Abstract
Cyclin D1 expression, usually absent in chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), has been described in the proliferation centers (PC) of some CLL/SLL. The prevalence of this finding is uncertain, as is the explanation for its occurrence and whether these cases have any other unique features. Cyclin D1 immunohistochemical staining was therefore investigated in 57 extramedullary CLL/SLL biopsies. In 6 cases, cyclin D1 immunofluorescence followed by CCND1 fluorescence in situ hybridization (FISH) and PC targeted analysis was performed using a Bioview Duet system. Excluding the prospectively selected cases that had the targeted FISH studies, cyclin D1+ PC were identified in 20% of cases. The cyclin D1+ CLL did not appear pathologically or phenotypically distinctive, though 46% had an interfollicular growth pattern. The cyclin D1+ PCs were SOX11− and lacked CCND1 translocations and gains in 5 of 5 informative cases. The recognition of cyclin D1 expression in PC of a significant minority of CLL/SLL can be a diagnostic aid and should not lead to the diagnosis of focal mantle cell lymphoma.
Keywords: Chronic lymphocytic leukemia, Small lymphocytic lymphoma, Cyclin D1, SOX11, Proliferation centers
Distinguishing chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) from mantle cell lymphoma (MCL) is considered important for clinical purposes, even though both entities show a clinical spectrum ranging from indolent to more aggressive.1 CLL/SLL is considered a “low grade” B-cell neoplasm and MCL has a reported median survival of only 3 to 5 years.2,3 Histologically, CLL/SLL is most confidently recognized in tissues when classic proliferation centers (PCs) are seen in the midst of a diffuse proliferation of small relatively round lymphocytes.2 The paler PCs are phenotypically distinct and reported to be sites of B-cell antigen receptor stimulation.4–9 Distinctive but nonspecific phenotypic features that have been reported include increased expression of proliferation-associated and activation-associated antigens.4,5 In addition, larger and more proliferative PCs have been associated with “atypical CLL”10 and with “accelerated” CLL, which has an aggressive clinical course.11
Unfortunately the distinction of CLL/SLL from MCL is not always straightforward because some MCLs may mimic CLL/SLL clinically, histologically, and/or phenotypically and vice versa. Specifically, some MCLs may be composed of small lymphoid cells with relatively round nuclei and even have foci mimicking PC,3,12 some present with blood and bone marrow involvement without adenopathy,13 and some may share even more phenotypic similarities than usual with CLL/SLL such as CD23 expression.3,14,15 Conversely, cases of CLL/SLL may manifest prominent adenopathy, have irregular nuclear contours,10 demonstrate a perifollicular growth pattern,16,17 and also show phenotypic variation, such as expression of FMC7 which is more commonly found in MCL.18 To aid in this distinction, one of the more important and heavily relied upon techniques when dealing with tissue sections is immunohistochemical staining for cyclin D1 because most MCLs express cyclin D1 whereas the vast majority of cases of CLL/SLL typically do not.
The cyclin D1 protooncogene is a regulator of cell cycle progression from the G1 to S phase, which is not expressed in normal lymphoid cells but is overexpressed in a subset of B-cell/plasma cell neoplasms, including MCL, plasma cell myeloma (PCM), and hairy cell leukemia (HCL).1 Cyclin D1 forms a complex with cyclin-dependent kinases 4 and 6, leading to phosphorylation and “inactivation” of the tumor suppressor protein retinoblastoma, releasing E2F, and culminating in cell cycle progression from the G1 to S phase.19–21 In MCL and a subset of cyclin D1+ PCM, the cyclin D1 is over-expressed because of a t(11;14)(q13;q32) translocation juxtaposing the immunoglobulin heavy chain gene (IGH@) and the CCND1 gene.22–25 Many of the other cyclin D1+, as well as some cyclin D1–, PCM show gains in the CCND1 gene, as detected with cytogenetic fluorescence in situ hybridization (FISH).25 The explanation for cyclin D1 expression in HCL is uncertain with only 1 unusual case reported to show a t(11;14) translocation.26 The cyclin D1+ HCLs, however, are reported to express the MCL-associated SOX11 transcription factor.27
The situation with cyclin D1 expression in CLL/SLL has become more complicated in recent years, perhaps because of the use of improved and more sensitive cyclin D1 immunohistochemistry.28 Cyclin D1+ CLL/SLL has traditionally been considered to be rare and in some reported cases the diagnosis is considered controversial. A recent series reported cyclin D1 positivity in PCs in 10% of CLLs including in 20% of lymph node biopsies, an observation noted in an earlier case report.28–32 The prevalence of this finding, however, is uncertain because the prior single case report of CLL/SLL with cyclin D1+ PC described 15 additional CLL/SLL cases and found no other cases with cyclin D1+ PC.30 Although cytogenetic studies have suggested a lack of the t(11;14) translocation in CLL/SLL with cyclin D1+ PC, a specific analysis of the PC has not been performed. Thus, it is unknown whether CCND1 abnormalities, like those in the dominant populations of MCL or PCM, are present specifically in the PC cells and whether, as in HCL,27 cyclin D1 expression might be correlated with expression of SOX11. Furthermore, data on the histopathologic and phenotypic features of the cyclin D1+ cases are limited.
To address these issues, 6 CLL/SLL with cyclin D1+ PC were characterized histopathologically and phenotypically. This included SOX11 staining in 5 cases which were then studied in detail using a targeted immunoFISH approach with cyclin D1 immunohistochemical study and a CCND1 “break-apart” probe. In addition, to assess the prevalence of this finding and its possible pathologic correlates, a contiguous series of 50 additional CLL/SLL cases with available cyclin D1 stain results were then reviewed.
Materials and Methods
Case Selection and Morphologic/Phenotypic Review
Seven cases that fulfilled the World Health Classification (WHO) classification criteria for CLL/SLL and had easily identified cyclin D1+ PCs were selected from the Division of Hematopathology files at University of Pittsburgh Medical Center (UPMC)–Presbyterian Hospital, Pittsburgh, PA. One case did not have additional available material and was excluded from further analysis. Two authors (J.F.G., S.H.S.) reviewed H&E-stained sections and the cyclin D1 immunohistochemical stains as well as findings of additional immunophenotypic studies such as flow cytometric studies. The size of the PCs, cellular components of the PCs, presence of residual reactive germinal centers, and patent sinuses were specifically assessed.
Immunohistochemical Stains for Cyclin D1 and SOX11
The cyclin D1 immunohistochemical staining had been previously performed using the Ventana BenchMark XT (Ventana, Tucson, AZ) and cyclin D1 rabbit monoclonal antibody (1:100 dilution, Clone SP4, 50-test dispenser, Cell Marque, Rocklin, CA). Prospective staining for SOX11 was performed using the Ventana BenchMark XT and the SOX11 rabbit polyclonal antibody at a 1:50 dilution (Sigma Aldrich, St Louis, MO).
Prevalence
To determine the prevalence of cyclin D1 positivity in the PC of CLL/SLL, extramedullary tissue biopsy specimens of CLL/SLL with an available H&E-stained section and a cyclin D1 immunohistochemical stain (performed as described earlier) diagnosed at UPMC from April 2005 through April 2010 were retrieved and reviewed. Cases were considered to have cyclin D1+ PC if discrete aggregates of cyclin D1+ lymphocyte nuclei were easily visible using a 10× objective. The pathologic and phenotypic features of these cases were also reviewed.
Immunofluorescence Staining/FISH (ImmunoFISH)
Immunofluorescence staining for cyclin D1 was performed on 5 cases. Briefly, 5-μm thick sections of formalin-fixed paraffin-embedded tissue were deparaffinized in xylene and ethanol, protein-blocked (DAKO X0909, DAKO, Carpinteria, CA) for 15 minutes, and then incubated with a cyclin D1 primary antibody (1:25 dilution, SC-753, Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. After washing with phosphate-buffered saline plus 0.05% polysorbate 20 (Igepal 630, Sigma Aldrich) slides were incubated at room temperature for 30 minutes in anti-rabbit IgG (Fl-1000, Vector Labs, Burlingame, CA) and then counterstained with DAPI I (30-804930 Abbott Molecular, Des Plaines, IL). Groups of positive staining cells with round nuclei were targeted and imaged using the Bioview Duet System (Bioview, Nes Ziona, Israel) equipped with an Olympus BX61 microscope (Olympus America, Center Valley, PA) using a 60× oil plan fluor objective and filter sets containing single band excitors for Texas red/rhodamine, fluorescein isothiocyanate (FITC), DAPI (UV 360 nm), a dual filter for SpectrumOrange/FITC, and a triple filter for SpectrumOrange, FITC, and DAPI. FISH analysis was then performed after removing the coverslips from the immunohistochemically imaged slides. The tissue was first digested for 28 minutes in protease solution (0.5 mg/mL) (Macron Chemical, Phillipsburg, NJ) at 37°C. The slides were washed in 2× saline sodium citrate (SSC, Abbott Molecular), followed by fixation in 10% buffered formalin phosphate (Fisher Scientific, Fair Lawn, NJ). Following fixation, the slides were again washed in 2× SSC. Dual-color FISH was performed using the LSI CCND1 direct-labeled break-apart rearrangement probe (1:10 dilution in LSI buffer #05J96-001, Abbott Molecular). The target tissue and probe were denatured at 90°C for 12 minutes and then incubated overnight at 37°C in a humidified chamber. The posthybridization wash was performed using 2× SSC/0.3% NP-40 at 73°C for 2 minutes. The slides were then air-dried in the dark, and counterstained with DAPI I. Areas with previously identified cyclin D1+ PC were identified using the Bioview Duet system and photographed for FISH analysis. Tissue sections from a lymph node involved by MCL were used as a positive control. It had numerous cells strongly positive to cyclin D1 and more than 50% of the cells had a CCND1 translocation.
Results
Clinicopathologic Features
The 6 patients (4 men, 2 women) included in the immunoFISH study were adults (range, 54–84 years) Table 1. All cases were nodal. Five cases had PCs of typical size and 1 case had larger PCs. All cases had identifiable paraimmunoblasts in the PCs and 2 cases had at least a moderate number of paraimmunoblasts. One case had scattered larger transformed cells in the PCs. All cases had at least some patent sinuses and 4 of 5 evaluable cases had reactive germinal centers and a pattern of “interfollicular SLL” Image 1. The sixth case was a needle core biopsy.
Table 1.
Clinical Features of Patients in ImmunoFISH Study
| Case | Age, y | Sex | Site | Lymphocytosis | Adenopathy | Splenomegaly | Residual GC | Patent Sinuses |
|---|---|---|---|---|---|---|---|---|
| 1 | 64 | M | Axillary LN | No | Diffuse | No | Yes | Yes |
| 2 | 54 | M | Neck LN | No | Diffuse | No | No | Yes |
| 3 | 60 | M | Cervical LN | Yes | Diffuse | Yes | Yes | Yes |
| 4 | 50 | F | Cervical LN | No | Diffuse | No | Yes | Yes |
| 5 | 70 | M | Supraclavicular LN | Yes | Diffuse | No | Yes | Yes |
| 6 | 84 | F | Axilla LN | Yes | Diffuse | Yes | NA, needle biopsy | Yes |
GC, germinal center; LN, lymph node; FISH, fluorescence in situ hybridization; NA, not available.
Image 1.
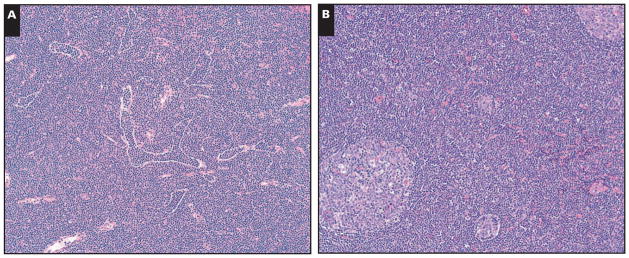
Partial preservation of lymph node architectural features. A (Case 2), Note the patent sinuses. B (Case 4), Prominent reactive germinal centers are present. (H&E, ×100)
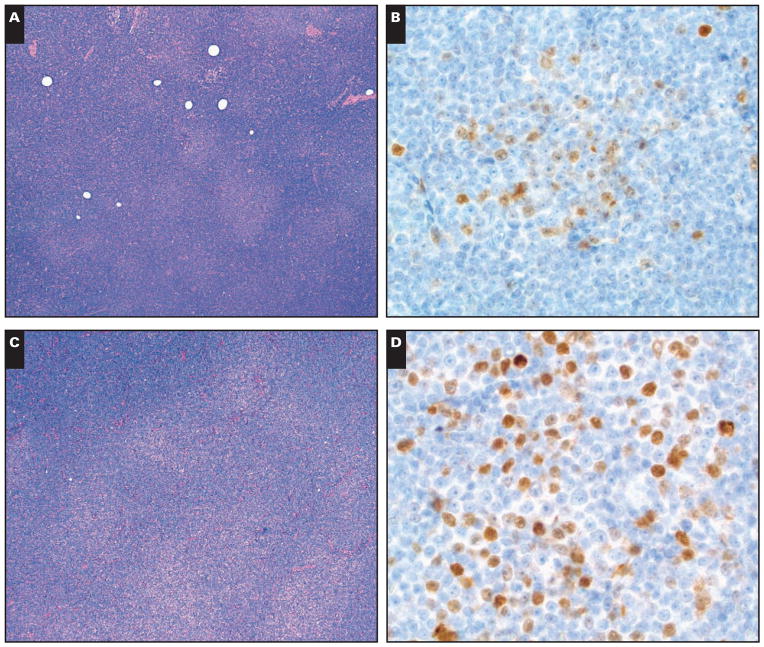
Five of six cases had a classic CLL/SLL phenotype that was CD5 positive, CD10 negative, CD23 positive, FMC7 negative, CD20 dim positive with dim surface light chain restriction. One case had a similar phenotype except it was CD5 negative and surface immunoglobulin (SIg) negative. Four of six cases were CD38 negative on either the diagnostic tissue specimen or a subsequent bone marrow evaluation. Five of the 6 cases had weak to moderate intensity cyclin D1 positivity in PC Image 2A and Image 2B and 1 case had stronger lymphoid staining in the PC Image 2C and Image 2D. Conventional FISH studies for CCND1 rearrangement were negative in 3 tested cases; however, 1 case had loss of the ATM tumor suppressor gene and a subset of cells with loss of the TP53 tumor suppressor gene. All 5 tested cases were negative for SOX11.
Image 2.
A and B (Case 1), The distinct proliferation centers showed weak to moderate intensity cyclin D1 staining. C and D (Case 2), In contrast, in this case, the proliferation centers are somewhat confluent and have moderate to strong cyclin D1 staining. (A and C, H&E, ×40; B and D, immunohistochemistry with hematoxylin counterstain, ×500.)
Prevalence
Review of the files of UPMC from April 2005 through April 2010 yielded an additional 50 cases that met the inclusion criteria for retrospective review. The patients (29 men, 21 women) were all adults (45–88 years). Of the 23 cases with concurrent flow cytometric immunophenotypic studies, 18 had a classic CLL/SLL type phenotype, 1 case had brighter than usual CD20 expression, 1 case was CD20 negative, 2 cases had brighter than usual SIg expression, and 1 had only partial CD23 expression. Cyclin D1+ PC was identified in 10 (20%) of 50 cases included in this retrospective review, in the 6 cases studied in more detail and 1 case initially chosen for more detailed study but which was not further pursued (because of insufficient material). All cases including the latter 7 cases were from the same period. Cyclin D1 staining intensity in the additional cases was variable even within the same PC, though it was unusual to find strongly staining cells. All cases had a majority of negative cells in the PCs. No cases had staining of lymphoid cells outside the PCs. Three additional cases with cyclin D1+ PC had an interfollicular growth pattern with germinal centers and intact sinuses, 2 had patent sinuses but no reactive germinal centers, and 5 had neither patent sinuses nor reactive germinal centers. Of the additional 5 cyclin D1+ cases with flow cytometric studies, 4 had a classic CLL/ SLL phenotype and 1 had a classic CLL/SLL phenotype except for brighter than usual SIg.
Immunofluorescence Staining/ImmunoFISH
Five cases had recognizable cyclin D1+ PC after staining with the fluorescently labeled cyclin D1 antibody. The positive cells in the PCs varied in intensity with few staining strongly. The number of PCs identified and analyzed per case ranged from 3 to 13 (median, 4). CCND1 rearrangements were not identified in either the PC or elsewhere in any of the CLL/SLL cases Image 3. No cases had extra copies suggesting CCND1 gains.
Image 3.
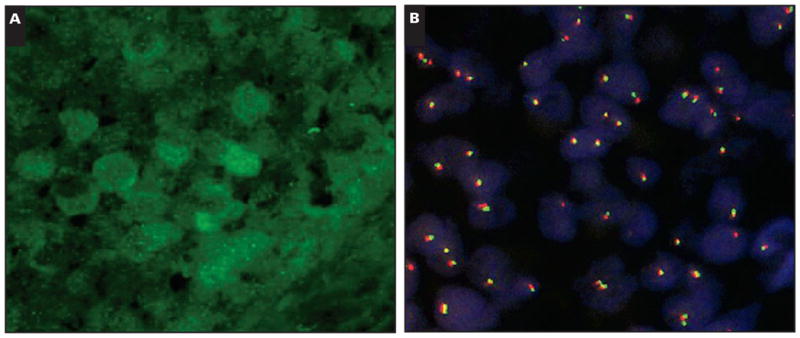
Immunofluorescence in situ hybridization analysis showing the green cyclin D1+ cells in the immunohistochemical stain (A) and the results of the CCND1 break-apart probe in this specific region (B). Cells with split signals indicating a CCND1 translocation are not seen. Note 2 fusion signals in all informative cells in the region of the cyclin D1 positive proliferation center. The cells with a single fusion signal or rare cells with a single red or green signal represent truncation. (A, fluorescence cyclin D1 stain, ×1,000; B, cyclin D1 break-apart probe, ×1,000.)
Discussion
It is important to distinguish CLL/SLL from MCL because, on average, CLL/SLL is considered a low-grade neoplasm even when the disease is nodal, whereas cases of MCL, especially those of nodal type, are clinically aggressive, with a median survival of 3 to 5 years.2,3,13 The distinction is based on well-characterized morphologic, phenotypic, and cytogenetic differences, including cyclin D1 expression in most MCLs and an absence of cyclin D1 in the vast majority of CLL/SLL. Given the overlapping clinical presentations,13 variability in the classic phenotypic features of both CLL/SLL and MCL,1,3,14,15,18,33 overlapping cytologic features with some MCLs composed of small round lymphocytes with even some foci resembling PC3,12,34 and some CLL/SLL with irregular nuclear contours10 and/or PC that are either not present or not readily apparent, the presence of cyclin D1 positivity is considered a very important diagnostic tool used to support the diagnosis of MCL when histologic material is available.
A small number of CLL/SLL with extensive cyclin D1 expression have been reported, though in the absence of an absolute gold standard for CLL/SLL, some might be considered controversial.32,35 Cyclin D1 positivity in CLL/ SLL, however, has also been reported to be restricted to the PC, initially seen in a single case report.30 This finding was reported to be “quite unusual” because a study of 15 additional cases failed to show any cyclin D1+ PC. Nevertheless, the importance of the finding as a diagnostic pitfall was emphasized, even if only found in 6% of the CLL/SLL cases analyzed.30 A more recent study found cyclin D1+ cells in the PC in about 10% of CLL/SLL and in 20% of nodal CLL/SLL29; however, whether the cyclin D1 positivity was the result of CCND1 translocation, gains, or another mechanism was not addressed. Although cytogenetic FISH studies performed in 3 of their cases failed to demonstrate a t(11;14) translocation, there is no comment about possible CCND1 gains that were presumed to be absent; more importantly, the PC cells were not specifically examined. It remains important to exclude the possibility of CCND1 gains restricted to PC, particularly because it was recently reported that the frequency of cells with varied genetic alterations in CLL is higher in PC than in the surrounding areas.36
The prevalence of at least 20% CLL with cyclin D1+ PC in the current extramedullary tissue-based study, with 7 additional cases identified during the same period (which, if included, would result in a prevalence of 30%) may be somewhat higher than previously reported. The recognition of this phenomenon in a significant minority of CLL/SLL may relate to the use of a better rabbit monoclonal anti-cyclin D1 antibody28 as seen in this study and the series reporting a 20% prevalence in lymph nodes with CLL/SLL. This antibody was not used in the past or in the study reporting only 1 of 16 positive cases.29,30
Although additional study is required to determine if there are specific clinical associations with the CLL/SLL having cyclin D1+ PC, no distinctive pathologic or immunophenotypic features have been suggested in these cases.29,30 Among the cases studied here, 46% did have a growth pattern like that reported in “interfollicular small lymphocytic lymphoma”16,37 and 69% had intact sinuses. Almost 80% of cases had a typical flow cytometric phenotype for CLL/SLL with the remaining cases showing common phenotypic variations. These cases do not appear to have features suggesting an overall more aggressive type of CLL/SLL, with most cases being predominantly CD38− and only 1 of the cases studied in greater detail showing larger than usual PCs. On the other hand, these cyclin D1+ PC cases do include some that have one or more adverse prognostic indicators such as the minority with CD38 expression or individual cases with ATM or TP53 deletion. Single cases have also been reported with trisomy 12,30 del(13)(q14.3), ins(14;19)(q32;p13p22), and t(X;17)(p11;q21).29
Cyclin D1 overexpression in MCL is caused by a CCND1 translocation, with classical and FISH cytogenetic studies being useful tools to document this characteristic abnormality in difficult cases or cases without tissue sections. Approximately 36% of PCMs that express cyclin D1 are also reported to have CCND1 translocations, while in the remaining cyclin D1+ cases, the overexpression is usually the result of increased copies of the CCND1 gene.25 The mechanism of cyclin D1 overexpression is unknown in HCL, though a recently reported unusual case had a CCND1 translocation.26 Neither CCND1 translocations nor gene amplifications were identified by FISH in the cyclin D1+ PC of the CLL/SLL studied here. This is consistent with the analyses in these cases that have not specifically looked at the PC and which report an absence of at least CCND1 translocations.29,30 Although our observations do not provide an explanation for the cyclin D1 expression in PC, they highlight how even FISH studies focused on the cyclin D1+ PC retain their usefulness in distinguishing MCL from CLL/SLL. These studies can be particularly useful in cases in which the cyclin D1 staining in CLL/SLL is stronger than usual and when a composite CLL/SLL and MCL38 is a consideration. Lymph nodes with MCL-like cells of uncertain significance/in situ MCL may also cause confusion because they might exhibit only focal cyclin D1 staining and the precise extent of the neoplasm may be uncertain.39,40 The cyclin D1 expression in these cases, however, would be expected to be in the mantle zones around reactive-appearing germinal centers. Rarely, PCs may be noted around germinal centers in CLL/SLL,16,17 thus in theory showing some cyclin D1 expression. However, the cases of MCL-like cells of uncertain significance/in situ MCL would show CCND1 translocations in the cyclin D1+ cells.
Another feature that distinguishes the cyclin D1 expression in CLL/SLL from other cyclin D1+ lymphoid/plasmacytic neoplasms is the lack of expression of the MCL-associated SOX11 transcription factor. SOX11 is expressed by about 90% of MCL as well as in many lymphoblastic lymphomas and some Burkitt lymphomas and HCLs.27,41–43 It should be recognized, however, that SOX11 negativity has been associated with indolent MCL that might look and behave like CLL/ SLL.43 Although CLL/SLL is known to be SOX11 negative, staining of scattered cells in PC might easily be missed if not specifically looked for. However, in contrast to HCL where cyclin D1 staining is reported to correlate with SOX11 expression,27 the cyclin D1+ PCs are SOX11 negative.
The fact that PCs have different expression of cyclin D1 compared with the surrounding CLL/SLL cells, while unexplained, is consistent with the observation that PCs are morphologically, phenotypically, and even sometimes cytogenetically distinct from the surrounding CLL/SLL. These paler foci in many CLL/SLL tissue infiltrates, where one finds larger neoplastic cells including paraimmunoblasts, are considered a critical component of CLL/SLL. The PC microenvironment of CLL/SLL both promotes the proliferation of the CLL/ SLL cells, perhaps also enhancing the likelihood of additional genetic abnormalities, and inhibits apoptosis.6–8,36 Antigenic drive with stimulation through the B-cell receptor is considered essential in the pathobiology of CLL/SLL, with the PC being a potential site where B-cell receptor stimulation occurs, with the presence of CD4+, CD40L+ T cells.6–9,44
Although one could speculate that the cyclin D1 expression in PC was simply related to the increased proliferation, many other highly proliferating lymphoid neoplasms are cyclin D1−. In addition to the increased cellular proliferation, other phenotypic changes seen in the PC, including their very frequent IRF4/MUM1 expression, are consistent with activation and potential B-cell receptor–mediated signaling at that site with changes that are similar to those that can be seen with mitogenic or immunologic stimulation of CLL/SLL cells.5 Some of the additional phenotypic alterations in PC include a higher frequency or greater expression of HLA-DC, HLA-DR, CD20, CD23, survivin (BIRC5), and possibly CD38 and diminished expression of IgD and CD9.4,5,44–51
In conclusion, approximately 20% to 30% of CLL/ SLL have cyclin D1 staining in the PC. It is important to recognize this finding and not to mistake these cases for MCL. The cyclin D1 positivity is not because of CCND1 translocation or gains and is not associated with SOX11 expression. The presence of a CCND1 translocation or gain, or SOX11 staining, therefore, should prompt one to consider MCL or one of the other cyclin D1+ lymphoid neoplasms. Given the importance of the microenvironment and PC in the pathobiology of CLL/SLL, further exploring the mechanism for cyclin D1 expression in these PCs might lead to further understanding of CLL/SLL and discovery of additional therapeutic targets.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. [Google Scholar]
- 2.Müller-Hermelink HK, Montserrat E, Catovsky D, et al. Chronic lymphocytic leukaemia/small lymphocytic lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. pp. 180–182. [Google Scholar]
- 3.Swerdlow SH, Campo E, Seto M, et al. Mantle cell lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. pp. 229–232. [Google Scholar]
- 4.Lampert IA, Wotherspoon A, Van Noorden S, et al. High expression of CD23 in the proliferation centers of chronic lymphocytic leukemia in lymph nodes and spleen. Hum Pathol. 1999;30:648–654. doi: 10.1016/s0046-8177(99)90089-8. [DOI] [PubMed] [Google Scholar]
- 5.Soma LA, Craig FE, Swerdlow SH. The proliferation center microenvironment and prognostic markers in chronic lymphocytic leukemia/small lymphocytic lymphoma. Hum Pathol. 2006;37:152–159. doi: 10.1016/j.humpath.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 6.Bertilaccio MT, Scielzo C, Muzio M, et al. An overview of chronic lymphocytic leukaemia biology. Best Pract Res Clin Haematol. 2010;23:21–32. doi: 10.1016/j.beha.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Caligaris-Cappio F, Chiorazzi N. Where is the biology of CLL leading us? Semin Cancer Biol. 2010;20:361–362. doi: 10.1016/j.semcancer.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Zenz T, Mertens D, Kuppers R, et al. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 9.Klein U, Dalla-Favera R. New insights into the pathogenesis of chronic lymphocytic leukemia. Semin Cancer Biol. 2010;20:377–383. doi: 10.1016/j.semcancer.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Bonato M, Pittaluga S, Tierens A, et al. Lymph node histology in typical and atypical chronic lymphocytic leukemia. Am J Surg Pathol. 1998;22:49–56. doi: 10.1097/00000478-199801000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Giné E, Martinez A, Villamor N, et al. Expanded and highly active proliferation centers identify a histological subtype of chronic lymphocytic leukemia (“accelerated” chronic lymphocytic leukemia) with aggressive clinical behavior. Haematologica. 2010;95:1526–1533. doi: 10.3324/haematol.2010.022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swerdlow SH, Zukerberg LR, Yang WI, et al. The morphologic spectrum of non-Hodgkin’s lymphomas with BCL1/cyclin D1 gene rearrangements. Am J Surg Pathol. 1996;20:627–640. doi: 10.1097/00000478-199605000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Orchard J, Garand R, Davis Z, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101:4975–4981. doi: 10.1182/blood-2002-06-1864. [DOI] [PubMed] [Google Scholar]
- 14.Kelemen K, Peterson LC, Helenowski I, et al. CD23+ mantle cell lymphoma: a clinical pathologic entity associated with superior outcome compared with CD23– disease. Am J Clin Pathol. 2008;130:166–177. doi: 10.1309/R94MAFJY5EA4A8C3. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, Peterson L, Nelson B, et al. Immunophenotypic variations in mantle cell lymphoma. Am J Clin Pathol. 2009;132:699–706. doi: 10.1309/AJCPV8LN5ENMZOVY. [DOI] [PubMed] [Google Scholar]
- 16.Bahler DW, Aguilera NS, Chen CC, et al. Histological and immunoglobulin VH gene analysis of interfollicular small lymphocytic lymphoma provides evidence for two types. Am J Pathol. 2000;157:1063–1070. doi: 10.1016/S0002-9440(10)64620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson SE, Swerdlow SH, Ferry JA, et al. Reassessment of small lymphocytic lymphoma in the era of monoclonal B-cell lymphocytosis. Haematologica. 2011;96:1148–1152. doi: 10.3324/haematol.2011.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Habib LK, Finn WG. Unsupervised immunophenotypic profiling of chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2006;70:124–135. doi: 10.1002/cyto.b.20091. [DOI] [PubMed] [Google Scholar]
- 19.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 20.Hunter T, Pines J. Cyclins and cancer: II, cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 21.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 22.Williams ME, Swerdlow SH, Rosenberg CL, et al. Chromosome 11 translocation breakpoints at the PRAD1/ cyclin D1 gene locus in centrocytic lymphoma. Leukemia. 1993;7:241–245. [PubMed] [Google Scholar]
- 23.Williams ME, Westermann CD, Swerdlow SH. Genotypic characterization of centrocytic lymphoma: frequent rearrangement of the chromosome 11 bcl-1 locus. Blood. 1990;76:1387–1391. [PubMed] [Google Scholar]
- 24.Hoyer JD, Hanson CA, Fonseca R, et al. The (11;14) (q13;q32) translocation in multiple myeloma: a morphologic and immunohistochemical study. Am J Clin Pathol. 2000;113:831–837. doi: 10.1309/4W8E-8F4K-BHUP-UBE7. [DOI] [PubMed] [Google Scholar]
- 25.Cook JR, Hsi ED, Worley S, et al. Immunohistochemical analysis identifies two cyclin D1+ subsets of plasma cell myeloma, each associated with favorable survival. Am J Clin Pathol. 2006;125:615–624. doi: 10.1309/BDR9-59TT-4JU6-388C. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Ketterling RP, Hanson CA, et al. A case of hairy cell leukemia with CCND1-IGH@ translocation: indolent non-nodal mantle cell lymphoma revisited. Am J Surg Pathol. 2011;35:1080–1084. doi: 10.1097/PAS.0b013e31821ddaec. [DOI] [PubMed] [Google Scholar]
- 27.Chen YH, Gao J, Fan G, Peterson LC. Nuclear expression of sox11 is highly associated with mantle cell lymphoma but is independent of t(11;14)(q13;q32) in non-mantle cell B-cell neoplasms. Mod Pathol. 2010;23:105–112. doi: 10.1038/modpathol.2009.140. [DOI] [PubMed] [Google Scholar]
- 28.Cheuk W, Wong KO, Wong CS, et al. Consistent immunostaining for cyclin D1 can be achieved on a routine basis using a newly available rabbit monoclonal antibody. Am J Surg Pathol. 2004;28:801–807. doi: 10.1097/01.pas.0000126054.95798.94. [DOI] [PubMed] [Google Scholar]
- 29.Abboudi Z, Patel K, Naresh KN. Cyclin D1 expression in typical chronic lymphocytic leukaemia. Eur J Haematol. 2009;83:203–207. doi: 10.1111/j.1600-0609.2009.01276.x. [DOI] [PubMed] [Google Scholar]
- 30.O’Malley DP, Vance GH, Orazi A. Chronic lymphocytic leukemia/small lymphocytic lymphoma with trisomy 12 and focal cyclin D1 expression: a potential diagnostic pitfall. Arch Pathol Lab Med. 2005;129:92–95. doi: 10.5858/2005-129-92-CLSLLW. [DOI] [PubMed] [Google Scholar]
- 31.Sola B, Roue G, Duquesne F, et al. Expression of cyclins D-type in B-chronic lymphoproliferative disorders. Leukemia. 2000;14:1318–1319. doi: 10.1038/sj.leu.2401805. [DOI] [PubMed] [Google Scholar]
- 32.Yang WI, Zukerberg LR, Motokura T, et al. Cyclin D1 (Bcl-1, PRAD1) protein expression in low-grade B-cell lymphomas and reactive hyperplasia. Am J Pathol. 1994;145:86–96. [PMC free article] [PubMed] [Google Scholar]
- 33.Matutes E, Owusu-Ankomah K, Morilla R, et al. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia. 1994;8:1640–1645. [PubMed] [Google Scholar]
- 34.Swerdlow SH, Habeshaw JA, Murray LJ, et al. Centrocytic lymphoma: a distinct clinicopathologic and immunologic entity—a multiparameter study of 18 cases at diagnosis and relapse. Am J Pathol. 1983;113:181–197. [PMC free article] [PubMed] [Google Scholar]
- 35.Bosch F, Campo E, Jares P, et al. Increased expression of the PRAD-1/CCND1 gene in hairy cell leukaemia. Br J Haematol. 1995;91:1025–1030. doi: 10.1111/j.1365-2141.1995.tb05429.x. [DOI] [PubMed] [Google Scholar]
- 36.Balogh Z, Reiniger L, Rajnai H, et al. High rate of neoplastic cells with genetic abnormalities in proliferation centers of chronic lymphocytic leukemia. Leuk Lymphoma. 2011;52:1080–1084. doi: 10.3109/10428194.2011.555889. [DOI] [PubMed] [Google Scholar]
- 37.Ellison DJ, Nathwani BN, Cho SY, et al. Interfollicular small lymphocytic lymphoma: the diagnostic significance of pseudofollicles. Hum Pathol. 1989;20:1108–1118. doi: 10.1016/0046-8177(89)90231-1. [DOI] [PubMed] [Google Scholar]
- 38.Fend F, Quintanilla-Martinez L, Kumar S, et al. Composite low grade B-cell lymphomas with two immunophenotypically distinct cell populations are true biclonal lymphomas: a molecular analysis using laser capture microdissection. Am J Pathol. 1999;154:1857–1866. doi: 10.1016/S0002-9440(10)65443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campo E, Swerdlow SH, Harris NL, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117:5019–5032. doi: 10.1182/blood-2011-01-293050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nodit L, Bahler DW, Jacobs SA, et al. Indolent mantle cell lymphoma with nodal involvement and mutated immunoglobulin heavy chain genes. Hum Pathol. 2003;34:1030–1034. doi: 10.1053/s0046-8177(03)00410-6. [DOI] [PubMed] [Google Scholar]
- 41.Dictor M, Ek S, Sundberg M, et al. Strong lymphoid nuclear expression of SOX11 transcription factor defines lymphoblastic neoplasms, mantle cell lymphoma and Burkitt’s lymphoma. Haematologica. 2009;94:1563–1568. doi: 10.3324/haematol.2009.008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica. 2009;94:1555–1562. doi: 10.3324/haematol.2009.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70:1408–1418. doi: 10.1158/0008-5472.CAN-09-3419. [DOI] [PubMed] [Google Scholar]
- 44.Malavasi F, Deaglio S, Damle R, et al. CD38 and chronic lymphocytic leukemia: a decade later. Blood. 2011;118:3470–3478. doi: 10.1182/blood-2011-06-275610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falini B, Fizzotti M, Pucciarini A, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–2092. [PubMed] [Google Scholar]
- 46.Granziero L, Ghia P, Circosta P, et al. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2777–2783. doi: 10.1182/blood.v97.9.2777. [DOI] [PubMed] [Google Scholar]
- 47.Stein H, Bonk A, Tolksdorf G, et al. Immunohistologic analysis of the organization of normal lymphoid tissue and non-Hodgkin’s lymphomas. J Histochem Cytochem. 1980;28:746–760. doi: 10.1177/28.8.7003001. [DOI] [PubMed] [Google Scholar]
- 48.Swerdlow SH, Murray LJ, Habeshaw JA, et al. Lymphocytic lymphoma/B-chronic lymphocytic leukaemia: an immunohistopathological study of peripheral B lymphocyte neoplasia. Br J Cancer. 1984;50:587–599. doi: 10.1038/bjc.1984.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuboi K, Iida S, Inagaki H, et al. MUM1/IRF4 expression as a frequent event in mature lymphoid malignancies. Leukemia. 2000;14:449–456. doi: 10.1038/sj.leu.2401696. [DOI] [PubMed] [Google Scholar]
- 50.Damle RN, Temburni S, Calissano C, et al. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood. 2007;110:3352–3359. doi: 10.1182/blood-2007-04-083832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patten PE, Buggins AG, Richards J, et al. CD38 expression in chronic lymphocytic leukemia is regulated by the tumor microenvironment. Blood. 2008;111:5173–5181. doi: 10.1182/blood-2007-08-108605. [DOI] [PubMed] [Google Scholar]