Abstract
KFase (kynurenine formamidase), also known as arylformamidase and formylkynurenine formamidase, efficiently catalyses the hydrolysis of NFK (N-formyl-L-kynurenine) to kynurenine. KFase is the second enzyme in the kynurenine pathway of tryptophan metabolism. A number of intermediates formed in the kynurenine pathway are biologically active and implicated in an assortment of medical conditions, including cancer, schizophrenia and neurodegenerative diseases. Consequently, enzymes involved in the kynurenine pathway have been considered potential regulatory targets. In the present study, we report, for the first time, the biochemical characterization and crystal structures of Drosophila melanogaster KFase conjugated with an inhibitor, PMSF. The protein architecture of KFase reveals that it belongs to the α/β hydrolase fold family. The PMSF-binding information of the solved conjugated crystal structure was used to obtain a KFase and NFK complex using molecular docking. The complex is useful for understanding the catalytic mechanism of KFase. The present study provides a molecular basis for future efforts in maintaining or regulating kynurenine metabolism through the molecular and biochemical regulation of KFase.
Keywords: α/β hydrolase, diazinon, kynurenine formamidase (KFase), N-formyl-L-kynurenine (NFK), PMSF, tryptophan metabolism
INTRODUCTION
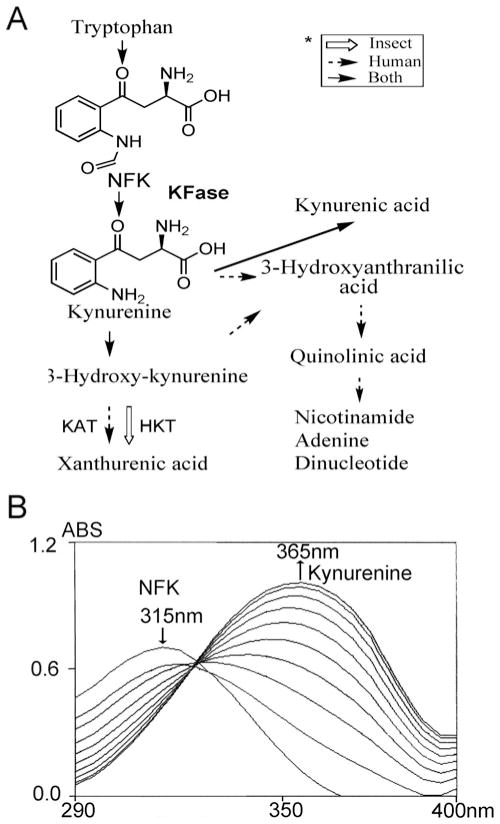
KFase (kynurenine formamidase; EC 3.5.1.9) catalyses the chemical reaction of NFK (N-formyl-L-kynurenine) and H2O to formic acid and kynurenine (Figure 1A). KFase is also capable of hydrolysing some other amides and esters [1]. Any tryptophan not used in protein synthesis is largely catabolized via the kynurenine pathway, a complex process involving a number of enzymes. In insects, the overall pathway includes oxidation of tryptophan to NFK, followed by hydrolysis of NFK to kynurenine. Kynurenine may then be either transaminated to kynurenic acid or hydroxylated to 3-hydroxykynurenine, which, in turn, is transaminated to xanthurenic acid [2]. In rodents and humans, kynurenines are further oxidized to nicotinamide adenine dinucleotide (Figure 1A).
Figure 1. Kynurenine pathway and KFase activity assay.
(A) Comparison of kynurenine pathways in insects and humans. Pathways specific to human are indicated by broken arrows and those specific to insects by open arrows. Shared pathways are indicated by normal arrows. HKT, 3-hydroxykynurenine transaminase; KAT, kynurenine aminotransferase. (B) Maximum absorbance for NFK and kynurenine is at 315 nm and 365 nm respectively. After adding KFase to the reaction mixture, the spectrum was scanned every 10 s. The production of kynurenine is indicated by the increase in absorbance at 365 nm and the decrease in absorbance at 315 nm. ABS, absorbance.
In mosquitoes, there is a significant load of tryptophan and its toxic tryptophan metabolite, 3-hydroxykynurenine after the proteolysis subsequent to a blood meal. However, 3-hydroxykynurenine is detoxified by 3-hydroxykynurenine transaminase, producing xanthurenic acid [3,4], which is known to play a key role in Plasmodium gametogenesis [5,6]. Thus it is possible that 3-hydroxykynurenine transaminase inhibitors could not only increase the levels of the toxic 3-hydroxykynurenine, but also reduce parasite reproduction in mosquitoes by reducing the concentrations of the critical Plasmodium developmental inducer molecule, xanthurenic acid. The crystal structures of 3-hydroxykynurenine transaminase and its homologue from mosquitoes have been solved, facilitating the development of novel insecticides to control mosquitoes and block malaria transmission [7,8].
In rodents and humans, several neuroactive intermediates are generated from the kynurenine pathway of tryptophan degradation. These include kynurenine, quinolinic acid, kynurenic acid and picolinic acid. Kynurenine is a potent vasodilator [9]; quinolinic acid is a potent endogenous excitant at amino acid receptors in the central nervous system [10]; kynurenic acid is an N-methyl-D-aspartate antagonist [11], and is implicated in the regulation of leucocyte binding to endothelium [12]; and picolinic acid is considered to be a neuroprotectant [13]. Interferon-γ and other cytokines, released during inflammation, stimulate the kynurenine pathway, resulting in the metabolism of tryptophan and the production of the kynurenine products [14,15]. This reduction in tryptophan and increase in kynurenines have been shown to modulate the immune response, largely by reduction of the T-cell population. It is not surprising that the kynurenine pathway has been implicated in several diseases and disorders having evidence of tryptophan and kynurenine imbalances, including acquired immune deficiency syndrome dementia complex, Alzheimer’s disease, Huntington’s disease, amyotrophic lateral sclerosis, neoplasia, malaria, depression, schizophrenia and cancer [16,17]. The involvement of the kynurenine pathway in these diseases suggests that research strategies targeting the kynurenine pathway may provide an alternative means of treatment. In fact, kynurenine metabolism enzymes are being studied for the development of cancer therapy [18], new antidepressant drugs that target the brain immune system [19,20], novel treatment of schizophrenia [21,22], Huntington’s disease [23] and others [24].
Because most metabolites from the kynurenine pathway are biologically active, their synthesis and further metabolism must be tightly controlled in humans. KFase, the second enzyme in the kynurenine pathway, may be just as important for kynurenine synthesis as the first enzyme, indoleamine 2,3-dioxygenase, which has already been targeted for the development of cancer therapy and antidepressant drugs [18–20]. KFase inhibitors may also reduce the metabolism of kynurenine, which is up-regulated in some diseases or disorders. For example, it has been found that inhibition of kynurenine pathway reduces central nervous system inflammation in a model of human African trypanosomiasis [25]. In mosquitoes, KFase inhibitors may potentially decrease the production of the signal molecule, xanthurenic acid, reducing the reproduction of the malaria parasite in mosquitoes.
Mouse KFase has been used as a model enzyme to study OP (organophosphorus) insecticide toxicity [26,27]. Several OP insecticides and their metabolites induce severe teratogenic effects, which seems due primarily to the inhibition of KFase activity, leading to lowered levels of nicotinamide adenine dinucleotide in developing chicken embryos [1,28]. It has been proposed that OP compounds may couple to the serine residue in the active site of KFases to inhibit the enzyme activity [1]. A crystal structure will validate how OP compounds inhibit KFase.
Although the crystal structure of an α/β serine hydrolase from yeast [29] has been identified as a KFase [30], it shares less than 10 % sequence identity with human KFase and Drosophila KFase. Drosophila KFase is much more similar to the human KFase than the yeast KFase, sharing more than 20 % sequence identity, and thus could be a better model to simulate the structural characteristics of its human counterpart. We now report, for the first time, the crystal structure of Drosophila KFase, and a complex structure with BME (2-mercaptoethanol) and a covalent adduct structure with PMSF. The determination of these tertiary structures may contribute to the rational design of selective inhibitors for the treatment of various human pathological conditions, as well as to the understanding of the toxicity of AChE (acetylcholinesterase)-based insecticides.
EXPERIMENTAL
Recombinant KFase production and KFase enzyme activity assay
KFase cDNA sequence (GenBank® accession number AAF52391.1) was amplified from a D. melanogaster larval cDNA pool using a forward primer (5′-AAAACATATGTAC-AATCCGAGGTGCAA-3′) and a reverse primer (5′-GAATTCA TTCAATTTCAATGTTGCGCAG-3′) containing NdeI and EcoRI restriction sites respectively. The amplified sequence was cloned into an Impact™-CN plasmid (New England Biolabs) for the expression of a fusion protein containing a chitin-binding domain. Transformed Escherichia coli cells were cultured at 37 °C. After induction with 0.2 mM IPTG (isopropyl β-D-thiogalactopyranoside) for recombinant protein expression, the cells were cultured at 16 °C for 24 h. Cells (12 l) were harvested as the starting material for affinity purification. To prevent serine and cysteine protease activity, 1 mM PMSF was added into the lysis buffer. Later on it was found that PMSF was covalently bound to the active site Ser157 of KFase, which may deactivate the enzyme and thus PMSF was not added in the cell lysis for characterization and crystallization of non-PMSF-bound KFase. The soluble fusion proteins were applied to a column packed with chitin beads and subsequently hydrolysed under reducing conditions. The affinity purification resulted in the isolation of KFase at around 85 %purity. Further purifications of the recombinant KFase were achieved by ion-exchange and gel-filtration chromatographies. The purified recombinant KFase was concentrated to 10 mg/ml in 20 mM Mops buffer (pH 7.5) using a Centricon YM-30 concentrator (Millipore). Protein concentration was determined by a protein assay kit from Bio-Rad Laboratories using BSA as a standard. NFK was synthesized as described previously [1,31]. Enzyme assays were performed based on the previously reported method with modifications [1]. The reaction mixture consisted of 50 mM potassium phosphate (pH 7.4), 0.45 mM NFK and 2 μg of KFase (only the batch of protein purified without adding PMSF) in a total volume of 600 μl. The absorbance increase at 365 nm resulting from the release of kynurenine was monitored for 90 s with a spectrophotometer (Hitachi) at room temperature (22°C). Blanks (reaction mixture without enzyme) were used to subtract any non-enzymatic activity. The tests were carried out in triplicate. At 365 nm, kynurenine has a molar extinction coefficient of 4530 M−1 · cm−1. Various concentrations of NFK (0.02–1.00 mM) were used in the kinetics study.
Kinetics of inhibition
It has been documented that KFase is inhibited by two major types of AChE-based insecticides, OP and carbamate [26,27,32]. The inhibition kinetics of some OP and carbamate insecticides and PMSF have been tested in chicken liver KFase [33]. In the present study we investigated the inhibition kinetics of OP compounds (diazinon and diazoxon) and PMSF for Drosophila KFase. PMSF and diazinon were purchased from Sigma–Aldrich; Diazoxon (a diazinon-O analogue) was purchased from Crescent Chemical Company. Inhibition of KFase was measured in the presence of 0.5 mM NFK by four or five concentrations of inhibitors, 4, 2, 1 and 0.5 mM for Diazinon, 0.9, 0.5, 0.25 and 0.13 mM for PMSF, and 188, 250, 375, 500 and 750 nM for diazoxon. A reaction without the inhibitor was included as a control. All of the assay conditions were the same as that used for the enzyme kinetics assay described above. The inhibitory data were analysed according to the reactions depicted in the following equation:
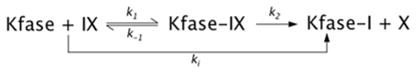 |
where IX is the inhibitor, X is the leaving group of the inhibitor and k2 is the phosphorylation or sulfonation constant.
Progressive inhibition of Kfase activity over time was continuously recorded for 60 s. The activity of Kfase at each 5 s interval was measured and the arithmetic average of three measurements for each time was used for fitting the inhibition curve. The apparent rate constant k (slope of the regression, the natural logarithm of residual activity compared with time) was determined according to pseudo-first order reaction. The 1/k was plotted against 1/[IX](1 − α) to obtain the dissociation constant Kd and k2 by the linear regression [34]:
where α = [S]/(Km + [S]) = 0.5/(0.32 + 0.5), therefore the equation for KFase can be expressed as:
The bimolecular reaction constant ki can be calculated by k2/Kd.
X-ray crystallography
The crystals were grown by a hanging-drop vapour diffusion method with the volume of reservoir solution at 500 μl and the drop volume at 2 μl, containing 1 μl of protein sample and 1 μl of reservoir solution. The optimized crystallization buffer for KFase-1.5 [1.5 Å (1 Å = 0.1 nm) resolution structure] consisted of 0.1 M Hepes (pH 7.5), 18 % PEG [poly(ethylene glycol)] 4000, and 8 % 2-propanol. Individual KFase crystals were cryogenized in the crystallization buffer containing 25 % ethylene glycol as a cryo-protectant solution. The KFase–PMSF covalent adduct was obtained by co-crystallization of the enzyme in the presence of 2 mM PMSF in 25 %ethylene glycol and 2 mM BME, and KFase-2.0 was crystallized under the same conditions excluding the PMSF. The protein used in the crystallization of KFase–PMSF and KFase-2.0 (2.0 Å resolution structure) was not treated with PMSF during the purification process. Diffraction data of KFase crystals were collected at the Brookhaven National Synchrotron Light Source beam line X29A (λ = 1.0750 Å). Data were collected using an ADSC Q315 CCD (charge-coupled device) detector. All of the data were indexed and integrated using HKL-2000 software; scaling and merging of diffraction data were performed using the program SCALEPACK [35]. The parameters of the crystals and data collection are listed in Table 1. The structure of KFase was determined by the molecular replacement method using a putative thioesterase (PDB code 2PBL, 22 % amino acid sequence identical) deposited by the Joint Center for Structural Genomics. The program MolRep [36] was employed to calculate both cross-rotation and translation of the model. The initial model was subjected to iterative cycles of crystallographic refinement with the Refmac 5.2 [37], Phenix.refine [38] and graphic sessions for model building using the program COOT [39]. The ligands were modelled based upon both the 2Fo − Fc and Fo − Fc electron density maps. Solvent molecules were added using COOT and refined with Refmac 5.2. PyMOL (http://www.pymol.org) was used to generate figures and analyse protein and substrate interaction.
Table 1.
Data collection and refinement statistics of KFase
| Crystal data | KFase-1.5 | KFase-2.0 | KFase–PMSF |
|---|---|---|---|
| Space group | C2221 | C2 | C2 |
| Unit cell | |||
| a (Å) | 71.6 | 75.8 | 75.5 |
| b (Å) | 76.0 | 47.1 | 47.0 |
| c (Å) | 210.4 | 85.7 | 85.3 |
| α = γ(°) | 90.0 | ||
| β (°) | 90.0 | 90.3 | 90.6 |
| Data collection | |||
| X-ray source | BNL-X29 | BNL-X29 | BNL-X29 |
| Wavelength (Å) | 1.0750 | 1.0750 | 1.0750 |
| Resolution (Å) | 1.50 (1.55–1.50) | 2.00 (2.07–2.00) | 1.64 (1.70–1.64) |
| Total number of reflections | 1 296 704 | 140 315 | 270 305 |
| Number of unique reflections | 91 964 | 20 558 | 36 922 |
| R-merge | 0.15 (0.34) | 0.07 (0.62) | 0.06 (0.68) |
| Redundancy | 12.2 (12.5) | 7.0 (6.7) | 7.5 (6.0) |
| Completeness (%) | 98.4 (86.9) | 97.0 (98.8) | 97.8 (88.9) |
| Refinement statistics | |||
| R-work (%) | 17.1 | 22.5 | 21.7 |
| R-free (%) | 19.8 | 27.3 | 24.8 |
| Rmsd bond lengths (Å) | 0.024 | 0.020 | 0.018 |
| Rmsd bond angles (°) | 2.070 | 1.663 | 1.525 |
| Number of ligand molecules | 2 SEB | 1 BME | 1 SEB |
| 5 EDO | 4 EDO | 4 EDO | |
| 1 Na | 1 Na | ||
| Number of water molecules | 515 | 83 | 155 |
| Average B overall (Å2) | 15.9 | 21.8 | 18.2 |
| Ramachandran plot (%) | |||
| Favoured regions | 98 | 97 | 97 |
| Allowed regions | 100 | 100 | 100 |
The values in parentheses are for the highest resolution shell. BNL, Brookhaven National Laboratory; EDO, ethylene glycol.
Molecular docking
To obtain insight into substrate binding and catalysis of the enzyme, AutoDock Vina was used for molecular docking to obtain KFase–NFK complex [41]. The NFK molecule for AutoDock Vina was prepared by using Marvin 5.3.7, 2010, ChemAxon (http://www.chemaxon.com). AutoDock Tools 1.5.4 (http://mgltools.scripps.edu/) was used to prepare the ligand and the receptor, after which docking was performed using AutoDock Vina. All of the protein side chains were immobilized and the rotatable bonds were left free to rotate for the ligand. The X, Y and Z dimensions of the grid were set to 48 Å×40 Å×44 Å around the benzyl ring of the SEB (o-benzylsulfonylserine) molecule in the KFase structure. The grid box covered the whole active-site cavity. The lowest energy configuration was considered the best docking pose for NFK.
RESULTS AND DISCUSSION
Enzyme purification and activity assay
Using affinity, ion-exchange and gel-filtration chromatography, KFase recombinant protein was purified to very high purity as judged by SDS/PAGE. KFase showed high activity towards chemically synthesized NFK (Figure 1B). The spectral changes in Figure 1(B) did not lead to an exact isosbestic point, which might be due to the formation of intermediates or an initial protein conformation change caused by addition of the substrate. The enzyme kinetics study determined its kinetic parameters towards NFK, with Km = 0.32 ± 0.06 mM, kcat = 1584 ± 267 min−1 and kcat/Km = 4950 min−1 · mM−1. The Km value is slightly higher than mouse recombinant KFase, which is 0.19 mM [42]. KFase has been previously studied in insects owing largely to the influence that the metabolites in the tryptophan oxidation pathway exerted on the colour of the compound eyes and wing colour pattern. In addition to the KFase activity shown to be present in D. melanogaster [43,44], a number of insect genes encoding KFase have been identified and cloned, including the KFase genes from Aedes aegypti mosquito [45], Heliconius butterfly [46], Tribolium castaneum and Bombyx mori [46]. The data shown in the present paper enabled the identification of D. melanogaster KFase for the first time at the biochemical level.
Overall structure
The first crystal structure of KFase (KFase-1.5) was determined by molecular replacement and refined to 1.5 Å resolution. The final model contains 605 amino acid residues (302 in chain A and 303 in chain B) and yields a crystallographic R value of 17.1 % and an Rfree value of 19.8 %. A total of 98 % of the residues are in favourable regions and 100 % of the residues are in allowed regions of the Ramachandran plot as defined with MolProbity [47]. Although there are two monomers in an asymmetric unit, its biological molecule is a monomer analysed by PISA (protein interfaces, surfaces and assemblies service) [48]. The second structure (KFase-2.0) was crystallized under a different condition without PMSF and refined to 2.0 Å resolution; the last structure (KFase–PMSF) was co-crystallized with 2 mM PMSF using the same conditions as for KFase-2.0 and refined to 1.64 Å resolution. The results of data collection and refinement are summarized in Table 1.
An overview of the structure model is shown in Figure 2(A). A structure homology search by Dali [49] revealed that, among those structures where the function is positively identified, KFase shows the most structural similarity with the carboxylesterases, which play an important role in the efficacy and detoxification of insecticides, and have been proposed as markers in environmental monitoring and toxicity identification evaluations [50]. The typical α/β hydrolase fold consists of an eight-stranded β-sheet surrounded by a number of α-helices [51]. KFase, as well as four different hydrolases which we superimposed on to the KFase-1.5 structure, all exhibited the distinctive α/β hydrolase fold structure or eight-stranded β-sheets surrounded by α-helices. The superimposed hydrolases include a carboxylesterase from Archaeoglobus fulgidus [PDB code 1JJI, rmsd (root mean square deviation) = 2.9, number of aligned positions = 250] [52], an acetylesterase from Rhodococcus sp. (PDB code 1LZL, rmsd = 2.8, number of aligned positions = 248) [53], a bacterial homologue of the mammalian hormone-sensitive lipase (PDB code 1JKM, rmsd = 3.2, number of aligned positions = 257) [54] and human AChE (PDB code 3LII, rmsd = 3.0, number of aligned positions = 232) [55] (Figure 2B). Close examination of the KFase structure reveals that residues Ser157, His276 and Asp244 line up perfectly (Supplementary Figure S1A at http://www.BiochemJ.org/bj/446/bj4460253add.htm). These three amino acid residues are important to KFase for its catalytic function as a hydrolase. They are conserved in most hydrolases and are known as the catalytic triad. These signature structural features place KFase in the α/β hydrolase fold group of the hydrolase family.
Figure 2. KFase fold.
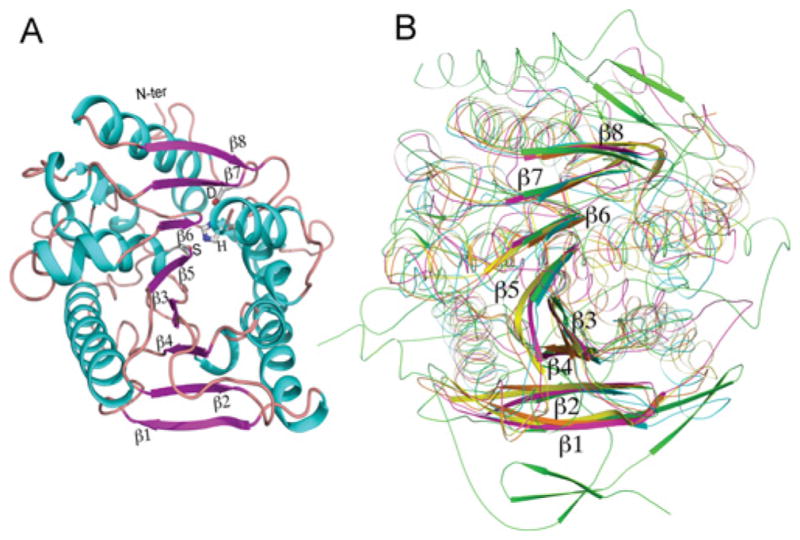
(A) Cartoon presentation of KFase showing an eight-stranded β-sheet surrounded by a number of helices. The catalytic triad is labelled as S (Ser157), H (His276), and D (Asp244) and the β-sheet strands were labelled as β1–β8. The α-helices are shown in cyan, the β-strands in purple and the loops in brown. (B) Superimposition of a carboxylesterase (yellow), an acetyl esterase (orange), a bacterial homologue of the mammalian hormone-sensitive lipase (pink) and human AChE (green) on to KFase (cyan) indicates that all of them have the α/β hydrolase fold signature structure. N-ter, N-terminus.
The active-site cavity
As shown in Figure 3, a well-defined cavity, approximately 11 Å deep, extends to the active-site residues Ser157, His276, and Asp244, via a wide passage (13 Å×7 Å). The active-site residues Ser157 and His276 are found at the bottom of the active-site cavity, Asp244 is nearby, but not on the surface of the cavity. The KFase active site is shallower than the active site of human AChE, which has been described as an ‘active-site gorge’ [55]. A total of 26 residues form the surface of the cavity, including residues Phe14, Pro15, Leu33, Phe36, Val37, Phe84, His86–Tyr89, Gln91–Met93, Met97, Ser98, Ile101, His156–Ala158, Val190, Val202, Phe248, His276, Phe277 and Ile280. Six of them are aromatic amino acid residues, whereas human AChE has 14 aromatic residues lining the active-site gorge surface. All of the hydrolases contain a catalytic triad, including amino acid residues serine, aspartic acid/glutamic acid/serine and histidine/aspartic acid/lysine (typically serine, aspartic acid/glutamic acid and histidine), which are important for catalytic function. In the tertiary structure of these hydrolases, the three amino acids must occupy the same positions relative to the active site. However, their positions in the primary sequence may vary widely among different types of hydrolases. For example, trypsin (a serine protease) has His57, Glu102 and Ser195 [56] (Supplementary Figure S1B) and AChE from Torpedo californica has Ser200, Glu327 and His 440 [57] as the catalytic triad (Supplementary Figure S1C). There are four groups of hydrolases which contain catalytic triads: the eukaryotic serine proteases, the cysteine proteases, subtilisins and the α/β hydrolase fold enzymes. The order of triad residues in KFase resembles α/β hydrolase fold arrangement, which is in three-dimensional space a mirror-image of the serine protease catalytic triad. The triad residues are spaced out in the primary structure in such a way that during protein folding they are all brought close together at the active site.
Figure 3. Active-site cavity.
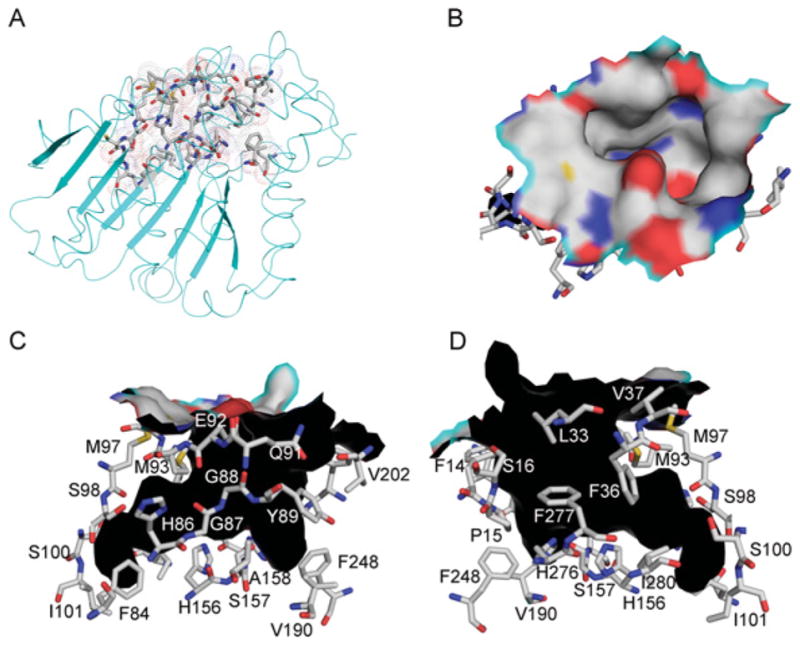
(A) KFase structure is shown as a cartoon diagram. The residues forming the active-site cavity are shown as sticks and a dot surface. (B) The top view of the surface of the active-site cavity. C, N, O and S atoms are coloured as silver, blue, red and yellow respectively. (C) One side view of the active-site cavity wall. The cavity shape is shown in black. The residues forming active-site cavity surface are shown as sticks. (D) The other side view of the active-site cavity wall.
Ligand binding and catalytic mechanism
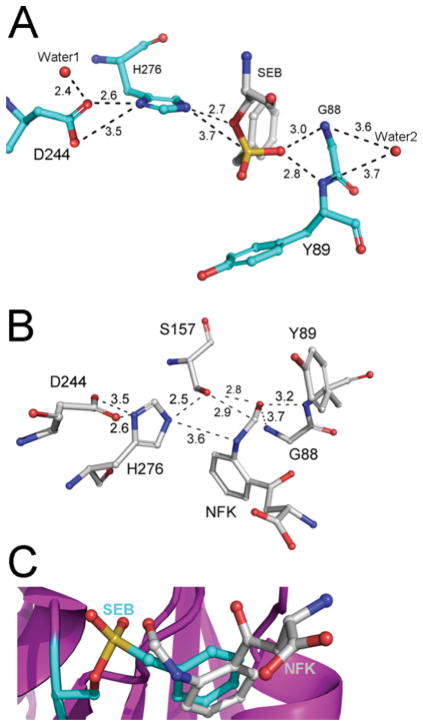
The residual electron density clearly revealed the presence of a ligand that forms a covalent bond with Ser157 in the active site of the KFase-1.5 structure. After diligently considering every compound added during the cell culture and purification process, we realized that the ligand could be PMSF, a hydrolase inhibitor added to the cell lysis buffer in the first step of protein purification. To confirm if PMSF was the ligand in the active site of KFase, we purified the protein without adding PMSF and crystallized it with and without PMSF under a new crystallization condition. The KFase–PMSF structure clearly shows that PMSF forms a covalent bond with Ser157 (Supplementary Figure S2B at http://www.BiochemJ.org/bj/446/bj4460253add.htm), whereas the KFase-2.0 structure crystallized without PMSF does not show electron density consistent with a covalent bond at Ser157. The sulfonyl group of PMSF is perfectly fitted into the omit map in the KFase-1.5 and KFase–PMSF structures. The benzyl ring seems disordered, which is due likely to the rotatable nature of the bond between the benzyl and sulfonyl groups of PMSF and also to the relatively large active site for a small PMSF molecule (Supplementary Figure S2).
To determine if ligand binding leads to any conformational change, we superimposed four chains from the three KFase structures. Significant conformational changes were revealed in two loop areas (Figure 4). However, these loop conformational changes are not associated with ligand binding because KFase-2.0 has the same conformation as KFase–PMSF. Instead, the conformational difference is probably associated with the crystallization conditions, because KFase-2.0 and KFase–PMSF structures crystallized in 25 % ethylene glycol have the same loop conformation, which is different from that of KFase-1.5, crystallized in Hepes buffer containing PEG 4000. A slight conformational difference between PMSF binding and non-PMSF structure was noticed, i.e. side chains of Met93, Lys33 and Tyr89 are tilted slightly away from the ligand-binding site in the non-PMSF structure (KFase-2.0).
Figure 4. KFase conformational changes.
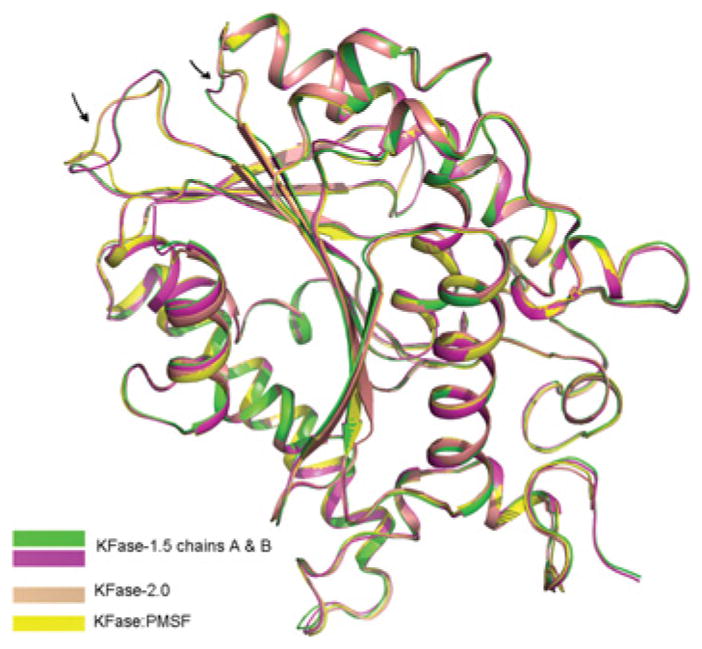
Superposition of all the subunits (three monomers) of KFase-1.5 and KFase-2.0 on to KFase–PMSF. Fragments with significant conformational changes are indicated by an arrow.
This newly formed structure by PMSF and Ser157 is named SEB, which is considered to be a sulfonate analogue of the first tetrahedral intermediate in the hydrolysis reaction (Figure 5). SEB is also similar to the tetrahedral intermediate formed by OP compounds and the active-site serine residue in an AChE [58]. This is because KFase has the same serine hydrolase signature structure as AChE; a catalytic triad and oxyanion hole formed by the main chain amide nitrogen atoms of Gly88 and Tyr89. Mouse KFase is a model enzyme to study OP insecticide toxicity [26,27]. The effect of OP diazinon/diazoxon on kynurenine pathway metabolites has been investigated [26,27]. Diazinon was able to increase NFK concentration and decrease kynurenine level in mouse liver in vivo [26]. Inhibition kinetics study of OP diazinon, diazoxon and PMSF against Drosophila KFase revealed that both diazinon and diazoxon inhibited the Drosophila enzyme, and the inhibition of PMSF against the enzyme was between diazinon and diazoxon (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/446/bj4460253add.htm showing the double reciprocal plot of rate of inhibition and Table 2 showing the inhibition parameters of Drosophila KFase). Note that diazinon (Supplementary Figure S3A) itself is not a strong AChE inhibitor, only showing inhibition up to 0.5 mM concentration in our test. In animals, diazinon is converted into diazoxon (Supplementary Figure S3C), a compound that is a strong AChE inhibitor [59,60]. A previous paper indicated that diazoxon was an approximately 40-fold better inhibitor of purified chicken liver KFase than PMSF and even more than diazinon [33]. In the present study, we showed that diazoxon was a 28 000- and 65 000-fold better inhibitor of Drosophila KFase than PMSF and diazinon respectively. Diazoxon is also a ~900-fold better inhibitor of Drosophila KFase than of chicken liver KFase. Note that the observed 900-fold cross-species difference for the diazoxon ki constant, whereas almost no difference was observed for PMSF is quite unusual. Further comparative studies of chicken KFase and Drosophila KFase structures and their diazoxon complexes might help to better elucidate the underlying mechanism. Alignment of Drosophila and mouse KFases verified that the two proteins share the same catalytic triad, the typical GHSAG motif seen in serine hydrolase (GXSXG) and the HGGYW sequence, from which the main chain amide nitrogen atoms of Gly88 and Tyr89 form the α/β hydrolase oxyanion hole. In addition, the active-site cavity of KFase is large enough to facilitate the binding of most OP compounds. This covalent association of PMSF with KFase suggests that inhibition of KFase by certain OP pesticides may be due to their interaction with the serine residue in the KFase’s active site.
Figure 5. Formation of SEB and enzyme reaction of NFK hydrolysis.
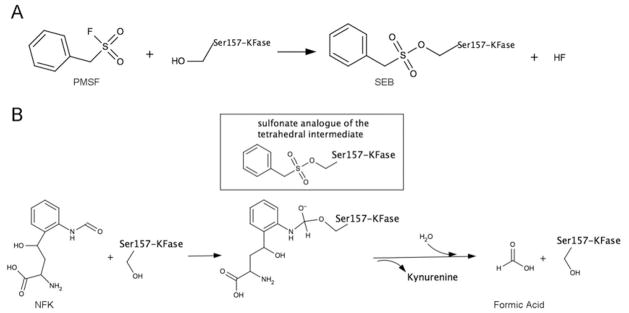
(A) The suggested molecular mechanism of covalent bonding of PMSF and Ser157. (B) Enzyme reaction of NFK hydrolysis. The first tetrahedral intermediate is shown in the Figure, which is mimicked by SEB, a sulfonate analogue of the tetrahedral intermediate.
Table 2.
Inhibition parameters for Drosophila KFase
| Inhibitor | Kd (mM) | k2 (min−1) | ki (min−1 · mM−1) |
|---|---|---|---|
| Diazinon | 1.8 ± 0.5 | 3.6 ± 0.02 | 2.0 |
| Diazoxon | 3.1×10−4 ± 1.2×10−4 | 40 ± 9 | 1.3×105 |
| PMSF | 0.3 ± 0.05 | 1.3 ± 0.1 | 4.6 |
Data are expressed as means ± S.E.M.
Using molecular docking and PMSF and KFase binding information, we modelled a KFase and NFK complex. The result indicated that the lowest predicted free energy of binding is − 6.9 kcal/mol (1 kcal = 4.184 kJ). The complex with the lowest binding energy was used for the analysis of the NFK and KFase interaction. The major interactions of PMSF and KFase are illustrated in Figure 6(A). It is worthwhile to note that two water molecules are involved in the hydrogen bond network in the active centre. ‘Water1’ has a close contact with Asp244 and ‘water2’ forms hydrogen bonds with Gly88 and Tyr89. The major interactions of KFase and NFK are shown in Figure 6(B). Briefly, the KFase–NFK complex obtained by molecular docking revealed that the substrate lies near the OG atom of Ser157, the NE2 atom of His267 and the N atoms of Gly88 and Tyr89. Ser157and His267 are directly involved in catalytic reaction and the main chain amide nitrogen atoms of Gly88 and Tyr89, which form the oxyanion hole, are used to stabilize the negatively charged transition state during hydrolysis. In addition to the aforementioned major interactions between NFK and KFase, NFK also connects to Pro15, Ser16 and Glu92 through hydrogen bonding and to Phe14, Leu33, Phe36, Met93, His156 and Phe277 via hydrophobic interactions. Interestingly, the binding sites of NFK and PMSF are well superposed, showing that their benzyl rings are essentially overlapping (Figure 6C).
Figure 6. The major interactions between ligand and KFase revealed by molecular docking.
The major residues involved in the catalytic reaction are shown in sticks. The distances between atoms directly involved in the reaction are labelled. (A) SEB and KFase interaction. (B) NFK and KFase interaction. (C) The superimposition of (A) and (B), showing that benzyl rings are well superimposed.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the NINDS (National Institute of Neurological Disorders and Stroke) [grant number NS062836].
This work was carried out in part at the National Synchrotron Light Source, Brookhaven National Laboratory. We are grateful to Haizhen Ding for helping with protein expression, to Elizabeth Watson for critically reading the paper, and to Dr Tao Cai and Dr Danilo Tagle from the National Institutes of Health for fruitful discussions at the early stage of the present study.
Abbreviations used
- AChE
acetylcholinesterase
- BME
2-mercaptoethanol
- KFase
kynurenine formamidase
- NFK
N-formyl-L-kynurenine
- OP
organophosphorus
- PEG
poly(ethylene glycol)
- rmsd
root mean square deviation
- SEB
o-benzylsulfonylserine
Footnotes
The atomic co-ordinates and structure factors have been deposited in the Protein Data Bank under codes 4E11, 4E14 and 4E15.
AUTHOR CONTRIBUTION
Qian Han and Jianyong Li conceived the general hypothesis, designed the experiments, carried out the enzyme purification, crystallization, enzymatic characterization and structure determination, analysed the results and wrote the paper. Howard Robinson carried out the diffraction data collection and processing, and helped with model building.
References
- 1.Pabarcus MK, Casida JE. Kynurenine formamidase: determination of primary structure and modeling-based prediction of tertiary structure and catalytic triad. Biochim Biophys Acta. 2002;1596:201–211. doi: 10.1016/s0167-4838(02)00232-7. [DOI] [PubMed] [Google Scholar]
- 2.Han Q, Beerntsen BT, Li J. The tryptophan oxidation pathway in mosquitoes with emphasis on xanthurenic acid biosynthesis. J Insect Physiol. 2007;53:254–263. doi: 10.1016/j.jinsphys.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han Q, Kim SR, Ding H, Li J. Evolution of two alanine glyoxylate aminotransferases in mosquito. Biochem J. 2006;397:473–481. doi: 10.1042/BJ20060469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Q, Fang J, Li J. 3-Hydroxykynurenine transaminase identity with alanine glyoxylate transaminase. A probable detoxification protein in Aedes aegypti. J Biol Chem. 2002;277:15781–15787. doi: 10.1074/jbc.M201202200. [DOI] [PubMed] [Google Scholar]
- 5.Garcia GE, Wirtz RA, Barr JR, Woolfitt A, Rosenberg R. Xanthurenic acid induces gametogenesis in Plasmodium, the malaria parasite. J Biol Chem. 1998;273:12003–12005. doi: 10.1074/jbc.273.20.12003. [DOI] [PubMed] [Google Scholar]
- 6.Billker O, Lindo V, Panico M, Etienne AE, Paxton T, Dell A, Rogers M, Sinden RE, Morris HR. Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature. 1998;392:289–292. doi: 10.1038/32667. [DOI] [PubMed] [Google Scholar]
- 7.Rossi F, Garavaglia S, Giovenzana GB, Arca B, Li J, Rizzi M. Crystal structure of the Anopheles gambiae 3-hydroxykynurenine transaminase. Proc Natl Acad Sci USA. 2006;103:5711–5716. doi: 10.1073/pnas.0510233103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han Q, Robinson H, Gao YG, Vogelaar N, Wilson SR, Rizzi M, Li J. Crystal structures of Aedes aegypti alanine glyoxylate aminotransferase. J Biol Chem. 2006;281:37175–37182. doi: 10.1074/jbc.M607032200. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Liu H, McKenzie G, Witting PK, Stasch JP, Hahn M, Changsirivathanathamrong D, Wu BJ, Ball HJ, Thomas SR, et al. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat Med. 2010;16:279–285. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone TW, Perkins MN. Quinolinic acid: a potent endogenous excitant at amino acid receptors in CNS. Eur J Pharmacol. 1981;72:411–412. doi: 10.1016/0014-2999(81)90587-2. [DOI] [PubMed] [Google Scholar]
- 11.Perkins MN, Stone TW. An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res. 1982;247:184–187. doi: 10.1016/0006-8993(82)91048-4. [DOI] [PubMed] [Google Scholar]
- 12.Barth MC, Ahluwalia N, Anderson TJ, Hardy GJ, Sinha S, Alvarez-Cardona JA, Pruitt IE, Rhee EP, Colvin RA, Gerszten RE. Kynurenic acid triggers firm arrest of leukocytes to vascular endothelium under flow conditions. J Biol Chem. 2009;284:19189–19195. doi: 10.1074/jbc.M109.024042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jhamandas K, Boegman RJ, Beninger RJ, Bialik M. Quinolinate-induced cortical cholinergic damage: modulation by tryptophan metabolites. Brain Res. 1990;529:185–191. doi: 10.1016/0006-8993(90)90826-w. [DOI] [PubMed] [Google Scholar]
- 14.Yasui H, Takai K, Yoshida R, Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2,3-dioxygenase: its possible occurrence in cancer patients. Proc Natl Acad Sci USA. 1986;83:6622–6626. doi: 10.1073/pnas.83.17.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor MW, Feng GS. Relationship between interferon-γ, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- 16.Chen Y, Guillemin GJ. Kynurenine pathway metabolites in humans: disease and healthy states. Int J Tryptophan Res. 2009;2:1–19. doi: 10.4137/ijtr.s2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Q, Cai T, Tagle DA, Li J. Structure, expression, and function of kynurenine aminotransferases in human and rodent brains. Cell Mol Life Sci. 2010;67:353–368. doi: 10.1007/s00018-009-0166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lob S, Konigsrainer A, Rammensee HG, Opelz G, Terness P. Inhibitors of indoleamine-2,3-dioxygenase for cancer therapy: can we see the wood for the trees? Nat Rev Cancer. 2009;9:445–452. doi: 10.1038/nrc2639. [DOI] [PubMed] [Google Scholar]
- 19.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piser TM. Linking the cytokine and neurocircuitry hypotheses of depression: a translational framework for discovery and development of novel anti-depressants. Brain Behav Immun. 2010;24:515–524. doi: 10.1016/j.bbi.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Erhardt S, Olsson SK, Engberg G. Pharmacological manipulation of kynurenic acid: potential in the treatment of psychiatric disorders. CNS Drugs. 2009;23:91–101. doi: 10.2165/00023210-200923020-00001. [DOI] [PubMed] [Google Scholar]
- 22.Han Q, Robinson H, Cai T, Tagle DA, Li J. Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV. Biosci Rep. 2011;31:323–332. doi: 10.1042/BSR20100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giorgini F, Guidetti P, Nguyen Q, Bennett SC, Muchowski PJ. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet. 2005;37:526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone TW, Darlington LG. Endogenous kynurenines as targets for drug discovery and development. Nat Rev Drug Discov. 2002;1:609–620. doi: 10.1038/nrd870. [DOI] [PubMed] [Google Scholar]
- 25.Rodgers J, Stone TW, Barrett MP, Bradley B, Kennedy PG. Kynurenine pathway inhibition reduces central nervous system inflammation in a model of human African trypanosomiasis. Brain. 2009;132:1259–1267. doi: 10.1093/brain/awp074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifert J, Pewnim T. Alteration of mice L-tryptophan metabolism by the organophosphorous acid triester diazinon. Biochem Pharmacol. 1992;44:2243–2250. doi: 10.1016/0006-2952(92)90353-k. [DOI] [PubMed] [Google Scholar]
- 27.Casida JE, Quistad GB. Serine hydrolase targets of organophosphorus toxicants. Chem Biol Interact. 2005;157–158:277–283. doi: 10.1016/j.cbi.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 28.Eto M, Seifert J, Engel JL, Casida JE. Organophosphorus and methylcarbamate teratogens: structural requirements for inducing embryonic abnormalities in chickens and kynurenine formamidase inhibition in mouse liver. Toxicol Appl Pharmacol. 1980;54:20–30. doi: 10.1016/0041-008x(80)90004-6. [DOI] [PubMed] [Google Scholar]
- 29.Arndt JW, Schwarzenbacher R, Page R, Abdubek P, Ambing E, Biorac T, Canaves JM, Chiu HJ, Dai X, Deacon AM, et al. Crystal structure of an α/β serine hydrolase (YDR428C) from Saccharomyces cerevisiae at 1.85 Å resolution. Proteins. 2005;58:755–758. doi: 10.1002/prot.20336. [DOI] [PubMed] [Google Scholar]
- 30.Wogulis M, Chew ER, Donohoue PD, Wilson DK. Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry. 2008;47:1608–1621. doi: 10.1021/bi701172v. [DOI] [PubMed] [Google Scholar]
- 31.Dalgliesh CE. Synthesis of N3-formyl-DL-kynurenine, Nα-acetyl-DL-kynurenine, and related compounds, and observation on the synthesis of kynurenine. J Chem Soc. 1952:137–141. [Google Scholar]
- 32.Pewnim T, Seifert J. Structural requirements for altering the L-tryptophan metabolism in mice by organophosphorous and methylcarbamate insecticides. Eur J Pharmacol. 1993;248:237–241. doi: 10.1016/0926-6917(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 33.Seifert J, Casida JE. Inhibition and reactivation of chicken kynurenine formamidase: in vitro studies with organophosphates, N-alkylcarbamates, and phenylmethanesulfonyl fluoride. Pestic Biochem Physiol. 1979;12:273–279. [Google Scholar]
- 34.Chen Z, Newcomb R, Forbes E, McKenzie J, Batterham P. The acetylcholinesterase gene and organophosphorus resistance in the Australian sheep blowfly, Lucilia cuprina. Insect Biochem Mol Biol. 2001;31:805–816. doi: 10.1016/s0965-1748(00)00186-7. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Reference deleted
- 41.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pabarcus MK, Casida JE. Cloning, expression, and catalytic triad of recombinant arylformamidase. Protein Expression Purif. 2005;44:39–44. doi: 10.1016/j.pep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Moore GP, Sullivan DT. The characterization of multiple forms of kynurenine formidase in Drosophila melanogaster. Biochim Biophys Acta. 1975;397:468–477. doi: 10.1016/0005-2744(75)90137-0. [DOI] [PubMed] [Google Scholar]
- 44.Moore GP, Sullivan DT. Biochemical and genetic characterization of kynurenine formamidase from Drosophila melanogaster. Biochem Genet. 1978;16:619–634. doi: 10.1007/BF00484718. [DOI] [PubMed] [Google Scholar]
- 45.Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferguson LC, Jiggins CD. Shared and divergent expression domains on mimetic Heliconius wings. Evol Dev. 2009;11:498–512. doi: 10.1111/j.1525-142X.2009.00358.x. [DOI] [PubMed] [Google Scholar]
- 47.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 49.Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics. 2006;14:5.5.1–5.5.24. doi: 10.1002/0471250953.bi0505s14. [DOI] [PubMed] [Google Scholar]
- 50.Wheelock CE, Phillips BM, Anderson BS, Miller JL, Miller MJ, Hammock BD. Applications of carboxylesterase activity in environmental monitoring and toxicity identification evaluations (TIEs) Rev Environ Contam Toxicol. 2008;195:117–178. doi: 10.1007/978-0-387-77030-7_5. [DOI] [PubMed] [Google Scholar]
- 51.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 52.De Simone G, Menchise V, Manco G, Mandrich L, Sorrentino N, Lang D, Rossi M, Pedone C. The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus. J Mol Biol. 2001;314:507–518. doi: 10.1006/jmbi.2001.5152. [DOI] [PubMed] [Google Scholar]
- 53.Zhu X, Larsen NA, Basran A, Bruce NC, Wilson IA. Observation of an arsenic adduct in an acetyl esterase crystal structure. J Biol Chem. 2003;278:2008–2014. doi: 10.1074/jbc.M210103200. [DOI] [PubMed] [Google Scholar]
- 54.Wei Y, Contreras JA, Sheffield P, Osterlund T, Derewenda U, Kneusel RE, Matern U, Holm C, Derewenda ZS. Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase. Nat Struct Biol. 1999;6:340–345. doi: 10.1038/7576. [DOI] [PubMed] [Google Scholar]
- 55.Dvir H, Silman I, Harel M, Rosenberry TL, Sussman JL. Acetylcholinesterase: from 3D structure to function. Chem Biol Interact. 2010;187:10–22. doi: 10.1016/j.cbi.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zakharova E, Horvath MP, Goldenberg DP. Structure of a serine protease poised to resynthesize a peptide bond. Proc Natl Acad Sci USA. 2009;106:11034–11039. doi: 10.1073/pnas.0902463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 58.Sanson B, Nachon F, Colletier JP, Froment MT, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, Weik M. Crystallographic snapshots of nonaged and aged conjugates of soman with acetylcholinesterase, and of a ternary complex of the aged conjugate with pralidoxime. J Med Chem. 2009;52:7593–7603. doi: 10.1021/jm900433t. [DOI] [PubMed] [Google Scholar]
- 59.Sparling DW, Fellers G. Comparative toxicity of chlorpyrifos, diazinon, malathion and their oxon derivatives to larval Rana boylii. Environ Pollut. 2007;147:535–539. doi: 10.1016/j.envpol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 60.Gallo MA, Lawryk NJ. Organic phosphorus pesticides. In: Hayes WJ Jr, Laws ER Jr, editors. Handbook of Pesticide Toxicology. Academic Press; New York: 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.