Summary
Storage granules are an important component of metabolism in many organisms spanning the bacterial, eukaryal and archaeal domains, but systematic analysis of their organization inside cells is lacking. In this study, we identify and characterize granulelike inclusion bodies in a methanogenic archaeon, Methanospirillum hungatei, an anaerobic microorganism that plays an important role in nutrient recycling in the ecosystem. Using cryo electron microscopy, we show that granules in mature M. hungatei are amorphous in structure with a uniform size. Energy dispersive X-ray spectroscopy analysis establishes that each granule is a polyphosphate body (PPB) that consists of high concentrations of phosphorous and oxygen, and increased levels of iron and magnesium. By scanning transmission electron tomography, we further estimate that the mass density within a PPB is a little less than metal titanium at room temperature and is about four times higher than that of the surrounding cytoplasm. Finally, three-dimensional cryo electron tomography reveals that PPBs are positioned off-centre in their radial locations relative to the cylindrical axis of the cell, and almost uniformly placed near cell ends. This positioning ability points to a genetic program that spatially and temporally directs the accumulation of polyphosphate into a storage granule, perhaps for energy-consuming activities, such as cell maintenance, division or motility.
Introduction
Archaea encompass one of the three domains of life and have been recognized to bear more similarity to eukaryotes than to bacteria, which is one of the main reasons behind the interest in studying this unique class of organisms. Believed to be among the oldest life forms known on earth, archaeal cells use special means to generate and store energy in order to thrive in harsh environments. The archaea Methanospirillum hungatei is a methanogen belonging to the Methanomicrobiales division of the Euryarchaeota. Initially isolated from an anaerobic benzoate degrading enrichment culture, it utilizes hydrogen plus carbon dioxide and/or formate as a source of energy and carbon for cell growth and methane production (Ferry et al., 1974; Ferry and Wolfe, 1977; Mountfort and Bryant, 1982). This organism is distinct from many other hydrogenotrophic methanogens in that it can form specialized and inter-dependent biochemical partnerships with a variety of syntrophic bacteria (Ferry and Wolfe, 1976; McInerney et al., 1981). When grown in co-culture, these methanogens aid in catalysing the breakdown of a variety of short-chain fatty acids, alcohols and aromatic compounds essential for the normal operation of the anaerobic food chains in nature (Stieb and Schink, 1985; McInerney et al., 2008). However, little is known concerning energy generation and storage or morphological determinants that favour the adaptation and survival of this physiologically distinct methanogen in many environmentally diverse habitats.
Methanospirillum hungatei species are morphologically distinct from other methanogen cell types. With a mildly curved surface, cells exhibit the characteristic ability to grow within a sheath-like structure containing one to more than ten cells along its length (Ferry et al., 1974; Patel et al., 1976; Beveridge et al., 1985). All cell types retain their outer sheath during all phases of growth where multiple flagella emanate from the sheath tips or ends. The recent sequencing of M. hungatei JF1 genome reveals it to possess the sixth largest (3.5 MB) of all archaea genomes sequenced to date (NC_007796.1). Whereas a core set of genes/proteins were identified for methane biosynthesis from hydrogen/carbon dioxide and/or formate, many other aspects of Methanosprillium cell energetics and physiology remain unexplored.
In this study, we employ a combination of biophysical methods, including cryo electron microscopy (cryoEM), cryo electron tomography, (cryoET), scanning transmission electron microscopy (STEM) tomography and energy dispersive X-ray (EDX) spectroscopy analyses, to identify and characterize high-density inclusion bodies (also called granules) distributed within the M. hungatei cell cytoplasm. We show that these structures, generally reaching 150 nm in diameter, are rich in phosphorous and oxygen and are typically positioned asymmetrically nearby each cell tip. With the likely function in phosphate and cellular energy storage, the presence of these granules may suggest an ancient origin of polyphosphate synthesis and accumulation in this model methanogenic archaean. Alternatively, they may have acquired this ability by lateral gene transfer from other organisms.
Results
Observation of high-density inclusion bodies within M. hungatei cells by cryoEM
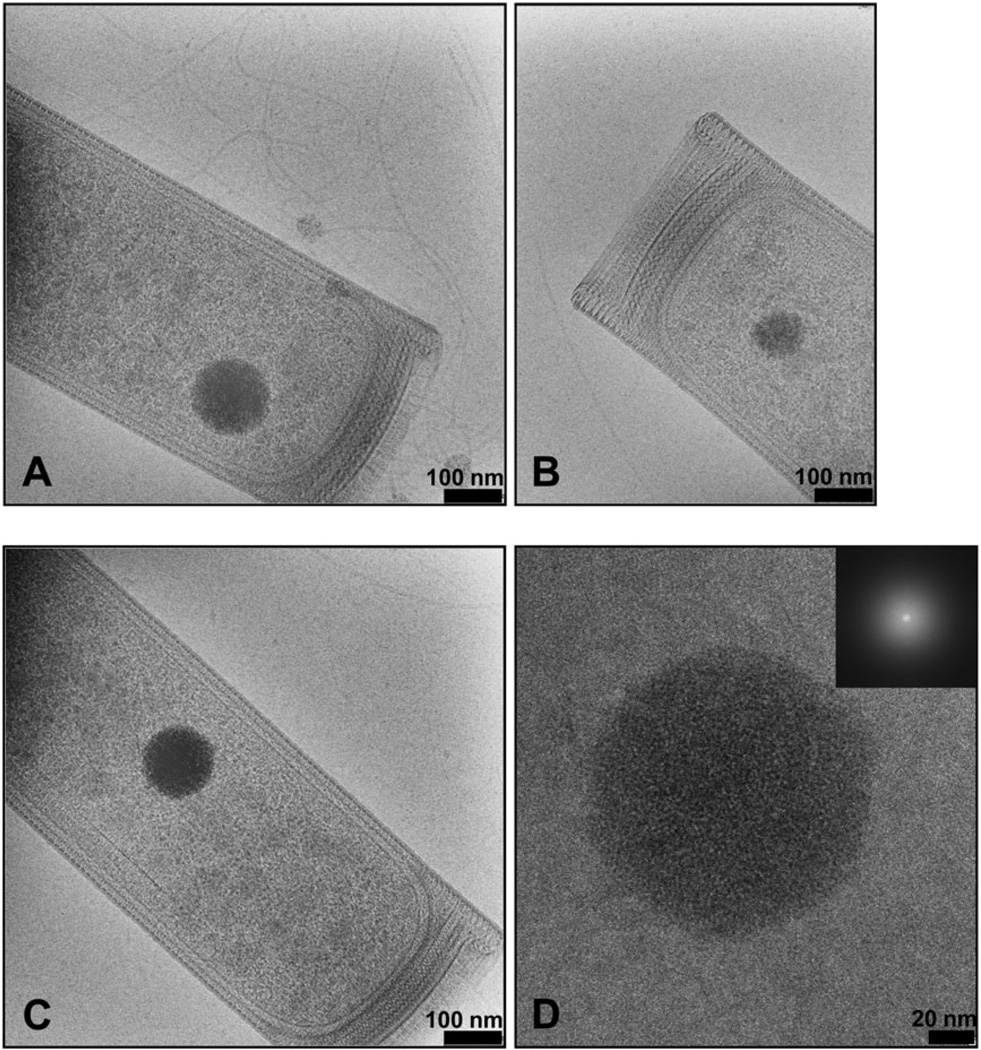
Previous ultrastructural studies of M. hungatei have relied on conventional transmission electron microscopy (TEM) of fixed cells using thin sectioning, followed by heavy metal staining (Beveridge et al., 1985; 1990; Southam and Beveridge, 1992; Firtel et al., 1993). Such approaches suffer from dehydration of the sample, staining artefacts, as well as missing information due to the impermeability of the outer sheath layer to the stain. CryoEM images of M. hungatei reveal that it has a multilayered envelope, which includes two proteinaceous surface layers, (a paracrystalline S-layer and a sheath layer) external to the cellular bi-layer membrane, as well as a multi-layered plug structure at the ends of each cell (Fig. 1A–C). The use of cryoEM allows us to visualize the whole cells embedded in vitreous ice thus preserving their structures in a natural state and eliminating artefacts associated with stain. The cryoEM images of M. hungatei cells show an elongated shape of each cell that is 5–10 µm in length and 0.4–0.5 mm in diameter with a characteristic multi-enveloped outer structure, with the layers of sheath, S-layer, and cellular membrane clearly visible. The sheath layer at both ends of the rod-shaped cell curls inward to form a circular lip and connects with the multi-layered plug structure (Fig. 1A–C). However, if there is a cell division occurring between two cells sharing one sheath, then the sheath is continuous between the two cells and there is a septum region separating the ends of each cell (Fig. 2, inset, see also Fig. 6A).
Fig. 1.
CryoEM of M. hungatei JF1 cells. A–C. Representative cryoEM images of cell ends showing characteristic high-density inclusion bodies.
D. Higher magnification cryoEM image of the same inclusion body shown in (C). The inset is the Fourier transform of the image, showing no pattern indicative of a crystalline lattice or ordered structure within the inclusion bodies.
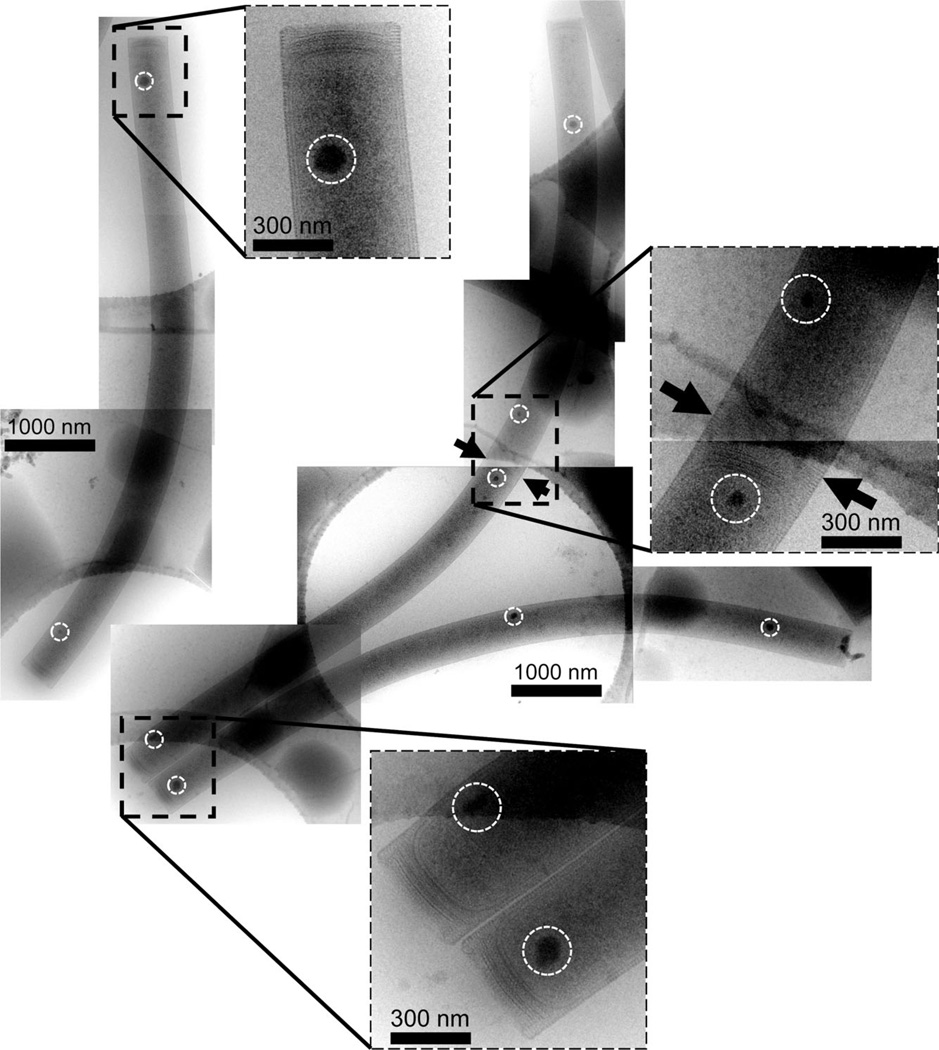
Fig. 2.
Composite cryoEM images showing whole M. hungatei JF1 cells and the relative longitudinal locations of the inclusion bodies. The location of a cell division/septum is indicated by the arrows and inclusion bodies are marked by circles.
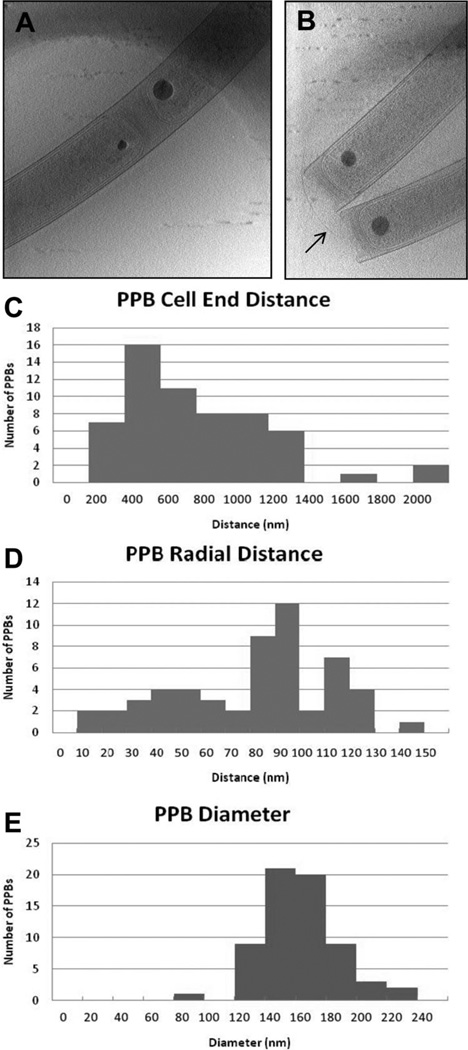
Fig. 6.
Location and size distributions of PPBs inside M. hungatei cells.
A and B. Central slices through 3D tomographic reconstructions of different M. hungatei cells. The slice shown in (A) contains the septum between two recently divided cells sharing a sheath. Note the size difference of the PPBs on the opposite sides of the septum. The slice shown in (B) contains many flagella (arrow) protruding from the ends of the cells.
C. Distribution of PPB distance from ends of cells.
D. Distribution of the PPB relative to the cylindrical axis of the cell.
E. Distribution of PPB diameters. These measurements are based on 65 PPBs in 3D cryoET reconstructions.
The most striking feature within the cytoplasm of the cell is an electron-dense inclusion body, which is similar to granules found in bacteria (Kornberg et al., 1999). To our best knowledge, this is the first time the high density inclusion bodies have been described in M. hungatei and no systematic analysis has been carried out previously. The granules varied from roughly spherical to an angular surfaced shape (Fig. 1A–C). Higher magnification images revealed that the bodies appeared to be made up of very small particles that have clustered into their characteristic spherical shape (Fig. 1D). No repeating or ordered arrangements were observed either visually as a crystalline pattern in the TEM image, or is obvious from the Fourier transform of the images of the inclusion bodies (Fig. 1D inset). Together, this evidence suggests that the bodies have an amorphous arrangement that would allow packing of smaller molecules into the structure (discussed below).
Elemental composition of inclusion bodies by EDX spectroscopy
In bacteria and other archaea, similar granules have been characterized utilizing both electron energy loss spectroscopy (EELS) and EDX spectroscopy in order to analyse the elemental composition of the inclusion bodies (Scherer and Bochem, 1983; Lechaire et al., 2002; Remonsellez et al., 2006). Here, we used an FEI Titan STEM instrument was employed to collect EDX spectra to analyse the elemental composition of the inclusion bodies within representative M. hungatei cells (Fig. 3). EDX spectroscopy was performed in the STEM microscope by utilizing the high-energy, focused electron probe to excite a specific region of the sample. The atoms being excited may eject electrons. If an inner shell electron is ejected, its position can be occupied by a higher energy electron from the outer shell, an event accompanied by energy release in the form of an emitted X-ray photon. The energy of these X-rays is characteristic of the specific atomic element from which they are emitted. Therefore, an energy-dispersive spectrometer can be used to measure the number and energy of the X-rays and give a quantitative description of the elemental composition of the specific region of the sample that is excited.
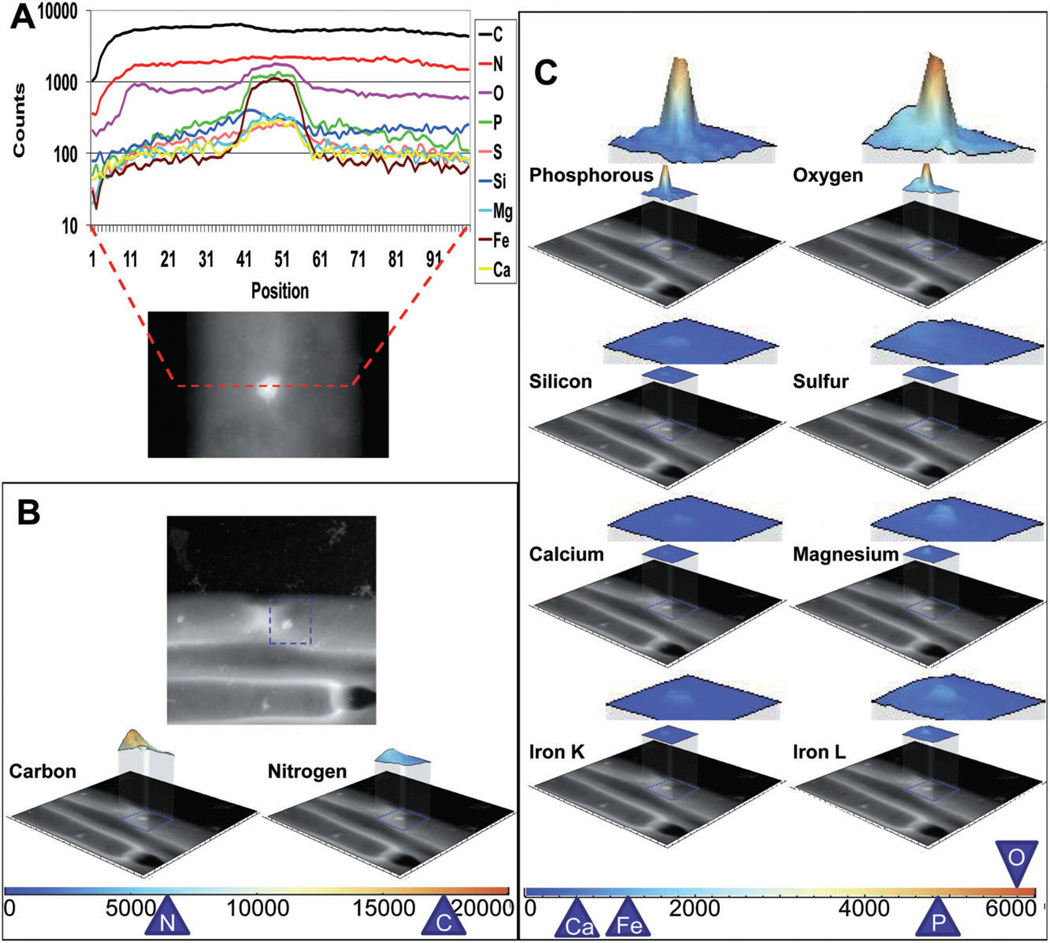
Fig. 3.
EDX analyses of the M. hungatei inclusion bodies. (A colour version of this figure is available online.)
A. A line spectrum across the width of a cell in the region with an inclusion body. The spectra of the most prevalent elements present in the cell are plotted using different colours (indicated in the box to the right).
B and C. Area spectra of the most prevalent elements identified through the EDX analyses. Top panel in (B): A STEM image of a region containing the ends of three cells. The blue box indicates the region analysed by EDX to produce an area spectrum (not shown). This area spectrum is then decomposed into individual ones corresponding to different elements. Those spectra with significant counts are displayed as three-dimensional colour maps with the colour-coding indicated below. Also shown below each colour spectrum is the STEM image to illustrate the location of the area scanned. Due to the use of carbon support film and the natural abundance of both carbon and nitrogen, the spectra for carbon and nitrogen have the highest counts and are shown in the bottom of (B) with a different scale. The spectra for the other prevalent element are shown in (C). Peak counts are also indicated on the colour scale bars at the bottom.
Using the EDX spectrum analysis tool integrated into FEI TIA software, we detected only the following elements in the sample: C, N, O, P, Fe, Mg, Ca, S and Si. The element line spectra across the M. hungatei bodies was obtained by scanning the electron probe along a line across the sample, and the spectra revealed a high abundance of phosphorous and oxygen within the granule region (Fig. 3A). Area scan spectra were also obtained by scanning specific areas of the cells that included one or more inclusion bodies, and the area scans also clearly established a high concentration of phosphorous and oxygen within the bodies (Fig. 3C). Among all the elements, carbon and nitrogen were found in highest abundance throughout the entire scanned region across the cells with a relative abundance of 3.5× and 1.3×, respectively, when compared with phosphorous, and a slight dip in carbon concentration was seen in the regions of the inclusion bodies (Fig. 3B), consistent with their being essential elements making up the cell envelope. After phosphorous and oxygen the next most prevalent elements, iron and magnesium, were found to be slightly concentrated (both about 0.2× phosphorous) within the granules relative to the cell cytoplasm. Lastly, calcium, silicon and sulfur were detected in small amounts within the granules at about 0.1× that of phosphorous.
The high concentration of phosphorous and oxygen within the inclusion bodies suggests that the granules are composed of concentrated phosphate, PO4−3. Previous studies have shown that phosphate concentrates into granules by polymerizing into long chains of poly-anionic phosphate, Cat3PO4(CatPO3)n, where Cat is a monovalent cation and n is the length of the polymer chain (Kulaev et al., 1999). The ratio of oxygen to phosphorous is approximately 5.2 (± 1.6) : 1 in the area of the granules. We reason that there are relatively more oxygen atoms than phosphorous atoms above and below the granules within the cell that also contributed to this ratio. Therefore, the actual oxygen to phosphorous ratio within the granules might be closer to 3:1, depending on the length of the phosphate chains. This high concentration of polyphosphate excludes proteins and other organic carbon, thus explaining the slight dip observed in the carbon count spectra across the area of the granules (Fig. 3A). The presence of low to intermediate concentrations of iron, magnesium, and calcium also suggests that the bodies are storing these elements as counter ions, and/or that the bodies are bound with enzymes or other structures with a higher concentration of these elements. It has also been previously shown that divalent cations, such as magnesium and calcium, can form ionic bonds between two separate phosphate groups. This cross-linking activity has been proven to allow phosphate chains to pack more tightly and thus would increase the density of the granule (Parsons et al., 2010). There was little apparent variation in packing or general elemental distribution within granules. This analysis allows us to conclude that the observed inclusion bodies are in fact polyphosphate bodies (PPBs).
Mass distribution of PPB by STEM tomography
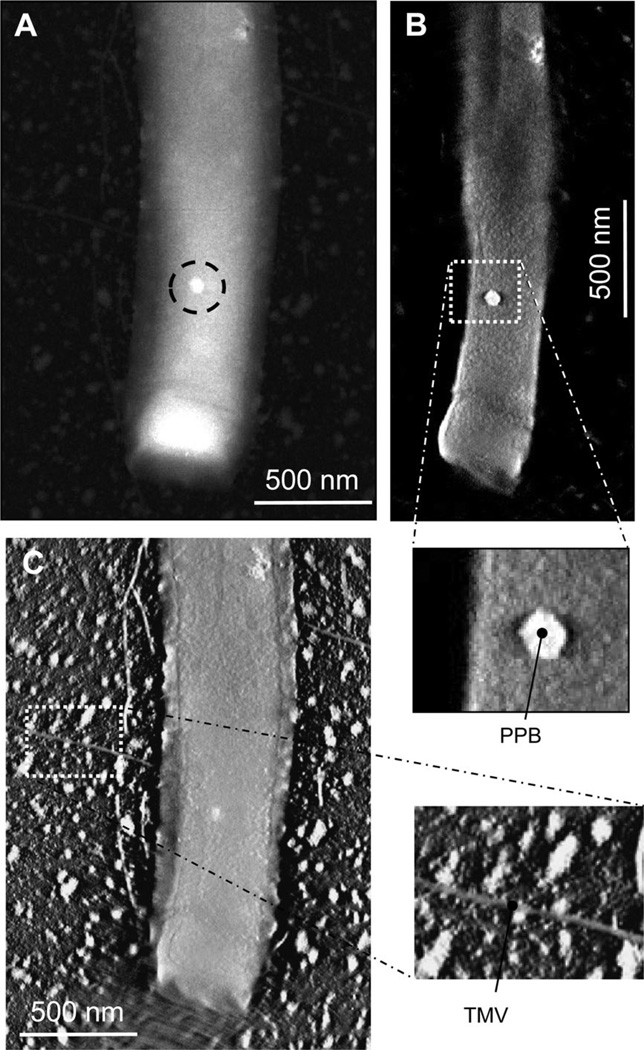
While cryoEM gives faithful representation of structural organization, it does not give accurate representation of mass distribution due to the phase contrast and noisy background perturbation intrinsic to the technique. One way to get a quantization of the mass distribution is to utilize STEM imaging, which allows the precise correlation of intensity to mass (Engel, 1978). In STEM imaging, an electron probe raster-scans over a square area of the sample grid divided into n2 elements or pixels, and at each pixel the electrons from the probe are scattered by the sample. An annular dark-field detector system capable of counting single electrons is used to collect most of the elastically scattered electrons. The resulting dark-field image can be used to directly measure the mass of the sample scanned by the electron probe, because, in unstained samples and in the absence of multiple scattering events, this annular detector signal is directly proportional to the number of atoms, weighted by atomic number (Z), irradiated by the electron probe. By utilizing theoretical data on elastic scattering it is possible to directly measure the mass, since the detector signal can be correlated directly to the number of electrons by dividing by the elastic scattering cross sections, the collection efficiency and the dose, determined by probe current, dwelling time, and pixel area (Muller and Engel, 1998). Alternatively, the mass can be estimated by comparing the signal of the sample to the signal of a mass standard, and in our case we utilized tobacco mosaic virus (TMV) particles with a known mass-per-unit-length (131.4 kDa nm−1).
We collected two STEM tomography tilt series of M. hungatei cells mixed with TMV particles and calculated the mass density of PPB using the known TMV mass as a reference (Fig. 4). Having the 3D reconstructions allowed us to separate PPB and TMV from the background. From the 3D STEM tomography reconstruction, we estimated that the average mass density of the PPB is 3.8 g cm−3, which is about four times higher than that of the surrounding cytoplasm. This value is larger than the density of most non-metal materials in their solid phases and is between densities of barium (3.51 g cm−3) and titanium (4.54 g cm−3).
Fig. 4.
Mass measurement of PPB inside M. hungatei cells by STEM tomography. Tobacco mosaic virus (TMV) is included to serve as a standard for mass measurements.
A. STEM image of the end of a cell containing a PPB (circled).
B and C. Slices extracted from the 3D STEM tomogram reconstructed from the tilt series of the cell shown in (A). Slice in (B) shows the PPB and that in (C) shows the TMV particles.
3D localization of PPBs inside cells
We also systematically examined the distribution of the M. hungatei PPBs within the cells by taking images of entire cells frozen in vitreous ice at lower magnification to establish locations and numbers of PPBs per cell. Two PPBs are present in every cell and within each field examined (Fig. 1A–C and Fig. 2), typically one near each of the two poles of every cell.
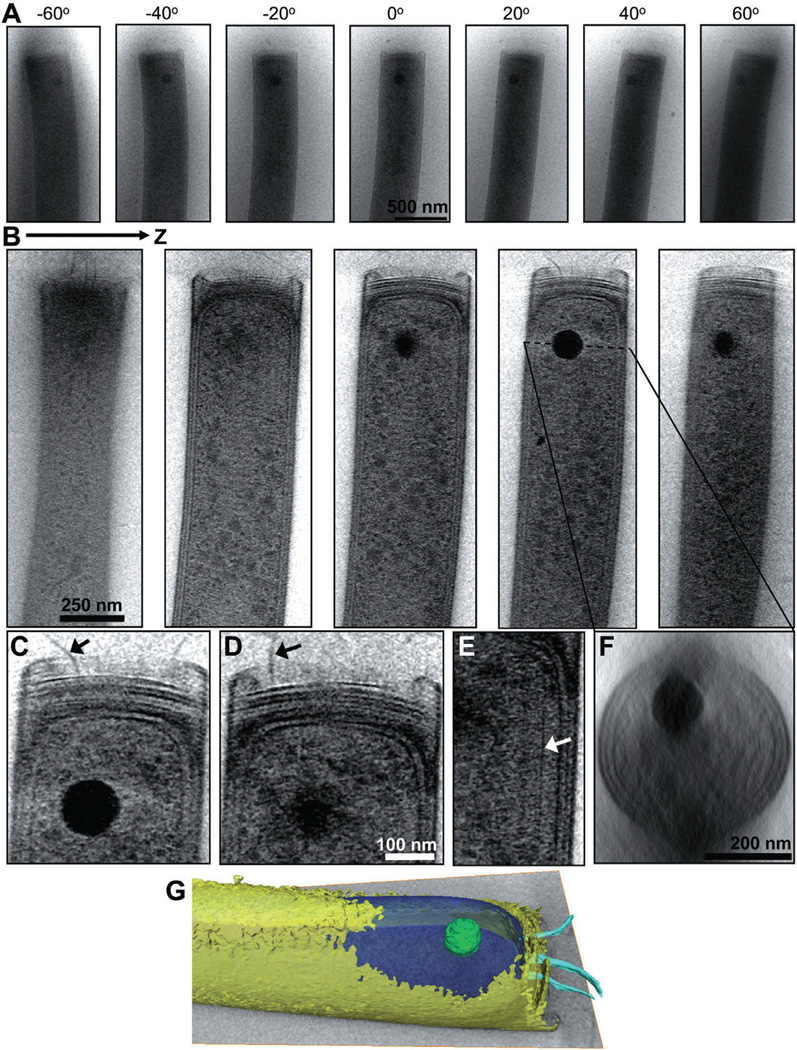
To precisely locate the PPB inside the M. hungatei cells, we employed cryoET to examine the methanogen in 3D. The 3D reconstructions of individual cells allowed us to examine the precise longitudinal and radial positions of each PPB within cell lumen (Fig. 5). Among the 65 cells analysed, we found that the average longitudinal distance of the PPB is 612 ± 52 nm from the cell end, measured from the plug (Fig. 6C). Perpendicular crosssectional views of the reconstructions through the centre of the PPB (Fig. 5F) also reveal the precise distance from the centre of the PPB to the radial centre of each cell. The PPBs are off-centred, on average, 74 ± 4 nm from the cylindrical axis of the cell (Fig. 6D). Interestingly, the offcentre distance is about the same as the radius of the PPB (described below) (i.e. 145/2 = 72.5 nm).
Fig. 5.
Three-dimensional localization of PPB inside M. hungatei cells by cryoET.
A. An example cryoET tilt series (± 70° tilts) of the end of an M. hungatei cell. Only seven of 71 images in the tilt series are shown for illustration and their tilt angles are indicated.
B. Density slices extracted from the 3D tomogram reconstructed from the tilt series shown in (A).
C-E. Higher magnification slices from the reconstruction show flagella and cytoskeletal structures, indicated by arrows.
F. A perpendicular cross-section through the 3D reconstruction showing the location of the PPB within the cell lumen.
G. Volume rendering of the tomogram with the PPB shown in green, the cytoplasm in blue, envelope in yellow and flagella in cyan. The cytoplasm and envelope are displayed semi-transparently so that the PPB volume is visible. Shown in greyscale is a slice from the 3D tomographic reconstruction. (A colour version of this figure is available online.)
From the 3D tomograms, we then measured the dimensions of the PPBs (Fig. 6E). The 65 bodies measured are 145 ± 3 nm in diameter, which reflects a fairly consistent size in fully mature cells (e.g. in stationary phase cells), but in actively growing and dividing cells the bodies are found in a range of diameters up to 200 nm. The approximate diameter of 150 nm would give a PPB volume of 1.8×106nm3 per body [i.e. V= (4/3)*π* 753]. For each PPB, this would be equivalent to approximately 0.2% of the total cell volume assuming cylindrical cell dimensions of 450 nm diameter by 5 µm length (cell volume V = 0.80 µm3). Calculating based on the volume of the body (1.8 × 106 nm3), the density of the body (3.8 g cm−3), and the molecular weight of each PO3 group of polyphosphate (79 g mol −1), we estimate that the maximum amount of phosphate that could be present in one PPB is about 8.7 × 10−5 pmol. If monovalent/divalent cations were to comprise 25% of the PPB, the value would be reduced to 6.5 × 10−5 pmol.
Distribution of PPBs in different stages of cell growth
In stationary phase cells, in which the cells have a more regular length and are not growing or dividing, the PPBs are always found near the ends of each cell and never intermediate along the length. Also, the PPBs in stationary phase cells are usually fully developed, with a mostly uniform size and shape in contrast to the smaller granules observed in newly dividing cells. Cell growth and division during exponential phase requires significant phosphate utilization for biosynthesis, which may limit phosphate accumulation and incorporation into the PPBs, and thus contribute to PPB size variability. In early stationary phase cells, the phosphate demand is reduced, allowing equilibration of the phosphate body development.
In exponential growth phase, as cells lengthen prior to cell division, one or two additional PPBs are often observed in the middle of the cell, and at roughly equal distance from the opposite end (Fig. 2). These PPBs appear to be in the formation process of with the smaller body at the mid-cell location in contrast to a larger body at the other end of the cell that has previously undergone cell division. This suggests that, during cell growth, two new PPBs are initiated and develop in the middle of the cell in preparation for cell division. When multiple M. hungatei cells share a common sheath with septum regions between adjacent cells (Fig. 2, arrows), the PPBs are present at both ends of adjacent cells (i.e. on both sides of the septum spacer regions). It is not known what guides PPB development during cell division but our observations of multiple cells suggest that, after sufficient cell elongation occurs, and before a new cell end forms, a new PPB initiates close by the pole adjacent to a newly forming septum. PPB migration from other cell locations was not evident. The newly formed PPB then gradually enlarges into a fully developed one as the cells further extend within the sheath and then divide. A similar and irregularly shaped small body also appears on the opposite side of the newly forming septum (Fig. 2, right inset indicated with arrows). These two small growing bodies, which would presumably be daughter bodies found at the new cell ends, appear to mature at similar rates. In some instances, one body is considerably larger (Fig. 6A) and it is unknown if this represents an existing body from an old cell, or if a development plan and/or cell metabolism allows more rapid PPB synthesis at this end.
Discussion
One of the first observations about the PPBs discovered in the frozen hydrated M. hungatei cells was their localization at the ends of each cell (Fig. 1). After further study, the PPB number, location and size revealed a relationship between the number of bodies and the length of the cells: as the cells grow, additional bodies develop along their length (Fig. 2). This may indicate a function for the bodies throughout the length of the cell, or they may be developing in preparation for a new division event after additional growth has occurred. The presence of PPBs in close proximity to the ends of nearly every cell indicates there may be some function required at the cell end. It is perhaps used as a phosphate-based energy source, for example, in flagellar/motor biogenesis or function, as the archaeal flagella protrude from the nearby cell ends and many can be seen in images of cells in motile phase (Fig. 6B). However, the fact that PPBs are also positioned at the ends of cells in the middle of chains of multiple cells within a single sheath would suggest other roles that are not tightly coupled to flagellar function, since the internal cells do not produce flagella.
PPBs in stationary, older cells are larger and more uniformly shaped than in pre-division or recently divided cells (e.g. Fig. 2). This indicates that as additional phosphate is taken up by the cell, PPBs grow as excess phosphate is incorporated. In a phosphate limited environment, it would be presumed that PPBs would fail to develop although this has not been tested experimentally. In addition, despite their amorphous composition, PPBs tend to round up into spherically shaped particles. This is most likely due to the need to minimize the surface energy of the PPB by achieving the maximum compaction and lowest surface area to volume ratio. It is known that short polyphosphate chains [e.g. two (pyrophosphate) or three (tripolyphosphate) phosphates] form crystalline structures naturally (Griffith and Grayson, 1977; Pritzker, 1998). The amorphous packing of PPBs inside M. hungatei observed in this study suggests that they contain longer phosphate chains. In other organisms these are reported to range from several hundred to over one thousand repeating units in length.
Is PPB development in M. hungatei programmed?
As noted above, PPBs in mature M. hungatei cells achieve a uniform size of about 150 nm in diameter and are positioned near cell ends (Fig. 1) but off-centre from the longitudinal cell axis. These PPBs are located about one radius in distance from the centre of the cell (Fig. 6D), and this would mean that the edge of each PPB is usually located in close proximity to the centre line of the cell (Fig. 5F). Therefore, if there are cytoskeletal structures that run along the length of the cells down their centres, the PPBs would be positioned to interact with such structures. It has yet to be observed, in M. hungatei, if there are any such intracellular structures contacting the PPB.
In contrast to the wealth of knowledge in bacterial systems (Rao et al., 2009), little is known about events of cell positioning and replication in the archaea. The ability to uniformly position PPBs within each cell in concert with the observed pattern for development of new bodies indicates that M. hungatei possesses a genetic program to spatially and temporally direct these events. This process would appear to be coupled to cell growth and division within the sheath since random cell locations were not observed. Whether polyphosphate is somehow directly involved in and essential for these events, or alternatively, is a by-product of them, is unknown.
Polyphosphate in other organisms
Related PPBs in bacteria and archaea have been reported to sequester heavy metals and other elements. In the archaeon, Sulfolobus metallicus, it was observed that PPBs decreased in number and size upon exposure to increasing copper concentration (Remonsellez et al., 2006). They concluded that the phosphate was utilized in metal ion detoxification by binding up the copper ions prior to being transported out of the cells. EDX and electron energy loss spectroscopy were used to verify the sequestration of these elements within the bodies of cells grown in an environment containing heavy metals. Considerably lower levels of polyphosphate were detected in Sulfolobus acidocaldarius and Sulfolobus solataricus cells and this was not altered by copper ion exposure (Remonsellez et al., 2006).
Few other archaeal species are reported to contain PPBs. Methanosarcina vacuolata as well as Methanosarcina barkeri strains Fusaro, 227, and Wiesmoor each contained a single round to oval shaped PPB-like granule of 0.15–0.30 µm in diameter with associated Ca and Fe ions (Scherer and Bochem, 1983). Presence of the bodies was variable depending on the nutritional conditions used, and they gradually disappeared as cells aged or starved. In contrast, addition of FeS appeared to stimulate body density which the authors suggested may be due to accumulation of polymeric iron phosphate. Interestingly, several of the above mentioned strains also contained up to a hundred very small irregular shaped inclusion bodies called ‘granula’ that were likely to be composed of polyphosphate based on their EDX spectra (Scherer and Bochem, 1983). The relationship of cyclic 2,3-diphosphoglycerate synthesis and ‘granula’ accumulation to 14% of the Methanosarcina mazei (formerly Methanosarcina frisia) cell dry weight is unknown, although the cellular phosphate accumulation was dependent on phosphate availability and the methanogenic substrate used (Rudnick et al., 1990). No related ‘granula’ bodies were observed in M. hungatei cells (this study).
Polyphosphate has been shown to be an important compound in the metabolic and regulatory pathways of many organisms (Kulaev and Kulakovskaya, 2000; Rao et al., 2009). It has very diverse roles in bacterial and eukaryotic cells, including phosphate reserves, cation sequestration and storage, cell envelope formation and function, modulating gene expression, regulation of enzyme activity, stress response and stationary phase adaptation, membrane channel function, etc. In E. coli the enzyme PPK (polyphosphate kinase) has been shown to be an important regulator of PPB biosynthesis (Ahn and Kornberg, 1990). The analogues for PPK in archaea species that exhibit PPB formation have not been reported, but a similar enzymatically regulated process is thought to occur. Inspection of the M. hungatei genome reveals two ppk-like genes, Mhun_0889 and Mhun_1431 that are candidates for this function (Ca. ~ 8.6 e-108 relative to E. coli b2501).
Polyphosphates were found to play a key role in regulatory control, in E. coli, during the transition from growth phase to stationary phase and to the survival of the cell in the stationary phase or under stress (Kornberg et al., 1999). Many archaea, including M. hungatei, exist in harsh environments that would be uninhabitable for many other organisms, therefore mechanisms to cope with environmental stress including phosphate limitation are expected. In this regard, the genome reveals both a high affinity ABC-type phosphate uptake system as well as a Na+ type low affinity phosphate symport system. The PPBs could certainly function to help M. hungatei handle other environmental stresses, such as metal toxicity as shown in Sulfolobus species (Remonsellez et al., 2006), or perhaps in aiding accumulation of iron as seen in Methanosarcina barkeri (Scherer and Bochem, 1983).
It was recently shown by cryo electron microscopy and tomography of carboxysomes in several bacterial cell types (i.e. in Halothiobacillus neapolitanus, Thiomonas intermedia and Thiomicrospira crunogena) (Iancu et al., 2010) that PPB-like granules were often associated with these shell structures. Although little is known about the properties or associated functions of these putative PPBlike granules, it was suggested that they may somehow provide cations needed for carboxysome synthesis and/or functioning. By analogy, M. hungatei might employ PPBs to sequester divalent cations (e.g. Fe) needed in great abundance for methane biosynthesis.
Other roles for PPBs are possible. For example, when methanogenic substrates (i.e. hydrogen or formate) are depleted in the cell’s immediate environment, cellular energy in the form of ion gradients and ATP becomes limited. Under these conditions, polyphosphate-based energy reserves in M. hungatei could then be employed to fuel essential cell functions, by re-synthesizing ATP using the anhydride bond energy stored in PPBs, and/or to restore electrochemical ion gradients. The latter would fuel motility and taxis to allow cells to find new substrate(s) and adapt accordingly. Based on the PPB volume and density values reported above, we have estimated that each granule contains about 6.5 × 10−5 pmol of phospho-anhydride repeating units. Were this stored bond energy in both PPB granules to be freely converted into ATP, it would be equivalent to 13 × 10−5 pmol of ATP or about 200 mMATP given the cell volume V = 0.80 µm3. Thus, the M. hungatei PPB appears to be a remarkably compact energy storage device! The two PPBs per cell require only ~ 0.4% of the cell volume and stores ~ 100-fold more energy than is contained in the cellular ATP pool within the other 99.6% of the cytoplasm (i.e. assuming an intracellular ATP concentration of 2 mM). These estimates of PPB density and energy storage in M. hungatei have significant implications for PPB in other archaea and bacteria. Regardless of PPB function(s), M. hungatei appears to be exquisitely adapted for life in low hydrogen environments.
Experimental procedures
Cell culture
Methanosprillum hungatei strain JF1 (DSM 864) was cultivated on a CO2/bicarbonate buffered mineral medium supplemented with trace metals, vitamins and sodium acetate with a vessel headspace containing a 80–20% atmosphere of hydrogen and carbon dioxide. The medium contained per 1000 ml: 0.54 g NaCl, 0.12 g MgSO4·7H2O, 5.0 g NH4Cl, 1.8 g KH2PO4, 2.9 g K2HPO4, 0.06 g CaCl2·2H2O, 2.72 g Na Acetate·3H2O, 10 ml of 100× trace metal solution and 1 ml of 1000× vitamin solution. Following sterilization, the medium was supplemented with 10 ml filter-sterilized solution of reducing reagent (2.5% Na2S·9H2O, 2.5% Cysteine HCl) and 20 ml of a 1 M NaHCO3 solution. Trace metals and vitamins were as previously described for M. acetivorans (Rohlin and Gunsalus, 2010). Following inoculation, anaerobe tubes containing 10 ml of medium were incubated at 37°C horizontally on a rotary drum shaker (60 RPM, New Brunswick). Cells were serially transferred at least three times with transfers made at mid-exponential phase to achieve 10 plus cell doublings prior to harvest and electron microcopy analysis.
Cryo electron microscopy
To optimize samples for cryoEM observation, we first performed negative stained electron microscopy of cells prepared on carbon-coated copper grids and stained with 1% uranyl acetate. To adjust the sample concentration, cells were centrifuged at low speed in a table-top microfuge, and re-suspended at the appropriate density using culture medium.
A droplet of 4 µl of the sample with optimized concentration was placed onto a glow discharged, 200 mesh Quantifoil grid (Ermantraut et al., 1998) with a spacing of 3.5 mm hole/1 µm edge. The sample was allowed to settle for 30 seconds, blotted by filter paper, and immediately plunged into liquid nitrogen cooled liquid ethane.
An FEI Tecnai F20 transmission cryo electron microscope with an accelerating voltage of 200 kV was used to image the M. hungatei cells imbedded in vitreous ice. Samples were imaged at a dosage of 33 and 6 electrons/Å2, for those shown in Figs 1 and 2, respectively. The targeted underfocus values were 2.5 µm.
Cryo electron tomography
For cryoET, we have used two cryo electron microscopes. For PPB location and size distribution, we used the abovementioned Tecnai F20 instrument with a 16 megapixel CCD camera. For the cryoET reconstructions shown in Fig. 5, we used an FEI Titan Krios instrument with a Gatan image filter (GIF) and a 4 mega pixel CCD camera, with the instrument operated at an accelerating voltage of 300 kV and a total dose of 100 electrons/−Å2 for each tilt series. During tilt series acquisition, we chose cells whose long axis was roughly parallel to the tilt axis of the sample holder. The cells were imaged at 29 000×. An underfocus value of 3–6 µm at zero degrees tilt was used. All tomography tilt series were recorded using the FEI Batch Tomography software with a tilt range from −70° to +70°.
STEM tomography and EDX analysis
We performed the STEM imaging/tomography and EDX analysis using an FEI Titan 80–300 kV scanning transmission electron microscope. Methanospirillum hungatei cells were placed on carbon coated copper grids, air-dried and imaged inside the Titan instrument.
For mass measurement using STEM tomography, we mixed TMV particles with the M. hungatei cells before placing the sample on grids. Samples were loaded on a Fischione 2020 tomography holder and FEI batch tomography software was used to collect STEM tomography tilt series at 300 kV accelerating voltage.
FEI TEM imaging and analysis (TIA) software package was used to acquire the line scan and area scan EDX spectra of different areas of the sample at 80 kV at a dosage of about 50 electrons/−Å2. The spectral data for the indicated elements were stored as counts over distance or area. The area spectra were output in 3D graphical format using Mathematica (Wolfram Research).
Electron tomography data analysis and 3D visualization
The tomography tilt series were processed with a suite of programs to generate a 3D reconstruction. Inspect3D (FEI Company) was used to quickly achieve an initial rough alignment (translation, rotation and tilt axis) of the tilt series using the graphical interface of the software. The image stacks were roughly aligned using cross-correlation with proper filtering. The high- and low-pass limits for the filtering were optimized so that a clearly distinguishable cross-correlation peak was obtained. After this alignment, the tilt axis was adjusted so that it was vertical and centred. This process was repeated 3–4 times until there were no further changes larger than one pixel in the x and y translational coordinates.
After the initial rough alignment, gold-bead tracking in Inspect3D was used to align the tilt series more precisely. Gold bead tracking started with finding several representative beads at zero degrees tilt to create a representative model, this model was used to find many other beads, bad beads were discarded, and the remainder were tracked throughout the whole tilt series. Errors in bead tracking were corrected and an alignment solution was calculated based on the bead positions. At this point the bead positions could be iteratively error adjusted to find a converged solution with the least amount of error. Once the converged alignment was found it was applied to the tilt series.
The aligned tilt series was then used to make a 3D reconstruction using GPU-based SIRT (Simultaneous Iterative Reconstruction Technique) reconstruction implemented in Inspect3D. The 3D reconstructions were saved as a stack of x–y plane images that are single pixel slices along the z-plane.
We also used the etomo tomography processing software from the Imod package (Kremer et al., 1996) to perform very similar alignment and reconstruction steps as those described above, including rough alignment by crosscorrelation, fine alignment by fiducial (gold-bead) tracking, and reconstruction by weighted back-projection. We further processed the reconstructions by applying a median filter to enhance the contrast.
Slices from the reconstructions were displayed using slicer within 3dmod from the Imod package. Amira (Visage Imaging GmbH, http://www.amira.com/) was used to create volume renderings of the 3D density maps of the cells.
Acknowledgements
We thank Feng Shen for technical assistance in STEM tomography, Matthew Mecklenburg for assistance in EDX data collection and presentation, and Bridget Carragher and Ruben Diaz-Avalos for the TMV sample used as a mass standard. This research was supported in part by the Department of Energy Biosciences Division grant award DE-FG03-86ER13498 and the UCLA-DOE Institute of Genomics and Proteomics to RPG, the Netherlands Organisation for Scientific Research (NWO) to AMH, and the National Institutes of Health (NIH; GM071940 and AI069015 to ZHZ). We acknowledge the use of electron microscopy facilities at the UCLA Electron Imaging Center for NanoMachines in California NanoSystems Institute (CNSI) supported by NIH (1S10RR23057 to ZHZ).
References
- Ahn K, Kornberg A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- Beveridge TJ, Stewart M, Doyle RJ, Sprott GD. Unusual stability of the Methanospirillum hungatei sheath. J Bacteriol. 1985;162:728–737. doi: 10.1128/jb.162.2.728-737.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ, Southam G, Jericho MH, Blackford BL. High-resolution topography of the S-layer sheath of the archaebacterium Methanospirillum hungatei provided by scanning tunneling microscopy. J Bacteriol. 1990;172:6589–6595. doi: 10.1128/jb.172.11.6589-6595.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A. Molecular weight determination by scanning transmission electron microscopy. Ultramicroscopy. 1978;3:273–281. doi: 10.1016/s0304-3991(78)80037-0. [DOI] [PubMed] [Google Scholar]
- Ermantraut E, Wohlfart K, Tichelaar W. Perforated support foils with pre-defined hole size shape and arrangement. Ultramicroscopy. 1998;74:75–81. [Google Scholar]
- Ferry JG, Wolfe RS. Anaerobic degradation of benzoate to methane by a microbial consortium. Arch Microbiol. 1976;107:33–40. doi: 10.1007/BF00427864. [DOI] [PubMed] [Google Scholar]
- Ferry JG, Wolfe RS. Nutritional and biochemical characterization of Methanospirillum hungatii . Appl Environ Microbiol. 1977;34:371–376. doi: 10.1128/aem.34.4.371-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry JG, Smith PH, Wolfe RS. Methanospirrillum, a new genus of methanogenic bacteria. Int J Syst Bacteriol. 1974;24:465–469. [Google Scholar]
- Firtel M, Southam G, Harauz G, Beveridge TJ. Characterization of the cell wall of the sheathed methanogen Methanospirillum hungatei GP1 as an S layer. J Bacteriol. 1993;175:7550–7560. doi: 10.1128/jb.175.23.7550-7560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith EJ, Grayson M. Topics in Phosphorous Chemistry. Ann Arbor, MI, USA: J. Wiley and Sons Interscience; 1977. [Google Scholar]
- Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol. 2010;396:105–117. doi: 10.1016/j.jmb.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kulaev I, Kulakovskaya T. Polyphosphate and phosphate pump. Annu Rev Microbiol. 2000;54:709–734. doi: 10.1146/annurev.micro.54.1.709. [DOI] [PubMed] [Google Scholar]
- Kulaev I, Vagabov V, Kulakovskaya T. New aspects of inorganic polyphosphate metabolism and function. J Biosci Bioeng. 1999;88:111–129. doi: 10.1016/s1389-1723(99)80189-3. [DOI] [PubMed] [Google Scholar]
- Lechaire JP, Shillito B, Frebourg G, Gaill F. Elemental characterization of microorganism granules by EFTEM in the tube wall of a deep-sea vent invertebrate. Biol Cell. 2002;94:243–249. doi: 10.1016/s0248-4900(02)01199-1. [DOI] [PubMed] [Google Scholar]
- McInerney MJ, Bryant MP, Hespell RB, Costerton JW. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl Environ Microbiol. 1981;41:1029–1039. doi: 10.1128/aem.41.4.1029-1039.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJ, Schink B, et al. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann NY Acad Sci. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- Mountfort DO, Bryant MP. Isolation and characterization of an anaerobic syntrophic benzoatedegrading bacterium from sewage sludge. Arch Microbiol. 1982;133:249–256. [Google Scholar]
- Muller SA, Engel A. Mass measurement in the scanning transmission electron microscope: a powerful tool for studying membrane proteins. J Struct Biol. 1998;121:219–230. doi: 10.1006/jsbi.1997.3953. [DOI] [PubMed] [Google Scholar]
- Parsons AJ, Ahmed I, Rudd CD, Cuello GJ, Pellegrini E, Richard D, Johnson MR. Neutron scattering and ab initio molecular dynamics study of crosslinking in biomedical phosphate glasses. J Phys Condens Matter. 2010;22:1–8. doi: 10.1088/0953-8984/22/48/485403. [DOI] [PubMed] [Google Scholar]
- Patel GB, Roth LA, van den Berg L, Clark DS. Characterization of a strain of Methanospirillum hungatti . Can J Microbiol. 1976;22:1404–1410. doi: 10.1139/m76-208. [DOI] [PubMed] [Google Scholar]
- Pritzker KPH. Calcium pyrophosphate crystal formation and dissolution. In: Amjad Z, editor. Calcium Phosphates in Biological and Industrial Systems. Norwell, MA, USA: Kluwer Academic Publishers; 1998. pp. 277–299. [Google Scholar]
- Rao NN, Gomez-Garcia MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- Remonsellez F, Orell A, Jerez CA. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology. 2006;152:59–66. doi: 10.1099/mic.0.28241-0. [DOI] [PubMed] [Google Scholar]
- Rohlin L, Gunsalus RP. Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol. 2010;10:62. doi: 10.1186/1471-2180-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick H, Hendrich S, Ulrich P, Biotevogel K-H. Phosphate accumulation and the occurrence of polyphosphates and cyclic 2,3-diphosphoglycerate in Methanocarcina frisia . Arch Microbiol. 1990;154:584–588. [Google Scholar]
- Scherer PA, Bochem H-P. Ultrastructural investigation of 12 Methanosarcinae and related species grown on methanol for occurence of polyphosphatelike inclusions. Can J Microbiol. 1983;29:1190–1199. [Google Scholar]
- Southam G, Beveridge TJ. Detection of growth sites in and protomer pools for the sheath of Methanospirillum hungatei GP1 by use of constituent organosulfur and immunogold labeling. J Bacteriol. 1992;174:6460–6470. doi: 10.1128/jb.174.20.6460-6470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieb M, Schink B. Anaerobic oxidation of fatty acids by Clostridium bryantii sp. nov., a sporeforming, obligately syntrophic bacterium. Arch Microbiol. 1985;140:387–390. [Google Scholar]