Abstract
A spider’s ability to store silk protein solutions at high concentration is believed to be related to the protein’s terminal domains. It has been suggested that a shift in salt concentration and pH can have a significant influence on the assembly process. Based on experimental data, a model has been proposed in which the N-terminal domain exists as a monomer during storage and assembles into a homodimer upon spinning. Here we perform a systematic computational study using atomistic, coarse-grained and well-tempered metadynamics simulation to understand how the NaCl concentration in the solution affects the N-terminal domain of the silk protein. Our results show that a high salt concentration, as found during storage, weakens key salt bridges between the monomers, inducing a loss in bond energy by 28.6% in a single salt bridge. As a result dimer formation is less likely as 35.5% less energy is required to unfold the dimer by mechanical force. Conversely, homodimer formation appears to be more likely at low salt concentrations as the salt bridge stays at the lower energy state. The link between salt concentration, structure and stability of the N-terminal domain provides a possible mechanism that prevents premature fiber formation during storage.
Keywords: Salt bridge, spidroin, assembly, metadynamics, free-energy landscape, coarse-grained model, nanomechanics, molecular dynamics
Introduction
Spider dragline silk stands out due to its exceptional mechanical properties such as high toughness and strength (1, 2). Nano-confined assemblies of hydrogen bonds have been proposed to yield this high mechanical performance (3, 4), maintain biodegradability and allow energy-efficient assembly of the material. In this sense, the in vivo fiber assembly of silk may inspire polymer processing towards a low-energy polymer fiber formation (5). Dragline silk mainly consists out of major ampullate spidroin 1 and 2. A highly repetitive core region, which is linked to the mechanical behavior of silk, makes up most of the spidroin, while the terminal domains are non-repetitive and related to the silk assembly process (6, 7). Before the major ampullate spidroin (MaSp) is drawn on demand into a fiber at ambient temperature, it is stored in a highly concentrated form in aqueous solution in the major ampullate gland (8). The production of silk strands is triggered by various processing variables, such as shear force, solvent removal and dope composition, that each impacts MaSp differently (8–10). The terminal domains of MaSp primary react on changes in the solvent condition (11–14), involving a shift from a situation with a strong presence of sodium chloride and near neutral condition in the gland, to predominant potassium and phosphate ions at a slight acidic condition in the spinning duct (9).
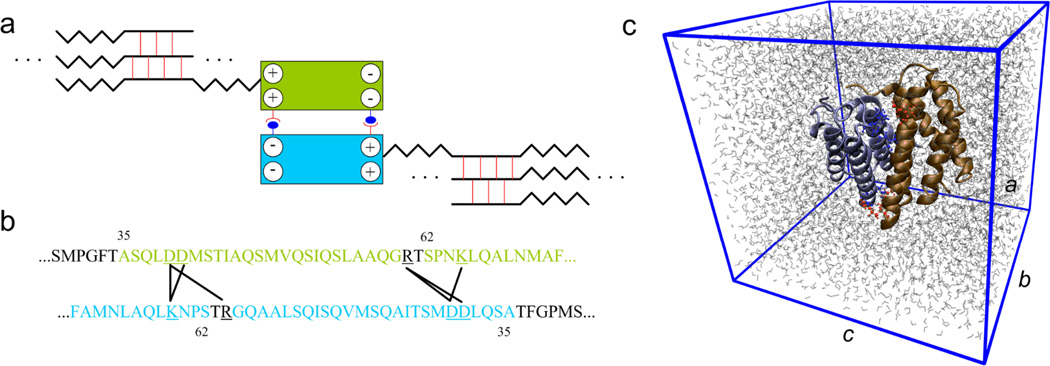
Specifically, the N-terminal domain of MaSp plays a critical role in delaying aggregation at neutral pH (11). At pH values of around 6 the N-terminal domain is found as a homodimer, while at high salt concentration and neutral pH it is found as a monomer (13–15). Several models based on these experimental observations have been proposed to describe the role of the terminal domains in the spinning process. Hagn et al. (13) proposed a model where the spidroin is stored in micelles with the terminal domains at the surface. The N-terminal domain is suggested to be present as a monomer while stored and later to assemble into a homodimer in the spinning duct (see Fig. 1a). However, how the formation process of the silk homodimer is governed by the solvent condition is not clear. This prevents us from understanding how spiders manage to store the silk in liquid crystalline phase and transition it into strong fibers during spinning.
Figure 1. Role of N-terminal domain in the silk assembly process.
Panel a: Schematic figure (not drawn to scale) of the association of the two monomers to form a dimer, which helps for the alignment of the spidroins and leads to very long spidroin strands. The two blocks represent the homodimer that is also depicted in Panel c. The zigzag line stands for the semi-amorphous region that alternates with β-sheet crystal forming parts, which connect different strands to a network. Panel b: Relevant domain within each of the monomer to form salt bridges. A black line connects the two amino acids involved in a salt bridge. These salt bridges are identified during the equilibrium molecular dynamic simulation by using the length criterion. Panel c: Simulation unit cell of the atomic structure of the homodimer of the N-terminal domain at equilibrium in water condition. The two monomers are depicted in different colors and each of them consists of five α-helices. The charged residues at the dimer interface are shown by red (for monomer 1) and blue (for monomer 2) in detail. The periodic box is given by the blue frame with lattice constants a, b and c in different directions. Water molecules are depicted by gray lines.
It is difficult for experimental studies to capture the details of certain molecular interactions. Computational approaches may give complementary insights into the detailed mechanisms of the events partially exposed by experimental observations. For example, Wallace and Shen (16) have investigated the effect of pH on the pKa values of the N-terminal domain using constant pH molecular dynamics. They revealed that glutamine residues are ionized at elevated pH and as a consequence water molecules enter the interface between the monomers and reduce the dimer stability. However, so far there has been no computational study that explains how the dimer interface is affected by high salt concentration, which is a condition essential for silk storage (13, 14). Here we use molecular dynamics simulation to elucidate the effect of sodium and chloride ions on the N-terminal domain and compare our results to experimental studies. The analysis on the molecular scale allows us to investigate the biophysical mechanism involved in destabilizing the homodimer. We focus on the mechanical stability of the assembled homodimer, and investigate the adhesion energy between the interfaces at two different salt conditions. Our result reveals how salt weakens the dimer bonding and alters its molecular conformation to prevent premature fiber formation while stored in the gland, which are consistent with experimental observations.
Materials and methods
Crystal structure of the N-terminal domain and molecular simulation setup
The crystal structure of the N-terminal domain from the spider Euprosthenops australis has been determined by X-ray crystallography with 1.7 Å resolution by Askarieh et al. (11) and is obtained from the Protein Data Bank (PDB identification code 3LR2). The initial structure is equilibrated in a solvent environment modeled by explicit TIP3P water molecules (17) using full-atomistic molecular dynamics. The molecular simulations are carried out using the CHARMM27 force field (18) and TIP3P water model (17) with the NAMD2.8 simulation package (19). We use a time step of 2 fs to simulate an isothermal-isobaric ensemble (NPT) at a temperature of T=300 K and pressure of P =1 atm using a Langevin-piston barostat and a friction coefficient of γ= 5 ps−1. We apply periodic boundary conditions in all directions and use the Particle Mesh Ewald method with 1 Å grid size to efficiently account for the full electrostatics. The simulated system is a rectangular unit cell with its size of 64 Å×61 Å×70 Å (a×b×c as shown in Fig. 1c). This edge length is chosen large enough to accommodate the protein structure. The minimum distance between mirroring structures is beyond 15 Å, which is the cutoff length of the van der Waals interaction. We neutralize the system by controlling the number of cations and anions added to the system. We equilibrate the homodimer separately for 20 ns at two different conditions. The environment of the spider’s spinning gland is represented by a NaCl concentration of 0.5 M, while the absence of NaCl in the spinning duct is modeled by a water box containing no salt. The two conditions are chosen to align with recent experiments that examined the influence of salt on the N-terminal domain (13, 14). During the equilibration process we monitor the root-mean-square deviation (RMSD, as shown in Fig. S1 in the Supporting Material) and the length of each cross-monomer salt bridge. A salt bridge is defined when the nitrogen and oxygen atoms of the charged residues pairs get closer than 4 Å during the equilibration (20).
Well-tempered metadynamics method
Free-energy calculations are carried out using the well-tempered metadynamics algorithm (21) implemented in the PLUMED1.3 (22) external plugin for the NAMD2.8 package. This method enhances the sampling speed in simulations by adding bias potentials during the simulation and has the advantage of ensuring convergence of the energy landscape without requiring an initial estimate. We chose the biased collective variable to be the distance between the center of mass of NH3 of Lys65 (in monomer 1) and COO of Asp40 (in monomer 2). The simulations are run with an initial configuration of the equilibrated homodimer obtained after equilibration for 20 ns. Other parameters include the enhanced temperature of 1,500 K where the collective variable is sampled, and Gaussian hills with the width of 0.35 Å are added to bias the energy of the collective variable at a rate of 0.1 kcal/mol for every 2 ps, corresponding to a deposition rate of 0.05 kcal/mol/ps. Each of the well-tempered metadynamics runs is 10 ns long and the convergence is confirmed (as shown in Fig. S2 in the Supporting Material).
Coarse-grained model with MARTINI force field
Additionally we carry out an equilibration and tensile test of the molecular structure using the residue-based coarse-grained MARTINI force field in explicit water. This allows us to explore the stability of the homodimer at time-scales of several hundreds of nanoseconds, and larger length-scales (23, 24). The initial coarse-grained geometry is generated from the full atomistic model of the homodimer (after the equilibration process, as shown in Fig. 1c). Each amino acid is represented by one to five beads, one of which is the backbone bead that is generated from the coordinate of the residue backbone and the rest of which are side-chain beads that are generated from the coordinate of the residue side chain. Every four water molecules are represented by a bead. The coarse-grained system includes 3,585 particles (503 for the protein plus 3,082 for water molecules), while the corresponding full atomistic system includes 40,635 particles (3,652 for the protein plus 36,984 for water molecules). We note that 59 ions (sodium plus chloride) are included in the system for the 0.5 M solution while 9 ions (sodium) are included as the counter ion for the non-salt solution. We use a 20 fs time step in this coarse-grained model, and apply the same ensemble (NPT) with the same constant temperature (300 K) and pressure (1 atm) as in the full atomistic model during the 200 ns equilibration phase. We ensure that the structures are fully equilibrated in different solutions by computing the RMSD of the structure and the potential energy (as shown in Fig. S3 and S4 in the Supporting Material), as well as the length of each of the salt bridge; each of which converges before 200 ns equilibrium.
Steered molecular dynamics: mechanical analysis
The simulations of the response of the homodimer under mechanical force are carried out with the steered molecular dynamics (SMD) method (25). The backbone bead at the C-terminal of one monomer is tethered to a spring with a force constant of 10 kcal/mol/Å2, being pulled in a constant rate, while the C-terminal of the other monomer is fixed. The pulling force is recorded versus the position of the tether point. The simulations are carried out at pulling velocity of 0.1 m/s. It is noted that for this pulling rate, the simulation time reaches 400 ns before the two monomers completely break apart. We record the force-displacement curve during the SMD simulation and compare the breaking force and external work for the homodimer in different solutions.
Results and discussion
We equilibrate the crystal structure of the N-terminal domain of E. australis (with PDB identification code 3LR2) via full-atomistic classic molecular dynamics simulations for 20 ns at two different solvent conditions, one non-salt and one with 0.5 M sodium chloride. The salt condition resembles the storing condition in the spider’s gland and parallel experiments that investigated the salt influence on the N-terminal domain (13–15). For the non-salt condition we align with the experiments that use a 10 mM or 20 mM phosphate buffer (13, 14), although in vivo there are potassium and phosphate ions prevalent in the spinning duct (9). The concentration of phosphate buffer used in the experiments is so low that we neglect the buffer in our simulations.
After simulating for 20 ns, there is no significant change for the structure as the RMSD stays within 1.9 Å for the non-salt condition and within 2.9 Å for the salt condition. The equilibrated structure for different solvent conditions maintains a homodimer composed of two monomers, each as a bundle of five α-helices as illustrated in Fig. 1c. The RMSD of the homodimer in both solutions converges by the end of those calculations, indicating that both structures reach their equilibrated states within the current setup of the simulations (as shown in Fig. S1).
For both conditions there are six residue pairs found to have the capacity to form intermolecular salt bridges to stabilize the dimer as shown in Fig. 1b and c. Those six residue pairs include:
Lys65(monomer 1)-Asp40(monomer 2) named SB1,
Arg60(monomer 1)-Asp40(monomer 2) named SB2,
Arg60(monomer 1)-Asp39(monomer 2) named SB3,
Asp40 (monomer 1)-Lys65(monomer 2) named SB4,
Asp39(monomer 1)-Arg60(monomer 2) named SB5 and
Asp39(monomer 1)-Lys65(monomer 2) named SB6.
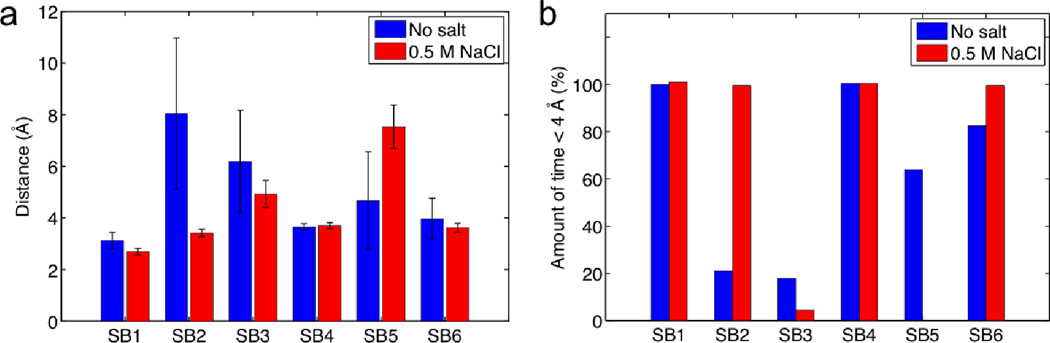
All residues that are involved in forming intermolecular salt bridges are located on helix 2 and helix 3 of each subunit. This observation is in agreement with the concept reported by Jaudzems et al. (26) that the two subunits dimerize via these two helices. It is interesting that SB4 and SB5 are in a symmetric position with regards to SB1 and SB3, respectively. We keep record of the length of the six salt bridges and statistically summarize the result in Fig. 2a. We find that SB1 and SB4 have the shortest length among all the salt bridges in both non-salt and salt conditions, indicating the strongest interaction to stabilize the dimer. We find more evidence as the importance of SB1 and SB4 as shown in Fig. 2b, which shows the distance between Lys65 and Asp40 (for SB1 and SB4) are permanently closer than 4 Å for all conditions and always form salt bridges with conserved length. We propose that these salt bridges represent the most relevant interactions to stabilize the two monomers since the rest of the salt bridges fluctuate more in length and maintain larger distances.
Figure 2. Distances between charged residues of the two molecules that are able to form intermolecular salt bridges.
Data is taken from the last 10 ns equilibration of the all-atom model. Panel a: Mean distance with standard deviation for salt and no salt condition. Panel b: Amount of time the residue pair has been in an effective salt bridge state (≤ 4 Å).
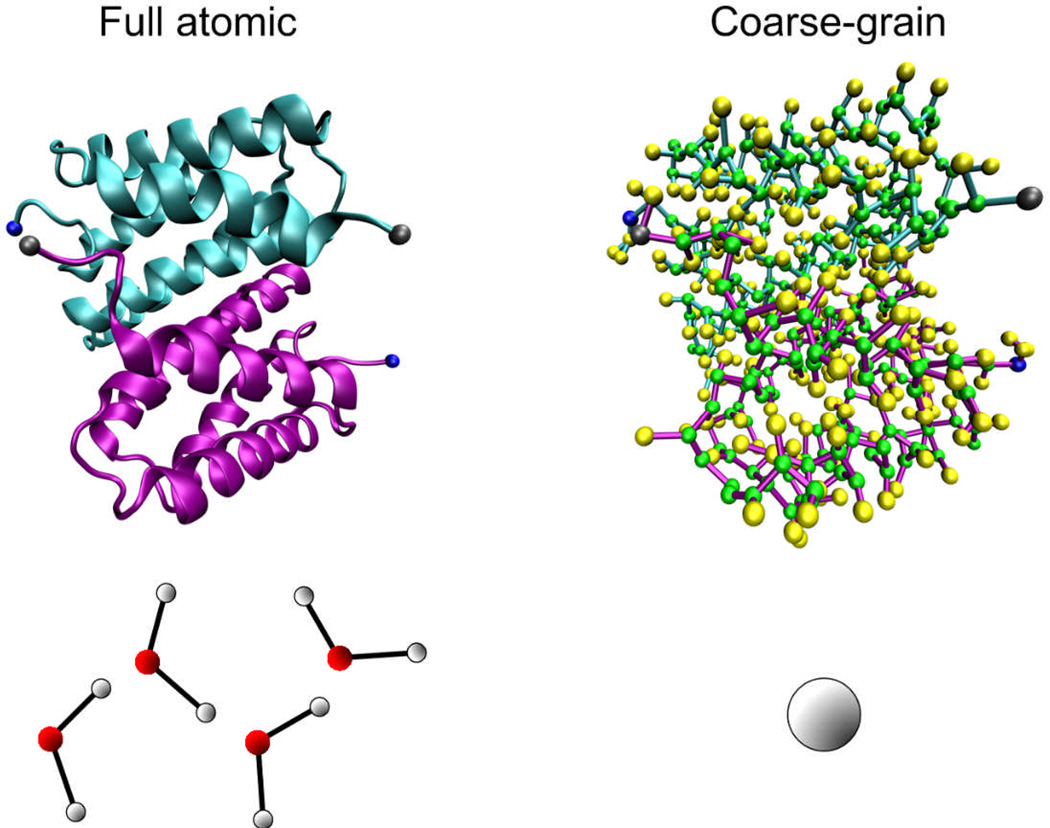
There is no direct evidence to show a difference in the stability of the homodimer in different solvent conditions via this classic molecular dynamics simulation. This issue most likely arises because the interaction between the two monomers is too strong and thermal fluctuations have no significant effect on the structure stability during the limited sampling time. To extend the sampling time we coarse-grain the model with the Martini force field to gain access to longer time scales. The coarse-grained model, as illustrated in Fig. 3, is set up based on the coordinates of each residue of the equilibrated atomic structure, and it preserves the secondary structure information as well as the non-bonded interactions between the residue pairs of the full atomic model.
Figure 3. Coarse-grained representation of the N-terminal dimer model.
By replacing the full atomistic representation (on left) of the protein and water with a residue-based bead model (on right, where several beads reflect one amino acid), much longer time-scales can be reached. In the coarse-grained model the backbone is represented by a green bead and the side chain is represented by yellow beads. We highlight the N-terminal and C-terminal of each monomer by coloring them in blue and gray, respectively. Four water molecules are represented by a single bead in the coarse-grained model.
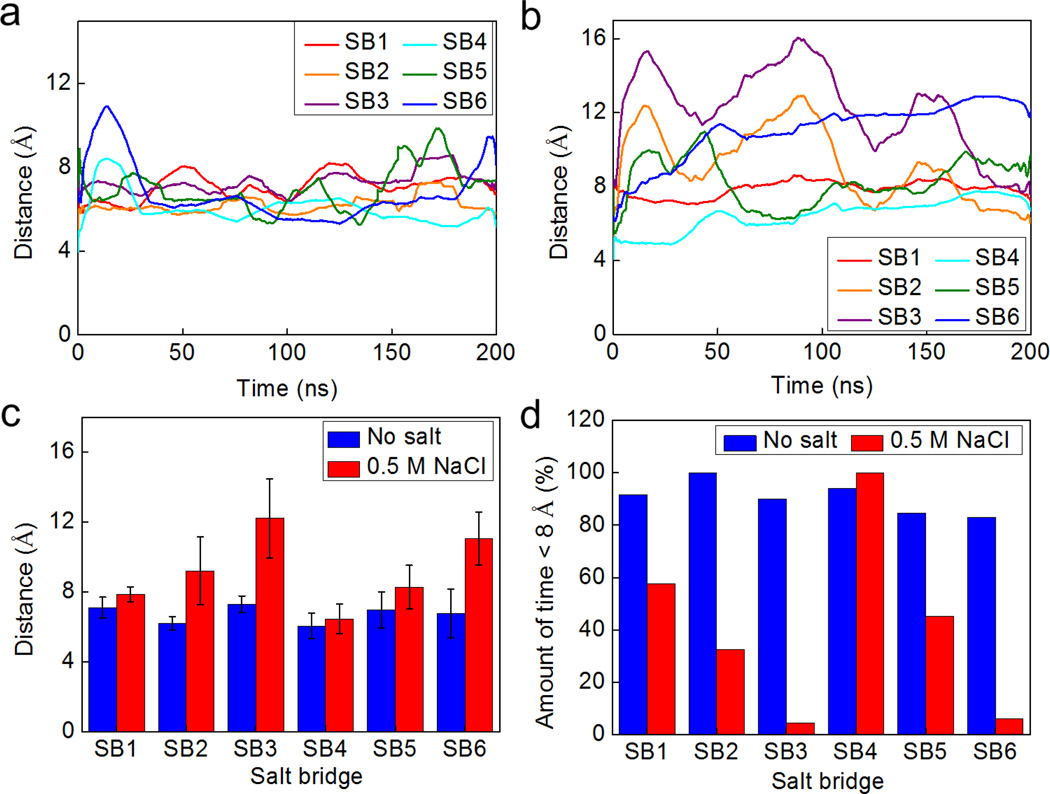
We equilibrate the coarse-grained model for 200 ns to ensure the structure is at equilibrium (convergence of RMSD as shown in Fig. S3 and potential energy as shown in Fig. S4 in the Supporting Material). Additionally we monitor the fluctuations of the distance of each salt bridge as shown in Fig. 4a and b for non-salt condition and salt condition, respectively. During the longer sampling time for the non-salt condition, all salt bridges have conserved lengths between 6 and 8 Å during the equilibration as shown in Fig. 4a. However, these salt bridges in the salt condition behave significant differently as shown in Fig. 4b, especially for SB2, SB3, and SB6, which fluctuate most significantly. We statistically summarize those results in Fig. 4c and find that all the salt bridges in the salt condition get longer than their counterparts in the non-salt condition; however, the extension is not as significant for SB1, SB4 and SB5. Moreover, SB1 and SB4 represent salt bridges with less fluctuation than all the others in the two solvent conditions, which agrees with the observation in full atomic simulations. It is noted that the distances for the salt bridges are much larger than the criterion we use in the full atomistic model (≤ 4 Å) because the coarse-grain scheme decreases the number of particles in the model, making the network sparser and thus leading to the increased salt bridge distance. A salt bridge of ~4 Å in the full atomistic model yields to ~8 Å once the model is converted to the coarse-grained form. We thereby increase the criterion to 8 Å to estimate the amount of time that each salt bridge is in effect as shown in Fig. 4d, where it is seen that in the salt condition almost all charged residue pairs stay most of the time during equilibrium at a distance to each other that does not meet the criterion of a salt bridge.
Figure 4. Fluctuation of the salt bridges within the dimer during the equilibrium at time-scales of hundreds of nanoseconds simulated by the coarse-grained model.
Panel a/b: Fluctuations of the distance between the mass center of the two amino acids involved in each of the salt bridge in non-salt condition (panel a) and for the solution with 0.5 M NaCl (panel b). Panel c: Statistical result (average ± standard deviation) for the distance of each of the salt bridge during the 200 ns equilibrium. Panel d: Amount of time the residue pair has been in an effective salt bridge state (< 8 Å for the coarse-grained model).
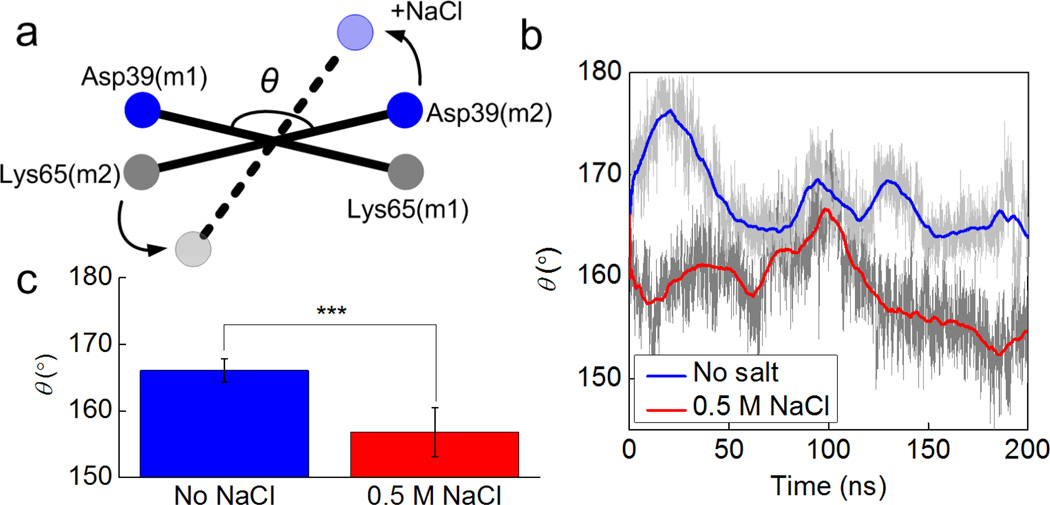
It is interesting to note that the solvent effect on the distance of salt bridges has a pattern as depicted in Fig. 4c and d (SB3>SB2>SB1 and SB6>SB5>SB4). We believe that this pattern can be explained by considering that the salt condition not only destabilizes each single salt bridge but also alters the overall conformation of the dimer as illustrated in Fig. 5a. We establish an end-to-end vector in each monomer and measure the angle between the two in forming the dimer. This angle is the same initially for the dimer in two solvent conditions but deviates from its original value during equilibration. The angle of the dimer in salt condition becomes much smaller than the angle of the dimer in non-salt condition, as shown in Fig. 5b. This difference is statistically significant during the last half of the equilibration process as shown in Fig. 5c, which means the salt condition is quite effective in altering the conformation of the homodimer. Also, the fluctuation of the measured angle is higher for the salt condition. This result provides evidence that the dimer in the salt condition is free to move while the no salt condition leads to a fixed, more aligned position. Landreh et al. (14) suggested a similar pH dependent mechanism where the dimer is able to rotate around the dimer interface at neutral pH (storing condition) and undergoes an electrostatic locking after a pH-drop as present in the spinning duct.
Figure 5. Fluctuations of the angle between two monomers in different solvent conditions.
Panel a: the schematic figure of the angle between two monomers, which is measured by the angle θ between two vectors as Asp39(m1)-Lys65(m1) and Asp39(m2)-Lys65(m2). Panel b: Fluctuations of the angle θ as a function of the simulation time. Panel c: Average and standard deviation of θ for different solvent conditions between 100 and 200 ns in the simulation as shown. We use 10 ns as the window to average θ and then perform the t-test for the 10 datasets, the result t-value=8.5 and P=0.00001, which suggests that the effect of salt on the overall dimer conformation is highly significant.
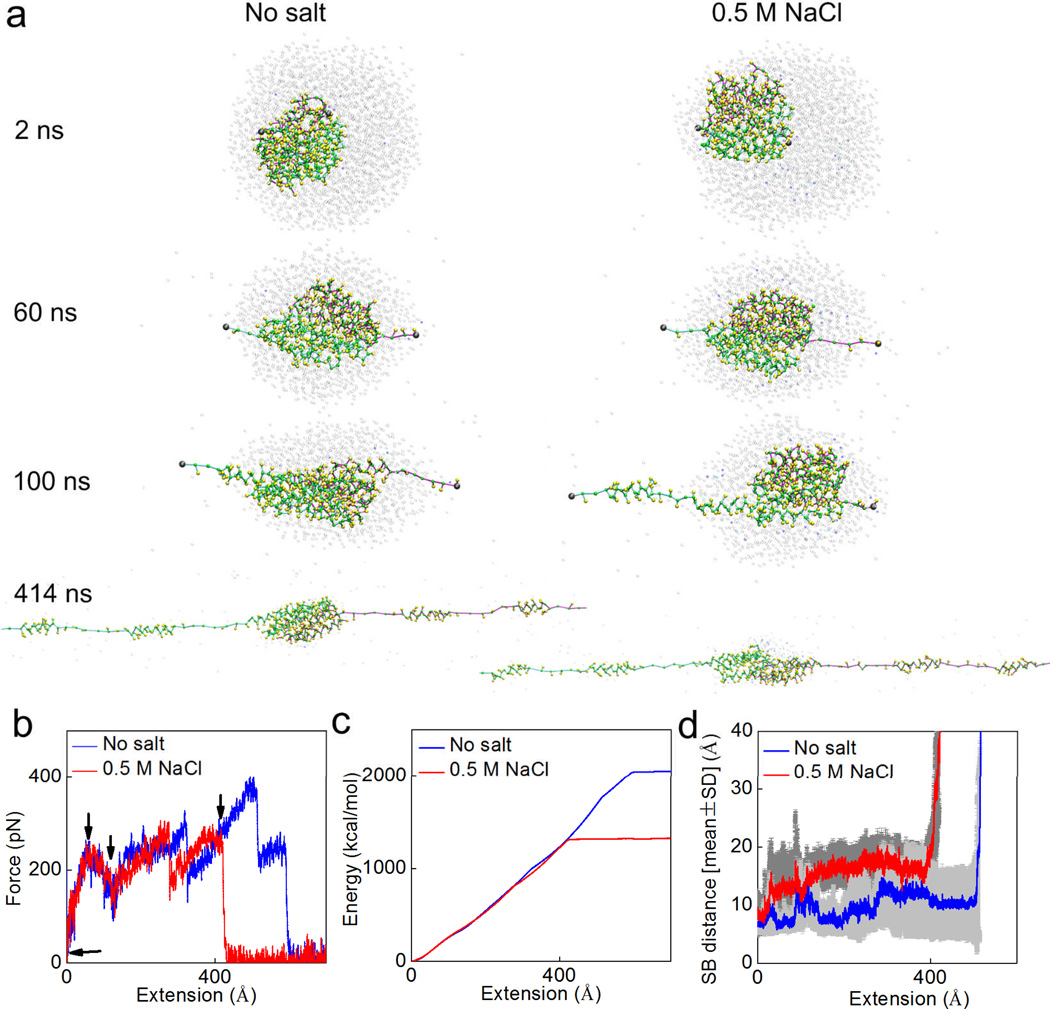
We further test the effect of salt by stretching the dimer via the SMD method in different solvent conditions. This method is similar to the tapping mode for atomic force microscopy in avoiding any direct force that instantly breaks the structure, and is thus promising to reveal a relation between mechanical and microstructural properties of proteins (25). The tensile tests, as shown in Fig. 6a, focus on the mechanical function of the N-terminal. They clearly show how strong is the assembly of the two monomers and how much energy is required to break the dimer. We apply force to the C-terminal of each monomer because it connects to the long MaSP strand that is subject to shear force during silk assembly. This test makes it clear how stable the two monomers are when they are subjected to force, which arises from the silk protein strands under shear deformation. The pulling rate in our simulation is 0.1 m/s, inspired by native spinning conditions. For instance, the vertical fall of a spider leads to physiological spinning rate ranging from 0.05 to 1.30 m/s (while the native spinning rate during web building can be lower, at ~0.02 m/s) (27–29). The results summarized in Fig. 6 show how a dimer under tension responds differently in different solvents. It is shown in Fig. 6a that for both the cases two α-helices of each monomer unfold from the bundle before the completely breaking apart of the dimer. This corresponds to the first two major force peaks. The third force peak corresponds to breaking apart as shown in Fig. 6b. It is noted that we only consider the major force peaks since the noisy peaks are caused by the oscillation of the spring applied in the SMD method to avoid large impact forces. Before breaking, there are more water molecules and ions observed at the interface of the dimer in the salt condition case, yielding to the larger salt-bridge distance before breaking as shown in Fig. 6d. The force for breaking apart of the dimer in the salt condition reaches 266 pN at a smaller extension level (415 Å versus 506 Å) and is 31.0% smaller than the breaking force obtained in the non-salt condition (383 pN). We extract the external energy applied to unfold the dimer by integrating the area below the force-extension curve as shown in Fig. 6c. The difference between those two energies is 35.5%, which only applies to the final stage before the dimer breaks apart, indicating that the salt condition destabilizes the interaction at the interface between the two monomers. The average distance of the six salt bridges during stretching as summarized in Fig. 6d clearly shows that the salt condition increases the salt bridge distance during the entire pulling process, making this interaction between the two monomers more vulnerable to the external force (or thermal fluctuations). We also point out that the simulation focuses primarily on fundamental questions at the microscopic scale. We are exploring the application of coarser models to investigate silk assembly at the macroscopic level.
Figure 6. Simulation snapshots for the deformation and mechanical response analysis during pulling apart of the dimer.
Panel a: Simulation snapshots taken as the applied extension is increased up to 400 Å. Panel b: Force-extension curve of the dimer under stretching with a rate of 0.1 m/s. Each of the arrow in this panel points to place where the corresponding snapshot in panel a is taken. Panel c: External energy applied to break the structure as a function of the extension, as the amount of area integrated below the force-extension curve. Panel d: the statistic result (average ± standard deviation for every 20 ps) of the distances of all the salt bridges (SB1–SB6) during the extension process. It is noted that the salt condition makes the dimer easier to break apart by breaking the salt bridges.
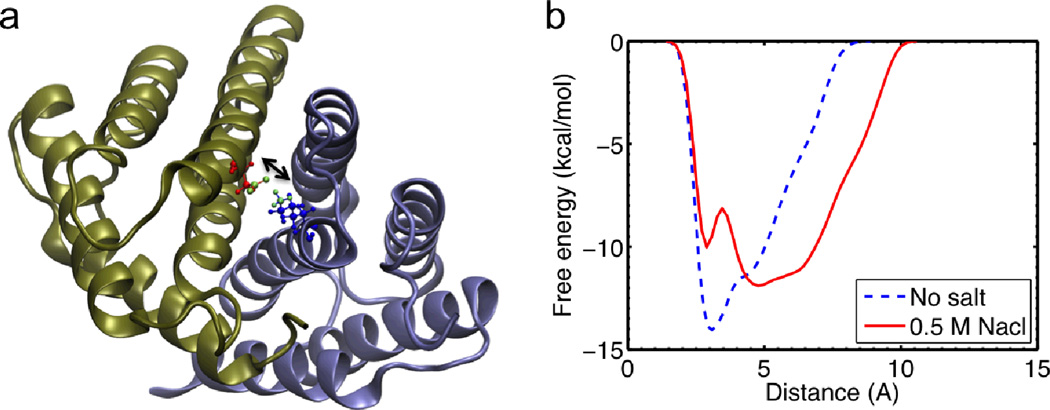
Finally, we carry out well-tempered metadynamics simulations for the full atomistic model to elucidate how a change in the salt condition leads to a decrease in the strength of the salt bridge. Metadynamics allows us to sample a much longer time-scale than it would be accessible with standard molecular dynamics. We focus on the interaction strength of SB1 as shown in Fig. 7a because it is shown to be the most relevant salt bridge at the interface to connect the two monomers within the dimer (Figs. 2 and 4). The free energy landscape as a function of the distance of SB1 is computed, and we find that the depth of the energy well keeps increasing as more bias potentials are added and it converges to the actual free-energy landscape before the end of the simulation (as shown in Fig. S2). We compare the free-energy results obtained in different solvent conditions. It is shown in Fig. 7b that for high salt condition, the initial structure is trapped in a local minimum at 4 Å, while the global energy minimum is shifted to a distance at around 5 Å. This increased distance is not counted as a salt bridge anymore, which explains the reason we obtain fewer salt bridges in the salt condition. Moreover, the height of the energy barrier for SB1 in salt condition to cross before breaking apart is 10 kcal/mol, which is 28.6% smaller than the energy barrier obtained for the non-salt condition. This energy difference agrees well with the difference in the tensile force and external energy to unfold the entire dimer (as discussed above). This salt bridge interaction is much larger than the van der Waals interaction (~1 kcal/mol) but much smaller than the covalent bond (~60 kcal/mol) and a little stronger than the hydrogen bond (9 kcal/mol). This explains why the interaction can affect the overall conformation of the dimer and act as a reversible switch to prevent or facilitate the assembly of spider silks. We believe that sodium chloride shields the electrostatic interactions between charged residues and thereby inhibits the formation of intermolecular salt bridges. Further investigation of additional collective variables would lead to a clearer picture of the free energy landscape between the charged residues at the dimer interface. Especially the inclusion of other charged residues that might have an effect on the residues of SB1 (Lys65 and Asp40) would be interesting. A likely candidate is the proximate Asp39 residue, as well as the Glu84 residue from the same subunit that is believed to form a “handshake interaction” with Asp40 (11).
Figure 7. The salt bridge pair SB1 Lys65(m1)-Asp40(m2), analysis with more detail.
Panel a: Visualization of the N-terminal domain with pertaining residues in more detail. The biased variable is the distance between NH3 of Lys65 and COO of Asp40 that are depicted in green. Panel b: Free energy profile of the distance between Lys65(monomer 1) and Asp40(monomer 2) with and without sodium chloride.
The energy landscape revealed by well-tempered metadynamics simulation in explicit solvent condition provides an accurate description of how a single salt bridge at the interface between two monomers reacts toward the state with lower free energy. The initial structure in the salt condition is trapped at a merely local energy minimum as shown in Fig. 7b while the global energy minimum is reached at longer distance between monomers. This fundamental mechanism causes the conformation change in the dimer as illustrated in Fig. 5. The existence of the energy barrier (~1.5 kcal/mol) between the initial state and the state with lower free energy and longer distance explains the difficulty to observe overall conformation change in classic molecular dynamics simulations with the full atomistic model for several tens of nanoseconds. The time span to observe an overall structure change can be theoretically estimated by τ0exp [ΔG/kBT] basing on the Bell model (30), where τ0 ≈ 0.1 ps is the reciprocal of a natural frequency of solids, ΔG = 6 × 1.5 kcal/mol is the total energy to cross for overall conformation change, kB represents the Boltzmann constant and T=300 K is the sampling temperature. It yields that at least 300 ns are necessary for overall conformation change. This explains why the overall structure change can only be observed in the simulation with the coarse-grained model over a long duration of time.
Our results demonstrate that an increased concentration of sodium chloride is important in destabilizing the homodimer. This confirms the experimental results that with a higher salt concentration the shift from monomeric state to the dimer state takes place at lower pH values. This has been seen in pull-down experiments (15) and electrospray-ionization mass spectrometry experiments (14). Divalent cations were found to destabilize the dimer state even further (15). This effect is in contrast to the role that ions play to build a bridge to crosslink protein molecules. For example, the calcium and magnesium ions (2+) in vimentin crosslink the tail domain and make the protein network more stable (31); ferric ions (3+) can stabilize many polymers into a highly crosslinked form (32). While other fibrillar molecules such as collagen and hard keratin provide strong intermolecular bonding through disulfide bonds (33, 34), dragline silk strands also form disulfide bonds at the C-terminal (12), but feature tunable bonding at the N-terminal controlled by the chemical and mechanical environment during the spinning process. This makes a reorientation of strands at the beginning of the spinning process possible, and might prevent premature assembly.
Conclusion
In this study, we modeled the dimer of the spider silk N-terminal domain in explicit solvent with different salt conditions. We use both an atomistic model and a coarse-grained model to understand the effect of salt on mechanical stability and the associated underlying molecular mechanisms. Recent experimental evidence shows that a lower salt concentration facilitates the formation of the dimers, whereas higher salt concentration favors formation of the monomers (13–15). These findings could be explained with the decreasing salt bridge strength for higher salt concentration. Furthermore, we detect a conformational change upon increasing ionic strength. This allows the monomers to rotate freely; an effect that also has been experimentally found to be linked to the pH (14). Our result implies that the interaction between the two monomers is tunable by altering the salt concentration. The crucial point for understanding silk fiber formation is that MaSp, which mainly consists of repetitive poly-Ala-rich and Gly-rich segments, quickly aggregates when passing the spinning duct without prior aggregation in the gland (which would be fatal for a spider). Our study shows that salt concentration is an important issue that strongly affects the physiological function of the solvent during the assembling process of spider silk fibers, together with the pH-value as revealed in earlier works (11, 13–16). The understanding of these key mechanisms may help us to improve synthetic silk spinning processes which have not yet been able to fully mimic the remarkable properties of natural silk. Often, the protein concentration used in synthetic approaches has to be significantly lower than seen in nature, owing to solubility problems. This and other related issues might partly explain the differences found when comparing synthetic silk to natural spider silk (35, 36). Some spinning devices are capable of mimicking the pH-drop (35, 36) and changes in ion concentrations (36), and point into the right direction of including appropriate solvent condition in the synthetic spinning process. The insights reported here may help to improve the design of these processes.
Supplementary Material
Acknowledgements
We acknowledge support from NIH (NIH/U01 EB014976), NSF and ONR, as well as the German National Academic Foundation (Studienstiftung des deutschen Volkes) (G.G.) and Dr.-Jürgen-Ulderup-Foundation (G.G.).
References
- 1.Agnarsson I, Kuntner M, Blackledge TA. Bioprospecting finds the toughest biological material: extraordinary silk from a giant riverine orb spider. Plos One. 2010;5(9):e11234. doi: 10.1371/journal.pone.0011234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410(6828):541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 3.Keten S, Buehler MJ. Geometric confinement governs the rupture strength of H-bond assemblies at a critical length scale. Nano Letters. 2008;8(2):743–748. doi: 10.1021/nl0731670. [DOI] [PubMed] [Google Scholar]
- 4.Qin Z, Buehler MJ. Cooperative deformation of hydrogen bonds in beta-strands and beta-sheet nanocrystals. Physical Review E. 2010;82(6) doi: 10.1103/PhysRevE.82.061906. 061906. [DOI] [PubMed] [Google Scholar]
- 5.Holland C, Vollrath F, Ryan AJ, Mykhaylyk OO. Silk and synthetic polymers: reconciling 100 degrees of separation. Adv Mater. 2012;24(1):105–109. doi: 10.1002/adma.201103664. [DOI] [PubMed] [Google Scholar]
- 6.Eisoldt L, Thamm C, Scheibel T. Review the role of terminal domains during storage and assembly of spider silk proteins. Biopolymers. 2012;97(6):355–361. doi: 10.1002/bip.22006. [DOI] [PubMed] [Google Scholar]
- 7.Hedhammar M, Rising A, Grip S, Martinez AS, Nordling K, Casals C, Stark M, Johansson J. Structural properties of recombinant nonrepetitive and repetitive parts of major ampullate spidroin 1 from Euprosthenops australis: Implications for fiber formation. Biochemistry. 2008;47(11):3407–3417. doi: 10.1021/bi702432y. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Knight DP, Vollrath F. Rheological characterization of Nephila spidroin solution. Biomacromolecules. 2002;3(4):644–648. doi: 10.1021/bm0156126. [DOI] [PubMed] [Google Scholar]
- 9.Knight DP, Vollrath F. Changes in element composition along the spinning duct in a Nephila spider. Naturwissenschaften. 2001;88(4):179–182. doi: 10.1007/s001140100220. [DOI] [PubMed] [Google Scholar]
- 10.Foo CWP, Bini E, Hensman J, Knight DP, Lewis RV, Kaplan DL. Role of pH and charge on silk protein assembly in insects and spiders. Applied Physics a-Materials Science & Processing. 2006;82(2):223–233. [Google Scholar]
- 11.Askarieh G, Hedhammar M, Nordling K, Saenz A, Casals C, Rising A, Johansson J, Knight SD. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature. 2010;465(7295):236–238. doi: 10.1038/nature08962. [DOI] [PubMed] [Google Scholar]
- 12.Hagn F, Eisoldt L, Hardy JG, Vendrely C, Coles M, Scheibel T, Kessler H. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature. 2010;465(7295):239–242. doi: 10.1038/nature08936. [DOI] [PubMed] [Google Scholar]
- 13.Hagn F, Thamm C, Scheibel T, Kessler H. pH-Dependent dimerization and saltdependent stabilization of the N-terminal domain of spider dragline silk-implications for fiber formation. Angew Chem Int Edit. 2011;50(1):310–313. doi: 10.1002/anie.201003795. [DOI] [PubMed] [Google Scholar]
- 14.Landreh M, Askarieh G, Nordling K, Hedhammar M, Rising A, Casals C, Astorga- Wells J, Alvelius G, Knight SD, Johansson J, Jornvall H, Bergman T. A pH-Dependent Dimer Lock in Spider Silk Protein. Journal of Molecular Biology. 2010;404(2):328–336. doi: 10.1016/j.jmb.2010.09.054. [DOI] [PubMed] [Google Scholar]
- 15.Gaines WA, Sehorn MG, Marcotte WR. Spidroin N-terminal domain promotes a pH-dependent association of silk proteins during self-assembly. Journal of Biological Chemistry. 2010;285(52):40745–40753. doi: 10.1074/jbc.M110.163121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace JA, Shen JK. Unraveling a trap-and-trigger mechanism in the pH-sensitive self-assembly of spider silk proteins. Journal of Physical Chemistry Letters. 2012;3(5):658–662. doi: 10.1021/jz2016846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 18.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 19.Nelson MT, Humphrey W, Gursoy A, Dalke A, Kale LV, Skeel RD, Schulten K. NAMD: A parallel, object oriented molecular dynamics program. International Journal of Supercomputer Applications and High Performance Computing. 1996;10(4):251–268. [Google Scholar]
- 20.Barlow DJ, Thornton JM. Ion-pairs in proteins. Journal of Molecular Biology. 1983;168(4):867–885. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 21.Barducci A, Bussi G, Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys Rev Lett. 2008;100(2) doi: 10.1103/PhysRevLett.100.020603. 020603. [DOI] [PubMed] [Google Scholar]
- 22.Bonomi M, Branduardi D, Bussi G, Camilloni C, Provasi D, Raiteri P, Donadio D, Marinelli F, Pietrucci F, Broglia RA, Parrinello M. PLUMED: a portable plugin for free-energy calculations with molecular dynamics. Computer Physics Communications. 2009;180(10):1961–1972. [Google Scholar]
- 23.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. The MARTINI coarse-grained force field: Extension to proteins. Journal of Chemical Theory and Computation. 2008;4(5):819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 24.Qin Z, Kreplak L, Buehler MJ. Hierarchical structure controls nanomechanical properties of vimentin intermediate filaments. PLoS ONE. 2009;4(10):e7294. doi: 10.1371/journal.pone.0007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316(5828):1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 26.Jaudzems K, Askarieh G, Landreh M, Nordling K, Hedhammar M, Jörnvall H, Rising A, Knight SD, Johansson J. pH-dependent dimerization of spider silk N-terminal domain requires relocation of a wedged tryptophan side chain. Journal of Molecular Biology. 2012;422(4):477–487. doi: 10.1016/j.jmb.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Breslauer DN, Lee LP, Muller SJ. Simulation of Flow in the Silk Gland. Biomacromolecules. 2009;10(1):49–57. doi: 10.1021/bm800752x. [DOI] [PubMed] [Google Scholar]
- 28.Ortlepp CS, Gosline JM. Consequences of forced silking. Biomacromolecules. 2004;5(3):727–731. doi: 10.1021/bm034269x. [DOI] [PubMed] [Google Scholar]
- 29.Vollrath F, Madsen B, Shao ZZ. The effect of spinning conditions on the mechanics of a spider's dragline silk. Proceedings of the Royal Society B-Biological Sciences. 2001;268(1483):2339–2346. doi: 10.1098/rspb.2001.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 31.Lin YC, Broedersz CP, Rowat AC, Wedig T, Herrmann H, MacKintosh FC, Weitz DA. Divalent Cations Crosslink Vimentin Intermediate Filament Tail Domains to Regulate Network Mechanics. Journal of Molecular Biology. 2010;399(4):637–644. doi: 10.1016/j.jmb.2010.04.054. [DOI] [PubMed] [Google Scholar]
- 32.Harrington MJ, Masic A, Holten-Andersen N, Waite JH, Fratzl P. Iron-Clad Fibers: A Metal-Based Biological Strategy for Hard Flexible Coatings. Science. 2010;328(5975):216–220. doi: 10.1126/science.1181044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keten S, Chou CC, van Duin ACT, Buehler MJ. Tunable nanomechanics of protein disulfide bonds in redox microenvironments. Journal of the Mechanical Behavior of Biomedical Materials. 2012;5(1):32–40. doi: 10.1016/j.jmbbm.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Buehler MJ. Nature designs tough collagen: Explaining the nanostructure of collagen fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12285–12290. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinahan ME, Filippidi E, Koster S, Hu X, Evans HM, Pfohl T, Kaplan DL, Wong J. Tunable silk: using microfluidics to fabricate silk fibers with controllable properties. Biomacromolecules. 2011;12(5):1504–1511. doi: 10.1021/bm1014624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rammensee S, Slotta U, Scheibel T, Bausch AR. Assembly mechanism of recombinant spider silk proteins. P Natl Acad Sci USA. 2008;105(18):6590–6595. doi: 10.1073/pnas.0709246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.