Abstract
Axon regeneration is a medically relevant process that can repair damaged neurons. This review describes current progress in understanding axon regeneration in the model organism Caenorhabditis elegans. Factors that regulate axon regeneration in C. elegans have broadly similar roles in vertebrate neurons. This means that using C. elegans as a tool to leverage discovery is a legitimate strategy for identifying conserved mechanisms of axon regeneration.
Keywords: axon, regeneration, Caenorhabditis elegans
INTRODUCTION
Caenorhabditis elegans is a comparatively new model for the study of axon regeneration. Here, we review progress in the field since its beginning in 2004 and discuss directions that future investigations might follow.
The Unrealized Potential of Axon Regeneration
Damaged axons can in some cases regenerate, restoring function to nervous systems after injury or disease. Regeneration of injured neurons is thought to be initiated by signals arising from the injury site (20, 66, 74). These injury signals promote remodeling of the cytoskeleton and plasma membrane at the site of injury, resulting in the generation of a new growth cone. Regulated changes in gene transcription and local and somatic protein synthesis are also required for successful axon extension during regeneration.
The response of a specific neuron to injury is the result of the interplay between various pathways that promote or suppress regeneration. In part, regeneration is regulated by signals from the neuronal environment. For example, myelin-derived signals [Nogo, MAG (myelin-associated glycoprotein), and OMgp (oligodendrocyte myelin glycoprotein)] and chondroitin-sulfate proteoglycans (CSPGs) inhibit regeneration in the vertebrate central nervous system (CNS) (97). However, regeneration is also regulated by intrinsic factors, and altering the intrinsic state of a neuron can improve regeneration, even in the presence of inhibitory factors (58, 59, 65). Thus, building a complete model of the mechanisms that regulate and mediate regeneration is critical to understanding how regeneration works and why it often fails.
Caenorhabditis elegans as a Regeneration Model
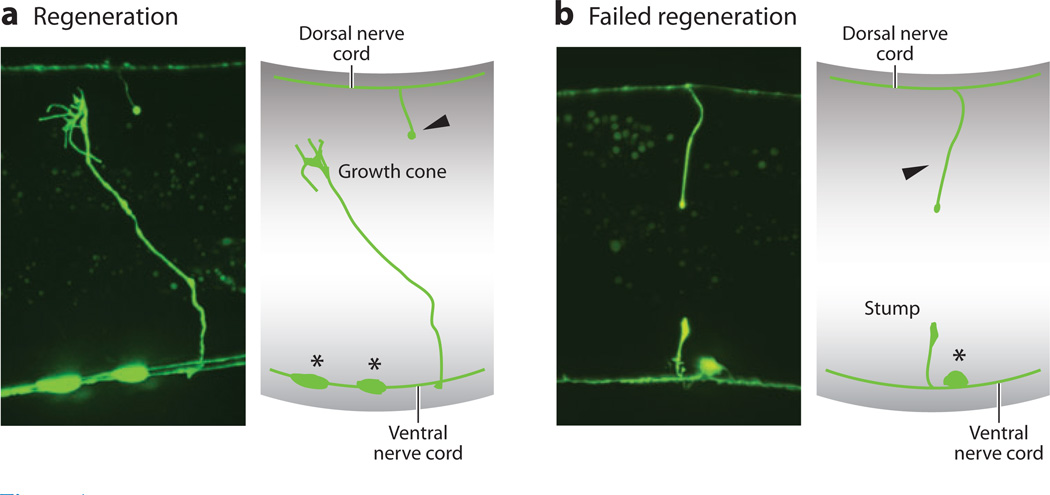
The nematode worm C. elegans has recently emerged as a model for axon regeneration. Axons can be severed through the transparent skin of the worm using a pulsed laser, triggering regeneration (96). Regeneration can then be studied in vivo at single-axon resolution (Figure 1). When this technique is combined with the powerful genetics and other tools available in C. elegans, it becomes possible to investigate the complex genetics and cell biology of axon regeneration more rapidly and in greater molecular detail than previously possible (27–29, 36, 57, 92, 95). An important limitation of C. elegans for these studies is the relative simplicity of the extrinsic neuronal environment. Worms have no myelin and no invading macrophages. Thus, the worm is a poor model for certain questions regarding the interaction between an injured neuron and its environment. In many cases, however, the simple environment surrounding C. elegans neurons can accelerate discovery by removing confounding factors.
Figure 1.
(a) A regenerating Caenorhabditis elegans GABA (γ-aminobutyric acid) neuron after laser surgery, and (b) a neuron that has failed to respond to injury (21).
An essential question of any biological model is its relevance to human biology. A precise comparison between the mechanisms of axon regeneration in C. elegans and those in humans (or at least vertebrate models) is not yet possible because these mechanisms are incompletely understood. However, results so far are encouraging: (a) In C. elegans, as in vertebrates, cAMP (cyclic adenosine monophosphate) acts to promote axon regeneration after injury (8, 28, 58); (b) the DLK-1 mitogen-activated protein kinase kinase kinase (MAPKKK) promotes regeneration in both C. elegans and mice (36, 43, 95); and (c) axon guidance molecules are important for regeneration in worms and mice (27, 37, 92). Thus, many factors that regulate axon regeneration in C. elegans have broadly similar roles in vertebrate neurons. In this review, we describe how C. elegans is used to study regeneration and summarize recent work that has begun to describe the mechanisms that regulate and execute regeneration.
INJURY MODELS IN CAENORHABDITIS ELEGANS
A major challenge in studying regeneration is that regeneration only occurs only in response to nerve injury. In mammals, injury is delivered by surgically cutting or crushing a nerve or a section of the spinal cord. These techniques are not easily applied to C. elegans because of the small size of the worm. Thus, the study of regeneration in C. elegans has required development of two novel injury models: laser axotomy and the β-spectrin mutant background.
Laser Axotomy
In 2004, it was found that laser pulses can sever GABA (γ-aminobutyric acid) neurons in C. elegans in intact, living animals and that regeneration can be observed after surgery (96). The first laser used to cut axons in C. elegans was a sophisticated, amplified Ti-sapphire laser (96). It is now clear that many varieties of pulsed laser can cut axons and elicit regeneration, including unamplified Ti-sapphire lasers (92), coumarin dye lasers (92), Nd:YAG (neodymium-doped yttrium aluminum garnet) lasers (68), and solidstate lasers (89). In addition to the GABA motor neurons, laser axotomy has been used to sever many other neurons, including the PHA phasmid sensory neurons (92), the PLM and ALM touch neurons (92), the sensory neuron AFD (18), the AWB chemosensory neuron (92), the AVM mechanosensory neuron (27), the DA/DB motor neurons (27), and the HSN neuron (27). There are no examples of neurons that cannot be cut, suggesting that laser axotomy can be used on any neuron in the worm.
Pulsed lasers sever axons by generating free electrons, by causing the formation (and collapse) of nanoscale bubbles, and by generating free plasma that results in a cavitation bubble (10). The precision of laser axotomy enables very accurate and controlled injuries at the single-neuron level. For example, Wu and colleagues (92) found that the success of regeneration in the ALM and PLM neurons depends on how close to the cell body the injury occurs: Injury close to the cell body results in increased regeneration. Similarly, Gabel and colleagues (27) showed that in the DA/DB motor neurons cutting closer than 20 µm to the cell body elicits growth from the cell body rather than the axon stump. A potential limitation of laser axotomy is the requirement to individually target and sever each individual axon that is to be studied. However, diligent application of this technique has yielded nearly all current data, including one study that analyzed hundreds of genotypes (15). One way to increase the speed of laser axotomy is by using a microfluidic platform to immobilize worms, combined with an automated axotomy system (3, 32, 73). A microfluidic and automated axotomy system has been used to screen for chemical modifiers of regeneration (75). Such systems hold the promise of significantly increasing experimental throughput.
β-Spectrin Mutants
β-spectrin is a major and essential component of the membrane skeleton, a dense two-dimensional protein mesh that is tightly associated with the plasma membrane in most metazoan cells (6, 7). In C. elegans, β-spectrin is encoded by the unc-70 gene (34). Neurons in animals lacking unc-70/β-spectrin develop with normal morphology. However, these neurons are fragile, and their axons break spontaneously when the mutant animals hatch and start to crawl (35). Broken axons regenerate, so regeneration can be studied in unc-70/β-spectrin mutant animals without the requirement for experimentally delivering nerve injuries.
Axon breaks in unc-70/β-spectrin mutant animals occur stochastically and may even recur in the same axon (35). Thus, it is more difficult to collect quantitative single-neuron data on regeneration using the unc-70/β-spectrin mutant background than using laser axotomy. However, because regeneration occurs spontaneously and constitutively, it is far easier to collect many animals that are regenerating. The unc-70/β-spectrin mutant background therefore may have advantages over laser surgery for experiments that require very large numbers of animals, such as genetic screens and genomic and proteomic studies. For example, this sensitized mutant background was successfully used in an unbiased RNA interference (RNAi) screen for genes affecting regeneration (36).
Differences Between Injury Models
An important and unresolved question is the extent to which different injury models—the use of different types of lasers, the study of different neurons, or the spontaneous breaks in unc-70/β-spectrin mutants—converge on common regeneration mechanisms. Detailed studies using femtosecond lasers to sever C. elegans axons show that dramatic changes in regeneration occur as a result of changing the number and energy of laser pulses, even though the total amount of energy applied is constant (10). Even the choice of fluorescent marker can influence regeneration (92). In some cases, such as the dlk-1 mitogen-activated protein kinase (MAPK) pathway, regeneration mechanisms show consistent effects across multiple neuron types, markers, and even injury models (36, 95). In other cases, however, the extent to which mechanisms remain consistent across different cells, markers, and injury models remains to be determined.
INTRINSIC REGENERATION PATHWAYS
Regeneration is regulated by intrinsic factors, and altering the intrinsic state of a neuron can improve regeneration, even in the presence of an inhibitory environment (58, 59, 64). Intrinsic pathways can also inhibit regeneration (17, 63, 81, 84). Both pro- and antiregeneration pathways have been described in C. elegans and are reviewed in this section.
Calcium/cAMP/PKA
Calcium and cAMP (cyclic adenosine monophosphate) signaling promote axon regeneration in a variety of systems (4, 13, 102). Increased calcium influx as a result of injury can result from entry of extracellular calcium through the breached axonal membrane, activation of voltage-gated calcium channels in response to depolarization, and release from intracellular stores (47, 101). Although increased intracellular calcium can result in neuronal cell death in some cases, calcium signaling can promote membrane resealing and regeneration in surviving neurons (71, 93). Increased intracellular calcium triggers multiple signaling events, including an increase in cAMP through modulation of adenylyl cyclase activity (19).
Calcium and cAMP also promote axon regeneration in C. elegans (28). Axotomy of the PLM touch neuron axons results in an acute increase in intracellular calcium. This increase is at least partially dependent on the voltagegated calcium channel subunit EGL-19 and the inositol triphosphate receptor ITR-1. Blocking the function of EGL-19 or ITR-1 reduces the acute increase in intracellular calcium and also reduces regeneration. Similarly, differences in the acute calcium increase in wild-type animals correlate with differences in the length of axon regeneration. Thus, intracellular calcium entry promotes regeneration in C. elegans. A major effector of calcium in regeneration is the adenylyl cyclase-cAMP-protein kinase A (PKA) signaling cassette. Interestingly, BAPTA (1,2-bis(o-aminophenoxy)ethane N, N, N′, N′ tetraacetic acid) treatment reduces the calcium baseline and reduces regeneration without affecting the size of the calcium transient. This suggests that total levels of calcium, rather than the relative size of the increase, are the final determinant of regeneration.
Channels, Transporters, and Neurotransmitters
Increased neuronal excitability results in more intracellular calcium, whereas decreased excitability results in less (reviewed above) (28). Consistent with the finding that calcium mediates regeneration, mutations that decrease excitability result in poor regrowth after axotomy, including the sodium pump nkb-1, the sodium channel unc-8, and the stomatins unc-1 and unc-24. Similarly, mutations that disrupt the synthesis of acetylcholine (cha-1) or acetylcholine transport (unc-17), or that disrupt the deg-3 subunit of the nicotinic acetylcholine receptor, result in decreased regeneration (15). Conversely, genetic lesions that make neurons more excitable, such as loss-of-function mutations affecting the slo-1 K+ channel or the K+ channel regulator mps-1, enhance regrowth (15).
MAP Kinase Signaling
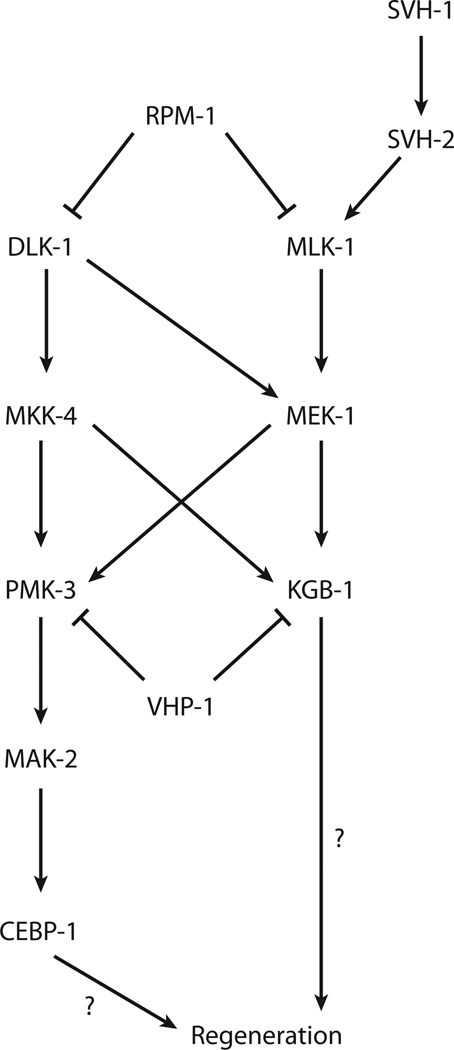
The dlk-1 MAPK regulates regeneration in both GABA motor neurons and sensory neurons (36, 95). An unbiased RNAi screen in unc-70/β-spectrin mutants for genes affecting regeneration identified dlk-1 as a candidate regeneration gene (36). dlk-1 is a MAPKKK that functions in a pathway with the MAP kinase kinase (MAPKK) mkk-4 and the p38 MAPK pmk-3 (56) (Figure 2). A major target of pmk-3 is the MAPKAPK (MAP kinase activated kinase) mak-2, which functions via the bZip-containing protein cebp-1 (95). This entire signaling pathway is required for regeneration. Loss of any component reduces regeneration, whereas activation of the pathway (by overexpression of dlk-1) increases regeneration (36, 95).
Figure 2.
Diagram of mitogen-activated protein kinase signaling during regeneration in Caenorhabditis elegans. Adapted from 48, 60.
The same RNAi screen in unc-70/β-spectrin mutants identified a second MAPKKK, mlk-1, as also having a significant role in regeneration (36). mlk-1 activates the MAPKK mek-1, which acts upstream of the JNK (c-Jun N-terminal kinase)-like kinase kgb-1 (52) (Figure 2). Again, this entire signaling pathway functions in regeneration. Further, the mlk-1 pathway exhibits significant cross talk with the dlk-1 pathway. In addition, the dlk-1 and mlk-1 MAPK pathways share common negative regulators. rpm-1, an E3 ubiquitin ligase that regulates synapse morphology and axon termination (30, 77, 100), mediates the degradation of mlk-1 as well as dlk-1 (56, 60). Hyperactivation of one or both of these pathways in rpm-1 mutants leads to increased regeneration (36). Both the dlk-1 and mlk-1 MAPK pathways are also regulated by the dual phosphatase vhp-1 (60).
In addition to these common regulators, the mlk-1 pathway is regulated by growth factor signaling. svh-1 is homologous to hepatocyte growth factor, macrophage stimulating protein, and plasminogen. svh-1 is a secreted factor that acts via the receptor-type tyrosine kinase svh-2 to activate regeneration via activation of the mlk-1 pathway. Although svh-2 functions in the injured neuron, svh-1 is expressed in the ADL neurons and functions as an extrinsic signal. Thus, the response of injured neurons is regulated in part by secreted factors deriving from other, uninjured neurons.
How does MAPK signaling facilitate regeneration? Activation of the dlk-1 pathway results in the stabilization of the cebp-1 messenger RNA (mRNA) through its 3′ untranslated region. Some mammalian MAPKAPKs (homologous to mak-2) can phosphorylate mRNA binding proteins, which may result in differential regulation of mRNA turnover (40). Thus, mak-2 may mediate cebp-1 mRNA stability by modifying RNA-binding proteins in response to axotomy. In the end, via the dlk-1 and mlk-1 pathways, axotomy results in localized translation of cebp-1 and also activation of kgb-1. However, the downstream effectors of cebp-1 are not known, nor are the effectors of kgb-1.
efa-6 and Microtubule Dynamics
Microtubules play a crucial role in axon regeneration. Destabilization of microtubules with nocodazole in regenerating rat sciatic nerve inhibits growth and transforms growth cones into retraction bulbs. Conversely, chemical stabilization of microtubules in the rat spinal cord by taxol treatment reduces the formation of retraction bulbs and enhances growth-cone formation. Retraction bulbs exhibit a disorganized microtubule structure and are static, whereas growth cones show a characteristic bundling of microtubules essential for growth-cone movement and axonal regrowth (22). Manipulation of microtubules results in enhanced regeneration in the mammalian CNS and may present novel therapeutic possibilities (39, 78, 82).
Microtubule dynamics are also important for C. elegans axon regeneration. efa-6 mutations result in increased PLM regrowth after axotomy (15). EFA-6 contains a variable N-terminal domain, a pleckstrin homology (PH) domain, and a Sec7 domain, which has a guanine exchange factor activity for ADPribosylation factor GTPases (26, 61). However, neither the PH domain nor the Sec7 domain of EFA-6 is required for inhibition of regeneration. Rather, the N terminus of EFA-6 inhibits regeneration by regulating microtubule organization. Although loss of efa-6 results in increased regeneration, overexpressing the N terminus of EFA-6 results in dysmorphic growth cones, less regeneration, and a reduction in dynamic microtubules (15). Stabilizing microtubules with taxol suppresses the inhibition of regeneration resulting from EFA-6 overexpression. This suggests that EFA-6 destabilizes microtubules in regenerating PLM axons and leads to failure of regrowth.
Notch Signaling
The lin-12 Notch receptor is a potent inhibitor of regeneration in C. elegans motor neurons (21). lin-12/Notch null animals regenerate significantly better than wild-type animals, whereas lin-12 gain-of-function alleles regenerate worse. Notch functions in the injured neuron to inhibit regeneration. Further, Notch activation after injury is necessary for Notch to inhibit regeneration: Blocking Notch activation with the γ-secretase inhibitor DAPT (N-[(3, 5-difluorophenyl)acetyl]-Lalanyl-2-phenyl]glycine-1,1-dimethylehyl ester) increases regeneration, even when injury has already occurred.
Notch activation typically involves sequential cleavage of Notch, first by a metalloprotease (site 2 cleavage) and then by the γ-secretase complex (site 3 cleavage). These cleavages release the Notch intracellular domain (NICD) into the cytoplasm. Notch inhibition of regeneration proceeds by this same activation mechanism: Both the sup-17/ADAM10 metalloprotease and presenilin (the catalytic component of γ-secretase) are required for Notch to inhibit regeneration, and NICD overexpression is sufficient to inhibit regeneration.
The activation mechanism for Notch inhibition of regeneration is not known. DSL (Delta/Serrate/LAG-2)-family Notch ligands are single-pass transmembrane proteins (87, 98). In addition to transmembrane DSL ligands, C. elegans expresses multiple secreted ligands thought to activate the receptor in cooperation with transmembrane ligands (16, 25, 45). However, loss of any single ligand does not improve regeneration. One possibility is that multiple ligands can redundantly activate Notch to inhibit regeneration. Alternatively, Notch activation might be ligand independent. In Drosophila, endocytosis of the Notch receptor can be mediated through Deltex in a ligand-independent manner and stabilized by Hif-α (41, 42, 55, 88, 94). Once endocytosed, Notch is cleaved by presenilin, and the NICD can be released into the cytoplasm. Calcium-dependent, ligand-independent Notch activation has also been described (67).
Notch activation is required within a short time frame (2 h) of axotomy to inhibit regeneration. This limited window of activation suggests that Notch may be directly activated by injury and that it signals immediately to inhibit regeneration. This immediate activation would allow enough time for an inhibitor of regeneration to be transcribed and translated to inhibit growth-cone initiation. Alternatively, Notch may be active at all times but may need to interact with other pathways that respond directly to injury or may need to be active continuously because its effectors are short lived. How Notch signaling affects regeneration remains an open question. One possibility that is consistent with our findings is that Notch could act by activating transcription of a direct inhibitor of regeneration. Some targets of Notch signaling have been identified, but their role in regeneration has not been examined.
Synaptic Vesicle Recycling
Synaptic vesicle exocytosis at nerve terminals results in the transfer of vesicle proteins and lipids to the plasma membrane; these are retrieved by endocytosis. Endocytosis— but not exocytosis—is also required for regeneration (15). Mutations in any of three key endocytosis genes (unc-26/synaptojanin, unc-57/endophilin, and unc-41/stonin) result in decreased regeneration. The requirement for synaptic vesicle endocytosis (but not exocytosis) genes in regeneration suggests that regeneration does not depend on synaptic vesicle cycling but on different cellular process. One potential mechanism is MAPK signaling, as the requirement for unc-57/endophilin during PLM regeneration can be bypassed by increased DLK-1 expression (15).
An Emerging Role for MicroRNAs
MicroRNAs function in many aspects of neural development and disease (46). Studies in cultured neurons, mice, and zebrafish suggest a role for microRNAs in central and peripheral regeneration (83, 91, 99). Because microRNAs are generally involved in the downregulation of their target genes, a regeneration-promoting role of microRNAs suggests an important role for the negative regulation of inhibitors of regeneration. In C. elegans, the Argonaute homolog alg-1 is required for PLM axon regeneration (15). alg-1 functions in the biogenesis of microRNAs (31), suggesting a role for microR-NAs in regulating regeneration in C. elegans. Although a general regeneration-promoting role for microRNAs has recently emerged, further study of the roles of individual microRNAs may reveal specific roles.
EXTRINSIC REGULATION OF REGENERATION IN CAENORHABDITIS ELEGANS
Regenerating axons navigate a very different environment than the one they encounter during development. Although some developmental growth and guidance cues may be maintained in adults, others are lost or replaced by inhibitory factors. Numerous studies in mammalian systems show that extrinsic factors from the glial scar and myelin act as potent inhibitors of regeneration in the CNS (reviewed in 11, 24, 50, 79, 97). C. elegans axons are not myelinated and navigate a very different extracellular environment than mammalian neurons. However, recent evidence suggests that axon guidance factors and elements of the C. elegans extracellular matrix (ECM) also play a role in the regulation of axon regeneration in worms.
Axon Guidance Factors
Regeneration in C. elegans is often characterized by guidance errors, premature termination, or branching (27, 92). By contrast, development of the nervous system is largely invariant. One mechanistic difference between regeneration and development is the function of axon guidance pathways. During development, ventral migration of the ALM mechanosensory neuron depends on the netrin and the Slit/Robo guidance pathways (27). These pathways depend on extracellular ligands (netrin and Slit) and their neuronal receptors (the netrin receptors unc-5 and unc-40 and the Slit receptor Robo). During regeneration, netrin and Slit also help to guide ALM ventral migration: Migration is defective in mutants for these genes. However, the neuronal receptors are dispensable for this process (27). Thus, axon guidance signals function differently in ALM during regeneration than during development.
A change in response to axon guidance molecules is also found in the PLM neurons. In these neurons, Slit/Robo signaling promotes outgrowth (49). During regeneration, however, Slit/Robo signaling inhibits axon extension (15). Further, accurate regeneration in the PLM neuron in adult animals is inhibited by the vab-1 Eph receptor (92). However, this function is independent of the kinase activity of vab-1 (92), whereas kinase activity is required for vab-1’s developmental function (53). Thus, differences in how axon guidance pathways function may account for some of the pathfinding errors during regeneration.
Extracellular Matrix
Many axons in C. elegans migrate along the basement membrane that runs between the body wall muscles and the epidermis. This basement membrane is required to maintain attachment of the muscle to the epidermis. Three genes—pxn-2, spon-1, and vab-19—that inhibit regeneration encode proteins that function to maintain muscle attachment. PXN-2 is a peroxidasin, which is a secreted protein that contains a catalytic peroxidase domain, immunoglobulin domains, and leucine-rich repeats. Peroxidases can inactivate metalloproteases, cross-link ECM components, and mediate cell-ECM adhesion (38, 70, 86). SPON-1 is spondin, a component of the ECM (44). VAB-19 is an ankyrin repeat protein, homologous to the human tumor suppressor Kank (76). VAB-19 is a cytoplasmic protein that localizes with components of epidermal attachment structures. Loss of any of these three genes causes defects in attachment of muscle to epidermis (29, 76, 90) and also results in increased regeneration (15, 29). These data suggest that muscle-epidermis attachment via the basement membrane inhibits regeneration. ECM proteins could act as a physical barrier to regeneration or could sequester essential soluble factors, rendering them unavailable to regrowing neurons. Alternatively, there may be receptor-mediated inhibitory mechanisms in C. elegans that are similar to inhibition of regeneration in vertebrates by CSPGs.
FUSION AND FUNCTIONAL REGENERATION
C. elegans neurons are capable of reconnecting directly to the severed distal fragment (14, 28, 57). Fusion restores membrane and cytoplasmic continuity between the two halves of the axon (28, 57). Fusion can also prevent degeneration of the distal fragment but only if it occurs relatively quickly after axotomy (57). If regenerating axons are given a choice of two distal fragments, one from the regenerating axon and one from another neuron, fusion occurs nearly exclusively with the correct fragment (57). Thus, reconnection after fusion can restore neuronal circuitry, at least in terms of morphology.
Fusion after axotomy requires the fusogen eff-1 (28). eff-1 also functions in homotypic fusions during normal development of epithelia, muscles, and sensory dendrites (54, 62). Fusion is also promoted by calcium and cAMP via PKA (28). Thus, fusion after axotomy is mediated by some of the same mechanisms as axon regeneration.
What of functional recovery? Regenerating axons in vertebrates can form de novo synapses and restore function providing they reach their target (1, 80). In C. elegans, loss of GABA neuron function (for example, in mutants that cannot synthesize GABA) leads to a characteristic shrinker behavioral phenotype (51). Severing all GABA commissures also results in the shrinker phenotype, and regeneration can restore GABA-mediated behavior (21, 96). Further, reduction of GABA regeneration (by increasing Notch signaling) also reduces behavioral recovery (21). Thus, regeneration can restore function to damaged neural circuits.
PERSPECTIVES
In a relatively short time, C. elegans has emerged as a highly tractable model to study axon regeneration. Current data suggest that many genes function similarly in worm and mammalian regenerating axons (see Introduction above). C. elegans offers the axon regeneration field a simple, highly invariant nervous system, genetic tractability, and the ability to study individual neurons in vivo. Although many novel signaling pathways that function in regeneration have been identified, many questions remain.
What Activates Regeneration Pathways?
MAPK pathways (48, 95) and probably also Notch signaling (21) are activated by injury. The cellular mechanism that links injury to activation is currently unknown. A variety of injury signaling mechanisms have been proposed, including calcium entry, electrical signals, and changes in trafficking of a regeneration factor. Of these, only calcium is known to function in C. elegans regeneration (28). One possibility is that calcium signaling is the single cue that activates all of the acute responses to injury. For example, modulation of calcium in vivo and at physiological conditions can activate Notch (69). Alternatively, other injury signals may exist. For example, disruption of the microtubule cytoskeleton in nerve injury may be an injury signal. In support of this idea, microtubule depolymerization can activate dlk-1 signaling (9). However, it remains to be determined whether this activation mechanism occurs in regenerating axons.
Answers to these questions and others may come via genetic screens. So far, two screens (an unbiased RNAi screen in unc-70/β-spectrin mutants and systematic screening of existing mutant alleles using laser axotomy) identified many genes involved in regeneration (15, 36). Additional genetic screens are likely to provide more details on the function of known pathways and also to identify additional factors that mediate regeneration.
What Are the Effectors of Regeneration Pathways?
In the end, regeneration signaling must converge on growth mechanisms that enable the injured neuron to generate a new growth cone, maneuver the growth cone to its target, and reconnect. These growth mechanisms are for the most part not understood in C. elegans. However, the recent description of microtubule dynamics in regenerating axons and the discovery of a novel regulator of these dynamics (see section on efa-6 and Microtubule Dynamics) suggest that it will be possible to analyze regeneration at the cell-biological level. Further application of cell-biological techniques, such as electron microscopy and super-resolution imaging, in combination with genetic analysis, should result in a better understanding of the growth mechanisms that mediate regeneration.
A second approach to cell biology that also has potential translational applications is the use of C. elegans as a platform to screen for drugs that are effective in improving regeneration after injury. A high-throughput screen using microfluidics identified a chemical enhancer of regeneration (75). Further, microinjecting compounds directly into the pseudocoelom of animals immediately after axotomy can enhance regeneration (15, 21). This technique may be used in the future to assess limited numbers of compounds for effects on regeneration. Such compounds may identify particular cell-biological processes that are important for regeneration.
How Good is Regeneration?
Most regeneration studies in C. elegans have used the morphology of the regenerating neuron as a measure of regenerative success. To date, two studies in the GABA motor neurons have shown that regeneration is accompanied by functional recovery at the level of whole-animal behavior (see section on Fusion and Functional Regeneration). However, such experiments are limited in their ability to accurately assess the function of individual regenerated neurons compared with their uninjured counterparts. C. elegans is a tractable model for a more detailed study of neuronal function using electrophysiological and optogenetic techniques (23, 33, 72). The future application of these techniques to the study of regeneration will yield information about how effective functional recovery is and may identify new pathways that are required for functional, rather than merely morphological, regeneration.
Why Inhibit Regeneration?
Axon regeneration can restore function, so why inhibit it? In the vertebrate CNS, pathways that inhibit regeneration also function in uninjured nervous systems and help maintain a stable, functional system by inhibiting aberrant growth and plasticity (2, 97). For example, CSPGs inhibit regeneration after injury in the CNS (11, 24, 50, 79, 97). In uninjured animals, enzymatic degradation of CSPGs results in ectopic growth and sprouting (5, 12, 20). These data suggest that inhibition of the injury response in the CNS is part of a broader program to limit plasticity. Similarly, it is possible that the C. elegans pathways that inhibit regeneration also function to stabilize the mature, uninjured nervous system.
Neurons in old C. elegans animals show a loss of stability and accumulate ectopic branches (85). Loss of jnk-1 causes an increase in ectopic neuronal branching in old animals, suggesting that jnk-1 contributes to nervous system stability (85). Consistent with the idea that stability pathways can also inhibit regeneration, jnk-1 mutants also display increased regeneration after nerve injury (36). In other ways, however, stability and regeneration seem to be separate processes. The dlk-1 pathway is required for regeneration but not for spontaneous branching in old animals. Loss of mlk-1 increases the incidence of age-dependent branching but decreases regeneration (36, 60, 85, 95). This suggests that in C. elegans, regeneration and spontaneous branching are promoted by different mechanisms. It remains to be seen whether other inhibitors of regeneration, such as Notch (21) and efa-6 (15), affect spontaneous branching and the overall stability of the nervous system.
Why Can Worms Regenerate at All?
Neurons in C. elegans can regenerate in response to injury. Is this ability due to a specialized regeneration mechanism that has evolved specifically to respond to nerve injury? Or is the response to injury part of a more general mechanism, such as homeostasis or stress response? Or is it even a pathological and undirected response to trauma?
During their life span in the wild, worms are subjected to desiccation, mechanical trauma, and predators. Thus, the ability to regenerate neurons quickly after they have been severed and to regain movement may pose a significant selective advantage. Alternatively, neuronal regeneration may be a particular manifestation of some broader biological process. A more complete understanding of the mechanisms that mediate regeneration might eventually help answer this question.
Conclusion
C. elegans has advanced the field of axon regeneration by providing a genetic system that is compatible with both high-throughput screening and single-neuron analysis. Additional forward genetics, a wider use of high-throughput techniques, functional regeneration assays, and drug validation will make C. elegans an even more robust model and further advance the study of axon regeneration.
Glossary
- MAG
myelin-associated glycoprotein
- Omgp
oligodendrocyte myelin glycoprotein
- CSPG
chondroitin-sulfate proteoglycan
- CNS
central nervous system
- cAMP
cyclic adenosine monophosphate
- MAPKKK
mitogen-activated protein kinase kinase kinase
- GABA
γ-aminobutyric acid
- RNAi
RNA interference
- MAPK
mitogen-activated protein kinase
- PKA
protein kinase A
- BAPTA
1,2-bis(o-aminophenoxy)ethane N, N, N′, N′,tetraacetic acid
- MAPKK
mitogen-activated protein kinase kinase
- N-[(3, 5-difluorophenyl)-acetyl]-Lalanyl-2-phenyl]glycine-1,1-dimethylehyl ester small (DAPT)
molecule inhibitor of γ-secretase
- NICD
Notch intracellular domain
- DSL
Delta/Serrate/LAG-2
- ECM
extracellular matrix
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aguayo AJ, Bray GM, Rasminsky M, Zwimpfer T, Carter D, Vidal-Sanz M. Synaptic connections made by axons regenerating in the central nervous system of adult mammals. J. Exp. Biol. 1990;153:199–224. doi: 10.1242/jeb.153.1.199. [DOI] [PubMed] [Google Scholar]
- 2.Akbik F, Cafferty WB, Strittmatter SM. Myelin associated inhibitors: a link between injury-induced and experience-dependent plasticity. Exp. Neurol. 2011;235:43–52. doi: 10.1016/j.expneurol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen PB, Chiu DT. Calcium-assisted glass-to-glass bonding for fabrication of glass microfluidic devices. Anal. Chem. 2008;80:7153–7157. doi: 10.1021/ac801059h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appenzeller O, Palmer G. The cyclic AMP (adenosine 3′,5′-phosphate) content of sciatic nerve: changes after nerve crush. Brain Res. 1972;42:521–524. doi: 10.1016/0006-8993(72)90553-7. [DOI] [PubMed] [Google Scholar]
- 5.Barritt AW, Davies M, Marchand F, Hartley R, Grist J, et al. Chondroitinase ABC promotes sprouting of intact and injured spinal systems after spinal cord injury. J. Neurosci. 2006;26:10856–10867. doi: 10.1523/JNEUROSCI.2980-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett V, Gilligan DM. The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane. Annu. Rev. Cell Biol. 1993;9:27–66. doi: 10.1146/annurev.cb.09.110193.000331. [DOI] [PubMed] [Google Scholar]
- 7.Bennett V, Stenbuck PJ. Identification and partial purification of ankyrin, the high affinity membrane attachment site for human erythrocyte spectrin. J. Biol. Chem. 1979;254:2533–2541. [PubMed] [Google Scholar]
- 8.Bhatt DH, Otto SJ, Depoister B, Fetcho JR. Cyclic AMP-induced repair of zebrafish spinal circuits. Science. 2004;305:254–258. doi: 10.1126/science.1098439. [DOI] [PubMed] [Google Scholar]
- 9.Bounoutas A, Kratz J, Emtage L, Ma C, Nguyen KC, Chalfie M. Microtubule depolymerization in Caenorhabditis elegans touch receptor neurons reduces gene expression through a p38 MAPK pathway. Proc. Natl. Acad. Sci. USA. 2011;108:3982–3987. doi: 10.1073/pnas.1101360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourgeois F, Ben-Yakar A. Femtosecond laser nanoaxotomy properties and their effect on axonal recovery in C. elegans. Opt. Expr. 2008;16:5963. doi: 10.1364/oe.16.005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cafferty WB, McGee AW, Strittmatter SM. Axonal growth therapeutics: regeneration or sprouting or plasticity? Trends Neurosci. 2008;31:215–220. doi: 10.1016/j.tins.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cafferty WB, Yang SH, Duffy PJ, Li S, Strittmatter SM. Functional axonal regeneration through astrocytic scar genetically modified to digest chondroitin sulfate proteoglycans. J. Neurosci. 2007;27:2176–2185. doi: 10.1523/JNEUROSCI.5176-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlsen RC. Comparison of adenylate cyclase activity in segments of rat sciatic nerve with a condition/test or test lesion. Exp. Neurol. 1982;77:254–265. doi: 10.1016/0014-4886(82)90243-6. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Chisholm AD. Axon regeneration mechanisms: insights from C. elegans. Trends Cell Biol. 2011;21:577–584. doi: 10.1016/j.tcb.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Wang Z, Ghosh-Roy A, Hubert T, Yan D, et al. Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron. 2011;71:1043–1057. doi: 10.1016/j.neuron.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen N, Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell. 2004;6:183–192. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- 17.Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J. Neurosci. 2010;30:9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung SH, Clark DA, Gabel CV, Mazur E, Samuel AD. The role of the AFD neuron in C. elegans thermotaxis analyzed using femtosecond laser ablation. BMC Neurosci. 2006;7:30. doi: 10.1186/1471-2202-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper DM, Mons N, Karpen JW. Adenylyl cyclases and the interaction between calcium and cAMP signalling. Nature. 1995;374:421–424. doi: 10.1038/374421a0. [DOI] [PubMed] [Google Scholar]
- 20.Corvetti L, Rossi F. Degradation of chondroitin sulfate proteoglycans induces sprouting of intact Purkinje axons in the cerebellum of the adult rat. J. Neurosci. 2005;25:7150–7158. doi: 10.1523/JNEUROSCI.0683-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Bejjani R, Hammarlund M. Notch signaling inhibits axon regeneration. Neuron. 2012;73:268–278. doi: 10.1016/j.neuron.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erturk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J. Neurosci. 2007;27:9169–9180. doi: 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filbin MT. Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat. Rev. Neurosci. 2003;4:703–713. doi: 10.1038/nrn1195. [DOI] [PubMed] [Google Scholar]
- 25.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, et al. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabel CV, Antoine F, Chuang CF, Samuel AD, Chang C. Distinct cellular and molecular mechanisms mediate initial axon development and adult-stage axon regeneration in C. elegans. Development. 2008;135:1129–1136. doi: 10.1242/dev.013995. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh-Roy A, Wu Z, Goncharov A, Jin Y, Chisholm AD. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. J. Neurosci. 2010;30:3175–3183. doi: 10.1523/JNEUROSCI.5464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotenstein JR, Swale RE, Fukuda T, Wu Z, Giurumescu CA, et al. The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development. 2010;137:3603–3613. doi: 10.1242/dev.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 32.Guo SX, Bourgeois F, Chokshi T, Durr NJ, Hilliard MA, et al. Femtosecond laser nanoaxotomy lab-on-a-chip for in vivo nerve regeneration studies. Nat. Methods. 2008;5:531–533. doi: 10.1038/nmeth.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat. Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammarlund M, Davis WS, Jorgensen EM. Mutations in β-spectrin disrupt axon outgrowth and sarcomere structure. J. Cell Biol. 2000;149:931–942. doi: 10.1083/jcb.149.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammarlund M, Jorgensen EM, Bastiani MJ. Axons break in animals lacking β-spectrin. J. Cell Biol. 2007;176:269–275. doi: 10.1083/jcb.200611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harel NY, Strittmatter SM. Can regenerating axons recapitulate developmental guidance during recovery from spinal cord injury? Nat. Rev. Neurosci. 2006;7:603–616. doi: 10.1038/nrn1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinecke JW, Li W, Daehnke HL, 3rd, Goldstein JA. Dityrosine, a specific marker of oxidation, is synthesized by the myeloperoxidase-hydrogen peroxide system of human neutrophils and macrophages. J. Biol. Chem. 1993;268:4069–4077. [PubMed] [Google Scholar]
- 39.Hellal F, Hurtado A, Ruschel J, Flynn KC, Laskowski CJ, et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science. 2011;331:928–931. doi: 10.1126/science.1201148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, et al. Mitogen-activated protein kinase–activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 2006;26:2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, et al. Drosophila Deltex mediates Suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- 42.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J. Cell Biol. 2011;195:1005–1015. doi: 10.1083/jcb.201104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh A, Horiuchi M, Bannerman P, Pleasure D, Itoh T. Impaired regenerative response of primary sensory neurons in ZPK/DLK gene-trap mice. Biochem. Biophys. Res. Commun. 2009;383:258–262. doi: 10.1016/j.bbrc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Klar A, Baldassare M, Jessell TM. F-spondin: a gene expressed at high levels in the floor plate encodes a secreted protein that promotes neural cell adhesion and neurite extension. Cell. 1992;69:95–110. doi: 10.1016/0092-8674(92)90121-r. [DOI] [PubMed] [Google Scholar]
- 45.Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, et al. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 2008;6:e196. doi: 10.1371/journal.pbio.0060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kosik KS. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 47.Kulbatski I, Cook DJ, Tator CH. Calcium entry through L-type calcium channels is essential for neurite regeneration in cultured sympathetic neurons. J. Neurotrauma. 2004;21:357–374. doi: 10.1089/089771504322972130. [DOI] [PubMed] [Google Scholar]
- 48.Li C, Hisamoto N, Nix P, Kanao S, Mizuno T, et al. The growth factor SVH-1 regulates axon regeneration in C. elegans via the JNK MAPK cascade. Nat. Neurosci. 2012;15:551–557. doi: 10.1038/nn.3052. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Kulkarni G, Wadsworth WG. RPM-1, a Caenorhabditis elegans protein that functions in presynaptic differentiation, negatively regulates axon outgrowth by controlling SAX-3/robo and UNC-5/UNC5 activity. J. Neurosci. 2008;28:3595–3603. doi: 10.1523/JNEUROSCI.5536-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGee AW, Strittmatter SM. The Nogo-66 receptor: focusing myelin inhibition of axon regeneration. Trends Neurosci. 2003;26:193–198. doi: 10.1016/S0166-2236(03)00062-6. [DOI] [PubMed] [Google Scholar]
- 51.McIntire SL, Jorgensen E, Horvitz HR. Genes required for GABA function in Caenorhabditis elegans. Nature. 1993;364:334–337. doi: 10.1038/364334a0. [DOI] [PubMed] [Google Scholar]
- 52.Mizuno T, Hisamoto N, Terada T, Kondo T, Adachi M, et al. The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 2004;23:2226–2234. doi: 10.1038/sj.emboj.7600226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohamed AM, Chin-Sang ID. Characterization of loss-of-function and gain-of-function Eph receptor tyrosine kinase signaling in C. elegans axon targeting and cell migration. Dev. Biol. 2006;290:164–176. doi: 10.1016/j.ydbio.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 54.Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 55.Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-α in development and survival of Drosophila blood cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakata K, Abrams B, Grill B, Goncharov A, Huang X, et al. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 57.Neumann B, Nguyen KC, Hall DH, Ben-Yakar A, Hilliard MA. Axonal regeneration proceeds through specific axonal fusion in transected C. elegans neurons. Dev. Dyn. 2011;240:1365–1372. doi: 10.1002/dvdy.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neumann S, Bradke F, Tessier-Lavigne M, Basbaum AI. Regeneration of sensory axons within the injured spinal cord induced by intraganglionic cAMP elevation. Neuron. 2002;34:885–893. doi: 10.1016/s0896-6273(02)00702-x. [DOI] [PubMed] [Google Scholar]
- 59.Neumann S, Woolf CJ. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron. 1999;23:83–91. doi: 10.1016/s0896-6273(00)80755-2. [DOI] [PubMed] [Google Scholar]
- 60.Nix P, Hisamoto N, Matsumoto K, Bastiani M. Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc Natl. Acad. Sci. USA. 2011;108:10738–10743. doi: 10.1073/pnas.1104830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Rourke SM, Christensen SN, Bowerman B. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat. Cell. Biol. 2010;12:1235–1241. doi: 10.1038/ncb2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oren-Suissa M, Hall DH, Treinin M, Shemer G, Podbilewicz B. The fusogen EFF-1 controls sculpting of mechanosensory dendrites. Science. 2010;328:1285–1288. doi: 10.1126/science.1189095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park KK, Liu K, Hu Y, Smith PD, Wang C, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu J, Cai D, Dai H, McAtee M, Hoffman PN, et al. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron. 2002;34:895–903. doi: 10.1016/s0896-6273(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 65.Qiu J, Cai D, Filbin MT. A role for cAMP in regeneration during development and after injury. Prog. Brain Res. 2002;137:381–387. doi: 10.1016/s0079-6123(02)37029-8. [DOI] [PubMed] [Google Scholar]
- 66.Raivich G, Makwana M. The making of successful axonal regeneration: genes, molecules and signal transduction pathways. Brain Res. Rev. 2007;53:287–311. doi: 10.1016/j.brainresrev.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 67.Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, et al. Calcium depletion dissociates and activates heterodimeric Notch receptors. Mol. Cell. Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao GN, Kulkarni SS, Koushika SP, Rau KR. In vivo nanosecond laser axotomy: cavitation dynamics and vesicle transport. Opt. Expr. 2008;16:9884–9894. doi: 10.1364/oe.16.009884. [DOI] [PubMed] [Google Scholar]
- 69.Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, et al. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature. 2004;427:121–128. doi: 10.1038/nature02190. [DOI] [PubMed] [Google Scholar]
- 70.Rees MD, Kennett EC, Whitelock JM, Davies MJ. Oxidative damage to extracellular matrix and its role in human pathologies. Free Radic. Biol. Med. 2008;44:1973–2001. doi: 10.1016/j.freeradbiomed.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 71.Rehder V, Kater SB. Regulation of neuronal growth cone filopodia by intracellular calcium. J. Neurosci. 1992;12:3175–3186. doi: 10.1523/JNEUROSCI.12-08-03175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richmond JE, Jorgensen EM. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat. Neurosci. 1999;2:791–797. doi: 10.1038/12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Microfluidic system for on-chip high-throughput whole-animal sorting and screening at subcellular resolution. Proc. Natl. Acad. Sci USA. 2007;104:13891–13895. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossi F, Gianola S, Corvetti L. Regulation of intrinsic neuronal properties for axon growth and regeneration. Prog. Neurobiol. 2007;81:1–28. doi: 10.1016/j.pneurobio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 75.Samara C, Rohde CB, Gilleland CL, Norton S, Haggarty SJ, Yanik MF. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc. Natl. Acad. Sci. USA. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarkar S, Roy BC, Hatano N, Aoyagi T, Gohji K, Kiyama R. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J. Biol. Chem. 2002;277:36585–36591. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- 77.Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 78.Sengottuvel V, Leibinger M, Pfreimer M, Andreadaki A, Fischer D. Taxol facilitates axon regeneration in the mature CNS. J. Neurosci. 2011;31:2688–2699. doi: 10.1523/JNEUROSCI.4885-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silver J, Miller JH. Regeneration beyond the glial scar. Nat. Rev. Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 80.Smith GM, Falone AE, Frank E. Sensory axon regeneration: rebuilding functional connections in the spinal cord. Trends Neurosci. 2011;35:156–163. doi: 10.1016/j.tins.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith PD, Sun F, Park KK, Cai B, Wang C, et al. SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron. 2009;64:617–623. doi: 10.1016/j.neuron.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stone MC, Nguyen MM, Tao J, Allender DL, Rolls MM. Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol. Biol. Cell. 2010;21:767–777. doi: 10.1091/mbc.E09-11-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strickland ER, Hook MA, Balaraman S, Huie JR, Grau JW, Miranda RC. MicroRNA dysregulation following spinal cord contusion: implications for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun F, Park KK, Belin S, Wang D, Lu T, et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tank EM, Rodgers KE, Kenyon C. Spontaneous age-related neurite branching in Caenorhabditis elegans. J. Neurosci. 2011;31:9279–9288. doi: 10.1523/JNEUROSCI.6606-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y, Rosen H, Madtes DK, Shao B, Martin TR, et al. Myeloperoxidase inactivates TIMP-1 by oxidizing its N-terminal cysteine residue: an oxidative mechanism for regulating proteolysis during inflammation. J. Biol. Chem. 2007;282:31826–31834. doi: 10.1074/jbc.M704894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 88.Wilkin M, Tongngok P, Gensch N, Clemence S, Motoki M, et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of Notch in the endosomal trafficking pathway. Dev. Cell. 2008;15:762–772. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 89.Williams W, Nix P, Bastiani M. Constructing a low-budget laser axotomy system to study axon regeneration in C. elegans. J. Vis. Exp. 2011;57:e3331. doi: 10.3791/3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Woo WM, Berry EC, Hudson ML, Swale RE, Goncharov A, Chisholm AD. The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development. 2008;135:2747–2756. doi: 10.1242/dev.015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu D, Raafat A, Pak E, Clemens S, Murashov AK. Dicer-microRNA pathway is critical for peripheral nerve regeneration and functional recovery in vivo and regenerative axonogenesis in vitro. Exp. Neurol. 2012;233:555–565. doi: 10.1016/j.expneurol.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu Z, Ghosh-Roy A, Yanik MF, Zhang JZ, Jin Y, Chisholm AD. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc. Natl. Acad. Sci. USA. 2007;104:15132–15137. doi: 10.1073/pnas.0707001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xie XY, Barrett JN. Membrane resealing in cultured rat septal neurons after neurite transection: evidence for enhancement by Ca2+-triggered protease activity and cytoskeletal disassembly. J. Neurosci. 1991;11:3257–3267. doi: 10.1523/JNEUROSCI.11-10-03257.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamada K, Fuwa TJ, Ayukawa T, Tanaka T, Nakamura A, et al. Roles of Drosophila Deltex in Notch receptor endocytic trafficking and activation. Genes Cells. 2011;16:261–272. doi: 10.1111/j.1365-2443.2011.01488.x. [DOI] [PubMed] [Google Scholar]
- 95.Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yanik MF, Cinar H, Cinar HN, Chisholm AD, Jin Y, Ben-Yakar A. Neurosurgery: functional regeneration after laser axotomy. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- 97.Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yochem J, Weston K, Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988;335:547–550. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]
- 99.Yu YM, Gibbs KM, Davila J, Campbell N, Sung S, et al. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur. J. Neurosci. 2011;33:1587–1597. doi: 10.1111/j.1460-9568.2011.07643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 101.Ziv NE, Spira ME. Spatiotemporal distribution of Ca2+ following axotomy and throughout the recovery process of cultured Aplysia neurons. Eur. J. Neurosci. 1993;5:657–668. doi: 10.1111/j.1460-9568.1993.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 102.Ziv NE, Spira ME. Axotomy induces a transient and localized elevation of the free intracellular calcium concentration to the millimolar range. J. Neurophysiol. 1995;74:2625–2637. doi: 10.1152/jn.1995.74.6.2625. [DOI] [PubMed] [Google Scholar]