Abstract
Fanconi anemia (FA) proteins function in maintaining genomic stability. Their major role is in the repair of DNA interstrand crosslinks, which by virtue of covalently binding the Watson and Crick strands of DNA, impede replication and transcription. Inappropriately repaired interstrand crosslinks cause genomic instability leading to cancer; conversely, their toxicity makes them a powerful chemotherapeutic. Here, we discuss how FA proteins promote stem cell function, prevent tumorigenesis, stabilize replication forks, and inhibit improper repair. We also review the most recent advances identifying endogenous aldehydes as possible culprits in DNA damage that induces the phenotypes seen in the FA patients.
Introduction
Fanconi anemia (FA) occurs in approximately one out of every 100,000 births 1. Biallelic mutations in FA genes lead to bone marrow failure and susceptibility to both acute myeloid leukemia and solid tumors, as well as congenital abnormalities and infertility 2. The function of the pathway, on the other hand, is anything but rarified: FA proteins participate in the repair of extraordinarily deleterious lesions, interstrand crosslinks (ICLs), and in maintaining genomic stability during DNA replication.
FA is genetically heterogeneous. To date, fifteen genes have been identified as mutated in patients (Figure 1, Table 1), many more interacting genes have been discovered, and there are still patients in which a mutation is yet to be identified. The known genes work together in ICL repair, coordinating the actions of multiple repair processes, in particular nucleases that are necessary for cutting the ICL out and for other nucleolytic processing necessary for the repair, translesion synthesis (TLS), a mode of damage tolerance that uses specialized polymerases to insert a base across from a lesion or abasic site, and homologous recombination (HR), the pathway best known for its role in repairing double-stranded breaks. The FA proteins may also counteract some of the activities of the non-homologous end joining (NHEJ) pathway, an error-prone repair pathway that is used to directly re-ligate DNA ends. Overall, FA interacts with many of the currently described genome maintenance pathways; as such, studying it provides a unique window into the elaborate interplay of multiple cellular networks.
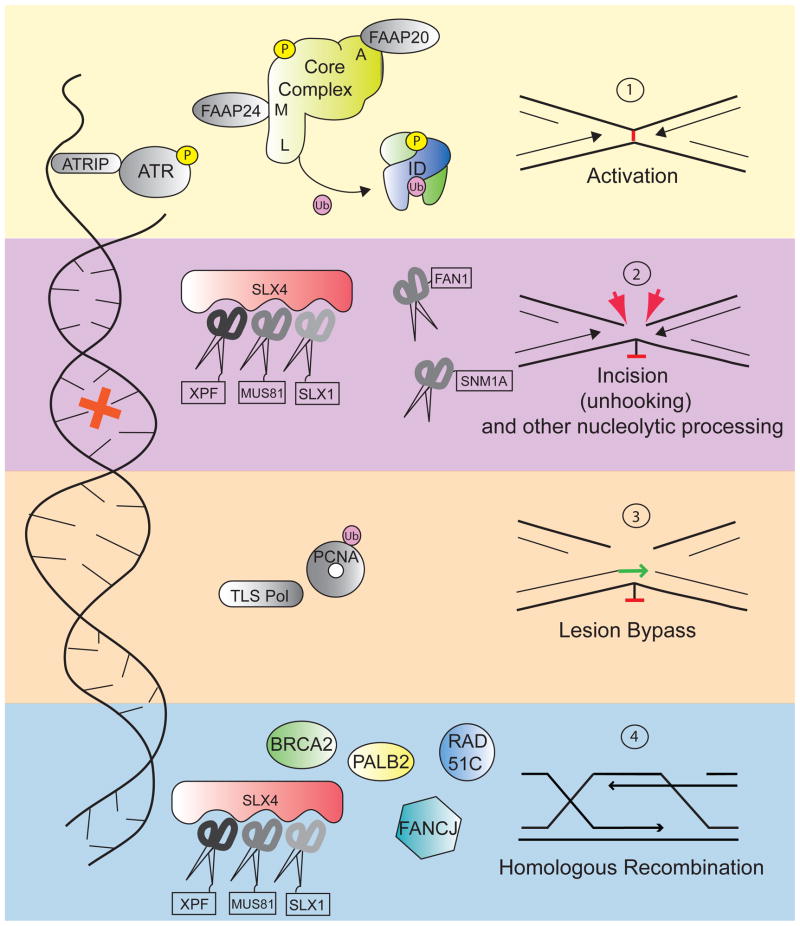
Figure 1. The FA pathway functions in ICL repair.
Upon detection of the crosslink, the core complex is activated and FANCL in the complex ubiquitylates the ID complex. The ID complex then coordinates the action of downstream repair factors, including nucleases and homologous recombination proteins, like the FA proteins BRCA2, FANCJ, PALB2, and RAD51C. Proteins that have been identified as mutated in FA patients are shown in color; factors known to participate in the pathway but are not clinically associated with FA are shown in grey. FANCJ, although not necessary for FANCD2 monoubiquitylation, may work at earlier steps than homologous recombination.
Table 1.
FA and FA-associated DNA repair factors and human disease
| Gene | Human phenotypes | Cancer Susceptibility in FA patients (bold) and in mutation carriers (italics) | Pathway and function | Link to FA |
|---|---|---|---|---|
| FANCA, B, C, D2, E, F, G, I, L1, M1 | BMF, growth retardation, thumb and radial ray defects, developmental abnormalities, hypogonadism, microcephaly, male infertility and female reduced fertility, hyperpigmentation |
AML, HNSCC, esophageal, gynecological, and liver cancers no cancers in mutation carriers for most groups2 |
FA;core complex; FANCL is a E3 ubiquitin ligase | Genes mutated in FA |
| FANCD1/BRCA2 | BMF, growth retardation, thumb and radial ray defects, microcephaly | AML, ALL, medulloblastoma, neuroblastoma Wilms tumor3; breast, ovarian and prostate cancer | FA, HR; loads RAD51 onto DNA | Gene mutated in FA |
| FANCJ/BRIP1 | BMF, growth retardation, thumb and radial ray defects, severe developmental abnormalities hyperpigmentation | AML, HNSCC; Breast and ovarian cancer | FA, HR; 3′-5′ helicase | Gene mutated in FA |
| FANCN/PALB2 | Anemia, growth retardation, thumb radial ray defects, hyperpigmentation, microcephaly | AML, medulloblastoma; neuroblastoma, Wilms tumor, hemangioendothelioma3; breast, ovarian, and pancreatic cancer | FA, HR; Promotes BRCA2 Function | Gene mutated in FA |
| FANCO4/RAD51C | Growth retardation thumb and radial ray defects, hypogonadism, cystic kidneys, renal failure | Breast and ovarian cancer | HR FA; coordinates | Gene mutated in FA-like syndrome |
| FANCP/SLX4 | BMF, Growth retardation, microcephaly, developmental defects including thumb and radial ray defects | HNSCC (one patient) | XPF-ERCC1, MUS81- EME1, and SLX1 nucleases | Gene mutated in FA |
| BLM | Growth retardation, sun sensitivity, chronic obstructive pulmonary disease, diabetes, recurrent infections, infertility | Broad predisposition including Non-Hodgkin’s lymphoma, AML, ALL, skin, colon, breast | 5′-3′ helicase, anti-recombinase | Interacts with core complex, FANCJ; genetic interaction with FANCP/SLX4 |
| BRCA1 | No patients with biallelic mutations reported yet | Breast and ovarian cancer | HR, ICL repair | Interacts with FANCJ, FANCD1/BRCA2; necessary for FA pathway activation Genetic interaction |
| LIGIV (DNA-PK, Ku)5 | Immune deficiency, pancytopenias, growth retardation, microcephaly, skin photosensitivity | T cell ALL (one report) | NHEJ | with FA proteins (see Table 2) |
| FAN1 | KIN/renal failure | None reported | ICL repair | Interacts with ID complex |
| XPF | Sun sensitivity, neurodegeneration, accelerated aging | Basal and squamous cell carcinoma of the skin | NER, FA nuclease | Interacts with FANCP/SLX4 |
Abbreviations used: ALL- acute lymphocytic leukemia; AML - acute myelogenous leukemia; BMF - bone marrow failure; CA congenital abnormalities; FA - Fanconi anemia; HNSCC - head and neck squamous cell carcinoma; HR - homologous recombination; ICL - interstrand crosslink; KIN - karyomegalic interstitial nephritis; NER - nucleotide excision repair; NHEJ - nonhomologous end joining.
As yet, no cancers have been reported in patients with FANCL or FANCM mutations, possibly due to their extreme rarity.
FANCC carriers show increased risk of breast cancer 75
In patients with FANCD1 and FANCN mutations, the growth, bone marrow, and developmental phenotypes are as in other FA complementation groups but the probability of malignancy is higher, the spectrum is different, and age of onset is lower, indicative of a more extreme subtype of FA.
FANCO is a provisional term, as patients with RAD51C mutations do not thus far display bone marrow failure or cancer, and chromosomal breakage is not as high as seen in FA individuals.
No mutations in Ku or DNA-PK have ever been observed in human patients.
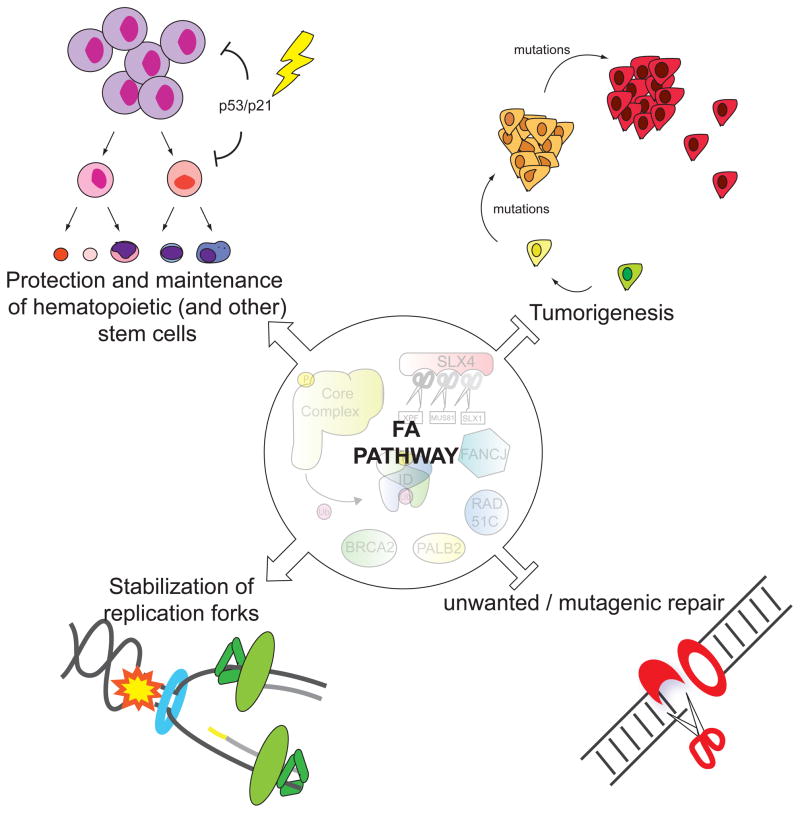
FA is also intimately involved with an expanding list of biological processes as new functions come to light (Figure 2). The bone marrow failure seen in patients suggests a function in stem cell biology, and indeed, recent work has begun to uncover tantalizing insights not only into FA pathogenesis, but also the intersection between DNA repair and stem cell maintenance. Several members of the FA pathway are also canonical breast and ovarian cancer predisposition genes, which together with the cancer susceptibility in FA patients points to an important role for FA proteins in suppressing tumorigenesis. Recent work has described an unanticipated role for FA proteins in replication fork stabilization. Finally, the identification of aldehydes as an endogenous genotoxin in FA provides an exciting glimpse into what challenges in the cellular environment are important in human disease. In this review, we highlight these topics and also give a primer on the current understanding of the molecular details of the FA pathway.
Figure 2. The diverse functions of the Fanconi anemia pathway.
The Fanconi pathway has many roles in human biology: as a regulator of the maintenance and proliferation of hematopoietic stem and progenitor cells, a tumor suppressor pathway, a preserver of replication fork stability during S phase, and a barrier against unwanted mutagenic repair processes. Besides the hematopoietic cells, other stem cells are most likely being affected by lack of FA pathway leading to stochastic developmental abnormalities and the infertility in FA individuals.
Stem cell function in the absence of the FA pathway
One of the clinical identifiers of FA is bone marrow failure which can be rescued by bone marrow transplantation, suggesting that disrupting the pathway leads to dysfunction of the hematopoietic stem and progenitor cells (HSPCs) themselves. While the bulk of FA research so far concentrates on the DNA repair functions of the pathway in terminally differentiated cells, this clinical manifestation points to a critical role in stem cell development that must be explored. A clear connection between the cellular and disease phenotypes in FA has been elusive, due to the difficulty in establishing mouse models that parallel human disease and because the limited proliferative potential of primary hematopoietic cells makes patient-derived in vitro systems infeasible. Understanding the interactions between FA pathway function and stem cell regulation and maintenance will likely prove to be key to elucidating the functional link between the pathway and the disease.
Recent research provides a glimpse into such a functional link between defects in genome maintenance and bone marrow failure in FA. Levels of p53, one of the central guardians of genome maintenance, and its transcriptional target p21, are elevated in primary blood, bone marrow, and even in the fetal FA livers during hematopoietic stem cell expansion in that organ. Perhaps due to this hyperactivation, the pool of available HSPCs is already compromised at birth in the FA individuals and the FA bone marrow is less proliferative than healthy bone marrow, with more cells in G0/G1 cell cycle stages and an increase in DNA damage signaling as indicated by the presence of phosphorylated H2AX (γH2AX). Not only are there fewer HSPCs in FA bone marrow, but those stem cells are also unable to produce progenitors in vitro 3.
Disrupting the FA pathway is itself sufficient to impair hematopoietic development in vitro. shRNA knockdown of either FANCA or FANCD2 in human embryonic stem cells leads to significantly reduced production of hematopoietic progenitor cells and their differentiated daughter populations 4. Exposing progenitor cells to crosslinking agents exacerbates the proliferation deficiency. These findings link the canonical ICL repair function of the FA pathway to its role in stem cell development. In murine hematopoetic stem cells and human bone marrow stromal cells, FA deficiency leads to an increase in binucleated cells, suggesting that cytokinesis failure also may contribute to bone marrow failure 5.
An important role for the FA pathway in stem cell function was uncovered when researchers attempted to derive induced pluripotent stem (iPS) cells from FA fibroblasts. They discovered that FA pathway deficient fibroblasts are refractory to reprogramming 6,7. It has been proposed, although not shown, that the FA proteins participate directly in reprogramming; alternatively, the increase in DNA damage signaling present in FA cells may preclude the normal cell cycle progression (division) necessary for reprogramming. It is also possible that the FA pathway is required to repair DNA damage that reprogramming itself induces. Discriminating between these possibilities will be key to understanding the role of the FA pathway in stem cells.
Taken together, recent data point to a connection between DNA repair defects and HSPC failure, though many mysteries remain. It is intriguing, for example, that FA deficiency leads to problems with both quiescent HSCs and their rapidly-dividing progenitors, two very different cellular landscapes. Understanding the underlying molecular functions of FA in hematopoietic stem cell populations will open up new avenues into understanding the pathophysiology of the disease. Furthermore, teasing out the importance of a pathway traditionally associated with DNA repair to the normal function of an important stem cell population will provide novel insight into stem cell biology in general.
FA pathway and tumor suppression
FA has a complicated relationship to tumorigenesis. While susceptibility to cancer, particularly AML and squamous cell carcinomas, is endemic to patients with homozygous mutations in the FA genes, four out of five downstream members of the pathway, which function in homology-directed repair of DNA ends, also confer susceptibility to breast cancer and ovarian cancer when mutated in only one copy 8–14 (Table 1). The famous breast cancer predisposition gene BRCA2 is itself an FA gene, FANCD1 15, and its partner PALB2 is FANCN 16. FANCJ is BRCA1-associated BRIP1 17,18 and RAD51C, one of the RAD51 paralogs important for HR, is provisionally named FANCO 19. In FA, patients with mutations in downstream pathway members evince more severe phenotypes, with more prevalent and earlier onset of tumors and leukemia. FANCD1 and FANCN patients also present with embryonal tumors at an extremely early age (< 2 years old).
The cancers that develop in FA patients also point to the complex landscape that is required for tumorigenesis to progress. Predisposition to AML may derive from the same hematopoietic stem cell instability that contributes to bone marrow failure. The type of squamous cell carcinomas of the head and neck common to FA are also associated with human papilloma virus (HPV), raising the possibility that these tumors develop in the context of infection with an oncogenic virus.
The correlation between downstream FA members and cancer is intuitive when one considers the mechanics of the pathway: while the upstream members belonging to the core and ID complexes are more self-contained, functioning in pathway activation and early coordination steps, the downstream components are intimately associated with homology directed repair (HR) which is used more broadly outside and independently of the FA pathway.
Basic functions of FA in ICL repair
Since crosslinked DNA impedes both transcription and replication, ICLs need to be removed during all stages of the cell cycle. Indeed, there is evidence for both replication-dependent and independent repair. In the absence of damage sensing by active replication forks and a homologous template, repair depends upon nucleotide excision repair, especially transcription-coupled nucleotide excision repair, and translesion synthesis proteins, including polymerase kappa 20,21. The involvement of Fanconi and Fanconi-associated proteins in G1 repair remains to be elucidated, although the core complex components and the ID complex are able to bind to ICLs independently of replication and their absence results in inefficient repair 22.
During S phase, the replication fork encounters the lesion and is forced to stall, activating a cascade of events leading to initiation of the DNA damage response. Current models favor a structure in which two replication forks converge upon a single lesion, which is most likely to occur in late S phase 23, although it is likely that a replication fork approaching the lesion from only one side would also elicit repair. Replication fork stalling at the site of damage is followed by an unhooking step, in which incisions are made on either side of the covalently linked nucleotides (Figure 1). On the strand that is incised, TLS, is recruited to bypass the unhooked crosslink. The incision event also effectively creates a double-stranded break, which is repaired by HR 23–25. The multi-step nature of ICL repair requires the tight coordination of several different repair pathways, and it is within this delicately balanced system that the FA pathway plays its regulatory role.
An important step in the FA pathway is the monoubiquitylation of two components, FANCI and FANCD2, which form a heterodimer called the ID complex. Monoubiquitiylation of FANCD2 at lysine 561, and to a lesser extent of FANCI at lysine 523, represents the activation step of the FA pathway 26,27. This step is dependent on the FA core complex, which is comprised of FANCA, B, C, E, F, G, L, and M, and accessory proteins including FAAP20, FAAP24 and FAAP100. With the exception of FANCM, each core complex member is absolutely required for successful monoubiquitylation of FANCD2-FANCI. The ubiquitin ligase function of the core complex depends upon the E3 ligase FANCL, with UBE2T lending its services as an E2 28–30. Ubiquitylation of FANCI and FANCD2 are mutually interdependent in mammalian cells, and also require discrete phosphorylation events: phosphorylation of FANCI at SQ/TQ sites close to the targeted lysine is necessary for specific ubiquitylation of the ID complex 31. Recent evidence that FANCD2 ubiquitylation is robustly stimulated by the presence of DNA suggests that this event may occur on chromatin 32. Ubiquitylated ID complex localized to chromatin orchestrates the actions of downstream repair proteins. Interestingly, the ID complex may also play a direct role in the regulation of nucleosome assembly at sites of damage: turnover of histone H3 after crosslinker treatment is markedly slowed after FANCD2 knockdown 33.
The downstream components, those that are dispensable for monoubiquitylation of the ID complex are FANCD1, FANCJ, FANCN, and FANCO - already introduced above as cancer predisposition genes necessary for homologous recombination - and FANCP (SLX4), which interacts with multiple nucleases, discussed in greater detail below. FANCD1 and FANCN have been shown to work in later stages of repair where they complete repair through HR. FANCJ largely remains a mystery and its role as an FA protein needs to be explored further.
Although the details of how the pathway is turned off are not fully elucidated, one player that clearly participates in the process is the deubiquitinating enzyme USP1, which removes the ubiquitin from FANCD2 34. In its absence, ubiquitinated FANCD2 is elevated but the cells are sensitive to crosslink damage 35,36. As such, it is clear that both appropriate activation and deactivation of the pathway are important for cell viability when challenged with ICL damage.
The FA Pathway Coordinates Nuclease Action
A key step in most repair pathways involves the physical removal of the damaged bases as well as processing of DNA by nucleases. In the case of ICLs, the lesion affects both Watson and Crick strands, and its repair requires the collaboration of several processing enzymes. Each incision event requires a nuclease with the correct structure specificity in order to yield a repair intermediate that can correctly feed into the subsequent step. For example, crosslink unhooking must precede the resection steps necessary to provide a substrate for HR. Therefore, the removal steps are a complex choreography of nuclease action. The FA pathway has been implicated in the recruitment and regulation of several different nucleases, among them XPF-ERCC1, MUS81-EME1, SLX1, and FAN1 37–42.
FANCP, or SLX4, is a recently identified FA protein that functions as a scaffold, modulator, and cofactor for three structure specific nucleases: XPF-ERCC1, MUS81-EME1, and SLX1 43–46. Essentially modular in nature, FANCP is unique in that it can bring a veritable toolbelt of DNA processing enzymes to the site of DNA damage, ensuring the presence of the best repair nuclease for the job. Among the three nucleases associated with SLX4, XPF-ERCC1 is the most important for the resistance to ICLs, with MUS81-EME1 and SLX1 playing less prominent roles 47,48. The exonuclease SNM1A is also biochemically active at sites of crosslinks. It is required for ICL resistance, consistent with an in vivo function in repair of these lesions. Although its exact interaction with the FA proteins remains largely unclear, it is thought to act in concert with SLX4-associated XPF-ERCC1 to process the crosslink after the initial incision 49. In vitro, SNM1A can digest ICL-containing DNA past the lesion, creating a preferred substrate for downstream TLS polymerases 49. Understanding precisely how SNM1A, as well as other nucleases, are coordinated and controlled by, or independent of, the FA proteins will be critical to understanding this multifaceted repair pathway.
FAN1, the Fanconi associated nuclease, was identified by multiple groups to be required for ICL resistance 38–41 implicating it in ICL repair. FAN1 can be recruited to the sites of DNA damage in a manner dependent upon the ubiquitylated ID complex and upon its own ubiquitin-binding domain. The precise mechanism of FAN1’s action upon crosslinked DNA remains elusive; however, it is clear that FAN1 participates in a separate, as-yet uncharacterized branch of crosslink repair in addition to the FA pathway.
This function of FAN1 was recently illuminated by studying another rare genetic disease called karyomegalic interstitial nephritis (KIN), which is characterized clinically by progressive renal failure and histologically by enlarged and hyperchromatic nuclei within the kidney tubular epithelium. A cohort of patients with KIN were seen to have mutations in FAN1. Interestingly, patients with KIN have no hallmarks of FA but cells from these patients exhibit significant sensitivity to crosslinking agents, which can be complemented with wild type FAN1, suggesting that an ICL repair defect conferred by FAN1 deficiency does, at least in part, underlie the syndrome 50. FAN1’s deficiency resulting in KIN and not FA, despite the fact that FAN1 and FANCD2 physically interact, suggests that FAN1 might be redundant with another nuclease in the FA pathway. It also implies that FAN1 has a function that is independent of the FA pathway in the kidney, where it may repair lesions that the FA pathway cannot. It is of interest that FAN1 expression is very high in the kidney. The deviance between KIN and FA phenotypes is a fascinating puzzle that might be a key to understanding different modes of ICL repair as well as a clue to the endogenous lesions that give rise to disease not only in these rare patients but also during normal aging.
FA pathway and replication stress
In addition to an indispensable role at DNA crosslinks, the FA pathway is required for the protection of replication fork stability under stress. The ID complex is monoubiquitylated during S phase even in unchallenged cells 15. The pathway is also activated by depletion of nucleotide pools with hydroxyurea (HU), and even by UV light 51. This has been puzzling since FA patient cell lines are not sensitive to either of these agents. However, recent data suggests that FANCA, ubiquitylated FANCD2, BRCA1, and BRCA2 are necessary for the protection of the stalled replication fork after HU treatment. When they are functionally absent, RAD51 is not deposited onto the fork under stress and the newly synthesized (nascent) DNA gets degraded in an MRE11-dependent manner 52,53. The outcome appears to be an increase in chromosomal instability, which, while not directly leading to immediate cellular demise, can still be mutagenic in the long term, thus contributing to the increase in tumorigenesis seen in FA patients. Importantly, the protective function of the FA/BRCA axis uncovered by these experiments appears to be separate from the canonical function of BRCA2 in HR. It will be important to address if any of the ICL sensitivity of FA cell lines might be secondary to a defect in fork protection function and whether the stem cell failure in FA can be mitigated by fork stabilization.
Cooperation with other DNA repair pathways
At its heart, FA pathway-mediated repair of ICLs is a highly regulated ensemble of other repair pathways: a nuclease borrowed from canonical NER makes incisions, translesion synthesis polymerases are responsible for filling in the gap across from the unhooked crosslink, and HR is coopted to resolve the double-stranded breaks that result when replication forks encounter the lesion. It is becoming clear that in addition to using other pathways for discrete repair steps, the FA pathway is intimately entwined with the DNA damage response as a whole, engaging in collaboration and antagonism with almost all of the classical repair pathways in the fight for survival after genotoxic insult. The most notable of these are the mismatch repair pathway components implicated in FANCJ 54 and FANCD2 function 55, and the BLM helicase, which appears to intersect with the FA pathway at multiple levels. BLM mutations are found in Bloom syndrome, an autosomal recessive disease of growth retardation, immunodeficiency, sun sensitivity and profound cancer predisposition. The BLM protein is responsible for unwinding a variety of DNA structures and together with topoisomerase IIIα is able to remove the double Holliday junctions (HJ) that form during HR 56. This process, called double HJ dissolution, prevents the inappropriate engagement of nucleolytic HJ resolvases whose action may lead to deleterious exchanges of DNA between sister chromatids.
There are two recently uncovered connections between FA and BLM. One pertains to ultrafine anaphase bridges (UFBs), thread-like DNA structures that form between masses of condensed DNA during anaphase, and are thought to arise from persistent, unresolved DNA catenanes or recombination intermediates. Both BLM and the ID complex localize to these microbridges, though FANCI and FANCD2 travel only to a subset of them, likely those caused by replication stress at fragile sites 57. These microbridges may underlie the cytokinesis failure in FA bone marrow cells discussed above. The second connection is with FANCP/SLX4, which collaborates with BLM during Holliday junction resolution. Co-depletion of these two proteins leads to a uniquely aberrant chromosome morphology, where sister chromatids appear to stay linked and the chromosomes themselves adopt an odd segmented appearance 58. As with many of the FA pathway’s apparent interactions with other repair pathways, the underlying mechanisms and relationships must still be elaborated, but taken together, the data reiterate how the players in the FA pathway have far-reaching influence in general genome maintenance.
Fanconi anemia and non-homologous end joining
The DNA repair pathway with a particularly fraught relationship with the FA pathway is that of non-homologous end joining (NHEJ). NHEJ is an error-prone mechanism for fixing double-stranded breaks when no sister chromatid is available to be the template for HR, in which DNA ends are ligated directly together without regard for homology. The Ku70-Ku80 heterodimer binds to DNA ends, where it recruits the catalytic subunit DNA-PK. LigIV is then brought in to carry out the ligation (reviewed in 59).
NHEJ is the default double-strand break (DSB) repair pathway in G0 and G1. In other phases of the cell cycle, the cell faces a choice of whether to use NHEJ or HR to restore the break. Recent work uncovers a dynamic shift between preferred pathways throughout the cell cycle with HR reaching a peak during late S phase 60. The decision also hinges on which factors access the free DNA end. The pattern of resection at the site of the double-stranded DNA break provides a particular substrate for each type of repair. HR requires extensive resection to create a long 3′ overhang that, once coated with RAD51, can initiate homology search, while NHEJ requires little resection if any. Competition between binding of HR and NHEJ factors influences the pattern, and only once the 3′ overhang has been successfully resected is the cell fully committed to HR 61.
The FA pathway is thought to play a role in pathway choice, funneling the DSBs created by ICL processing or other forms of replication stress into high-fidelity HR repair. There has therefore been speculation that NHEJ-mediated repair in the absence of a functional FA pathway is an underlying cause of the gross chromosomal abnormalities observed in FA cell lines. Without a functional FA pathway, perhaps the breaks become subject to the profligate end-pairing of NHEJ and thereby generate chromosomal rearrangements. Animal and human cell models have been developed to test this hypothesis and so far are giving contradictory findings (Table 2).
Table 2. Summary of genetic interactions between FA and NHEJ.
Different combinations of FA and NHEJ genes that have been shown to genetically interact, but offer conflicting views on the nature of those interactions and their relevance to human disease. Experimental outcomes are classified by model organism and the method of gene loss or depletion. Color coding indicates full rescue (green), partial rescue (blue), or no rescue (red)
| Model System | FA gene | NHEJ gene | Outcome |
|---|---|---|---|
| C. elegans | fcd-2 (FANCD2) (germline mutation) | lig-4 (germline mutation) | Rescue of crosslink sensitivity and aberrant meiotic crossovers 63 |
| DT40 cells | ΔFANCC (knockout) | ΔKu70 (knockout) | Partial rescue of crosslink sensitivity 64 |
| ΔFANCC (knockout) | ΔLIGIV or ΔDNA- PKcs (knockout) | No rescue 64 | |
| Mouse embryonic fibroblasts | Fanca−/−, Fancc −/− (germline mutation) | DNA-PKcs (Nu7026 inhibitor) | Partial rescue of crosslink sensitivity 63 |
| Fancd2 −/− (germline mutation) | Prkdcsc/sc (scid mouse) | No rescue 65 | |
| Fancd2−/− (germline mutation) | Ku80, 53BP1 (germline mutations) | No rescue Increase in chromosomal instability and sensitivity 66 | |
| Human cells | MO59J cells FANCD2 siRNA | DNA-PKcs deficient cell line | Rescue of crosslink (MMC) sensitivity 63 |
| HeLa cells FANCA siRNA |
DNA-PKcs (Nu7026 inhibitor) | Partial rescue of crosslink (MMC) sensitivity 63 | |
| HeLa cells FANCD2 siRNA |
DNA-PKcs inhibitor | Rescue of crosslink sensitivity 63 | |
| PD773(FANCD2) PD331 (FANCC) FA patient mutations |
KU80 siRNA | Partial rescue of crosslink (MMC) sensitivity 63 |
In the nematode Caenorhabditis elegans, the basic components of the FA pathway are conserved, including FANCD2 (FCD-2), FANCI (FNCI-1), FANCM (FCM-1) and FANCJ (DOG-1) 62. The C. elegans ID complex is ubiquitinated and travels to sites of damage like its human counterpart, and knockout of all four members leads to hypersensitivity to crosslinking agents. The worm is an excellent model system to observe the effects of knocking out NHEJ components in a FA-deficient background, as it mimics relevant phenotypes with a pared-down pathway. When LIG-4, the DNA ligase important for NHEJ, is mutated in the fcd-2 knockout, the sensitivity to crosslinking agents is corrected to near-wild type levels. These data suggest that NHEJ does in fact have a deleterious effect when the FA pathway is deficient 63.
The C. elegans model is also useful for studying the interplay between FA and NHEJ during meiosis. While FA patients and knockout mice exhibit fertility defects, the mechanisms underlying this loss of fertility are poorly understood. The regulation and resolution of meiotic crossovers derived from recombination of programmed double-stranded breaks is mediated by many proteins that also serve in a DNA repair capacity, raising the possibility that the FA pathway might also play a role. In the fcd-2 mutant, RAD51 foci persist during meiosis, indicative of unresolved HR intermediates, and inappropriate nonhomologous chromosome engagements are observed after diakinesis 63. Similar to what is seen in somatic cells, the deletion of LIG-4 ameliorates the aberrant crossovers observed in the fcd-2 mutant.
In chicken DT40 cells, the ICL hypersensitivity and chromosome breakage endemic to FANCC cell lines can be suppressed by also knocking out Ku70 64. Interestingly, knocking out either DNA-PK or LigIV in the DT40 cells effects no such rescue, in contrast to the observations in C. elegans. In this case, the specificity of Ku rescue may point to a direct role for FA proteins, possibly the ID complex, in antagonizing Ku attachment at DNA ends.
The implication that NHEJ underlies the hypersensitivity and chromosomal breakage phenotypes in FA is of great clinical interest, as it provides a putative pharmaceutical target. If the cellular phenotypes that underlie the disease are caused not by ICLs themselves but by aberrant repair processes that occur in the absence of the FA pathway, perhaps NHEJ inhibition could serve as a highly specific treatment. It is therefore critical to investigate the relationship between these two pathways in mammalian cell lines. RNAi knockdown of Ku80 in FANCC and FANCD2-deficient patient cell lines partially suppresses sensitivity to the killing effects of crosslinking agents. Furthermore, the addition of a DNA-PK inhibitor mitigates the hypersensitivity resulting from knockdown of FANCA or FANCD2 by RNAi in HeLa cells, or by knockout of FancA or FancC in mouse embryonic fibroblasts 63. This again is in contrast to genetic experiments that show that the crosslink sensitivity of the Fancd2 knockout is not changed in the setting of decreased DNA-PK activity (Prkdc sc/sc mouse)65 and that loss of 53BP1 or Ku80 renders Fancd2-deficient cells even more sensitive to DNA crosslinking agents than Fancd2 deficiency alone66.
The inconsistencies between these studies raise questions about how to parse data from different organisms, genetic backgrounds, and experimental setups. As hinted by these experiments, different components of the NHEJ pathway may play different roles in different organisms and even in different cells of the same organism; indeed, fibroblasts may not accurately reflect how cells more relevant to FA, like HSPCs, might behave. Furthermore, interaction between components of the two pathways may be exquisitely sensitive to the spatial and temporal relationships between them. When thinking about possible therapeutic implications of these findings, it is important to remember that the full inhibition of the Ku proteins is not feasible in human cells since they have essential function at human telomeres 67. Overall, well-controlled work, especially in human hematopoietic cells, will be required to figure out the interplay of FA and NHEJ pathways. It will also be necessary to determine how these extremely toxic ICLs are repaired if both FA and NHEJ are compromised.
Alcohol metabolism poses a challenge for FA-deficient organisms
The average human is not exposed to the clastogenic agents used in the lab to model DNA repair defects. The FA pathway is required for dealing with replication stresses that can arise during unchallenged cell cycle progression, as shown by the activation of the pathway during normal S-phase. It is likely that the pathway also evolved in order to maintain genome integrity in the face of mutagenic agents that arise endogenously from processes of cellular metabolism. Oxidative stress within the cell can yield nucleophilic crosslinking agents like malondialdehyde and acrolein, which derive from lipid peroxidation; nitric oxide, another endogenous genotoxin that can promote the formation of crosslinks, is generated independently of lipids 68. Aldehydes, discussed in greater detail below, are also a major source of endogenous crosslink damage. Understanding how damage from these endogenous agents feeds into the FA pathway is critical to understanding the disease.
Ethanol metabolism has long been known to produce reactive aldehydes that can function as carcinogens: acetaldehyde, an intermediate in the metabolic processing of ethanol, is in particular implicated as a genotoxic agent. Acetaldehyde, which is generated from alcohol metabolism, was shown to stimulate monoubiquitylation of FANCD2 in vitro, and FA cells demonstrated sensitivity to formaldehyde 69,70. Until recently, though, no in vivo studies functionally linked these cellular byproducts to concrete phenotypes in an FA model. The first indication that such cellular byproducts may indeed pose a challenge to FA-deficient systems comes from a genetic interaction between FA genes and superoxide dismutase (SOD), an enzyme which decreases the load of oxidative damage in the cell as a whole. Fancc−/−;Sod−/− mice display bone marrow hypocellularity, which is not present in either of the single mutants 71. Studies investigating the effects of interfering with acetaldehyde catabolism has uncovered a strong connection to the FA pathway. Chicken DT40 cells knocked out for several FA components, including FANCB, FANCC, FANCL, and FANCJ, are sensitive to treatment with acetaldehyde 72. In addition, FA DT40 knockouts are synthetic lethal with mutations in the formaldehyde catabolism gene ADH5, suggesting that toxicity is generated when the cells permit natural toxic aldehydes to build up 73.
A double mutant for FANCD2 and aldehyde dehydrogenase 2 (ALDH2), an important enzyme in acetaldehyde catabolism, was created to test the relevance of acetaldehyde sensitivity in the developing mouse. The double mutant did not survive, with embryos dying between embryonic day 9.5 and 13.5, if and only if the mother was homozygous for the ALDH2 mutation. In contrast, pregnant females with one wild-type ALDH2 allele could carry pups to term. When heterozygous mothers were fed ethanol during pregnancy, the double mutant pups exhibited drastic developmental abnormalities, such as exencephaly and eye defects 72. These data suggest that the developing mouse is exquisitely sensitive to toxic aldehydes, casting light onto the significance of aldehyde catabolism in FA deficient backgrounds.
A more important question is whether toxic aldehydes can produce symptoms in the mouse that mimic the human disease phenotype. Chronic alcohol abuse is correlated with bone marrow problems, reminiscent of the bone marrow failure observed in FA. Indeed, Fancd2−/−;Aldh2−/− double mutants mice displayed increased levels of ethanol-induced bone marrow failure compared to wild-type or single mutants. Even without administration of ethanol, the double mutants die early, due to abnormalities that closely resemble acute lymphoblastic leukemia.
Most tellingly, the double mutant mice that do not suffer from leukemia develop aplastic anemia that is similar to that seen in human FA patients. The bone marrow failure in these mice can be traced back to a DNA repair deficiency in the HSPC pool, suggesting that endogenous aldehyde may be particularly harmful to hematopoetic stem cells, and providing the first direct link between acetaldehyde related toxicity and bone marrow failure phenotypes in FA 74.
Animal (particularly mouse) models of FA allow us to explore what genetic and environmental factors contribute to disease pathogenesis, and as such can lead to better disease management and treatment plans. Especially as sequencing becomes more affordable, understanding what genetic factors modify and shape the progress of FA symptoms will offer better diagnostics and tailored treatment. Discovering what commonly encountered genotoxic agents contribute to the clinical manifestations of FA will provide therapeutic targets and lifestyle modifications, not only for FA but for many diseases that derive from DNA repair defects.
Looking Forward
As a multi-functional, highly connected pathway, the FA pathway is well poised to be a very important topic in DNA repair and in biology. There is evidence of functional relationships between the FA pathway and virtually all of the cellular DNA repair pathways, although many of the details remain murky. Understanding the mechanisms behind these connections will foster understanding of DNA repair as a whole, rather than a grab-bag of disparate pathways. The genetic link between FA and breast cancer will also be an important topic as research progresses, shedding light onto the nature of tumor suppressors, and the enigma of genotype-phenotype correlations. As technologies make it easier to manipulate stem cells, the function of FA in hematopoietic lineages and other stem cells will become a growing subfield within FA research. We may finally gain insight into the developmental abnormalities that are seen in patients. Combining studies of FA function and stem cell development will allow us to create a powerful model of DNA repair within stem and progenitor cell populations. Another pressing issue is the identification of the endogenous damage that the FA pathway responds to. It is likely that the lesions arise from multiple sources and that there will be nuances to how the FA pathway is responding to each of them. Finally, there are many details about the molecular mechanism of ICL repair that are still missing. Coordination and regulation of the nucleases identified as necessary for repair is a topic of intense interest. With the identification of new players in the pathway we can only expect that the pathway will become more complex but also more revealing. Deciphering the mechanics of FA pathway function will help efforts to understand both a fascinating disease, and basic human biology.
Acknowledgments
We thank Arleen Auerbach, Siobhan Gregg, Elizabeth Garner, Yonghwan Kim, Supawat Thongthip and Anderson Wang for their comments. Work in our lab is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists, the Starr Center Consortium grant, and by grant # 8 UL1 TR000043 from the National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health. A.S. is Rita Allen Foundation, Irma T. Hirschl, the Alexandrine and Alexander Sinsheimer Foundation scholar, and is a recipient of the Doris Duke Clinical Scientist Development Award. M.C.K. is supported by an American Cancer Society-J.T. Tai Postdoctoral Fellowship.
Footnotes
AUTHOR INFORMATION
The authors declare no competing financial interests.
References
- 1.Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A. 2011;155A:1877–1883. doi: 10.1002/ajmg.a.34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. S0027-5107(09)00053-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceccaldi R, et al. Bone Marrow Failure in Fanconi Anemia Is Triggered by an Exacerbated p53/p21 DNA Damage Response that Impairs Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. S1934-5909(12)00247-0 [pii] This study identifies activation of the p53/p21 axis as a major contributing factor to loss of hematopoeitic stem cells in Fanconi anemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tulpule A, et al. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115:3453–3462. doi: 10.1182/blood-2009-10-246694. blood-2009-10-246694 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinciguerra P, Godinho SA, Parmar K, Pellman D, D’Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest. 2010;120:3834–3842. doi: 10.1172/JCI43391. 43391 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller LU, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. blood-2012-02-408674 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. nature08129 [pii] This paper demonstrates that human fibroblasts deficient in the FA pathway are refractory to reprogramming into iPS cells unless they are first corrected with an appropriate gene, and that FA-corrected cells can be reprogrammed into iPS cells that give rise to phenotypically normal lineages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. nrc3088 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meindl A, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–414. doi: 10.1038/ng.569. ng.569 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Rafnar T, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43:1104–1107. doi: 10.1038/ng.955. ng.955 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. ng1959 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. ng1902 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Tischkowitz M, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. 0701724104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh T, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. 1115052108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. 1073834 [pii] This paper identifies the breast cancer predisposition gene BRCA2 as mutated in a very severe form of Fanconi anemia, firmly establishing that Fanconi anemia is a DNA repair deficiency disease. [DOI] [PubMed] [Google Scholar]
- 16.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. ng1942 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Levran O, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. ng1624 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Levitus M, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. ng1625 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Vaz F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. ng.570 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Enoiu M, Jiricny J, Scharer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks670. gks670 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, et al. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, et al. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. S1097-2765(09)00475-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. science.1182372 [pii] This work uses a cell-free system to demonstrate the requirement for FANCD2 and FANCI in the replication-dependent repair of interstrand crosslinks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. S0092-8674(08)01075-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. 333/6038/84 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. S0092-8674(07)00320-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi T, et al. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 28.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. S1097-2765(08)00844-7 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. S1097-2765(06)00452-7 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241ng1241[pii]. [DOI] [PubMed] [Google Scholar]
- 31.Ishiai M, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. nsmb.1504 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 2012;40:4553–4561. doi: 10.1093/nar/gks053. gks053 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato K, et al. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 2012;31:3524–3536. doi: 10.1038/emboj.2012.197. emboj2012197 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nijman SM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. S1097276505010415 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Kim JM, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. S1534-5807(09)00003-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oestergaard VH, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. S1097-2765(07)00632-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y, et al. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. ng.750 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. S0092-8674(10)00676-8 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. science.1192656 [pii] [DOI] [PubMed] [Google Scholar]
- 40.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. S0092-8674(10)00675-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. S1097-2765(10)00464-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoepker C, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. ng.751 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Fekairi S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. S0092-8674(09)00776-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. S0092-8674(09)00777-6 [pii] This work, along with references 43, 45, and 46, identifies SLX4 as a partner to multiple DNA repair nucleases necessary for ICL repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersen SL, et al. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. S1097-2765(09)00432-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munoz IM, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. S1097-2765(09)00433-X [pii] [DOI] [PubMed] [Google Scholar]
- 47.Bhagwat N, et al. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol Cell Biol. 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. MCB.00086-09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim Y, et al. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. doi: 10.1182/blood-2012-07-441212. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang AT, et al. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011;25:1859–1870. doi: 10.1101/gad.15699211. 25/17/1859 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012 doi: 10.1038/ng.2347. ng.2347 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. ddi065 [pii] [DOI] [PubMed] [Google Scholar]
- 52.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. S0092-8674(11)00375-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. S1535-6108(12)00214-0 [pii] This work identifies an important role for FANCD2 and BRCA1/2 in protecting the stability of replication forks, illuminating a repair-independent function for these proteins in genome maintenance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng M, et al. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007;26:3238–3249. doi: 10.1038/sj.emboj.7601754. 7601754 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams SA, et al. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum Mol Genet. 2011;20:4395–4410. doi: 10.1093/hmg/ddr366. ddr366 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. nature02253 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. ncb1882 [pii] [DOI] [PubMed] [Google Scholar]
- 58.Wechsler T, Newman S, West SC. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. nature09790 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. S1097-2765(12)00554-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 62.Youds JL, Barber LJ, Boulton SJC. elegans: a model of Fanconi anemia and ICL repair. Mutat Res. 2009;668:103–116. doi: 10.1016/j.mrfmmm.2008.11.007. S0027-5107(08)00301-1 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Adamo A, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. S1097-2765(10)00493-4 [pii] This work investigates the relationship between the FA pathway and nonhomologous end joining (NHEJ), suggesting that the FA cellular phenotypes derive from aberrant repair by NHEJ. [DOI] [PubMed] [Google Scholar]
- 64.Pace P, et al. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. science.1192277 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Houghtaling S, et al. Fancd2 functions in a double strand break repair pathway that is distinct from non-homologous end joining. Hum Mol Genet. 2005;14:3027–3033. doi: 10.1093/hmg/ddi334. ddi334 [pii] [DOI] [PubMed] [Google Scholar]
- 66.Bunting SF, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. S1097-2765(12)00174-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. 0903362106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pang Q, Andreassen PR. Fanconi anemia proteins and endogenous stresses. Mutat Res. 2009;668:42–53. doi: 10.1016/j.mrfmmm.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marietta C, Thompson LH, Lamerdin JE, Brooks PJ. Acetaldehyde stimulates FANCD2 monoubiquitination, H2AX phosphorylation, and BRCA1 phosphorylation in human cells in vitro: implications for alcohol-related carcinogenesis. Mutat Res. 2009;664:77–83. doi: 10.1016/j.mrfmmm.2009.03.011. S0027-5107(09)00115-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ridpath JR, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. 67/23/11117 [pii] [DOI] [PubMed] [Google Scholar]
- 71.Hadjur S, et al. Defective hematopoiesis and hepatic steatosis in mice with combined deficiencies of the genes encoding Fancc and Cu/Zn superoxide dismutase. Blood. 2001;98:1003–1011. doi: 10.1182/blood.v98.4.1003. [DOI] [PubMed] [Google Scholar]
- 72.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. nature10192 [pii] [DOI] [PubMed] [Google Scholar]
- 73.Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. nsmb.2173 [pii] [DOI] [PubMed] [Google Scholar]
- 74.Garaycoechea JI, et al. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. nature11368 [pii] This paper, along with reference 71, identifies the aldehydes resulting from ethanol metabolism as important endogenous genotoxins contributing to FA pathogenesis. Together, they demonstrate that increasing aldehyde load in FA-mutant mice leads to phenotypes strikingly similar to the human disease, including developmental abnormalities, leukemia and bone marrow failure. [DOI] [PubMed] [Google Scholar]
- 75.Berwick M, et al. Genetic heterogeneity among Fanconi anemia heterozygotes and risk of cancer. Cancer Res. 2007;67:9591–9596. doi: 10.1158/0008-5472.CAN-07-1501. 67/19/9591 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]