Abstract
Five genotypes (GI–V) of Japanese encephalitis virus (JEV) have been identified, all of which have distinct geographical distributions and epidemiologies. It is thought that JEV originated in the Indonesia-Malaysia region from an ancestral virus. From that ancestral virus GV diverged, followed by GIV, GIII, GII, and GI. Genotype IV appears to be confined to the Indonesia-Malaysia region, as GIV has been isolated in Indonesia from mosquitoes only, while GV has been isolated on three occasions only from a human in Malaysia and mosquitoes in China and South Korea. In contrast, GI–III viruses have been isolated throughout Asia and Australasia from a variety of hosts. Prior to this study only 13 JEV isolates collected from the Indonesian archipelago had been studied genetically. Therefore the sequences of the envelope (E) gene of 24 additional Indonesian JEV isolates, collected throughout the archipelago between 1974 and 1987, were determined and a series of molecular adaptation analyses were performed. Phylogenetic analysis indicated that over a 14-year time span three genotypes of JEV circulated throughout Indonesia, and a statistically significant association between the year of virus collection and genotype was revealed: isolates collected between 1974 and 1980 belonged to GII, isolates collected between 1980 and 1981 belonged to GIV, and isolates collected in 1987 belonged to GIII. Interestingly, three of the GII Indonesian isolates grouped with an isolate that was collected during the JE outbreak that occurred in Australia in 1995, two of the GIII Indonesian isolates were closely related to a Japanese isolate collected 40 years previously, and two Javanese GIV isolates possessed six amino acid substitutions within the E protein when compared to a previously sequenced GIV isolate collected in Flores. Several amino acids within the E protein of the Indonesian isolates were found to be under directional evolution and/or co-evolution. Conceivably, the tropical climate of the Indonesia/Malaysia region, together with its plethora of distinct fauna and flora, may have driven the emergence and evolution of JEV. This is consistent with the extensive genetic diversity seen among the JEV isolates observed in this study, and further substantiates the hypothesis that JEV originated in the Indonesia-Malaysia region.
Key Words: Indonesia, Japanese encephalitis virus, Molecular epidemiology
Introduction
Japanese encephalitis virus (JEV) is a flavivirus with a single-stranded, positive-sense RNA genome that is approximately 11 kilobases in length. The genome contains 5′ and 3′ untranslated regions and a single open reading frame that encodes a polyprotein that is cleaved into three structural proteins: the capsid (C), the precursor of the membrane (prM), and the envelope (E), followed by seven non-structural proteins (NS): NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5 (Chambers et al. 1990). The surface of the flavivirus particle is comprised of 90 E protein dimers (Kuhn et al. 2002), each of which contains three structurally distinct domains: domain I is the structurally central domain and acts as a hinge between domains I and III, domain II is the dimerization domain and contains the hydrophobic fusion peptide at its distal end, and domain III is the receptor-binding domain (Rey et al. 1995).
The enzootic transmission cycle of JEV involves rice-paddy-breeding culicine mosquitoes, primarily Culex tritaeniorhynchus, ardeid wading birds, and/or domestic swine. Humans and other non-avian vertebrates are considered dead-end hosts.
Five genotypes of JEV have been identified. GI is comprised of isolates collected in northern Thailand, Cambodia, Malaysia, Korea, China, Japan, Vietnam, Taiwan, northern Australia, and India between 1967 and the present; GII includes isolates that have been collected sporadically between 1951 and 1999 in Korea, southern Thailand, Malaysia, Indonesia, Papua New Guinea, and northern Australia; GIII is composed of isolates collected in mostly temperate regions of Asia between 1935 and the present; GIV consists of isolates collected only from mosquitoes on three islands encompassing the Indonesian archipelago between 1980 and 1981 (Chen et al. 1992); and GV consists of only three isolates, one from a human in Malaysia in 1952 (Mohammed et al. 2011), a second from a mosquito in China in 2009 (Li et al. 2011), and a third from a mosquito in South Korea in 2010 (Takhampunya et al. 2011).
Previous studies have proposed that JEV originated in the Indonesia-Malaysia region from an ancestral virus common to JEV and Murray Valley encephalitis virus (MVEV) (Solomon et al. 2003). From this ancestral virus, GV of JEV diverged, followed by GIV, GIII, GII, and GI (Mohammed et al. 2011). Genotypes I–III have spread throughout most of Asia and Australasia, while GIV seems to have remained confined to the Indonesia-Malaysia region (Solomon et al. 2003; Mohammed et al. 2011). To date, only 13 Indonesian JEV isolates collected between 1978 and 1981 have been genetically characterized (Chen et al. 1992), consequently limiting our understanding of the genetic variation and evolution of the virus. We hypothesized that genetic characterization of additional Indonesian JEV isolates would reveal that the extent of genetic diversity is greater than currently recognized. Therefore, we determined the nucleotide sequence of the E gene of 24 JEV isolates that were obtained from mosquitoes collected throughout the Indonesian archipelago from 1974 to 1987, and then performed a series of phylogenetic and molecular adaption analyses.
Materials and Methods
Virus isolates
The 24 JEV isolates sequenced in this study were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch (Table 1). Most of the viruses were originally isolated by scientists at the United States Naval Medical Research Unit No. 2, and the Indonesian Ministry of Health.
Table 1.
Details of the Japanese Encephalitis Virus Isolates Used in This Study
| Isolate | Origin | Year | Host | Genotype | GenBank accession no. |
|---|---|---|---|---|---|
| Ishikawab | Japan | 1998 | Mosquito | GI | ABO51292 |
| KV1899b | Korea | 1999 | Swine | GI | AY316157 |
| JEV/sw/Mie/40/2004b | Japan | 2004 | Swine | GI | AB241118 |
| XJ69b | China | 2007 | Mosquito | GI | EU880214 |
| DjAr703a | West Java | 1974 | C. quiquefasciatus | GII | JQ429287 |
| Bennettb | Korea | 1951 | Human | GII | FJ872376 |
| WTP-70-22b | Malaysia | 1970 | Mosquito | GII | HQ223286 |
| JKT654c | Kapuk, Java | 1978 | C. tritaeniorhynchus | GII | HQ223287 |
| JKT657c | Kapuk, Java | 1978 | C. tritaeniorhynchus | GII | L42157 |
| JKT1105c | Kapuk, Java | 1979 | C. gelidus | GII | L42155 |
| JKT2219c | Kapuk, Java | 1979 | C. tritaeniorhynchus | GII | L42165 |
| JKT2363c | Kapuk, Java | 1979 | C. tritaeniorhynchus | GII | L42163 |
| JKT5441 (U70406)b | Indonesia | 1981 | Mosquito | GII | U70406 |
| FUb | Australia | 1995 | Human | GII | AF217620 |
| JKT220507a | Jakarta, Java | January-1979 | Mosquito | GII | JQ429291 |
| JKT811a | Kapuk, Java | January-1979 | C. tritaeniorhynchus | GII | JQ429303 |
| JKT1110a | Kapuk, Java | February-1979 | C. tritaeniorhynchus | GII | JQ429288 |
| JKT1724a | Lombok | March-1979 | C. tritaeniorhynchus | GII | JQ429304 |
| JKT1749a | Kapuk, Java | March-1979 | C. tritaeniorhynchus | GII | JQ429305 |
| JKT2254a | Lombok | March-1979 | A. annularis | GII | JQ429293 |
| JKT2267a | Lombok | March-1979 | A. vagus | GII | JQ429294 |
| JKT1729a | Kapuk, Java | April-1979 | C. tritaeniorhynchus | GII | JQ429289 |
| JKT1754a | Kapuk, Java | April-1979 | C. tritaeniorhynchus | GII | JQ429290 |
| JKT2212a | Kapuk, Java | October-1979 | C. tritaeniorhynchus | GII | JQ429292 |
| JKT2303a | Kapuk, Java | November-1979 | C. tritaeniorhynchus | GII | JQ429295 |
| JKT2329a | Kapuk, Java | November-1979 | C. tritaeniorhynchus | GII | JQ429298 |
| JKT2352a | Kapuk, Java | November-1979 | C. tritaeniorhynchus | GII | JQ429299 |
| JKT2362a | Kapuk, Java | November-1979 | C. tritaeniorhynchus | GII | JQ429296 |
| JKT2380a | Kapuk, Java | November-1979 | C. vishnui | GII | JQ429297 |
| JKT4312a | Kapuk, Java | December-1979 | C. gelidus | GII | JQ429300 |
| JKT4331a | Kapuk, Java | December-1979 | C. tritaeniorhynchus | GII | JQ429301 |
| JKT4332a | Kapuk, Java | December-1979 | C. tritaeniorhynchus | GII | JQ429302 |
| JKT5441a | Indonesia | June-1980 | Mosquito | GII | JQ429306 |
| Matsunagab | Japan | 1939 | Human | GIII | FJ872381 |
| Roumb | Korea | 1946 | Human | GIII | FJ872377 |
| Equineb | Japan | 1947 | Equid | GIII | FJ872378 |
| Tairab | Japan | 1948 | Human | GIII | FJ515933 |
| Beijing-1b | China | 1949 | Mosquito | GIII | L48961 |
| SA14b | China | 1954 | Mosquito | GIII | JEU14163 |
| HV1b | Taiwan | 1958 | Human | GIII | AF098735 |
| JaGAr01b | Japan | 1959 | Mosquito | GIII | AF069076 |
| JaTH160b | Japan | 1960 | Human | GIII | AB269326 |
| Lingb | Taiwan | 1965 | Human | GIII | L78128 |
| JaOH0566b | Japan | 1966 | Human | GIII | AY508813 |
| GP78b | India | 1978 | Human | GIII | AF075723 |
| JKT1724 (U70404)b | Lombok | 1979 | Mosquito | GIII | U70404 |
| JKT1749 (U70405)b | Kapuk, Java | 1979 | Mosquito | GIII | U70405 |
| JaOArS982b | Japan | 1982 | Mosquito | GIII | NC_001437 |
| K87P39b | Korea | 1987 | Mosquito | GIII | AY585242 |
| CH1392b | Taiwan | 1990 | Mosquito | GIII | AF254452 |
| T1P1b | Taiwan | 1997 | Mosquito | GIII | AF254453 |
| 05734b | India | 2005 | Human | GIII | EF623988 |
| JKT27-085a | Central Java | January-1987 | Mosquito | GIII | JQ429307 |
| JKT27-087a | Central Java | January-1987 | Mosquito | GIII | JQ429308 |
| “Korea Jap B”b | Korea | circa 1950 | Unknown | GIII | FJ872379 |
| V9-3901b | Japan | circa 1950 | Human | GIII | FJ872382 |
| V9-3902b | Japan | circa 1950 | Human | GIII | FJ872383 |
| V9-4399b | Japan | circa 1950 | Human | GIII | FJ872380 |
| JKT8442c | Bali | 1980 | C. tritaeniorhynchus | GIV | L42159 |
| JKT6468b | Flores | 1981 | C. tritaeniorhynchus | GIV | AY184212 |
| JKT7003c | Java | 1981 | Mosquito | GIV | L42161 |
| JKT7887c | Java | 1981 | Mosquito | GIV | L42160 |
| JKT9092c | Bali | 1981 | Mosquito | GIV | L42158 |
| JKT7089a | Bantul, Java | June-1981 | C. vishnui | GIV | JQ429309 |
| JKT7180a | Central Java | July-1981 | C. tritaeniorhynchus | GIV | JQ429310 |
| MVE-1-51d | Australia | 1951 | Human | AF161266 |
Indonesian viruses sequenced in this study.
The nucleotide sequences of these viruses were retrieved from GenBank to generate phylogenies.
The nucleotide sequences of these Indonesian JEV isolates were downloaded from GenBank to determine the overall distribution of Indonesian JEV isolates according to year of collection and genotype.
All viruses are JEV except for MVE-1-51, which is a strain of Murray Valley encephalitis virus.
C., Culex; A., Anopheles.
Cell culture, virus growth, RNA extraction, RT-PCR of the E gene, DNA purification, and sequencing
These were performed as described previously (Schuh et al. 2010, 2011).
Phylogenetic analyses
Phylogenetic relationships were inferred using the neighbor-joining (NJ) and maximum-parsimony (MP) methods within the PHYLIP package (Felsenstein, 1989), as well as with the maximum-likelihood (ML) method implemented in the PHYML program (Guindon et al. 2003). The robustness of the phylogenies was evaluated by bootstrap resampling with 1000 replicates. Trees were visualized using FigTree v1.1.2 (Rambaut, 2008).
Distribution of Indonesian JEV isolates according to year of collection and genotype
To test the null hypothesis that there is no association between the year of collection of the Indonesian JEV isolates and their genotype, a list of all Indonesian JEV isolates for which nucleotide sequence information were available was created, in addition to the isolates that were sequenced in this study (Table 1). The relationships between: (1) the year of collection of all of the Indonesian isolates and genotype, and (2) the year of collection of Java isolates only and genotype, was evaluated using the Pearson's chi-square test with α=0.05, that was calculated based on the exact distribution of the test statistic (IBM SPSS Statistics version 19). Post-hoc analyses were then performed to determine which cells in the table of year of collection versus genotype contributed most to the statistically significant Pearson's chi-square value. Residuals (the difference between the observed and the expected frequency), and standardized residuals/z-scores (SR) were calculated, and then the standardized residuals were compared against the critical z-value (±1.96) for p=0.05 (IBM SPSS Statistics version 19).
Recombination and molecular adaptation analyses
The E gene sequence alignment was examined for evidence of recombination as described previously (Schuh et al. 2011).
All molecular adaptation analyses were performed using methods available on the Datamonkey webserver (Pond et al. 2005). Either the E gene sequence alignment (six duplicate sequences were removed), or the E protein sequence alignment (16 duplicate sequences were removed), was used to perform the analyses.
The E gene sequence alignment was analyzed for selective pressures by calculating the global ratio of non-synonymous (dN) to synonymous (dS) nucleotide substitutions. Additionally, evidence of positive selection (dN:dS>1) was evaluated using the single-likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), internal FEL (IFEL), and relative effects likelihood (REL) methods, using a NJ phylogeny and the reversible nucleotide substitution model (Kosakovsky Pond et al. 2005). Statistically significant evidence of positive selection was guided by a p value<0.05 for the SLAC, FEL, and IFEL methods, and a BF>100 for the REL method.
Evidence of directional selection within the E protein alignment was assessed using the directional evolution in protein sequences (DEPS) method (Kosakovsky Pond et al. 2008). Statistically significant shifts in amino acid residue frequencies (p<0.05), and/or a significantly large number of substitutions towards a particular residue (empirical Bayes factor >100), were examined over the phylogeny using the Jones, Taylor, Thorton (JTT) amino acid substitution model and an NJ phylogeny.
Identification of co-evolving sites within the E protein alignment was performed using the Spidermonkey method (Poon et al. 2007). The JTT amino acid substitution model was used to estimate substitutions over a NJ phylogeny. Reconstructed ancestral sequences were resampled, a two-parent directed network was used, and sites were filtered based on a minimum number of substitutions across the phylogeny (threshold ≥2). Sites with a posterior probability ≥0.90 were reported.
Results
Geographical distribution of the Indonesian JEV isolates
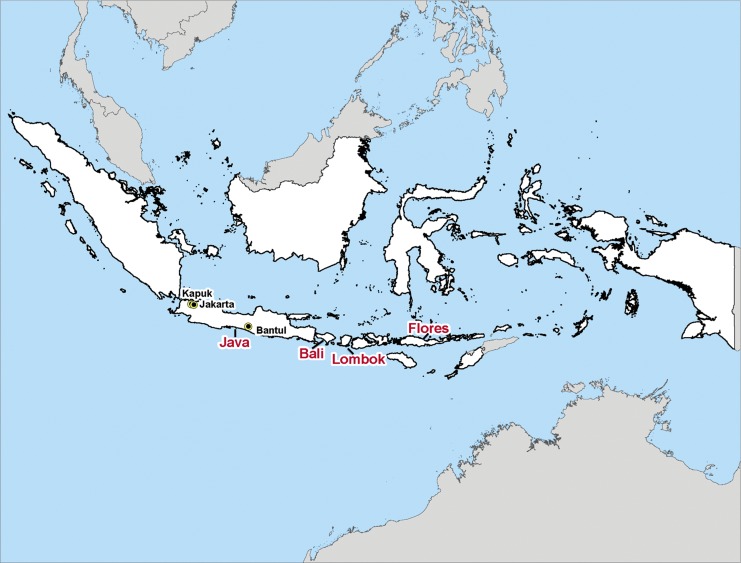
Figure 1 depicts the geographical distribution of the 24 Indonesian JEV isolates sequenced in this study, as well as 10 others that were retrieved from GenBank (Table 1).
FIG. 1.
Map of the Indonesian archipelago. The islands comprising Indonesia are shown in white, large text indicates the islands where Japanese encephalitis virus (JEV) isolates included in this study were collected, and small text indicates the cities in Java where JEV isolates included in this study were collected. Twenty-seven of the Indonesian isolates were from the island of Java (one from Jakarta, 19 from Kapuk, one from West Java, three from Central Java, one from Bantul, and two from unspecified locations in Java), two were from the island of Bali, three were from the island of Lombok, one was from the island of Flores, and one was from an unspecified location in Indonesia. The map was created using the ArcView GIS version 9.1 (Environmental Research Systems Institute 2004) from geographic boundary files downloaded from the DIVA-GIS, Geodatabase (DIVA-GIS, 2011). (Color image available online at www.liebertpub.com/vbz).
Genetic relationships among the Indonesian JEV isolates
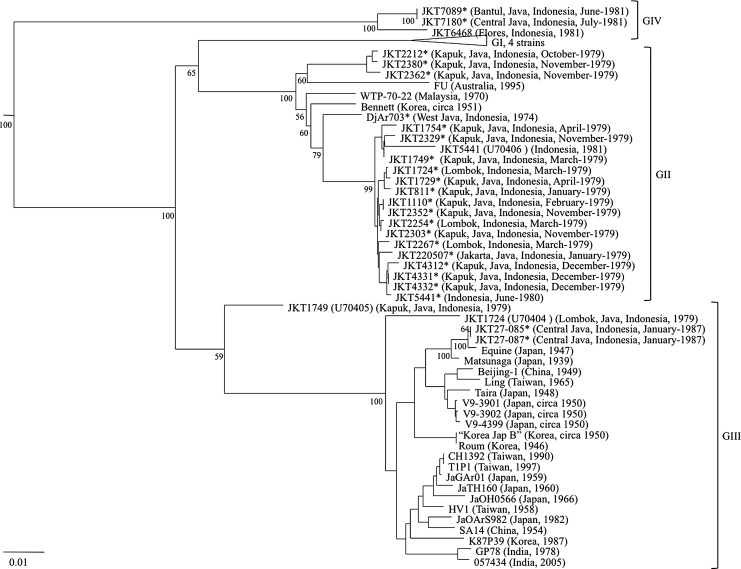
Phylogenies generated using all three methods of phylogenetic inference sequence (NJ, MP, and ML) identified four major clades (representing GI–IV; Fig. 2). Twenty of the 24 Indonesian JEV isolates sequenced in this study belonged to GII (collected between 1974 and 1979 in Jakarta, Kapuk, West Java, and Lombok), two isolates belonged to GIII (collected in 1987 in Central Java), and two isolates belonged to GIV (collected in 1981 in Bantul and Central Java).
FIG. 2.
Neighbor-joining phylogeny based on nucleotide sequence information derived from the E gene of the Japanese encephalitis virus (JEV) isolates. The tree was rooted using the MVE-1-51 isolate of Murray Valley encephalitis virus, which is a member of the Japanese encephalitis serogroup, but has been removed to allow for better visualization of branch lengths. Horizontal branch lengths are proportional to the genetic distance between strains, and the scale at the lower-left of the tree indicates the number of nucleotide substitutions per site. Genotype (GI–IV) is represented to the right of the tree. Bootstrap percentages based on 1000 replicates are indicated to the lower-left of the genotype-defining nodes within the phylogeny, as well as additional selected nodes. The Indonesian isolates sequenced in this study are indicated by asterisks.
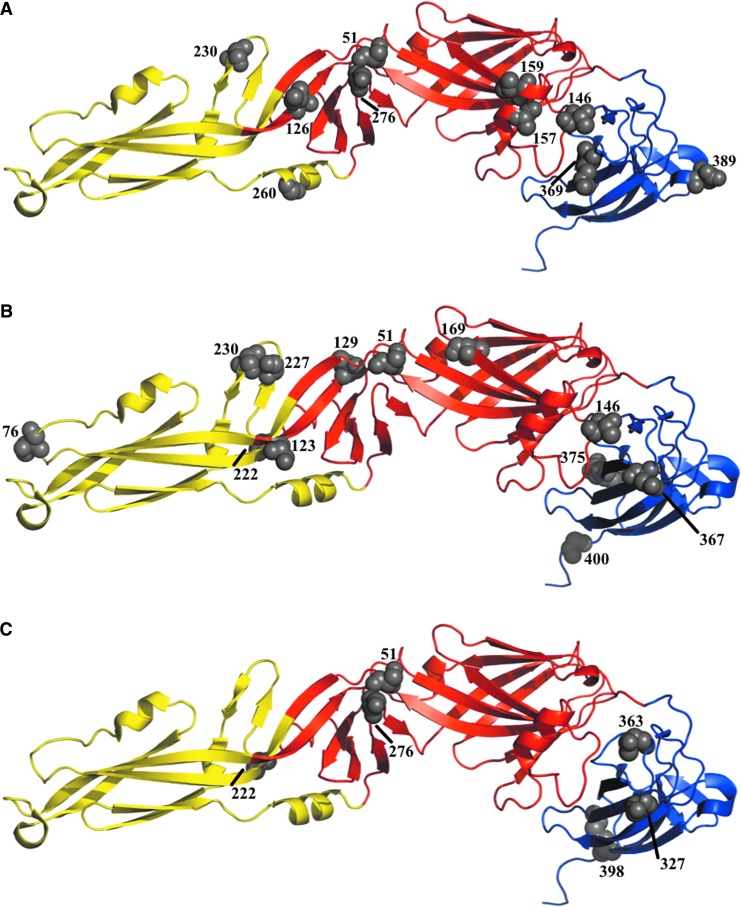
The topology of the phylogeny was compared to the deduced amino acid alignment to identify coding substitutions that could be involved in phylogenetic divergence and potentially result in phenotypic changes. Node-defining amino acid substitutions within the E protein that were specific to the Indonesian viruses sequenced in this study were determined and modeled onto the predicted three-dimensional crystal structure of the E protein of JEV (Fig. 3A).
FIG. 3.
The location of important amino acids sites mapped onto the three-dimensional E protein structure of Japanese encephalitis virus (JEV) (PBD ID: 3P54, MMDB ID: 87213): (A) Node-defining amino acid substitutions specific to the Indonesian viruses sequenced in this study. (B) Sites identified by the directional evolution in protein sequences (DEP)S analysis to exhibit evidence of directional evolution. (C) Sites detected by the Spidermonkey algorithm to display evidence of co-evolution. Domain I is indicated in red, domain II in yellow, and domain III in blue. The image was created using MacPyMOL version 1.3. (Color image available online at www.liebertpub.com/vbz).
Genotype II of the phylogeny is composed of two major groups of viruses. The first group is comprised of 3 Indonesian isolates (JKT2212, JKT2380, and JKT2362 [Kapuk, Java, and October/November 1979]), that were sequenced in this study and share the A146T node-defining amino acid substitution, as well as the FU isolate (Australia, 1995). The second group is comprised of the remaining 17 GII Indonesian isolates sequenced here: WTP-70-22 (Malaysia, 1970) diverged first, followed by the Bennett (Korea, circa 1951), and then the DjAr703 isolate (West Java, 1974). The DjAr703 isolate is the oldest virus and is ancestral to the other 16 GII isolates that were all collected in 1979 in Java, Lombok, or undefined locations within Indonesia. Four of these viruses (JKT4312, JKT4331, JKT4332, and JKT5441) share the S230N node-defining amino acid substitution.
Two of the viruses sequenced in this study grouped together within GIII (JKT27-085 and JKT27-087 [Central Java, January 1987]), and share the A157T node-defining amino acid substitution. These two viruses grouped closely with the equine isolate (Japan, 1947), and these three viruses are defined by two node-defining amino acid substitutions (A51V and S276N).
The JKT7089 (Bantul, Java, June 1981), and JKT7180 (Central Java, July 1981) isolates sequenced here belonged to GIV, along with the JKT6468 isolate (Flores, 1981). When the JKT7089 and JKT7180 isolates were compared with the JKT6468 isolate, six amino acid substitutions were noted: T126I, I159V, R260G, Q369K, E389D, and T490I.
Re-examination of three previously sequenced Indonesian JEV strains
The phylogeny revealed that three previously sequenced viruses (JKT1749 [U70405], JKT1724 [U70404], and JKT5441 [U70406]) were not phylogenetically distributed according to the pattern observed in this study (year of collection versus genotype; Fig. 2). Given these discrepancies we re-sequenced the E gene of these three viruses, and found that the JKT1724, JKT1749, and JKT5441 isolates belonged to GII (Fig. 2).
Distribution of Indonesian JEV isolates according to year of collection and genotype
Indonesian isolates of JEV that were sequenced in this study, and isolates for which nucleotide sequence information was retrieved from GenBank, were categorized according to their year of collection and genotype (Table 2).
Table 2.
Distribution of Indonesian Japanese Encephalitis Virus Isolates According to Year of Collection and Genotypea
| |
Year |
||||||
|---|---|---|---|---|---|---|---|
| Genotype | 1974 | 1978 | 1979 | 1980 | 1981 | 1987 | Total |
| II | 1 | 1 | 22 | 1 | 0 | 0 | 25 (73.5%) |
| III | 0 | 0 | 0 | 0 | 0 | 2 | 2 (5.9%) |
| IV | 0 | 0 | 0 | 1 | 6 | 0 | 7 (20.6%) |
| Total | 1 (2.9%) | 1 (2.9%) | 22 (64.7%) | 2 (5.9%) | 6 (17.6%) | 2 (5.9%) | 34 (100.0%) |
JEV isolates for which there is nucleotide sequence information available, including isolates sequenced in this study and isolates for which the nucleotide sequence was retrieved from GenBank.
A Pearson's chi-square test was used to test the null hypothesis that there is no association between the year of virus collection and genotype. Based on an alpha level <0.05, we rejected the null hypothesis and concluded that there is a relationship between the year of virus collection and genotype (Pearson's chi-square=64.891, exact p value=0.000). Post-hoc analysis revealed that among virus isolates collected in 1979 and 1981, there were fewer GIV (SR=−2.1) and GII (SR=−2.1) isolates than expected, respectively. Furthermore, among virus isolates collected in 1981 and 1987, there were more GIV (SR=4.3) and GIII (SR=5.5) isolates than expected, respectively.
Given that the majority of the Indonesian JEV isolates were collected in Java, the hypothesis was re-tested using Java isolates only. Again, we rejected the null hypothesis and concluded that there was a relationship between the year of virus collection and genotype in Java (Pearson's chi square=50.000, exact p value=0.000). Post-hoc analysis indicated that among virus isolates collected in 1981 and 1987, in Java there were more GIV (SR=4.2) and GIII (SR=4.6) isolates than expected, respectively.
Recombination and molecular adaptation analyses
No evidence of recombination was obtained.
The global dN:dS ratio across the E gene alignment was 0.055 (95% CI: 0.045,0.066), which suggested the occurrence of predominantly purifying selection (virus-encoded proteins are conserved over time due to the selective pressure against deleterious variants). No sites were identified to be under positive selection using the SLAC, FEL, IFEL, and REL methods.
The DEPS analysis revealed elevated amino acid substitution rates towards seven residues: A, I, M, N, P, T, and V (Table 3). Of these seven residues, directional evolution towards P was subjected to the strongest bias (83.26), but affected the smallest proportion of sites (1.35%), whereas evolution towards T was subjected to a weak bias (4.31), but affected the largest proportion of sites (16.36%; Table 3). Fifteen sites were found to be involved in this directional evolution: 51, 76, 123, 129, 146, 169, 222, 227, 230, 367, 375, 400, 474, 483, and 484 (Table 4). The locations of several of these sites are indicated on the predicted three-dimensional crystal structure of the E protein of JEV (Fig. 3B).
Table 3.
Directional Selection Analysis of the E Protein Sequences
| Residuea | p Valueb | Biasc | Proportion (%)d | No. of sitese |
|---|---|---|---|---|
| A | 0.0000 | 3.09 | 12.5 | 3 |
| I | 0.0000 | 23.05 | 3.07 | 3 |
| M | 0.0006 | 51.29 | 1.75 | 3 |
| N | 0.0000 | 34.30 | 2.17 | 2 |
| P | 0.0000 | 83.26 | 1.35 | 2 |
| T | 0.0000 | 4.31 | 16.63 | 2 |
| V | 0.0025 | 20.97 | 3.34 | 2 |
The target residue for directional selection.
The p value associated with the test of directional versus non-directional selection.
The relative rate of amino acid substitution across the alignment towards the target site.
The percentage of sites that evolve under a directional model versus a non-directional model.
The number of sites that show evidence of directional selection for the particular site.
Table 4.
Amino Acid Sites within the Envelope Protein That Were Found to Be Under Directional Selection
| Site | Compositiona | MRCAb | Target | Inferred substitutionsc | DEPS EBF |
|---|---|---|---|---|---|
| 51 | S35V2T1 | S | V | S0↔1T, S0↔2V | 298.3 |
| 76 | T36M2 | T | M | M2↔0T | 692.7 |
| 123 | S33N3R2 | S | N | N3↔0S, R2↔0S | 6656.3 |
| 129 | T33M4L1 | T | M/T | L1↔0T, M4↔1T | >105/>105 |
| 146 | T32A5S1 | T | A/T | A5↔0T, S1↔0T | 200.6/122.6 |
| 169 | V34I4 | V | I | I4↔0V | >105 |
| 222 | A19S18L1 | A | A | A0↔1L, A14↔6S | 2558.1 |
| 227 | S34P4 | S | P | P4↔0S | >105 |
| 230 | S34N4 | S | N | N4↔0S | >105 |
| 367 | A19S19 | A | A | A11↔36S | 1637.4 |
| 375 | M36I2 | M | I | I2↔0M | 126.2 |
| 400 | A35P2E1 | A | P | A0↔1E, A0↔2P | 1594.9 |
| 474 | V33I5 | V | I | I5↔0V | >105 |
| 483 | L36M2 | L | M | L0↔2M | 318.8 |
| 484 | A35V3 | A | V | A0↔3V | 1654.1 |
Amino acid composition at the site.
Reconstructed most recent common ancestor (MRCA) at the site.
Amino acid substitutions inferred when Ab↔cD indicates b substitutions from A to D and c substitutions from D to A.
Empirical Bayes factor for evidence in favor of a directional selection model at the site for the target residue.
DEPS, directional evolution in protein sequences.
The Spidermonkey method detected three sets of sites that exhibited evidence of co-evolution: (1) S51V and S276N, (2) S222A and T327S, and (3) T363A and K398R (Table 5). These sites are indicated on the predicted three-dimensional crystal structure of the E protein of JEV (Fig. 3C).
Table 5.
Co-evolving Sites Within the E Protein
| Site 1a | Site 2a | No. of isolatesb | Posterior probabilityc | Supporting replicates (%)d |
|---|---|---|---|---|
| S51V | S276N | 2 | 0.90 | 99 |
| S222A | T327S | 19 | 0.98 | 99 |
| K398R | T363A | 2 | 0.94 | 99 |
The amino acid substitution pattern.
The number of isolates in the analysis that possessed the given pattern of co-evolution.
The posterior probability for site 1 and site 2 being conditionally dependent.
The percentage of ancestral replicates that have a posterior probability≥to that of the maximum likelihood ancestral state.
Discussion
Although it has been suggested that the Indonesia-Malaysia region is the origin of JEV evolution, prior to this study only 13 isolates collected within a 4-year time span from Indonesia had been studied genetically. Therefore, the objective of the current study was to determine the extent of genetic variation among Indonesian JEV isolates by expanding the phylogenetic analysis of Chen and associates (1992), through the incorporation of 24 newly sequenced Indonesian isolates collected between 1974 and 1987. This study revealed a statistically significant association between the year of virus collection and genotype: isolates collected between 1974 and 1979 belonged to GII, isolates collected between 1980 and 1981 belonged to GIV, and isolates collected in 1987 belonged to GIII. Further analysis indicated that among virus isolates collected in 1979 there were fewer GIV isolates than expected, among virus isolates collected in 1981 there were fewer GII isolates and more GIV isolates than expected, and among virus isolates collected in 1987 there were more GIII isolates than expected. It is interesting to note that three genotypes of JEV circulated throughout Indonesia over a time-span of 14 years, whereas with the exception of one instance (Schuh et al. 2010), GIII circulated exclusively throughout northern and western Asia for at least 45 years (collection of the prototype GIII isolate in Japan in 1935 to the collection of the GI YN79-Bao83 GI isolate in China in 1979). Although GI has recently emerged and displaced GIII as the most frequently isolated virus genotype throughout most of Asia (Nga et al. 2004; Nitatpattana et al. 2008), a GI virus has never been isolated in Indonesia. However, several GI isolates have been collected in neighboring northern Malaysia (Tsuchie et al. 1997), and isolates representative of genotypes II, III, and V have been collected throughout Malaysia (Tsuchie et al. 1994, 1997; Mohammed et al. 2011). The tropical climate of the Indonesia-Malaysia region coupled with the vast array of distinct fauna and flora present throughout the region may have facilitated the emergence and evolution of JEV. This is consistent with the extensive genetic diversity observed among isolates collected from this region, as well as with the previously proposed hypothesis that JEV originated from an ancestral virus in the Indonesia-Malaysia region (Solomon et al. 2003; Mohammed et al. 2011).
Three previously sequenced Indonesian isolates (JKT1749 [U70405], JKT1724 [U70404], and JKT5441 [U70406]) were re-sequenced here because they were not phylogenetically distributed according to the pattern observed in this study (year of collection versus genotype), and a previous study revealed that the JKT1749 [U70405] and JKT1724 [U70404] isolates belonged to GIII in an E gene phylogeny, but belonged to GII in a prM gene phylogeny (Williams et al. 2000). Consistent with the findings of Williams and colleagues, re-sequencing of the E gene of JKT1749 and JKT1724 revealed that these two isolates did indeed belong to GII.
Twenty of the 24 Indonesian JEV isolates sequenced in this study belonged to GII, three of which (JKT2212, JKT2380, and JKT2362 [all from Kapuk, Java, October/November 1979]) group together and share the A146T node-defining amino acid substitution. Site 146 was also found to be under directional evolution. The JKT2212, JKT2380, and JKT2362 isolates share a most recent common ancestor with the FU isolate, from an asymptomatic human during the 1995 JE outbreak that occurred on Badu Island, which is located in the Torres Strait between Queensland, Australia and Papua New Guinea (Hanna et al. 1996). A subsequent study found that the JEV isolates from Papua New Guinea were >99% identical to virus isolates collected during the 1995 outbreak on Badu Island, and therefore concluded that JEV may have been introduced from Papua New Guinea to Australia (Johansen et al. 2000). The close phylogenetic grouping between the three Indonesian isolates collected in 1979 and the FU isolate suggest that JEV may have been introduced to the Papua New Guinea-Australia region from Indonesia sometime between 1979 and 1995. A previous hypothesis suggests that JEV gradually moved eastward along the Indonesian archipelago, through natural mosquito-bird and mosquito-pig transmission cycles, until it arrived undetected in Papua New Guinea (Mackenzie et al. 2002). In fact, strong serological evidence suggests that JEV arrived in Papua New Guinea in the late 1980s (Mackenzie et al. 2002). Although flaviviral serological results are difficult to interpret due to heterotypic reactivity, it is noteworthy that JEV-seropositive swine were observed in Timor and Irian Jaya between 1989 and 1991 (Mackenzie et al. 2002). It is also plausible that JEV may have been introduced into Papua New Guinea and subsequently into Australia through wind-blown mosquitoes (Ritchie et al. 2001), black flying foxes (Pteropus spp.; van den Hurk et al. 2009), and/or wind-blown biting midges (Culicoides spp.; Mackenzie et al. 2002).
The remaining 17 GII Indonesian isolates sequenced here (collected in Java, Lombok, and unknown locations in Indonesia) were descendants of the WTP-70-22 (Malaysia, 1970) and Bennett (Korea, circa 1951) isolates, and four of these viruses (JKT4312, JKT4331, JKT4332, and JKT5441) share the S230N node-defining amino acid substitution. Site 230 was also found to be under directional evolution. Between 1972 and 1974, the time period coinciding with the collection of the DjAr703 isolate (West Java, 1974), the United States Naval Medical Research Unit No. 2 and the Indonesian Ministry of Health collected mosquitoes from Kapuk, Java, ultimately resulting in 12 isolates of JEV (Dirk Van Peenen et al. 1975). During the study period, Kapuk was an area of Jakarta reserved for the raising and slaughter of swine, and 11 of the 12 JEV isolates that were made came from pools of C. tritaeniorhynchus, thereby implicating domestic swine and C. tritaeniorhynchus in the enzootic transmission cycle of JEV (Dirk Van Peenen et al. 1975). An additional 19 isolates of JEV were made from mosquitoes (C. tritaeniorhynchus, geldius, vishnui, and fuscocephala), captured between October 1978 and April 1980 in another study that took place in Kapuk, Java, and 14 of these isolates were sequenced in this study (11 of these isolates were descendants of the DjAr703 isolate, and 3 of these isolates share a most recent common ancestor with the FU isolate mentioned above; Olson et al. 1985b). Three of the 17 GII Indonesian viruses (JKT1724, JKT2254, and JKT2267) were collected in villages near Gerung, West Lombok during March of 1979 (Olson et al. 1985a), where a previous study indicated a low human seroprevalence of JEV (Olson et al. 1983). The JKT1724 isolate was obtained from a pool of C. tritaeniorhynchus feeding on domestic buffalo or resting in buffalo stables, the JKT2254 isolate was made from a pool of Anopheles annularis collected from buffalo stables, and the JKT2267 isolate was made from a pool of Anopheles vagus collected by CDC light traps (Olson et al. 1985a). Interestingly, the minimum frequencies of JEV infection in zoophilic Anopheles spp. were higher than in C. tritaeniorhynchus (Olson et al. 1985a). It has been previously suggested that the relatively low minimum frequency of JEV infection in C. tritaeniorhynchus, coupled with the absence of swine from Lombok, may have been responsible for the low seroprevalence of JEV among humans residing in this area (Olson et al. 1985a).
Two of the 24 Indonesian JEV isolates sequenced in this study belonged to GIII (JKT27-085 and JKT27-087 [both Central Java, January 1987]), and share the A157T node-defining amino acid substitution. These two isolates grouped closely with the equine isolate (Japan, 1947), and together these three isolates share the S51V and S276N node-defining amino acid substitutions. Sites 51 and 276 also exhibited evidence of co-evolution. It is interesting to note that they lie adjacent to each other within domain I of the three-dimensional structure of the E protein of JEV. The close phylogenetic and evolutionary relationships between the JKT27-085, JKT27-087, and equine isolates may indicate that the viruses responsible for the major JE epidemics occurring in Japan in 1924, 1935, and 1948 (Innis, 1995) evolved in Indonesia, and were subsequently transported by migratory birds traveling along the East Asian-Australasian flyway to Japan.
Two of the 24 Indonesian JEV isolates sequenced in this study belonged to GIV (JKT7089 [Bantul, Java, June 1981] and JKT7180 [Central Java, July 1981]). These two isolates possessed six amino acid substitutions in the E protein when compared to the previously sequenced JKT6468 (Flores, 1981) isolate (T126I, I159V, R260G, Q369K, E389D, and T490I). Notably, these two GIV isolates, as well as the other five GIV isolates, were all collected from mosquitoes (three isolates were made from mosquito pools, three isolates were made from C. tritaeniorhynchus mosquitoes, and one isolate was made from a Culex vishnui mosquito) in Indonesia (four isolates were collected in Java, two isolates were collected in Bali, and one isolate was collected in Flores) between 1980 and 1981 (one isolate was collected in 1980 and six isolates were collected in 1981). As GIV has not been collected since 1981, it remains unknown whether this genotype is still circulating in Indonesia or if it has become extinct. The reasons underlying the failure of GIV to spread beyond the Indonesian archipelago are unknown, but could be due to one or more factors. The vector competence of C. tritaeniorhynchus, the primary vector for JEV throughout Asia, for GIV of JEV may be lower compared to the other three virus genotypes; GIV may preferentially infect and disseminate to the salivary glands in a mosquito species that does not exist throughout Asia; the replicative ability of GIV in birds may be poor, implying that birds infected with this genotype may not produce sufficient viremias to transport the virus to mainland Asia; or the GIV transmission cycle may involve an amplifying host that is a non-migratory bird, and/or the GIV transmission cycle may involve a non-avian amplifying host that is either non-migratory or is geographically confined to Indonesia.
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grant AI 067847 and contract HHSN2722010000401/HHSN27200004/D04. A.J.S. is supported by NIH T32 training grant AI 60549.
Author Disclosure Statement
No competing financial interests exist.
References
- Chambers TJ. Hahn CS. Galler R, et al. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chen WR. Rico-Hesse R. Tesh RB. A new genotype of Japanese encephalitis virus from Indonesia. Am J Trop Med Hyg. 1992;47:61–69. doi: 10.4269/ajtmh.1992.47.61. [DOI] [PubMed] [Google Scholar]
- Dirk Van Peenen PF. Joseph PL. Atmosoedjono S, et al. Japanese encephalitis virus from pigs and mosquitoes in Jakarta, Indonesia. Trans R Soc Trop Med Hyg. 1975;69:477–479. doi: 10.1016/0035-9203(75)90102-9. [DOI] [PubMed] [Google Scholar]
- Environmental Research Systems Institute I. ArcView GIS. 9.1. Redlands; California: 2004. [Google Scholar]
- Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Free spatial data [database on the Internet] 2011. Mar 10, 2011. http://www.diva-gis.org/Data http://www.diva-gis.org/Data
- Guindon S. Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hanna JN. Ritchie SA. Phillips DA, et al. An outbreak of Japanese encephalitis in the Torres Strait, Australia, 1995. Med J Aust. 1996;165:256–260. doi: 10.5694/j.1326-5377.1996.tb124960.x. [DOI] [PubMed] [Google Scholar]
- Innis B. Japanese Encephalitis. In: Porterfield J, editor. Exotic Viral Infections. London: Chapman & Hall; 1995. [Google Scholar]
- Johansen CA. van den Hurk AF. Ritchie SA, et al. Isolation of Japanese encephalitis virus from mosquitoes (Diptera: Culicidae) collected in the Western Province of Papua New Guinea, 1997–1998. Am J Trop Med Hyg. 2000;62:631–638. doi: 10.4269/ajtmh.2000.62.631. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL. Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL. Poon AF. Leigh Brown AJ, et al. A maximum likelihood method for detecting directional evolution in protein sequences and its application to influenza A virus. Mol Biol Evol. 2008;25:1809–1824. doi: 10.1093/molbev/msn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ. Zhang W. Rossmann MG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MH. Fu SH. Chen WX, et al. Genotype v Japanese encephalitis virus is emerging. PLoS Negl Trop Dis. 2011;5:e1231. doi: 10.1371/journal.pntd.0001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS. Johansen CA. Ritchie SA, et al. Japanese encephalitis as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia. Curr Top Microbiol Immunol. 2002;267:49–73. doi: 10.1007/978-3-642-59403-8_3. [DOI] [PubMed] [Google Scholar]
- Mohammed MA. Galbraith SE. Radford AD, et al. Molecular phylogenetic and evolutionary analyses of Muar strain of Japanese encephalitis virus reveal it is the missing fifth genotype. Infect Genet Evol. 2011;11:855–862. doi: 10.1016/j.meegid.2011.01.020. [DOI] [PubMed] [Google Scholar]
- Nga PT. del Carmen Parquet M. Cuong VD, et al. Shift in Japanese encephalitis virus (JEV) genotype circulating in northern Vietnam: implications for frequent introductions of JEV from Southeast Asia to East Asia. J Gen Virol. 2004;85(Pt 6):1625–1631. doi: 10.1099/vir.0.79797-0. [DOI] [PubMed] [Google Scholar]
- Nitatpattana N. Dubot-Peres A. Gouilh MA, et al. Change in Japanese encephalitis virus distribution, Thailand. Emerg Infect Dis. 2008;14:1762–1765. doi: 10.3201/eid1411.080542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JG. Ksiazek TG. Gubler DJ, et al. A survey for arboviral antibodies in sera of humans and animals in Lombok, Republic of Indonesia. Ann Trop Med Parasitol. 1983;77:131–137. doi: 10.1080/00034983.1983.11811687. [DOI] [PubMed] [Google Scholar]
- Olson JG. Ksiazek TG. Lee VH, et al. Isolation of Japanese encephalitis virus from Anopheles annularis and Anopheles vagus in Lombok, Indonesia. Trans R Soc Trop Med Hyg. 1985a;79:845–847. doi: 10.1016/0035-9203(85)90135-x. [DOI] [PubMed] [Google Scholar]
- Olson JG. Ksiazek TG. Tan R, et al. Correlation of population indices of female Culex tritaeniorhynchus with Japanese encephalitis viral activity in Kapuk, Indonesia. Southeast Asian J Trop Med Public Health. 1985b;16:337–342. [PubMed] [Google Scholar]
- Pond SL. Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Poon AFY. Lewis FI. Pond SLK, et al. An evolutionary-network model reveals stratified interactions in the V3 loop of the HIV-1 envelope. Plos Comput Biol. 2007;3:2279–2290. doi: 10.1371/journal.pcbi.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2008. FigTree v1.1.2.
- Rey FA. Heinz FX. Mandl C, et al. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Ritchie SA. Rochester W. Wind-blown mosquitoes and introduction of Japanese encephalitis into Australia. Emerg Infect Dis. 2001;7:900–903. doi: 10.3201/eid0705.017524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AJ. Li L. Tesh RB, et al. Genetic characterization of early isolates of Japanese encephalitis virus: genotype II has been circulating since at least 1951. J Gen Virol. 2010;91(Pt 1):95–102. doi: 10.1099/vir.0.013631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AJ. Tesh RB. Barrett AD. Genetic characterization of Japanese encephalitis virus genotype II strains isolated from 1951 to 1978. J Gen Virol. 2011;92(Pt 3):516–527. doi: 10.1099/vir.0.027110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T. Ni H. Beasley DW, et al. Origin and evolution of Japanese encephalitis virus in southeast Asia. J Virol. 2003;77:3091–3098. doi: 10.1128/JVI.77.5.3091-3098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhampunya R. Kim HC. Tippayachai B, et al. Emergence of Japanese encephalitis virus genotype V in the Republic of Korea. Virol J. 2011;8:449. doi: 10.1186/1743-422X-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchie H. Oda K. Vythilingam I, et al. Genetic study of Japanese encephalitis viruses isolated in Malaysia. Jpn J Med Sci Biol. 1994;47:101–107. doi: 10.7883/yoken1952.47.101. [DOI] [PubMed] [Google Scholar]
- Tsuchie H. Oda K. Vythilingam I, et al. Genotypes of Japanese encephalitis virus isolated in three states in Malaysia. Am J Trop Med Hyg. 1997;56:153–158. doi: 10.4269/ajtmh.1997.56.153. [DOI] [PubMed] [Google Scholar]
- van den Hurk AF. Smith CS. Field HE, et al. Transmission of Japanese encephalitis virus from the black flying fox, Pteropus alecto, to Culex annulirostris mosquitoes, despite the absence of detectable viremia. Am J Trop Med Hyg. 2009;81:457–462. [PubMed] [Google Scholar]
- Williams DT. Wang LF. Daniels PW, et al. Molecular characterization of the first Australian isolate of Japanese encephalitis virus, the FU strain. J Gen Virol. 2000;81(Pt 10):2471–2480. doi: 10.1099/0022-1317-81-10-2471. [DOI] [PubMed] [Google Scholar]