Abstract
The emergence of high-throughput technologies for measuring biological systems has introduced problems for data interpretation that must be addressed for proper inference. First, analysis techniques need to be matched to the biological system, reflecting in their mathematical structure the underlying behavior being studied. When this is not done, mathematical techniques will generate answers, but the values and reliability estimates may not accurately reflect the biology. Second, analysis approaches must address the vast excess in variables measured (e.g. transcript levels of genes) over the number of samples (e.g. tumors, time points), known as the ‘large-p, small-n’ problem. In large-p, small-n paradigms, standard statistical techniques generally fail, and computational learning algorithms are prone to overfit the data. Here we review the emergence of techniques that match mathematical structure to the biology, the use of integrated data and prior knowledge to guide statistical analysis, and the recent emergence of analysis approaches utilizing simple biological models. We show that novel biological insights have been gained using these techniques.
Keywords: Bayesian analysis, computational molecular biology, signal pathways, metabolic pathways, databases
INTRODUCTION
In the 1980s, sequencing technology advanced rapidly leading to the creation of GenBank [1] and the need for data analysis tools that permitted researchers to compare large numbers of sequences [2]. Until quite recently, sequence data has been gathered with low error rates on stable genomes, which simplifies analysis. The advent of microarrays in biological research [3, 4] resulted in a new dynamic type of data, where error levels were initially quite high and remain in the 5 – 10% range, perhaps reflecting not technical variation but inherent ‘biological’ noise. In fact, such biological noise has now been shown to have significant phenotypic consequences and to be an aspect driving some biological behaviors, such as cell death [5]. Dynamic data with high error rates raised new issues for analysis, and we are only beginning to fully address the difficulties of such analyses.
In addition, with the emergence of SNPchips, microarrays focused on measurement of single nucleotide polymorphisms (SNPs), statistical geneticists raised a new issue that had been ignored by most researchers focused on expression microarrays—the problem of high dimensionality [6]. In essence the problem is that standard statistical techniques, when limited to relatively small numbers of samples, lack power in the face of thousands or millions of variables of potential interest. Genetic interactions, where one SNP or allele influences the effect on phenotype of another, lead to a further combinatorial explosion, essentially eliminating any hope of making meaningful statistical statements on genetic interactions from the data alone, even if millions of individuals were to be included in a study.
Presently, high-throughput data is routinely generated for genome sequences (next generation sequencing), polymorphisms (SNPchips), transcripts (expression microarrays), miRNAs (microRNA arrays), proteins (mass spectrometry and protein microarrays) and metabolites (mass spectrometry and NMR). Genome-wide association studies (GWAS) have now identified ∼50 disease susceptibility loci [7], and extremely large studies with multiple institutional collaborators and diverse populations are being undertaken [8]. For proteomics, the overwhelming complexity of the proteome coupled to the dominance of a small set of high abundance proteins in terms of total protein mass creates unique problems for analysis. Early work focused on 2D gel separation, which limited data volume, however gel-free separation techniques were developed leading to greatly increased throughput [9]. In addition, antibody arrays, where protein- and phosphoprotein-specific antibodies are spotted onto a substrate have been developed [10], permitting studies similar to expression microarrays though more limited in coverage. With the development of mass spectrometric methods [11], metabolomic studies have become more common as well. For all of these technologies, experiments involving integration with other molecular data types have been performed and are discussed below.
One path forward for the analysis of integrated data was identified early in the development of statistics by Bayes [12] and rediscovered and extended by Laplace [13]. The standard Bayesian paradigm relies on knowledge that is independent of but related to the data to guide the potential inferences to those that are most probable. This is summarized in Bayes’ equation,
where the left-hand side is known as the posterior distribution and represents the goal of the analysis, the probability that the model (M) is correct given the data (D). The right-hand side includes the probability that the data arises from the model (the likelihood), the probability of the model itself (the prior), and the probability of the data itself (the evidence or marginal likelihood of the data). The prior, p(M), is the source of additional information that can guide Bayesian statistical techniques to focus only or primarily on model distributions that agree with existing biological knowledge. The data and model may in fact be complex, comprising multiple interdependent models and integrated data sets, which results in a more complex form of Bayes’ equation. However, the modification of the posterior probability distribution by the prior remains the key feature of any Bayesian approach. It is worth noting that all data analysis makes assumptions about the underlying model to be fit, such as linear relationships in linear regression or mathematical models of planetary motion in Newtonian or Einsteinian mechanics. Bayesian methods specifically modify these models by including prior information on the distribution of the parameters in the form of the prior.
MICROARRAY ANALYSIS: WHERE IT BEGAN
With microarrays biologists suddenly had data sets comprising thousands of simultaneous measurements of transcript levels. Early analysis methods, beyond simple statistical tests between classes, were borrowed from other fields, such as hierarchical clustering from phylogenetics [14]. These methods, which were primarily clustering approaches, introduced the concept of ‘guilt by association’ (GBA), where similar expression profiles were taken to indicate similar function. There has been ongoing debate about the validity of GBA. One reason for differences in interpretation of the validity probably lies in the thresholds applied in different applications. In receiver operating characteristic (ROC) analysis [15], clustering tends to deviate from the expectation of random classification only at high specificity (for example, see the results for three clustering metrics compared to a gold standard in ref. [16]). This portion of the ROC curve can be considered to contain the ‘low hanging fruit’, where the genes showing the highest similarity across all samples group together. This suggests that GBA may work well for estimating gene-gene relationships, such as shared gene ontology terms, when expression profiles are highly similar, but that it could fail as expression profiles become only moderately similar (i.e. at the edges of clusters). One way dissimilarity can arise between expression profiles of genes sharing a function is for one of the genes to be involved in a second biological function activated in a different subset of samples.
The earliest methods to address biological knowledge during analysis focused on two main issues that clustering ignored—multiple-regulation of genes due to gene reuse in different biological processes and non-orthogonality of biological process activity arising from the natural simultaneity of biological behaviors. We modified our Bayesian Decomposition (BD) algorithm, a Markov chain Monte Carlo algorithm for medical spectroscopic imaging [17], to address these two issues in microarray studies [18]. Kim and Tudor, and Brunet and colleagues independently extended non-negative matrix factorization (NMF), introduced by Lee and Seung for image analysis [19], to microarray analysis [20,21]. Subsequently it was realized that sparseness is critical to the capture of patterns strongly tied to biological processes in such matrix factorization methods, and Gao and Church provided a sparse NMF method [22]. Fortuitously, sparseness was already a feature of BD through its atomic prior [23], as its initial use was in spectroscopy where peaks tend to be isolated features on a noisy background. More recently, Carvalho and colleagues introduced Bayesian Factor Regression Modeling (BFRM), an additional Markov chain Monte Carlo method for microarray data analysis [24]. While BD identifies patterns that provide a non-orthogonal basis for the data, BFRM first isolates the mean behavior of a gene and then performs an analysis that focuses on differences in transcript levels between samples. BD is therefore useful for systems modeling where projection onto biological functions is desired, while BFRM provides discrimination between samples in terms of gene signatures, as is desired in biomarker discovery [25].
While these methods address some of the underlying issues in the initial approaches to the analysis of high-throughput data, they cannot alone overcome the curse of dimensionality. It is conceivable and perhaps likely that distinct distributions of genes into patterns, especially as the patterns cannot generally be known a priori, can reproduce the measured data. An example is shown in Figure 1, where a small set of genes show cell cycle related behaviors, and two different sets of patterns are shown (Figure 1B and C), both of which can equally well explain all the observed behavior. In this case, we have purposely included no genes that appear upregulated only in G1 phase, so that the difference between the two sets of patterns is that one has a pattern related only to G1 and the other a pattern showing G1 and G2 simultaneously. Mathematically, there is no way to choose between these two sets of patterns, as each fit the data equally well and are parsimonious. However, biologically we prefer G1 and G2 to be separate phases of the cell cycle, and such a preference can be encoded in an algorithm as a prior, reducing the probability of the solution shown in Figure 1C. While this is an artificial and highly simplified construct, it shows, at its most basic, the idea of allowing biological knowledge to guide analysis.
Figure 1:
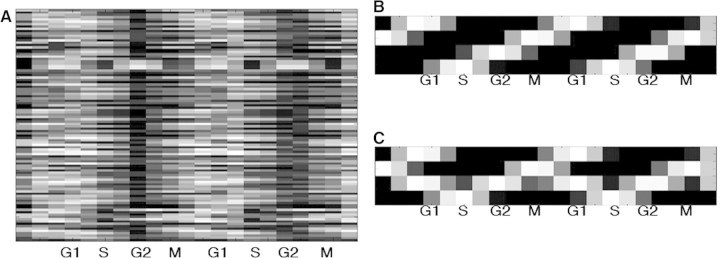
An example of two mathematical solutions to matrix factorization. The 100 genes show regulation in each of the four cell cycle phases (A). The logical set of patterns to explain the data show upregulation in each cycle (B). However, in this artificial example, G2 is always coexpressed with G1, permitting another equally valid mathematical solution (C). Prior knowledge can be used to inform an algorithm to prefer B over C.
INTEGRATED DATA
Prior knowledge requires information from outside the measured data, and this can come in two forms. The first is knowledge gained in previous experiments and codified in the literature or databases. The second is data from different domains [e.g. transcription factor (TF) binding from chromatin immunoprecipitation on microarray (ChIP-chip) experiments] that can be used to provide prior probabilities for analysis (e.g. the probability that a gene is transcribed depends on the transcript levels of other genes with the same TF binding site).
However, it is also often useful to simply integrate all data into a single data set for analysis. Daemen and colleagues demonstrated that integrating global microarray and targeted proteomics data together provided a more accurate signature for the response to cetuximab in rectal cancer patients [26], even though the proteins measured were not even mapped to their corresponding genes but instead served as independent data points. An approach that links a gene to its encoded protein was introduced by Wabnik and colleagues, who applied rough sets to integrated data [27]. They integrated gene ontology data, protein feature data derived from amino acid sequence, and transcript data in a supervised learning approach to generate rules that predicted gene ontology annotations for proteins of unknown function. English and Butte brought together 49 obesity related data sets comprising microarray, genetics, proteomics and knock-down experiments in human, mouse, rat and worm, which required linking of molecular types across different species [28]. This improved discovery of obesity related genes, as confirmed by ROC analysis of gold-standard genes.
Kim and colleagues integrated array-CGH (aCGH) and expression data in a study of prostate cancer progression [29]. Here copy number alterations in sets of patients were compared to expression changes in the genes related to the identified aCGH regions to rank genes related to progression. Essentially this method looked for overlap in lists of genes generated from analyses of different types of data. Use of overlap in lists of genes can be done using ChIP-chip data as well. Yu and colleagues focused on the key oncogenic transcription factor EZH2 in a study of prostate cancer [30]. This study integrated transcript data from a focused cell line study of EZH2 disregulation, Oncomine tumor signatures, and PRC2 (polycomb repressor complex containing EZH2) ChIP-chip data to identify candidate genes directly regulated by EZH2.
While studies like these are powerful, we expect that statistical techniques that go beyond mere overlap of gene lists will increase power. In some ways, the overlapping gene list approach is similar to GBA in that the genes with the strongest signatures will be found; here those signatures are coordinated changes in different data domains (e.g. TF binding and expression) rather than similarity of expression profile. As we remain in the early days of systems biology, such ‘low-powered’ approaches are likely to continue to yield important discoveries, however, reanalysis of the data with more statistical and model-oriented approaches may provide further insights. Carey and Gentleman introduced an R/Bioconductor framework that integrates genomic and transcriptomic data, and extensions should permit application of the plethora of R tools to integrated data [31]. Naturally, this structure permits integrated data to be used as prior information as well.
PRIOR KNOWLEDGE IN MICROARRAY ANALYSIS
Not surprisingly, microarray analysis, as the oldest high-throughput technology in biology, has seen the most activity in the use of prior information to guide analysis. Part of the drive for the use of prior information arose from attempts to reconstruct genetic networks, or transcriptional regulatory networks (TRNs), from time series microarray data. An early success using a Bayesian Network (BN) in yeast [32] generated excitement in the field, but the approach did not succeed in more complex organisms. It was realized that for TRNs additional information from TF binding could reduce the number of genes that potentially were responding directly to the expression of a TF, and in a seminal paper, Lee and colleagues reproduced the regulatory network of S. cerevisiae during the cell cycle using ChiP-chip and expression arrays [33].
Ucar and colleagues extended this approach to include nucleosome occupancy measurements in order to refine the TF binding prediction to include the concept of functional binding, where the TF is not only bound but is active, which correlates with nucleosome occupancy [34]. The FANTOM and Riken consortiums used a different approach to address the issue of active TFs in regulatory networks [35]. Using deepCAGE sequencing of mRNA transcripts coupled to TF binding site motifs from the JASPAR [36] and TRANSFAC [37] databases, they created a time series prediction of active TF binding sites. These were then used to interpret expression data and reproduce a TRN in time series data. Alternatively, curated data on validated regulation of genes by TFs in TRANSFAC can be used as prior knowledge to enhance the statistical power of Bayesian methods, as done by Kossenkov and colleagues with BD [38]. A similar approach based on experiments that modify receptor signaling has been used to generate summaries of transcript changes downstream from these receptors [39].
Recently, BNs, which relate all gene pairs in a graphical structure with an edge defining a probability of relationship, have returned to popularity. Djebbari and Quackenbush used literature review and protein–protein interaction (PPI) data to seed a BN with probabilities of gene–gene expression relationships [40]. This seeded BN served as a prior structure that was then modified based on the expression data, improving recovery of true gene–gene interactions. In this work, the BN could encode phenomenological relationships in addition to direct interactions, which is discussed in detail in the section on pathway-focused analysis below. Ulitsky and Shamir used a similar approach in their algorithm, CEZANNE, where they seeded a graphical model of gene-gene interactions from PPI data [41]. The analysis then proceeded from the expression data and improved recovery of functional modules.
Mani and colleagues took a different approach to the use of prior information from network or graphical relationships in their study of B cell lymphomas [42]. Beginning with the curated B cell interactome database, which provides a network of probable interactions, they looked for changes in mutual information between genes in terms of lymphoma subtypes. They referred to these as gain of correlation (GoC) and loss of correlation (LoC) relationships, and they showed that these are related to gain or loss of regulatory behaviors that distinguish the different types of cancerous B cells.
An interesting recent paper reversed the typical information flow and used microarray data as prior information for identification of proteins from shotgun proteomics [43]. Ramakrishnan and colleagues identified proteins likely to be present in a sample based on mRNA levels under similar experimental conditions and used this information to improve their ability to identify proteins from MS/MS peaks.
Alternatively, one can set up a two-way information flow between transcript and protein levels during clustering. Rogers and colleagues used coupled clustering, where the data itself was used to adjust the strength of the coupling between transcript level and protein level driving the clustering [44]. This approach led to recovery of clusters of proteins and transcripts that were linked functionally. Interestingly, the relationship between transcript and protein clusters was rarely one-to-one, suggesting that strong functional coupling between biological processes is typical.
GENOME-WIDE ASSOCIATION STUDIES
As noted in the Introduction section, statistical geneticists were among the first to raise the alarm over the inability of standard analytical methods to address the large number of variables in high-throughput studies. One of the primary platforms leading to this concern was the SNPchip, capable of measuring hundreds of thousands to millions of genetic variants, which simply overwhelms the ability of traditional statistical tests to obtain significance in any potential study sample size. Not surprisingly, therefore, knowledge-based analysis methods are beginning to appear to address the problem of identifying SNPs that will have an impact on disease or treatment response.
One approach is to focus on the effects of SNPs on gene expression. Huang and colleagues in two studies identified SNPs associated with growth inhibition by chemotherapeutic agents in cell lines [45,46]. These SNPs were then linked to gene expression changes in the HapMap cell lines, and a statistical model identified SNPs associated with gene expression changes that correlated with chemotherapeutic response. This effectively nominated candidate SNPs to explain differences in sensitivity to chemotherapy through integration of expression and genotype data.
Protein structure and promoter structure provide additional points of leverage for prior knowledge in a SNP-based study. While it is now clear that non-coding and non-regulatory regions of the genome have impacts on disease in ways not fully understood, it seems reasonable that a SNP that changes an amino acid or disrupts of TF binding site may be more likely to impact disease. Lee and Shatkay determined SNP location in terms of exons, splice sites, TF binding sites, microRNAs and amino acids that have post-translational modification sites to score the likely SNP effect [47]. Carter and colleagues focused on differentiating driver mutations from passenger mutations within missense mutations occurring in cancer [48]. This work, based on random forest classification trained on known driver and passenger mutations, showed excellent area under the ROC curve (AUC = 0.91) and predicted 8% of mutations identified in a recent glioblastoma study are driver mutations [49]. Other drivers of protein function are protein interaction domains. Chen and Jeong created a random forest classifier based on protein physicochemical features, amino acid composition, and amino acid substitution rates to predict protein interaction sites [50].
To date, these protein-focused methods have been used to study SNPs that have been ranked in statistical tests. Now that these methods exist, the logical next step will be to apply them during statistical analysis, allowing them to create prior distributions based on the probability that a mutation may be deleterious. Methods for utilizing such prior knowledge have been created for expression-QTL (eQTL), where microarray data and genotype data are integrated to improve QTL studies. For instance, Degnan and colleagues recently demonstrated the use of microarray data as a phenotypic measure to focus on SNPs that might drive expression changes [51].
THE EMERGENCE OF PATHWAY-FOCUSED ANALYSIS
Throughout the last decade there has been a growing realization that it may be more useful to focus on pathways rather than on individual genes. This has been strongly validated recently for signaling pathways in glioblastoma multiforme [49], where the PI3K, RAS and P53 pathways are almost always modified, but in each individual tumor different genes in these pathways can be affected. This was further validated in a larger data set by the Cancer Genome Atlas Research Network [52]. These studies followed earlier work showing that there were limited numbers of driver mutations in breast and colorectal cancer, but that the driver mutations shared functional assignments [53]. This pathway-centric view of disease provides an impetus for the development of integrated analysis methods to discover drug targets [54].
In biological studies, the term ‘pathway’ is not highly specific. It can describe a specific set of metabolic reactions, the enzymes (i.e. proteins) involved, and the small molecules generated. This ‘metabolic’ pathway is a well-defined set of molecular entities and each molecular modification and component is believed to be included in the pathway description. Alternatively, ‘pathway’ can refer to a set of protein–protein interactions that lead to conformational changes in proteins and transduce a signal through the cell, known as a ‘signaling’ pathway, where some steps and components remain unknown. However, ‘pathway’ often describes a series of changes, in which many intermediate events remain unknown, such as in TRNs where many cellular reactions may be ignored, or genetic pathways in which ‘downstream’ and ‘upstream’ are defined by epistasis in deletion experiments. Genetic networks can involve multiple types of molecular networks, including signaling and transcriptional, as both types of pathways can lead to changes in phenotypes.
Data analysis has utilized these different pathway definitions to link data together. Both the connectivity map [55] and molecular concepts analysis [56] treated pathways as conceptual links that integrated gene expression signatures with other data types. The connectivity map identified correlations between changes in gene expression and small molecules, then related these to possible disease states, linking targets of small molecules to disease. Molecular concepts analysis added pathways and gene ontology functions and generated networks linking genes, drugs, pathways, transcription factor binding sites and expression changes.
Since many of our insights into biological pathways come from model organisms, it is not surprising that we have detailed phylogenetic and orthological information for human pathways in organisms from yeast to mouse. Liu and colleagues leveraged this orthology by comparing mouse lung development at 10 stages to human lung cancer staging [57]. Using principal component analysis, they identified signatures in the mouse transcript data and through the use of orthologs were able to project human tumor transcript data onto these. The results predicted outcome, suggesting lung cancer development mirrored certain aspects of natural lung development. Alexeyenko and Sonnhammer fully exploited the use of orthologs and data integration in FunCoup [58], a Bayesian framework that integrates microarray, miRNA target prediction, localization, protein–protein interactions (PPI), protein expression, phylogenetic profiling, TF binding site and protein domain data from plant through human. A Naïve Bayes Network (NBN) trained on known interactions is used to predict novel networks. By holding out data on one organism, they demonstrated its ability to recover known networks from orthologous data alone.
As noted for microarray analysis, BNs are popular as they permit probabilistic links between nodes (in that case, genes). For BNs, the networks are often conceptual, allowing diverse data to be linked, with a node representing expression of a gene connected to a node representing a protein by an edge with a defined probability (e.g. a gene's expression to an upstream signaling protein that drives its expression indirectly). As these ‘pathways’ in a BN are abstractions, they do not necessarily represent direct molecular interactions, although a BN can be limited to direct molecular interactions if desired. One very successful use of an NBN was the creation of a genome-wide functional network for the mouse, MouseNET, by Guan and colleagues [59]. This network integrated PPI including homologous interactions, phenotype and disease data from MGI and OMIM, phylogenetic profiles, functional relationship predictions, and tissue specific expression data. Importantly, because NBNs assume independence, Guan and colleagues tested data for such independence, allowing them to exclude data that was effectively derivative to avoid skewing statistics.
A number of studies have relied on PPI data, where the pathway is a representation of direct molecular interactions, to guide analysis of microarray data in human disease. The concept in these studies is that PPI networks provide a logical point to increase the probability of coordinated expression a priori. Unlike the studies in glioblastoma, where different genes show aberrations within a pathway, the target here is coordinated changes in genes linked together through PPI. This could represent the result of coordinated regulation by biological processes that have been disregulated, affecting a full pathway, or feedback where variation in one component leads to variations in linked molecules. Chuang and colleagues identified statistically significant transcript differences between breast cancer metastatic states and identified subnetworks in PPI data with coordinated expression changes [60], using permutation testing for significance estimation. The analysis recovered a number of key pathways, including the P53, SMAD4, ERBB2, RAS and MYC pathways. Liu and colleagues performed a similar analysis in type 2 diabetes, relying on the number of samples showing a subnetwork to be coordinately changed instead of permutation tests for ranking subnetworks [61]. The analysis recovered insulin signaling and nuclear receptor networks as consistently differentially expressed as expected.
Heiser and colleagues applied a different approach for identifying the networks driving disease using breast cancer cell line data [62]. They established an initial signaling network model based on prior pathway knowledge. Initial states involved cell line status information, such as mutation status in key proteins. This initial network than evolved based on the data and sets of rules on binary states, such as [if kinase HRAS is active, kinase BRAF is active]. Working from mutation status, copy number variation, protein levels, and microarray data, they identified key nodes that were context dependent, suggesting potential personalized treatment targets.
ANALYSIS USING BIOLOGICAL MODELS
The last study suggests a transition from prior information through linking (e.g. protein isoform to gene as in PPI subnetworks) to a view of a complex system. A systems view allows more detailed modeling, including relationships such as a protein isoform being related not only to the gene encoding it but also to a different protein isoform or a set of genes through a transcription factor. The relationships are more definite than in a general BN, as they now involve both probabilities of interaction but also definitive rules. Bidaut and colleagues used knowledge of TF factor regulation to interpret results of BD analysis of the Rosetta compendium of yeast deletion mutants [63], identifying transcriptional signatures related to MAPK signaling [64]. Chang and colleagues utilized a pathway model to identify core sets of genes showing transcriptional responses to changes in signaling, enlarged this set with BFRM, and then identified these signatures in tumor samples [65].
Enlarging this approach to include metabolite levels, metabolic pathways, and expression has led to a substantial novel insight. Sreekumar and colleagues utilized prior knowledge on gene expression and TF binding in prostate cancer to identify a change in a key metabolite associated with prostate cancer progression [66]. Sarcosine was one of many metabolites to show substantial changes in levels during prostate cancer progression, however it is produced by the enzyme GNMT, a methyl transferase with an androgen receptor binding site upstream. As androgen is known to play an important role in prostate cancer aggressiveness, this allowed prediction that sarcosine might serve as a marker of aggressiveness and potentially even be a driver of such aggressiveness, which was validated in cell line studies. This has led to a follow-up study of sarcosine as a biomarker for prostate cancer detection.
While limited in scope, this last study shows the potential of knowledge-based analysis, even when only a very limited biological model can be created. While biological systems are so complex that an overarching theory remains in the distant future, limited models that encompass adequate complexity to allow deep insight are now possible due to the vast gathering of information that began with Mendel and continues to accelerate. Importantly, biological systems are highly non-linear with highly interconnected subsystems, which guarantees that non-mathematical models will fail to capture all reliable predictions [67]. Integrating the prior knowledge with appropriate non-linear models should greatly reduce the risk of overlooking valuable insights into the processes underlying disease and treatment.
CONCLUSION
The last decade has seen rapid advances in the use of biological knowledge to guide analysis. At its simplest, this has been through integration of data from different molecular domains; for instance recognizing that a gene encodes a specific protein, thus allowing data from microarrays and PPI networks to be analyzed together. The integration has become more powerful with increasing numbers of data types, which now include pathway models, disease phenotypes, genetic aberrations, protein structural information, and gene expression data. The statistical power is greatest when we can formalize our knowledge, as in a Bayesian framework, and is most predictive when we can create a full mathematical model permitting simulation and hypothesis generation.
Integration of data for analysis often requires retrieval of data from a variety of databases and knowledge-bases. Unfortunately, this is not a trivial matter in most cases, as the knowledge-bases often use incompatible standards making automated retrieval of information extremely difficult. A number of efforts aim to improve this through use of controlled vocabularies and ontologies, including the cancer biomedical informatics grid (caBIG®) initiative [68] and the open biological ontologies effort [69]. For medical data, the situation is somewhat better, with the unified medical language system well established [70], although not always incorporated into existing systems. While critical to the use of knowledge as prior information for analysis, a review of the issues and technologies for data retrieval is beyond the scope of this work.
Two often overlooked issues play a critical role in knowledge-based analysis—data provenance and covariance within high-throughput data. Data provenance is crucial to encoding the probability of prior knowledge, which relies on knowing when multiple sources of evidence are independent. Unfortunately, through data sharing between databases and limited traceability of information, it is often impossible to verify independence. This affects our ability to assign prior probabilities. Covariance within data is equally problematic. Most statistical techniques assume independence, and as biological processes naturally lead to correlations in measurements of biological entities, such as transcripts, the statistics can become highly skewed. These two problems both lead to the same result—an incorrect statistical estimate of differences or similarities within a biological system. One must keep both these issues in mind when performing analyses using knowledge from databases or high-throughput data, and, as was done for MouseNET, performing tests of data independence can often be useful.
Looking forward, the goal for knowledge-based analysis should be the creation of a full mathematical models of biological processes. Looking back, we can note that one of the greatest achievements of the last century, our ability to model quantum processes, led to technologies unthinkable in 1910, including instantaneous worldwide communications, medical imaging and laser treatments. It is important to remember that quantum theory did not emerge instantly with Newton's brilliance. Instead, simpler models were first created and tested against experimental data. The models were refined over hundreds of years, until deep insights suitable to the development of new areas of engineering emerged. However, none of these developments would have been possible with a purely phenomenological, non-mathematical approach to physics. Biology will require an equivalent evolution in mathematical methods, from the development of specialized data analysis techniques to test biological hypotheses to the creation of models suitable for mathematical exploration.
Key Points.
Biological systems are complex, and the mathematical structure of statistical and computational analysis techniques should be matched to the underlying biological structure.
High-throughput data platforms generate thousands to millions of simultaneous measurements while sample sizes remain limited, leading to the ‘curse of dimensionality’, which limits the power of statistical and computational learning approaches.
Leveraging existing biological knowledge from decades of traditional research and integrating data from different molecular domains can increase the power of mathematical methods, if they are designed to exploit this information.
Bayesian statistics provides a proven and well-developed framework for inclusion of existing knowledge through the use of prior distributions.
The greatest insights have arisen from the analysis of data in light of models of the biological process being studied, even though these models remain extremely limited.
Biography
Michael Ochs has been trained in astrophysics with a focus on the structure of quasars. His present research at Johns Hopkins University is on the use of Bayesian statistical approaches and computational modeling in cancer research.
References
- Burks C, Fickett JW, Goad WB, et al. The GenBank nucleic acid sequence database. Comput Appl Biosci. 1985;1:225–33. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–80. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, et al. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–32. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B. Finding genes influencing susceptibility to complex diseases in the post-genome era. Am J Pharmacogenomics. 2001;1:203–21. doi: 10.2165/00129785-200101030-00005. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Manolio TA. Collaborative genome-wide association studies of diverse diseases: programs of the NHGRI's office of population genomics. Pharmacogenomics. 2009;10:235–41. doi: 10.2217/14622416.10.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe MR, Griffin TJ. Gel-free mass spectrometry-based high throughput proteomics: tools for studying biological response of proteins and proteomes. Proteomics. 2006;6:4678–87. doi: 10.1002/pmic.200500876. [DOI] [PubMed] [Google Scholar]
- Borrebaeck CA, Wingren C. High-throughput proteomics using antibody microarrays: an update. Expert Rev Mol Diagn. 2007;7:673–86. doi: 10.1586/14737159.7.5.673. [DOI] [PubMed] [Google Scholar]
- Han J, Danell RM, Patel JR, et al. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics. 2008;4:128–40. doi: 10.1007/s11306-008-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes T. An essay towards solving a problem in the doctrine of chances. Philos Trans Royal Soc. 1763;53:370–418. [PubMed] [Google Scholar]
- Laplace P-S. Mémoire sur la probabilité des causes par les événements, Memoires de mathématique et de physique présentés à l'Académie Royale des Sciences, par divers savants, et lûs dans ses assemblées 6 1774. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko TA, Bhagwat JG, Zou KH, et al. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform. 2005;38:404–15. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Cherepinsky V, Feng J, Rejali M, et al. Shrinkage-based similarity metric for cluster analysis of microarray data. Proc Natl Acad Sci USA. 2003;100:9668–73. doi: 10.1073/pnas.1633770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs MF, Stoyanova RS, Arias-Mendoza F, et al. A new method for spectral decomposition using a bilinear Bayesian approach. J Magn Reson. 1999;137:161–76. doi: 10.1006/jmre.1998.1639. [DOI] [PubMed] [Google Scholar]
- Moloshok TD, Klevecz RR, Grant JD, et al. Application of Bayesian Decomposition for analysing microarray data. Bioinformatics. 2002;18:566–75. doi: 10.1093/bioinformatics/18.4.566. [DOI] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–91. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- Brunet JP, Tamayo P, Golub TR, et al. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 2004;101:4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Tidor B. Subsystem identification through dimensionality reduction of large-scale gene expression data. Genome Res. 2003;13:1706–18. doi: 10.1101/gr.903503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Church G. Improving molecular cancer class discovery through sparse non-negative matrix factorization. Bioinformatics. 2005;21:3970–5. doi: 10.1093/bioinformatics/bti653. [DOI] [PubMed] [Google Scholar]
- Sibisi S, Skilling J. Prior distributions on measure space. J Royal Stat Soc B. 1997;59:217–35. [Google Scholar]
- Carvalho CM, Chang J, Lucas J, et al. High-dimensional sparse factor modelling: applications in gene expression genomics. J Am Stat Assoc. 2008;103:1438–56. doi: 10.1198/016214508000000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossenkov A, Ochs MF. Matrix factorization for recovery of biological processes from microarray data. In: Johnson M, Brand L, editors. Methods Enzymology, Computer Methods B. New York: Elsevier-Academic Press; 2009. pp. 59–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daemen A, Gevaert O, De Bie T, et al. Integrating microarray and proteomics data to predict the response on cetuximab in patients with rectal cancer. Pac Symp Biocomput. 2008;13:166–77. [PubMed] [Google Scholar]
- Wabnik K, Hvidsten TR, Kedzierska A, et al. Gene expression trends and protein features effectively complement each other in gene function prediction. Bioinformatics. 2009;25:322–30. doi: 10.1093/bioinformatics/btn625. [DOI] [PubMed] [Google Scholar]
- English SB, Butte AJ. Evaluation and integration of 49 genome-wide experiments and the prediction of previously unknown obesity-related genes. Bioinformatics. 2007;23:2910–7. doi: 10.1093/bioinformatics/btm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Dhanasekaran SM, Mehra R, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–39. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- Yu J, Cao Q, Mehra R, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–31. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Carey VJ, Davis AR, Lawrence MF, et al. Data structures and algorithms for analysis of genetics of gene expression with Bioconductor: GGtools 3.x. Bioinformatics. 2009;25:1447–8. doi: 10.1093/bioinformatics/btp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, et al. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–20. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Ucar D, Beyer A, Parthasarathy S, et al. Predicting functionality of protein-DNA interactions by integrating diverse evidence. Bioinformatics. 2009;25:i137–44. doi: 10.1093/bioinformatics/btp213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Forrest AR, van Nimwegen E, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–62. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–6. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossenkov AV, Peterson AJ, Ochs MF. Determining transcription factor activity from microarray data using Bayesian Markov Chain Monte Carlo sampling. Stud Health Technol Inform. 2007;129:1250–4. [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebbari A, Quackenbush J. Seeded Bayesian Networks: constructing genetic networks from microarray data. BMC Syst Biol. 2008;2:57. doi: 10.1186/1752-0509-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shamir R. Identifying functional modules using expression profiles and confidence-scored protein interactions. Bioinformatics. 2009;25:1158–64. doi: 10.1093/bioinformatics/btp118. [DOI] [PubMed] [Google Scholar]
- Mani KM, Lefebvre C, Wang K, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan SR, Vogel C, Prince JT, et al. Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics. 2009;25:1397–403. doi: 10.1093/bioinformatics/btp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Girolami M, Kolch W, et al. Investigating the correspondence between transcriptomic and proteomic expression profiles using coupled cluster models. Bioinformatics. 2008;24:2894–900. doi: 10.1093/bioinformatics/btn553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Kistner EO, et al. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol Cancer Ther. 2008;7:3038–46. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81:427–37. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, Shatkay H. An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics. 2009;25:1048–55. doi: 10.1093/bioinformatics/btp103. [DOI] [PubMed] [Google Scholar]
- Carter H, Chen S, Isik L, et al. Cancer-specific High-throughput Annotation of Somatic Mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–7. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XW, Jeong JC. Sequence-based prediction of protein interaction sites with an integrative method. Bioinformatics. 2009;25:585–91. doi: 10.1093/bioinformatics/btp039. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Lasky-Su J, Raby BA, et al. Genomics and genome-wide association studies: an integrative approach to expression QTL mapping. Genomics. 2008;92:129–33. doi: 10.1016/j.ygeno.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Gan CM, Zhang X, et al. A multidimensional analysis of genes mutated in breast and colorectal cancers. Genome Res. 2007;17:1304–18. doi: 10.1101/gr.6431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Tomlins SA, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–54. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kho AT, Kohane IS, et al. Predicting survival within the lung cancer histopathological hierarchy using a multi-scale genomic model of development. PLoS Med. 2006;3:e232. doi: 10.1371/journal.pmed.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyenko A, Sonnhammer EL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107–16. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Myers CL, Lu R, et al. A genomewide functional network for the laboratory mouse. PLoS Comput Biol. 2008;4:e1000165. doi: 10.1371/journal.pcbi.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, et al. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Liberzon A, Kong SW, et al. Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genet. 2007;3:e96. doi: 10.1371/journal.pgen.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser LM, Wang NJ, Talcott CL, et al. Integrated analysis of breast cancer cell lines reveals unique signaling pathways. Genome Biol. 2009;10:R31. doi: 10.1186/gb-2009-10-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–26. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Bidaut G, Suhre K, Claverie JM, et al. Determination of strongly overlapping signaling activity from microarray data. BMC Bioinformatics. 2006;7:99. doi: 10.1186/1471-2105-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Carvalho C, Mori S, et al. A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Mol Cell. 2009;34:104–14. doi: 10.1016/j.molcel.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Strogatz SH. Exploring complex networks. Nature. 2001;410:268–76. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- Cimino JJ, Hayamizu TF, Bodenreider O, et al. The caBIG terminology review process. J Biomed Inform. 2009;42:571–80. doi: 10.1016/j.jbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37:W170–3. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Libr Assoc. 1993;81:170–7. [PMC free article] [PubMed] [Google Scholar]


