Abstract
The RV144 HIV-1 vaccine trial (Thailand, 2003 to 2009), using immunogens genetically matched to the regional epidemic, demonstrated the first evidence of efficacy for an HIV-1 vaccine. Here we studied the molecular evolution of the HIV-1 epidemic from the time of immunogen selection to the execution of the efficacy trial. We studied HIV-1 genetic diversity among 390 volunteers who were deferred from enrollment in RV144 due to preexisting HIV-1 infection using a multiregion hybridization assay, full-genome sequencing, and phylogenetic analyses. The subtype distribution was 91.7% CRF01_AE, 3.5% subtype B, 4.3% B/CRF01_AE recombinants, and 0.5% dual infections. CRF01_AE strains were 31% more diverse than the ones from the 1990s Thai epidemic. Sixty-nine percent of subtype B strains clustered with the cosmopolitan Western B strains. Ninety-three percent of B/CRF01_AE recombinants were unique; recombination breakpoint analysis showed that these strains were highly embedded within the larger network that integrates recombinants from East/Southeast Asia. Compared to Thai sequences from the early 1990s, the distance to the RV144 immunogens increased 52% to 68% for CRF01_AE Env immunogens and 12% to 29% for subtype B immunogens. Forty-three percent to 48% of CRF01_AE sequences differed from the sequence of the vaccine insert in Env variable region 2 positions 169 and 181, which were implicated in vaccine sieve effects in RV144. In conclusion, compared to the molecular picture at the early stages of vaccine development, our results show an overall increase in the genetic complexity of viruses in the Thai epidemic and in the distance to vaccine immunogens, which should be considered at the time of the analysis of the trial results.
INTRODUCTION
A major obstacle for the development of a safe and globally effective HIV vaccine is the extensive genetic diversity of the virus (1, 2). Most infections in the pandemic are due to HIV type 1 (HIV-1) group M, which is classified into nine subtypes (A to D, F to H, J, and K) differing by roughly 15% in their full-genome nucleotide sequence and up to 35% in the sequences of the gp120 envelope protein. Numerous recombinants have been characterized in the pandemic (3). Some of them have achieved wide dissemination (i.e., circulating recombinant forms [CRFs]), while others have been retrieved from only a single individual (i.e., unique recombinant forms [URFs]). HIV-1 subtypes and recombinants have distinct geographic distributions (4); for example, subtype B is the predominant strain in North America and Western Europe, while infections in South, East, and Southeast Asia are mostly due to subtypes B, C, and CRF01_AE and their recombinants.
Due to the early and continuous participation in the planning and execution of HIV-1 vaccine trials, the Thai epidemic is one of the best studied (5). The early 1990s saw a great expansion of the HIV-1 epidemic, with a nationwide seroprevalence among young military conscripts of 3.7%, reaching 12% among those from northern Thailand (6, 7). At that time, HIV-1 strains segregated on the basis of risk factors, with subtype B predominating among injecting drug users (IDUs) and CRF01_AE being the main clade among those with heterosexual exposure risk (8–12), though through ensuing years, CRF01_AE has come to predominate the epidemic among IDUs as well (13–21).
From the mid-1990s to the mid-2000s, a series of molecular processes (e.g., the expansion of the genetic diversity within circulating subtypes, shifts in the proportions of different molecular forms, the emergence of intersubtype recombinants, and the influx of new strains from other epidemics) played major roles in the evolution of this epidemic. First, it was noted that the within-subtype interhost genetic divergence had increased significantly compared to that noted in earlier surveys (9, 10, 17, 22–25). Second, the barriers that maintained the segregation of subtypes on the basis of risk factors started to efface; the representation of CRF01_AE among IDUs increased, especially among new infections (26), to the point where CRF01_AE accounted for 80 to 90% of the cases in Bangkok and the northern province of Chiang Mai (27, 28). Coinfections (29) and superinfections (30) with subtype B and CRF01_AE were documented, and intersubtype recombinants were detected among individuals with either IDU (28, 31) or heterosexual (32, 33) exposure. While most of the recombinants were URFs, a new CRF, CRF15_01B, was found throughout the country among individuals with either risk factor, reinforcing the linkage between the different epidemics in Thailand (31, 32). Finally, the connection of the Thai HIV-1 epidemic with other epidemics in the region, through travel and trafficking of drugs and persons, has been reflected in the genetic relatedness of Thai strains to those from other countries in the Golden Triangle (i.e., a major illicit opium-producing area spanning Myanmar [Burma], Vietnam, Laos, and Thailand) (34, 35) and China (36). Subtypes A, C, D, and CRF02_AG—which are more common in African epidemics—have been detected sporadically (37–39); however, to date there have been no indications that these strains have spread locally.
In preparation for the RV144 trial, community cohort surveys were conducted from 1998 to 2001 in Rayong and Chon Buri Provinces (i.e., the areas where the trial was later to be executed) and found that 90.5% of prevalent infections were due to CRF01_AE, 2.4% were due to subtype B, and 7.2% were due to recombinants (including CRF15_01B) or dual infections (40). Prospective volunteers for RV144 were enrolled in screening protocol RV148 to assess eligibility (41); those with preexisting HIV infection were deferred from enrollment in the RV144 trial and are the subject of the current study. This study attempts (i) to provide a molecular snapshot of the HIV-1 epidemic in eastern Thailand between 2003 and 2005, (ii) to correlate the viral genetic information in this cohort with the epidemiological characteristics of the volunteers for RV144, and (iii) to determine the genetic relationship between the immunogens incorporated in the vaccine and the viral strains that challenged the trial participants. The HIV-1 strains characterized in RV148 were drawn at the same time, from the same location, and from the same social networks as those in the RV144 trial, and they represent one of the closest descriptions we may have to the HIV-1 strains to which the RV144 participants might have been exposed during the trial. This study is complementary to the recent work of Rolland et al. (42) that describes a variable region 2 (V2)-delimited sieve analysis of HIV-1 strains infecting RV144 trial participants, as it describes viral strains that are the nearest possible approximations to the viral strains to which the placebo and vaccine recipients in RV144 might have been exposed. The thorough analysis of near-full-length RV144 breakthrough viruses is the focus of a separate report (M. Rolland, unpublished data).
MATERIALS AND METHODS
Population under study.
From September 2003 to December 2005, 26,676 volunteers in Rayong and Chon Buri Provinces in East Thailand who consented for the RV144 trial were assessed for eligibility in the setting of the screening protocol RV148. One hundred twenty-eight individuals were not referred for protocol-defined HIV testing because they met at least one exclusion criterion and/or failed to meet at least one inclusion criterion. Out of the 26,548 volunteers who were tested for HIV, 418 had HIV infection (see reference 41 for details on the testing reagents and algorithms), were counseled and referred to designated Ministry of Public Health (MOPH) hospitals for evaluation of the status of their infection, and were considered for standard treatment and care on the basis of the published Thai MOPH guidelines (43; see reference 41 for details on counseling). Samples from 390/418 individuals were available for HIV-1 genotyping. CD4+ T cell enumeration was performed using standard flow cytometry (FACSCalibur; Becton, Dickinson, Palo Alto, CA), and HIV-1 RNA quantification was performed using reverse transcription-PCR technology (Roche Amplicor version 1.5 test; Roche Diagnostics, Indianapolis, IN).
Ethical review.
The screening protocol RV148 was reviewed and approved by the ethical committees of the Thai MOPH, the Royal Thai Army, Mahidol University, and the Human Subjects Research Review Board of the U.S. Army Medical Research and Materiel Command. The study was conducted in accordance with U.S. and Thai FDA regulations as well as the International Conference on Harmonisation (ICH) technical requirements for registration of pharmaceuticals for human use guideline on good clinical practice (GCP) for clinical trials, guideline E6 (http://www.ich.org/products/guidelines/efficacy/article/efficacy-guidelines.html).
HIV-1 genotyping.
Viral RNA was extracted from serum samples by robotic extraction (MagnaPure; Roche Diagnostics), and the determination of HIV-1 subtype and recombinants was achieved through a multiregion hybridization assay (MHAbce) of the whole available sample set and near-full-genome amplification and sequencing of a subset of samples.
(i) MHAbce version 2.
The presence of infections with subtypes B, C, and CRF01_AE, and recombinants and dual infections combining these strains was evaluated using MHAbce version 2, as previously published (32). Briefly, 8 regions throughout the HIV-1 genome (p17 in gag; protease, reverse transcriptase, and integrase in pol; the first exon of tat; gp120 and gp41 in env; and nef) were reverse transcribed and PCR amplified with universal primers. Subsequently, each region was subject to real-time PCRs with respective fluorescent TaqMan probes specific for subtypes B, C, and CRF01_AE. Samples with concordant patterns of probe reactivity throughout the genome were assigned to the corresponding subtype or CRF, while those samples showing discordant patterns of probe reactivity were considered intersubtype recombinants. Patients with samples exhibiting dual-probe reactivity in the same genome region were considered dually infected after verification by cloning and real-time PCR subtyping of at least 20 clones per sample. Using typeability criteria determined at assay validation, only samples showing probe reactivity in 4 or more genome regions were considered typed (32).
(ii) Near-full-genome amplification and sequencing.
Near-full-genome HIV-1 sequences were obtained as either a complete HIV-1 genome or two half genomes overlapping by 1.5 kb, as previously described (44).
Phylogenetic analyses.
Alignments of RV148 near-full-genome HIV-1 sequences and subtype and CRF reference sequences (3) were obtained using the HIVAlign program (http://www.hiv.lanl.gov/content/sequence/VIRALIGN/viralign.html) and were manually edited with BioEdit version 7.0.9.0 software (45). RV148 sequences were assigned to the corresponding subtype or CRF through phylogenetic analyses of nucleotide sequences conducted in the MEGA5 program (46) using maximum likelihood trees; bootstrap values supporting relevant clusters were computed through the Kimura 2-parameter model (47) and neighbor-joining trees (48) with 1,000 iterations. The structure of recombinant forms was determined using the jumping profile hidden Markov model (jpHMM; http://jphmm.gobics.de/) (49) and was confirmed through subgenomic phylogenetic analysis. The location of all the recombination breakpoints is reported in the HXB2 coordinate system.
Nucleotide sequence diversity was determined through a maximum likelihood estimation using the general reversible model, as implemented in Hypothesis Testing Using Phylogenies (HyPhy) (50). The distance of the deduced protein sequences of RV148 immunogens and published sequences from East and Southeast Asia to RV144 immunogens was computed using the p-distance, Poisson (51), Jones-Taylor-Thornton (JTT) (52), Dayhoff (53), and HIV-Between (54) models implemented in MEGA5 and HyPhy. None of these models was specifically tailored to weigh the impact of amino acid substitution on immune recognition. For the current analysis, we selected the p-distance model, which is the one that makes the fewest a priori assumptions, as we found that it had a very strong linear correlation with the more complex models (range of r2 values, 0.9557 to 0.9999) (data not shown).
Network analysis.
The sharing of common recombination breakpoints between RV148 intersubtype recombinants and published HIV-1 strains from East and Southeast Asia was studied using the RecDraw program (55), allowing 27 bp of mapping imprecision (i.e., the average distance between the jpHMM-predicted and previously published breakpoints, as determined by Schultz et al. [49]). Network diagrams were built using the UCINET version 6.275/NetDraw version 2.091 (Analytic Technologies, Lexington, KY) and NodeXL (Social Media Research Foundation; http://www.smrfoundation.org) programs.
Statistics.
Differences in the distribution of gender, HIV-1 subtypes, recombinants, and dual infections across sociodemographic characteristics were assessed using the chi-square test and Fisher's exact test (if the expected cell count was less than 5) for categorical variables and through the Wilcoxon rank sum test for continuous variables; crude odds ratios and 95% confidence intervals (CIs) were calculated using JMP version 10.0 (SAS Institute, Cary, NC). JMP was also employed to assess differences in genetic diversity among time intervals and country of sampling using the Wilcoxon rank sum test and to assess the linear fit between genetic distances, computed by least-squares regression using different models. All of the presented P values are unadjusted for multiple comparisons.
Nucleotide sequence accession numbers.
The HIV-1 sequences obtained in this study have been deposited in GenBank under accession numbers JN248316 to JN248357.
RESULTS
Sociodemographic characteristics of the population under study.
In order to determine eligibility for enrollment in RV144, volunteers were first enrolled in the screening protocol RV148. Out of 26,548 volunteers who were tested for HIV, 418 individuals (1.57%) had a confirmed HIV infection and were deferred from enrollment in RV144 (see Materials and Methods) (56). The sociodemographic risk factors for HIV seropositivity have been previously reported (41) and included female sex, older age, lower education, widowed/divorced/separated marital status, and fisherman/sex worker/entertainment worker self-reported occupation.
Samples from 390/418 volunteers who were ineligible for RV144 because HIV infection was detected during screening were available for HIV-1 genotyping. Table 1 shows their sociodemographic and clinical characteristics. Among the seropositive volunteers, comparable numbers of female (n = 192) and male (n = 198) individuals were represented. Statistically significant differences in sociodemographic characteristics between male and female volunteers were observed. Male seropositive volunteers were enriched for transgender individuals (P < 0.001), volunteers who were single (P < 0.001), volunteers who had completed at least senior high school (P = 0.02), or volunteers who were office/factory workers (P = 0.02). Among females there was enrichment in volunteers who had not completed primary school (P = 0.03) or who were married (P < 0.001), divorced (P < 0.001), entertainment workers (P = 0.04), or housewives (P < 0.001) (data not shown). The distribution of plasma viral load (pVL) and CD4 cell counts did not differ significantly between males and females (data not shown).
Table 1.
Sociodemographic and clinical characteristics of screened volunteers who were deferred from enrollment in the RV144 vaccine trial due to preexisting HIV infection
| Characteristic | Female (n = 192) | Male (n = 198) | Total (n = 390) |
|---|---|---|---|
| No. (%) of volunteers by gender | |||
| Female | 192 (100) | 10 (5.1) | 202 (51.8) |
| Male | 0 (0) | 187 (94.4) | 187 (48.0) |
| Missing | 0 (0) | 1 (0.5) | 1 (0.2) |
| Age (yr) | |||
| Median | 25 | 26 | 26 |
| Interquartile interval | 22–28 | 23–27 | 22–28 |
| No. (%) of volunteers by the following characteristic: | |||
| Birthplace | |||
| Chon Buri | 44 (22.9) | 52 (26.3) | 96 (24.6) |
| Rayong | 45 (23.4) | 44 (22.2) | 89 (22.8) |
| Other | 103 (53.7) | 102 (51.5) | 205 (52.6) |
| Screening site | |||
| Chon Buri | |||
| Panthong | 8 (4.2) | 16 (8.1) | 24 (6.2) |
| Sri Racha | 11 (5.7) | 17 (8.6) | 28 (7.2) |
| Bang Lamung | 48 (25.0) | 54 (27.3) | 102 (26.1) |
| Sattahip | 30 (15.6) | 23 (11.6) | 53 (13.6) |
| Rayong | |||
| Ban Chang | 12 (6.3) | 20 (10.1) | 32 (8.2) |
| Mueang | 23 (12.0) | 29 (14.6) | 52 (13.3) |
| Ban Khai | 23 (12.0) | 19 (9.6) | 42 (10.8) |
| Klaeng | 37 (19.2) | 20 (10.1) | 57 (14.6) |
| Marital status | |||
| Single | 21 (10.9) | 97 (49.0) | 118 (30.2) |
| Married | 123 (64.2) | 89 (44.9) | 212 (54.4) |
| Separated | 2 (1.0) | 0 (0.0) | 2 (0.5) |
| Divorced | 40 (20.8) | 11 (5.6) | 51 (13.1) |
| Widowed | 6 (3.1) | 1 (0.5) | 7 (1.8) |
| Education | |||
| Did not complete primary school | 13 (6.8) | 4 (2.0) | 17 (4.3) |
| Primary school | 107 (55.8) | 99 (50.0) | 206 (52.9) |
| Junior high school | 54 (28.1) | 60 (30.3) | 114 (29.2) |
| Senior high school/vocational school | 15 (7.8) | 28 (14.1) | 43 (11.0) |
| Vocation school | 2 (1.0) | 5 (2.5) | 7 (1.8) |
| Bachelor's degree or higher | 1 (0.5) | 2 (1.0) | 3 (0.8) |
| Occupation | |||
| Laborer | 56 (29.1) | 62 (31.3) | 118 (30.2) |
| Office/factory | 35 (18.2) | 57 (28.8) | 92 (23.6) |
| Merchant/business | 15 (7.8) | 17 (8.6) | 32 (8.2) |
| Housewife | 26 (13.5) | 0 (0.0) | 26 (6.7) |
| Entertainment | 15 (7.8) | 6 (3.0) | 21 (5.4) |
| Agriculture | 8 (4.2) | 10 (5.1) | 18 (4.6) |
| CSW | 8 (4.1) | 9 (4.5) | 17 (4.4) |
| Fishery | 3 (1.6) | 5 (2.5) | 8 (2.0) |
| Government officer | 1 (0.5) | 4 (2.0) | 5 (1.3) |
| Military/police | 0 (0) | 4 (2.0) | 4 (1.0) |
| Student | 0 (0) | 3 (1.5) | 3 (0.8) |
| Other | 0 (0) | 2 (1.0) | 2 (0.5) |
| Unemployed | 25 (13.0) | 19 (9.6) | 44 (11.3) |
| Plasma viral load (no. of RNA copies/ml) | |||
| Median | 30,371 | 33,841.5 | 32,919 |
| Interquartile interval | 7,137–121,999 | 14,518.8–127,640 | 9,629–124,361 |
| No. (%) of patients with VLs of <400 copies/ml | 8 (4.1) | 6 (3.0) | 14 (3.6) |
| CD4+ cell count (no. of cells/μl) | |||
| Median | 401 | 397.5 | 399 |
| Interquartile interval | 260–568.75 | 276.75–583.25 | 272–577.5 |
| No. (%) of patients with CD4 counts of: | |||
| <200 cells/μl | 33 (17.1) | 27 (13.6) | 60 (15.3) |
| <350 cells/μl | 73 (37.8) | 77 (38.9) | 150 (38.4) |
HIV-1 subtype distribution among RV148 seropositive individuals.
Out of the 390 specimens tested with MHAbce, 375 samples (96.2%) showed probe reactivity in 4 or more regions, which was the preestablished typeability requirement (32) (see Materials and Methods). The main reason for untypeability was undetectable pVL (data not shown). Results from MHAbce showed a predominance of infections due to CRF01_AE (90.2%), followed by B/CRF01_AE recombinants (6.1%) and subtype B (3.2%) (Table 2). Two cases showed dual-probe reactivity for subtype B and CRF01_AE. Additionally, among the untyped samples, there was one specimen showing probe reactivity with subtype C probes in two regions and with CRF01_AE probes in one region. A near-full-genome sequence was retrieved, revealing that the sample corresponded to subtype C, clustering within the African radiation (data not shown).
Table 2.
HIV clade distribution among 375 volunteers deferred from enrollment in the RV144 vaccine trial due to preexisting HIV infection, genotyped by MHAbce and full-genome sequencing
| Clade | No. (%) of volunteers |
|
|---|---|---|
| MHAbce only | MHAbce with sequencing confirmation | |
| CRF01_AE | 338 (90.2) | 344 (91.7) |
| Subtype B | 12 (3.2) | 13 (3.5) |
| B/CRF01_AE recombinants | 23 (6.1) | 16 (4.3) |
| B/CRF01_AE dual infection | 2 (0.5) | 2 (0.5) |
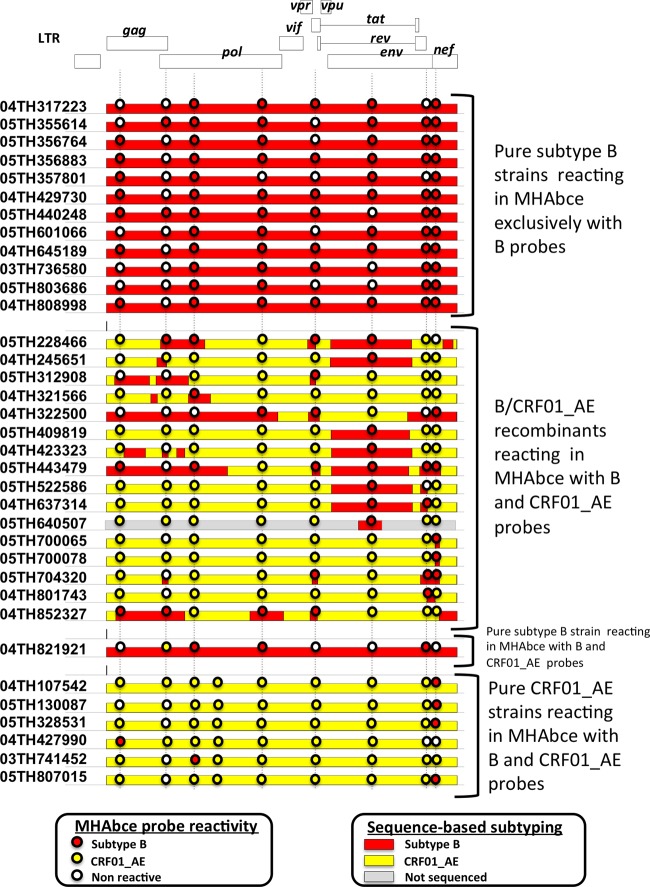
All typeable samples showing reactivity with subtype B probes in at least one region were subject to near-full-genome sequencing (Fig. 1). All 12 samples identified as pure subtype B by MHAbce showed a concordant subtype designation by near-full-genome sequencing. Out of the 23 samples identified as B/CRF01_AE recombinants by MHAbce, 16 were confirmed to be recombinants by sequencing, while 1 and 6 strains were determined by near-full-genome sequencing to be pure subtype B and CRF01_AE strains, respectively. Detailed inspection of near-full-genome sequences revealed that the main reason for heterotypic probe reactivity in MHAbce was the presence of infrequent polymorphisms in the probe binding sites, which had not been anticipated in the design of the assay (32).
Fig 1.
MHAbce reactivity pattern and genomic structure of typed samples showing reactivity with subtype B probes in one or more regions. Color-coded horizontal bars represent the genomic structure based on near-full-genome sequencing, and the circles depict the corresponding MHAbce probe reactivity. Vertical dotted lines indicate the genomic locations of the MHAbce probes. For sample 05TH640507, only a subgenomic sequence was retrieved (HXB2 residues 6991 to 7573). LTR, long terminal repeat.
The subtype distribution combining MHAbce- and near-full-genome sequencing-based genotyping is shown in Table 2. Overall, CRF01_AE strains accounted for 91.7% of the infections, and pure subtype B strains were only slightly outnumbered by B/CRF01_AE intersubtype recombinants (3.5% and 4.3%, respectively). While B/CRF01_AE dual infections were detected, they were very infrequent (0.5%).
Analysis of near-full-genome sequences. (i) Subtype B.
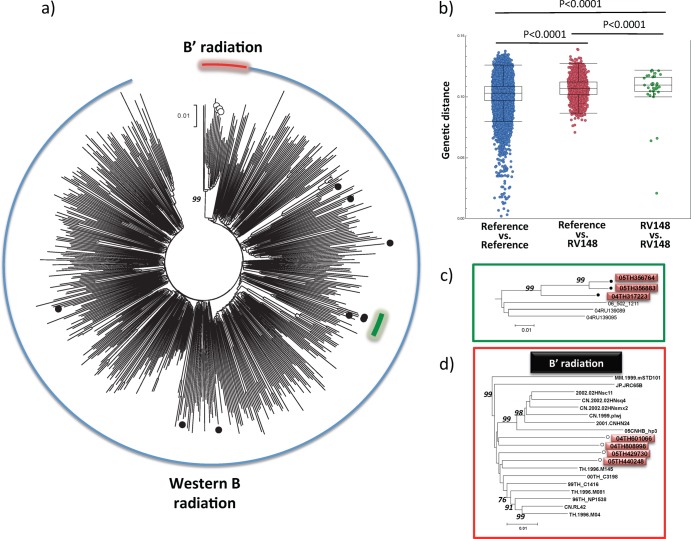
Sequences from the 13 subtype B samples were analyzed (Fig. 2). The subtype B strains circulating in Southeast Asia can be subclassified into two different clades on the basis of their genetic sequences and especially their motif at the crown of the V3 loop in gp120: sequences clustering with strains from the Americas and Western Europe carrying the GPGR motif in V3 (which are designated Western B throughout the text) and a separate cluster of sequences identified primarily in Southeast Asia and China and carrying the GPGQ motif in V3, referred to in the literature as B′ or Thai B (57, 58). In RV148, 9 sequences clustered with strains from the Western B radiation, while the remaining 4 sequences clustered within the B′ radiation (Fig. 2a). There were no significant differences in the distribution of sociodemographic characteristics between carriers of Western B and B′; however, there was a trend toward a higher prevalence of Western B strains in the Bang Lamung area of Chon Buri Province that borders on Pattaya (5/9 for Western B versus 0/4 for B′, P = 0.10), an international tourist destination.
Fig 2.
Location of RV148 subtype B strains in the global subtype B phylogeny. (a) Black circles depict RV148 B strains from the Western B radiation, and hollow circles depict RV148 strains from the B′ radiation. (b) Distribution of pairwise genetic distance within the cosmopolitan Western B epidemic (blue circles), among RV148 sequences from the Western B radiation (green circles), and between these two groups (red circles). This analysis was based on near-full-genome sequences from 344 cosmopolitan Western B strains that were sampled between 2000 and 2007. (c) Detail of a trio of RV148 strains with low genetic diversity (red boxes). (d) Subtree of the B′ radiation, where the RV148 strains are highlighted (red boxes). The phylogenetic trees were built using near-full-genome sequences. Numbers on the branches of the phylogenetic trees represent the percentage of bootstrap replicates supporting the relevant clusters.
The genetic diversity among RV148 Western B strains (median, 0.11; interquartile range [IQR], 0.105 to 0.116) was only slightly higher than the genetic diversity between RV148 and contemporaneous reference Western B strains (median, 0.107; IQR, 0.102 to 0.112; P < 0.0001) or among contemporaneous reference Western B strains (median, 0.103; IQR, 0.097 to 0.109; P < 0.0001) (Fig. 2b). Overall, these results support the hypothesis of multiple introductions of Western B strains in the area. The genetic diversity observed among RV148 B′ strains (median, 0.069; IQR, 0.068 to 0.070; data not shown) was significantly lower than the one observed among Western B RV148 strains (P = 0.001).
Among the Western B strains, there was a pair of sequences (05TH356764 and 05TH356883) that showed a high level of sequence identity (distance, 0.021) (Fig. 2c). They were both sampled in the same site 2 weeks apart. Neither the remaining Western B RV148 strains nor the four strains from the B′ radiation showed a high level of sequence identity to other strains from the same cohort or to reference strains (Fig. 2d).
(ii) CRF01_AE.
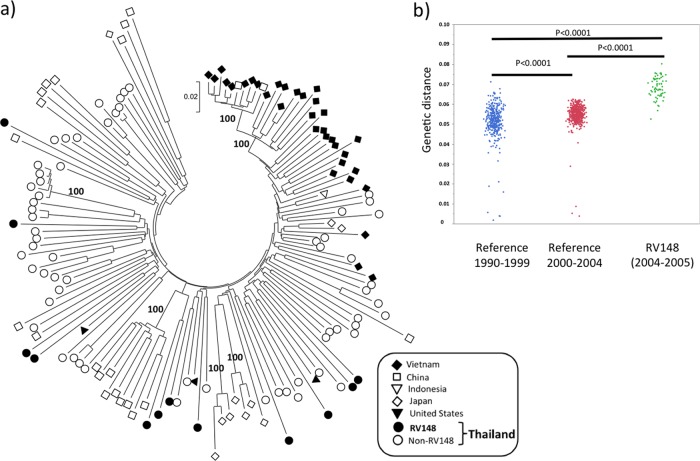
CRF01_AE sequences from 12 individuals were analyzed, including the 6 pure CRF01_AE strains depicted in Fig. 1 and 6 randomly selected specimens (Fig. 3a). The median genetic distance among these sequences was 0.068 (IQR, 0.065 to 0.073). The genetic diversity among CRF01_AE RV148 strains was significantly higher than the genetic diversity in Thailand from 1990 to 1999 (median, 0.052; IQR, 0.049 to 0.056; P < 0.0001) and 2000 to 2004 (median, 0.055; IQR, 0.053 to 0.058; P < 0.0001), indicating that the Thai CRF01_AE epidemic has undergone a significant expansion in the past 2 decades (Fig. 3b); no contemporaneous reference full-genome CRF01_AE sequences from Thailand were available for comparison. None of the sequenced RV148 strains showed high similarity with either reference or other RV148 sequences. Overall, the RV148 sequences were distributed throughout the East and Southeast Asian CRF01_AE radiation, intermingling with the sequences sampled throughout the region.
Fig 3.
Genetic diversity of near-full-genome CRF01_AE sequences in RV148. (a) Location of CRF01_AE RV148 strains in the context of the Asian CRF01_AE radiation. The different symbols depict the country of sampling of each sequence, as indicated in the box. Sequences sampled in the United States, but likely acquired in Southeast Asia (85), were also included. Numbers on the branches of the phylogenetic tree represent the percentage of bootstrap replicates supporting the relevant clusters. (b) Distribution of pairwise nucleotide genetic distances among CRF01_AE strains in different time periods in the history of the Thai HIV-1 epidemic: 1990 to 1999 (n = 30) and 2000 to 2004 (n = 32).
(iii) B/CRF01_AE recombinants.
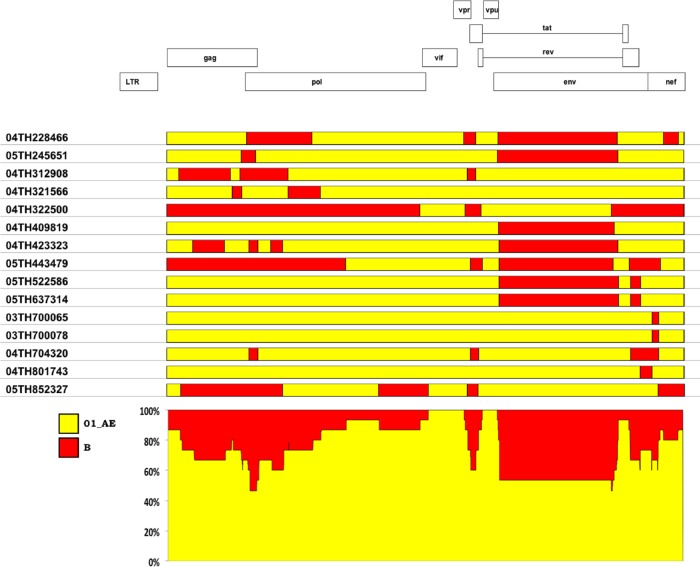
In RV148, by combining MHAbce and sequencing, we were able to identify 16 recombinants, all of them between subtype B and CRF01_AE; near-full-genome sequences were generated for 15 of them (Fig. 1). CRF01_AE comprised the majority of the genomic content of the RV148 intersubtype recombinants (Fig. 4); 13/15 sequences had a CRF01_AE genomic content greater than 50% (range, 59.0% to 98.8%). Some genomic areas were CRF01_AE in all of the recombinants (e.g., vif and vpu), while in other genomic areas (e.g., gp120 in env) there were equivalent numbers of strains carrying either clade. In 14/15 recombinants, the subtype B components belonged to the B′ radiation. Strain 04TH322500 was the only intersubtype recombinant whose subtype B content was contributed by the Western B radiation (data not shown). The number of breakpoints observed among RV148 B/CRF01_AE recombinants did not differ significantly from the number observed among previously described B/CRF01_AE recombinants (data not shown).
Fig 4.
CRF01_AE comprises the majority of the genomic content of B/CRF01_AE recombinants in RV148. On the basis of recombination breakpoint mapping, the subtype composition of all the recombinants was added at each genomic position. Some areas showed equivalent contributions from CRF01_AE and subtype B (e.g., gp120 in env), while other areas were predominantly CRF01_AE (e.g., vif).
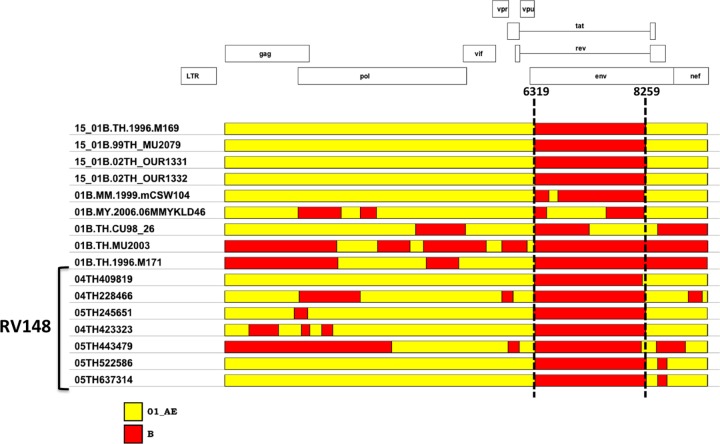
The genomic structures of intersubtype recombinants isolated in RV148 were compared to those of recombinants containing subtypes B and CRF01_AE from the published literature. All of the RV148 recombinants, with the exception of strain 04TH801743, shared at least one breakpoint with previously reported CRFs and URFs. Strain 04TH409819 was the only B/CRF01_AE recombinant that shared a full pattern of recombination breakpoints with a previously described CRF, CRF15_01B, which has been found in Thailand in individuals attending antenatal/family planning clinics in Lampang and Rayong Provinces (32, 40), an IDU attending an opiate treatment clinic in Chiang Mai (59), and a female commercial sex worker (CSW) in Bangkok (59) (Fig. 5). There was a group of six additional strains (i.e., 04TH228266, 05TH245651, 04TH423323, 04TH443479, 05TH522586, and 05TH637314) that shared both of the CRF15_01B recombination breakpoints in the envelope region; all of these recombinants incorporated additional breakpoints in other genomic areas.
Fig 5.
Seven RV148 B/CRF01_AE intersubtype recombinants share common recombination breakpoints with CRF15_01B and with other published Southeast Asian recombinants. The vertical dashed lines indicate the location of CRF15_01B breakpoints in the HXB2 coordinate system. See the text for details.
There were two pairs of B/CRF01_AE recombinants that shared their complete recombination patterns. Strains 05TH522586 and 05TH637314 were mostly CRF01_AE and contained two subtype B segments in env (Fig. 1). These two segments corresponded, respectively, to the gp120 segment present in CRF15_01B and the gp41/second exon of the rev segment present in CRF34_01B. The genetic distance between these two strains was 0.039. The volunteers from whom these strains were recovered were screened at different sites located 12 km apart in Rayong Province, and their visits occurred 6 months apart. The other pair of strains sharing their complete recombination pattern was 03TH700065and 03TH700078, which were composed of a single 109-bp subtype B fragment in nef embedded in a CRF01_AE background. The genetic distance between these two strains was 0.026. This recombinant structure was fully shared with strain 02TH.OUR840I, which had been retrieved from a 2002 incident infection within an IDU cohort in Chiang Mai, Thailand (28); the genetic distances from 02TH.OUR840I to 03TH700065 and 03TH700078 were 0.055 and 0.058, respectively. Individuals 03TH700065and 03TH700078 were screened on the same day at the same site in Rayong Province, and the possibility of a direct epidemiological linkage between them cannot be excluded. For this reason, and despite the shared recombination pattern with 02TH.OUR840I, this recombinant should not be designated a CRF at this time.
Location of RV148 intersubtype recombinants in the context of the East and Southeast Asian HIV-1 recombinant network.
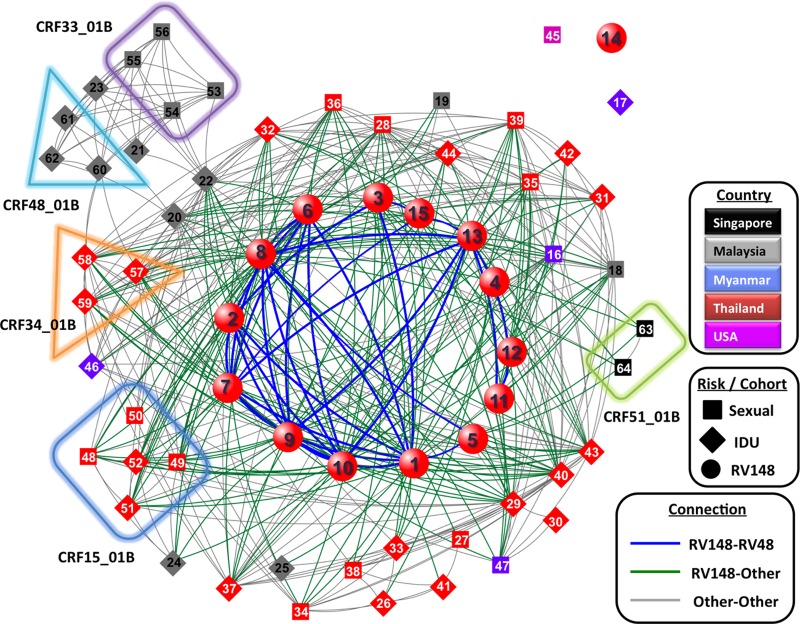
The irreversible nature of HIV-1 recombination makes recombination breakpoints extremely useful markers in molecular epidemiology studies; recombinants are generated within multiply infected individuals, and when transmitted to new individuals, their genomic structure is preserved as a stable form. Additionally, transmitted recombinants may further recombine with other strains in subsequent multiply infected individuals, and some of the breakpoints from the parental strains may be preserved. We have previously shown that if shared recombination breakpoints are used to compare different sequences and strains that share common recombination breakpoints are grouped together, the vast majority of the recombinants sampled in East and Southeast Asia are part of a single, structured, and intricate network (55). Here we analyzed the RV148 recombinants in the backdrop of the network composed of all the reported recombinants containing subtypes B and CRF01_AE (Fig. 6). RV148 strains were an integral part of the larger East/Southeast Asian network. With the exception of strain 04TH801743, all of the RV148 strains were linked to at least one reported strain. RV148 recombinants were directly linked to reported recombinants sampled in diverse countries (i.e., Malaysia, Myanmar, Singapore, and Thailand) and from individuals with diverse reported risk factors for HIV-1 infection (i.e., IDU and sexual contact). While there were clear connections linking RV148 strains to other strains from the same cohort, RV148 strains showed a higher level of connection to reference strains (P = 0.001; data not shown).
Fig 6.
RV148 recombinants are embedded in the larger network of Southeast Asian HIV-1 recombinants. In the network diagram, each node represents a recombinant color coded by country of sampling and shape coded by risk factor for HIV-1 acquisition (risk factors in RV148 were not recorded and thus are depicted as a separate category). Lines connecting nodes link strains that share common recombination breakpoints, and they are color coded to help distinguish relationships within RV148, within published strains, and between these two groups. Boxes group together strains that belong to the same CRF. Numbers within the nodes identify the strains, as follows: 1, 04TH228466; 2, 05TH245651; 3, 04TH312908; 4, 04TH321566; 5, 04TH322500; 6, 04TH409819; 7, 04TH423323; 8, 05TH443479; 9, 05TH522586; 10, 05TH637314; 11, 03TH700065; 12, 03TH700078; 13, 04TH704320; 14, 04TH801743; 15, 05TH852327; 16, 01B.MM.1999.mCSW104; 17, 01B.MM.2000.mIDU502; 18, 01B.MY.2003.03MYKL018_1; 19, 01B.MY.2004.04MYKL016_1; 20, 01B.MY.2004.04MYKL019_1; 21, 01B.MY.2005.05MYKL043_1; 22, 01B.MY.2006.06MMYKLD46; 23, 01B.MY.2007.07MYKLD47; 24, 01B.MY.2007.07MYKLD48; 25, 01B.MY.2007.07MYKLD49; 26, 01B.TH.-0.1269 12_69; 27, 01B.TH.-.CM237; 28, 01B.TH.-.CU98_26; 29, 01B.TH.-.MU2003; 30, 01B.TH.-.NP1623; 31, 01B.TH.-.TH1326; 32, 01B.TH.-.TH283; 33, 01B.TH.-.TH9_95; 34, 01B.TH.1996.M005; 35, 01B.TH.1996.M043; 36, 01B.TH.1996.M171; 37, 01B.TH.1999.OUR2574; 38, 01B.TH.2000.00TH_C2254_BE; 39, 01B.TH.2000.00TH_R1741; 40, 01B.TH.2001.OUR033I; 41, 01B.TH.2002.OUR740I; 42, 01B.TH.2002.OUR840I; 43, 01B.TH.2002.OUR846I; 44, 01B.TH.2002.OUR847I; 45, 01B.US.1998.99US_MSC5043; 46, 01BC.MM.1999.mIDU107; 47, 01BC.MM.2000.mCSW503; 48, 15_01B.TH.1996.M169; 49, 15_01B.TH.1999.99TH_MU2079; 50, 15_01B.TH.1999.99TH_R2399; 51, 15_01B.TH.2002.02TH_OUR1331; 52, 15_01B.TH.2002.02TH_OUR1332; 53, 33_01B.MY.05.05MYKL045_1; 54, 33_01B.MY.2005.05MYKL007_1; 55, 33_01B.MY.2005.05MYKL015_2; 56, 33_01B.MY.2005.05MYKL031_1; 57, 34_01B.TH.1999.OUR1969P; 58, 34_01B.TH.1999.OUR2275P; 59, 34_01B.TH.1999.OUR2478P; 60, 48_01B.MY.2007.07MYKT014; 61, 48_01B.MY.2007.07MYKT016; 62, 48_01B.MY.2007.07MYKT021; 63, 51_01B.SG.2011.11SG_HM021; 64, 51_01B.SG.2011.11SG_HM091. See the text for details.
HIV-1 subtype distribution and its association with sociodemographic variables.
In East Africa and Thailand, it has previously been reported that sociodemographic characteristics can be associated with more genetically complex HIV-1 epidemics (60–66). In the current study, we explored the association between recorded sociodemographic variables and the infecting HIV-1 strain. There were no statistically significant differences in the distribution of subtypes and recombinants on the basis of sex, gender, age, birthplace, marital status, education level, or occupation (Table 3).
Table 3.
Comparison of sociodemographic variables across the different genotypes detected among 375 HIV-infected volunteers deferred from enrollment in RV144
| Characteristic | No. (%) of volunteers with the following genotype: |
Odds ratio for non-CRF01_AE (95% CI) | ||||
|---|---|---|---|---|---|---|
| Total | CRF01_AE | Subtype B | B/CRF01_AE recombinants | B/CRF01_AE dual infection | ||
| Sex | ||||||
| Female | 183 | 172 (94.0) | 5 (2.7) | 6 (3.3) | 0 (0.0) | Reference |
| Male | 192 | 172 (89.6) | 8 (4.2) | 10 (5.2) | 2 (1.0) | 1.82 (0.85–3.91) |
| Gender | ||||||
| Female | 192 | 181 (94.3) | 5 (2.6) | 6 (3.1) | 0 (0.0) | Reference |
| Male | 182 | 162 (89.0) | 8 (4.4) | 10 (5.5) | 2 (1.1) | 2.03 (0.94–4.4) |
| Missing | 1 | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NAa |
| Age (yr) | ||||||
| 18–25 | 185 | 170 (91.9) | 8 (4.3) | 6 (3.2) | 1 (0.5) | Reference |
| 26–30 | 190 | 175 (91.6) | 5 (2.6) | 10 (5.3) | 1 (0.5) | 1.04 (0.50–2.17) |
| Birthplace | ||||||
| Rayong | 85 | 80 (94.1) | 1 (1.2) | 4 (4.7) | 0 (0.0) | Reference |
| Chon Buri | 92 | 82 (89.1) | 6 (6.5) | 3 (3.3) | 1 (1.1) | 1.95 (0.66–6.50) |
| Other | 198 | 182 (91.9) | 6 (3.0) | 9 (4.6) | 1 (0.5) | 1.40 (0.53–4.42) |
| Marital status | ||||||
| Single | 112 | 103 (92.0) | 4 (3.6) | 4 (3.6) | 1 (0.9) | Reference |
| Married | 206 | 187 (90.8) | 7 (3.4) | 11 (5.3) | 1 (0.5) | 1.16 (0.52–2.78) |
| Separated | 2 | 2 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Divorced | 49 | 46 (93.9) | 2 (4.1) | 1 (2.0) | 0 (0.0) | 0.75 (0.16–2.63) |
| Widowed | 6 | 6 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Education | ||||||
| Primary | 196 | 177 (90.3) | 7 (3.6) | 11 (5.6) | 1 (0.5) | Reference |
| Did not complete primary school | 17 | 15 (88.2) | 1 (5.9) | 1 (5.9) | 0 (0.0) | 1.24 (0.19–4.87) |
| Junior high school | 111 | 103 (92.8) | 5 (4.5) | 2 (1.8) | 1 (0.9) | 0.72 (0.29–1.66) |
| Senior high school/vocational school | 41 | 39 (95.1) | 0 (0.0) | 2 (4.9) | 0 (0.0) | 0.48 (0.07–1.74) |
| Vocational school | 7 | 7 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Bachelor's degree or higher | 3 | 3 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Occupation | ||||||
| Office/factory | 87 | 80 (92.0) | 1 (1.1) | 5 (5.8) | 1 (1.1) | Reference |
| Laborer | 117 | 108 (92.3) | 2 (1.7) | 6 (5.1) | 1 (0.9) | 0.95 (0.34–2.77) |
| Merchant/business | 31 | 30 (96.8) | 1 (3.2) | 0 (0.0) | 0 (0.0) | 0.38 (0.02–2.27) |
| Housewife | 24 | 24 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Entertainment | 20 | 19 (95.0) | 1 (5.0) | 0 (0.0) | 0 (0.0) | 0.60 (0.03–3.68) |
| Agriculture | 16 | 14 (87.5) | 1 (6.3) | 1 (6.3) | 0 (0.0) | 1.63 (0.23–7.63) |
| Male/female CSW | 15 | 13 (86.7) | 1 (6.7) | 1 (6.7) | 0 (0.0) | 1.76 (0.24–8.28) |
| Fishery | 8 | 7 (87.5) | 1 (12.5) | 0 (0.0) | 0 (0.0) | 1.63 (0.08–11.3) |
| Government officer | 5 | 5 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Military/police | 4 | 3 (75.0) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 3.81 (0.18–34.7) |
| Student | 3 | 3 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Other | 2 | 2 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Unemployed | 43 | 36 (83.7) | 4 (9.3) | 3 (7.0) | 0 (0.0) | 2.22 (0.71–6.95) |
NA, not applicable.
Genetic distance of RV148 strains to immunogens from RV144 regimen.
The cocirculation in Thailand of subtype B, CRF01_AE, and their recombinants poses an added level of difficulty to the process of HIV-1 vaccine development, and the incorporation of subtype B and CRF01_AE immunogens into vaccine candidates was implemented early on (reviewed in reference 67). However, genetic changes in the local epidemic occurring in the time span elapsing from immunogen selection to vaccine testing might have had profound implications for the success of vaccine trials and need to be considered in the analysis of vaccine efficacy. Here we assessed the genetic distance of the RV148 protein sequences to the different immunogens, and we analyze these results in the context of the evolving HIV-1 epidemics in East and Southeast Asia (Fig. 7).
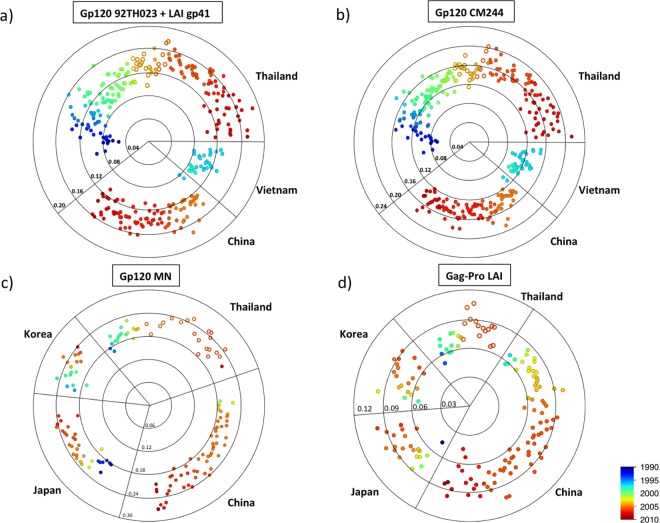
Fig 7.
Temporal and geographical differences in the distance of southeast Asian strains to RV144 immunogens. (a) Protein distance of RV148 CRF01_AE and B/CRF01_AE recombinants whose gp120 region is CRF01_AE and published HIV-1 CRF01_AE strains sampled in East and Southeast Asia to the 92TH023 gp120 linked to the LAI gp41 immunogen presented in the ALVAC-HIV. (b) Protein distance of RV148 CRF01_AE and B/CRF01_AE recombinants whose gp120 region is CRF01_AE and published HIV-1 CRF01_AE strains sampled in East and Southeast Asia to the CM244 gp120 immunogen presented in the AIDSVAX B/E. (c) Protein distance of RV148 subtype B and B/CRF01_AE recombinants whose gp120 region is subtype B and published HIV-1 subtype B strains sampled in East and Southeast Asia to the MN gp120 immunogen presented in the AIDSVAX B/E. (d) Protein distance of RV148 strains to subtype B and B/CRF01_AE recombinants whose gag-pro region is subtype B and published HIV-1 subtype B strains sampled in East and Southeast Asia to the LAI Gag immunogen presented in the ALVAC-HIV. Each point in the radar chart depicts an RV148 (hollow circles) or published strain (full circle), and its radial distance to the center represents the protein p-distance to the immunogen. Concentric circles show the distance scale. The country and year of sampling are indicated.
(i) Distance to CRF01_AE Env immunogens: 92TH023 gp120 linked to LAI gp41 (ALVAC-HIV) and CM244 gp120 (AIDSVAX B/E).
Figures 7a and b depict the genetic distance of 21 RV148 strains—13 CRF01_AE B and 8 B/CRF01_AE recombinants whose corresponding genomic region is CRF01_AE—along with those of 284 CRF01_AE reference sequences sampled in Thailand, China, and Vietnam between 1990 and 2009 to the CRF01_AE Env immunogens. The median distances of RV148 strains to the CRF01_AE ALVAC-HIV and AIDSVAX B/E Env immunogens were 0.136 (IQR, 0.127 to 0.145) and 0.155 (IQR, 0.142 to 0.163), respectively. Interestingly, when analyzing sequences sampled in Thailand from 1990 to date, there is a clear, progressive, and significant increase in the distance to both CRF01_AE Env immunogens: the median distances of sequences from 1990 to 1994 to ALVAC-HIV and AIDSVAX B/E were 0.081 (IQR, 0.071 to 0.108) and 0.102 (IQR, 0.081 to 0.120), respectively (see Fig. S1a and b in the supplemental material). These represent significant increases in the distance to immunogens of 68% and 52% (P < 0.0001), respectively. Sequences sampled in Thailand and Vietnam between 1995 and 1999 fit in this progression, and those sampled in Thailand between 2005 and 2009 were farther away from the immunogens than the RV148 samples (median distance to ALVAC-HIV, 0.146 [IQR, 0.138 to 0.158; P = 0.01]; median distance to AIDSVAX B/E Env, 0.166 [IQR, 0.152 to 0.179; P = 0.03]).
(ii) Distance to gp120 MN (AIDSVAX B/E).
Figure 7c depicts the genetic distance of 20 RV148 strains—13 subtype B and 7 B/CRF01_AE recombinants whose corresponding genomic region is subtype B—along with those of 128 subtype B reference sequences sampled in Thailand, China, Japan, and South Korea between 1986 and 2009 to the subtype B Env immunogen. The median distance of RV148 strains to the MN immunogen in AIDSVAX B/E was 0.220 (IQR, 0.201 to 0.239). This reflected only a 12% increase from the measurement evidenced on sequences sampled in Thailand from 1990 to 2002 (median, 0.196; IQR, 0.186 to 0.203; P = 0.0002) (see Fig. S1c in the supplemental material); too few reference sequences were available to conduct a comparison with shorter temporal intervals. The median distance from sequences from China, Japan, and South Korea to the immunogen fell within the same range as the RV148 measurement (0.204, 0.218, and 0.221, respectively).
(iii) Distance to HIV-1 Gag and Pro of LAI (ALVAC-HIV).
Figure 7d depicts the genetic distance of 15 RV148 strains—13 subtype B and 2 B/CRF01_AE recombinants whose corresponding genomic region is subtype B—along with those of 140 subtype B reference sequences sampled in Thailand, China, Japan, and South Korea between 1986 and 2009 to the subtype B Gag/Pro immunogen. The median distance of RV148 strains to LAI was 0.084 (IQR, 0.066 to 0.088), which is 29% greater than the distance from sequences retrieved in Thailand from 1990 to 2002 (median, 0.065; IQR, 0.060 to 0.074; P = 0.0005) (see Fig. S1d in the supplemental material). The distance of subtype B strains sampled in East Asia—most of them contemporaries of RV148 strains—to the LAI immunogen was equivalent to that of RV148.
Variation in V1 and V2 of HIV-1 Env.
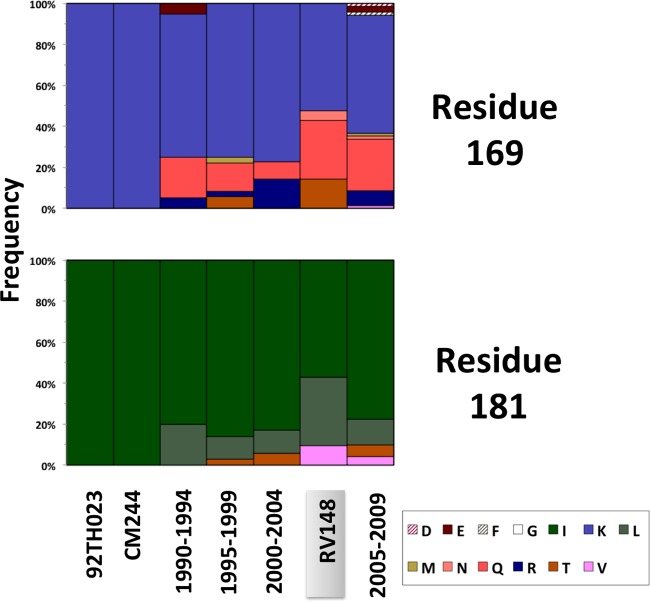
Previous studies have shown that sequence variation in variable regions 1 and 2 (V1V2) can dramatically impact antibody binding and neutralization (68–72). Most recently, analyses in RV144 revealed that the level of binding of IgG antibodies to V1V2 correlated inversely with the rate of HIV-1 infection (73) and that there was an increased vaccine efficacy against viruses with genetic signatures in V2 (42). Given the importance of this variable region, we compared the V1V2 sequences from RV148 and reference isolates to those of the vaccine inserts. We focused our analysis on CRF01_AE sequences from Thailand (1990 to 2009) and the corresponding CRF01_AE immunogens (i.e., 92TH023 presented in ALVAC-HIV and CM244 presented in AIDSVAX B/E), as this constituted the largest data set. After excluding areas with extensive length variation, there were 18 positions where more than 20% of the RV148 sequences presented amino acid residues different from those of RV144 immunogens (i.e., residues 130, 133, 148, 152, 154, 155, 158, 161, 165, 166, 169, 170, 171, 173, 178, 181, 183, and 192 in the HXB2 numbering system). In all of these sites, sequence variation was not restricted to RV148 but could be evidenced throughout the 2 decades that spanned molecular epidemiology studies in Thailand, with no consistent temporal trend (data not shown). Also, the amino acid residues involved in toggling at each position coincided among the compared periods, probably reflecting convergent elicitation of immune responses and pathways of viral escape during natural HIV-1 infection. The sieve analysis conducted on RV144 breakthrough cases revealed significant differences between vaccine and placebo recipients in the amino acid residues represented at positions 169 and 181; the vaccine efficacy was higher against viruses carrying a lysine at position 169 (169K) and against viruses not carrying an isoleucine at position 181 (181X) (42). Figure 8 shows that, in RV148, 11/21 individuals carried 169K, which was less than the proportion evidenced among viruses sampled in Thailand from 1995 to 1999 and 2000 to 2004 (unadjusted P = 0.07 and 0.053, respectively). Similarly, 9/21 individuals carried the 181X signature in RV148, which represented a proportion greater than the proportions evidenced from 1990 to 1995, 2000 to 2004, and 2005 to 2009 (unadjusted P = 0.02, 0.04, and 0.07, respectively). Due to the large numbers of comparisons carried out, these unadjusted P values should be interpreted as trends.
Fig 8.
Amino acid variation in V1V2 positions 169 and 181 among CRF01_AE sequences sampled in Thailand between 1990 and 2009 and their relation to the RV144 vaccine immunogens. The frequency of each amino acid residue at the corresponding position is depicted for published Thai CRF01_AE gp120 sequences sampled between 1990 and 1994 (n = 20), 1995 and 1999 (n = 36), 2000 and 2004 (n = 35), and 2005 and 2009 (n = 71), as well as CRF01_AE and B/CRF01_AE recombinants whose gp120 region is CRF01_AE from RV148 (n = 21). The amino acid residues presented in 92TH023 and CM244 RV144 immunogens are depicted for comparison.
V1V2 is notable for its high level of N-glycosylation; RV148 sequences presented a median of 8 potential N-glycosylated sites (IQR, 7 to 10), which did not differ significantly from that for reference sequences (data not shown). Despite V1V2 sequence variation, the N-glycosylation site 160 and the cationic nature of the C strand, which have been shown to be critical for neutralization by PG9 and related antibodies, were conserved among RV148 and reference sequences (data not shown). Overall, this analysis shows that there have been no notable changes in the patterns of V1V2 sequence variation among CRF01_AE strains in Thailand in the past 2 decades.
DISCUSSION
RV144 was the first vaccine trial to demonstrate a significant, though modest, protection from HIV-1 acquisition (modified intent-to-treat vaccine efficacy, 31.2% [95% CI, 1.1 to 52.1%]) (56). Subsequent studies have defined correlates of risk, importantly, vaccine-induced IgG to the V1V2 region (73), and analysis of breakthrough infections (42) has further identified genetic signatures in Env, particularly at HXB2 amino acid positions 169 and 181, which are associated with vaccine efficacy. In the broader context, understanding the molecular epidemiology of volunteers deferred from enrollment in RV144 due to preexisting HIV-1 infection (i) provides a cross-sectional representation of the HIV-1 epidemic in Thailand at the time of recruitment for RV144, (ii) helps better define who volunteered for the trial, and (iii) is an important approximation to the viral strains that challenged the vaccine and placebo recipients in RV144 and their relationship to the immunogens.
Cross-sectional representation of the HIV epidemic in Thailand at the time of recruitment for RV144.
In RV148, we observed that the predominant strain, with a prevalence of over 90%, was CRF01_AE, followed by equivalent proportions of subtype B and B/CRF01_AE recombinants and only minor occurrences of dual infections. These results are in high concordance with the findings of the community cohort studies performed in the same area between 1998 and 2001 in preparation for RV144 (40). Regarding the level of genetic diversity among full-genome CRF01_AE sequences in RV148, we noticed increases of 31% and 24% compared to the measurements from Thailand from 1990 to 1999 and 2000 to 2004, respectively. This observation supports previous observations indicating that in the past 2 decades the CRF01_AE Thai epidemic has experienced a continued genetic expansion (25).
The proportion of RV148 strains that were subtype B was 3.5%. An interesting aspect of the East and Southeast Asian HIV-1 epidemics is the cocirculation of two different clusters within subtype B: Western B and B′ (57, 58). The latter is widespread throughout the region and has been incorporated by the vast majority of subtype B-containing recombinants (e.g., CRF07_BC, CRF08_BC, and CRF15_01B). In RV148, the Western B strains outnumbered the B′ strains in a 2:1 ratio. Our phylogenetic analyses indicate that the Western B strains did not proceed from a single source but seemed to be part of the larger, cosmopolitan, and contemporary Western B epidemic. The fact that more than half of the Western B strains in RV148 were sampled near Pattaya, a hub for international tourism, suggests that this area may be an important port of entry for these strains. While B′ strains continue to be detected in Thailand, they do not seem to constitute the majority of the subtype B strains in Rayong and Chon Buri Provinces. It is possible that the populations that used to bear B′ strains were high-risk groups (e.g., IDUs) and became superinfected with CRF01_AE strains, which led to the emergence of numerous B/CRF01_AE URFs (28). It can be expected that if the process of absorption of B′ strains into B/CRF01_AE recombinants persists and the influx of Western B strains continues, pure B′ strains may eventually be replaced by Western B strains in the Thai HIV-1 epidemic. In contrast to the evolution of B′, the incorporation of Western B strains in B/CRF01_AE recombinants has been restricted to a few examples (e.g., CRF51_01B [74], CM237 [75], and strain 04TH322500 in the current study), which suggests that Western B circulates in social networks where dual infection and dissemination of recombinants is not frequent.
The cocirculation of subtype B and CRF01_AE in Southeast Asia has led to the emergence of intersubtype recombinants. While some of them expanded in the population (e.g., CRF15_01B and CRF34_01B) (59, 76), the majority of recombinants have been sampled only on single occasions (i.e., URFs). An interesting feature of the URFs in Thailand is that most of them share common recombination breakpoints. Molecular epidemiological studies support the hypothesis that these similarities reflect common ancestry (28, 77–79), though some common breakpoints may represent independent and convergent events (80). In RV148, 1 out of 15 sequenced B/CRF01_AE recombinants belonged to a CRF, CRF15_01B, which is the CRF that has spread among different provinces and social networks throughout Thailand (32, 59). Additionally, 6 URFs shared with CRF15_01B both of the signature recombination breakpoints in env, suggesting that this CRF plays a central role in the Thai HIV-1 epidemic, not only by spreading to some extent as a stable form but also as a major contributor of genetic material to new recombinants. The genomic structure of CRF15_01B can be compared to a pseudotype virion (81), as it is composed of a single B′ segment extending from the beginning of gp120 (excluding the leader peptide, which is CRF01_AE) to the end of the ectodomain of gp41 in a CRF01_AE background. The retention of these motifs in the CRF15_01B-derived URFs reiterates a phenomenon observed in other subtype B-containing recombinants sampled worldwide (e.g., CRF14_BG [82]) and may indicate that these motifs interact better in this configuration, which warrants further structural and functional analyses (A.-L. Chenine et al., unpublished data).
It has previously been shown that the sharing of recombination breakpoints among CRFs and URFs in East and Southeast Asia links virtually all of these recombinants in a single network, which connects strains from different geographic locations and strains sampled from individuals with different routes for HIV-1 acquisition (55). Here we have shown that all but one of the RV148 recombinants are fully integrated within this larger network. The fact that the direct connectedness of RV148 to other reference recombinants was higher than the direct connectedness to other members of RV148 indicates that recombinants in RV148 did not proceed from a single pocket or subpopulation but, rather, that they represent a more diverse pool of sequences. This may shed light on the social networks from which the RV148 volunteers were drawn (see below).
Who volunteered for the trial?
The information retrieved from RV148 volunteers included basic sociodemographic characteristics, but the volunteers were not asked questions regarding behaviors that may increase the risk for HIV acquisition (41). In cohorts in Thailand and worldwide, it has been shown that there were associations between the complexity of HIV-1 strains and sociodemographic characteristics and behaviors that may increase the risk of HIV infection (e.g., more frequent needle sharing among IDUs [61], the presence of sexually transmitted infections [63], exchanging sex for money [65, 66), and having high numbers of sex partners [66]). We compared the frequency of different molecular forms—pure subtypes and recombinants—between RV148 and other low-risk cohorts (i.e., RV109, a mother-to-child transmission [MTCT] study conducted in Lampang between 1996 and 1997 [32]) and high-risk cohorts (i.e., a study conducted among IDUs seeking treatment in Chiang Mai between 1999 and 2000 [61] and the Thai Red Cross [TRC] study conducted among high-risk men who have sex with men [MSM] and heterosexuals in Bangkok between 2006 and 2007 [83]) from Thailand that have also been genotyped using MHAbce. In RV148, the frequency of non-CRF01_AE strains (9.8%) was higher than that in the low-risk MTCT study (5.1%) but lower than the ones observed in higher-risk cohorts: IDUs (18.7%) and high-risk MSM and heterosexuals (20%). In RV148, the combined frequency of complex molecular forms (i.e., recombinants and dual infections) was 6.6%; likewise, these complex strains were more frequent in RV148 than in the low-risk MTCT cohort (2.3%) but less frequent in RV148 than in the high-risk Opiate-Users Research (14.7%) and TRC (13%) cohorts. Overall, these results are consistent with the idea that the level of risk for HIV-1 infection among RV148 HIV-infected individuals was at the level of what is expected from a community-based cohort, which was the aim of RV144 (56). While individuals belonging to groups with high-risk behaviors may also have volunteered for the trial, their representation in RV148 seems to be only minor. This is an important point, as the level of risk of vaccine trial populations can impact the observed vaccine efficacy (84).
The fact that in RV148 we observed a large number of URFs along with a limited number of multiply infected individuals can be explained through the model through which, in the region, there are small groups with high levels of exposure to HIV-1, where recombinants are being generated in multiply infected individuals. The linkage of these groups with the larger, lower-risk community then leads to the dissemination of URFs. Following this model, in order to limit the expansion in HIV-1 genetic diversity, public health efforts should be directed at identifying these high-risk groups and strengthening effective intervention campaigns.
An approximation to the viral strains that challenged the vaccine and placebo recipients in RV144 and their relationship to the immunogens.
The complex activities involving vaccine development can take more than a decade from the time of immunogen selection until testing in efficacy trials. Even if the immunogens were chosen to minimize the mismatch to the circulating strains at the time of vaccine development, the molecular characteristics of the local epidemic can dramatically change by the time of vaccine testing in efficacy trials. Here we showed that the distance to the immunogens increased 52% to 68% for CRF01_AE Env, 12% for subtype B Env, and 29% for subtype B Gag-Pro from the early 1990s to the mid-2000s. The CRF01_AE Thai epidemic in the early 1990s was very compact and continued expanding throughout the decade. Considering the data for the period from 2005 to 2009 from Thailand, the increase in genetic diversity had not plateaued at the time of RV148. On the other hand, it seems that the subtype B epidemic is not expanding at such a fast pace. It is important to continue the molecular characterization of communities where vaccine trials are planned, as this phenomenon can impact the analysis of vaccine efficacy in future trials. At the time of RV148, the distance of CRF01_AE and subtype B strains from other countries in East and Southeast Asia to the immunogens was equivalent to that measured in Thailand. This means that the distance of circulating HIV-1 strains to the immunogen should not be a major concern at the time of extending the testing/usage of the RV144 vaccine regimen to other countries in the region.
Of note, the above-mentioned protein distance calculations were based on the entirety of the immunogen sequence. The analysis of immunological correlates of protection from acquisition in RV144 has revealed a significant inverse association between binding of IgG antibodies to V1V2 and infection risk (73). This region of gp120 is recognized as one of the most variable areas of the HIV-1 proteome. Our analysis of CRF01_AE strains indicates that the nature and level of sequence variation in highly polymorphic sites in V1V2 have been maintained since the early 1990s to date, in contrast to the results from the analyses based on the whole immunogen. Some V1V2 sites showed an extremely high level of mismatch between RV148 sequences and immunogens; 48% and 43% of the sequences differed from the sequence of the vaccine insert in positions 169 and 181, respectively, with some trends suggesting differences with previous time points. Interestingly, these are two sites where recent analyses by Rolland et al. have detected significant sieve effects in RV144 (42), further supporting the need for continuing molecular epidemiological surveys in communities where vaccine efficacy trials are planned or under way.
Limitations of the study.
The current study, involving nearly 400 individuals, constitutes one of the largest HIV-1 molecular surveys conducted in Southeast Asia to date; however, it has several limitations. The use of high-throughput MHAbce allowed the cost-effective genotyping of virtually the entire cohort and allowed the detection of recombinants that would have been missed by partial-genome sequencing; nevertheless, it is possible that some of the samples typed as pure CRF01_AE may in fact be recombinants incorporating other subtypes in genomic areas not interrogated by MHAbce. Additionally, MHAbce is not capable of detecting multiple infections due to the same clade (e.g., coinfection with two distinct CRF01_AE strains). Thus, the current tallies of recombinants and multiple infections should be considered lower-limit estimates. Another limitation of the current study arises from the fact that near-full-genome sequences were retrieved from only 10% of the individuals, with special emphasis on subtype B and intersubtype recombinants. For this reason, the study was not powered to systematically address the presence of direct epidemiological linkage among HIV-1 infected individuals, especially among those infected with CRF01_AE.
Finally, the current work focused exclusively on the RV144 volunteers deferred from enrollment in the vaccine trial due to preexisting HIV-1 infection. The thorough analysis of RV144 breakthrough cases is the focus of a separate report (Rolland, personal communication) and will provide wider context to the current observations.
Conclusion.
In summary, our results show that at the time of enrollment for RV144, the HIV-1 epidemic in Rayong and Chon Buri Provinces was dominated by CRF01_AE strains. Some representation of subtype B was noted, with an increase in the Western B frequency due to multiple entries in the region and waning of B′ being detected. Most of the intersubtype recombinants were URFs embedded within the larger regional network of recombinants. The level of genetic complexity observed in this study strongly indicates that the RV148 volunteers presented risk levels consistent with those for a community-based cohort. Compared to the molecular picture at the early stages of the process of vaccine development, we evidenced substantial changes in the epidemic (i.e., the expansion of the genetic diversity within CRF01_AE, the increase in the frequency and complexity of B/CRF01_AE recombinants, and shifts in the Western B/B′ balance) which had the potential to impact the efficacy of the vaccine and should be considered at the time of the analysis of the trial results.
Supplementary Material
ACKNOWLEDGMENTS
The opinions expressed in this article are those of the authors and do not represent the official views of the U.S. Department of Health and Human Services, the National Institute of Allergy and Infectious Diseases, the U.S. Department of Defense, or the Department of the Army.
This work was supported in part by an Interagency Agreement (Y1-AI-2642-12) between the U.S. Army Medical Research and Materiel Command and the National Institute of Allergy and Infectious Diseases. This work was also supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense.
We report that we do not have any potential conflict of interest relevant to this article.
We thank the volunteers who participated in this study; Sonchai Watanna and Wiwat Wiriyakiija; the staff members of the Ministry of Public Health at health centers and district hospitals in Rayong Province (Ban Chang, Ampur Mueang, Ban Khai, Klaeng), Chon Buri Province (Panthong, Sri Racha, Bang Lamung, Sattahip), and surrounding provinces; and Christiana Gammon-Richardson and Nasheed Moqueet for technical assistance.
Footnotes
Published ahead of print 10 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03070-12.
REFERENCES
- 1. Kijak GH, McCutchan FE. 2005. HIV diversity, molecular epidemiology, and the role of recombination. Curr. Infect. Dis. Rep. 7:480–488 [DOI] [PubMed] [Google Scholar]
- 2. Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 11:45–56 [DOI] [PubMed] [Google Scholar]
- 3. Kuiken C, Leithner T, Hahn B, Mullins J, Wolinsky S, Foley B, Apetrei C, Mizrahi I, Rambaut A, Korber B. 2012. HIV sequence compendium 2012. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 4. Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. 2008. The challenge of HIV-1 subtype diversity. N. Engl. J. Med. 358:1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tovanabutra S, Birx D, McCutchan F. 2004. Molecular epidemiology of HIV in Asia and the Pacific. In Lu Y, Essex M. (ed), AIDS in Asia. Kluwer Academic, New York, NY [Google Scholar]
- 6. Nelson KE, Celentano DD, Suprasert S, Wright N, Eiumtrakul S, Tulvatana S, Matanasarawoot A, Akarasewi P, Kuntolbutra S, Romyen S, Sirisopana N, Theetranont C. 1993. Risk factors for HIV infection among young adult men in northern Thailand. JAMA 270:955–960 [PubMed] [Google Scholar]
- 7. Sirisopana N, Torugsa K, Mason CJ, Markowitz LE, Jugsudee A, Supapongse T, Chuenchitra C, Michael RA, Burke DS, Singharaj P, Johnson AE, McNeil JG, McCutchan FE, Carr JK. 1996. Correlates of HIV-1 seropositivity among young men in Thailand. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 11:492–498 [DOI] [PubMed] [Google Scholar]
- 8. Weniger BG, Limpakarnjanarat K, Ungchusak K, Thanprasertsuk S, Choopanya K, Vanichseni S, Uneklabh T, Thongcharoen P, Wasi C. 1991. The epidemiology of HIV infection and AIDS in Thailand. AIDS 5(Suppl 2):S71–S85 [DOI] [PubMed] [Google Scholar]
- 9. Ou CY, Takebe Y, Luo CC, Kalish M, Auwanit W, Bandea C, de la Torre N, Moore JL, Schochetman G, Yamazaki S, Gayle HD, Young NL, Weniger BG. 1992. Wide distribution of two subtypes of HIV-1 in Thailand. AIDS Res. Hum. Retroviruses 8:1471–1472 [DOI] [PubMed] [Google Scholar]
- 10. McCutchan FE, Hegerich PA, Brennan TP, Phanuphak P, Singharaj P, Jugsudee A, Berman PW, Gray AM, Fowler AK, Burke DS. 1992. Genetic variants of HIV-1 in Thailand. AIDS Res. Hum. Retroviruses 8:1887–1895 [DOI] [PubMed] [Google Scholar]
- 11. Ou CY, Takebe Y, Weniger BG, Luo CC, Kalish ML, Auwanit W, Yamazaki S, Gayle HD, Young NL, Schochetman G. 1993. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet 341:1171–1174 [DOI] [PubMed] [Google Scholar]
- 12. Weniger BG, Takebe Y, Ou CY, Yamazaki S. 1994. The molecular epidemiology of HIV in Asia. AIDS 8(Suppl 2):S13–S28 [PubMed] [Google Scholar]
- 13. Mason CJ, Kitsiripornchai S, Markowitz LE, Chanbancherd P, Supapongse T, Jugsudee A, Sirisopana N, Chuenchitra C, Torugsa K, VanCott TC, Michael RA, Nitayaphan S. 1998. Nationwide surveillance of HIV-1 prevalence and subtype in young Thai men. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:165–173 [DOI] [PubMed] [Google Scholar]
- 14. Entz AT, Ruffolo VP, Chinveschakitvanich V, Soskolne V, van Griensven GJ. 2000. HIV-1 prevalence, HIV-1 subtypes and risk factors among fishermen in the Gulf of Thailand and the Andaman Sea. AIDS 14:1027–1034 [DOI] [PubMed] [Google Scholar]
- 15. Limpakarnjanarat K, Mastro TD, Saisorn S, Uthaivoravit W, Kaewkungwal J, Korattana S, Young NL, Morse SA, Schmid DS, Weniger BG, Nieburg P. 1999. HIV-1 and other sexually transmitted infections in a cohort of female sex workers in Chiang Rai, Thailand. Sex. Transm. Infect. 75:30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beyrer C, Artenstein A, Kunawararak P, VanCott T, Mason C, Rungreungthanakit K, Hegerich P, Nelson KE, Khamboonruang C, Natpratan C. 1997. The molecular epidemiology of HIV-1 among male sex workers in northern Thailand. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:304–307 [DOI] [PubMed] [Google Scholar]
- 17. Subbarao S, Limpakarnjanarat K, Mastro TD, Bhumisawasdi J, Warachit P, Jayavasu C, Young NL, Luo CC, Shaffer N, Kalish ML, Schochetman G. 1998. HIV type 1 in Thailand, 1994-1995: persistence of two subtypes with low genetic diversity. AIDS Res. Hum. Retroviruses 14:319–327 [DOI] [PubMed] [Google Scholar]
- 18. Tovanabutra S, Robison V, Wongtrakul J, Sennum S, Suriyanon V, Kingkeow D, Kawichai S, Tanan P, Duerr A, Nelson KE. 2002. Male viral load and heterosexual transmission of HIV-1 subtype E in northern Thailand. J. Acquir. Immune Defic. Syndr. 29:275–283 [DOI] [PubMed] [Google Scholar]
- 19. Kunanusont C, Foy HM, Kreiss JK, Rerks-Ngarm S, Phanuphak P, Raktham S, Pau CP, Young NL. 1995. HIV-1 subtypes and male-to-female transmission in Thailand. Lancet 345:1078–1083 [DOI] [PubMed] [Google Scholar]
- 20. Kitayaporn D, Vanichseni S, Mastro TD, Raktham S, Vaniyapongs T, Des Jarlais DC, Wasi C, Young NL, Sujarita S, Heyward WL, Esparza J. 1998. Infection with HIV-1 subtypes B and E in injecting drug users screened for enrollment into a prospective cohort in Bangkok, Thailand. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:289–295 [DOI] [PubMed] [Google Scholar]
- 21. Vongsheree S, Phutiprawan T, Sri-ngam P, Thaisri H, Puangtabtim W, Sawanpanyalert P. 2002. Co-existence of HIV-1 subtypes B′ and E infections among Thai injecting drug users. Asian Pac. J. Allergy Immunol. 20:29–35 [PubMed] [Google Scholar]
- 22. Ichimura H, Kliks SC, Visrutaratna S, Ou CY, Kalish ML, Levy JA. 1994. Biological, serological, and genetic characterization of HIV-1 subtype E isolates from northern Thailand. AIDS Res. Hum. Retroviruses 10:263–269 [DOI] [PubMed] [Google Scholar]
- 23. Wang Z, Lyles CM, Beyrer C, Celentano DD, Vlahov D, Natpratan C, Markham R, Khamboonruang C, Nelson K, Yu XF. 1998. Diversification of subtype E human immunodeficiency virus type 1 env in heterosexual seroconverters from Northern Thailand. J. Infect. Dis. 178:1507–1511 [DOI] [PubMed] [Google Scholar]
- 24. Auwanit W, Ayuthaya PI, Duangchanda S, Mukai T, Kurata T, Ikuta K. 2000. Highly variable sequences in the env V3 region of HIV type 1 distributing among Thai carriers from 1995 to 1997. AIDS Res. Hum. Retroviruses 16:283–289 [DOI] [PubMed] [Google Scholar]
- 25. Utachee P, Jinnopat P, Isarangkura-Na-Ayuthaya P, de Silva UC, Nakamura S, Siripanyaphinyo U, Wichukchinda N, Tokunaga K, Yasunaga T, Sawanpanyalert P, Ikuta K, Auwanit W, Kameoka M. 2009. Genotypic characterization of CRF01_AE env genes derived from human immunodeficiency virus type 1-infected patients residing in central Thailand. AIDS Res. Hum. Retroviruses 25:229–236 [DOI] [PubMed] [Google Scholar]
- 26. Wasi C, Herring B, Raktham S, Vanichseni S, Mastro TD, Young NL, Rubsamen-Waigmann H, von Briesen H, Kalish ML, Luo CC, Pau C-P, Baldwin A, Mullins JI, Delwart EL, Esparza J, Heyward WL, Osmanov S. 1995. Determination of HIV-1 subtypes in injecting drug users in Bangkok, Thailand, using peptide-binding enzyme immunoassay and heteroduplex mobility assay: evidence of increasing infection with HIV-1 subtype E. AIDS 9:843–849 [DOI] [PubMed] [Google Scholar]
- 27. Kalish ML, Baldwin A, Raktham S, Wasi C, Luo CC, Schochetman G, Mastro TD, Young N, Vanichseni S, Rubsamen-Waigmann H, Vonbriesen H, Mullins JI, Delwart E, Herring B, Esparza J, Heyward WL, Osmanov S. 1995. The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: implications for HIV vaccine trials. AIDS 9:851–857 [DOI] [PubMed] [Google Scholar]
- 28. Tovanabutra S, Beyrer C, Sakkhachornphop S, Razak MH, Ramos GL, Vongchak T, Rungruengthanakit K, Saokhieo P, Tejafong K, Kim B, De Souza M, Robb ML, Birx DL, Jittiwutikarn J, Suriyanon V, Celentano DD, McCutchan FE. 2004. The changing molecular epidemiology of HIV type 1 among northern Thai drug users, 1999 to 2002. AIDS Res. Hum. Retroviruses 20:465–475 [DOI] [PubMed] [Google Scholar]
- 29. Artenstein AW, VanCott TC, Mascola JR, Carr JK, Hegerich PA, Gaywee J, Sanders-Buell E, Robb ML, Dayhoff DE, Thitivichianlert S, Nitayaphan S, McNeil JG, Birx DL, Michael RA, Burke DS, McCutchan FE. 1995. Dual infection with human immunodeficiency virus type 1 of distinct envelope subtypes in humans. J. Infect. Dis. 171:805–810 [DOI] [PubMed] [Google Scholar]
- 30. Ramos A, Hu DJ, Nguyen L, Phan KO, Vanichseni S, Promadej N, Choopanya K, Callahan M, Young NL, McNicholl J, Mastro TD, Folks TM, Subbarao S. 2002. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J. Virol. 76:7444–7452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tovanabutra S, Polonis V, De Souza M, Trichavaroj R, Chanbancherd P, Kim B, Sanders-Buell E, Nitayaphan S, Brown A, Robb MR, Birx DL, McCutchan FE, Carr JK. 2001. First CRF01_AE/B recombinant of HIV-1 is found in Thailand. AIDS 15:1063–1065 [DOI] [PubMed] [Google Scholar]
- 32. Kijak GH, Tovanabutra S, Sanders-Buell E, Watanaveeradej V, de Souza MS, Nelson KE, Ketsararat V, Gulgolgarn V, Wera-arpachai M, Sriplienchan S, Khamboonrueng C, Birx DL, Robb ML, McCutchan FE. 2007. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v. 2. Virology 358:178–191 [DOI] [PubMed] [Google Scholar]
- 33. Viputtijul K, de Souza M, Trichavaroj R, Carr JK, Tovanabutra S, McCutchan FE, Sriplienchan S, Buapunth P, Chuenchitra C, McNeil JG, Birx DL, Brown AE, Nitayaphan S. 2002. Heterosexually acquired CRF01_AE/B recombinant HIV type 1 found in Thailand. AIDS Res. Hum. Retroviruses 18:1235–1237 [DOI] [PubMed] [Google Scholar]
- 34. Takebe Y, Motomura K, Tatsumi M, Lwin HH, Zaw M, Kusagawa S. 2003. High prevalence of diverse forms of HIV-1 intersubtype recombinants in central Myanmar: geographical hot spot of extensive recombination. AIDS 17:2077–2087 [DOI] [PubMed] [Google Scholar]
- 35. Liao H, Tee KK, Hase S, Uenishi R, Li XJ, Kusagawa S, Thang PH, Hien NT, Pybus OG, Takebe Y. 2009. Phylodynamic analysis of the dissemination of HIV-1 CRF01_AE in Vietnam. Virology 391:51–56 [DOI] [PubMed] [Google Scholar]
- 36. Piyasirisilp S, McCutchan FE, Carr JK, Sanders-Buell E, Liu W, Chen J, Wagner R, Wolf H, Shao Y, Lai S, Beyrer C, Yu XF. 2000. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J. Virol. 74:11286–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sukasem C, Churdboonchart V, Chasombat S, Kohreanudom S, Watitpun C, Pasomsub E, Piroj W, Tiensuwan M, Chantratita W. 2007. Surveillance of genotypic resistance mutations in chronic HIV-1 treated individuals after completion of the National Access to Antiretroviral Program in Thailand. Infection 35:81–88 [DOI] [PubMed] [Google Scholar]
- 38. Praparattanapan J, Tragoolpua Y, Pathom-aree W, Kotarathitithum W, Chaiwarith R, Nuntachit N, Sirisanthana T, Supparatpinyo K. 2011. Current molecular epidemiology and recombination of HIV type 1 subtypes in northern Thailand. AIDS Res. Hum. Retroviruses 27:1201–1206 [DOI] [PubMed] [Google Scholar]
- 39. Sirivichayakul S, Phanuphak P, Pankam T, O-Charoen R, Sutherland D, Ruxrungtham K. 2008. HIV drug resistance transmission threshold survey in Bangkok, Thailand. Antivir. Ther. 13(Suppl 2):109–113 [PubMed] [Google Scholar]
- 40. Watanaveeradej V, Benenson MW, Souza MD, Sirisopana N, Nitayaphan S, Tontichaivanich C, Amphaipit R, Renzullo PO, Brown AE, McNeil JG, Robb ML, Birx DL, Tovanabutra S, Carr JK, McCutchan FE. 2006. Molecular epidemiology of HIV type 1 in preparation for a phase III prime-boost vaccine trial in Thailand and a new approach to HIV type 1 genotyping. AIDS Res. Hum. Retroviruses 22:801–807 [DOI] [PubMed] [Google Scholar]
- 41. Ministry of Public Health-Thai AIDS Vaccine Evaluation Group 2011. Screening and evaluation of potential volunteers for a phase III trial in Thailand of a candidate preventive HIV vaccine (RV148). Vaccine 29:4285–4292 [DOI] [PubMed] [Google Scholar]
- 42. Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, Decamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O'Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O'Connell RJ, Desouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490:417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ministry of Public Health of Thailand 2000. National guidelines for clinical management of HIV/AIDS in adults and children. Ministry of Public Health, Bangkok, Thailand [Google Scholar]
- 44. Tovanabutra S, Sanders EJ, Graham SM, Mwangome M, Peshu N, McClelland RS, Muhaari A, Crossler J, Price MA, Gilmour J, Michael NL, McCutchan FM. 2010. Evaluation of HIV type 1 strains in men having sex with men and in female sex workers in Mombasa, Kenya. AIDS Res. Hum. Retroviruses 26:123–131 [DOI] [PubMed] [Google Scholar]
- 45. Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 46. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 48. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 49. Schultz AK, Zhang M, Bulla I, Leitner T, Korber B, Morgenstern B, Stanke M. 2009. jpHMM: improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 37:W647–W651. 10.1093/nar/gkp371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679 [DOI] [PubMed] [Google Scholar]
- 51. Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166 In Bryson V, Vogel H. (ed), Evolving genes and proteins. Academic Press, New York, NY [Google Scholar]
- 52. Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275–282 [DOI] [PubMed] [Google Scholar]
- 53. Schwartz RM, Dayhoff MO. 1979. Protein and nucleic acid sequence data and phylogeny. Science 205:1038–1039 [DOI] [PubMed] [Google Scholar]
- 54. Nickle DC, Heath L, Jensen MA, Gilbert PB, Mullins JI, Kosakovsky Pond SL. 2007. HIV-specific probabilistic models of protein evolution. PLoS One 2:e503. 10.1371/journal.pone.0000503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kijak GH, Tovanabutra S, Beyrer C, Sanders-Buell EE, Arroyo MA, Robb ML, Michael NL, McCutchan FE, Kim JH. 2010. RecDraw: a software package for the representation of HIV-1 recombinant structures. AIDS Res. Hum. Retroviruses 26:1317–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 57. Kalish ML, Luo CC, Weniger BG, Limpakarnjanarat K, Young N, Ou CY, Schochetman G. 1994. Early HIV type 1 strains in Thailand were not responsible for the current epidemic. AIDS Res. Hum. Retroviruses 10:1573–1575 [DOI] [PubMed] [Google Scholar]
- 58. Graf M, Shao Y, Zhao Q, Seidl T, Kostler J, Wolf H, Wagner R. 1998. Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B′-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res. Hum. Retroviruses 14:285–288 [DOI] [PubMed] [Google Scholar]
- 59. Tovanabutra S, Watanaveeradej V, Viputtikul K, De Souza M, Razak MH, Suriyanon V, Jittiwutikarn J, Sriplienchan S, Nitayaphan S, Benenson MW, Sirisopana N, Renzullo PO, Brown AE, Robb ML, Beyrer C, Celentano DD, McNeil JG, Birx DL, Carr JK, McCutchan FE. 2003. A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res. Hum. Retroviruses 19:561–567 [DOI] [PubMed] [Google Scholar]
- 60. Herbinger KH, Gerhardt M, Piyasirisilp S, Mloka D, Arroyo MA, Hoffmann O, Maboko L, Birx DL, Mmbando D, McCutchan FE, Hoelscher M. 2006. Frequency of HIV type 1 dual infection and HIV diversity: analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res. Hum. Retroviruses 22:599–606 [DOI] [PubMed] [Google Scholar]
- 61. Kijak GH, Beyrer C, Tovanabutra S, Sripaipan T, Suriyanon V, Moqueet N, Sanders-Buell E, Saokhieo P, Timpan U, Jittiwutikarn J, Robb ML, Birx DL, Celentano DD, McCutchan FE. 2011. Socio-demographic and drug use factors associated with HIV-1 recombinants and dual infections in northern Thai drug users: associations of risk with genetic complexity. Drug Alcohol Depend. 116:24–30 [DOI] [PubMed] [Google Scholar]
- 62. Arroyo MA, Hoelscher M, Sateren W, Samky E, Maboko L, Hoffmann O, Kijak G, Robb M, Birx DL, McCutchan FE. 2005. HIV-1 diversity and prevalence differ between urban and rural areas in the Mbeya region of Tanzania. AIDS 19:1517–1524 [DOI] [PubMed] [Google Scholar]
- 63. Arroyo MA, Sateren WB, Foglia G, Kibaya R, Langat L, Wasunna M, Bautista CT, Scott PT, Shaffer DN, Robb ML, Michael NL, Birx DL, McCutchan FE. 2009. Short communication: HIV type 1 genetic diversity among tea plantation workers in Kericho, Kenya. AIDS Res. Hum. Retroviruses 25:1061–1064 [DOI] [PubMed] [Google Scholar]
- 64. Arroyo MA, Sateren WB, Serwadda D, Gray RH, Wawer MJ, Sewankambo NK, Kiwanuka N, Kigozi G, Wabwire-Mangen F, Eller M, Eller LA, Birx DL, Robb ML, McCutchan FE. 2006. Higher HIV-1 incidence and genetic complexity along main roads in Rakai District, Uganda. J. Acquir. Immune Defic. Syndr. 43:440–445 [DOI] [PubMed] [Google Scholar]
- 65. Land AM, Luo M, Pilon R, Sandstrom P, Embree J, Wachihi C, Kimani J, Plummer FA, Ball TB. 2008. High prevalence of genetically similar HIV-1 recombinants among infected sex workers in Nairobi, Kenya. AIDS Res. Hum. Retroviruses 24:1455–1460 [DOI] [PubMed] [Google Scholar]
- 66. Khawcharoenporn T, Apisarnthanarak A, Gesprasert G, Jaiyen Y, Mundy LM, Thitithanyanont A. 2011. Predictors for recombinant HIV infection in a Thai cohort. Sex. Transm. Dis. 38:1046–1049 [DOI] [PubMed] [Google Scholar]
- 67. Rerks-Ngarm S, Brown AE, Khamboonruang C, Thongcharoen P, Kunasol P. 2006. HIV/AIDS preventive vaccine ‘prime-boost’ phase III trial: foundations and initial lessons learned from Thailand. AIDS 20:1471–1479 [DOI] [PubMed] [Google Scholar]
- 68. Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, Decker J, Wibmer CK, Gao F, Alam SM, Easterbrook P, Abdool Karim S, Kamanga G, Crump JA, Cohen M, Shaw GM, Mascola JR, Haynes BF, Montefiori DC, Morris L. 2011. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol. 85:11502–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moore PL, Gray ES, Sheward D, Madiga M, Ranchobe N, Lai Z, Honnen WJ, Nonyane M, Tumba N, Hermanus T, Sibeko S, Mlisana K, Abdool Karim SS, Williamson C, Pinter A, Morris L. 2011. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J. Virol. 85:3128–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. van Gils MJ, Bunnik EM, Boeser-Nunnink BD, Burger JA, Terlouw-Klein M, Verwer N, Schuitemaker H. 2011. Longer V1V2 region with increased number of potential N-linked glycosylation sites in the HIV-1 envelope glycoprotein protects against HIV-specific neutralizing antibodies. J. Virol. 85:6986–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chaillon A, Braibant M, Moreau T, Thenin S, Moreau A, Autran B, Barin F. 2011. The V1V2 domain and an N-linked glycosylation site in the V3 loop of the HIV-1 envelope glycoprotein modulate neutralization sensitivity to the human broadly neutralizing antibody 2G12. J. Virol. 85:3642–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Saunders CJ, McCaffrey RA, Zharkikh I, Kraft Z, Malenbaum SE, Burke B, Cheng-Mayer C, Stamatatos L. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069–9080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ng OT, Eyzaguirre LM, Carr JK, Chew KK, Lin L, Chua A, Leo YS, Redd AD, Quinn TC, Laeyendecker O. 2012. Identification of new CRF51_01B in Singapore using full genome analysis of three HIV type 1 isolates. AIDS Res. Hum. Retroviruses 28:527–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Swanson P, Devare SG, Hackett J., Jr 2003. Full-length sequence analysis of HIV-1 isolate CM237: a CRF01_AE/B intersubtype recombinant from Thailand. AIDS Res. Hum. Retroviruses 19:707–712 [DOI] [PubMed] [Google Scholar]
- 76. Tovanabutra S, Kijak GH, Beyrer C, Gammon-Richardson C, Sakkhachornphop S, Vongchak T, Jittiwutikarn J, Razak MH, Sanders-Buell E, Robb ML, Suriyanon V, Birx DL, Michael NL, Celentano DD, McCutchan FE. 2007. Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in northern Thailand. AIDS Res. Hum. Retroviruses 23:829–833 [DOI] [PubMed] [Google Scholar]
- 77. McCutchan FE, Hoelscher M, Tovanabutra S, Piyasirisilp S, Sanders-Buell E, Ramos G, Jagodzinski L, Polonis V, Maboko L, Mmbando D, Hoffmann O, Riedner G, von Sonnenburg F, Robb M, Birx DL. 2005. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J. Virol. 79:11693–11704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Carr JK, Avila M, Gomez Carrillo M, Salomon H, Hierholzer J, Watanaveeradej V, Pando MA, Negrete M, Russell KL, Sanchez J, Birx DL, Andrade R, Vinoles J, McCutchan FE. 2001. Diverse BF recombinants have spread widely since the introduction of HIV-1 into South America. AIDS 15:F41–F47 [DOI] [PubMed] [Google Scholar]
- 79. McCutchan FE, Carr JK, Murphy D, Piyasirisilp S, Gao F, Hahn B, Yu XF, Beyrer C, Birx DL. 2002. Precise mapping of recombination breakpoints suggests a common parent of two BC recombinant HIV type 1 strains circulating in China. AIDS Res. Hum. Retroviruses 18:1135–1140 [DOI] [PubMed] [Google Scholar]
- 80. Fan J, Negroni M, Robertson DL. 2007. The distribution of HIV-1 recombination breakpoints. Infect. Genet. Evol. 7:717–723 [DOI] [PubMed] [Google Scholar]
- 81. Levy JA. 2007. HIV and the pathogenesis of AIDS, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 82. Delgado E, Thomson MM, Villahermosa ML, Sierra M, Ocampo A, Miralles C, Rodriguez-Perez R, Diz-Aren J, Ojea-de Castro R, Losada E, Cuevas MT, Vazquez-de Parga E, Carmona R, Perez-Alvarez L, Medrano L, Cuevas L, Taboada JA, Najera R. 2002. Identification of a newly characterized HIV-1 BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J. Acquir. Immune Defic. Syndr. 29:536–543 [DOI] [PubMed] [Google Scholar]
- 83. Arroyo MA, Phanuphak N, Krasaesub S, Sirivichayakul S, Assawadarachai V, Poltavee K, Pankam T, Ananworanich J, Paris R, Tovanabutra S, Kijak GH, McCutchan FE, Phanuphak P, Kim JH, de Souza M. 2010. HIV type 1 molecular epidemiology among high-risk clients attending the Thai Red Cross anonymous clinic in Bangkok, Thailand. AIDS Res. Hum. Retroviruses 26:5–12 [DOI] [PubMed] [Google Scholar]
- 84. Robb ML, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Kunasol P, Khamboonruang C, Thongcharoen P, Morgan P, Benenson M, Paris RM, Chiu J, Adams E, Francis D, Gurunathan S, Tartaglia J, Gilbert P, Stablein D, Michael NL, Kim JH. 2012. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect. Dis. 12:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tovanabutra S, Brodine SK, Mascola JR, Sankale JL, Sanders-Buell E, Kim B, Birx DL, McCutchan FE. 2005. Characterization of complete HIV type 1 genomes from non-B subtype infections in U.S. military personnel. AIDS Res. Hum. Retroviruses 21:424–429 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.