Abstract
Septic synovitis is a potentially debilitating and life-threatening disorder in horses. We hypothesized that a universal bacterial real-time PCR (RT-PCR) assay would have improved sensitivity and decreased turn-around time for detection of bacteria in synovial fluid (SF) samples. Forty-eight SF samples were collected from 36 horses that presented to two referral institutions with suspected septic synovitis. Universal RT-PCR, bacterial culture and SF analysis were performed on all samples, and an interpretation on the sample being septic or not was derived by three board certified specialists from the history, clinical assessment and SF characteristics. RT-PCR results were compared to a composite standard comprised of positive culture and interpretation by all three specialists of samples as “septic”. For 41 of 48 samples (85%), culture and RT-PCR results were concordant. Compared to the composite standard, 83% of samples were correctly classified by RT-PCR (turn-around time of approximately 4 hours). Relative sensitivity and specificity of RT-PCR were 87% and 72% respectively, and 56% and 86% for culture. Hence, universal RT-PCR was a rapid and highly sensitive test, which may accelerate diagnosis and improve outcome for horses with septic synovitis.
Résumé
La synovite septique est une condition potentiellement débilitante et mortelle chez les chevaux. Nous avons émis l’hypothèse qu’une épreuve universelle d’amplification en chaîne par la polymérase en temps réel (RT-PCR) pourrait avoir une sensibilité augmentée et une diminution du délai d’obtention des résultats pour la détection de bactérie dans des échantillons de liquide synovial (SF). Quarante-huit échantillons de SF ont été prélevés à partir de 36 chevaux présentés à deux centres de référence avec une synovite septique suspectée. L’épreuve RT-PCR universelle, la culture bactérienne et l’analyse du SF ont été effectuées sur tous les échantillons et une interprétation à savoir si l’échantillon était septique ou non était obtenue de trois spécialistes certifiés à partir de l’histoire du cas, de l’évaluation clinique et des caractéristiques du SF. Les résultats du RT-PCR ont été comparés à un composite standard consistant en une culture et une interprétation positives par les trois spécialistes d’échantillons comme étant «septique». Pour 41 des 48 échantillons (85 %), les résultats de la culture et du RT-PCR concordaient. Comparativement au composite standard, 83 % des échantillons ont été classés correctement par le RT-PCR (délai d’obtention des résultats approximatif de 4 heures). La sensibilité et la spécificité relatives du RT-PCR étaient de 87 % et 72 %, respectivement, 56 % et 86 % pour la culture. Ainsi, l’épreuve RT-PCR universelle était un test rapide et hautement sensible, ce qui pourrait accélérer le diagnostic et améliorer le sort des chevaux avec une synovite septique.
(Traduit par Docteur Serge Messier)
Introduction
Septic synovitis is a common and potentially life-threatening orthopedic condition in horses that can be difficult and costly to treat. It is often associated with negative impacts on athletic performance and even death (1,2). The combination of history, clinical signs, and synovial fluid (SF) analysis is routinely used for clinical diagnosis of septic synovitis. A prompt and accurate diagnosis is crucial to initiate early treatment, as this can reduce the likelihood of cartilage damage and other intrasynovial changes, improve the likelihood of successful treatment, and minimize long-term effects.
The current “gold standard” for diagnosis of septic synovitis consists of a positive bacterial culture from SF in a horse with clinical signs consistent with septic synovitis (3). However, SF from up to 69% of clinical cases with suspected septic synovitis may yield negative culture results, hampering diagnosis and therapeutic measures (1,2,4). Culture of the synovial membrane, in addition to synovial fluid, increases the likelihood of detecting a microorganism (5). Gram staining of SF smears can also be done, but the sensitivity is low and false positives occur (1). Enrichment culture methods increase the sensitivity of culture, but negative results are still common (6,7). Possible explanations include the intrinsic bactericidal properties of SF, prior antimicrobial therapy having eliminated live organisms, or the presence of only a very small number of organisms (1,5,8). These limitations have fueled the search for a more sensitive test, which would also detect bacteria after an antimicrobial therapy has been installed.
Molecular (culture-independent) diagnostic techniques, such as polymerase chain reaction (PCR), are based on the detection of bacterial DNA and are currently widely used in human diagnostic microbiology laboratories (9–14). The 16S (also named broad range or universal) primers amplify specific 16S ribosomal-DNA (16S rDNA) regions that are present in essentially all bacteria. The 16S PCR assay has been used to test synovial fluid and tissue samples from human patients with various forms of septic and non-septic arthritis and has greatly improved detection rate, speed, and accuracy of bacterial identification (12,13). When using real-time PCR (quantitative or qPCR) technology, amplification and detection of DNA products via continuous fluorescence measurements are coupled in a single reaction vessel. This process is typically more sensitive than conventional PCR, easier to perform, reduces turn-around time and decreases risk of contamination because less handling of samples is required (11,14). The qPCR will also allow for quantification of the amount of bacterial DNA present (15).
In veterinary medicine, minimal data are available regarding the use of molecular methods for the diagnosis of septic synovitis and in horses, only conventional PCR techniques with 16S primers have been described (16,17). An overall sensitivity and specificity of 89% and 93%, respectively, was obtained when using a conventional 16S PCR assay on 57 SF samples from horses clinically suspected of having septic synovitis (17). While the data from both equine and human studies are encouraging, a major limitation of most 16S rDNA PCR studies is failure to report quality control aspects of assays, such as limit of detection and scrutiny of cut-off points in the studies using real time technology (11,12,14). Evaluation of results is often arbitrary, as accurate interpretation of a clinical sample being septic or not is inherently limited by the absence of a true “gold standard.” The 16S PCR could detect bacterial infections in the face of antimicrobial therapy and possibly also allow for monitoring of bacterial clearance from the synovial cavity and guide antimicrobial therapy (18).
The objective of this study was to compare a 16S qPCR assay with standard clinical, clinico-pathological, and microbiological methods for diagnosis of septic synovitis in horses.
Materials and methods
Animals
Horses with clinically suspected synovial infection admitted to the Veterinary Teaching Hospital of the Ontario Veterinary College, University of Guelph and to Milton Equine Hospital, were included in the study. Inclusion criteria comprised of history and clinical signs leading to suspicion of septic synovitis, as determined by the attending clinician. Samples obtained on admission from different synovial structures in the same horse were included in the analysis; however, samples taken from the same structure at a later date were excluded.
A total of 36 horses were used. History and clinical data were gathered for each horse. Synovial fluid from clinically normal adult horses with no history of lameness or clinical signs of synovitis was used as negative control in the qPCR assays. These horses had been euthanized for reasons other than musculoskeletal disorders and the SF samples had no growth on aerobic and anaerobic culture.
Synovial fluid collection, cytological analysis, and microbial culture
All clinical and control SF samples were collected in the same manner. Hair overlying the joint/tendon sheath was clipped, and the skin was aseptically prepared with 4% chlorhexidine gluconate soap, followed by 70% isopropyl alcohol and a 0.5% chlorhexidine gluconate solution in 70% isopropyl alcohol. Sterile gloves were worn to perform the synoviocentesis. A SF sample of a minimum of 1 mL was collected into a syringe by needle aspiration. In cases where less synovial fluid was obtained, a direct smear of the synovial fluid was submitted for cytology. Aliquots for SF analysis and qPCR were transferred into separate EDTA tubes (Vacutainer systems, Becton Dickinson, Franklin Lakes, New York, USA). Conventional enrichment methods were used to maintain consistency between the 2 locations, and SF for microbial culture was placed in sterile serum collection tubes and in trypticase soy broth (TSB; Oxoid Microbiology Products, Hampshire, United Kingdom).
Synovial fluid for cytological analysis, and microbial culture was delivered to the Animal Health Laboratory (AHL) of the University of Guelph. Microbial culture was done as follows: blood agar plates were directly inoculated and incubated at 35°C for 48 h under both aerobic and anaerobic (80% nitrogen, 10% carbon dioxide, and 10% hydrogen) conditions. For enrichment, the inoculated TSB were incubated aerobically at 35°C for 72 h and sub-cultured onto blood agar after 24, 48, and 72 h for further incubation, as described above. Bacterial isolates were subsequently identified by using Gram staining and conventional biochemical methods (19). The SF samples for the qPCR assay were stored on ice until freezing at −20°C in 400 μL aliquots within 24 h of collection.
Laboratory handling and automated DNA extraction
All laboratory handling involving qPCR was done in a biological safety cabinet in a designated room. The cabinet was cleaned with 5% NaOH solution and irradiated with 254 nm UV light for 10 min prior to use. Laboratory coats, disposable gloves, and a dedicated set of pipettes were used. All plastic ware (e.g., reaction tubes, filter tips) was sterilized prior to use and reagents were divided into aliquots to prevent frequent handling.
Prior to extraction, 100 μL of 25 mg/mL lysozyme (Lysozyme; Sigma-Aldrich, St. Louis, Missouri, USA) in sterile water were added to 400 μL of SF. The mixture was vigorously shaken by hand and incubated in a heating bath at 37°C for 30 min. The DNA was then extracted using a robotic magnetic bead-based system (Maxwell 16 Instrument; Promega, Madison, Wisconsin, USA) and a DNA purification kit (Maxwell 16 DNA Purification Kit; Promega), in accordance with the manufacturer’s instructions.
Broad-range RT-PCR amplification and sequence analysis of the products
Broad-range primers targeting nucleotides 750–769 and 964–979, located at the V5–V6 region of the bacterial 16S rRNA gene were used, resulting in an amplicon length of 195bp (20, 21). The forward and reverse primer sequences were: 5′-GGA TTA GAT ACC CTG GTAGT-3′ and 5′-GGT AAG GTT CTT CGCG-3′, respectively (20,21).
The qPCR mixture used consisted of 4 μL of Master mix (LightCycler FastStart DNA MasterPlus SYBR green I; Roche Diagnostics, Laval, Quebec), 13.2 μL of DNA-free PCR-grade water, and 0.4 μM of each primer per reaction tube. Two microliters of template DNA extract was added to the reaction mixture, resulting in a final reaction volume of 20 μL. Prior to qPCR, the Master mix and DNA-free PCR-grade water were incubated with deoxyribonuclease (2U/μL, DNAse I; Applied Biosystems, Foster City, California, USA), at 1 μL per 50 μL of Master mix/water. The mixture was placed in a heating block for 10 min at 37°C, then the deoxyribonuclease was inactivated by heating to 75°C for 30 min, and cooled on ice prior to addition of the primers. The qPCR was done using a (LightCycler 1.5 Instrument, Roche Diagnostics) as follows: initial denaturation at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 5 s, primer annealing at 57°C for 5 s, and extension at 72°C for another 15 s. A melting cycle was done prior to cooling down, which included heating to 95°C with a rate of 0.2°C/s.
The positive control consisted of 15 ng/μL of DNA extracted from a pure Staphylococcus aureus culture suspended in normal equine synovial fluid (NSF). The negative control consisted of NSF and both controls were handled in the same manner as the clinical samples. Additionally, each qPCR assay included a negative control consisting of DNA-free PCR-grade water. Analysis and management of the qPCR data (Cycle threshold Ct and melting curves) were done using the appropriate software (LightCycler Software 3.3; Roche Diagnostics).
Samples were considered positive if the threshold cycle crossed at least 2.0 cycles before NSF within the same assay [determined by receiver operating characteristic (ROC) curve analysis], and if the melting curve showed a single melting peak within the temperature range of the primers. The expected melting temperature for the primers was 87.73°C ± 0.86°C (mean ± SD), as based on 52 positive control samples consisting of genomic bacterial DNA. By means of serial dilutions, the analytical sensitivity or limit of detection (LOD) of the assay was determined to be 103 CFU/mL of starting concentration for Staphylococcus aureus and Escherichia coli.
The PCR products were purified with a purification kit (QiaQuick Polymerase Chain Reaction Purification Kit; QIAGEN, Valencia, California, USA), and sequencing of the amplicons was done in both directions, with a commercial sequencer (ABI 3730xl automated DNA Analyzer; Applied Biosystems, Carlsbad, California, USA). Low quality sequences were trimmed manually at the ends while evaluating the trace data with sequence viewer software (4Peaks, Version 1.7.2; Mekentosj B.V, Aalsmeer, The Netherlands), making sure to include only tall distinct peaks with little overlap. Sequences were evaluated using BLAST (Basic Local Alignment Search Tool) from the National Center for Biotechnology Information database, or software from the Ribosomal Database Project (22). A percent identity score of ≥ 95 and ≥ 97 was used to classify an organism to genus and species, respectively. Ultimate species identification was assigned when there was > 95% and > 97% identity with bacterial genus and species, respectively, in the genomic databases (23).
Agreement between culture and RT-PCR
Assay agreement was defined as samples having identical qPCR and culture results, i.e., positive or negative, otherwise samples with conflicting results were considered discordant. Agreement of organism was assessed by corresponding species identification or identification at a higher taxonomic level between the organism cultured and the organism obtained by sequencing of the product of a positive qPCR assay. The qPCR assays were repeated once on samples with discordant assay results or identification of dissimilar organisms.
Statistical analysis
The data were compiled using an electronic database (Excel, Mac 2011; Microsoft Corporation, Redmond, Washington, USA) and subsequently exported and re-formatted in another program (STATA 10/MP; Stata Corp, College Station, Texas, USA) for analysis. The history, clinical data and SF analysis results were assessed by 2 ACVS-board certified surgeons (JBK and NCC) and 1 ACVP-board certified clinical pathologist (DB). All 3 observers were blinded to the culture and qPCR results. The samples were categorized as septic or non-septic by the observers and inter-observer agreement beyond chance between the 3 observers was investigated using Cohen’s kappa-statistic and McNemar’s Chi-square test. The degree of agreement was interpreted based on the following scale described by Landis and Koch: < 0 poor; 0.01–0.20 slight; 0.21–0.40 fair; 0.41–0.60 moderate; 0.61–0.80 substantial; and 0.81–1.00 almost perfect agreement (24).
The qPCR results were measured on a continuous scale (i.e., number of cycles). To establish the optimal cut-off value of the qPCR assay, the cycle numbers were compared with positive culture results and required agreement of all 3 observers to interpret the sample as septic. Based on different threshold cycle cut-off values, an ROC curve analysis was done to determine the area under the curve (AUC) and the optimal cycle cut-off value to maximize sensitivity and specificity of the qPCR test by plotting the results of the qPCR test against 1-Specificity (25).
A Fisher’s exact test was used to compare the proportion of samples with bacterial growth on culture in horses that had received antimicrobial therapy versus the proportion of samples with bacterial growth on culture in horses without antimicrobial therapy. Agreement beyond chance between the culture and qPCR results was investigated using Cohen’s kappa-statistic. Relative sensitivity and specificity of culture, qPCR, and the combination of culture and RT-PCR were estimated, and compared with agreement on sepsis of all 3 observers or only for the 2 surgeons. P-values < 0.05 were considered statistically significant.
Results
Signalment and clinical data
A total of 48 SF samples were obtained from 36 horses. In 8 horses, 6 of which were foals, SF samples were obtained from more than one synovial structure. Mean age ± SD of horses was 3.1 ± 4.1 y, ranging from 1 d to 15 y old. Thoroughbreds (17/36; 47%) and Standardbreds (10/36; 28%) were most frequently represented, followed by Warmbloods (3/36; 8.3%), American Quarter Horses (3/36; 8.3%), and horses of mixed breed (3/36; 8.3%). Seventeen females (47%), 15 intact males (42%), and 4 geldings (11%) were included.
The tarsocrural joint was most frequently involved (23/48; 48%), followed by the fetlock joint (10/48; 21%), carpal joints (7/48; 15%), and stifle joints (4/48; 8%). Tendon sheath involvement was rare (4/48; 8%) with 2 digital flexor tendon sheaths, 1 carpal and 1 tarsal sheath, being affected. Twenty-one of the 36 horses (58%) had received antimicrobial therapy prior to sample collection, 8 horses had not received any antimicrobial therapy, and in 7 cases the history of antimicrobial drug use was unknown. Route of antimicrobial delivery was not consistently reported. Four of the 10 foals that presented with sepsis were recumbent, and the remaining 6 foals exhibited lameness at a walk. In 2 of the 26 adult horses, degree of lameness was not reported. Twenty of the remaining 24 adults (83%) exhibited lameness at a walk. In all but 3 records, the presence of synovial effusion was recorded.
Gross appearance of the SF was considered abnormal in 43 out of 48 samples, with an increase in turbidity (87%) and decrease in viscosity (75%) being the most frequently reported anomalies. The mean ± SD total nucleated cell count of the SF samples was 45.3 ± 44.9 × 109 cells/L, whereas the median and range were 34.2 × 109 cells/L and 0.5 to 152 × 109 cells/L, respectively. Mean ± SD total protein was 39 ± 38 g/L, and median and range were 39 g/L and 20 to 68 g/L, respectively. Less than 1 mL of SF was obtained in 4 samples, and a direct smear of SF was sent to the laboratory for differential analysis. All samples were evaluated cytologically; 26 had greater than 80% neutrophils, and degenerative changes were identified in 16 of these 26 samples. Neutrophil degeneration was only observed in synovial samples with more than 80% neutrophils. In 9 of the 48 cytology slides evaluated (20%), intra-cellular bacteria were observed.
Based on clinical data and cytological analysis, all 3 observers agreed in the interpretation of 36 out of 48 SF samples (75%); 27 samples were interpreted as septic and 9 as non-septic. There was moderate to substantial inter-observer agreement beyond chance for all pair-wise combinations of raters (range κ = 0.44 to 0.78), and when all 3 observers were considered together (κ = 0.59, Table I). Agreement on the samples being septic by the 2 surgeons only turned out to be identical to agreement on sepsis by all 3 observers.
Table I.
Cohen’s kappa and McNemar values for agreement between 3 observers on the diagnosis of septic or non-septic synovitis status of 48 synovial fluid (SF) samples with corresponding 95% confidence intervals
| Kappa | 95% CI | P-value | McNemar’s χ2 | Prob > χ2 | |
|---|---|---|---|---|---|
| Clinical pathologist and surgeon 1 | 0.57 | 0.30 to 0.83 | 0.0001 | 5.44 | 0.02 |
| Clinical pathologist and surgeon 2 | 0.44 | 0.17 to 0.70 | 0.0006 | 5.33 | 0.02 |
| Surgeon 1 and surgeon 2 | 0.78 | 0.50 to 1.0 | 0.0001 | 0.20 | 0.65 |
| All 3 observers | 0.59 | 0.0001 |
χ2 — Chi-square test; CI — Confidence Interval.
Microbial culture results
Eighteen of the 48 SF samples (38%) yielded bacterial growth on culture, of which one with mixed growth. Seven of these (39%) grew organisms only after culture enrichment. Organisms cultured from SF samples consisted of Actinobacillus equuli (n = 6), Staphylococcus aureus (n = 5), coagulase negative Staphylococcus sp. (n = 2), Salmonella Hadar (n = 1), Enterobacter cloacae (n = 1), Enterobacter agglomerans (n = 1), Enterococcus sp. (n = 1), Pseudomonas aeruginosa (n = 1), and Streptococcus zooepidemicus (n = 1). Bacteria were isolated from 7/28 SF samples (25%) from 21 horses known to have received antimicrobial therapy prior to sample collection compared to 5/13 (38%) that had not been treated (P = 0.055). Bacteria were isolated from 15 of the 27 samples (56%) that had been interpreted as septic by all 3 observers.
RT-PCR assay results
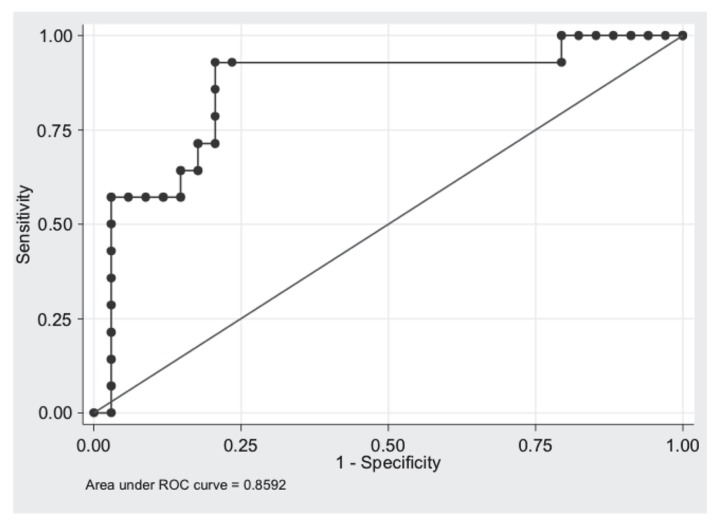
Except for the additional deoxyribonuclease step, the qPCR methodology used in this study was adopted from a previous assay (20,21). Because of day-to-day variability in qPCR parameters of the negative control samples, only intra-run comparisons between clinical samples and negative controls were done. The ROC curve analysis revealed an optimal Ct cut-off of 2.0 cycles between clinical samples and normal synovial fluid samples. The AUC was 0.86, consistent with moderate accuracy (Figure 1). Microbial culture and qPCR results are demonstrated in Table II. Time needed between sample collection and obtaining results from the qPCR assay was approximately 4 h.
Figure 1.
Receiver operating characteristics (ROC) curve analysis of broad range RT-PCR assay’ cycle threshold values versus bacterial growth on culture and agreement of all 3 observers on the sample being septic.
Table II.
Results of the microbial culture and 16S RT-PCR of synovial fluid (SF) samples to detect septic synovitis in horses. Number of SF samples (number of SF samples after repeat RT-PCR)
| RT-PCR | Culture | Total number of samples | |
|---|---|---|---|
|
| |||
| Positive | Negative | ||
| Positive | 13 (15) | 7 (4) | 20 (19) |
| Negative | 5 (3) | 23 (26) | 28 (29) |
Agreement between culture and qPCR results
Using the Ct cut-off of 2 cycles, a 75% (36/48) agreement between results from culture and qPCR assay was identified, which represented moderate agreement beyond chance between the 2 assays (κ = 0.45, Sχ̄ = 0.14, P = 0.0007). A second qPCR assay was done in the remaining 12 samples with discordant assay results, and results subsequently increased agreement to 85% (41/48). Four of the 7 remaining discordant results did not have bacterial growth on culture, but their RT-PCR assay results were positive. Three of these samples were from the same horse and an additional sample from this horse yielded mixed growth (Staphylococcus aureus and an Acinetobacter sp.) on culture, and was positive with the qPCR assay. The 4th sample, which was bacterial culture negative and qPCR assay positive twice, belonged to a septic foal with multiple joints involved and had been assigned as “septic” by the observers. Eighteen of the 27 samples (67%) interpreted as septic by all 3 observers, were positive for the qPCR assay.
Three samples with bacterial growth on culture remained negative by the qPCR assay. The observers had interpreted 2 as non-septic, of which 1 had grown a coagulase negative Staphylococcus sp. after enrichment. The 3rd sample, designated as septic, had a total nucleated cell count (TNCC) of 70.4 × 109 cells/L and Streptococcus zooepidemicus was isolated.
Relative sensitivity and specificity for qPCR compared to agreement of the observers was 87% and 72%, respectively (Table III). Moreover, the sensitivity and specificity of culture and qPCR results were combined relatively to the agreement of the observers were estimated 85% and 81%, respectively.
Table III.
Test performance of RT-PCR assay relative to observers’ assessment (all 3 observers, clinical diagnosis) and bacteriological culture as “gold” standard for diagnosing septic synovitis
| Sensitivity | Specificity | PPV | NPV | P-value | |
|---|---|---|---|---|---|
| Culture results vs clinical diagnosis | 56% | 86% | 83% | 60% | 0.007 |
| RT-PCR results vs clinical diagnosis | 87% | 72% | 74% | 86% | 0.001 |
| Culture and RT-PCR results combined vs clinical diagnosis | 85% | 81% | 85% | 81% | 0.008 |
| RT-PCR results vs culture results | 61% | 84% | 78% | 70% | 0.001 |
PPV — Positive predictive value; NPV — Negative predictive value.
Species identification
Of 18 culture positive samples, 9 samples had a similar organism at species or genus level identified after sequencing of the qPCR product. Repetition of the sequencing assay resulted in 3 more samples identified concordant with culture, resulting in 72% (13/18) SF samples that had similar organisms identified by culture and qPCR. The remaining 5 sequences were considered to be erroneous, as the amplicon obtained showed no sequence similarity with any of those in the databases.
Discussion
Compared with the currently available diagnostic tests for septic synovitis in horses, the 16S qPCR assay performed well in detecting septic synovitis from SF samples in this population of horses. Addition of the qPCR assay resulted in a large increase in relative sensitivity (56% to 85%), with minimal impact on specificity (86% to 81%). The minimal turn-around time for qPCR (i.e., 4 h) was shorter than for culture, which required a minimum of 24 to 48 h.
Despite the use of enrichment culture, bacteria were only isolated from 56% of samples that had been interpreted as septic, which is within the range of 31% to 76% reported in some previous studies (1,2,4,17). Longer transportation times for samples received from another referral hospital or antimicrobial therapy administered prior to collection of the samples could have also contributed to the lower bacterial culture yield observed, since over 55% of horses had received such therapy. More specialized enrichment media, as used in blood culture bottles, contain agents that absorb antimicrobials and inactivate growth inhibitors potentially present in SF (7). A limitation of this study is that a conventional enrichment medium was used, and using specialized media could possibly have increased isolation rate (7). Interestingly, both in humans and horses, studies have shown that prior antimicrobial therapy did not influence the chance of obtaining bacterial growth (2,26). In this study, there was not a significant impact of antimicrobials on culture outcome; however, the P-value (0.055) and relatively small sample size suggest that further study of the impact of antimicrobial therapy on SF culture in horses is required.
The lack of a true “gold standard“ for diagnosis of septic synovitis limits accurate interpretation of a SF sample as septic or not. Bacterial culture is considered the “gold standard” for diagnosing septic synovitis in the majority of published literature (3). Frequently, no growth is obtained from bacterial culture but the clinician still makes the diagnosis of septic synovitis based on clinical data and SF analysis results. We compared both culture and the qPCR assay results to the same standard (observers’ agreement), allowing us to make a true comparison between the 2 different tests. Only one clinical study has described the use of a conventional 16S PCR for detection of bacterial DNA in SF samples of horses (17). That study reported a higher sensitivity and specificity than the present study, but all horses included were considered to be septic, which might be an over-interpreted status since there was lack of a true “gold standard.” Also, DNA extraction and purification were done overnight, and subsequent amplicon analysis is required, increasing the time needed to obtain assay results (17).
The analytical sensitivity of our assay was in the same range as in previous studies evaluating contamination of human platelet concentrates by means of a 16S rDNA qPCR assay, where an LOD of 102 to 103 CFU/mL was reported (27,28). However, it is unknown what concentration of bacteria is clinically relevant. As a result, it is also unknown what sensitivity is desirable for a test to detect bacteria in SF samples of horses with suspected septic synovitis.
One major disadvantage of 16S PCR is the high risk of obtaining false-positive results due to contamination during collection of the sample or execution of the assay. To minimize contamination, strict aseptic techniques were used for sample collection. Sample handling in the laboratory was done in a dedicated PCR room, inside a biological safety cabinet. In a human study, skin biopsy was done prior to SF sampling in order to decrease the risk of contamination with skin bacteria; however, this is impractical in a clinical setting in horses (29). Contamination of PCR reagents like Taq DNA polymerase with recombinant bacterial proteins or DNA is also a major problem when using 16S primers (30). In this study, the qPCR Master mix was pre-treated with deoxyribonuclease to decrease the amount of “background” DNA.
The increased risk of detecting background DNA due to high analytical sensitivity of real time technology compared to conventional PCR most likely contributed to the lower specificity in this study (12,17). Additionally, SYBR Green, a non-specific fluorescent dye was used as detection agent in the qPCR assay. The SYBR Green binds to any double-stranded DNA, potentially decreasing specificity of the assay (31). The SYBR Green is used frequently due to low cost and ease of assay development. Nevertheless, a probe-based assay may be more appropriate when working with 16S primers (31).
Thirteen percent of samples (4/30) were deemed to yield false negative results by culture compared to the qPCR assay, which is similar to the proportion identified in another study (12). All 4 of these qPCR assay samples were identified as septic by all 3 observers, and we believe lack of sensitivity of bacterial culture was responsible for the discordant results. Conversely, 2 of 3 “false negative” qPCR assay samples (positive for culture) had been interpreted by the observers as non-septic. One of these had yielded a coagulase negative Staphylococcus sp. after enrichment, which was most likely a skin contaminant (1,32). We suspect contamination occurred during culture sample handling, as the qPCR assay did not detect any bacterial DNA. One “false negative” qPCR assay sample was designated as septic by all observers. The TNCC of this sample was 70.4 × 109 cells/L, of which 95% were degenerate neutrophils and intracellular bacteria observed on cytology. Therefore, we suspect this was a truly false negative qPCR result, which could have been related to inadequate DNA extraction, the small volume used for the qPCR assay or a sample handling error.
Sequencing of qPCR amplicons from positive samples did not always yield quality data for organism identification. Potential explanations could be the presence of multiple species (contaminants or etiologic organisms) or low-copy-number sequences (14). Difficulty with bacterial identification using 16S rRNA gene sequencing has previously been described (33,34). Without obtaining genus identification, it is challenging and probably impossible to determine whether these samples are true or false positives. Therefore, sensitivity and specificity of the assay were based strictly on qualitative data, and samples were diagnosed either as infected or non-infected based on a minimum of 2-cycle difference between the clinical sample and negative control.
A limitation of PCR is that a positive result only indicates the presence of bacterial DNA, inherently resulting in a lack of antimicrobial susceptibility data. It is also unknown if the DNA is from a living pathogen, which challenges the clinical relevance of a PCR positive result. On the other hand, this could be an advantage when patients have received antimicrobial therapy prior to sample collection and live organisms cannot be obtained from the sample. The use of 16S rRNA reverse transcriptase PCR assay could be promising, as it detects the more labile mRNA, which is suggestive of live organisms (35). Clearance of bacterial DNA from the synovial cavity could be monitored, and the appropriate time for change or discontinuation of antimicrobial therapy could be determined (18). This could possibly decrease costs, limit potential antimicrobial-related diarrhea, and prevent the development of antimicrobial resistance when unnecessary treatment is installed or continued after resolution of infection.
Further standardized research addressing the risk of contamination and analyzing amplicon sequences is needed prior to clinical application of the assay. The main advantage of the use of 16S qPCR for diagnosis of septic synovitis is that it is a rapid diagnostic test with a higher sensitivity than bacterial culture. The assay can complement the more specific bacteriological culture results, which also provides the equine clinician with antimicrobial susceptibility data. Ideally, both tests would be available in clinical settings.
References
- 1.Schneider RK, Bramlage LR, Moore RM, Mecklenburg LM, Kohn CW, Gabel AA. A retrospective study of 192 horses affected with septic arthritis/tenosynovitis. Equine Vet J. 1992;24:436–442. doi: 10.1111/j.2042-3306.1992.tb02873.x. [DOI] [PubMed] [Google Scholar]
- 2.Taylor AH, Mair TS, Smith LJ, Perkins JD. Bacterial culture of septic synovial structures of horses: Does a positive bacterial culture influence prognosis? Equine Vet J. 2010;42:213–218. doi: 10.2746/042516409X480403. [DOI] [PubMed] [Google Scholar]
- 3.Steel CM. Equine synovial fluid analysis. Vet Clin Equine. 2008;24:437–454. doi: 10.1016/j.cveq.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Lescun TB, Vasey JR, Ward MP, Adams SB. Treatment with continuous intrasynovial antimicrobial infusion for septic synovitis in horses: 31 cases (2000–2003) J Am Vet Med Assoc. 2006;228:1922–1929. doi: 10.2460/javma.228.12.1922. [DOI] [PubMed] [Google Scholar]
- 5.Madison J, Sommer M, Spencer P. Relations among synovial membrane histopathologic findings, synovial fluid cytologic findings, and bacterial culture results in horses with suspected infectious arthritis: 64 cases (1979–1987) J Am Vet Med Assoc. 1991;198:1655–1661. [PubMed] [Google Scholar]
- 6.Montgomery RD, Long IR, Milton JL, DiPinto MN, Hunt J. Comparison of aerobic culture, synovial membrane biopsy, and blood culture medium in detection of canine bacterial arthritis. Vet Surg. 1989;18:300–303. doi: 10.1111/j.1532-950x.1989.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 7.Dumoulin M, Pille F, van den Abeele A, et al. Use of blood culture medium enrichment for synovial fluid culture in horses: A comparison of different culture methods. Equine Vet J. 2010;42:541–546. doi: 10.1111/j.2042-3306.2010.00091.x. [DOI] [PubMed] [Google Scholar]
- 8.Gruber BF, Miller BS, Onnen J, Welling R, Wojtys EM. Antibacterial properties of synovial fluid in the knee. J Knee Surg. 2008;21:180–185. doi: 10.1055/s-0030-1247816. [DOI] [PubMed] [Google Scholar]
- 9.Marin M, Munoz P, Sanchez M, et al. Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine. 2007;86:195–202. doi: 10.1097/MD.0b013e31811f44ec. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Millar BC, Moore JE, et al. Employment of broad-range 16S rRNA PCR to detect aetiological agents of infection from clinical specimens in patients with acute meningitis — rapid separation of 16S rRNA PCR amplicons without the need for cloning. J Appl Microbiol. 2003;94:197–206. doi: 10.1046/j.1365-2672.2003.01839.x. [DOI] [PubMed] [Google Scholar]
- 11.Rosey A, Abachin E, Quesnes G, et al. Development of a broad-range 16S rDNA real-time PCR for the diagnosis of septic arthritis in children. J Microbiol Methods. 2007;68:88–93. doi: 10.1016/j.mimet.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol. 2006;44:1018–1028. doi: 10.1128/JCM.44.3.1018-1028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariani BD, Levine MJ, Booth RE, Tuan RS. Development of a novel, rapid processing protocol for polymerase chain reaction-based detection of bacterial infections in synovial fluids. Mol Biotech. 1995;4:227–237. doi: 10.1007/BF02779016. [DOI] [PubMed] [Google Scholar]
- 14.Yang S, Ramachandran P, Hardwick A, et al. Rapid PCR-based diagnosis of septic arthritis by early gram-type classification and pathogen identification. J Clin Microbiol. 2008;46:1386–1390. doi: 10.1128/JCM.02305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkbright S, Fatovich D, Kee C, et al. Quantitative rt-PCR holds promise as a screening tool for patients with severe sepsis. Emerg Med Australas. 2011;23:502–506. doi: 10.1111/j.1742-6723.2011.01445.x. [DOI] [PubMed] [Google Scholar]
- 16.Pille F, Martens A, Schouls LM, et al. Detection of bacterial DNA in synovial fluid from horses with infectious synovitis. Res Vet Sci. 2004;77:189–195. doi: 10.1016/j.rvsc.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Pille F, Martens A, Schouls L, et al. Broad range 16S rRNA gene PCR compared to bacterial culture to confirm presumed synovial infection in horses. Vet J. 2007;173:73–78. doi: 10.1016/j.tvjl.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 18.van der Heijden I, Wilbrink B, Vije A, Schouls L, Breedveld F, Tak P. Detection of bacterial DNA in serial synovial samples obtained during antibiotic treatment from patients with septic arthritis. Arthritis Rheum. 1999;42:2198–2203. doi: 10.1002/1529-0131(199910)42:10<2198::AID-ANR23>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 19.Cowan ST, Steel KJ. Bacterial characters and characterization. In: Barrow GI, Feltham RKA, editors. Manual for the Identification of Medical Bacteria. Cambridge, United Kingdom: Cambridge University Press; 2004. pp. 21–45. [Google Scholar]
- 20.Kobayashi N, Bauer TW, Togawa D, et al. A molecular gram stain using broad range PCR and pyrosequencing technology. Diagn Mol Pathol. 2005;14:83–89. doi: 10.1097/01.pas.0000162753.38284.1a. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi N, Procop GW, Krebs V, Kobayashi H, Bauer TW. Molecular identification of bacteria from aseptically loose implants. Clin Orthop Rel Res. 2008;466:1716–1725. doi: 10.1007/s11999-008-0263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petti CA. Detection and identification of microorganisms by gene amplification and sequencing. Clin Infect Dis. 2007;44:1108–1114. doi: 10.1086/512818. [DOI] [PubMed] [Google Scholar]
- 24.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 25.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 26.Ghanem E, Parvizi J, Clohisy J, Burnett S, Sharkey PF, Barrack R. Perioperative antibiotics should not be withheld in proven cases of periprosthetic infection. Clin Orthop Relat Res. 2007;461:44–47. doi: 10.1097/BLO.0b013e318065b780. [DOI] [PubMed] [Google Scholar]
- 27.Mohammadi T, Reesink HW, Vandenbroucke-Grauls CMJE, Savelkoul PHM. Optimization of real time PCR assay for rapid and sensitive detection of eubacterial 16S ribosomal DNA in platelet concentrates. J Clin Microbiol. 2003;41:4796–4798. doi: 10.1128/JCM.41.10.4796-4798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rood IGH, Koppelman MHGM, Pettersson A, Savelkoul PHM. Development of an internally controlled PCR assay for broad range detection of bacteria in platelet concentrates. J Microbiol Meth. 2008;75:64–69. doi: 10.1016/j.mimet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Wilbrink B, van der Heijden IM, Schouls LM, et al. Detection of bacterial DNA in joint samples from patients with undifferentiated arthritis and reactive arthritis, using polymerase chain reaction with universal 16S ribosomal RNA primers. Arthritis Rheum. 1998;41:535–543. doi: 10.1002/1529-0131(199803)41:3<535::AID-ART20>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol. 2000;38:1747–1752. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shipley GL. An introduction to real-time PCR. In: Dorak MT, editor. Real-time PCR. New York: Taylor & Francis Group; 2006. pp. 1–38. [Google Scholar]
- 32.Piette A, Verschraegen G. Role of coagulase-negative staphylococci in human disease. Vet Microbiol. 2009;134:45–54. doi: 10.1016/j.vetmic.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mignard S, Flandrois JP. 16S rRNA sequencing in routine bacterial identification: A 30-month experiment. J Mircobiol Meth. 2006;67:574–581. doi: 10.1016/j.mimet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Kempsell KE, Cox CJ, Hurle M. Reverse Transcriptase-PCR Analysis of Bacterial rRNA for Detection and Characterization of Bacterial Species in Arthritis Synovial Tissue. Infect Imm. 2000;68:6012–6026. doi: 10.1128/iai.68.10.6012-6026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]