Before going to the gym for a workout or after indulging in cake at the office party, people with diabetes can use a portable monitor to take a quick blood glucose measurement and adjust their food or insulin intake to prevent extreme dips or spikes in blood sugar. The inexpensive finger-prick blood testing devices that allow diabetics to check their blood sugar throughout the day may sound like small conveniences. That is unless you are diabetic and can remember back a decade or more, when having that disease came with far more fear and guessing and far less control over your own wellbeing.
The quality of life afforded to diabetics by technologies that easily and inexpensively extract information from the body offers a glimpse of what all medicine could be like: more predictive and preventive, more personalized to the individual’s needs and enabling more participation in maintaining one’s own health. In fact, we believe that medicine is already headed in that direction, largely because of new technologies that make it possible to acquire and analyze biological information quickly and cheaply.
One of the keys to this evolution in medicine is the extreme miniaturization of technologies for making diagnostic measurements from miniscule amounts of blood or even single cells taken from diseased tissues. These emerging tools, constructed at the scale of microns and nanometers (one billionth of a meter), can manipulate and measure large numbers of biological molecules rapidly, precisely, and, eventually, at a cost of pennies or less per measurement. That combination of cost and performance opens up new avenues for studying and treating disease by permitting the human body to be viewed as a dynamic system of molecular interactions. Such systems-level measurements are then integrated into computational models, which, in turn, can reveal early indicators of a problem. When these insights are combined with new nanotechnology-based therapies, the treatment can be targeted to the problem and only the problem, thereby avoiding serious side effects.
Although we anticipate that all medicine will eventually operate by these principles, cancer research offers current examples of how technology at the ultra-small scale is providing the data needed to paint a big-picture systems view of disease.
Systems Medicine
Modeling a system requires vast amounts of data, and living organisms are full of information that could be described as digital—it can be measured and quantified and programmed into the model. Such biological information starts with an organism’s genetic code. Every cell in the human body carries a full copy of the human genome, which is made up of three billion pairs of DNA bases, the letters of the genetic alphabet. Those “letters” encode some 25,000 genes, representing instructions for operating cells and tissues. Inside each cell, genes are transcribed into a more portable form, discrete snippets of messenger RNA that carry those instructions to cellular equipment that reads the RNA and churns out chains of amino acids according to the encoded instructions. Those amino acid chains, in turn, fold themselves into proteins, the three-dimensional molecular machines that execute most of the functions of life.
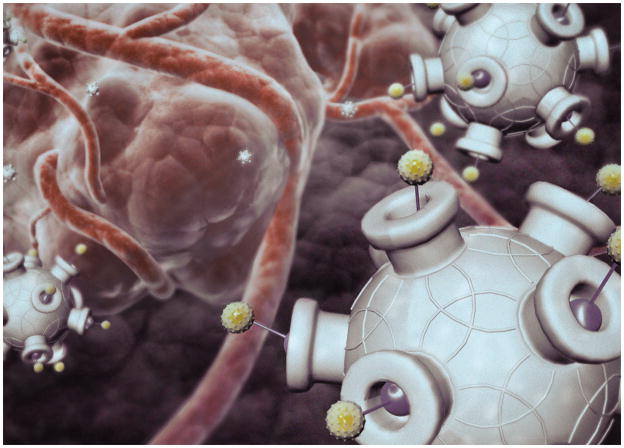
NANOPARTICLES CONSTRUCTED to carry a therapeutic payload are studded with proteins that act as keys for gaining entry to tumor cells.
Within a biological system, such as a person, all of this “data” is transmitted, processed, integrated and ultimately executed through networks of proteins interacting with one another and with other biologically relevant molecules inside cells. And when the entire system is viewed as a network of these interrelated events, disease can be seen as a consequence of something perturbing the network’s normal programmed patterns of information. The initial cause could be a flaw within the system, such as a random change in DNA that alters an encoded instruction, or even some environmental influence impinging on the system from outside, such as the ultraviolet radiation in sunlight that can trigger DNA damage that eventually leads to melanoma. As an initial disruption produces ripple effects and feedback, the information patterns continue to change, and these dynamically altered patterns explain the nature of the disease mechanistically [see illustration on page 46].
Of course, building an accurate computer model of this kind of biological network is a staggering effort. The task can require computational integration of millions or more measurements of messenger RNA and protein levels to comprehensively capture the dynamics of the system’s transition from health to disease. Nevertheless, an accurate model—meaning one capable of correctly predicting the effects of perturbations—can be the foundation for dramatic change in the way illness and health are understood and how they are treated medically.
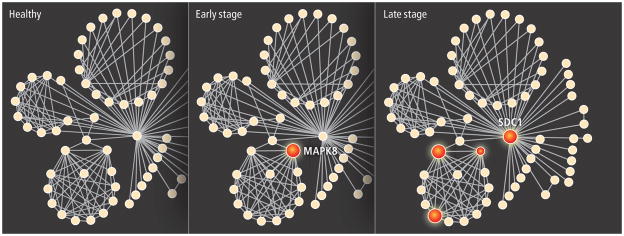
PROSTATE CELLS contain groups of proteins (solid circles) that interact (lines) with one another in small networks; changes in cellular levels of certain proteins accompany a shift from health to disease. Early-stage prostate cancer cells show a rise in levels of MAPK8, a protein known to regulate cell movement. In late-stage cancer cells, levels of SDC1 are 16 times higher than in early-stage cells. Relative amounts of these two proteins can offer diagnostic clues to the presence and progression of disease.
Over the past several decades, for example, cancer has been the most intensively studied of all diseases, yet tumors have traditionally been characterized by fairly coarse features that include their size, their location in a particular organ or tissue, and whether malignant cells have spread from the primary tumor. The more advanced the cancer according to such diagnostic “stages,” the more bleak the prognosis for the patient. But even that conventional wisdom offers plenty of contradictions. Patients diagnosed with identical cancers and given similar treatments from the standard repertoire of radiation and chemotherapies often respond very differently—one group of patients may experience full recovery, whereas a second group succumbs rapidly.
Large-scale measurements of messenger RNA and protein concentrations within biopsied tumors have revealed the inadequacy of such traditional approaches by showing how two patients’ cancers that are seemingly the same really contain networks perturbed in dramatically different ways. Based upon such molecular analysis, many cancers that were at one time considered to be a single disease are now identified as separate diseases.
About 80 percent of human prostate tumors grow so slowly that they will not ever harm their hosts, for instance. The remaining 20 percent will grow more quickly, invading surrounding tissues and even spreading (metastasizing) to distant organs, eventually killing the patient. Our research group is now attempting to identify the disease-perturbed networks in prostate cells that characterize both these major cancer types so that a doctor could identify from the outset which kind a patient has. That information could spare 80 percent of patients unnecessary surgery, irradiation or chemotherapy, along with the pain, incontinence and impotence that accompany those treatments.
We are also analyzing the networks within the prostate that distinguish subtypes among the more aggressive 20 percent of cases that might require distinct treatment regimens. For example, by analyzing the networks characteristic of early-stage and metastatic prostate cancers, we have identified a protein secreted into the blood that appears to be an excellent identifying marker for metastatic cancer. Tools of this kind that can stratify a given disease such as prostate cancer into its precise subtype would allow a clinician to make a rational choice of the appropriate therapy for each individual.
Detecting Disease
Although such analyses of mRNAs and proteins from tumor tissues can be informative about the nature of a known cancer, the systems approach can also be applied to distinguishing between health and illness. Blood bathes every organ in the body, carrying away proteins and other molecules, so it provides an excellent window into the entire body system. The ability to detect an imbalance in particular proteins or messenger RNAs could therefore serve to signal the presence of disease and pinpoint its location and its nature.
Our research group has addressed the challenge of using blood to assess the status of the whole-body system by comparing messenger RNA populations produced in the 50 or so individual organs, and we have found that each human organ has 50 or more messenger RNA types that are made primarily in just that organ. Certain of these RNAs encode organ-specific proteins that are secreted into the bloodstream, and the levels of each will reflect the operation of the networks that control their production within the organ. When those networks are disease-perturbed, corresponding protein levels will be altered. Those changes should make it possible to identify the disease because each disease in the organ will disturb distinct biological networks in unique ways.
If the levels of about 25 proteins from each of these organ-specific fingerprints can be assessed, computational analysis should make it possible to detect all diseases by determining which networks are perturbed—just from blood measurements. Beyond early detection—which is so important in cancer—this approach would provide the ability to stratify a patient’s disease into its different subtypes, to follow its progression and to follow its response to therapy. We have shown an initial proof of this principle by following the evolution of prion disease in mice.
We injected the mice with infectious prion proteins that lead to a degenerative brain disease akin to “mad cow disease,” then we analyzed the complete populations of brain messenger RNAs in the infected and control animals at 10 different time points during the onset of the disease. From these data we identified 300 changing messenger RNAs that encoded the core prion disease response. About 200 of those RNAs belonged to four biological networks that explained virtually every known aspect of the disease, and about 100 other RNAs described previously unknown aspects of prion disease. Study of these disease-perturbed networks also allowed us to identify four blood proteins that predicted the presence of prion disease before any apparent symptoms and could therefore serve as presymptomatic diagnostic markers, with obvious benefits for preventive medicine. These studies required about 30 million measurements, and we developed a series of software programs for analyzing, integrating and finally modeling these enormous amounts of data. Constructing predictive network models of disease, and translating those models into medically useful tools will require rapid, sensitive and—most importantly—cheap methods for DNA sequencing and for measuring mRNA and protein concentrations.
[DIAGNOSTICS]: PENNIES PER PROTEIN.
Information is the most valuable commodity in a systems approach to medicine, so diagnostic tests will have to easily and accurately measure large numbers of biological molecules for a few cents or less per measurement. Extreme miniaturization allowed the authors and their colleagues to produce a prototype chip that can measure concentrations of a panel of cancer-associated proteins in a droplet of blood in 10 minutes, at a cost of five to 10 cents per protein.
![[DIAGNOSTICS]: PENNIES PER PROTEIN](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/1403/3700418/3323d3a6c18c/nihms478906u1.jpg)
[THE BASICS]: NANOTECH IN MEDICINE.
At the scale of one nanometer—one billionth of a meter—materials and devices can interact with cells and biological molecules in unique ways. The nanoscale technologies already used in research or therapies are generally between 10 nanometers, the size of an antibody protein, and 100 nanometers, the size of a virus. These devices and particles are being applied as sensors to detect molecules such as proteins or DNA, as imaging enhancers, and as a means to target specific tissues and deliver therapeutic agents.
![[THE BASICS]: NANOTECH IN MEDICINE](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/1403/3700418/16679363ab47/nihms478906u2.jpg)
| NANOTECHNOLOGY | USE | HOW IT WORKS |
|---|---|---|
NANOWIRES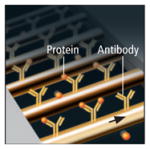
|
Sensor | Conductive wire, 10 to 20 nanometers thick, is strung across a channel through which a sample will pass. To detect proteins or DNA, probes made of complementary antibodies or DNA are attached to each wire. When a protein meets its matching antibody, it binds to the probe and changes the conductive properties of the wire, allowing the event to be detected electronically. |
CANTILEVERS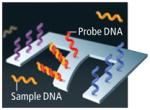
|
Sensor | Molecular probes, such as single-stranded DNA, can also be attached to beams just a few thick. When exposed to a DNA sample, complimentary strands will bind to the probes on the cantilever, causing the beam to bend slightly. That response can be detected visually or by a change in the beam’s electrical conductivity. |
QUANTUM DOTS
|
Imaging | Nanocrystals made of inorganic elements such as cadmium or mercury encased in latex or metal respond to light by emitting fluorescence at different wave-lengths and intensities depending on their composition. Antibodies attached to the crystals can cause the dots to bind to a select tissue, such as a tumor, which can then be more easily seen with conventional imaging devices. |
NANOSHELLS
|
Imaging Tissue targeting | Solid silica nanospheres, sometimes encased in a thin layer of gold, will travel through the bloodstream without entering most healthy tissues, but they tend to pass through the “leaky” walls of tumor blood vessels into the tumor tissue. Once a large number of the nanoshells accumulate in a tumor, heat delivered to the tumor will be absorbed by the spheres, killing the tissue. Depending on their composition, nanoshells can also absorb or scatter light, enhancing tumor images made with certain forms of spectroscopy. |
NANOPARTICLES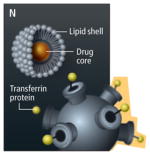
|
Tissue targeting Delivery | Particles composed of a variety of materials can be constructed to contain therapeutic molecules in their core and to release them at a desirable time and location. Such delivery vehicles include simple lipid shells that passively leak through tumor blood vessel walls, then slowly release a traditional chemotherapy drug into the tissue. Newer nanoparticles are more complexly designed, including exterior elements such as antibodies to target tumor-specific proteins, and materials that minimize the particles’ interaction with healthy tissues. |
Measuring Molecules
Many scientists have noted that technological advances in DNA sequencing have mirrored Moore’s Law for microprocessors: namely, that the number of functional elements that can be placed on a chip per unit cost has doubled every 18 months for the past several decades. In fact, next-generation DNA sequencing machines are increasing the speed of reading DNA at a pace that is much faster than Moore’s Law. For example, the first human genome sequenced probably took about three to four years to complete and cost perhaps $300 million. We believe that within five to 10 years an individual human genome sequence will cost less that $1,000—a 300,000-fold reduction in price—and be done in a day. Over the next decade, similar advances in other relevant biomedical technologies will permit predictive and personalized medicine to emerge.
At present, a test to measure levels of a single diagnostic cancer protein, such as prostate-specific antigen, in a patient’s blood costs a hospital about $50 to perform. Given that systems-based medicine will require measurements of large numbers of such proteins, this price must fall dramatically. Measurement time is also a cost. A blood test today could take a few hours to a few days, in part because of the many steps needed to separate blood components—cells, plasma, proteins and other molecules—before they can each be measured using tests of varying accuracy.
Extreme miniaturization can offer enhanced precision and significantly faster measurements than can be achieved with current technologies. Several microscale and nanoscale technologies are already proving their value as research tools for gathering the data needed to construct a systems view of biological information. For use in patient care, though, the demands of a systems approach to medicine will mandate that each measurement of a protein should only cost a few pennies—a target that is unlikely to be met by many emerging nanotechnologies.
Two of us (Heath and Hood) have developed a four-centimeter-wide chip that tests protein levels in a droplet of blood, employing a highly miniaturized variant of conventional protein-detection strategies [see box on page 47]. The chip is made only of glass, plastic and reagents, so it is very inexpensive to produce. Our device takes about two microliters of blood, separates the cells from the plasma, then measures a panel of a dozen plasma proteins, all within a few minutes of blood collection. The projected cost of using the prototype version is perhaps five to 10 cents per protein tested, but when fully developed, this technology should be able to meet the cost demands of systems medicine.
Extending the capabilities of the chip to measure hundreds of thousands of proteins will take time, but advances in microfluidics design, surface chemistry and measurement science are quickly bridging the gap between what is possible today and what will be required to fully realize a new predictive and personalized medicine. Our colleagues Stephen R. Quake and Axel Scherer, for instance, have developed a microfluidic system that directly integrates valves and pumps on a chip. Their miniaturized plumbing permits chemical reagents, biomolecules, and biological samples to be precisely directed into any one of a large number of individual chambers on the chip, with each chamber representing a separate and independent measurement. Their concept thus turns a lab-on-a-chip into many labs-on-a-chip, providing avenues for further reducing the costs of biological measurements.
Extremely miniaturized technology has similarly important implications for therapies and prevention. Insights into diseased networks can ultimately provide new targets for novel therapies that restore network dynamics to normalcy. In the shorter term, the systems view can help to target existing drugs far more effectively by matching the optimal drug combination to each patient. Nanotechnology, moreover, can also radically reduce the amount of each drug that is required to treat the cancer.
Tiny and Targeted
Nanoparticle therapeutics are small relative to most things but large compared to a molecule, and operating at this scale provides an unprecedented level of control over the therapeutic particles’ behavior within the body. Nanoparticles can range between one and 100 nanometers (nm) in size and can be assembled from a variety of existing therapeutic agents, such as chemotherapy drugs or gene-silencing RNA (siRNA) strands. These payloads may be encapsulated in synthetic materials such as polymers or lipidlike molecules, and targeting agents such as antibodies and other molecules designed to bind with specic cellular proteins can be added to the particle’s surface. This modularity makes nanotherapeutics especially versatile and capable of performing complex functions at the right place and the right time inside a patient.
ON TARGET.
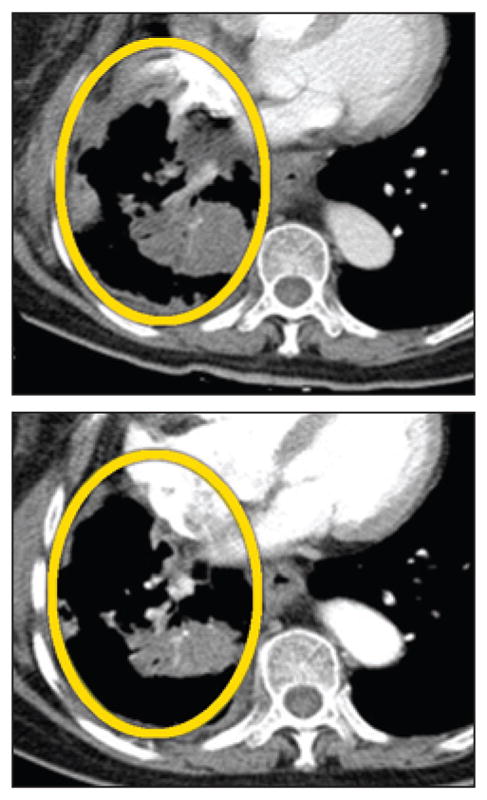
An experimental nanotherapy, IT-101, encapsulates a chemotherapy drug, camptothecin, inside a nanoparticle designed to circulate for an extended period in the bloodstream and to accumulate in tumors. In a human safety trial, evidence of the treatment’s efficacy was seen in some patients with advanced cancers. In the CT scans below, views of a patient’s midsection show a large lung tumor (top, circled) before treatment with IT-101 and after six months of treatment (bottom), when the tumor had shrunk considerably.
One of the greatest challenges in developing and using cancer drugs is delivering them to the diseased tissues without poisoning the patient’s entire body. Size alone gives even simple nanoparticle therapies special properties that determine their movement into and throughout tumors. Nanoparticles smaller than 10 nm are, like so-called small-molecule drugs, rapidly eliminated through the kidney, whereas particles larger than 100 nm have a difficult time moving through a tumor. Particles within the 10- to 100-nm range travel throughout the bloodstream to seek out tumors, although they are unable to escape into most healthy tissues through blood vessel walls. Because tumors, in contrast, have abnormal blood vessels whose walls are riddled with large pores, nanoparticles can leak into the surrounding tumor tissue. As a result, nanoparticles have a tendency to accumulate in tumors while minimizing effects on other parts of the body and avoiding the traditional ravishing side effects of cancer drugs.
Even when a standard drug manages to get to tumor cells, cellular pump proteins may eject it from the cell before it has a chance to work, a common mechanism of drug resistance. Nanoparticles enter a cell by endocytosis, a natural process that creates a pocket of cell membrane around a foreign object to draw it inside the cell, protecting the particle’s payload from the cellular pumps [see box on next page].
Certain cancer therapeutics that are now re-classified as nanoparticles have existed for some time and illustrate some of these basic advantages of nanoparticles in reaching tumor cells while minimizing effects on healthy tissue. Liposomal doxorubicin, for example, is a traditional chemotherapy compound encapsulated in a lipid shell that has been used to treat ovarian cancer and multiple myeloma. The lipid-encased version of the drug has far lower heart toxicity than doxorubicin alone, although a new side effect, skin toxicity, has been observed.
Newer nanoparticles—for example one known as IT-101, which has already undergone human safety testing in phase I trials, have more complex designs that provide multiple functions. IT-101 is a 30-nm particle assembled from polymers joined to the small-molecule drug camptothecin, which is closely related to two FDA-approved chemotherapy drugs, irinotecan and topotecan. The IT-101 particles are designed to circulate in the patient’s blood and they remain there for more than 40 hours, whereas camptothecin by itself would circulate for only a few minutes. This long circulation period allows time for IT-101 to escape into tumors and accumulate there. The particles then enter tumor cells and slowly release the camptothecin to enhance its effects. As the drug is released, the rest of the nanoparticle components disassemble and the small individual polymer molecules harmlessly exit the body through the kidney.
In the clinical trials, dosages of the drug were achieved that provided high quality of life without the side effects, such as vomiting, diarrhea and hair loss typical of chemotherapeutics, and with no new side effects. The general high quality of life while on treatment is exciting, and although phase I trials focus on establishing safety, the tests also provided evidence that the drug was active in patients [see sidebar on preceding page]. That is encouraging because patients in phase I cancer trials have had numerous courses of standard therapy that failed before entering the trial. After completing the six-month trial, several of these patients have remained on the drug on a compassionate use basis, and long-term survivors of approximately one year or more include patients with advanced lung, renal and pancreatic cancers.
[CASE STUDY]: DESIGNED TO DELIVER.
An experimental nanoparticle therapeutic called CALAA-01 illustrates some of the advantages such agents can offer. In addition to having a natural tendency to accumulate in tumors, nanoparticles can be designed to home to one or more receptors commonly found on cancer cells. The particles’ mode of cell entry also allows them to evade cellular pumps that eject some drugs.
Customized Structure.
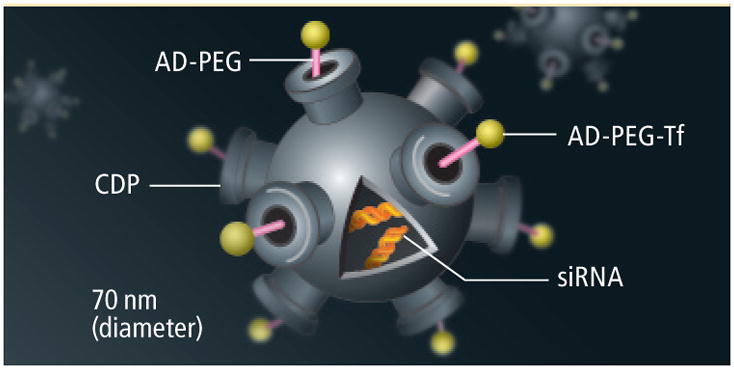
The particle is built with biocompatible materials: a cyclodextrin-containing polymer (CDP) with polyethylene glycol (PEG) stalks to which transferrin proteins (Tf) are attached. Inside, as many as 2,000 siRNA molecules—the therapeutic agents—are stored.
Passive Tumor Targeting.
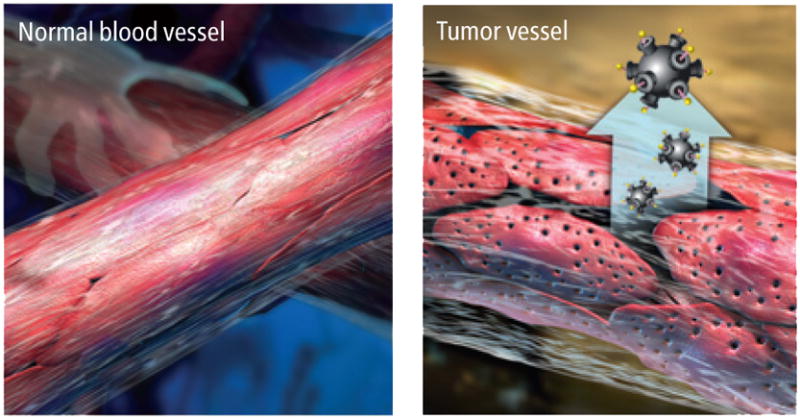
When the particles enter a patient’s bloodstream, they circulate freely but cannot penetrate most blood vessel walls. Tumor vessels are abnormally leaky, with large pores that allow nanoparticles to pass through and accumulate in the tumor tissue.
Active Tumor Targeting.
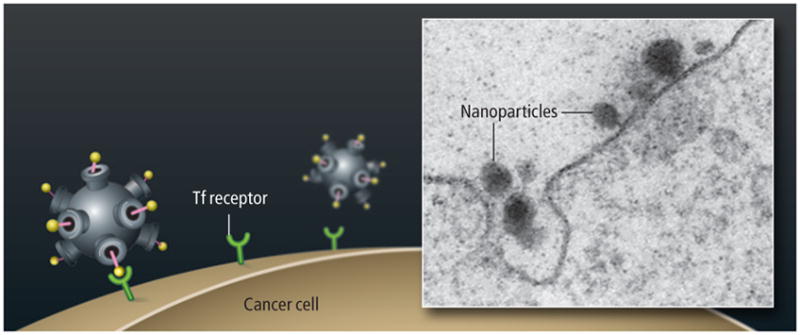
Transferrin receptors on the surface of a cancer cell bind to the transferrin protein on the nanoparticle, causing the cell to internalize the nanoparticle by endocytosis.
Controlled release.
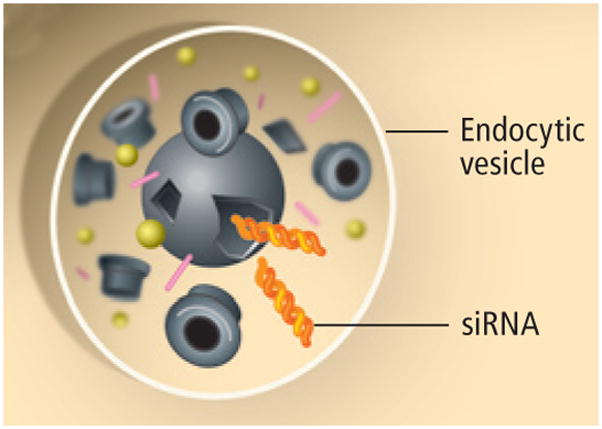
Once inside the cell, a chemical sensor within the nanoparticle responds to the low pH within the endocytic vesicle by simultaneously triggering disassembly of the nanoparticle and release of the siRNA molecules that will block a gene’s instructions from being translated into a protein the cancer cell needs to survive.
Because the side-effect profile of this drug is so low, it will next be tested in a phase II (efficacy) trial in women diagnosed with ovarian cancer who have undergone chemotherapy. Instead of simply “waiting and watching” for the cancer to progress, IT-101 will be given as maintenance therapy in the hope of preventing disease progression. These observations from IT-101 testing and similarly encouraging news from trials of other new nanoparticle-based treatments are beginning to provide a picture of what may be possible with well-designed nanoparticle therapeutics. Indeed, the next generation of synthetic nanoparticles, which are far more sophisticated, offer a glimpse of the real potential of this technology and the importance these drugs will hold for the systems-based view of disease and treatment.
Calando Pharmaceuticals in Pasadena, Calif., began trials in 2008 of a siRNA delivery system invented by one of us (Davis) that illustrates the newer approach. Proteins on the surface of the particles target specific receptors that occur in high concentrations on the surface of cancer cells. Once inside the cells, the particles release siRNA molecules, which are tailored to match a specific gene of interest and inhibit the manufacture of the gene’s encoded protein.
This multifunctional nanotherapeutic is just the beginning of the story, though. Once the principles of nanoparticle function in humans are more fully established, the concept can be applied to create a therapeutic system that can carry combinations of drugs, each with its own customized release rates. For example, if one wished to inhibit a protein that causes a drug to be ineffective, then an option would be to create a nanoparticle that first releases siRNA to inhibit the gene for that protein before releasing the drug molecule. As greater understanding of the molecular transitions from health to disease and vice versa is gained, the nanoparticle approach is likely to play an increasing role in the treatment of disease at the molecular level.
The Big Picture
A systems approach to disease is predicated on the idea that the analysis of dynamic disease-perturbed networks and the detailed mechanistic understanding of disease that it provides can transform every aspect of how we practice medicine—better diagnostics, effective new approaches to therapy and even prevention. This systems biology approach to disease is driving the development of many new technologies, including microfluidics, nanotechnologies, new measurement and visualization instrumentation, and computational advances that can analyze, integrate and model large amounts of biological information.
In the next 10 to 20 years, predictive and personalized medicine will be revolutionized by at least two new approaches. The sequence of individual human genome will permit us to determine with ever increasing accuracy the probable future health of an individual. Inexpensive measurements of blood proteins will permit us to assess, regularly and comprehensively, how that individual’s health is evolving.
Preventive medicine starts with the identification of proteins within a diseased network that, if perturbed, will restore network behavior to normalcy, and will eventually lead to prophylactic drugs that prevent disease. For instance, a woman at increased risk for ovarian cancer, who at age 30 starts taking a nanotherapeutic that is specially designed to offset the molecular source of the risk, might lower her lifetime chance of developing ovarian cancer from 40 percent to 2 percent.
With this kind of knowledge about the causes of health and disease available, people will also be able to participate more effectively in their own health decisions, much as today’s diabetics have tools and information that help them manage their own daily well being.
The realization of a form of medicine that is predictive, personalized, preventive and participatory will have broad implications for society. The health care industry will have to fundamentally alter its business plans, which are currently failing to produce cost effective and highly efficacious drugs. Emerging technologies will also lead to the digitization of medicine—meaning the ability to extract disease-relevant information from single molecules, single cells or single individuals—just as information technologies and communications have been digitized over the past 15 years. As a result of the new high-throughput, low-cost technologies, the cost of health care should fall sharply making it widely accessible, even in the developing world.
NANOSCALE THERAPIES.
Nanoscale particles designed to treat cancer include drugs already in use, such as a liposome-encased version of the chemotherapy doxorubicin, as well as a variety of experimental polymer drug combinations in which the drug and polymer molecules are meshed or chemically joined into untargeted nanoparticles (composites, conjugates, micelles, dendrimers). Newer targeted particles bear additional features that increase their affinity for, and facilitate entry into, cancer cells.
| PARTICLE TYPE | DEVELOPMENT STAGE | EXAMPLES |
|---|---|---|
| Liposome | FDA approved | DaunoXome, Doxil |
| Albumin-based | FDA approved | Abraxane |
| Polymeric micelle | Clinical trials | Genexol-PM, SP1049C, NK911, NK012, NK105, NC-6004 |
| Polymer-drug conjugate | Clinical trials | XYOTAX (CT-2103), CT-2106, IT-101, AP5280, AP5346, FCE28068 (PK1), FCE28069 (PK2), PNU166148, PNU166945, MAG-CPT, DE-310, Pegamotecan, NKTR-102, EZN-2208 |
| Targeted liposome | Clinical trials | MCC-465, MBP-426, SGT-53 |
| Targeted polymer-based particle | Clinical trials | FCE28069(PK2), CALAA-01 |
| Solid inorganic or metal particle | Clinical trials (gold) and preclinical | Carbon nanotubes, silica particles, gold particles (CYT-6091) |
| Dendrimer | Preclinical | Polyamidoamine (PAMAM) |
For cancer, the very exciting promises that should be realized over the next 10 years are, first, that presymptomatic blood diagnoses will catch fledgling cancers that can be cured by conventional therapy; second, that cancers of a particular tissue (for example, breast or prostate) will be stratified into distinct types that can be matched with drugs that provide very high cure rates; and third, the identification of disease-perturbed networks will allow more rapid development of drugs that are less expensive and far more effective. This new approach to medicine therefore has the potential to transform health care for virtually everyone living today.
KEY CONCEPTS.
A “systems” approach to medicine views the body as a complex network of molecular interactions that can be measured and modeled, revealing causes of disease such as cancer.
Extremely miniaturized tools can inexpensively measure and manipulate molecules for systems medicine.
Nanoscale therapies deliver precisely targeted treatments to tumors while avoiding healthy tissues.
—The Editors