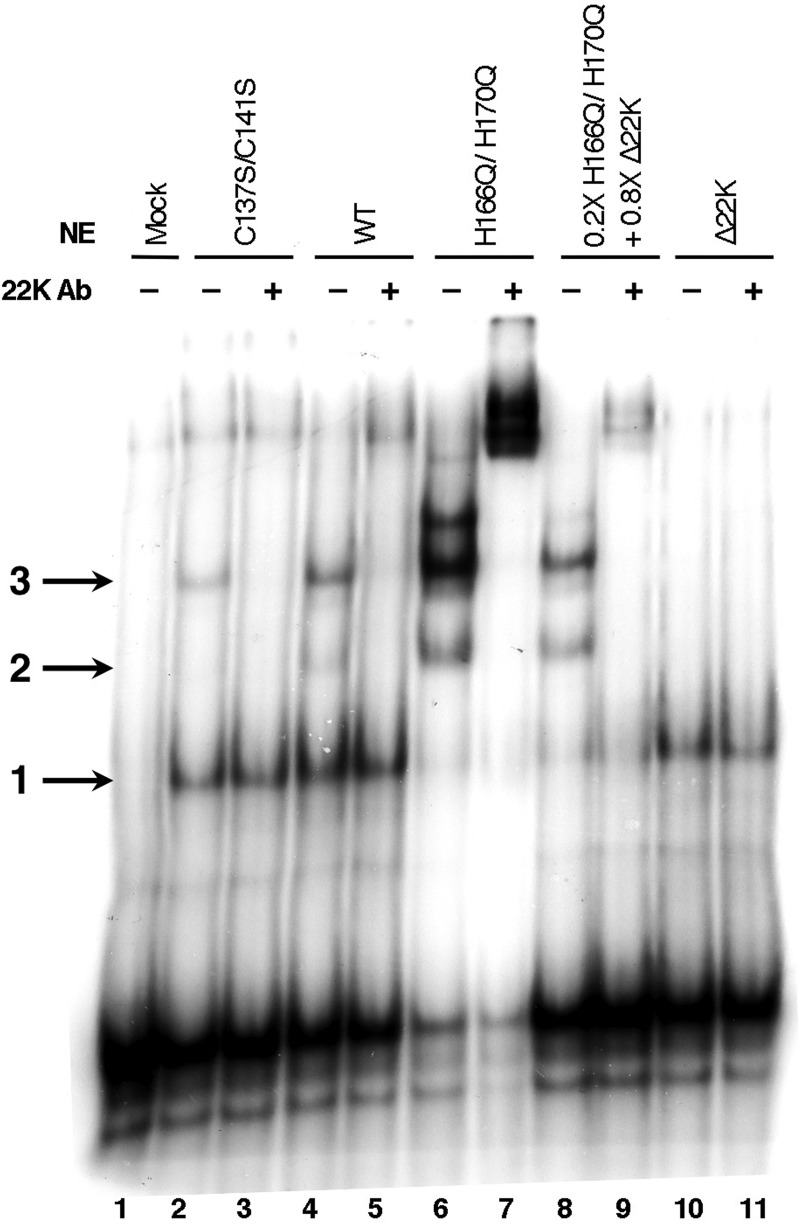
To examine if the conserved cysteine and histidine residues constitute a DNA binding motif involved in the DNA-protein interaction of L4-22K with packaging sequences, we analyzed DNA binding in in vitro gel mobility shift assays. Nuclear extracts were prepared at late times after infection of cells with viruses Ad5 WT, C137S/C141S, or H166Q/H170Q, and the formation of complexes specific to Ad5 packaging sequences was assessed by using a probe containing A repeats 1 and 2. Consistent with previous results (11), three complexes were observed with extracts form WT-infected cells (Fig. 4, lane 4). Complex 1 corresponds to the binding of IVa2 to the CG motif in the A1 repeat; complex 2 is formed with the L4-22K protein, in addition to IVa2, binding to the TTTG motif of the A2 repeat; complex 3 is due to the binding of an additional IVa2 protein to the CG sequence in the A2 repeat (11, 20, 21). Note that only complex 1 was formed with extracts from Δ22K virus-infected cells, since functional L4-22K protein was not present (Fig. 4, lanes 10 and 11). The amounts of complexes 1, 2, and 3 formed relative to each other varied somewhat from experiment to experiment. With extracts from C137S/C141S mutant virus-infected cells, complexes 1 and 3 were detected (lane 2). Although complex 2 was weak and difficult to detect in this assay, we postulate that binding to the packaging sequence exists, since addition of an antibody directed against the unique coding region of the L4-22K protein eliminated complex 3 (lane 3), similar to that observed with Ad5 WT-infected extract (lane 5).
Fig 4.
The L4-22K-C137S/C141S and L4-22K-H166Q/H170Q mutant proteins bind to Ad5 packaging sequences in vitro. Gel mobility shift assays were done with a 32P-labeled probe containing Ad5 A repeats 1 and 2. Six micrograms of nuclear extract from N52.E6-Cre cells infected with WT, C137S/C141S, H166Q/H170Q, or Δ22K or mock infected and harvested at 24 hpi were used for the assays. In lanes 8 and 9, 1.2 μg (0.2× of total sample) of nuclear extract from H166Q/H170Q-infected cells was mixed with 4.8 μg (0.8× of total sample) of nuclear extract from Δ22K-infected cells prior to incubation with probe. In lanes 3, 5, 7, 9, and 11, anti-L4-22K protein antibody was added to the binding reaction mixture prior to incubation with probe. The positions of complexes 1, 2, and 3 are indicated by the arrows to the left of the gel.
In addition to complexes 1, 2, and 3, additional complexes formed with nuclear extracts prepared from H166Q/H170Q mutant virus-infected cells (Fig. 4, lane 6). As shown in Fig. 3C and discussed again below, L4-22K expression with the H166Q/H170Q mutant virus was elevated compared to Ad5 WT, which may explain the formation of these additional complexes. Nuclear extracts from cells infected with Ad5 WT, C137S/C141S, or H166Q/H170Q were also analyzed for L4-22K and IVa2 protein expression, and the levels of both proteins were comparable to those observed with whole-cell extracts (data not shown). The additional complexes observed with the H166Q/H170Q mutant contained the L4-22K protein, as observed by supershifts when antibody against L4-22K protein was added (lane 7). Biochemical studies have provided evidence that L4-22K binding promotes cooperative assembly of IVa2 onto packaging sequences, increasing affinity for the loading of additional IVa2 monomers to complexes with an existing IVa2 and L4-22K monomer already present (33, 34). To determine if the observed complexes were due solely to elevated amounts of L4-22K protein, we normalized the levels of nuclear extract from H166Q/H170Q virus-infected cells to protein levels from Ad5 WT-infected nuclear extracts (Ad5 WT L4-22K protein levels corresponded to 0.2× of H166Q/H170Q mutant L4-22K protein levels [data not shown]). To supplement IVa2 in the reaction mixture, we added the remaining amount of nuclear extract (0.8× of the total amount of nuclear extract) from Δ22K virus-infected cells. As shown in lane 8, complexes 1, 2, and 3 formed, and the upper complexes were reduced in intensity; the addition of an antibody against L4-22K resulted in a supershift of complexes 2 and 3 (lane 9). We conclude that the conserved cysteine and histidine residues in the C-terminal region of the L4-22K protein are not critical for the formation of protein-DNA complexes on the packaging sequences. We also found further evidence of the role of L4-22K in promoting cooperative assembly of protein-protein and protein-DNA interactions onto packaging sequences, as observed by the additional complexes formed with elevated levels of L4-22K protein beyond the normal levels observed during a WT infection.
L4-22K regulates Ad late gene expression and acts via a mechanism different from that of its role in viral genome packaging.
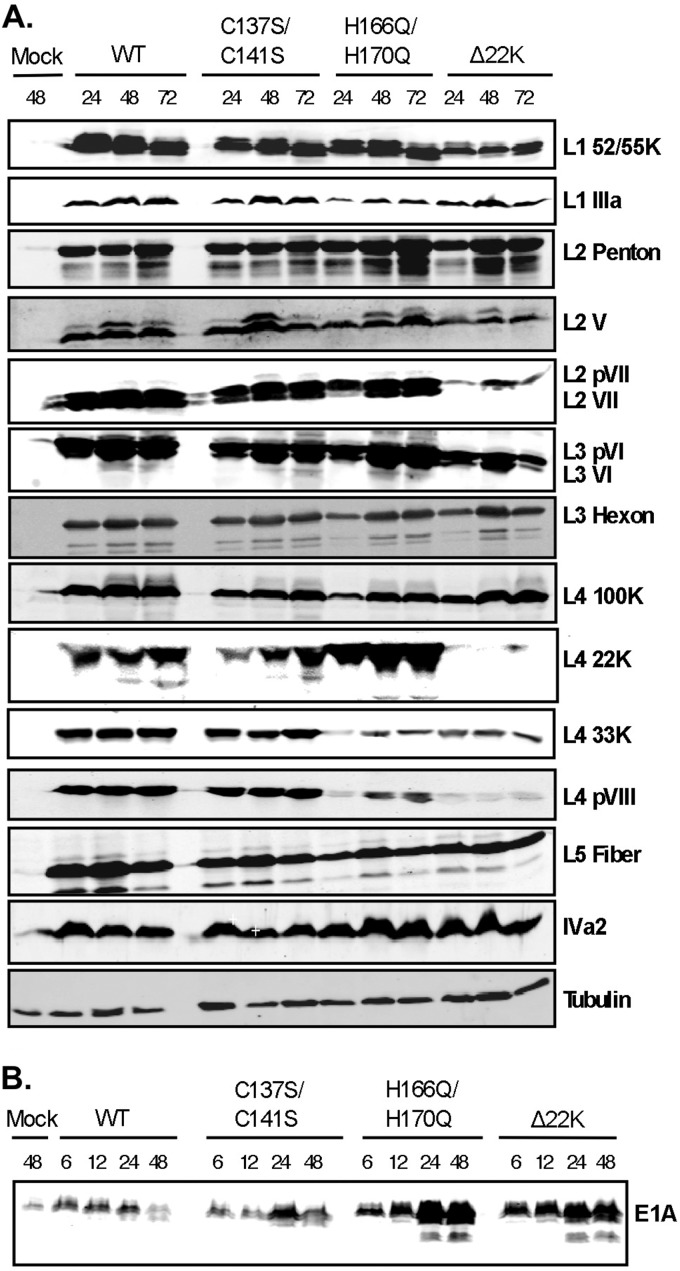
To examine if viral gene expression was affected with the L4-22K mutant viruses, N52.E6-Cre cells were infected with viruses Ad5 WT, C137S/C141S, H166Q/H170Q, or Δ22K, and whole-cell extracts were prepared at different times after infection and analyzed by Western blotting for levels of different viral proteins. Late gene expression (regions L1 to L5) with the C137S/C141S mutant was comparable to that of WT (Fig. 5A). Therefore, the defect in production of infectious progeny shown in Fig. 3 with the C137S/C141S mutant cannot be attributed to decreases in late gene products required for virus assembly and DNA packaging. Consistent with previous data (12), minor capsid proteins IIIa, VI, and VIII, core proteins V and VII, and the viral splicing factor L4-33K exhibited a decrease in expression with the Δ22K mutant compared to Ad5 WT (Fig. 5A). All other products were only modestly delayed (hexon, penton, and fiber) or not affected (L4-100K). Analysis of late proteins expressed with the H166Q/H170Q mutant showed that, similar to the Δ22K mutant, proteins IIIa, L4-33K, and VIII were decreased, while the levels of other Ad late proteins were only modestly affected. The level of L4-22K protein expression was increased with the H166Q/H170Q mutant compared to Ad5 WT and the C137S/C141S mutant. Intermediate protein IVa2 was also analyzed, and no significant changes were observed with the mutant viruses. Previous studies described a role for L4-22K, in addition to regulating late gene expression, in suppressing early gene expression (12). To determine if the conserved cysteine and histidine residues in L4-22K are key to this function, we analyzed E1A expression with the C137S/C141S and H166Q/H170Q mutant viruses and compared the results to those with Ad5 WT and the Δ22K mutant (Fig. 5B). Both Δ22K and H166Q/H170Q mutant viruses had elevated E1A expression levels compared to both Ad5 WT and the C137S/C141S mutant as early as 6 hpi. These results indicated that the L4-22K conserved cysteine residues are not important for suppression of early gene expression but suggested the possibility that the conserved histidine residues may be involved in this function.
Fig 5.
Western blot analyses of viral gene expression. (A) N52.E6-Cre cells were infected with WT, C137S/C141S, H166Q/H170Q, or Δ22K or mock infected, and whole-cell extracts were isolated at 6, 12, 24, 48, or 72 hpi as indicated at the top. Late proteins from regions L1 to L5 and intermediate protein IVa2 (A) and early region E1A protein (B) are shown with designations on the right. Tubulin was used as the loading control.
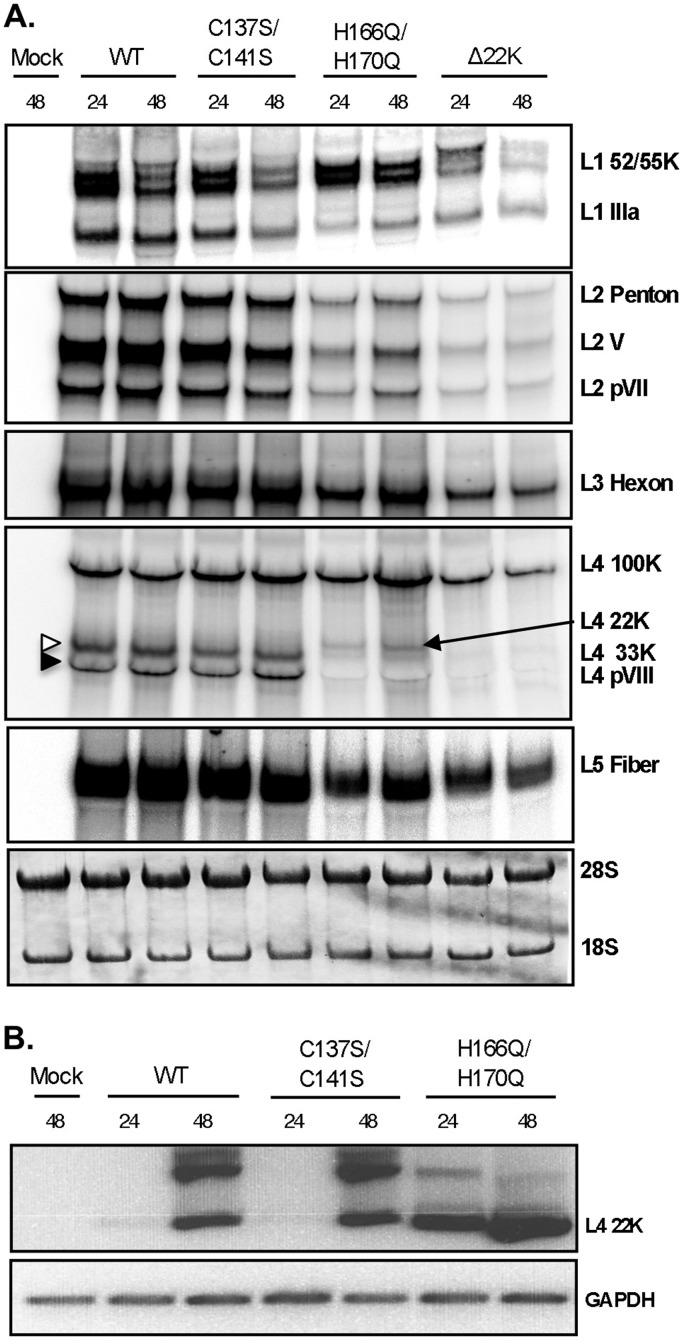
Previous studies presented evidence that L4-22K regulates gene expression and drives the early-to-late phase switch during Ad infection by acting at the level of late gene mRNA production and/or stability (12, 13). To determine whether the observed delays and decreases in late gene expression are due to L4-22K exerting its posttranscriptional effect at the level of protein or mRNA production, Northern blot analyses were performed to measure levels of Ad late transcripts (Fig. 6A). Consistent with the Western blot analyses, late mRNA levels were not affected with the C137S/C141S mutant compared to Ad5 WT. The mRNA levels for most of the major and minor capsid proteins were decreased with the H166Q/H170Q and Δ22K mutants, suggesting that the corresponding reduction in late protein expression (Fig. 5A) is at the level of mRNA production and/or stability. Note that this was not a global effect and that the posttranscriptional role of L4-22K is selective for a particular set of mRNAs. The reduction in IIIa mRNA levels is consistent with the decreased levels of L4-33K protein, since previous studies have shown that L4-33K regulates alternative splicing of the L1 unit by activating the IIIa splice site during late times of infection (5, 10, 15). Since the levels of L4-22K mRNA were undetectable for Ad5 WT and the C137S/C141S mutant by Northern blotting, we used RT-PCR to measure L4-22K mRNA levels (Fig. 6B). Consistent with the levels of protein expression (Fig. 5A), elevated levels of L4-22K mRNA were observed with the H166Q/H170Q mutant as early as 24 hpi, compared to Ad5 WT and the C137S/C141S mutant. L4-22K mRNA levels for the H166Q/H170Q mutant were elevated enough to be visible by Northern blotting (Fig. 6A, arrow).
Fig 6.
Analysis of Ad late mRNA levels. N52.E6-Cre cells were infected with Ad5 WT, C137S/C141S, H166Q/H170Q, or Δ22K, or mock infected, and total cytoplasmic mRNAs were isolated at 24 and 48 hpi. (A) Cytoplasmic RNA was analyzed by Northern blotting using 32P-labeled probes designed around the coterminal 3′ ends of each mRNA family (L1 to L5) with the designated mRNAs labeled to the right. The blot was stained with methylene blue as a loading control (bottom panel). (B) L4-22K mRNA was detected by RT-PCR using a forward primer specific for tripartite leader 3 and reverse primer directed to nucleotides 26682-26703 (corresponding to the intron of L4-33K). Equivalent amounts of cDNA derived from the reverse transcriptase reaction mixture were used in a PCR mixture with primers directed against GAPDH (bottom panel).
We conclude that the observed changes in protein levels with the L4-22K mutants are reproduced at the mRNA level, confirming a posttranscriptional mechanism for L4-22K in regulating viral gene expression. In addition, the C137S/C141S mutations in the L4-22K protein had no effect on gene expression, suggesting that the L4-22K protein exploits a mechanism to exert its posttranscriptional role that is distinct from that of genome packaging. Finally, we found that the H166Q/H170Q mutant had a similar phenotype with respect to mRNA production and changes in protein expression as an L4-22K null mutant.
The L4-22K protein acts at the level of processing of the L4-33K mRNA.
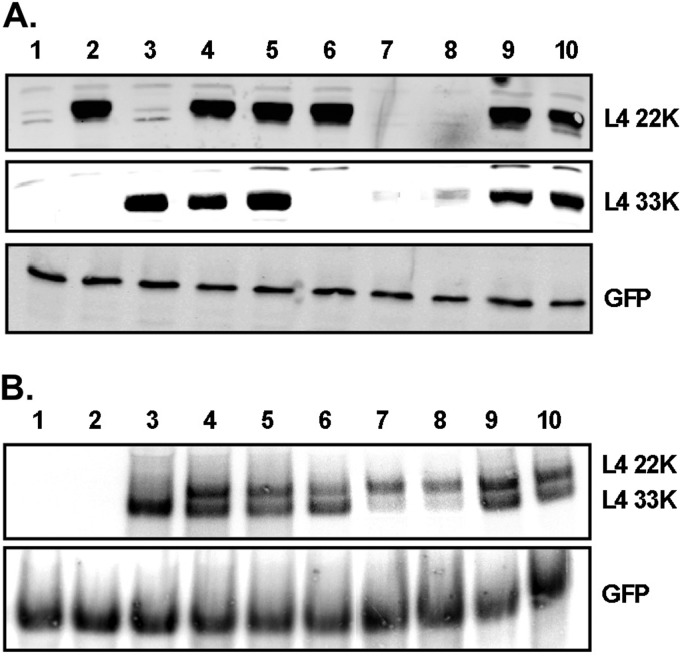
To investigate whether the posttranscriptional role of L4-22K is at the level of alternative mRNA splicing, we analyzed the mRNA and protein levels of L4-33K in an in vivo splicing assay with and without L4-22K protein expression (Fig. 7). N52.E6-Cre cells were cotransfected with plasmids that expressed GFP and WT or mutant L4 proteins, protein levels were analyzed by Western blotting, and mRNA levels were analyzed by Northern blotting. To compensate for any differences in transfection efficiencies, all samples were normalized to GFP mRNA and GFP protein levels. Expression plasmids pcDNA3-L4-22K and pcDNA3-L4-33K contain the coding sequences for the L4-22K and L4-33K proteins, respectively (Fig. 7A and B, lanes 2 and 3). Note that the mRNA transcribed with transfection of pcDNA3-L4-22K was not visible with the probe used in the Northern blot assay (Ad5 nt 26769-27590) (Fig. 7B, lane 2). Plasmid pcDNA3-L4 contains the genomic sequences encoding both L4-22K and L4-33K proteins, including the intron interrupting the protein reading frame of L4-33K (Fig. 1A); hence, production of the coding L4-33K mRNA requires splicing of the initial transcript (Fig. 7B, lane 4). When this plasmid was transfected into N52.E6-Cre cells, both L4-22K and L4-33K proteins were expressed to similar levels (Fig. 7A, lane 4). Plasmids pcDNA3-L4-22K− and pcDNA3-L4-33K− have a stop codon in the L4-22K and L4-33K reading frames, respectively, in the background of the pcDNA3-L4 plasmid, and the L4 pre-mRNA is still transcribed regardless of protein expression. Transfection of pcDNA3-L4-33K− resulted in normal expression levels of the L4-22K protein, while L4-33K was not expressed (Fig. 7A, lane 6). Transfection of pcDNA3-L4-22K− resulted in a significant decrease in L4-33K protein expression (Fig. 7A, compare lanes 4 and 7) due to inefficient splicing of the L4-33K pre-mRNA (Fig. 7B, compare lanes 4 and 7).
Fig 7.
In vivo splicing assay for the L4-33K pre-mRNA. N52.E6-Cre cells were cotransfected with expression plasmids for WT or mutant L4 proteins and an expression plasmid for GFP and harvested 48 h after transfection. Cells were used for Western blot analysis (A) and Northern blot analysis (B); all samples were normalized to GFP protein or mRNA levels. Lane 1, pcDNA3; lane 2, pcDNA3-22K; lane 3, pcDNA3-33K; lane 4, pcDNA3-L4; lane 5, pcDNA3-L4-C137S/C141S; lane 6, pcDNA3-L4-33K−; lane 7, pcDNA3-L4-22K−; lane 8, pcDNA3-L4-22K− cotransfected with pcDNA3 empty vector; lane 9, pcDNA3-L4-22K− cotransfected with pcDNA3-22K; lane 10, pcDNA3-L4-22K− cotransfected with pcDNA3-22K-C137S/C141S.
To further probe if the L4-22K protein is required for efficient splicing of the L4-33K transcript, we cotransfected equal amounts of pcDNA3-L4-22K− and pcDNA3-L4-22K and analyzed protein expression and mRNA levels. Expression of L4-33K was complemented when the L4-22K protein was expressed in trans (Fig. 7A, lane 9), consistent with restoration of L4-33K pre-mRNA splicing (Fig. 7B, lane 9). This result was not observed when pcDNA3-L4-22K− was cotransfected with empty vector pcDNA3 (Fig. 7A and B, lane 8), demonstrating that L4-22K, and not any other sequence in the vector background, is required for efficient splicing of the L4-33K mRNA. The results from Fig. 5 and 6 suggest that the conserved cysteine residues in the C terminus of L4-22K are not required for the regulation of viral gene expression, and so we analyzed if substitution mutations of these residues had any effect on the in vivo splicing of L4-33K mRNA. Transfection of plasmid pcDNA3-L4-C137S/C141S had no effect on the protein or mRNA levels of either the L4-22K or L4-33K genes (Fig. 7A and B, lane 5). Coexpression of plasmid pcDNA3-L4-C137S/C141S with plasmid pDNA3-L4-22K− complemented the L4-22K plasmid to restore L4-33K protein expression, consistent with an increase in L4-33K pre-mRNA accumulation (Fig. 7A and B, lane 10).
We conclude that the posttranscriptional effects observed on specific Ad mRNAs when functional L4-22K is not present during infection (Fig. 6) can be attributed to a role of the L4-22K protein in regulating alternative splicing of specific transcripts. The presence of full-length L4-22K protein in an in vivo splicing assay resulted in an increase in L4-33K mRNA accumulation and a subsequent increase in L4-33K protein levels. This effect was observed even with the C137S/C141S L4-22K mutant, suggesting that the conserved cysteine residues are not essential for the splicing activity of L4-22K.
DISCUSSION
We previously demonstrated that the Ad5 L4-22K protein is an integral component at key steps in the viral infectious cycle. First, L4-22K binds to Ad packaging sequences in vitro and in vivo and is essential for viral genome packaging and, therefore, for the production of infectious progeny (11, 12). Additionally, L4-22K has a distinct function in the temporal control of viral gene expression by activating MLP transcription (7), and more importantly, by regulating the accumulation of specific Ad late transcripts and suppressing early gene expression (12, 13). To further explore the functions of L4-22K, we generated and characterized two new L4-22K mutant viruses containing point mutations in conserved pairs of cysteine and histidine residues in the C-terminal region of L4-22K (C137S/C141S and H166Q/H170Q) (Fig. 1). We proposed that the primary sequence and configuration of these amino acids was consistent with a DNA binding motif and speculated that the DNA binding activity of L4-22K to the packaging sequences may be mediated by this putative zinc finger motif. Our results showed that the conserved cysteine residues are essential for efficient generation of infectious progeny, while no changes in gene expression patterns were observed with the C137S/C141S mutant virus. The H166Q/H170Q mutant recapitulated the phenotype of an L4-22K null mutant virus, suggesting an important role for these histidine residues in both Ad genome packaging and viral gene expression, although the possibility of an indirect effect on L4 mRNA splicing cannot be ruled out (discussed below). Both mutant proteins bound to packaging sequences in vitro, suggesting that the tandem CXXXC and HXXXH motifs do not comprise a protein domain critical for DNA binding. Our results confirm in vivo the essential role of L4-22K as a posttranscriptional regulator of late gene expression of specific Ad mRNAs with the primary targets the L4-33K and L4-VIII mRNAs and proteins. Our findings show that the cysteine mutations did not affect the ability of L4-22K to regulate gene expression (early or late) at the mRNA or protein levels (Fig. 5 and 6). These results uncouple the functions of L4-22K in regulating viral gene expression and genome packaging and suggest that the conserved cysteine residues are unique to the molecular mechanism behind Ad genome packaging. Finally, we confirmed the posttranscriptional effect of L4-22K on L4-33K expression and revealed that this occurs at the level of mRNA splicing.
Both the C137S/C141S and H166Q/H170Q mutant viruses had infectivity and viral genome replication comparable to Ad5 WT (Fig. 2A and B), demonstrating that events leading up to the late phase of infection do not account for the observed effects on late protein production and viral genome packaging. With respect to plaque formation, the C137S/C141S mutant virus behaved similarly to WT, while the H166Q/H170Q mutant virus had a severe defect in plaque formation on A549 cells (Fig. 2C). The apparent discrepancy in the results with the H166Q/H170Q mutant virus likely reflects the different assays used. The FFU assay (Fig. 2B) is a direct measure of how many cells are infected by the virus at an earlier time of infection (18 hpi), while the plaque assay (Fig. 2C) is a measure of virus production and spread, a late event during the infectious cycle. With the H166Q/H170Q mutant virus, infectivity (the particle/FFU ratio) was similar comparable to WT, consistent with the DNA replication data. However, the H166Q/H170Q mutant virus had a significant defect in virus spread. When supplemented with L4-22K in trans, plaque formation with the H166Q/H170Q mutant virus was partially restored (Fig. 2D). A role for L4-22K in Ad-induced cell death by regulating expression of ADP was previously proposed (12). ADP is known to facilitate cell lysis, and its deletion results in a small plaque phenotype that is due to impaired virus release (3, 35). Western blot analysis showed that ADP protein levels were significantly reduced with the H166Q/H170Q mutant and comparable to the low levels observed with L4-Δ22K (data not shown). This may explain the severe defect in plaque formation with the H166Q/H170Q mutant in noncomplementing cells. Additionally, the fact that we did not observe complete rescue in plaque formation when L4-22K was supplemented in trans, whereas the L4-Δ22K null mutant was rescued to WT levels, suggests that the H166Q/H170Q mutant protein may act as a dominant negative effector (discussed below).
The conserved cysteine residues of L4-22K are important for virus growth and the production of mature virions (Fig. 3A and D). The infectious virus yield and mature particle production with the C137S/C141S mutant was restored when the L4-22K protein was supplemented in trans (Fig. 3B and D). To determine if the reduction in genome packaging in vivo correlated with a defect in binding of the C137S/C141S mutant protein to packaging sequences, we analyzed DNA binding in vitro by using a gel mobility shift assay (Fig. 4). The data showed that the conserved cysteine residues were not critical for binding of the L4-22K protein to the packaging sequences in vitro, indicating that these amino acids are important for a different function of the protein in viral genome packaging. The conserved histidine residues also were not required for DNA binding, indicating that it is highly unlikely that the cysteine and histidine residues are part of a zinc finger motif involved in binding of L4-22K to the packaging domain. A role for L4-22K as a transcription factor stimulating major late transcription through direct binding to DNA sequences located downstream of the MLP start site was previously proposed (7, 11, 13). We cannot rule out the possibility that the conserved cysteine and histidine residues are part of a DNA binding motif necessary for L4-22K to bind to the DE and activate the MLP. However, this seems unlikely, since the C137S/C141S mutations had no effect on expression of genes transcribed from the major late promoter (Fig. 5). The possibility that mutation of the conserved cysteine residues leads to a disruption in the overall protein folding also seems unlikely, since there was no observed phenotype with respect to mRNA processing and viral protein expression (Fig. 5 and 6). However, we cannot exclude the idea that the cysteine residues could be involved in a disulfide bond formation. Additionally, zinc finger motifs also mediate protein-protein interactions in addition to protein-nucleic acid interactions (32). Although no published data exist for any viral or cellular factors that directly interact with the L4-22K protein, this possibility is an open question.
The phenotype of the H166Q/H170Q mutant virus suggests that the mutations severely disrupt the major functions of the L4-22K protein. Infectious virus yield and the production of mature virions were impaired (Fig. 3 and data not shown). Alterations in both early and late gene expression were also observed with similar targets and levels as a nonfunctional L4-22K mutant, and the observed results correlated well with mRNA levels (Fig. 5 and 6). Of significance, the mRNA and protein expression levels of L4-33K and L4-pVIII were decreased with both the H166Q/H170Q and Δ22K mutant viruses, with a large increase in mRNA and protein levels of L4-22K with the H166Q/H170Q mutant virus as early as 24 hpi, when mRNA levels were undetectable with WT and the C137S/C141S mutant (Fig. 5 and 6). The increase of L4-22K transcripts inversely correlated with the decreased levels of L4-33K mRNA (Fig. 6). Both transcripts are posttranscriptionally processed from the same pre-mRNA, but the L4-33K mRNA contains an additional splice of its internal intron (Fig. 1A). If the additional splicing event required for appropriate translation of the L4-33K protein was disrupted, this would increase the pool of L4-22K mRNAs. Our data demonstrated that L4-22K regulates splicing of the L4-33K pre-mRNA (Fig. 7), and we suggest that the conserved histidine residues are critical for this activity. However, we cannot rule out the possibility that the proximity of the two point mutations introduced to change the histidine residues interfered with the function of the internal splice acceptor site of L4-33K to affect the processing of the L4-33K pre-mRNA. In order to examine the contribution of reduced L4-33K gene expression to the H166Q/H170Q mutant virus phenotype, we investigated if virus yield was altered when L4-33K was supplemented in trans. The results showed that this did not augment overall virus yield or mature virion production (Fig. 3B and data not shown), although both L4-33K and L4-22K protein expression levels were comparable to WT (Fig. 3C). Interestingly, supplementation of L4-22K to H166Q/H170Q mutant infections also had no significant effect on virus yield or mature virion production. This result could be due to a dominant negative effect of the H166Q/H170Q mutant protein on the activity of WT L4-22K and correlates with the inability of L4-22K to restore plaque formation with the H166Q/H170Q mutant (Fig. 2D).
Morris and Leppard showed that the L4-22K protein is important for the temporal control of viral gene expression (13). Consistent with this report, we recently showed that L4-22K activates Ad late gene expression and suppresses Ad early gene expression and is involved in the transition from the early to late stages of infection (12). Our results reveal that in vivo one mechanism by which the L4-22K protein influences late gene expression is at the level of pre-mRNA processing (Fig. 7). In the absence of functional L4-22K protein, the levels of L4-33K protein expression were significantly decreased, which correlated with a decrease in L4-33K mRNA levels (Fig. 7). When L4-22K was supplied by transient transfection, we observed an increase in L4-33K mRNA accumulation and a corresponding increase in L4-33K protein levels (Fig. 7). Our data demonstrate that L4-22K indeed functions at the level of pre-mRNA splicing and activates splicing of the internal intron of L4-33K. The role of L4-22K as a regulator of pre-mRNA splicing, as opposed to other posttranscriptional mechanisms, is not surprising given that the L4-22K mRNA is nearly identical to the L4-33K mRNA, with the exception of the internal intron of L4-33K. The L4-33K protein is an RNA splicing factor that preferentially activates L1-IIIa splicing (10). We recently showed that L4-33K is required for the accumulation of L1-IIIa and pVI, but other late mRNAs are not primary targets of L4-33K in regulating late gene expression (37). It is therefore likely that L4-33K is not the only viral factor acting as an activator of pre-mRNA splicing and that L4-22K is a strong candidate due to shared sequences between the two proteins that may be critical for their function as mRNA splicing activators. It is interesting that the splice acceptor site for the second exon of L4-33K is common to the acceptor site of the pVIII gene, as previously published (36) and as we confirmed by RT-PCR and nucleotide sequence analysis (data not shown). L4-pVIII is the other gene that was significantly decreased both at the mRNA and protein level with H166Q/H170Q and Δ22K mutant virus infections (Fig. 5 and 6). We anticipate that expression of L4-22K in an in vivo splicing assay would stimulate pVIII mRNA accumulation.
It is interesting that L4-22K mRNA levels were almost undetectable for WT infection compared to the other highly abundant Ad late mRNAs (Fig. 6). By using an antibody against the common regions of L4-22K and L4-33K proteins, we detected significantly larger amounts of L4-33K protein compared to L4-22K protein in WT infection, and these proportionally corresponded to the mRNA levels (unpublished data). These observations suggest that although L4-22K is multifunctional and critical for Ad infection, it is not found in abundance compared to the total pool of Ad late viral proteins. We propose a feedback loop, wherein L4-22K regulates accumulation of different L4 transcripts, including its own, thereby regulating the levels of L4 gene products. As L4-22K protein levels increase, L4-33K pre-mRNA splicing will be stimulated, which would effectively reduce the accumulation of L4-22K mRNAs. Since L4-33K itself regulates Ad late mRNA splicing (5, 10, 15), this fine-tunes the temporal switch of gene expression patterns during the late phase of infection. Finally, the results of gel mobility shift assays with extracts prepared from H166Q/H170Q mutant-infected cells demonstrated that overexpression of L4-22K alters the binding of packaging proteins to the packaging sequences (Fig. 4). We speculate that L4-22K protein levels are low and precisely regulated to promote the appropriate formation of a packaging assembly complex on the viral packaging domain.
ACKNOWLEDGMENTS
We thank our colleagues, including Gudrin Schiedner, Stefan Kochanek, Arnold Levine, Carl Anderson, Daniel Engel, David Matthews, Christopher Wiethoff, Maxim Balakirev, Ann Tollefson, and William Wold, for providing important reagents used in this study. We thank current laboratory members and Kai Wu for informative discussions throughout the project. We thank Mary Anderson and Ilana Shoshani for excellent technical assistance.
This work was supported by NIH grant AI041636. D.G. was supported by a SUNY Doctoral Diversity Fellowship, a W. Burghardt Turner Fellowship, and NIH training grant T32AI007539 from the National Institute of Allergy and Infectious Diseases.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print 1 May 2013
REFERENCES
- 1. Berk AJ. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685 [DOI] [PubMed] [Google Scholar]
- 2. Evans JD, Hearing P. 2002. Adenovirus replication, p 39–70 In Curiel DT, Douglass JT. (ed), Adenoviral vectors for gene therapy. Elsevier, New York, NY [Google Scholar]
- 3. Lichtenstein DL, Toth K, Doronin K, Tollefson AE, Wold WSM. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 23:75–111 [DOI] [PubMed] [Google Scholar]
- 4. Weitzman MD, Ornelles DA. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686–7696 [DOI] [PubMed] [Google Scholar]
- 5. Akusjärvi G. 2008. Temporal regulation of adenovirus major late alternative RNA splicing. Front. Biosci. 13:5006–5015 [DOI] [PubMed] [Google Scholar]
- 6. Young CSY. 2003. The structure and function of the adenovirus major late promoter. Curr. Topics Microbiol. Immunol. 272:213–249 [DOI] [PubMed] [Google Scholar]
- 7. Backström E, B KK, Lan X, Akusjarvi G. 2010. Adenovirus L4-22K stimulates major late transcription by a mechanism requiring the intragenic late-specific transcription factor-binding site. Virus Res. 151:220–228 [DOI] [PubMed] [Google Scholar]
- 8. Hayes BW, Telling GC, Myat MM, Williams JF, Flint SJ. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xi Q, Cuesta R, Schneider RJ. 2005. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k viral mRNAs. J. Virol. 79:5676–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Törmänen H, Backström E, Carlsson A, Akusjärvi G. 2006. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 281:36510–36517 [DOI] [PubMed] [Google Scholar]
- 11. Ostapchuk P, Anderson ME, Chandrasekhar S, Hearing P. 2006. The L4-22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu K, Orozco D, Hearing P. 2012. The adenovirus L4-22K protein is multifunctional and is an integral component of crucial aspects of infection. J. Virol. 86:10474–10483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morris SJ, Leppard KN. 2009. Adenovirus serotype 5 L4-22K and L4-33K proteins have distinct functions in regulating late gene expression. J. Virol. 83:3049–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsson S, Svensson C, Akusjärvi G. 1992. Control of adenovirus major late gene expression at multiple levels. J. Mol. Biol. 225:287–298 [DOI] [PubMed] [Google Scholar]
- 15. Farley DC, Brown JL, Leppard KN. 2004. Activation of the early-late switch in adenovirus type 5 major late transcription unit expression by L4 gene products. J. Virol. 78:1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris SJ, Scott GE, Leppard KN. 2010. Adenovirus late-phase infection is controlled by a novel L4 promoter. J. Virol. 84:7096–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang W, Flint SJ. 1998. The tripartite leader sequence of subgroup C adenovirus major late mRNAs can increase the efficiency of mRNA export. J. Virol. 72:225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Logan J, Shenk T. 1984. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc. Natl. Acad. Sci. U. S. A. 81:3655–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ostapchuk P, Hearing P. 2005. Control of adenovirus packaging. J. Cell. Biochem. 96:25–35 [DOI] [PubMed] [Google Scholar]
- 20. Ewing SG, Byrd SA, Christensen JB, Tyler RE, Imperiale M. 2007. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J. Virol. 81:12450–12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang W, Imperiale M. 2000. interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ostapchuk P, Almond M, Hearing P. 2011. Characterization of empty adenovirus particles assembled in the absence of a functional adenovirus IVa2 protein. J. Virol. 85:5524–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gustin KE, Imperiale M. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma H-C, Hearing P. 2011. Adenovirus structural protein IIIa is involved in the serotype specificity of viral DNA packaging. J. Virol. 85:7849–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schiedner G, Hertel S, Kochanek S. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105–2116 [DOI] [PubMed] [Google Scholar]
- 26. Evans J, Hearing P. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replicaiton and inhibition of genome concatenation. J. Virol. 77:5295–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maizel JV, White DO, Scharff MD. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115–125 [DOI] [PubMed] [Google Scholar]
- 29. Brown T, Mackey K, Du T. 2004. Analysis of RNA by Northern and slot blot hybridization. Curr. Protoc. Microbiol. 4:1–19 [DOI] [PubMed] [Google Scholar]
- 30. Larkin M, Blackshields G, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm F, Lopez R, Thompson J, Gibson T, Higgins D. 2007. ClustalW and ClustalX verison 2. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 31. Krishna SS, Majumdar I, Grishin NV. 2003. Structural classification of zinc fingers. Nucleic Acids Res. 31:532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brayer KJ, Segal DJ. 2008. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell. Biochem. Biophys. 50:111–131 [DOI] [PubMed] [Google Scholar]
- 33. Yang T-C, Maluf NK. 2010. Self-association of the adenoviral L4-22K protein. Biochemistry 49:9830–9838 [DOI] [PubMed] [Google Scholar]
- 34. Yang T-C, Maluf MN. 2012. Cooperative heteroassembly of the adenoviral L4-22K and IVa2 proteins onto the viral packaging sequence DNA. Biochemistry 51:1357–1368 [DOI] [PubMed] [Google Scholar]
- 35. Tollefson AE, Scaria A, Hermiston Tw, Ryerse JS, Wold LJ, Wold WSM. 1996. The adenovirus death protein (E3-11.6k) is required at very late stages of infection for efficient cell lysis and release of adenvirus from infected cells. J. Virol. 70:2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vrati S, Boyle D, Kocherhans R, Both GW. 1995. Sequence of ovine adenovirus homologs for 100K hexon assembly, 33K, pVIII, and fiber genes: early region E3 is not in the expected location. Virology 209:400–408 [DOI] [PubMed] [Google Scholar]
- 37. Wu K, Guimet D, Hearing P. 3 April 2013. The adenovirus L4-33K protein regulates both late gene expression patterns and viral DNA packaging. J. Virol. 10.1128/JVI.00652-13 [DOI] [PMC free article] [PubMed] [Google Scholar]