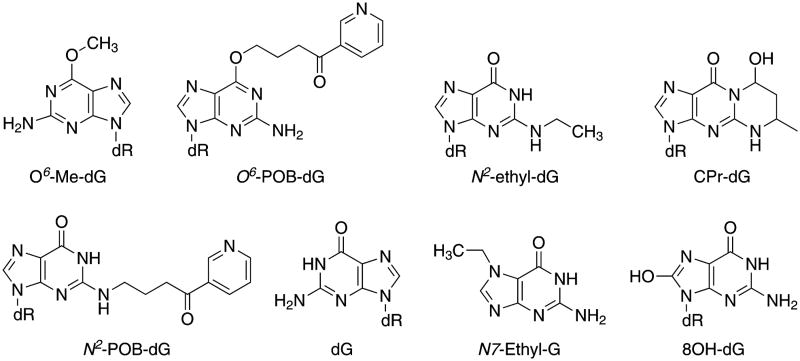
1. Introduction
Structural modifications of nucleobases within the deoxyribonucleic acid (DNA) of living cells can be induced as a result of actions of specialized DNA-modifying enzymes (creating epigenetic DNA modifications) or may result from exposure to reactive endogenous and exogenous electrophiles and oxidants (creating DNA adducts). Epigenetic DNA modifications such as 5-methylcytosine (MeC) are important regulators of cell function that influence chromatin structure and levels of gene expression. DNA methyltransferases (DMTs) catalyze the addition of the C-5 methyl group to cytosine nucleobases.1 DMTs preferentially recognize hypomethylated 5′-CG-3′ sequences, producing epigenetic modifications which preserve DNA methylation patterns. C-5 cytosine methylation controls gene expression by mediating the binding of specific proteins (methyl-CpG binding proteins) to MeCG sites, followed by the recruitment of histone-modifying enzymes that promote chromatin remodeling.2 Recent studies have discovered additional cytosine modifications, e.g. 5-hydroxymethyl-C, and 5-formyl-C, and 5-carboxyl-C; these modifications have been hypothesized to be demethylation intermediates or they may possess their own epigenetic functions within cells.3-5
In contrast to epigenetic modifications, chemical DNA damage including nucleobase alkylation, oxidation, deamination, and cross-linking occurs at a variety of sites, including the N-7, O-6, C-8, and N-2 of guanine; the N-1, N-3, and N-7 of adenine; the O-2 and O-4 of thymine; and the O-2 and N-4 of cytosine (Scheme 1 and Chart 1).6 Some carcinogens are inherently reactive towards DNA, while others must first be metabolically activated to electrophilic intermediates (e.g. epoxides, quinone methides, diazonium ions, and nitrenium ions), which subsequently bind to DNA producing nucleobase adducts (Figure 1).6 All living cells contain extensive DNA repair systems responsible for removing nucleobase lesions. If structurally modified DNA bases escape repair, they may induce base mispairing during DNA replication; thus, the chemical damage would be converted into permanent genetic damage (mutations).6 Accumulation of mutations in genes controlling cell growth, proliferation, programmed cell death, and cell differentiation is likely to cause cancer.7-10
Scheme 1.
DNA sites frequently modified by carcinogens and their metabolites.
Chart 1. Structures of representative DNA adducts.
a1,N6-etheno-2′-deoxyadenosine (εAdo); 3,N4-etheno-2′-deoxycytosine (εdCyd); O6-methyl-2′-deoxyguanosine (O6-Me-dG); N7-ethylguanine (N7-Me-G); 8-oxo-7,8-dihydro-2′deoxyguanosine (8-oxo-dG); 8-oxo-7,8-dihydro-2′deoxyadenosine (8-oxo-dA); N7-ethylguanine (N7-Ethyl-G); 1, 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2′-deoxyadenosine (1,N6 -αHMHP-dA); 1,N6-(2-hydroxy-3-hydroxymethyl-propan-1,3-diyl)-2′-deoxyadenosine (1,N6 -γHMHP-dA)
Figure 1.
Central role of DNA adducts in chemical carcinogenesis.
Due to their central role in chemical carcinogenesis, DNA adducts are considered the true mechanism-based biomarkers of carcinogen exposure. The presence of DNA adducts within a given tissue can be correlated to the formation of reactive intermediates available for binding to DNA and other biomolecules.11 Unlike hemoglobin adducts that reflect a cumulative exposure to carcinogens over time, DNA adducts provide information on the burden of DNA damage within a given tissue at a specific time. Adducts can be used to quantify the capacity of DNA repair systems and to assess the potential for genetic damage as a result of faulty replication. By employing these measurements, DNA adduct levels have been utilized to set human exposure limits for industrial and environmental chemicals and also to identify individuals and populations at risk for developing cancer.11-14
The concentrations of epigenetically modified DNA bases in vivo are relatively high (e.g. four MeC per 100 of total nucleobases), however, the amounts of chemically induced DNA adducts in animal and human tissues can be quite low, in the range of 0.01 - 10 adducts per 108 normal nucleotides. Therefore, analytical methods used for quantifying carcinogen-DNA adducts must be ultra-sensitive, accurate, and specific, allowing the quantitation of low abundance DNA lesions in the presence of a large molar excess of normal nucleosides. Early studies of DNA damage utilized radiolabel-based assays such as 32P-postlabeling methods to measure adduct levels.15-18 Recent developments in mass spectrometry instrumentation have offered an alternative approach that provides both accurate and sensitive quantitation and structural information for the damaged bases, without the need for radioactivity.12 DNA adduct structure can be established using tandem mass spectrometry experiments, while mass spectrometry in combination with stable isotope labeled internal standards (isotope dilution HPLC-ESI-MS-MS or IDMS) is considered a golden standard for DNA adduct analysis due to its high specificity, sensitivity, and accurate quantification.14 Furthermore, mass spectrometry can be used for sequencing native and structurally modified DNA.
The present review is devoted to the applications of mass spectrometry to DNA adduct and epigenetic DNA modification identification, screening, and quantitation. We will also discuss the use of MS based approaches to map the distribution of DNA modifications along DNA duplexes and to establish the biological consequences of DNA adduct formation in cells. Taken together, this article provides an overview of the contributions of mass spectrometry to the field of chemical carcinogenesis and epigenetics, with a primary focus on the new developments and recent advances in the field.
2. Overview of mass spectrometry instrumentation employed in the analysis of DNA modifications
In mass spectrometry (MS) methodology, analyte ions are separated according to their mass to charge (m/z) ratios, providing a very accurate and selective mode of detection of specific molecular species. The selectivity of MS analysis improves as the mass resolution and mass accuracy of the MS instrument is increased. Additional specificity is provided by using tandem mass spectrometry monitoring for both intact ions and mass fragments. In spite of the high selectivity provided by mass spectrometry, the extreme complexity of biological samples requires that the analyte be separated from the bulk of the sample matrix in order to achieve trace level detection of DNA adducts (e.g. 1 adduct per 109 normal nucleosides). This separation is accomplished by coupling chromatography to mass spectrometry. A direct coupling of liquid chromatography (LC) and MS became a routine and extremely powerful analytical tool with the development of the electrospray ion source. Electrospray ionization provides a “soft”, robust, and efficient interface enabling the ionization of analytes directly from mostly aqueous solutions at atmospheric pressure. The successful coupling of LC and MS techniques has led to an explosion in the capabilities of existing mass spectrometry instrumentation and catalyzed the development of new technologies available for the analysis of DNA modifications. Many types of mass analyzers are commercially available including quadrupoles, ion traps, orbital traps (Thermo Scientific Orbitrap™ technology), and time of flight (TOF) mass spectrometers19;20 and these mass analyzers can be combined forming “hybrid” instruments with further increased capabilities. New LC-MS approaches are being developed including column switching, nanospray operation, and chip based methodologies. These approaches are increasing the sensitivity and specificity of MS analyses enabling the detection of DNA adducts in occupationally exposed populations, in cancer patients treated with alkylating drugs, and in individuals with no known carcinogen exposure.
2.1 Liquid chromatography
Efficient separation of DNA adducts from normal/unmodified nucleosides/nucleobases and other components of biological samples is essential for their sensitive detection by mass spectrometry. Such separation can be achieved online using LC-MS systems. A wide variety of liquid chromatography stationary phases with differing selectivity are available, including non-polar reverse phase packing with and without modifiers imbedded within the stationary phase, polar hydrophilic interactions phases (HILIC), as well as the less common ion exchanger and mixed-mode packing materials. The majority of normal and structurally modified nucleosides/nucleobases can be resolved from normal bases and other sample components on reverse phase chromatographic columns using C18 stationary phases, although particularly polar DNA lesions may require the use of more retentive porous graphitic carbon phases (e.g. Hypercarb, available from Thermo Scientific).21 In addition, normal phase chromatography (HILIC) has been successfully used for the analysis of some DNA adducts,22-25 although sample solubility in the typically highly organic solvents used in this technique may be problematic, especially for complex biological samples.
The choice of column length, diameter, and the particle size of the stationary phase can substantially influence the efficiency and throughput of chromatographic separations. Narrower columns typically providing improved sensitivity by generating sharper chromatographic bands. Longer columns generally provide an increased ability to separate compounds, but at the expense of longer HPLC runs and potentially reduced sensitivity as chromatographic peaks widen. Smaller particles within the column provide a greater surface area of the stationary phase and subsequently improve the efficiency of the chromatographic separation and sensitivity, but at the expense of increased column back pressure, which may limit the throughput of the system. A wide range of column dimensions have been used for DNA adduct analysis, with typical diameters of 0.5 – 2 mm, lengths of 50 – 150 mm, and particle sizes of 1.8 – 5 μm. HPLC flow rates are dependent primarily on column dimensions, with flows ranging from 250 nL/min to 1 mL/min used for the analysis of DNA adducts. As described below, more sophisticated HPLC systems incorporating on-line analyte enrichment via column switching techniques have been recently introduced.20;26-30 In some cases, this approach allows for direct injection of unpurified samples, minimizing or eliminating sample preparation steps.
Capillary electrophoresis (CE)31 has been used as an alternative to liquid chromatography for the in-line mass spectrometric analysis of modified DNA. Both unmodified and modified nucleotides16;32-36 and oligonucleotides37-40 have been analyzed, the entire field has been thoroughly reviewed.41 Typical analysis is performed using negative ion electrospray ionization and with sample stacking35;42 for maximum sensitivity. The coupling of capillary electrophoresis with electrospray is typically done with one of three different source geometries,41 namely with and without sheath liquid and with a liquid junction. The most commonly used types of mass spectrometers are triple quadrupole16;33;35;43;44 and Q-TOF16;37;40 instruments operated in both MS and MS2 mode, however in theory any mass spectrometer with an electrospray ion source could be used for capillary electrophoresis-mass spectrometric analysis.
2.2 Electrospray and atmospheric pressure chemical ionization
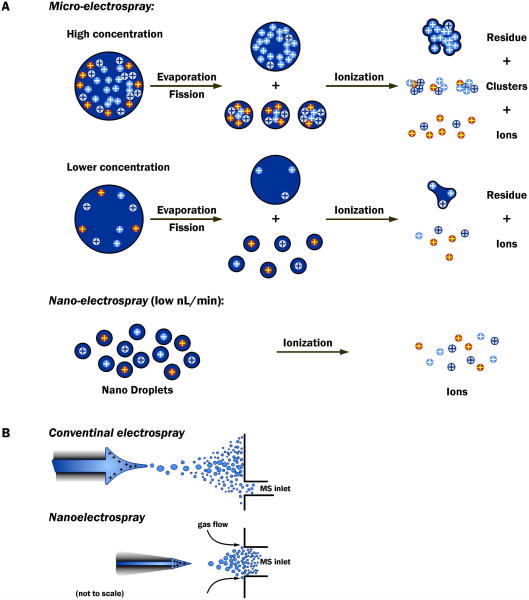
The development of electrospray ionization (ESI)45 enabled the direct coupling of mass spectrometry and liquid chromatography, and allowed for direct mass spectrometric analysis of polar molecules such as DNA nucleosides, nucleotides, and oligonucleotides.46 In an electrospray ion source, liquid is passed through an narrow orifice with a voltage (1 - 5 kV) applied either to the conductive tip of the orifice or directly to the liquid through a liquid-liquid junction.47;48 This results in the production of a mist of finely charged aerosol particles. Within the mist, the size of charged droplets gradually decreases, ultimately resulting in the production of gas phase ions under atmospheric pressure conditions. The actual mechanism by which the charged aerosol particles produce “naked” gas phase ions is complex and not fully understood, but has been proposed to involve ion desorption and/or droplet fission.49;50
The conversion ions within the liquid phase to gas phase ions is a competitive process due to the finite amount of space and charge available on the charged droplets.51 Thus other components of the sample can interfere with, or overpower the ionization of the analyte of interest. The phenomenon of background materials interfering with analyte ionization is commonly observed in trace ESI analysis and is referred to as ion suppression.52,53 As ion suppression will reduce the sensitivity and reproducibility of an assay, the phenomenon must be taken into consideration when developing LC-MS methods for analyzing DNA modifications. Sample clean up procedures and high efficiency HPLC separations are effective means of separating the analyte of interest from the bulk of the components within the sample, thereby assuring optimal electrospray ionization efficiency of the analyte and subsequently sensitive mass spectrometry.
Depending on the polarity of the voltage applied to the electrospray source, the technique generates either protonated or deprotonated ions and therefore requires that the analyte of interest contains either basic or acidic functionalities. DNA molecules are polyanions due to their negatively charged phosphodiester backbone and are thus typically analyzed in negative ion mode. Nucleoside monophosphates are similarly often analyzed using ESI- -MS.54 In contrast, free DNA nucleobases and nucleosides are readily protonated at pH < 5, and are therefore typically detected in positive ion mode.12;55 In some rare cases when the adducted base contains a strongly electron withdrawing substituents such as a nitro group, negative ion mode is preferred.56
The ionization efficiency of ESI is inversely proportional to the HPLC flow rate. Therefore, improved sensitivity is realized by reducing HPLC flow rates and subsequently decreasing the HPLC column diameter to maintain optimal chromatographic performance at reduced flows. 57-59 Column miniaturization is commonly used to improve assay sensitivity and allows for the analysis from the limited size of typical biological samples. HPLC flow rates of 140 - 600 nL/min and column diameters of 0.075 mm have been used to maximize the sensitivity of quantitative DNA adduct assays58;60-69 (see Section 5.5.4) and to identify and characterize novel adducts.70-73
Atmospheric pressure chemical ionization (APCI) is another common ionization technique utilized to detect DNA modifications.27;28 The APCI source volatilizes the HPLC effluent in a heated nebulizer with a help of drying gas (typically nitrogen). Molecules within the effluent are ionized as they pass through a corona discharge generated by a high voltage applied to a metal needle.74 The resulting plasma produces ions of the mobile phase solvent, which in turn ionize analyte molecules usually by protonation (M+H)+ in the positive ion mode or deprotonation (M-H)- in the negative ion mode.75 The formation of solvent adduct ions is also possible. Charge transfer and electron capture can additionally occur for some analytes, resulting in very high sensitivities.76 The mean free path between charge transferring collision events is short and quickly reaches equilibrium, allowing most thermodynamically ions to be observed.57 Unlike electrospray ionization that occurs in solution, APCI is a gas phase ionization process capable of ionizing molecules which are otherwise not easily ionizable. The gas phase ionization process is additionally amenable to higher HPLC flow rates (up to 3 mL/min). Since DNA nucleosides and nucleobases contain basic functional groups and are easily and efficiently ionized by ESI, APCI is rarely used for their analysis, with the exception of rare cases when high HPLC flow rates must be employed.
2.3 Matrix Assisted Laser Desorption Ionization (MALDI)
Matrix assisted laser desorption ionization (MALDI) is a powerful tool for mass spectrometric analysis of oligonucleotides and nucleic acids.77 MALDI is a “soft” ionization process producing either positively or negatively charges ions. Samples are prepared for MALDI analysis by mixing and co-crystallizing with a large excess of UV or IR-absorbing organic matrix, which is deposited onto a MALDI target. A pulsed UV or infrared laser then strikes the dried analyte/matrix solid, resulting in desorption/ablation of the sample and analyte ionization. MALDI matrices are most commonly organic acids containing a chromophore that absorbs the wavelength of the laser. The role of matrix is to absorb radiation from the laser pulse, to facilitate analyte ionization, and also to protect the analyte from radiation damage.77 The preferred matrix for the analysis of DNA is 3-hydroxypicolinic acid (3-HPA)78 while for RNA it is a mixture of 2,3,4- and 2,4,6-trihydroxy-acetophenone or 3-HPA.79 MALDI is well-suited for coupling to time-of-flight (TOF) mass spectrometry since both operate in a pulsed fashion. Furthermore, the mass ranges of ions which can be generated using MALDI matches up well with the mass range possible with TOF instrumentation. MALDI-MS of nucleic acids has been performed almost exclusively with TOF instruments with delayed ion extraction.77
2.4 Mass analyzers
Our ability to detect and characterize structural modifications of DNA is largely dependent upon the current state-of-the-art MS instrumentation and techniques; as MS instrumentation has become increasingly more powerful, our understanding of endogenous and exogenous modifications present in cellular DNA has rapidly evolved. Many types of mass analyzers are available commercially, including quadrupoles, ion traps, orbital traps, and time of flight (TOF) mass spectrometers.19 The different instruments vary in their resolution power, duty cycle, dynamic range, and MS/MS capabilities, as well as their cost and their ease of use. In this section, we will describe the main types of MS instruments available commercially and their advantages and disadvantages for the qualitative and quantitative analyses of DNA modifications.
2.4.1 Quadrupole mass analyzers
A quadrupole mass analyzer (Q) consists of four parallel conductive circular or hyperbolic rods. Asymmetric RF and DC voltages are applied to the two sets of opposing rods, forming an electromagnetic field allowing the passage (or filtering) of specific ions through the center space of the apparatus. The voltages applied to the rod pairs can be selected to enable only ions which fall within a narrow range of mass to charge ratio (m/z) to have a stable trajectory;13;19 these selected ions are able to pass through the quadrupole mass filter. Alternatively, quadrupoles can be operated in the “RF only mode” allowing all ions entering the quadrupole to be transmitted. Triple quadrupoles (Q1q2Q3) contain two mass filtering quadrupoles (Q1 and Q3) and a third quadrupole which serves as a collision cell/ion guide (q2) positioned between the two mass filters. Triple quadrupole mass analyzers are well suited for tandem mass spectrometry (MS/MS) experiments. The collision cell is filled with an inert collision gas (typically Ar or N2) at a pressure of 1-2 mTorr and is operated in RF only mode, allowing transmission of all ions. Ions within the specified m/z range exit the first quadrupole Q1 and are accelerated to 10-50 V. Upon entering the collision cell (q2), they collide with the inert gas, resulting in cleavage of weaker bonds, a process called collision induced dissociation (CID). The resulting fragment ions exit the collision cell and are then analyzed in the third quadrupole (Q3). Common modes of operating triple quadrupole instruments are discussed in Section 2.5. Triple quadrupoles are the main type of mass analyzer used for trace quantitative analysis due to their high selectivity (especially when operated in selected reaction monitoring mode - see Section 2.5), high sensitivity, large dynamic range, high precision, relatively low cost, and ease of operation.
2.4.2 Ion trap mass analyzers
Ion trap mass analyzers work by trapping and accumulating ions within a large m/z range (e.g. 15 – 4000) within a confined space. Following the trapping period, the accumulated ions can be selectively ejected out of the trap and detected based on their m/z value, allowing for mass analysis.80 Alternatively, ion fragmentation spectra (MS2) can be acquired by ejection of all ions other than the m/z of interest, and then fragmenting the selected ions via collision induced fragmentation with background He gas, and the subsequent mass analysis of resulting fragment ions. This process of collecting and fragmenting target ions can be repeated multiple times, allowing for the acquisition of MSn spectra (n ≥ 3). Ion traps are powerful tools for acquiring structural information for novel DNA adducts, DNA sequencing, and other qualitative analysis. Although ion traps have not been traditionally used for trace quantitative analysis due to their lower duty cycle as compared to that of triple quadrupole instruments,19 they offer an additional level of selectivity when operated in MS3 mode and have been recently used for sensitive detection of minor DNA adducts in human tissues.81
2.4.3 Time of flight mass analyzers
Time of flight (TOF) mass analyzers are extended m/z range mass analyzers with a resolving power of 10,000 or better and typical mass accuracies of 2 - 5 ppm. TOF analyzers determine the m/z of ions by measuring the length of time it takes for ions to migrate through a field-free flight tube. Ions accelerated to the same kinetic energy (25-50 kV) enter a 1-2 m long drift tube and travel towards the ion detector. Since ions of higher m/z travel slower than the ions of lower mass, they separate in space according to their velocities. The flight time is inversely proportional to ion velocity, and the m/z values can be calculated from the observed flight time. TOF mass analyzers can be coupled to MALDI or electrospray ion sources.19 TOF technology has improved in recent years, with significant advances in their sampling speed, resolving power, and sensitivity.
TOF mass analyzers are powerful tools due to their ability to determine the accurate mass of detected ions, while obtaining full spectral data. A hybrid instrument consisting of a quadrupole mass filter, a collision cell, and a TOF mass analyzer (QqTOF) is popular in the field of proteomics.82 Traditionally, TOF instruments have been limited to qualitative analyses; however, QqTOF instrument have been employed for quantitation, since their high resolution capabilities can eliminate chemical noise.83 However, the typical sensitivity of QqTOF instruments is lower than that of triple quadrupoles.83
2.4.4 Orbital trap (Orbitrap) instruments
A new type of mass analyzer, the Orbitrap,84 has recently become available commercially. The Orbitrap offers excellent resolution as high as >240,000 and mass accuracy of 1-3 ppm. 85 The Orbitrap technology was originally sold solely as a hybrid instrument with the device coupled to an ion trap instrument, under the brand name of LTQ Orbitrap™ (Thermo Scientific, San Jose, CA). In addition to the original ion trap-orbital trap configuration, the Orbitrap technology is now available as a standalone device (Exactive Plus Orbitrap™) and as a hybrid instrument coupled to a quadrupole (Q Exactive™ Hybrid Quadrupole-Orbitrap). To avoid confusion between these different configurations, the mass analyzing device will be referred to here as an orbital trap.
The orbital trap is an ion trapping device 84 consisting of a central “spindle” electrode and an outer “barrel” electrode. Discrete packets of ions are injected into the region between the electrodes at a specific kinetic energy and are trapped between the central “spindle” electrode and the outer “barrel” electrode by application of a static electrostatic field. Ions of all m/z values follow a circular orbit around the z axis, and the frequency of their oscillation (ω) is dependent on their m/z values: ω = [(z/m) × k]1/2, where z is the ion charge, m is the ion mass, and k is dependent on the field strength. Ion oscillations are detected as image current and are transformed into mass spectra using a Fourier transformation (FT).
The original orbital trap mass analyzers were the hybrid ion trap-orbital trap instruments, and were used almost exclusively for proteomics analyses.85-87 The standalone orbital trap instruments introduced more recently (Exactive Plus Orbitrap™) can be used for a variety of applications, including small molecule analysis.88-92 The quadrupole-orbital trap hybrid instrument which was introduced in 2011 (Q Exactive™ Hybrid Quadrupole-Orbitrap Mass Spectrometer) is well suited for proteomics and small molecule quantitation.92-96 Our research group has successfully employed orbital trap technology to improve the sensitivity of DNA adduct detection in human samples.66 Excellent sensitivity and greatly improved signal to noise ratios have been achieved for complex samples containing trace amounts of DNA adducts in the presence of a large excess of normal nucleosides (see Section 6.4).
2.4.5 Hybrid instruments
Several mass analyzers of different types can be combined together in to a single system to form “hybrid” instruments with expanded capabilities. For example, quadrupoles and ion traps have been coupled to TOF, ion trap, and orbital trap analyzers. Another example is the use of two TOF analyzers (TOF-TOF) in sequence. Often mass analyzers (quadrupole, TOF, orbital trap) which are not capable of MS2 operation individually are coupled together to attain that capability as a hybrid instrument. The commercially available instruments and their capabilities have been recently reviewed.55;97
2.5 MS Scanning Methods
Several ion scanning modes can be used to analyze DNA modifications, including selected ion monitoring (SIM), selected reaction monitoring (SRM, also MRM), constant neutral loss (CNL), precursor scanning, product ion scanning, and data dependent scanning (Scheme 2) SIM and SRM methods offer the greatest sensitivity and are used primarily for targeted detection and quantitative analysis of DNA adducts, while the other three modes provide structural information and are useful for identification and characterization of novel DNA modifications.
Scheme 2.
Tandem mass spectrometry scanning modes.
SIM is the simplest way to improve ion detection sensitivity when using a single stage mass spectrometer unable to perform tandem mass spectrometry experiments. SIM adds selectivity by monitoring ions within a narrow m/z range, increasing sensitivity 100-fold due to an improved duty cycle. However, no structural information beyond molecular weight is obtained from SIM analyses.
Many of the most sensitive scanning methods require the use of instruments capable of performing MS/MS experiments, such as triple quadrupoles (Q1q2Q3) and the quadrupole linear ion trap (QqQtrap). Product ion mode (Scheme 2A) involves the isolation of the analyte ions in Q1, followed by their fragmentation in q2 and mass analysis of the fragment ions in Q3. This mode is useful for characterizing novel DNA modifications, confirming analyte identity, and providing information regarding their chemical structure.
In the precursor ion scanning mode (Scheme 2B), the Q3 is set to pass only the fragment ions of interest, while Q1 is scanned within a broader range, so that only analyte ions producing fragment ions of interest are recorded. Precursor ion scanning mode can be used to identify adducts based upon the common chemical structure of DNA, e.g. guanine, adenine, cytosine, or thymine adducts.
The best sensitivity is typically achieved using the selected reaction monitoring (SRM) mode (Scheme 2C). In SRM mode with a triple quadrupole mass spectrometer (Section 2.4.1), Q1 selects ions within a user-specified narrow m/z range, which then enter the collision cell (q2) and are fragmented by CID. The resulting fragment ions (product ions) enter Q3, and only product ions of the specified m/z pass through Q3 and are recorded at the detector. In SRM experiments, only ions of the user-specified mass that produce specific fragments under CID conditions are detected. These multi-step criteria add selectivity and improve signal to noise ratios of the assay, since it is unlikely that sample components will have both the same precursor and fragment masses as the analyte of interest.
Constant neutral loss (CNL) scanning mode (Scheme 2D) allows the user to specify a mass difference between the precursor and product ions (fragment mass). This method is typically used when unknowns or multiple analytes with similar structures are being analyzed, assuming all of the targeted compounds lose the same neutral fragment under CID conditions (e.g., the neutral loss of deoxyribose sugar from DNA nucleosides or the loss of a phosphate group from nucleotides).98 In the CNL mode, Q1 scans and transmits all m/z ions within a broad range, which are then dissociated in q2 before passing to Q3, where the resulting product ions are scanned. Q1 and Q3 scan are synchronized but offset from each other by the user-defined mass difference.
Data dependent scanning is based on repeated acquisition of full scan spectra and MSn spectra (Scheme 2E). This scanning mode requires a real time decision making by the mass spectrometry software based upon initial programming of the data dependent method. The simplest data dependent methods involve acquisition of a full scan spectrum, followed by MS2 fragmentation of the most abundant ions. The masses of ions selected for fragmentation are then put into an exclusion list and these masses are no longer eligible for fragmentation upon observation in subsequent full scan spectra. However, the exclusion list has a time limit, so that ions of a given mass once again become eligible for MS2 analysis later on during the chromatographic run. The data acquisition cycle of full scan data acquisition and MS2 analysis of the most abundant ions is repeated for the entire HPLC-MS/MS run. This approach is commonly used in proteomics analyses and can be applied to unknown adduct screening (adductomics, Section 4.7). Alternatively, an inclusion list containing masses of interest can be used to trigger MS2 analysis of targeted analytes. Overall, data dependent acquisition is a powerful approach useful for screening and identifying novel DNA modifications.
3. Sample Preparation
Sample preparation for DNA adduct analysis by mass spectrometry typically involves several standard steps, e.g. DNA hydrolysis, analyte enrichment, and fortifying with an appropriate internal standard (Scheme 3). Genomic DNA can be obtained from a variety of sources of differing complexity. Most methods include DNA hydrolysis and a purification step that removes the bulk of sample matrix in order to avoid ion suppression and to achieve optimal sensitivity.
Scheme 3.
Sample processing scheme for HPLC-ESI-MS/MS analysis of DNA adducts.
3.1 DNA Sources for adduct analyses
Common biological sources of DNA for adduct analysis include tissues, blood, oral buccal cells, and urine. Depending on lesion abundance and analytical method sensitivity, quantitative analysis of DNA modifications requires between 1 and 200 μg of DNA. DNA isolated from organ tissues including liver, lungs, kidney, and brain are often used in laboratory animal studies.67;68;99 Since these tissues are not available from human biomonitoring studies, DNA can be isolated from human blood, saliva, and oral buccal cells in these cases.62;100 Purified DNA (e.g. calf thymus DNA) and synthetically produced oligonucleotides are commercially available and can be used for method development, in vitro experiments, and mechanistic/structural experiments.
Some DNA adducts are excreted as free nucleobases and nucleosides in urine due to their spontaneous hydrolysis and/or active repair.101-103 Human urine is a complex matrix containing high concentrations of salts and other polar compounds which can interfere with mass spectrometric analysis of polar DNA adducts. Careful consideration of these potential interfering constituents is required when developing effective sample preparation and on-line liquid chromatography methods necessary for the analysis of urinary adducts. Several laboratories have successfully analyzed urinary nucleosides in humans by HPLC-ESI-MS/MS.24;104-107
3.2 DNA hydrolysis
Genomic DNA strands are very long, linear biopolymers consisting of a heterogeneous sequence of four nucleosides (deoxyadenosine, deoxyguanosine, deoxycytidine, and thymidine monophosphates) connected by phosphodiester bonds. With the exception of experiments with pure synthetic oligodeoxynucleotides of a specified sequence, MS analysis of DNA modifications requires DNA hydrolysis to the corresponding monomers (nucleobases, nucleosides, or nucleotides). This is because mass spectrometric analysis of DNA monomers is characterized by an increased sensitivity, accuracy and precision as compared to the analysis of DNA polynucleotides. Some DNA adducts, including 7-alkylguanines, 3-alkyladenines, and 2-alkylcytosines, can be readily and selectively released from a DNA backbone as free bases upon heating (thermal hydrolysis).6;108 Hydrolytically stable adducts may be analyzed by enzymatic digestion of DNA by nucleases and phosphatases to release deoxynucleosides. (Care should be taken when digesting chemically modified DNA as many adducts are capable of blocking nuclease enzymes, leading to incomplete enzymatic hydrolysis).109 Alternatively, heating in the presence of acid (mild acid hydrolysis) releases all purines as free bases, including adenine and guanine adducts.6,110
As a word of caution, some DNA lesions can be artificially generated during enzymatic digestion and other sample processing steps. For example, 8-oxoguanine (8-oxo-dG) is readily produced from dG in an aerobic environment.111-113 The artifactual formation of 8-oxo-dG during DNA isolation and sample processing can be minimized by adding antioxidants, metal chelators, and free radical trapping agents.114 Artifactual deamination of cytosine and adenine can be caused by deaminases present as contaminants in commercial nuclease and phosphatase enzymes used for enzymatic DNA digestion. dC and dA deaminase inhibitors can be used to avoid adventitious formation of 2′-deoxyuridine (dU) and 2′-deoxyinosine (dI), respectively, during DNA isolation and processing.115-117
3.3 Sample enrichment strategies
DNA hydrolysates require several cleanup steps to enrich DNA adducts and to remove the bulk of unmodified nucleosides, proteins, inorganic salts, and other sample components that can interfere with MS analysis (Scheme 3).13;14 Some examples of sample preparation methods used in DNA adduct analyses include ultrafiltration, liquid/liquid extraction, solid phase extraction, immunoaffinity purification, and off-line HPLC.
Ultrafiltration can be used to remove partially depurinated DNA remaining following thermal or acid hydrolysis of DNA.108;110 This method is also useful for removing proteins following enzymatic hydrolysis of DNA. Disposable ultrafiltration centrifugation devices are available commercially with a typical molecular weight cutoff used to remove the DNA backbone of 3-10 kDa.
Liquid/liquid extraction with organic solvents can be used to isolate hydrophobic DNA adducts, while polar components of the biological sample remain in the aqueous phase.118 This method is effective and economical, but is limited to hydrophobic lesions such as those of polycyclic aromatic hydrocarbons and aromatic amines.
Solid phase extraction (SPE) has much broader application than liquid/liquid extraction due to the large variety of SPE cartridges available commercially. In SPE, samples are loaded on a small disposable column packed with chromatographic stationary phase under conditions that facilitate analyte binding. Following several washing steps to remove unwanted components, the analytes of interest are eluted with a stronger solvent. Depending on the structure of the analyte, SPE separations can employ cation exchange, anion exchange, or mixed mode liquid chromatography packing to isolate specific DNA lesions from complex mixtures.66;108;119;120 Immunoaffinity purification is based on a similar principle, with the exception that the sample enrichment is achieved via analyte binding to a monoclonal or polyclonal antibody attached to a solid support.121
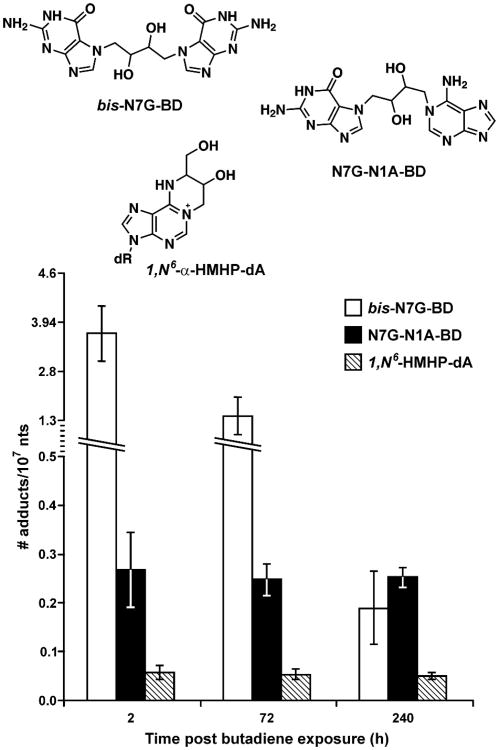
Off-line HPLC separation can be effectively used to remove the bulk of impurities and to enrich the analyte prior to MS analysis. HPLC fractions containing the compounds of interest are selected, concentrated, and injected onto an HPLC column for HPLC-MS analysis. Retention time markers can be used during offline HPLC cleanup to control for any variations in analyte retention time between samples, e.g. due to small temperature changes and the influence of sample matrix. Our laboratory has successfully employed offline HPLC cleanup to enrich guanine-guanine adducts of 1,2,3,4-diepoxybutane (bis-N7G-BD) prior to their quantitative analysis by nanoHPLC-nanospray MS/MS.68;69 In general, offline HPLC purification prior to HPLC-MS/MS provides better sensitivity than methodologies employing SPE cleanup, but the technique is more time consuming and susceptible to carryover problems.
3.4 MS Detection of Nucleoside and Nucleobase Adducts
The formation of DNA adducts can occur via modification of the nucleic base or phosphate moieties of DNA. Structural modifications of DNA nucleobases are directly related to chemical carcinogenesis8 and are more frequently studied than phosphate group modifications. Therefore, discussions within this section are focused on detecting nucleobase DNA adducts by mass spectrometry.
Chromatographic separation of DNA adducts can be accomplished using gas or liquid chromatography. DNA nucleosides, nucleotides, and nucleobases are polar species with limited volatility, requiring extensive derivatization steps in order to make them amenable to GC separation.122 In contrast, liquid chromatography can be used to separate modified nucleosides, nucleotides, nucleobases, and oligonucleotides directly, with no need for derivatization. Therefore, LC-MS has become the preferred approach for DNA adduct analysis. Typically DNA adduct analyses involves HPLC coupled with atmospheric pressure ionization methods such as electrospray and atmospheric pressure chemical ionization. DNA nucleosides and nucleobases are usually analyzed as positive ions, since they are readily protonated. The resulting [M+H]+ ions are available for detection in full scan or selected ion monitoring (SIM) mode (Section 2.5). MS/MS fragmentation can be utilized to obtain structural information.
4. DNA adduct identification and screening
4.1 Molecular formula determination
High resolution mass spectra can be used to determine the elemental composition of structurally modified nucleotides, nucleosides, and nucleobases, facilitating the structural characterization of these targets. Recent developments in mass spectrometry instrumentation has provided a number of different systems capable of routinely acquiring high resolution mass spectra.55 Mass accuracy of 2 - 5 ppm is routinely achieved when using orbital trap (Orbitrap™) and time-of-flight technologies (Sections 2.3.3 and 2.3.4). Both instruments, when combined with a quadrupole or an ion trap mass analyzer into a hybrid instrument, can be used to determine the accurate mass of product ions generated from CID. High resolution mass measurements (resolving powers of > 30,000) can be useful when analyzing complex biological samples, especially when the analyte is present at trace levels. Low abundance analytes can be detected in the presence of a complex sample matrix because accurate mass measurements enable distinguishing analyte ions from potentially interfering compounds of similar mass. The typical resolving power of time-of-flight mass analyzers is 15,000 – 50,000, while the resolving power of orbital trap (Orbitrap™ technology) instruments is as high as > 240,000. FT-ICR instruments are capable of even higher resolving power (≥750,000), depending upon the size of the magnet employed. For orbital traps and FT-ICR mass spectrometers, there is an inverse relationship between resolving power and scan speed, which hinders the application of these instruments to analytical problems requiring rapid analysis. Among the three types of mass analyzers capable of high resolution mass measurements, FT-ICR instruments are the most expensive and require regular maintenance of the superconducting magnet and therefore are rarely used for DNA adduct detection.
MS resolving power required to conclusively determine the molecular formula of a given analyte is dependent upon its molecular mass and the number of possible elements considered.123 This is illustrated in Table 1, where the numbers of possible molecular formulas were calculated at 2 ppm and 5 ppm mass accuracy and considering various combinations of biologically relevant elements for hypothetical analytes of increasing molecular mass: m/z 152.05669 (e.g. guanine), 268.10403 (e.g. dG), 299.12510 (e.g. POB-Gua); 415.17244 (POB-dG); and 570.19832 (BPDE-dG). It is clear from this representative example that as the molecular size increases, it becomes more difficult to assign the elemental composition of an unknown nucleoside, although further increasing the resolution will reduce the number of possibilities. Simple approaches can be applied to constrain the number of potential molecular formulas use “rings-and-double-bond equivalents”124 and “nitrogen rule”125 approaches, although they should be used with care due to certain limitations.126
Table 1.
Comparison of possible molecular formulas for a selection of nucleosides and nucleobases at different mass accuracies and possible elemental components illustrating the relationship between analyte size, elements considered, and number of possible molecular formulas.
| 2 ppm | 5 ppm | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| M·H+ (amu) | C,H,N,O | C,H,N,O,P | C,H,N,O,P,S | C,H,N,O | C,H,N,O,P | C,H,N,O,P,S |
| 152.05669a | 1 | 1 | 1 | 1 | 2 | 3 |
| 268.10403b | 1 | 2 | 3 | 3 | 5 | 11 |
| 299.12510c | 1 | 3 | 5 | 4 | 11 | 16 |
| 415.17244d | 1 | 5 | 15 | 5 | 17 | 45 |
| 570.19832e | 2 | 22 | 72 | 6 | 46 | 116 |
In addition to constraining the types of elements considered when determining elemental composition of an unknown, filters can be applied to constrain the number of possible molecular formulas. A list of heuristic filtering methods has been carefully examined for determining molecular formulas by accurate mass measurement.126 One of these filters, which can be particularly powerful, includes the comparison of the observed and theoretical isotope patterns. Careful theoretical analysis of the interplay between mass accuracy and isotopic pattern measurements for molecular formula determination has been conducted by Kind and Fiehn.123 Calculation of theoretical isotope patterns can be conducted using commercial data analysis software.
For example, consider the isotopic pattern (the relative ratio of M and M+1 peaks) of an unknown at m/z 415.17244. If one includes C,H,N,O, and P as possible elements present, there are 5 possible molecular formulas when using a 2 ppm mass accuracy tolerance (Table 1), with 12Cv-1 13CHwNzOyPz/CvHwNzOyPz ratios ranging between 0.12 to 0.20. The observed CvHwNzOyPz ·H+ /12Cv-113CHwNzOyPz·H+ signal ratio can be used to determine the molecular formula of the unknown analyte (Figure 2). In addition, the number of nitrogen atoms in the molecular formula could be determined by comparing the signal from the isotopologue with a single 13C atom (12Cv-113CHw14NzOyPz·H+) to the signal from the isotopologue with a single 15N atom (CvHw15N14Nz-1OyPz·H+). A more sophisticated isotopic pattern analysis can be performed where the M+2 signal abundance is considered along with the M and M+1 isotope pattern. When this type of analysis is conducted, the best match for the compound shown in Figure 2 is C19H22O5N6·H+ (O6-POB-dG, Chart 2).
Figure 2.
Observed (top) and theoretical (bottom) accurate mass spectra of O6-pyridyloxobutyl-2-deoxyguanosine [O6-POB-dG+H]+. The top spectrum was obtained with an LTQ Orbitrap Velos (Thermo Scientific) operated at a resolution of 60,000. Shown on the bottom is the predicted spectrum generated with Xcalibur Qualbrowser software (Thermo Scientific).
Chart 2. Structures of DNA adducts used for discussion of molecular formula determination.a.
aguanine (G); 2′-deoxyguanosine (dG); O6-[4-(3-pyridyl)-4-oxobut-1-yl]guanine (O6-POB-G); O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine (O6-POB-dG); 7,8,9-trihydroxy-10-(N2-deoxyguanosyl)-7,8,9,10-tretrahydrobenzo[a]pyrene (N2-BPDE-dG)
High resolution mass spectra are useful for resolving the analyte of interest from background chemical noise and co-eluting species of very similar mass, and also for identifying and distinguishing isotopologues. In the case of O6-POB-dG, the mass of the 13C isotopologue is approximately 20 ppm higher than the mass of the 15N isotopologue, and this difference can be detected by mass spectrometer with a resolving power of ≥ 40,000 (10% definition) or ≥ 80,000 (FWHM definition). It should be noted that signals of interest must exhibit good signal to noise ratios to enable this type of analyses. Furthermore, the ion signal being evaluated should be within the linear dynamic range of the signal intensity.123
4.2 Types of MS/MS Fragmentation
Valuable structural information for unknown nucleosides, nucleotides, and nucleobases can be obtained using tandem mass spectrometry (see Section 2.5). In addition to providing fragmentation patterns for novel DNA modifications and confirming the identity of known DNA lesions, MS/MS and MSn experiments can dramatically increase the selectivity and sensitivity of assays quantifying DNA adducts.
DNA adduct assays typically utilize fragmentation reactions under low-energy collision-activated dissociation conditions (CAD), commonly referred to as collision-induced dissociation (CID). Two different types of CAD exist, namely “ion-trap-type CAD” and “beam-type CAD”.127 While ion-trap-type CAD is available on ion traps and ion trap-time-of-flight instruments, beam-type CAD is available with triple quadrupole, quadrupole-orbital trap, and quadrupole-time-of-flight instruments.127 Quadrupole ion traps and quadrupole-orbital trap instruments have both types of fragmentation available. The two fragmentation processes differ in the mechanism by which the energy is imparted to the precursor ions, forcing their fragmentation. In the case of the beam-type CAD mechanism, fragmentation occurs as a result of the energy imparted to the precursor ion as it is accelerated and subjected to multiple collisions with the inert gas within the collision cell.128 In the case of the ion-trap-type CAD mechanism, analyte ions undergo resonance excitation, leading to their energetic collisions with background gas (typically He) to produce fragment ions.128 The fragment ions which initially form are no longer in resonance with the excitation frequency used to excite the precursor ions and therefore the fragments will not undergo further fragmentation. The two different types of CAD fragmentation can result in significantly different product spectra for the same analyte (Figure 3).
Figure 3.
Comparison of “beam-type CAD” (triple quadrupole) and “ion-trap-type CAD” MS/MS spectra of N2-ethyl-2′-deoxyguanosine obtained at increasing collision energy.
4.3 MS/MS Fragmentation Pathways of Modified Nucleosides
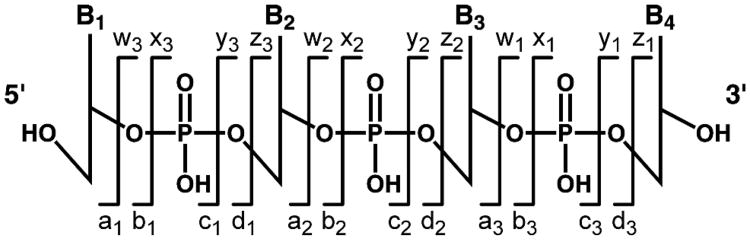
MS/MS fragmentation pathways of native and structurally unmodified nucleosides have recently been reviewed.129 All DNA nucleosides have a common structural feature, namely the presence of a deoxyribose moiety bound to one of the four nucleobases or their derivatives through a glycosidic bond. Therefore, low energy CAD spectra of structurally modified DNA nucleosides (B-X-dR, where B = nucleobase, X = modification, and dR is the deoxyribose sugar) is typically dominated by the cleavage of the glycosidic bond and a neutral loss of dR (116 amu), leading to protonated nucleobase ions ([B-X] +H)+ (Figure 3). An interesting exception has been reported by Farmer et al. for the DB[a,I]P dihydroepoxide adduct of dA, which produced mostly fragments corresponding to the DB[a,I]P moiety.30
The MS/MS fragmentation pathway corresponding to the neutral loss of dR is often used for quantitation of nucleoside adducts or for confirmation of adduct identity. Depending on the nature of the modified nucleoside, instrumentation used, and most importantly, the amount of energy used for fragmentation, other types of mass fragments may be observed. For example, Figure 3 contains product ion spectra of N2-ethyl-2′-deoxyguanosine using triple quadrupole (beam-type CAD) and ion trap-like fragmentation. For CID experiment conducted on a triple quadrupole system at a collision energy of 10 volts, fragment ions corresponding to the neutral loss of dR (m/z 180) are observed exclusively. Increasing the collision energy to 40 volts leads to the formation of additional fragment ions at m/z 163 (60%), 135 (100%), and 110 (50%) (Figure 3, left panel). In contrast, fragmentation of the same analyte using an ion trap (ion-trap CAD) leads almost exclusively to the loss of the dR group, regardless of the amount of collisional energy applied (Figure 3, right panel). As the intensity of new fragments increase with the increased collision energy in the triple quadrupole mass spectrometer, the intensity of the ion signal at m/z 180 decreases (Figure 3, left panel). In contrast, the amount of total MS/MS signal and m/z 180 signal in particular is independent of collisional activation for the similar experiment conducted using an ion trap (ion-trap-CAD) fragmentation (Figure 3, right panel).
Other MS/MS fragmentation pathways observed for structurally modified nucleosides include the loss of the substituent producing protonated nucleoside ions and the ions corresponding to carcinogen-derived portion of the molecule. In most cases, fragment ions corresponding to the protonated base of origin [B+H]+ are observed and can be used to identify the origin of a specific nucleobase modification. MS/MS spectra of a series of representative dG adducts obtained using a beam-type CAD are given in Figure 4. All modified nucleosides shown in Figure 4 produce [Gua+H]+ (m/z 152) and [Gua+H-NH3]+ ions (m/z 135) characteristic of dG adducts. The same two fragments are observed upon MS/MS fragmentation of protonated guanine (see Section 4.4).129;130 Similar results were reported by Inagaki and coworkers for the fragmentation of N7-Et-Gua (Chart 3), dG, CPr-dG, and N2-ethyl-dG. 131 In contrast, fragments m/z 135 and m/z 152 were not observed upon fragmentation of 8-oxo-dG (Chart 1), probably because of the difficulty of cleaving the oxygen-carbon bond.
Figure 4.
MS/MS spectra of unmodified dG and representative dG adducts: N2-ethyl-2′-deoxyguanosine (N2-ethyl-dG), O6-methyl-2-deoxyguanosine (O6-methyl-dG), O6-pyridyloxobutyl-2-deoxyguanosine (O6-POB-dG), and exocyclic crotonaldehyde adduct (CPr-dG).
Chart 3. Structures of DNA adducts used for discussion of nucleobase fragmentation.a.
a2′-deoxyguanosine (dG); N2- ethyl-2′deoxyguanosine (N2-Ethyl-dG); N2-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine (N2-POB-dG); O6-methyl-2′-deoxyguanosine (O6-Methyl-dG); O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine (O6-POB-dG); (CPr-dG); N7-ethylguanine (7-Ethyl-G); 8-Hydroxy-2′-deoxyguanosine (8OH-dG)
4.4 Fragmentation of Modified Nucleobases
The MS/MS fragmentation of protonated guanine has been carefully studied.130 For example, Tuytten and coworkers have reported a rigorous analysis of the MS2-4 fragmentation of dG .130;132 They observed the loss of the ribose moiety upon MS2 analysis and the subsequent fragmentation of [Gua+H]+ produces the m/z 135 ion referred to above as well as [Gua-HNCNH+H]+ (m/z 110), and a minor product ion of [Gua-HNCO+H]+ (m/z 109). Gregson and McCloskey determined that the ion at m/z 135 [G-NH3·H]+ can result from the loss of the exocyclic N2-amino group or the N-1 nitrogen following a Dimroth rearrangement-like ring-opening process.130
Fragmentation pathways of unmodified cytosine have been reviewed recently.133 The main fragments observed are at m/z 95 [M+H-NH3]+, m/z 69 [M+H-NHCO]+, with a minor peak at m/z 94 [M+H- H2O]+.
The MS/MS fragmentation of adenine is more complex, revealing four major pathways:134-136 (i) neutral loss of NH3 (m/z 119) followed by the loss of two molecules of HCN (or HNC)) (m/z 92, 65); (ii) elimination of NH2CN (m/z 94) and two molecules of HCN (m/z 67,40); (iii) the sequential loss of three molecules of HCN (m/z 109,82,55); and (iv) the formation of NH4+ (m/z 18).
Unlike nucleoside adducts (Section 4.3), modified nucleobases typically do not fragment by a common pathway. One exception is N-7 substituted guanine adducts which undergo common fragmentation: the alkyl-guanine bond is cleaved and a proton is transferred to guanine, resulting in protonated guanine ions at m/z 152.34 A notable exception is N7-methylguanine, which preferentially loses ammonia [M+H-NH3]+.137
4.5 Identification of DNA adducts using fragmentation information and accurate mass
Structural identification of an unknown DNA adduct frequently starts with the determination of the molecular formula based on accurate mass measurements (Section 4.1). Any formula under consideration should be consistent with the presence of a specific nucleobase (adenine, guanine, cytosine, or thymine) within the adduct structure. For example, the molecular formula of any deoxyguanosine-derived nucleoside should contain at least 10 C, 13 H, 4 O and 5 N atoms (dG, C10H13O4N5). As discussed in Section 4.1 and shown in Table 1, as the molecular size of an unknown increases, unambiguous assignment of its molecular formula from accurate mass measurements becomes more difficult. In these cases, accurate measurements of fragment mass obtained from CID experiments can be helpful, since fragment ions are smaller in size than the parent molecule, and their molecular formula can be more easily and unambiguously assigned.
Product ion spectra can be very helpful in characterizing unknown adduct structures. As discussed above (Section 4.3), most modified DNA nucleosides readily lose deoxyribose under CID conditions, with a characteristic neutral loss of m/z 116. Product spectra containing accurate mass data can be useful since the neutral loss of the dR (m/z 116.04735) can be used as evidence of nucleoside identity. Higher energy spectra can be obtained with a triple quadrupole mass spectrometer to reveal additional fragments such as those corresponding to unsubstituted nucleobases: m/z 152 (G), 136 (A), 127 (T), 112 (C), or a characteristic fragment ion of the protonated base (m/z 135 for guanine). Alternatively, the nucleobase ions generated from CID in an ion trap can be subjected to further fragmentation (MS3 experiment) to generate characteristic fragments.138
4.6 DNA Adduct Structure Determination
Since each DNA nucleobase can in theory be alkylated at several alternative sites (Scheme 1), the best way to identify the substitution site of the observed DNA adduct is to compare the HPLC retention time and MS/MS fragmentation of the unknown adduct to that of a synthetic standard.138,139 In general, mass spectral data alone cannot determine the structural identity of an unknown DNA adduct, although MS/MS fragmentation data can provide a wealth of structural information. Efforts are underway to develop methods to elucidate unknown analyte structures using MSn data by automated searching140-142 (e.g. by comparing the experimental spectra to those found in MSn spectral databases), or in silico by comparing the observed spectra to theoretical fragments generated from candidate structures selected based on experimental accurate mass measurements.143 Although databases for small molecule fragmentation are being developed,144-146 they are not yet sufficiently comprehensive to be of much use in identifying unknown DNA adducts. Currently available software for processing MSn data140 is either not capable of follow up data processing or is not specifically designed to deal with MSn data.146
Chiarelli and coworkers investigated using MS/MS fragmentation patterns to determine the structure of thirteen C8-substituted alkylaniline adducts of guanine and deoxyguanosine.147;148 The authors147;148 reported that a visual inspection of fragmentation patterns alone could not distinguish between isomeric adducts. However, isomeric structures could be differentiated based upon a calculated similarity index. It was concluded that a database of product ion spectra could be generated allowing the identification of unknown guanine adducts produced by aromatic amines.147;148
MS/MS fragmentation patterns can also be useful in determining the site of the modification on guanine or deoxyguanosine. As mentioned above (section 4.4), guanine adducts with substitution at the N-7 position undergo a common fragmentation to protonated guanine (m/z 152), with the exception of N7-methylguanine, which fragments to [G+H-NH3]+ (m/z 135).137 Therefore, the presence of a fragment ion at m/z 152 is suggestive of an N-7 guanine substitution, although modifications at other positions cannot be ruled out. Turesky and Vouros have made an interesting observation that isomeric C8-dG and N2-dG adducts of aromatic amines, 2-amino-3-methylimidazo [4,5-f]quinoline and 2-amino-3,4-dimethylimidazo[4,5-f]quinoxaline have distinct MS3 fragmentation patterns. While the N2 adducts lose NH3 (17 amu) from the guanine base [BH2-NH3]+, the isomeric C8 adducts lose CONH3 (45 Da) from the guanine base [BH2-CONH3]+.149 It has been proposed that this fragmentation pattern may be a general way to distinguish between C8 and N2-substituted derivatives of dG using MS3, since the same differences in fragmentation have been observed for dG adducts induced by 2-aminofluorene.147
4.7 Unknown adduct screening (Adductomics)
Screening DNA samples for unknown or unanticipated adducts is a relatively new field referred to as “adductomics”. The motivation for undertaking this field of study has been recently reviewed.150 In brief, the global investigation of DNA adducts independent of a priori assumptions regarding the formation of specific adducts is critical, because the complexity of the adducts produced in vivo from endogenous sources or as a result of exposure to complex mixtures of chemicals cannot be anticipated or predicted. For this reason, several groups have been actively engaged in developing the field of adductomics.30;100;131;151-160
The use of LC-MS/MS for adductomics experiments relies on the general observation discussed in Section 4.3 that the fragmentation of protonated modified nucleosides result in the formation of the corresponding protonated modified nucleobases, with a mass difference corresponding to the loss of a deoxyribose moiety (-116 amu). In a typical experiment, DNA samples are enzymatically hydrolyzed to free nucleosides and analyzed by tandem mass spectrometry.
The most commonly used approach is to use a triple quadrupole instrument operated in a constant neutral loss mode (CNL) or “pseudo” CNL mode (Scheme 2D) detecting any molecules undergoing a neutral loss of 116 (dR). In a typical CNL experiment, The Q1 and the Q3 simultaneously scan with a constant offset of -116 amu. This approach has been successfully used by Gangl, Turesky, and Vouros,156;157 Compagnone and coworkers,158 and Farmer et al.30 The pseudo-CNL approach is similar to CNL. However, instead of actually scanning the quadrupoles, the system is set to monitor a number of contiguous selected reaction monitoring transitions (Scheme 2C and section 2.5), all of which involve a loss of 116 amu. Multiple analyses are performed using SRM methods covering different mass ranges enabling a large range to be ultimately covered for a given sample.152-155;159 It is expected that the “pseudo” CNL approach should provide greater sensitivity than the traditional CNL approach since SRM scanning offers additional sensitivity. However, as the procedure requires multiple HPLC-MS/MS runs for each sample (typically 7-15), it is more time consuming and is also susceptible to increased instrument variability.
Another useful approach in the adductomics field is data dependent scanning (Section 2.5, Scheme 2E). In brief, full scan MS analysis is followed by fragmentation of the most abundant ion(s) observed at a given point of the LC-MS/MS run. The data dependent full scan/MS2 process is continuously repeated throughout the entire chromatographic run. Van den Driessche and coworkers151 adopted this approach to quadrupole-TOF instrumentation. Turesky and coworkers used the data-dependent approach on an ion trap instrument, with the addition of MS3 fragmentation of all those ions which lost 116 amu upon MS2.100;160
Recently, Inagaki and coworkers suggested an alternative approach to adductomics analyses.131 These authors postulate that guanine adducts can be identified by characteristic fragments at m/z 152 and 135, respectively, corresponding to protonated guanine and protonated [Gua+H-NH3]+ (Section 4.4). This new approach was employed to study acrylamide-DNA adducts.131 A similar approach has been suggested for adenine adducts, with a characteristic fragment at m/z 136 [Ade +H]+. One possible benefit of this approach is that it employs a simple thermal or acid hydrolysis of DNA, releasing free nucleobases, rather than using enzymatic hydrolysis to nucleosides. However, this approach appears to be limited to adducts of Gua and Ade bases.
Since adductomics is a relatively new field, a limited number of analyses have been conducted to date. There is a great room for advances in the field, especially considering the rapidly improving capabilities of mass spectrometry instrumentation. One capability which could facilitate the identification of novel or unanticipated adducts in the future is the use of high resolution mass spectrometry. To our knowledge, the only study to date which employed instrumentation capable of providing accurate mass of unknown adducts was the one by Van den Driessche and coworkers,151 who used a quadrupole-TOF instrument. However, accurate mass data was not reported in that study.151 The new generation of orbital trap and quadrupole-TOF instruments are capable of providing both molecular formula and fragmentation information to facilitate adduct identification. In addition, HPLC flow rates could be reduced to capillary or nano HPLC flow rates, providing a significant increase in sensitivity. The increased sensitivity enables testing samples of limited size. To our knowledge, none of the adductomics studies reported to date have operated using nanoflow techniques, which are standard in the field of proteomics. Turesky and coworkers100;160 have employed 6 μL/min flow rates with a CaptiveSpray™ ion source (Bruker, Billerica, MA), which is reported to offer sensitivity similar to that of nanospray ionization. Finally, data analysis software approaches tailored to adductomics data analysis, especially for experiments conducted in a dependent scan mode, would be very valuable in advancing the field. The ongoing comprehensive studies of the MS/MS fragmentation of known DNA adducts should assist in the detection and identification of novel DNA modifications.
5. Quantitative analysis of DNA adducts
Quantitative analysis of DNA adducts by mass spectrometry can be performed in combination with gas chromatography (GC) or liquid chromatography (LC) separations. Traditional methods of DNA adduct quantitation have employed GC-MS.161 However, gas chromatography has fallen out of favor with the recent developments of LC-MS techniques allowing for direct analysis of DNA nucleobases, nucleosides, and nucleotides from aqueous solutions. In contrast, derivatization is required for GC analysis of DNA monomers, which are otherwise insufficiently volatile to be analyzed directly by this technique.162 Nevertheless, many sensitive and accurate GC-MS methodologies have been reported for quantifying DNA adducts; many of these techniques are comparable to the corresponding HPLC-MS methodologies.163
HPLC-ESI-MS/MS has become the standard approach to the quantifying DNA adducts with the advent of the atmospheric pressure ionization sources, electrospray (ESI) and atmospheric pressure chemical ionization (APCI). As described above, electrospray ionization is particularly well suited for analyzing protonated DNA nucleobases and nucleosides, especially when coupled with reverse phase liquid chromatography.164 While electrospray ionization is currently the dominant ion source for LC-MS analysis of DNA adducts, APCI can be a more sensitive technique when judicious derivatization is employed.165
The sensitivity of electrospray ionization increases as the flow rate is decreased. LC-MS analysis at analytical flow rates of 0.2 - 1.0 mL/min does not allow the sensitivity necessary to detect DNA adducts at low levels. The use of capillary flow rates (5-15 μL/min) with 3-5 mm ID columns increases the inherent sensitivity of detecting DNA adducts by HPLC-ESI MS. Operating LC-MS systems within this flow range can be performed with minimal modifications to the standard commercial electrospray ion sources.
In some cases when DNA adduct concentrations are particularly low and/or a limited amount of DNA is available for analysis, the sensitivity of capillary HPLC-ESI-MS/MS is insufficient. The use of nanoHPLC (250 – 500 nL/min) provides an additional increase in sensitivity. Operating in this flow range, however, requires the use of specialized nanospray ion sources and very narrow HPLC columns (usually 75-100 um ID). NanoHPLC columns can be self-packed using homemade column/emitters pulled from fused silica tubing or commercially available empty fused silica tubing with built-in frit/emitters (e.g. New Objective, Woburn, MA or Thermo Scientific, West Palm Beach, FL). Pre-packed nanoHPLC columns are also available commercially. Originally, the low flow used for nanoHPLC (typically 200 – 500 nL/min) was generated by splitting the flow from a higher flow generated by a conventional HPLC. The flow could be split either before61 or after64 the injection port. Currently available systems are designed to directly and accurately deliver nano flow rates, facilitating nano HPLC-nanospray MS analysis of DNA adducts.66-69
The sensitivity of LC-MS-based DNA adduct analysis can be limited by the amount of sample entering the mass spectrometer for detection, in which case the miniaturization of HPLC separations will improve limits of detection and quantitation. Alternatively, adduct detection in biological samples can be limited by chemical noise from the matrix. In the latter case, the selectivity of MS detection can be improved adding an additional level of fragmentation (MS3)100;166 or by using high resolution MS to reduce or eliminate chemical noise.66
5.1 Sample preparation for quantitative analysis
Varying degrees of sample cleanup are required to remove the bulk of unmodified nucleosides, proteins, inorganic salts, and other sample components prior to quantitative HPLC-ESI-MS/MS analysis (Scheme 3).13;14 Some examples include ultrafiltration, liquid/liquid extraction, solid phase extraction (SPE), immunoaffinity purification, and off-line HPLC (Section 3.3). The extent and the type of sample cleanup is highly variable depending upon the sample source, analyte concentration, the type of adduct targeted (nucleobase, nucleoside, nucleotide), and specificity/selectivity of the mass spectrometric method. Insufficient sample cleanup can result in excessive chemical noise, co-eluting HPLC peaks, and analyte signal suppression (Section 2.2).
5.2 Quantitative data acquisition
Quantifying DNA adducts by mass spectrometry is typically performed in the selected ion monitoring mode (SIM) or selected reaction monitoring mode (SRM, also referred to as multiple reaction monitoring (MRM)) (Section 2.5). The benefit of using SIM is the simplicity of method development and the ability to use inexpensive single stage quadrupole instrumentation. However, SRM based methods offer improved sensitivity over SIM due to the additional criteria required prior to detecting an MS/MS signal. The use of tandem mass spectrometry and reaction monitoring reduces chemical noise, improving method detection limits, assay accuracy and precision, and also generates fewer false positive measurements.
SRM experiments are conducted almost exclusively using triple quadrupole instrumentation. However, ion traps, quadrupole-ion traps, quadrupole-TOF, ion trap-orbital traps, or quadrupole-orbital trap instruments are also capable of operating in a reaction monitoring mode. Ion traps can also perform additional fragmentation reactions (MSn).81 An example of how an MS3 technique can be utilized to improve the performance of an assay is shown in Figure 5. Here a UPLC-ESI/MS3 method was developed166 using a linear quadrupole ion-trap operating in MS3 mode to measure 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AL-I) and 7-(deoxyguanosin-N2-yl) aristolactam I (dG-AL-I) adducts (Chart 4); this method attained significantly lower limits of quantitation (0.2 and 1.0 adducts per 108 DNA bases, respectively). The method was used to quantify adduct levels in upper urinary tract tissues of patients with urothelial carcinomas; results suggested this methodology could supplement currently employed 32P-postlabeling techniques for biomonitoring DNA adducts in human tissues.
Figure 5.
Chromatograms obtained upon UPLC-MS3 analysis of renal cortex DNA from human subject with a carcinoma of the upper urinary tract along with MS3 fragmentation spectra of dA-AL-II and dA-AL-I. Reprinted with permission from Reference 166. Copyright 2012 American Chemical Society.
Chart 4. Structures of DNA adducts highlighted in Section 5.5.a.
aN7-(2′-hydroxyethyl)guanine (N7-HEG); 5-hydroxymethyl-2′-deoxyuridine (HmdU); 3,N4-ethenocytosine (εCyt); 7-(1′,2′-dihydroxyheptyl)-3H-imidozo(2,1-i)purine (DHH- εAde); 1,N6-ethenoadenine (εAde); N2,3-ethenoguanine (N2,3-εGua); 1,N2-ethenoguanine (1,N2- εGua); 4-hydroxyestrogen-1-N3-adenine (4-OH-E-1-N3Ade); 1,N6-etheno-2′-deoxyadenosine (εdAdo); 3,N4-etheno-2′-deoxycytidine (εdCyt); 1,N2- etheno-2′-deoxyguanosine (1,N2- εdGuo); O2-ethylthymidine (O2-edT); O4-ethylthymidine (O4-edT); 1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol (N7G-N1A-BD); 1,4-bis-(guan-7-yl)-2,3-butanediol (bis-N7G-BD); N2-hydroxymethyl-2′-deoxyguanosine (N2-HOMe-dG); N7-ethylguanine (N7-Ethyl-G); N-(deoxyguanosin-8-yl)-PhIP (C8-dG-PhIP); 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene (dG-N2-B[a]P); 7-(deoxyadenosin-N6-yl)aristolactam I (dA-AL-I); 7-deoxyguanosin-N2-yl aristolactam I (dG-AL-I)
Traditional methods for quantify DNA adducts has been performed on instrumentation capable of unit mass resolution and accuracy. More recently, mass spectrometers capable of high resolution accurate mass measurements have become available both in the form of improved TOF instruments as well as the orbital trap (Thermo Scientific's Orbitrap technology) instruments. High resolution mass measurements, especially when used in conjunction with tandem mass spectrometry experiments, provide enhanced selectivity and sensitivity for quantitative analyses of low abundance DNA adducts (see example in Figure 6).66 This approach was used in the analysis66 of 7-ethyl-Gua (Chart 4) in human leukocyte DNA to investigate the exposure of smokers to an unknown ethylating agent. Nanoelectrospray-high resolution tandem mass spectrometry using an orbital trap (LTQ Orbitrap Velos) with a resolution of 55,000 and 5 ppm mass tolerance was used and attained a detection limit of 10 amol on column and a limit of quantitation of 8 fmol/μmol Gua starting with 180 μg DNA (corresponding to 36 μg DNA on-column). This method allowed for the measurement of 7-ethyl-Gua in the DNA of both smokers and non-smokers, which was not possible using the existing, lower resolution triple quadrupole techniques.119
Figure 6.
Chromatograms obtained upon LC-NSI-HRMS/MS analysis of human leukocyte DNA (129 μg, 12.9 μg on column) containing 59.4 fmol N7-ethyl-Gua /μmol Gua. The relatively higher amount of analyte in this sample allowed for the confirmation of its identity by additional monitoring of the accurate mass of the molecular ion of N7-ethyl-Gua and the internal standard. Panel A shows the result from monitoring of the accurate mass of 7-ethyl-Gua (m/z 180.08799). Panel B shows the result from the monitoring of the accurate mass of [15N5]7-ethyl-Gua (m/z 185.07317). Panel C shows the results from the transition at m/z 180 [M + H]+ → m/z 152.05669 [Gua + H]+ for 7-ethyl-Gua, and panel D shows the corresponding transition m/z 185 [M + H]+ → m/z 157.04187 [Gua + H]+ for the internal standard. Results are shown with a 5 ppm mass tolerance. Reprinted with permission from Reference 66. Copyright 2011 American Chemical Society.
5.3 Stable isotope dilution
DNA adducts present in biological samples are typically subjected to several processing/enrichment steps prior to MS analysis to remove the bulk of unwanted materials within the sample and thereby minimize the potential for ion suppression. All sample clean-up procedures inherently lose a certain amount of the analyte of interest, and recovery can vary significantly from sample to sample. The sometimes necessary derivatization steps122,165 can add additional variability to sample processing.
To accommodate the inherent variability of sample processing while maintaining a reproducible and accurate quantitative assay, the technique of isotope dilution mass spectrometry (IDMS) is commonly used. The technique of IDMS utilizes a stable isotope-labeled analog of the analyte as an internal standard for quantitation. When spiked into the sample early in the analysis, stable isotope tagged internal standards increase the reproducibility and accuracy of the analysis. Because the stable isotope-labeled internal standards are chemically identical to the analyte, the behavior of the isotope mimics the analyte throughout sample extraction / processing procedures (Scheme 3) as well as compensating for ion suppression within the ESI source. For a given sample, recovery and ionization differences are accounted for by normalizing the detected analyte signal to that of the internal standard (Figure 5, 6) prior to quantitation against a calibration curve.
Internal standards possessing 13C or 15N labels are preferable, since the chromatographic retention time of these internal standards are virtually identical to that of the analyte containing natural isotopes. Deuterated standards are also useful, although a slight shift in retention times is commonly observed, and there is the possibility of small but non-negligible isotope effects upon sample recovery and MS/MS fragmentation pathways. Care also must be taken to assure that the deuteriums are not exchangeable under the conditions used for sample processing and analysis. Ideally, multiple labels (at least 3) should be used, to negate contribution to the internal standard signal from natural occurring isotopomers of the analyte (especially 13C). Finally, care should be taken when deciding on the amount of internal standard added. Sufficient amounts should be used to assure good ion signal quantitation and levels should be within the dynamic range of the instrument such that a linear relationship between the measured analyte and internal standard signals is assured.
5.4 Method validation
To ensure accurate and reliable assay HPLC-MS/MS methods must be validated prior to running true samples. In its simplest form, the validation is performed by fortifying sample of the same matrix type with known amounts of analyte and then processing these known samples through the entire assay procedure. The results are typically expressed as a plot of measured analyte amounts versus the amounts that were spiked into the matrix (Figure 7). Spiking experiments can also be used to determine the limit of detection (LOD) (S/N ≥ 3) and the limit of quantitation (LOQ) (S/N ≥ 10) for the analyte of interest in a given matrix. Method accuracy and precision (intraday and interday) is calculated by repeated analyses of spiked samples. To ensure that sample differences do not impact the performance of the assay, it is advisable that matrices from at least 6 different, unique sources and tested within the method validation.
Figure 7.
Relationship between detected and added 7-ethyl-Gua. Various amounts of 7-ethyl-Gua were added to calf thymus DNA (0.3 mg, 30 μg on column) and analyzed by the method described in the text; R2 = 0.99. 7-Ethyl-Gua present in the calf thymus DNA was subtracted from each value. Reprinted with permission from Reference 66. Copyright 2011 American Chemical Society.
5.5 Recent examples of Mass Spectrometric Quantitation of DNA adducts
The application of ESI-MS/MS methods for the quantitative analysis of DNA adduct has been recently reviewed.55 Table 2 provides representative examples of DNA adduct which have been quantified by mass spectrometry, and also highlights technical differences in the methodologies.
Table 2.
Examples of published mass spectrometry based detection methods for DNA adducts.
| Adduct | Adduct Type | LOD | Sample Preparation | Details | Species | Source | Ref. |
|---|---|---|---|---|---|---|---|
| GC-MS | |||||||
|
| |||||||
| N7-HEG* | Epoxide derived | 1-butanol extraction, HPLC, derivatization (HONO, pentafluorobenzyl bromide, pivalic anhydride), silica SPE, HPLC | NICI, SIM | human | blood | 167;169 | |
| 1,N6-ethenoadenine | Exocyclic | 0.030,0. 400 fmol(on-column) 6.3, 36 fmol (total) | SPE, derivatization (pentafluorobenzyl bromide), Si SPE, Acetonide Formation | NICI, SIM | human | urine, placenta | 106;172-174 |
| 3,N6-ethenocytosine | Exocyclic | 7.4 fmol (LOQ - Total) | SPE, derivatization (pentafluorobenzyl bromide), Si SPE | NICI, SIM | human | urine | 171 |
| HmdU* | Oxidation | 50 fmol (starting material) 500 amol (on-column) | Derivatization (3,5-bis(trifluoromethyl)benzyl Bromide), isooctane extraction | NICI, SIM | human | cultured cells | 170 |
| 1,N2-ethenoguanine N2,3-ethenoguanine | Exocyclic | LOQ = 15fmol (1,N2- εGua) LOQ = 7.6 fmol (N2,3-εGua) | Immunoaffinity, Derivatization (pentafluorobenzyl bromide), Si SPE | immunoaffinity NICI, SIM high res. | rat | liver | 170,175 |
|
| |||||||
| LC-MS/MS (Conventional Electrospray) | |||||||
|
| |||||||
| 4-OHE2-N7G* | Natural Product | 70 pg | MCX SPE, BondElut SPE | human | urine | 102 | |
| Mel-dGuo (Melphalan)* | Alkylating Drug | 900 fg | Column Switching | rat | liver, mammary | 312 | |
| AFB1-N7Gua (Aflatoxin)* | Natural Product | 0.02 pg | OasisMCX SPE, immunoaffinity column, BondElut LRC SPE | human | urine | 103 | |
| ES-3′-N6-dA, ES-3′-N2-dG,ES-3′-C8-dG (Estragole) | Natural Product | 0.2-0.5 fmol | SPE | SIM | rats | liver | 99 |
| O6-methyl*/O6-ethyl-dG* | Nitrosamine | 0.03 O6-Me-dG/108 nts 0.05 O6-Et-dG/108 nts | Centrifugation | Column Switching | mouse | liver | 313 |
| DHP-dA,* DHP-dG* (Pyrrolizidine alkaloids) |
Natural Product | Enzymatic Digest | rat | liver | 314 | ||
| dG-C8 -IQ* | Aromatic Amine | 6 fmol | C18 SPE | rat | liver | 315 | |
| dG-C8-4-ABP* | Aromatic Amine | 5 fmol/300 μg DNA | C18 isolute SPE | human | pancreas | 316 | |
| dG-C8-4-ABP* | Aromatic Amine | 0.72/107 nts | DNA digest w/on-line column switching | Column Switching | mouse | liver | 317 |
| C8-dG-MeIQx* | Aromatic Amine | 500 fg | SepPak C18 SPE | rat | liver | 318 | |
| N2-dG-MeIQx* | Aromatic Amine | 750 fg | SepPak C18 SPE | rat | liver | 318 | |
| N8-Ade-benzidine* | Aromatic Amine | 22 pg | SepPak C18 SPE | 319 | |||
| N8-Ade-2-aminofluorene* | Aromatic Amine | 51 pg | SepPak C18 SPE | 319 | |||
| 1,N2-ethenoguanine | Exocyclic | 1 pg | SCX, C18-OH SPE | human | urine | 107 | |
| εA* | Exocyclic | 2-3 pg | C18-OH SPE | human | urine | 106 | |
| 1,N2-propano-dG* | Exocyclic | 0.015 fmol/μg DNA | Centrifugation (YM-10), Oasis HLB SPE | rat | liver | 320 | |
| PdG, * Et-dG* | Exocyclic | 250 fg? | DNA digest w/Liquid extraction | human | blood | 321 | |
| 1,A/2-εdGuo* | Exocyclic | 20 fmol | Chloroform extraction | Column Switching | rat | liver | 322 |
| N7-ethylyguanine | Ethanol/acetaldehyde | 0.59 pg/mL urine | SepPak C18 SPE | Column Switching | human | urine | 105 |
| C8-HEG* | Ethanol/acetaldehyde | 50 fmol | HPLC | rat | liver | 323 | |
| MP-dGuo, * MP-dAdo* | Polyaromatic hydrocarbon | 10 fmol/2 fmol in 100 μg DNA | OasisHLB SPE | rat | liver | 324 | |
| N2-BPDE-dG* | Polyaromatic hydrocarbon | mouse | liver,lung, kidney | 325 | |||
| N2-BPDE-dG* | Polyaromatic hydrocarbon | Oasis HLB | mouse | liver | 326 | ||
| N2-BPDE-dG* | Polyaromatic hydrocarbon | 1/108 nts | DNA digest | Column Switching | mouse | liver | 327 |
| BP-6-N7Gua* | Polyaromatic hydrocarbon | 2.5 fmol/1 mL urine | SepPak C8 and Strata-SCX SPE | Column Switching | human | urine | 328 |
| DB[a,l]P-N6-dA dibenzo-[a,l]pyrene | Polyaromatic Hydrocarbon | 8 fmol/100 μg DNA | Enzymatic digestion ADD | mice | oral tissue | 329 | |
| THBG* | Epoxide derived | 3.5 fmol | YM-10 filtration | mouse/rat | liver/lung | 330 | |
| HMVK-dGuo* | Epoxide derived | 5 fmol/200 μg DNA | Centricon-10, HPLC | rat | liver | 331 | |
| N7-GA-Gua, * N3-GA-Ade* | Epoxide derived | 1.5-2 adducts/108 nts | Centrifugal filtration | mouse | liver/lung/kidney | 332 | |
| N7-HEG* | Epoxide derived | 0.25 ng/mL urine | Centrifugation | Column Switching | human | urine | 333 |
| N7-HEG* | Epoxide derived | 0.1 fmol | YM-10 filtration | rat | liver, heart, spleen, kidney, colon, stomach, lung | 334 | |
| N7-HEG, * N1-HedA* | Epoxide derived | 0.5 fmol ea. | YM-3 filtration, HPLC | rat | liver | 335 | |
| 8-oxo-dG* | Oxidation | 2 fmol | Immunoaffinity column purification | 1mm column, 50 μl/min | mouse | prostate | 336 |
| 8-oxo-dG/A* | Oxidation | 10 fmol 8-oxodA | Oasis HLB SPE | human | urine | 337 | |
| 8-OH-dG* | Oxidation | 0.024 ng/mL | SepPak C18 SPE | 338 | |||
| CEdG* | Oxidation | 0.2 pg | Strata-X-C | rat | urine | 339 | |
| N6-FFM-dAdo,*N2-FFM-dGuo* | Food Contaminant | LOD = 1.3 fmol (N2-FFM-dG) LOD= 0.3 fmol (N6-FFM-dA) |
Oasis HLB SPE | UPLC | human | cells | 340 |
| Luc-N2-dG and N6-dA | Food Additive | LOD = 4 pM, LOQ = 1 pM 0 (Luc-N2-dG) LOD = 0.6 pM, LOQ =2 pM Luc-N6-dA |
On-line SPE | Column switching | rat | liver, kidney | 341 |
|
O6-methylguanine O6-carboxymethyl guanine |
Endogenous (food) | LOQ = 0.03 pmol/mg DNA (O6-MeG) LOQ = 0.05 pmol/mg DNA (O6-CMG) |
Oasis HLB SPE | UPLC | human | cells | 342 |
| N7-(3-benzo[1,3]dioxol-5-yl-2-hydroxypropyl)guanine | Food Additive | mouse | urine | 343 | |||
| 7-(2-oxoethyl)guanine | From vinyl chloride (Industrial) | LOD = 1 fmol, LOQ = 1.5 fmol | YM-10 filtration | Derivatization (O-t-butyl hydroxylamine) | rat | Liver, lung, kidney, spleen, testis, brain | 344 |
|
| |||||||
| LC-MS/MS (APCI) | |||||||
|
| |||||||
| εdA* | Exocyclic | 8 pM | Oasis HLB SPE | Column Switching | human | urine | 27 |
| εdC* εdA* εAde* | Exocyclic | 70 pM dAde, 100 pM dC, 17 pM dA | OasisHLB | Column Switching | human | urine | 28 |
|
| |||||||
| LC-MS/MS (Capillary Flow Electrospray) | |||||||
|
| |||||||
| G-NOR-G cross-links* (Cyclophosphamide) | Alkylating Drug | 5 fmol | Thermal hydrolysis, SPE (Oasis Max) | human | blood | 310 | |
| Hydroxyethyl-dG* | Nitrosamine | 100-150 fmol | 4mm syringe filter | rat | liver | 345 | |
| O6-POB-dG* | Nitrosamine | 50 fmol | Strata-X C18 SPE | mouse | liver | 346 | |
| dG-C8-MeIQx,* dG-C8-PhIP*, dG-C8-4-ABP* | Aromatic Amine | 5-10 adducts per 109 nts | Enzymatic digest, SPE (HyperSep) | linear ion trap-MS3 | human | saliva | 100 |
| POB adducts* | Nitrosamine | 3fmol G, 1 fmol dGuo, 100 amol Thd, 2 fmol Cyt | StrataX SPE | rat | liver/lung | 347 | |
| 7-POB-Gua* | Nitrosamine | Strata-X SPE | rat | liver/lung | 348 | ||
| O6-mdGuo* | Nitrosamine | 24 fmol O6-mdGuo | Column Switching | rat | liver | 20 | |
| C-εdC*, C-εdG*, H-εdA*, H-εdC*, H-εdG* | Exocyclic | Not specified | 0.2 um filter, Supelclean LC-C18 SPE | mouse | intestine | 349 | |
| gdG* | Exocyclic | gdG: 1.5-2pmol | 4 mm syringe filter | rat | liver | 345 | |
| Propano-dG* | Exocyclic | 4 adducts/109 nts | Strata-X SPE | human | lung | 350 | |
| Cro-dGuo* | Exocyclic | 0.2 fmol | Strata-X SPE | human | liver/lung | 351 | |
| N7-ethylguanine | Ethanol/acetaldehyde | 0.25 fmol | Ym-30, Strata-X SPE | human | liver | 119 | |
| N2-ethyl-dGuo* | Ethanol/acetaldehyde | 0.4 fmol | StrataX, OasisMAX | human | liver,lung, leukocytes | 352 | |
| N2-ethyl-dGuo* | Ethanol/acetaldehyde | 0.05 fmol on column, 3 adducts/109 nts | Strata-X, OasisMAX SPE | human | leukocytes | 353 | |
| Bis-N7G-BD* | Epoxide derived | 6 adducts/109 nts | YM-10 filtration, offline HPLC | mouse/rat | liver, lung, kidney, brain, thymus | 67 | |
| 1,N6-HMHP-dA* | Epoxide derived | 1.5 adducts/109 nts | Enxymatic hydrolysis, Clean Carbo SPE | Column switching | mouse | liver | 26 |
|
| |||||||
| LC-MS/MS (Nanospray) | |||||||
|
| |||||||
| 4-OH-E1-N3Ade* | Alkylating drug/Natural Product | SPE, offline HPLC | Not IDMS, Capillary flow | human | breast | 64 | |
| εdC*, εdA*, εdG* | Exocyclic | LOQ = 0.18 fmol (εdC) LOQ = 4.0 fmol (εdA) LOQ = 3.4 fmol (εdG) |
SPE | human | saliva, blood, placenta | 60-62 | |
|
O2-ethylthymidine N3-ethylthymidine O4-ethylthymidine |
Ethylation (tobacco smoke) | LOQ = 50 fg (O2-edT) LOQ = 100 fg (N3-edT) LOQ = 100 fg (O4-edT) LODs 10× lower |
SPE | human | leukocytes | 63 | |
| N2-ethylguanine | Ethanol and acetaldehyde induced | 10 amol standard, 2 adducts per 109 nts | Thermal hydrolysis, ultrafiltration, Strata-X SPE | HRMS (Orbitrap) | human | leukocytes | 66 |
| N2-hydroxymethyl-dG* | 20 amol | Offline HPLC | UPLC | rat | nasal tissue | 65 | |
| N7G-N6A-BD* | Epoxide derived | 3 adducts/109 nts | YM-10 filtration, SepPak C18 SPE | mouse/rat | liver | 67 | |
| Bis-N7G-BD* N7G-N6A-BD* | Epoxide derived | 0.5 fmol/100 μg DNA | Ultrafiltration, off-line HPLC | mouse | liver; lung, kidney | 68;69 | |
|
| |||||||
| Other (High Resolution/Accurate Mass and MS3) | |||||||
|
| |||||||
| dG-C8-MeIQx*, dG-C8-PhIP*, dG-C8-4-ABP* | Aromatic amine | 5-10 adducts per 109 nts | Enzymatic digest, SPE (HyperSep) | MS3 (Linear Ion Trap) Capillary Flow, CaptivaSpray Source | human | saliva; mammary tissue | 81;100;189 |
| 7(deoxyadenosin-N6-yl) aristolactam (dA-AL-I), 7-(deoxyguanosin-N2-yl) aristolactam (dG-AL-I) | Food contaminant | LOQ = 0.3 adducts/108 nts (dA-AL-I) LOQ = 1 adducts/108 nts (dG-AL-I), 10 μg DNA |
Protein precipitation, On-line SPE | MS3 (Linear Ion Trap) Capillary UPLC, CaptivaSpray Source, Column Switching | human | renal cortex | 166 |
| N2-ethylguanine | Ethanol and acetaldehyde induced | 10 amol standard 2 adducts per 109 nts | Thermal hydrolysis, ultrafiltration, Strata-X SPE | HRMS (Orbitrap), Nanoflow | human | leukocytes | 66 |
Abbreviations: (mel-dGuo), melphalan-deoxyguanosine; (O6-Me-dG) O6-methyl-2′-deoxyguanosine; C8-dG-4-ABP, N-(deoxyguanosine-8-yl)-4-ABP adducts; C8-dG-4-ABP, N-(deoxyguanosine-8-yl)-4-ABP adducts; C8-dG-MeIQx, N-(deoxyguanosine-8-yl) MeIQx; N2-dG-MeIQx, 5-(deoxyguanosin-N2-yl) MeIQx; (1,N2-propano-dG)1,N2-propanodeoxyguanosine; (N8-Ade-benzidine) N-(adenin-8-yl)-benzidine-adenine; (N8-Ade-2-aminofluorene) N-(adenin-8-yl)-aminofluorene adenine; (N2-BPDE-dG) 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene; (BP-6-N7Gua) N7-(benzo[a]pyrene-6-yl)guanine; (THBG) N7-(2,3,4-trihydroxybut-1-yl) guanine; (N3-GA-Ade) N3-(2-carbamoyl-2-hydroxyethyl)adenine; (N7-GA-Gua) N7-(2-carbamoyl-2-hydroxyethyl)guanine; (N7-HEG) N7-(2′-hydroxyethyl)guanine; (dG-C8-IQ) N-(deoxyguanosine-8-yl)-2-amino-3-methylimidazo[4,5-f]quinoline; (DHP-dG) ±6,7-dihydro-7-hydroxy-1-hydroxymethyl-5H-pyrrolizine-2′deoxyguanosine; (DHP-dA) ±6,7-dihydro-7-hydroxy-1-hydroxymethyl-5H-pyrrolizine-2′deoxyadenosine; (εdG) 1,N2-etheno-2′-deoxyguanosine; (εdA) exocyclic 1,N6-etheno-dA adducts; (1,N2-εdGuo)1,N2-etheno-2′-deoxyguanosine; (Et-dG) N2-ethyl-2′-deoxyguanosine; (PdG) α-S- and α-R-methyl-γ-hydroxy-1,N2-propano-2′-deoxyguanosine; (7-POB-Gua) 7-[4-(3-pyridyl)-4-oxobut-1-yl]guanine; (POB adducts) 7-[4-(3-pyridyl)-4-oxobut-1-yl] adducts; (gdG) glyoxal 2′-deoxyguanosine; (N2-ethyl-dGuo) N2-ethyldeoxyguanosine; (bis-N7G-BD) bis-(guan-7-yl)-2,3-butanediol; (Cro-dGuo) 1,N2-propanodeoxyguanosine adducts; (Propano-dG)1,N2-propanodeoxyguanosine adducts; (H-εdG) heptanone-etheno-2′-deoxyguanosine adducts; (H-εdC) heptanone-etheno-2′-deoxycytidine adducts; (H-εdA) heptanone-etheno-2′-deoxyadenosine adducts; (N1-HedA) N1-hydroxyethyl-deoxyadenosine; (8-oxo-dG) 8-oxo-7,8-dihydro-2′deoxyguanosine; (8-oxo-dA) 8-oxo-7,8-dihydro-2′deoxyadenosine; (8-OH-dG) 8-hydroxy-2′-deoxyguanosine; (CEdG) N2-(1-carboxyethyl)-2′-deoxyguanosine; (N6-FFM-dAdo) N6-((2-formylfuran-5-yl)methyl)-2′-deoxyadenosine; (N2-FFM-dGuo) N2-((2-formylfuran-5-yl)methyl)-2′-deoxyguanosine; (εdC) 3,N4-etheno-2′-deoxycytidine; (εAde) 1,N6-ethenoadenine; (εdA) 1,N6-etheno-2′-deoxyadenosine; (G-NOR-G) N,N-bis[2-(N7-guaninyl) ethyl] amine DNA-DNA cross-links; (Hydroxyethyl-dG) O6-2-hydroxyethyldeoxyguanosine; (O6-POB-dG) O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine; (C8-dG-PhIP) N-(deoxyguanosin-8-yl)-PhIP; (C8-dG-MeIQx) N-(deoxyguanosine-8-yl)MeIQx; (C8-dG-4-ABP) N-(deoxyguanosine-8-yl)-4-ABP adducts; (1, N6 -αHMHP-dA) 1, N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2′-deoxyadenosine; (4-OH-E1-N3Ade) 4-hydroxyestrogen-1-N3-adenine (4-OH-E-1-N3Ade); (N7G-N6A-BD) 1-(guan-7-yl)-4-(aden-6-yl)-2,3-butanediol; (HmdU) 5-hydroxymethyl-2′-deoxyuridine; (N7-HEG) N7-(2′-hydroxyethyl)guanine; (AFB1-N7-Gua) aflatoxin B1-N7-guanine
5.5.1 GC-MS
GC-MS quantitation of DNA adducts was reviewed by Giese and co-authors in 1997.161 Since then, there have been a number of papers describing the quantitation of DNA adduct using GC-MS techniques; some representative examples are included in Table 2. All of the GC-MS methods listed here employed a single quadrupole instrument operated in the SIM mode.
Giese and workers developed a quantitative method for the analysis of N7-(2′-hydroxyethyl)guanine (N7-HEG) (Chart 4) in DNA using GC-MS with negative ion chemical ionization. N7-HEG-d4 was used as an internal standard. Targeted adducts were released from DNA using neutral thermal hydrolysis. N7-HEG adducts were extracted using 1-butanol, purified by reverse phase HPLC, and derivatized with HONO, pentafluorobenzyl bromide and pivalic anhydride to add electron withdrawing pentafluorobenzyl groups.167;168 The derivatized samples were purified by silica solid phase extraction and reverse phase HPLC and analyzed by GC-MS. This assay was used to determine N7-HEG levels in the granulocyte DNA (0.1-11.5 μg) of hospital workers resulting from exposure to ethylene oxide.169 A considerable inter-individual variability was observed, with adduct concentrations ranging between 1.6 and 241.3 adducts/107 nucleotides.169
Sowers and coworkers170 developed a method for the quantitation of 5-hydroxymethyl-2′-deoxyuridine (HmdU) (Chart 4) using isotope dilution with 15N2,d2-HmU or 15N2,d2-HmdU internal standard. GC-MS analyses were conducted using negative ion chemical ionization following derivatization with 3,5-bis(trifluoromethyl)benzyl bromide. Samples were purified by an isooctane extraction procedure. The reported detection limit was 50 fmol of HmdU per sample, which corresponds to 500 amol HmdU on-column. The method was used to analyze DNA from cultured cells. The authors observed a correlation between toxicity and exogenous HmdU adduct levels. Furthermore the kinetics of HmU repair was investigated using stable isotope-labeled uridine as a metabolic tracer.170
Chen and coworkers developed a GC-MS assay for the quantitation of potentially promutagenic 3,N4-ethenocytosine (εCyt) adducts in human urine (Chart 4).171 The analysis employed GC-MS with negative ion chemical ionization, using [13C4,15N3] εCyt as an internal standard. Human urine samples were fractionated by C18 SPE, followed by derivatization with pentafluorobenzyl bromide. GC-MS analysis was conducted in the selective ion monitoring mode using [M - 181]− - fragments of pentafluorobenzylated εCyt and its internal standard. One fg of pentafluorobenzylated εCyt standard was detectable (S/N > 40), and the holistic limit of quantification for the assay was 7.4 fmol. This method was approximately 1000-fold more sensitive than the previous existing HPLC/fluorescence assay for 3,N4-etheno-2′-deoxycytidine in DNA. The method was verified by comparison of the results of the GC/NICI/MS analysis of chloroacetaldehyde-treated calf thymus DNA to those obtained using the existing HPLC/fluorescence methods. The GC-MS method was used for analysis of εCyt in urine samples from two smokers, and the average levels of εCyt were determined to be 101±17 pg/mL/g of creatinine.171
Chen, Chung, and coworkers developed a quantitative GC-MS method for 7-(1′,2′-dihydroxyheptyl)-3H-imidozo[2,1-i]purine (DHH-εAde) (Chart 4) and 1,N6-ethenoadenine (εAde) (Chart 4).172 These adducts form endogenously upon DNA alkylation by the epoxide metabolite of trans-4-hydoxy-2-nonenal, a peroxidation product of ω-6 polyunsaturated fatty acids. The quantitative GC-MS analysis was performed in a negative ion chemical ionization mode, with [15N5] εAde and [15N5]DHH- εAde as internal standards. The [M – PFB]- ions at m/z 158 and m/z 328 were monitored for PFB εAde and ACT-PFB-DHH- εAde, respectively. Sample preparation involved acid hydrolysis of DNA, C18 solid phase extraction (SPE), derivatization by pentafluorobenzylation (PFB), a silica-based SPE step, and acetonide (ACT) formation. The on-column limits of detection for PFB-εAde and ACT-PFB-DHH-εAde were 30 amol and 0.4 fmol, respectively. The detection limits for the entire assay were 6.3 fmol for εAde and 36 fmol for DHH-εAde, respectively. The validated GC-MS method was used to quantify εAde in human placental DNA samples, and 2.3 εAde adducts per 106 Ade bases were detected.173 The method was also used to measure εAde concentrations in human urine.174 A significant correlation was observed between excreted εAde levels and cigarette smoking in males.174 Finally, the established GC-MS method to verify a new LC-ESI-MS/MS assay for εAde.106 Similar levels were observed when the same samples were re-analyzed by LC/ESI/MS/MS.
Swenberg et al. have developed an immunoaffinity purification-high resolution GC/NICI/MS method for N2,3-ethenoguanine (N2,3-εGua) and 1,N2-ethenoguanine (1,N2-εGua) (Chart 4) in vivo.175 [13C4,15N2]-N2,3-εGua and [13C3]-1,N2-εGua were used as internal standards. 1,N2-εGua and N2,3-εN2Gua adducts were purified by immunoaffinity chromatography using immobilized polyclonal antibodies, derivatized to their 3,5-bis(pentafluorobenzyl) (PFB) derivatives, and quantified using GC/NICI-MS. The instrument was operated in the selected ion monitoring mode, with high resolution data acquisition of the [M - 181]- fragments. The limits of quantitation for 1,N2-εGua and N2,3-εN2Gua in hydrolyzed DNA were 7.6 fmol and 15 fmol, respectively, which corresponds to 5.0 N2,3-εGua and 8.7 1,N2-εGua adducts/108 unmodified Gua bases. 1,N2-εGua was the predominant ethenoguanine adduct formed when lipid peroxidation products react with DNA. N2,3-εGua was detected in DNA treated with the vinyl chloride (VC) metabolite 2-chloroethylene oxide and in hepatocyte DNA from rats exposed to 1100 ppm VC for 4 weeks (6 h/day for 5 days/week).176
5.5.2 HPLC-MS/MS (APCI and Conventional ESI)
The rapid growth in the availability and technology of HPLC-MS instrumentation has led to a shift from GC-MS based methods to HPLC-MS based methods for quantifying DNA adducts. As discussed above, an important advantage of HPLC-MS over GC-MS techniques is eliminating lengthy and labor intensive derivatization steps. Traditionally, HPLC-MS was operated under analytical flow rate conditions (0.2-1 ml/min), and some representative examples are listed in Table 2.
5.5.3 LC-MS/MS (Capillary Flow Electrospray)
In an effort to increase HPLC-MS sensitivity, capillary HPLC ESI-MS methods have been developed. As discussed above (section 2.3), electrospray ionization efficiency increases with decreasing flow rate. A variety of methods have been developed utilizing a decreased flow rate of between 5 - 15 μL/min (capillary or micro flow); these flow rates are typically achieved using commercially available HPLC systems specifically designed to operate at this flow range. Small injection volumes (< 10 μL) are required when using capillary HPLC to ensure good chromatographic peak shape. . Conventional electrospray sources can operate in this flow range used with little or no modification. Examples of this type of analysis were recently discussed in a detailed review of LC-MS/MS quantitation,55 and some examples are listed in Table 2.
5.5.4 Nanoflow HPLC-nanospray MS/MS
The sensitivity of ESI LC-MS/MS methods can be further increased by reducing the chromatographic flow to sub-μL/min levels (typically 250 – 500 nl/min, nanoHPLC). These slower flow rates used for nanospray mass spectrometry further increase the sensitivity of the analysis by improving ionization efficiency and ion transport from the source into the mass analyzer. It has also been reported that nanoflow ionization is less susceptible to ion suppression. The sensitivity of electrospray-based MS techniques is inversely proportional to the HPLC flow rate.177;178 due to the following two considerations. First, as the flow decreases the ionization of the analyte becomes more efficient, as illustrated in Figure 9A.59 Secondly, nanospray flow rates allow the electrospray plume to be directly aimed into the mass spectrometer increasing the number of analyte ions entering the mass spectrometer, as shown in Figure 9B.59 The practical operation of electrospray ion sources at nl/min range was accomplished177;179 and has been used to great effect in the field of proteomics.59 More recently, nanoflow rates of 250-500 nL/min and column dimensions of 0.075-0.10 mm which were typically used in proteomics have been adopted for highly sensitive analysis of DNA adducts. Applications of nanospray ionization for quantifying DNA adducts are discussed in detail in Section 5.5.4.
Figure 9.
(A) Simplified illustration showing concentration and flow rate effects on the ESI process. For larger flow rates, which produce larger droplets, analyte surface activity, concentration, and competition from other species can affect overall ionization efficiency, the extent of ionization “suppression”, and quantitation. At sufficiently low flow rates and analyte concentrations, each droplet contains on average less than one analyte molecule, ionization efficiency is 100%, and suppression/matrix effects are eliminated. Reprinted with permission from Reference 59. Copyright 2004 American Chemical Society. (B) Normal flow rate electrospray (top) vs a lower flow rate electrospray (bottom) that produces smaller droplets. By allowing closer proximity to the MS inlet, the lower flow rate electrospray affords more efficient ion introduction. Reprinted with permission from Reference 59. Copyright 2004 American Chemical Society.
Delivering the optimal flow rates for nano ESI requires either flow splitting from a conventional HPLC or using specialized nanoflow pump; specific care must be taken to avoid significant void volumes. Consideration must also be given to injection volumes since the size of the injection is limited due to the low flow rates; chromatographic peak shape will suffer if the injection volume is too large for the flow rate. Also, a specialized nanospray ion source is necessary with varying degrees of complexity depending upon the application. This approach has been taken in a number of recent studies, where maximum assay sensitivity was necessary to successfully quantify DNA adducts. These are listed in Table 2 and are briefly discussed below.
Gross and coworkers have developed an assay for the measurement of estrone-metabolite-modified N3-adenine adduct (4-OH-1-N3Ade) (Chart 4) in breast tissue of women with breast cancer. This study employed an LCQ Deca (Thermo Scientific, San Jose, CA) quadrupole ion-trap with a capillary HPLC. Samples (5 μL) were loaded onto the column (75 μm ID × 12 cm C18 column self-packed into a 15 μm PicoFrit nano tip form New Objective Inc., Woburn, MA). The HPLC flow was delivered by a standard pump (8 μl/min) and split to achieve 270 nl/min flow rates through the column.15N5 labeled 4-OH-1-N3Ade was used as an internal standard. 4-OH-1-N3Ade concentrations in breast tissue from five women were in the range of 20-70 fmol/g, while it was undetectable in another female subject. The authors concluded that the sensitivity of the method was not sufficient to permit distinctions between cancer and non-cancer patients.
Etheno DNA adducts (1,N6- etheno-2′-deoxyadenosine (εdAdo), 3,N4-etheno-2′-deoxycytidine (εdCyt), and 1,N2-etheno-2′-deoxyguanosine (1,N2 -εdGuo) (Chart 4) and nucleoside adducts derived from acrolein (AdG) and crotonaldehyde (CdG) can be induced by exogenous exposure (industrial chemicals) and endogenous sources (lipid peroxidation). A nano HPLC-nanospray MS/MS method was developed for quantitation of these adducts levels in human saliva,62 placenta,60 and leukocyte DNA samples.61 Chen and coworkers used a TSQ Quantum Ultra EMR triple quadrupole mass spectrometer (Thermo Scientific, San Jose, CA), equipped with a nanospray ionization (NSI) interface. The triple quadrupole was operated with a Q1 mass width of 0.2 amu (lower than standard 0.7 amu) to conduct highly selected reaction monitoring (H-SRM) experiments. They used a hand pulled 75 μm ID × 120 cm column hand packed with C18 stationary phase. An UltiMate 3000 Nano LC system (Dionex, Amsterdam) was used with a 30 μL/min flow split before the injection port to achieve a flow rate of 300 nL/min. The injection port had a 2 μL loop. The detection limits were reported as 0.73, 160, and 34 amol on-column for εdAdo, εdCyd, and 1,N2 -εdGuo standards, respectively and limits of quantitation for the entire assay were 0.18, 4.0, and 3.4 fmol, respectively. εdAdo, εdCyd, and 1,N2 -εdGuo were detected in human placental DNA, with the concentrations of 28.2, 44.1, and 8.5 adducts per 108 normal nucleosides, respectively. The amounts of εdAdo, εdCyd, and 1,N2-εdGuo in leukocyte DNA samples from 11 human volunteers were 16.2 ± 5.2, 11.1 ± 5.8, and 8.6 ± 9.1 (mean ± S.D.) in 108 normal nucleotides, respectively. DNA (30 μg) was isolated from 1-1.5 mL of blood. The relative standard deviations were within 10%. An aliquot equivalent to 6 μg of DNA hydrolysate was used. The authors concluded that the method will allow for these adducts to be used as noninvasive biomarkers in clinical studies for cancer risk assessment and for evaluation of cancer chemopreventative agents.
Chen and coworkers used a very similar approach to that described above to measure levels of the ethylated DNA adducts, O2-ethylthymidine (O2 -εdT) and O4 -ethylthymidine(O4-εdT) (Chart 4), which accumulate in the body due to their slow repair and are thought to have promutagenic properties.63 These authors employed nanoLC-NSI/MS/MS methodology H-SRM to attain impressive detection limits for O2-edT,N3-edT, and O4 -edT: 5.0, 10, and 10 fg, respectively, injected on-column. The limits of quantification for the entire assay were reported as 50, 100, and 100 fg, respectively, corresponding to 1.1, 2.3, and 2.3 adducts in 109 normal nucleotides, respectively. The assay requires 50 μg of DNA obtained from 1.5-2.0 mL of blood. Adduct concentrations in leukocyte DNA from 20 smokers were 44.8 ± 52.0, 41.1 ± 43.8, 48.3 ± 53.9 per 108 normal nucleotides for O2-εdT, N3-εdT, and O4-εdT, respectively. The corresponding adduct levels in 20 nonsmokers were 0.19 ± 0.87, 4.1 ± 13.3, and 1.0 ± 2.9, respectively. Adduct concentrations in human leukocyte DNA were significantly higher in smokers than in nonsmokers. Chen et al. suggested that this assay will be clinically feasible for simultaneous quantification of the three ethylated thymidine adducts as potential biomarkers for exposure to ethylating agents and for cancer risk assessment.
Our laboratory has employed nanospray ionization for quantitation of DNA-DNA crosslinks resulting from exposure to 1,3-butadiene, an important industrial chemical compound used in rubber and plastic manufacturing and also present in automobile exhaust and cigarette smoke. A nano HPLC-nanospray MS/MS method was initially developed for guanine-adenine crosslinks, 1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol (N7G-N1A-BD) (Chart 4).67 The liquid chromatography method utilized a nanoAcquity UPLC (Waters, Milford, MA) system. Samples (8 μL) were injected onto a 0.18 × 20 mm Symmetry C18 (Waters) trapping column eluted at a flow rate of 10 μL/min for 1 min. The flow rate was then reduced to 350 nL/min and re-directed from the trapping column to a 75 μm ID × 100 mm Atlantis C18 analytical column. The effluent from the second column entered the TSQ Quantum Ultra AM (Thermo Scientific, Waltham, MA) triple quadrupole mass spectrometer with a nanospray ion source.67 Excellent sensitivity was achieved, allowing the quantitation of this minor adduct in laboratory rats and mice treated with 62.2 – 625 ppm BD by inhalation.51 A similar nanospray ionization method was developed for the analysis of BD-induced guanine-guanine cross-links, 1,4-bis-(guan-7-yl)-2,3-butanediol (bis-N7G-BD) (Chart 4).68;69 For this work, the analytical column employed was a self-packed Zorbax SB-C18 column created using a 15 μm PicoFrit nano tip (75 μm ID × 18 cm, from New Objective Inc., Woburn, MA). The LOD values achieved in this assay were 0.5 fmol bis-N7G-BD/100 μg DNA, with the LOQ of 1.0 fmol/100 μg DNA allowing for the adduct to be quantified in vivo at 3 adducts per 109 nucleotides. This method enabled the investigation of the formation, persistence and repair of these adducts in laboratory animals treated with low ppm and sub-ppm concentrations of BD;52 test levels which approach the exposure limits for occupationally exposed workers.
Swenberg and coworkers have employed nano HPLC-nanospray MS/MS to quantify endogenous and exogenous N2 -hydroxymethyl-dG adducts (N2 -HOMe-dG in Chart 4) in nasal and bone marrow DNA of laboratory rats exposed to 0.7-15.2 ppm [13CD2]-formaldehyde by inhalation.65 Deuterated formaldehyde was used in order to distinguish between exogenous and endogenously formed adducts. These authors employed a nano-UPLC system (Waters (Milford, MA) interfaced to a Quantum Ultra (Thermo Electron, Waltham, MA) triple quadrupole mass spectrometer operated in the SRM mode. Samples were loaded onto a trapping column at a flow rate of 5 μL/min, followed by transfer to an analytical column eluted at 600 nL/min. The method accuracy (∼85%) and precision (∼10%) was verified by fortifying blank DNA (50 μg) with known amounts of the analyte (4 fmol, 8 fmol, or 16 fmol, in triplicate). The reported limit of detection of the new method was 20 amol on column. These authors65 reported that endogenous DNA adducts dominated at low formaldehyde exposures, comprising more than 99% of total N2-HOMe-dG. Exogenous N2-HOMe-dG adducts were not detectable in the bone marrow of rats exposed to less than 15.2 ppm [13CD2] formaldehyde and were formed in a highly nonlinear fashion at higher exposures.65
6. New Approaches to MS analysis of DNA modifications
A number of promising new liquid chromatography-mass spectrometry approaches have been developed in the recent years with the goal of improving the sensitivity and specificity of analyzing DNA modifications in vivo. Several such approaches will be discussed in the present section: on-line sample purification (sample switching), chip-MS, MSn, and high resolution MS.
6.1 On-line sample purification
As discussed above, HPLC-MS analysis of DNA adducts in biological samples can be challenging due to ionization suppression resulting from other components within the sample matrix (salts, proteins, unmodified nucleosides). Therefore, extensive sample processing is required prior to loading the sample on an analytical column for HPLC-MS analysis; these procedures are often time-consuming and can result in significant losses of the analyte of interest. Column switching methods couple on-line sample clean-up with HPLC separation and MS analysis, potentially eliminating offline cleanup procedures, preventing sample loss, and increasing sample throughput.
Column switching (or 2D chromatography) involves loading the sample onto a trapping column and then washing away salts and other non-retained contaminants. After flushing the column, the flow of the mobile phase is switched and the trapping column is back-flushed, transferring the analyte onto a second, analytical column for final separation and MS analysis (Figure 8). Column switching has been successfully used by several groups to quantify DNA adducts in biological samples.20;27;28;180-184 For example, Doerge et al. have developed a column switching HPLC-ESI-MS/MS method to quantify exocyclic εdAdo and εdCyd adducts (Chart 1) in liver and lung DNA of mice exposed to urethane.183 The method employed a reverse phase trapping column (Luna C18, 2 mm × 30 mm, 3 μm) and a Luna C18, 2 mm × 150 mm, 3 μm analytical column coupled to a Quattro LC-triple quadrupole operated in the ESI+ mode. SRM transitions corresponding to a loss of deoxyribose from protonated molecules of εdAdo and εdCyd were monitored. The limits of detection (LOD) of their method were 0.3 – 0.9 εdA adducts per 108 normal nucleotides and 0.7 – 1.8 εdC adducts per 108 normal nucleotides. Liver DNA from untreated and exposed mice contained 1.0 and 2.2 ε-dA adducts per 108 normal nucleotides and 1.0 and 2.7 εdC adducts in 108 normal nucleotides, respectively. The method sensitivity for εdC was lower than that for εdA, and the authors suggested that immunoaffinity online cleanup could be used to increase the sensitivity of εdC detection in the future.183
Figure 8.
Diagram of valve positions employed in column switching HPLC-ESI-MS/MS. The sample is loaded onto the trapping column in position A and washed to remove contaminants. In position B, the second pump backflushes the analyte from the trapping column onto the analytical column and into the mass spectrometer.
Poulson et al. have developed a column switching method for analyzing 1,N6-etheno-2′-deoxyadenosine (εdAdo) adducts (Chart 1) present in urine.27 The 2D separation was performed with a Luna trapping LC column (75 × 4.6 mm, 5 μm) and a Synergi Polar-RP analytical column (150 × 4.6 mm, 4 μm). An API3000 triple quadrupole with an APCI source was used for the analysis. Urine (3 mL) was initially purified by Oasis HLB solid phase extraction prior to online column switching –ESI-MS/MS analysis. The LOQ value for εdAdo was 600 amol injected, or 0.7 pmol/L urine; however, no increase in εdAdo levels were observed in smoker's urine compared to healthy non-smoker's urine.27 The same group has modified the above method to measure εdCyd adducts in human urine with an LOQ of 100 pM for ε-dC.28
Brink et al.20 have developed a column switching method for the simultaneous determination of O6-methyl-2′-deoxyguanosine (O6-Me-dG, Chart 1), 8-oxo-7,8-dihydro-2′deoxyguanosine (8-oxo-dG, Chart 1), and εdAdo (Chart 1) in rat liver DNA. These three adducts can be formed endogenously or from exposure to exogenous chemicals. The LOQ values for these three adducts ranged between 24 and 48 fmol, although only one internal standard, isotopically labeled O6-Me-dG, was used for all three analytes. All three adducts were observed in rats dosed with dimethylnitrosamine. A major advantage of this method was that much lower levels of artifact 8-oxo-dG (Chart 1) were observed as compared to other LC-MS/MS methods that use off-line sample cleanup methods.20
Chao et al. reported another column switching method for 8-oxo-dG.182 These authors reported the removal of more than 99% of dG from biological samples using an ODS-3 trap column (Intersil) and PolyaminII endcapped analytical HPLC column (150 × 4.6 mm, YMC). The LOD value was 0.13 8-oxo-dG adducts/106 dG (1.8 fmol on column) when using 20 μg mouse liver DNA per analysis, which is lower than background levels of 8-oxo-dG in human lymphocytes (0.3 adducts/106 dG). The low LOD coupled with short analysis time makes this method ideal for high throughput human analysis.182 The most recent column switching method for 8-oxo-dG quantitation simultaneously analyzes 8-oxo-dG and 8-oxo-dA (Chart 1) in rat liver DNA.184 The LOD for both adducts using this method was 5 fmol. While 8-oxo-dA was roughly 30 fold less abundant than 8-oxo-dG, there was less artifact formation of 8-oxo-dA. It was concluded that the combination of 8-oxo-dG and 8-oxo-dA provides accurate quantitation of DNA oxidation.
Column switching has also been used for quantifying alkylguanine DNA adducts.181 Chao et al. have developed a column switching method for the simultaneous analysis of N7-methylguanine (N7-Me-G) and N7-ethylguanine (N7-Et-G) (Chart 1).181 The method employs a Nucleosil NH2 trap column (35 mm × 4.6 mm, 10 μm) and a Polyamine-II endcapped HPLC column (150 mm × 4.6 mm, 5 μm, YMC). The HPLC method was only 15 min long, and the LODs were 0.42 fmol N7MeG and 0.17 fmol N7EtG on column. The column switching method was used to develop dose response curves in liver DNA of mosquito fish exposed to N-nitrosodiethylamine (NDEA) and N-nitrosodimethylamine (NDMA).181 These authors reported that their column switching method had greater sensitivity than previously developed LC-MS/MS methods.181
Our laboratory has developed column switching methods for two exocyclic DNA adducts of 1,3-butadine, 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2′-deoxyadenosine (1, N6-αHMHP-dA) and 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2′-deoxyadenosine (1, N6-γHMHP-dA) (Chart 1).26 The limit of detection of our method is 1.11,N6-HMHP-dA/109 nts, sensitive enough to obtain dose response curves for mice exposed to 62.5 to 625 ppm BD (Figure 10).
Figure 10.
Column switching capLC-ESI+-MS/MS analysis of 1,N6-HMHP-dA in liver DNA from a control mouse (A) and a mouse exposed to 200 ppm BD for 2 weeks (B). Dose-dependent formation of 1,N6-HMHP-dA in liver of laboratory mice exposed to increasing concentration of BD by inhalation (C). Reprinted with permission from Reference 26. Copyright 2010 American Chemical Society.
6.2 Chip/LC-MS
Establishing and maintaining good chromatographic and electrospray performance in the nanospray mode can be challenging; this has led to efforts to simplify nanospray operation as well as making the technique more robust. Chip-MS is the use of a microchip with electrospray needles and/or nano columns embedded in it. This approach facilitates MS operation in the nanoflow regime, potentially making for more robust operation.185 This design makes it easier for “non-experts” to operate in the nanospray mode and in some cases improves system performance. Some recent examples include the Agilent CHIP-MS, the Thermo Scientific (Waltham, MA) EASY-Spray source, which employs pre-packed nanoflow columns (0.075 mm ID, 2-3 μm) up to 50 cm in length, and the New Objective (Woburn, MA) PicoChip source, which allows for simple setup of an integrated nanoflow column and nanospray emitter. Conversely, the Bruker Daltonics (Billerica, MA) CaptiveSpray source is designed to provide nanospray-like sensitivity at higher HPLC flow rates (1-5 μL/min).
More dramatic modifications to the traditional column/emitter arrangement involve the incorporation of microfabricated devices in the form of a “chip”, with various designs incorporating chromatography, nanospray emitters, or both. A versatile device which contains a fraction collector and silicon-based integrated chip with an array of ESI nozzles is offered by Advion (Ithaca, NY). This device allows for coupling to chromatography or direct infusion of a sample without an HPLC column,34 as well as fraction collection during mass spectrometry data acquisition. A simplified version of this device has been recently introduced (American Society for Mass Spectrometry National Meeting, 2012). This latest version allows for direct coupling of the chip ESI nozzle device to chromatography to allow for easier nanospray operation, with an advantage of instantaneous switching of nozzles in the case of the emitter blockage. Applied Biosystems (Framingham, MA) offers a chip-based chromatography device (cHiPLC Nanoflex) designed for making nanoflow chromatography easier and more robust. Agilent Technologies (Santa Clara, CA) has a device (HPLC-Chip Cube) which incorporates both the chromatography and the nanospray emitter into a single chip for the most user-friendly configuration of nanospray operation.
The majority of previous chip-MS applications have been limited to the analysis of peptides or drugs/drug metabolites.185;186 However, the Vouros group at Northeastern recently employed chip-MS analysis for quantitation of DNA adducts. Glick et al. first reported a chip-based nano-LC/MS ion trap method for the quantitation of 2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine (PhIP) induced DNA adducts (e.g. C8-dG-PhIP in Chart 4).187 The same laboratory has recently used a small molecule chip for quantitative analysis of guanine adducts in DNA extracted from bladder tissue of rats exposed to 4-aminobiphenyl (4-ABP).188 Their methods employ an Agilent LC/MSD XCT Ultra Ion Trap configured with an Agilent small molecule Chip Zorbax 80SB-C18, 5 μm particle size. The Agilent Chip contains an enrichment column (40 nL) and analytical column (75 μm i.d., 43 μm in length). For analyses of 4-ABP induced dG adducts (Chart 4), two SRM transitions were monitored corresponding to dR loss from dG-ABP and its internal standard, dG-ABP-d9 (m/z 435.4 → 319 and m/z 444 → 328, respectively). The limit of detection of this method was reported as 2 dG-ABP adducts/108 nts, requiring only 1.25 μg DNA. The method was used to quantify dG-ABP adducts in bladder DNA from rats dosed with ABP by IP injection; in these samples, dG-ABP was detected at levels starting at 80 adducts/108 nts.188
6.3 MSn
As discussed above, the use of tandem mass spectrometry can dramatically increase LC-MS method selectivity for minor DNA lesions due to improved signal to noise ratios. Since ion trap instruments are capable of MS3 and MS4 experiments, additional selectivity can be achieved by including additional MS/MS transitions. For example, Turesky and coworkers have developed81;81 an MS3 method for several dG adducts of heterocyclic aromatic amines present in tobacco smoke and cooked meat.64 Their HPLC-ESI-MS/MSn approach employs consecutive reaction monitoring in the MS3 scan stage mode using a linear quadrupole ion trap mass spectrometer. The method was employed to analyze C8-dG adducts of the heterocyclic aromatic amines, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), 2-amino-9H-pyrido[2,3-b]indole (ARC), 2-amino-3,8-dimethylmidazo[4,5-f]quinoxaline (MeIQx), 4-aminobiphenyl (4-ABP), and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) (see C8-dG-PhIP, Chart 4).64 The chromatographic separation was achieved at a flow rate of 6 μL/min using a 0.32 mm ID C18 column. An LTQ linear ion trap (ThermoElectron, San Jose, CA) was interfaced with an Advance CaptiveSpray ion source from Michrom Bioresource Inc. (Auburn, CA). Fragment ions corresponding to the loss of deoxyribose ([M + H - 116]+), followed by MS3 full scanning to characterize the product ions of the aglycone adducts [BH2]+ and the two or three most abundant ions, were used for quantitative measurements. For DNA adducts of HAAs and 4-ABP, the isolation widths were set at 3.0 and 1.0 amu for the MS2 and MS3 scan modes, respectively. For N2-BPDE-dG (Chart 4), the isolation widths were 6 amu and 5 amu for the MS2 and MS3 scan modes, respectively. One μscan was used for data acquisition, and about 12 scans were acquired for each adduct and its corresponding internal standard.
DNA was isolated from saliva of 37 human volunteers on restricted diets. The concentrations of PhIP, MeIQx, and 4-ABP-dG adducts in these samples ranged from 1 to 9 adducts per 108 nucleotides. This study demonstrated that PhIP exposure is significant in humans and that saliva is a promising biological fluid for DNA adducts induced by tobacco and dietary carcinogens in humans. The same methodology was used to quantify DNA adducts of PhIP and 4-ABP in tumor-adjacent breast tissue of breast cancer patients.189 The authors found that only 1 of 70 biopsy samples contained measureable level of PhIP-DNA adduct (3 adducts/109 nucleotides) and no measureable amounts of 4-ABP-DNA adducts.189 This is in contrast to immunohistochemistry (IHC) and 32P-post-labeling results, which reported the same adducts at concentrations that should have been measureable by the MS-based method. Therefore, exact identities of the adducts measured by IHC or 32P-post-labeling are uncertain.
This same research group has developed an assay for 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AL-I) and 7-(deoxyguanosin-N2 -yl) aristolactam I (dG-AL-I) (Chart 4) resulting from exposure to aritolochic acids (AL).166 Consumption of aritolochic acids has been implicated in the etiology of so-called Chinese herbs nephropathy and of Balkan endemic nephropathy, both caused by carcinomas of the upper urinary tract.166 Turesky et al. previously employed HPLC-ESI-MS/MSn methodology to identify AL-DNA adducts in the renal cortex of a woman who developed end-stage renal failure after using a herbal remedy containing aristolochic acid139 as also from patients in the Balkan countries190 and Taiwan.191 In their new method,166 Turesky et al. employed a trapping column to enable on-line sample clean-up. The LOQ values for dA-AL-I and dG-AL-I were reported as 0.3 and 1.0 adducts per 108 DNA bases, respectively, using 10 μg of DNA (Figure 5). The new method was used to quantify AL-DNA adducts in tissues of rodents exposed to AA and in the renal cortex of Taiwanese patients with carcinomas of the upper urinary track. In human tissues, dA-AL-I was detected at levels ranging from 9 to 338 adducts per 108 DNA bases, whereas dG-AL-I was not found. The researchers concluded that HPLC-ESI-MS/MSn methods is complementary to 32P-postlabeling techniques previously used for biomonitoring of AL-DNA adducts in human tissues.
6.4 High Resolution/Accurate Mass Mass Spectrometry
The use of high resolution mass spectrometry is a promising approach to maximize signal to noise ratios for DNA adduct analyses in complex samples. For example, Balbo and coworkers have developed a nano HPLC – accurate mass MS/MS based method to quantify N7-ethylguanine (Chart 4) in human leukocyte DNA. Cigarette smoking has been proposed to be a source of an unknown ethylating agent that cam modify genomic DNA. The authors66 have developed a nanospray based tandem mass spectrometry method for quantifying N7-ethylguanine in vivo (7-ethyl-G, Chart 4). [15N5]7-Ethyl-Gua was used as the internal standard. High resolution/accurate mass data acquisition utilizing an LTQ Orbitrap Velos instrument (Thermo Scientific, Waltham, MA) was used in order to reduce chemical noise. A Nano2D-LC HPLC (Eksigent, Dublin, CA) system equipped with a 1 μL injection loop was utilized, with a flow rate of 300 nL/min and one microliter of sample was injected onto a C18 capillary column (75 μmID, 10 cm length, 15 μm orifice). The column was created by hand packing a commercially available fused-silica emitter (New Objective, Woburn MA). The LC-NSI-HRMS/MS method used LTQ Orbitrap Velos instrument (Thermo Scientific, Waltham, MA) and nanospray source (Thermo Scientific) and a Nano2D-LC HPLC (Eksigent, Dublin, CA). Isolations of 1 amu mass windows and for the analyte (m/z 180) and internal standard (m/z 185) were performed sequentially followed by transport to the HCD collision cell for fragmentation. The transitions of m/z 180 [M + H]+ → m/z 152.05669 [Gua + H]+ for 7-ethyl-Gua and m/z 185 → m/z 157.04187 for the internal standard were monitored in the Orbitrap detector set at a resolution of 30,000 (at 400) amu with an actual resolution of 55,000 at the detected masses. Accurate mass monitoring of the fragment ions at 5 ppm (152.05669 ± 0.0008 and 157.04187 ± 0.0008, respectively) was used to generate extracted mass chromatograms for quantitation. A full scan event was also performed over 100-500 amu mass range at a resolution setting of 30,000 to monitor for the accurate mass at 5 ppm of the molecular ion of the analyte m/z 180.08799 and of the internal standard m/z 185.07317, to confirm the identity of the analyte. A detection limit of approximately 10 amol on column was achieved, while the limit of quantitation was about 8 fmol/μmol Gua starting with 180 μg DNA (corresponding to 36 μg DNA on-column).
The limit of quantitation of the new method was defined by identifying the lowest measurement with a coefficient of variation lower than 5%. A measure of the signal-to-noise could not be used for determining the LOQ because there was virtually no observable noise. The method was used to determine the concentrations of N7-ethylguanine in leukocyte DNA samples from 30 smokers and 30 nonsmokers. Adduct concentrations in smokers were 49.6 ± 43.3 (range 14.6-181) fmol/μmol Gua, while the corresponding amounts in nonsmokers were 41.3 ± 34.9 (range 9.64-157) fmol/μmol Gua, with no significant difference between smokers and non-smokers.66 This study for the first time established the presence of N7-ethylguanine in human leukocyte DNA.
7. Mass spectrometry-based mapping of nucleobase damage along DNA sequences
The ability of DNA lesions to induce mutations and cancer is strongly dependent on their chemical structure, their ability to be recognized by specialized DNA repair proteins, and their positions within the genome.192;193 Local DNA sequence context plays a major role in mediating the rate of DNA adduct repair and their mispairing potency.194 Furthermore, the exact location of a DNA lesion within the gene will determine whether it results in mutations affecting the structure and function of the corresponding protein.
Most carcinogens and drugs exhibit distinct DNA sequence preferences.195;196 Some possible mechanisms of small molecule-DNA recognition include pre-covalent interactions between the DNA modifying agent and its target sequence within DNA (e.g. electrostatic or hydrophobic interactions, π-π stacking), as well as electronic and steric factors that influence the rates of the chemical reaction between carcinogen and its target base. For example, GC-rich sites and poly G runs have long been recognized as targets for DNA oxidation and alkylation.197-199 Furthermore, the introduction of the C5-methyl group on cytosine as observed physiologically200 increases the reactivity of CG dinucleotides towards carcinogen metabolites and drugs, e.g. mitomycin C, esperamicins, and benzo[a]pyrene diolepoxide.199;201-203
Traditional methods of mapping nucleobase damage along DNA sequences have involved gel electrophoresis (PAGE) analysis of DNA strand breaks generated at the sites of nucleobase damage by the action of repair endonucleases or hot piperidine treatment.204 Unfortunately, many important biologically relevant DNA adducts, e.g. O6-Me-dG, O6-POB-dG, and N2-ethyl-dG, cannot be converted to DNA strand breaks and thus cannot be detected by gel electrophoresis methods, furthermore, the structures and the optical identities of carcinogen-modified bases cannot be determined by PAGE. Mass spectrometry based approaches described below are applicable to any DNA adduct and are capable of providing additional structural information, facilitating mechanistic studies of the origins of DNA sequence specificity.
7.1 DNA Sequencing by Tandem Mass Spectrometry
The most direct way to sequence structurally modified DNA is to employ tandem mass spectrometry (MS/MS). MS/MS has been extensively used in sequencing native (unmodified) oligonucleotides. Initial efforts in this area have employed fast atom bombardment (FAB)205;206 and plasma desorption (PD)207;208 coupled with single stage mass spectrometry to achieve “bidirectional” sequencing of oligonucleotides. These techniques resulted in the cleavage of phosphodiester bonds, thereby leading to the formation of fragment ions (5′-P and 3′-P sequence ions) which can be used to determine the sequence.205 However, matrix interferences and surface fragmentation interfered with accurate sequencing by FAB-MS, a problem solved by employing tandem mass spectrometry.209 Cerny et. al. were among the first to sequence a short oligonucleotide (less than 6 bases in length) using fast atom bombardment (FAB) coupled with tandem mass spectrometry (FAB-MS/MS).209;210 FAB-MS/MS sequencing of oligonucleotides soon fell out of favor due to its high detection limit (10 nmol for oligonucleotides of MW>1200).211 Instead, MALDI-MS methods using a variety of matrices have been developed and successfully applied to oligonucleotide sequencing.212-215 The use of a delayed extraction (DE) methodology216 has improved the resolution of MALDI-MS, while post source decay (PSD)211;217;218 provided useful fragmentation information during oligonucleotide sequencing. Recently, the use of 1,5-diaminonapthalene as a matrix has been reported to improve in source dissociation (ISD) of oligonucleotides during MALDI-MS.219 More spectral fragment ions have been observed with this technique which is useful for accurate sequencing.219
McLuckey et. al.220 employed ESI-MS/MS to determine the sequence of short oligonucleotides (4-8 nucleotides in length) and proposed a comprehensive nomenclature for fragment ions, which is analogous to the nomenclature of peptide fragments (Scheme 4). Under most CID conditions, the phosphodiester bond can be cleaved at four different positions, giving rise to several types of 5′-terminus containing fragments (a, b, c, and d) and several types of 3′-terminus group containing fragments (w, x, y, and z).220 Each of these ions is assigned a numerical subscript (an, zn etc.), where n represents the cleavage position from the respective termini. In addition, “a” series of ions that underwent the neutral loss of a DNA base are commonly observed due to the facile cleavage of N-glycosydic bonds. The corresponding ions are designated as an-Bn, where upper case B denotes the cleaved base, and subscript n stands for the position of the base from the 5′ terminus. Finally, d and w series ions can lose water molecules to form ions with the same m/z as fragment ions c (d-H2O) and x (w-H2O), respectively. Structurally modified oligomers exhibit similar fragmentation patterns, and the characteristic mass shifts observed for modification-containing fragments can be used to determine the positions of structurally altered bases within the sequence.221
Scheme 4.
Nomenclature of common ion series observed upon MS/MS sequencing of DNA.
Negative ion ESI-MS/MS is especially suitable to oligonucleotide sequencing due to its high sensitivity and the production of multiply charged ions.220;222-226 However, CID spectra of oligonucleotides can be quite complex, especially for longer oligonucleotides (> 10-mers), making data interpretation challenging.224;226;227 Computer algorithms have been developed to facilitate MS/MS data interpretation for oligonucleotides, and some of these are available online free of charge. The recent introduction of high resolution/mass accuracy capabilities of orbital trap (Orbitrap™ technology) and Q-TOF instruments for use in oligonucleotide sequencing (Figure 11) facilitates accurate sequence determination of both structurally modified and unmodified oligonucleotides.228
Figure 11.
Examples of representative MS/MS spectra of oligonucleotides obtained using an Orbitrap Velos mass spectrometer.
Iannitti and coworkers were among the first to employ ESI-MS/MS for sequencing and characterizing the oligodeoxynucleotides covalently modified by alkylating agents.229 Sequence selectivity of nucleobase alkylation by hedamycin was investigated by incubating the drug with different 6 mer oligonucleotide sequences, followed by offline HPLC purification and desalting of the differentially adducted oligomers. The individual purified and desalted oligonucleotides were then injected onto a triple quadrupole mass spectrometer, and the adduct position within oligonucleotide sequence was determined by negative ion ESI-MS/MS.229 Marzilli et. al. employed a similar strategy to map aflatoxin-B1 (AFB1) guanine adducts along the 5′-CCGGAGGCC-3′ sequence.230 These authors have isolated three isomeric oligonucleotides resulting from AFB1 epoxide adduction at three different guanines, and the adduct position was established by HPLC-ITMS. Identification of the exact site of modification was possible by CID, which yielded characteristic fragmentation patterns.230 In a separate study, Glover et. al. employed ESI-MS/MS to confirm that para-benzoquinone preferentially alkylates cytidine residues of DNA by comparing the MS/MS spectra of unmodified 7-mer oligonucleotide (5′- GTTCTTG-3′) with the corresponding spectra of para-benzoquinone-modified 5′- GTTCTTG-3′.231 Iannitti et. al. used negative ion ESI-MS/MS to identify the sequence specificity of n-bromohexylphenanthridinium bromide and ESI+-MS/MS to obtain structural information of the adduct formed by this intercalating agent.232 However, sequence specificity of cisplatin could not be determined by ESI--MS/MS. Instead, positive ion ESI-MS/MS was employed, which revealed the formation of intra strand cross link by cisplatin between the G4 and G6 guanines in the oligonucleotide sequence 5′-CACGTG-3′.232
Harsch et. al. investigated the chemoselectivity of the reaction of carcinogenic bay region diol epoxide (-)-(1R,2S,3S,4R)-1,2-epoxy-3,4-dihydroxy-1,2,3,4-tetrahydrobenzo-[c]phenanthrene with 12-mer DNA oligonucleotide (5′-TAGTCAAGGGCA-3′).233 Reactions were performed by incubating the carcinogen with both single stranded and double stranded dodecamers. These authors233 were able to successfully resolve positional isomers of adducted oligonucleotides using a polystyrene divinylbenzene column. Harsch et. al. found that in reactions of (-)-(1R,2S,3S,4R)-1,2-epoxy-3,4-dihydroxy-1,2,3,4-tetrahydrobenzo[c]phenanthrene with double stranded dodecamer, the diol epoxide preferentially alkylated adenine rather than guanine nucleobases and produces 3-fold more adducts at A7 as compared to A6. On the other hand, reaction of the bay region diol epoxide with single stranded dodecamer yielded oligonucleotides with more dG modifications than dA.233 Colgrave et. al. applied the methodology developed by Iannitti et. al. to study the site and sequence specificity of alkylation of 14 mer oligonucleotide sequences by minor groove binding alkylating agents duocarmycin C2 and duocarmycin C1.234
Wang et. al. employed sustained off-resonance irradiation collisionally-activated dissociation (SORI-CAD) on a FT-ICR instrument to map the sites of interstrand cross linking by mitomycin C and 4,5′,8-trimethylpsoralen.235 This was achieved by comparing the fragmentation patterns of unmodified single-stranded oligonucleotides to those containing interstrand cross-links.235 Xiong et. al. developed a ion-pair reversed-phase nano-high-performance liquid chromatography coupled with nano-ESI-MS/MS to map the preferential sites of adduction by ± anti-BPDE on a double stranded oligonucleotide sequence.236 Using this highly sensitive method, they were able to resolve and characterize four positional isomeric ± anti-BPDE adducted oligonucleotides. Interestingly, several diastereoisomers were also resolved, although their stereochemistry could not be identified.236 Chan et. al. employed negative ion ESI-MS/MS for mapping the site and sequence specificity of aristocholic acids (AA-I and AA-II).237 5′-TTTATT-3′, 5′-TTTGTT-3′, 5′-TTTCTT-3′, and 5′-TACATGTGT-3′ oligonucleotide sequences were incubated in the presence of AA-I and AA-II. The reaction mixtures were purified by SPE or offline HPLC and subsequently analyzed by quadrupole time-of-flight MS. These experiments confirmed that aristocholic acids form covalent adducts with A and G bases of DNA.237
Chowdhury et. al. have developed an elegant and direct method for mapping the sites of base modification in oligonucleotide sequences.238 The advantage of this method was that incubation mixtures were directly analyzed by HPLC-MS/MS, avoiding the need for SPE or offline HPLC purification. This method was applied successfully to study the differences in the alkylation of multiple sites in a double stranded DNA 15-mer (representing codons 156, 157, 158, 159 of the p53 tumor suppressor gene) following reactions with BPDE or N-hydroxy-4-aminobiphenyl.238 These authors have confirmed that cytosine methylation in CpG dinucleotides led to an increase in the guanine alkylation by BPDE and N-hydroxy-4-aminobiphenyl.238
Jamin et. al. have utilized ESI-MS/MS to study the effect of different bases flanking on the 3′ or 5′ side of a guanine base that can be potentially alkylated by heterocyclic aromatic amines, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and 2-amino-3-methylimidazo[4,5-f]quinoline (IQ).239 Model oligonucleotides (5′-TTTTTXGYTTTT-3′, where X and Y can be either C/A/G/T) were incubated with activated HAAs, and the reaction mixtures were directly analyzed using HPLC-MS/MS. It was observed that IQ preferred 5′-GGG-3′ sequences, while PhIP preferred 5′-GGA/G/T-3′ trinucleotides, with the site of adduction being the center guanine base.239
Anichina et. al. employed ESI-MS/MS as a screening tool to test the reactivity of structurally related bromobenzoquinones with single stranded and double stranded oligonucleotides.240 They applied their methodology for testing of five compounds namely, 1,4-benzoquinone, 2,5-dibromo-1,4-benzoquinone, 2,5-dimethyl-3,6-dibromo-1,4-benzoquinone, 2,3,5,6-tetrabromo-1,4-benzoquinone, and 3,4,5,6-tetrabromo-1,2-benzoquinone and found that the reactivity of these closely related compounds with DNA varies depending on the extent of their bromination and methylation.240 More recently, Sharma et. al. developed a novel reversed phase-ion pairing-liquid chromatography electrospray ionization tandem mass spectrometry using a monolithic poly(styrene-divinylbenzene) capillary column eluted with a gradient of triethylammonium bicarbonate and methanol.241 This method was initially applied to map the sites of adduction by N-acetylaminofluorene (AAF) in a 15-base pair oligonucleotide fragments containing codon 135 of the p53 gene. A complex mixture of 13 different oligonucleotide products was obtained including the singly, bis-, tris-, and tetra AAF adducted oligonucleotides and isomeric olignucleotides that were successfully characterized in a single HPLC-ESI-MS/MS run.241 Additionally, this method was successfully applied to profile a complex oligonucleotide mixture including seven unmodified 15 mer oligonucleotides, an unmodified 17 mer, AAF modified 12 mer, two AAF modified 15 mer oligonucleotides, and a AAF modified 18 mer oligonucleotide.241
An alternate approach to map the sites of adduction in an oligonucleotide sequence is to digest the modified oligonucleotide and analyze the digested fragments by ESI-MS. Schrader et. al. have coupled capillary zone electrophoresis with ESI-MS to study the reactivity and sequence selectivity of styrene oxide with DNA.242;243 Experiments were performed with both oligonucleotides and calf thymus DNA, which were then enzymatically digested to random fragments. The digested oligonucleotide fragments were resolved by capillary zone electrophoresis and detected by MS. Since the adducted oligonucleotides have different mass and mobility than unmodified oligonucleotide fragments, it is possible to identify the site of adduction by looking at the molecular ions of adducted oligonucleotide fragments.242;243 Wang et. al. determined the sites of interstrand cross link formed by mitomycin C or a 4,5′,8-trimethylpsoralen by treating double stranded oligonucleotides with the drugs and digesting it with nuclease P1 and analyzing the digested products including the bearing the cross-linked nucleobase moiety by both ESI-MS and ESI-MS/MS.244 Gao et. al. subjected the dodeca-oligonucleotides modified by N-acetoxy-N-(trifluoroacetyl)-2-aminofluorene to 3′ and 5′ exonuclease digestion, and the digests were analyzed by LC-MS to determine the sites of adduction.245
Structural analysis of a modified oligonucleotide by tandem mass spectrometry results in a large amount of complex mass spectral data, which needs to be interpreted in order to map the sites of modification of oligonucleotides by carcinogens. Manual interpretation of this data can be cumbersome, but can be facilitated by computational interpretation. Several useful computer algorithms have been developed in the last 10 years for interpreting the MS/MS spectra. McCloskey and coworkers developed a program called Mongo oligo mass calculator.246 When the user inputs a known oligonucleotide sequence, the program can perform multiple functionalities including molecular mass calculations, predicting electrospray series, potential MS/MS fragments obtained by collision-induced dissociation (CID), and fragments of enzymatic digests by endonuclease and exonuclease. Simple oligonucleotide sequencer (SOS) was the first automated sequencing algorithm launched in 2002 by Rozenski and McCloskey.247 This algorithm could be successfully used for de novo sequencing of oligonucleotides 20 bases long or lesser but can't be used for base or sugar modified oligonucleotides of greater than 10 bases.247 The program identifies the 5′ (a–B-ions)- and 3′ (w-ions)-end ion series and extends the series by addition of masses of one of the four bases to derive different combinations of m/z values which will be compared with the experimental spectra.247
Oberascher et. al. started with comparative sequencing algorithm (COMPAS)248 for sequencing oligonucleotides and later developed a global de novo sequencing algorithm that starts with a known nucleotide composition and generates a theoretical mass spectra by extending it with different base combinations leading to increased run times.249 More recently, Oberascher et. al. improved their previous global de novo sequencing algorithm by usage of simulated annealing for stochastic optimization.250 Briefly, the nucleotide composition of a given oligonucleotide is determined from accurate molecular mass measurements to yield an initial sequence. The algorithm predicts the fragmentation spectra for this reference sequence and compares it with the experimental fragmentation spectra and a parameter called fitness (FS) which measures the difference between the predicted and observed m/z.250 If the FS value is smaller, a simulated annealing algorithm is applied where in two bases of the initially predicted oligonucleotide sequence are randomly changed and again the FS value is calculated. If the FS value is greater than the first iteration, the new sequence will become the reference sequence and this process continues until the optimal sequence is predicted. None of the software mentioned above have the ability to predict the sequence of oligonucleotide from the MS and MS/MS data of digested modified oligonucleotide.250
In 2008, Vouros and co-workers developed a novel software called GenoMass that uses ‘reversed pseudo-combinatorial” approach to predict the sequence of an oligonucleotide from the mass spectral data of its digest.251 The software creates a combinatorial isomer library consisting of 2-12 mer fragments in different combinations of the four bases. When the raw data from the mass spectrometry is submitted, the program scans through the data to match with the above computational data. If a match is found, it lists the mass, ion intensity and shows the extracted ion chromatogram.251 The initial functionality of the software was only identifying the adducted oligonucleotide fragments after enzymatic digestion and predicting the MS and CID spectrum of digested oligonucleotide fragments generated after digestion of modified or unmodified oligonucleotides.251 The GenoMass software was developed further by Vouros and co-workers and they recently reported a newer version252 which has the ability to map the exact site of modification in an oligonucleotide sequence by matching the combinatorial library with the experimental tandem mass spectrometry data.252 Using the newer version of the software, they mapped the position of N-acetylaminofluorene (AAF) adduct on a 17-mer (5′-CCTACCCCTTCCTTGTA-3′) oligonucleotide (5′-CCTACCCCTTCCTTGTA-3′).252
7.2 Exonuclease ladder sequencing
DNA sequence information, including the presence of endogenously and exogenously modified bases, can be obtained by mass spectral analysis of exonuclease ladders.253 Depending on their identity, DNA phosphodiesterases (PDEs) sequentially remove mononucleotides in either the 5′→ 3′ or the 3′→ 5′ direction. Since time controlled exonuclease digests contain a mixture of DNA fragments differing from each other by one or more mononucleotides (DNA “ladders”), mass differences between adjacent peaks in the resulting mass spectra correspond to individual nucleotides, making it possible to determine the complete DNA sequence (Scheme 5).109;253
Scheme 5.
Exonuclease ladder sequencing of DNA. In separate experiments, DNA is partially digested with phosphodiesterases I and II to generate two series of DNA fragments, which are analyzed by MALDI-TOF MS. Mass differences between adjacent signals in MALDI-TOF spectra can be used to identify DNA sequence and to detect the presence of endogenous or chemical modifications.
Pieles et. al. have developed an enzymatic ladder sequencing method for oligonucleotide sequencing using MALDI-TOF spectrometry.254 The exonuclease ladder approach is applicable to many DNA adduct types, e.g. N2-BPDE-dG, 8-oxo-dG, oxazolone,O6-Me-dG (Figure 12), and O6-POB-dG.253 Complete sequences can be deduced from as little as 50 pmol DNA. No a priori information on DNA sequence or adduct type is required. However, important limitations of this approach are the requirement for pure DNA samples and potential resistance of bulky adducts such as O6-POB-dG to exonuclease digestion.255
Figure 12.
Exonuclease ladder sequencing of a synthetic DNA 18-mer containing site specific O6-Me-dG. Reprinted with permission from Reference 253. Copyright 2010 American Chemical Society.
7.3 Stable isotope labeling of DNA (IDL-MS)
While ESI-MS/MS and exonuclease ladder sequencing described above are powerful approaches to locate a variety of nucleobase modifications, they are limited by their requirement of available pure oligomers containing the adducted base at a specific site. Furthermore, the length of the oligomers that can be sequenced is limited. This is not practical for chemical reactivity studies where DNA duplexes containing many potential reactive sites are treated with carcinogens. Our laboratory has developed a methodology based on stable isotope labeling of DNA and mass spectral analysis (ILD-MS) enabling accurate quantification of DNA adducts originating form specific sites within a DNA sequence.256 The ILD-MS (mass tagging) approach involves placing15N- or 13C-labeled nucleobases at specific positions within DNA oligodeoxynucleotides representing gene sequences of interest, followed by carcinogen exposure, enzymatic or acid hydrolysis of alkylated DNA, and mass spectral analysis of the resulting nucleoside adducts (Scheme 6). DNA adducts formed at the isotopically tagged position can be distinguished from the lesions originating at other sites by their molecular weight, which is increased due to the presence of15N and or 13C atoms in the molecule. The extent of reaction at the labeled nucleobase is directly calculated from the peak areas (A) in the extracted ion chromatograms corresponding to unlabeled and stable isotope-labeled adducts (Scheme 6): % reaction at X = A15N-X × 100/(AX + A15N- X). By preparing a series of oligomers of the same sequence, but with different label positions, the extent of the adduct formation at each site within the sequence can be quantified. Since the stable isotope-labeled nucleobase is chemically identical to the unlabeled base, its reactivity will represent the extent of reaction in unlabeled DNA.
Scheme 6.
Strategy for quantitation of dG adducts at specific sites within DNA by stable isotope labeling of DNA – mass spectrometry (ILD-MS) approach.
Over the past decade, the ILD-MS approach has been successfully employed to analyze the distributions of structurally defined nucleobase lesions along DNA duplexes following treatment with various carcinogens and reactive oxygen species.257;258 For example, we employed this methodology to map the formation of benzo[a]pyrene diolepoxide derived N2-BPDE-dG adducts along frequently mutated regions of the p53 tumor suppressor gene.201 For each sequence, a series of strands was prepared containing a [1,7,NH2-15N3] isotope label at one of the highlighted guanines. This label served as an isotope “tag” to enable the quantification of guanine lesions originating from that specific position (Scheme 6). 5-Methylcytosine (MeC) was incorporated at all physiologically methylated sites. Each isotopically labeled oligomer was carefully purified, structurally characterized, and annealed to the complementary unlabeled strand. Following treatment with (±) anti BPDE (10 μM), the adducted DNA was enzymatically hydrolyzed to 2′-deoxy-nucleosides in the presence of DNAse I, phosphodiesterase I, and alkaline phosphatase. Capillary HPLC-ESI+-MS/MS was used to establish the relative amounts of N2-BPDE-dG lesions originating from the stable isotope labeled dG and from unlabeled dGs elsewhere in the sequence. For example, quantitative analysis of N2-BPDE-dG and 15N3-N2-BPDE-dG was based on selected reaction monitoring of mass transitions m/z 570.2 → 454.1 and m/z 573.2 → 457.1, respectively (Figure 13), and percent reaction at the labeled site was established from HPLC-ESI+-MS/MS areas as shown in Scheme 6. A TSQ Quantum mass spectrometer (Thermo Scientific, Waltham, MA) was interfaced with an Agilent 1100 capillary HPLC operated at a flow rate of 15 μL/min. All four N2-BPDE-dG diastereomers, (+) trans, (−) trans, (+) cis, and (−) cis N2-BPDE-dG, were resolved and quantified separately (Figure 13A).201
Figure 13.
ILD-MS based mapping of diastereomeric N2-BPDE –dG adducts along DNA sequence: HPLC-ESI-MS/MS traces of N2-BPDE –dG adducts originating from 15N3-labeled guanine (m/z 573.1) and elsewhere in the sequence (m/z 570.1) (A). N2-BPDE –dG distribution along p53 gene-derived DNA duplex as determined by ILD-MS (B). Reprinted with permission from Reference 201. Copyright 2004 American Chemical Society.
We found that BPDE adducts were formed preferentially at the endogenously methylated CG dinucleotides, including those within p53 codons 157, 158, 245, 248, and 273 (Figure 13B). Many of these sites coincide with known hotspots for G → T transversions detected in smoking induced lung cancer. Targeted formation of N2-BPDE-dG at MeCG dinucleotides within the p53 gene (Figure 13B) has been previously detected by endonuclease incision-ligation mediated PCR assay and is consistent with the prevalence of G →T transversions at these sites.
8. Mass spectrometry of epigenetic modifications
8.1 Cytosine modifications occurring naturally
Endogenous cytosine methylation is involved in many physiological processes, including gene expression, host defense, genomic imprinting, and X chromosome inactivation.1 5-Methylcytosine (MeC) bases are formed by the enzymatic methylation of the C5 position of cytosine in CG dinucleotides (CpG sites) and represent about 1% of total bases in the mammalian genome.259 DNA methylation patterns are maintained by the action of methyltransferase enzymes. If present in gene promoter sequences, MeC typically reduce the levels of gene expression.260 While the reverse process of removing the 5-methyl group is known to occur, the mechanism of demethylation has not yet been identified. Many tumor types, including smoking-induced lung cancer, are characterized by aberrant DNA methylation patterns, resulting in the silencing of tumor suppressor genes, activating oncogenes, and decreasing chromosome stability.200
Recent studies have discovered several related epigenetic modifications of DNA in addition to MeC, including 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC).261;262 These modifications are thought to arise from the Tet (ten eleven translocation protein)-mediated oxidation of MeC.18 5hmC, 5fC, and 5caC are present in genomic DNA of various organs and are hypothesized to play a yet unidentified role in epigenetic signaling or possibly serve as demethylation intermediates.
8.2 MS methods to quantify 5-methylcytosine and related epigenetic modifications
Among the various bases involved in epigenetic changes, 5-methylcytosine (MeC) has been widely quantified using various mass spectrometric methods. HPLC-ESI-MS/MS analysis was used as a tool for quantification of methylated nucleoside (5-methyl-dC) or its free base (MeC) in DNA from various sources to determine the levels of global DNA methylation.263-271 Most of the earlier methods did not employ isotopically labeled internal standards for quantitation, but rather reported the relative amounts of methylated base with respect to either dC or dG. For example, Thuc Le et al. developed a mass spectrometric method using selected reaction monitoring for simultaneous relative quantitation of MeC and 5hmC.272 These authors compared the levels of MeC and 5hmC in human pluripotent stem cells versus differentiated parental fibroblast cells where, the pluripotent stem cells had increased levels of 5meC and 5hmeC.272
5-Methylcytosine was also quantified in urine of smokers and non-smokers using an online solid-phase extraction (SPE) coupled to an isotope dilution HPLC-ESI-MS/MS (LC) procedure.267 This method employed deuterated internal standards for absolute quantitation of MeC and 5-Me-dC. In another study, 5-medC was quantified in human urine samples using HPLC-MS/MS analysis.273
The less abundant epigenetically modified bases such as 5hmC have been detected using immunological,4;5;274;275 enzymatic,276-278 and 32P-radioactive labeling 1D or 2D thin layer chromatography methods.3 More recently, mass spectrometry methods have played a major role in detecting and quantifying minor epigenetic modifications in experimental systems and in studies of their tissue distribution in vivo. For example, Globisch et al.261 used HPLC-ESI-MS/MS methodology to determine tissue distribution of 5hmC and other active demethylation intermediates. These authors found that while the concentrations of mC in all tissues were 4.3 % of dG in all tissues, hmC concentrations were highly variable, with 0.4-0.7 % hmC detected in CNS tissues, 0.2-0.4% in heart, muscle, kidney, and < 0.1% in the lung, spleen, and testes.261 In contrast, 5–formylcytosine (fC), 5-carboxylcytosine (caC), and 5–hydroxymethyluracil (hmU) were not detected in that study. Global DNA levels of 5hmC were analyzed in the DNA from cerebrums of rats using isotope dilution liquid chromatography-tandem mass spectrometry (LC-MS/MS) and were found to be 0.66%.279 There are very few mass spectrometric methods in literature for the quantification of 5fC and 5caC. Among them, Ito et al. 262 employed HPLC-ESI-MS/MS to quantify 5fC and 5caC in genomic DNA of mouse embryonic stem cells.
9. Mass spectrometry to identify the biological consequences of DNA damage
In addition to determining DNA adduct structures and quantifying their levels in biological samples, mass spectrometry has played a major role in determining the biological consequences of DNA damage. For example, the instrumentation can be used to determine the identity of nucleotides inserted opposite DNA adducts upon in vitro and in vivo DNA replication and to investigate DNA repair mechanisms and adduct persistence in tissues. Several examples of such applications are briefly described in this section.
9.1 Primer extension
As discussed in the Introduction, DNA can be modified by endogenous and exogenous agents leading to the formation of nucleobase adducts (Scheme 1).280 The resulting structurally modified nucleobases may block the normal DNA replication process due to their size or their effects on DNA structure.281 The specialized translesion synthesis polymerases (TLS polymerases) can copy across bulky or distorting DNA lesions, avoiding replicative crisis.282-284 In general, lesion bypass polymerases have large active sites and lack endonuclease proofreading activity, which enables them to bypass DNA lesions. As a result, these specialized DNA polymerases have lower fidelity and are more likely to introduce errors, leading to mutations.285-287 Mass spectrometry along with gel electrophoresis is an important tool employed in studying the mutagenic ability of structurally defined DNA adducts in vitro.288-292 These in vitro translesion synthesis experiments are generally performed by annealing a DNA template strand containing a site specific DNA adduct to a complementary DNA strand (primer). The length of the primer strand is shorter than the template, so that the 3′ terminus of the primer is positioned prior to the lesion290;292 (Scheme 7). The template-primer duplexes are incubated in the presence of a TLS polymerase and all four dNTPs allowing for primer extension. The primer extension products have been traditionally visualized by gel electrophoresis using a 32P-end labeled primer.293-295 However, the exact sequence of primer extension products cannot be determined by gel electrophoresis, and multiple time consuming experiments involving individual dNTPs must be conducted to evaluate each incorporation step.292 More importantly, gel electrophoresis fails to detect deletion products occurring when a polymerase skips the modified base, resulting in frameshift mutations. HPLC-ESI-MS/MS can be used to accurately sequence primer extension products in a single experiment, without the use of radioactivity.292
Scheme 7.
MS/MS identification of in vitro replication products of adduct-containing DNA following site-specific cleavage with UDG/piperidine.
Because of a size limitations for sequencing oligonucleotide by MS/MS, Zang et. al have developed a methodology in which the primer is engineered to contain a uracil residue close to its 3′ terminus (e.g. Scheme 7).292 Following primer extension, the extension products are cleaned with uracil DNA glycosylase (UDG) hot piperidine to generate shorter oligonucleotides which can be readily sequenced by tandem mass spectrometry.
For the specific template-primer complex shown in Scheme 7, 7 mer products are generated when the primer extension is complete. Depending on the specific base incorporated opposite the adduct (X) and assuming the enzyme does not make any additional errors while copying beyond the adduct, the sequence of the extension product can be any of the following: 5′p-TCAATGA-3′, 5′p-TCGATGA-3′, 5′p-TCCATGA-3′, or 5′p-TCTATGA-3′. The preliminary identity of the product can be obtained by determining the m/z values of the observed oligonucleotide products using full scan HPLC-ESI- MS. MS/MS spectra of each product are then obtained using ion trap or triple quadrupole instrumentation, and are compared to the predicted spectra (Mongo Oligo Mass Calculator v2.06) (Example in Table 3).
Table 3.
Predicted CID fragments for DNA 7-mer, 5′p-TCTATGA-3′.
| n | ch | a-B | w | y | d-H2O |
|---|---|---|---|---|---|
| 1 | -1 | 330.217 | 250.237 | 303.189 | |
| -2 | 151.090 | ||||
| 2 | -1 | 481.274 | 659.427 | 579.447 | 592.374 |
| -2 | 240.133 | 329.209 | 289.219 | 295.683 | |
| -3 | 196.786 | ||||
| 3 | -1 | 770.459 | 963.624 | 883.644 | 896.571 |
| -2 | 384.725 | 481.308 | 441.318 | 447.781 | |
| -3 | 256.147 | 320.536 | 293.876 | 298.185 | |
| -4 | 223.386 | ||||
| 4 | -1 | 1074.656 | 1276.834 | 1196.854 | 1209.781 |
| -2 | 536.824 | 637.913 | 597.923 | 604.386 | |
| -3 | 357.546 | 424.939 | 398.279 | 402.588 | |
| -4 | 267.908 | 318.452 | 298.457 | 301.689 | |
| -5 | 241.149 | ||||
| 5 | -1 | 1387.866 | 1581.031 | 1501.051 | 1513.978 |
| -2 | 693.429 | 790.011 | 750.021 | 756.485 | |
| -3 | 461.950 | 526.338 | 499.678 | 503.987 | |
| -4 | 346.210 | 394.501 | 374.506 | 377.738 | |
| -5 | 276.766 | 315.399 | 299.403 | 301.989 | |
| -6 | |||||
| 6 | -1 | 1692.063 | 1870.216 | 1790.236 | 1843.188 |
| -2 | 845.527 | 934.604 | 894.614 | 921.090 | |
| -3 | 563.349 | 622.733 | 596.073 | 613.724 | |
| -4 | 422.259 | 466.798 | 446.803 | 460.041 | |
| -5 | 337.606 | 373.236 | 357.240 | 367.831 | |
| -6 | 281.170 | 310.862 | 297.532 | 306.358 | |
| -7 | 262.448 |
The methodology developed by Zang et. al. has been successfully used in translesion synthesis studies of several important DNA adducts including 1,N2 -etheno guanine,288;292 malondialdehyde-deoxyguanosine (M1-dG),290;296 7-8-dihydro-8-oxodeoxyguanosine,297;298 O6-benzylguanine,299;300 O6-methylguanine,300;301 O6-[4-oxo-4-(3-pyridyl)butyl] (O6-Pob-G)300,N2-alkyl guanine adducts,302 C8- and N2-deoxyguanosine adducts of the dietary mutagen 2-amino-3-methylimidazo]4,5-f[quinolone,291 N6-deoxyadenosine-BPDE adducts,303 2,6-diamino-4-hydroxy-N5 -(methyl)-formamidopyrimidine (MeFapy-dGuo),304 and 7-(2-oxoheptyl)-1,N2 -etheno-2′-deoxyguanosine.289
Our laboratory has recently utilized an HPLC-ESI-MS/MS methodology for studying translesion synthesis across 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2′-deoxyadenosine (1,N6-γ-HMHP-dA) adducts formed by the alkylation of adenine by 1,2,3,4-diepoxybutane (genotoxic metabolite of 1,3-butadiene). 305 The primer/template sequences were as shown in Scheme 7.305 Briefly, an 18-mer oligonucleotide template strand (5′-TCATXGA ATCCTTCCCCC-3′) where X=1,N6-γ-HMHP-dA was annealed to a 13 mer primer (5′-GGGGGAAGGAUTC-3′) and incubated in the presence of individual TLS polymerases (hPol η, hPol κ) and all the four dNTPs. At the end of the polymerase reaction, the primer extension products were cleaved with UDG/piperidine, and the resulting short oligonucleotide products were sequenced by HPLC-ESI--MS/MS on an Orbitrap Velos (Thermo Scientific). A representative extracted ion chromatogram revealing the presence of six different primer extension products following hPol κ catalyzed primer extension are shown in Figure 14. The relative yields of individual products were calculated by comparing peak areas to that of a known internal standard (14-mer). As evident from Figure 14, incorporation of incorrect bases (G, A) along with the correct base (T) was observed. The error-free product 5′p-TCTATGA-3′ constituted only 18% of all the observed products. We also observed single and double deletion/frameshift mutation products.305. Based on these results, we proposed that 1,N6-γ-HMHP-dA adducts could lead to A→T and A→C transversions in vivo.305
Figure 14.
HPLC-ESI-MS detection of primers extension products following in vitro replication of DNA template containing 1,N6-HMHP-dA. MS analysis reveals multiple mutated products and -1 deletions.
9.2 Site-specific mutagenesis
The Wang group recently developed a new mass spectrometry-based methodology allowing for the determining the mutagenicity of specific DNA lesions in vivo, e.g. in living cells.306 This is a modification of the site specific mutagenesis approach developed by Essigmann et al.307 In short, double-stranded plasmids are constructed containing site-specific DNA lesions. The plasmids are introduced into bacterial or mammalian cells and are allowed to replicate. The products formed from replication or transcription are amplified by PCR, digested with restriction enzymes, and the resulting restriction fragments are sequenced by HPLC-ESI--MS/MS. These experiments can determine the influence of specific DNA lesions on the efficiency and fidelity of DNA replication. This approach has been applied to several important DNA adducts, including N2-carboxyalkyl-2′-deoxyguanosine lesions produced by glyoxal.306
9.3 MS studies of DNA repair
Mass spectrometry provides an accurate and specific tool for investigating DNA adduct repair in vitro and in vivo. For example, MALDI-TOF MS in combination with hot piperidine cleavage was used to detect base excision repair of oxidative dG adducts in synthetic DNA duplexes.308 DNA duplexes containing site specific 8-oxo-G, oxaluric acid, oxazolone, or cyanuric acid lesions were incubated with recombinant formamidopyrimidine glycosylase (Fpg enzyme), and the resulting abasic sites were converted to strand breaks in the presence of hot piperidine. The cleavage products were detected by MALDI-TOF using 3-hydroxypicolinic acid matrix. Fpg efficiently excised 8-oxo-G and oxaluric acid and to some extent oxazolone, but not cyanuric acid. These data suggest that some DNA lesions formed via ONOO− exposures (cyanuric acid) are not repaired by Fpg and are not uncovered by assays based on piperidine cleavage at the site of lesion.308
Guza et al. have employed isotope dilution HPLC-ESI-MS/MS to investigate the influence of DNA sequence on the kinetics of repair of O6-alkylguanine adducts by the specialized repair protein,O6-alkylguanine-DNA alkyltransferase (AGT).309 O6-Me-dG adducts were placed at different sites (G5, G6, G7, G8, G9, G10, or G11) within the synthetic double stranded DNA sequences representing codons 8-17 of the K-ras protooncogene (5′-G1TA G2TT G3G4A G5CT G6G7T G8G9C G10TA G11G12C AAG13 AG14T-3′). The second guanine of K-ras codon 12 is a major mutational hotspot for G → A transitions observed in lung tumors of smokers and in neoplasms induced in laboratory animals by exposure to methylating agents.O6-Me-dG containing double stranded DNA oligomers were incubated with human recombinant AGT protein at conditions favoring second-order kinetics, and the reactions were quenched at varying times. Following acid hydrolysis, isotope dilution HPLC-ESI-MS/MS was used to determine the amounts of unrepaired O6-Me-dG remaining in DNA. The relative extent of repair for O6-methyl-dG located at G5, G6, G7, G8, G9, G10, or G11 following a 10 s incubation with AGT varied by 5-30% depending on sequence position. A similar strategy was recently used to analyze the kinetics of AGT-mediated repair of O6-pyridyloxobutylguanine lesions.120
In vivo investigations of DNA adduct repair in various tissues can be achieved in laboratory animal studies. Following exposure period, animals are allowed to recover for specified periods of time. DNA is extracted from tissues, followed by standard sample preparation techniques and HPLC-MS/MS analysis as described in Sections 3 and 5. This strategy allows for determining the kinetics of DNA adduct removal in various tissues of a living animal, to observe tissue differences in DNA repair, and to identify DNA lesions that are resistant to repair and thus persist in vivo, increasing their chances of inducing genetic damage. For example, Goggin et al.68 employed capillary HPLC-ESI-MS/MS to study the in vivo persistence of DNA-DNA adducts and exocyclic adducts of 1,2,3,4-diepoxybutane in tissues of laboratory mice exposed to 1,3-butadiene (Figure 15).68
Figure 15.
Persistence of bifunctional DEB-DNA adducts in mouse and rat liver DNA. (A) Female B6C3F1 mice were exposed to 625 ppm BD for 2 weeks, and tissues were collected 2, 72, or 240 h post exposure. (B) Female F344 rats were exposed to 1250 ppm BD for 2 weeks, and tissues were collected 2, 24, 72, or 144 h post exposure. Bis-N7G-BD, N7G-N1A-BD, and 1,N6-HMHP-dA, were quantified by isotope dilution HPLC-ESI-MS/MS analysis of DNA hydrolysates.
More recently, our laboratory has developed sensitive capillary HPLC-ESI-MS/MS methodologies to monitor the kinetics of N,N-bis[2-(N7-guaninyl) ethyl] amine DNA-DNA cross-links (G-NOR-G) cross-link repair in peripheral blood lymphocytes of cancer patients undergoing chemotherapy with cyclophosphamide (CPA).310 DNA was extracted from peripheral blood lymphoblasts of cancer patients receiving 50-60 mg/kg CPA intravenously. Extracted DNA samples (5-20 μg) were spiked with an internal standard of [15N10]-G-NOR-G and subjected to neutral thermal hydrolysis, releasing G-NOR-G nucleobase adducts from the DNA backbone. Following solid phase extraction, G-NOR-G conjugates were quantified by capillary HPLC-ESI-MS/MS in the selected reaction monitoring mode. The highest numbers of G-NOR-G adducts (up to 18 adducts per 106 normal nucleotides) were observed 4-8 h following CPA administration; adduct levels gradually decreased over time, probably due to their spontaneous depurination, active repair, and white blood cell turnover. Interestingly, CPA-DNA adduct concentrations were elevated in patients with Fanconi Anemia as compared to non-Fanconi Anemia cancer patients; this is consistent with compromised interstrand DNA cross-link repair in Fanconi Anemia.311
10. Conclusions
Over the past 30 years, mass spectrometry has played a central role in characterizing endogenous and exogenous DNA modifications. It has also become the method of choice for quantitative analyses of DNA adducts in vitro and in vivo. Mass spectrometry provides valuable structural information used to unambiguously identify DNA nucleosides, nucleotides, and nucleobases. Furthermore, the use of isotope dilution mass spectrometry enables accurate, precise, and reproducible quantitation of DNA damage in tissues and cells. A variety of possibilities are available for chromatographic separation of DNA adducts, and a wide range of mass spectrometry systems with various capabilities can be used for both qualitative and quantitative analysis of DNA modifications. The field of DNA adduct analysis by mass spectrometry is expanding rapidly, as novel instrumentation and approaches are being introduced, including online sample preparation, nanoflow LC-MS, high resolution HPLC-MS/MS, and chip based methology. These rapid developments in chromatography and mass spectrometry instrumentation and methodologies are enabling the analysis of low abundance DNA adducts and epigenetic modifications in human samples. Furthermore, mass spectrometry based methods are widely used for sequencing of carcinogen-modified DNA and in combination with biotechnology tools, to determine the biological consequences of DNA damage. Mass spectrometry will undoubtedly remain on the forefront of chemical carcinogenesis and epigenetics research, continuing to provide valuable information about structural modifications of nucleic acids in cells.
Acknowledgments
We are thankful to Bob Carlson (University of Minnesota Masonic Cancer Center) for preparing figures and tables for this manuscript, Dewakar Sangaraju for his help with several sections, and Gregory Janis (MedTox Laboratories) for his valuable comments and suggestions on the manuscript. This work is supported by grants from the National Cancer Institute (CA 100670, CA 095039, CA 138338).
Biographies
Srikanth Kotapati completed his Bachelor's and Master's degrees in Pharmacy in 2007 at the Birla Institute of Technology and Sciene, Pilani (BITS, Pilani), India. He is currently a Ph.D. candidate in the Tretyakova laboratory at the Department of Medicinal Chemistry and the Masonic Cancer Center -University of Minnesota, Twin Cities. His thesis project involves the development of HPLC-ESI-MS/MS methods for quantification of urinary biomarkers of human exposure to 1,3-butadiene and application of these methods in identifying ethnic/racial differences in lung cancer risk. Additionally, he is investigating the effects of 1,3-butadiene-DNA adducts on DNA replication using gel electrophoresis and tandem mass spectrometry techniques.

Natalia Tretyakova was born in Moscow, Russia and obtained her B.Sc. and M.S. degrees in Chemistry from Moscow State University. In 1992, she moved to the United States and received her Ph.D. degree in 1997 at the University of North Carolina, Chapel Hill under the direction of Professor James Swenberg. She spent 3 years as a Postdoctoral Associate at the Massachusetts Institute of Technology (advisor, Professor Steven Tannenbaum). In 2000, she started her independent academic career at the Department of Medicinal Chemistry and the Masonic Cancer Center at the University of Minnesota-Twin Cities, where she is currently a Professor of Medicinal Chemistry. She teaches drug metabolism, nucleic acid chemistry, and mass spectrometry courses.
Dr. Tretyakova has authored 68 publications in peer-reviewed journals and books. Her research interests include chemical carcinogenesis, mass spectrometry of structurally modified nucleic acids, quantitative analysis of DNA adducts and epigenetic modifications in humans, and the use of mass spectrometry to sequence carcinogen-modified DNA.

Peter Villalta was born in 1965 in Fort Frances, Ontario and obtained a B.Sc. degree in 1987 from St. John's University in Collegeville, Minnesota. In 1993, he received a Ph.D. degree in Chemistry from the University of Minnesota under the direction of Professor Doreen Leopold. He spent 2 years as a Postdoctoral Associate at the National Oceanic and Atmospheric Administration in Boulder, Colorado (advisor, Dr. Carl Howard). In 1995, he took a position as a senior research scientist at Aerodyne Research in Billerica, Massachusetts. In 1999, he moved to his current position as the coordinator of mass spectrometry services at the Masonic Cancer Center of the University of Minnesota. His research interests include developing methodologies for screening unknown DNA adducts (Adductomics) and improving in vivo DNA adduct quantitation through advanced mass spectrometry and chromatography methodologies.

Reference List
- 1.Riggs AD, Jones PA. Adv Cancer Res. 1983;40:1. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- 2.Widschwendter M. Dis Markers. 2007;23:1. doi: 10.1155/2007/860404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriaucionis S, Heintz N. Science. 2009;324:929. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Nature. 2011;473:398. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 5.Jin SG, Wu X, Li AX, Pfeifer GP. Nucleic Acids Res. 2011;39:5015. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. Plenum Press; New York and London: 1983. [Google Scholar]
- 7.Weinberg RA. The Biology of Cancer. Garland Science; New York, NY: 2006. [Google Scholar]
- 8.Harris CC. Cancer Res. 1991;51:5023s. [PubMed] [Google Scholar]
- 9.Hecht SS. J Natl Cancer Inst. 1999;91:1194. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 10.Rajski SR, Williams RM. Chem Rev. 1998;98:2723. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 11.Swenberg JA, Fryar-Tita E, Jeong YC, Boysen G, Starr T, Walker VE, Albertini RJ. Chem Res Toxicol. 2008;21:253. doi: 10.1021/tx700408t. [DOI] [PubMed] [Google Scholar]
- 12.Farmer PB, Brown K, Tompkins E, Emms VL, Jones DJ, Singh R, Phillips DH. Toxicol Appl Pharmacol. 2005;207:293. doi: 10.1016/j.taap.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 13.Farmer PB, Singh R. Mutat Res. 2008;659:68. doi: 10.1016/j.mrrev.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Koc H, Swenberg JA. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:323. doi: 10.1016/s1570-0232(02)00135-6. [DOI] [PubMed] [Google Scholar]
- 15.Beach AC, Gupta RC. Carcinogenesis. 1992;13:1053. doi: 10.1093/carcin/13.7.1053. [DOI] [PubMed] [Google Scholar]
- 16.Gennaro LA, Vadhanam M, Gupta RC, Vouros P. Rapid Commun Mass Spectrom. 2004;18:1541. doi: 10.1002/rcm.1516. [DOI] [PubMed] [Google Scholar]
- 17.Randerath K, Reddy MV, Gupta RC. Proc Natl Acad Sci USA. 1981;78:6126. doi: 10.1073/pnas.78.10.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeppky RN, Ye Q, Goelzer P, Chen Y. Chem Res Toxicol. 2002;15:470. doi: 10.1021/tx0101393. [DOI] [PubMed] [Google Scholar]
- 19.Boyd RK, Basic C, Bethem RA. Tools of the Trade V Mass Analyzers for Quantitation: Separation of Ions by m/z Values. John Wiley & Sons; West Sussex, England: 2008. Trace Quantitative Analysis by Mass Spectrometry; pp. 266–301. [Google Scholar]
- 20.Brink A, Lutz U, Volkel W, Lutz WK. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;830:255. doi: 10.1016/j.jchromb.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 21.Matter B, Malejka-Giganti D, Csallany AS, Tretyakova N. Nucleic Acids Res. 2006;34:5449. doi: 10.1093/nar/gkl596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong LZ, McCullagh JSO. J Chromatogr A. 2011;1218:5480. doi: 10.1016/j.chroma.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 23.Pucci V, Giuliano C, Zhang RA, Koeplinger KA, Leone JF, Monteagudo E, Bonelli F. J Sep Sci. 2009;32:1275. doi: 10.1002/jssc.200800722. [DOI] [PubMed] [Google Scholar]
- 24.Tuytten R, Lemiere F, Van Dongen W, Witters E, Esmans EL, Newton RP, Dudley E. Anal Chem. 2008;80:1263. doi: 10.1021/ac702057u. [DOI] [PubMed] [Google Scholar]
- 25.Zhao HQ, Chen JH, Shi Q, Li X, Cao W, Li JX, Wang XR. Chem J Chinese U. 2012;33:44. [Google Scholar]
- 26.Goggin M, Seneviratne U, Swenberg JA, Walker VE, Tretyakova N. Chem Res Toxicol. 2010;23:808. doi: 10.1021/tx900439w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillestrom PR, Hoberg AM, Weimann A, Poulsen HE. Free Radic Biol Med. 2004;36:1383. doi: 10.1016/j.freeradbiomed.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 28.Hillestrom PR, Weimann A, Poulsen HE. J Am Soc Mass Spectrom. 2006;17:605. doi: 10.1016/j.jasms.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Singh R, Arlt VM, Henderson CJ, Phillips DH, Farmer PB, Gamboa dC. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:2155. doi: 10.1016/j.jchromb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R, Teichert F, Seidel A, Roach J, Cordell R, Cheng MK, Frank H, Steward WP, Manson MM, Farmer PB. Rapid Commun Mass Spectrom. 2010;24:2329. doi: 10.1002/rcm.4645. [DOI] [PubMed] [Google Scholar]
- 31.Quigley WW, Dovichi NJ. Anal Chem. 2004;76:4645. doi: 10.1021/ac040100d. [DOI] [PubMed] [Google Scholar]
- 32.Bhowmik SK, Jung BH. Rapid Commun Mass Spectrom. 2012;26:1426. doi: 10.1002/rcm.6245. [DOI] [PubMed] [Google Scholar]
- 33.Ding J, Vouros P. J Chromatogr A. 2000;887:103. doi: 10.1016/s0021-9673(00)00469-6. [DOI] [PubMed] [Google Scholar]
- 34.Singh R, Farmer PB. Carcinogenesis. 2006;27:178. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- 35.Apruzzese WA, Vouros P. J Chromatogr A. 1998;794:97. doi: 10.1016/s0021-9673(97)00820-0. [DOI] [PubMed] [Google Scholar]
- 36.Willems AV, Deforce DL, Van den Eeckhout EG, Lambert WE, Van Peteghem CH, De Leenheer AP, Van Bocxlaer JF. Electrophoresis. 2002;23:4092. doi: 10.1002/elps.200290026. [DOI] [PubMed] [Google Scholar]
- 37.Deforce DL, Raymackers J, Meheus L, Van Wijnendaele F, De Leenheer A, Van den Eeckhout EG. Anal Chem. 1998;70:3060. [Google Scholar]
- 38.Harsch A, Vouros P. Anal Chem. 1998;70:3021. doi: 10.1021/ac9713823. [DOI] [PubMed] [Google Scholar]
- 39.Chen B, Bartlett MG. Biomed Chromatogr. 2012;26:409. doi: 10.1002/bmc.1696. [DOI] [PubMed] [Google Scholar]
- 40.Willems A, Deforce DL, Van Bocxlaer J. Methods Mol Biol. 2008;384:401. doi: 10.1007/978-1-59745-376-9_14. [DOI] [PubMed] [Google Scholar]
- 41.Willems AV, Deforce DL, Van Peteghem CH, Van Bocxlaer JF. Electrophoresis. 2005;26:1221. doi: 10.1002/elps.200410278. [DOI] [PubMed] [Google Scholar]
- 42.Wolf SM, Vouros P. Anal Chem. 1995;67:891. doi: 10.1021/ac00101a016. [DOI] [PubMed] [Google Scholar]
- 43.Barry JP, Norwood C, Vouros P. Anal Chem. 1996;68:1432. doi: 10.1021/ac9510245. [DOI] [PubMed] [Google Scholar]
- 44.Deforce DL, Lemiere F, Hoes I, Millecamps RE, Esmans EL, De Leenheer A, Van den Eeckhout EG. Carcinogenesis. 1998;19:1077. doi: 10.1093/carcin/19.6.1077. [DOI] [PubMed] [Google Scholar]
- 45.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Mass Spectrom Rev. 1990;9:37. [Google Scholar]
- 46.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 47.Blades AT, Ikonomou MG, Kebarle P. Anal Chem. 1991;63:2109. [Google Scholar]
- 48.Bruins AP. ESI source design. Wiley; 2010. Electrospray and MALDI mass spectrometry: fundamentals, instrumentation, practicalities, and biological applications; pp. 123–147. [Google Scholar]
- 49.Bruins AP, Covey TR, Henion JD. Anal Chem. 1987;59:2642. [Google Scholar]
- 50.Kebarle P, Verkerk UH. On the mechanism of electrospray ionization mass spectrometry. Wiley; 2010. Electrospray and MALDI mass spectrometry: fundamentals, instrumentation, practicalities, and biological applications. [Google Scholar]
- 51.Cech NB, Enke CG. Mass Spectrom Rev. 2001;20:362. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 52.Kebarle P, Tang L. Anal Chem. 1993;65:972A. [Google Scholar]
- 53.Buhrman DL, Price PI, Rudewicz PJ. J Am Soc Mass Spectrom. 1996;7:1099. doi: 10.1016/S1044-0305(96)00072-4. [DOI] [PubMed] [Google Scholar]
- 54.Kim J, Park S, Tretyakova NY, Wagner CR. Mol Pharm. 2005;2:233. doi: 10.1021/mp0500162. [DOI] [PubMed] [Google Scholar]
- 55.Tretyakova NY, Goggin M, Sangaraju D, Janis G. Chem Res Toxicol. 2012;25:2007. doi: 10.1021/tx3002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tretyakova NY, Burney S, Pamir B, Wishnok JS, Dedon PC, Wogan GN, Tannenbaum SR. Mutat Res. 2000;447:287. doi: 10.1016/s0027-5107(99)00221-3. [DOI] [PubMed] [Google Scholar]
- 57.Covey TR, Schneider BB, Javaheri H, Yves LeBlanc JC, Ivosev G, Corr JJ, Kovarik P. ESI, APCI, and MALDI a comparison of the central analytical figures of merit: sensitivity, reproducibility, and speed. Wiley; 2010. Electrospray and MALDI mass spectrometry: fundamentals, instrumentation, practicalities, and biological applications; pp. 443–490. [Google Scholar]
- 58.Hoes I, Van Dongen W, Lemiere F, Esmans EL, Van Bockstaele D, Berneman ZN. J Chromatogr B Biomed Sci Appl. 2000;748:197. doi: 10.1016/s0378-4347(00)00400-x. [DOI] [PubMed] [Google Scholar]
- 59.Smith RD, Shen Y, Tang K. Acc Chem Res. 2004;37:269. doi: 10.1021/ar0301330. [DOI] [PubMed] [Google Scholar]
- 60.Chen HJ, Lin WP. Anal Chem. 2009;81:9812. doi: 10.1021/ac9019472. [DOI] [PubMed] [Google Scholar]
- 61.Chen HJ, Lin GJ, Lin WP. Anal Chem. 2010;82:4486. doi: 10.1021/ac100391f. [DOI] [PubMed] [Google Scholar]
- 62.Chen HJ, Lin WP. Anal Chem. 2011;83:8543. doi: 10.1021/ac201874d. [DOI] [PubMed] [Google Scholar]
- 63.Chen HJ, Wang YC, Lin WP. Anal Chem. 2012;84:2521. doi: 10.1021/ac203405y. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Q, Aft RL, Gross ML. Chem Res Toxicol. 2008;21:1509. doi: 10.1021/tx8001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu K, Moeller B, Doyle-Eisele M, McDonald J, Swenberg JA. Chem Res Toxicol. 2011;24:159. doi: 10.1021/tx1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balbo S, Villalta PW, Hecht SS. Chem Res Toxicol. 2011;24:1729. doi: 10.1021/tx200262d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goggin M, Swenberg JA, Walker VE, Tretyakova N. Cancer Res. 2009;69:2479. doi: 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goggin M, Sangaraju D, Walker VE, Wickliffe J, Swenberg JA, Tretyakova N. Chem Res Toxicol. 2011;24:809. doi: 10.1021/tx200009b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sangaraju D, Goggin M, Walker V, Swenberg J, Tretyakova N. Anal Chem. 2012;84:1732. doi: 10.1021/ac203079c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Embrechts J, Lemiere F, Van DW, Esmans EL, Buytaert P, Van ME, Kockx M, Makar A. J Am Soc Mass Spectrom. 2003;14:482. doi: 10.1016/S1044-0305(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 71.Embrechts J, Lemiere F, Van Dongen W, Esmans EL. J Mass Spectrom. 2001;36:317. doi: 10.1002/jms.136. [DOI] [PubMed] [Google Scholar]
- 72.Wang JJ, Marshall WD, Frazer DG, Law B, Lewis DM. Anal Biochem. 2003;322:79. doi: 10.1016/j.ab.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 73.Wang JJ, Marshall WD, Law B, Lewis DM. Int J Mass Spectrom Ion Proc. 2003;230:45. [Google Scholar]
- 74.Thomson BA. The heated nebulizer LC-MS interface. Elsevier Ltd; Oxford: 2007. Encyclopedia of Mass Spectrometry; pp. 366–370. [Google Scholar]
- 75.Bruins AP. Mass Spectrom Rev. 1991;10:53. [Google Scholar]
- 76.Singh G, Gutierrez A, Xu K, Blair IA. Anal Chem. 2000;72:3007. doi: 10.1021/ac000374a. [DOI] [PubMed] [Google Scholar]
- 77.van den Boom D, Berkenkamp S. MALDI-MS of Nucleic Acids and Practical Implementations in Genomics and Genetics. Wiley-VCH Verlag GmbH & Co KGaA; Weinheim, Germany: 2007. MALDI MS: A Practical Guide to Instrumentation, Methods and Applications; pp. 131–179. [Google Scholar]
- 78.Nordhoff E, Kirpekar F, Karas M, Cramer R, Hahner S, Hillenkamp F, Kristiansen K, Roepstroff P, Lezius A. Nucleic Acids Res. 1994;22:2460. doi: 10.1093/nar/22.13.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu YF, Chung CN, Taranenko NI, Allman SL, Martin SA, Haff L, Chen CH. Rapid Commun Mass Spectrom. 1996;10:383. doi: 10.1002/(SICI)1097-0231(199602)10:3<383::AID-RCM485>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 80.Dass C. Mass Analysis and Ion Detection. Wiley-Interscience; New York: 2001. Principles and Practice of Biological Mass Spectrometry; pp. 59–93. [Google Scholar]
- 81.Goodenough AK, Schut HA, Turesky RJ. Chem Res Toxicol. 2007;20:263. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kristensen DB, Imamura K, Miyamoto Y, Yoshizato K. Electrophoresis. 2000;21:430. doi: 10.1002/(SICI)1522-2683(20000101)21:2<430::AID-ELPS430>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 83.Lacorte S, Fernandez-Alba AR. Mass Spectrom Rev. 2006;25:866. doi: 10.1002/mas.20094. [DOI] [PubMed] [Google Scholar]
- 84.Makarov A. Anal Chem. 2000;72:1156. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 85.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach JJ, Cox J, Horning S, Mann M, Makarov A. Mol Cell Proteomics. 2012;11:O111. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kalli A, Hess S. Proteomics. 2012;12:21. doi: 10.1002/pmic.201100464. [DOI] [PubMed] [Google Scholar]
- 87.Frese CK, Altelaar AF, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJ, Mohammed S. J Proteome Res. 2011;10:2377. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- 88.Bernsmann T, Furst P, Godula M. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2011;28:1352. doi: 10.1080/19440049.2011.619504. [DOI] [PubMed] [Google Scholar]
- 89.Moulard Y, Bailly-Chouriberry L, Boyer S, Garcia P, Popot MA, Bonnaire Y. Anal Chim Acta. 2011;700:126. doi: 10.1016/j.aca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 90.Chitescu CL, Oosterink E, de Jong J, Linda Stolker AA. Anal Bioanal Chem. 2012;403:2997. doi: 10.1007/s00216-012-5888-8. [DOI] [PubMed] [Google Scholar]
- 91.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Anal Chem. 2010;82:3212. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He X, Kozak M, Nimkar S. Anal Chem. 2012;84:7643. doi: 10.1021/ac3019476. [DOI] [PubMed] [Google Scholar]
- 94.Moller I, Thomas A, Delahaut P, Geyer H, Schanzer W, Thevis M. J Pharm Biomed Anal. 2012;70:512. doi: 10.1016/j.jpba.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 95.Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. Mol Cell Proteomics. 2012:1709. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holcapek M, Jirasko R, Lisa M. J Chromatogr A. 2012;1259:3. doi: 10.1016/j.chroma.2012.08.072. [DOI] [PubMed] [Google Scholar]
- 98.Bryant MS, Lay JO., Jr J Am Soc Mass Spectrom. 1992;3:360. doi: 10.1016/1044-0305(92)87064-6. [DOI] [PubMed] [Google Scholar]
- 99.Ishii Y, Suzuki Y, Hibi D, Jin M, Fukuhara K, Umemura T, Nishikawa A. Chem Res Toxicol. 2011;24:532. doi: 10.1021/tx100410y. [DOI] [PubMed] [Google Scholar]
- 100.Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, Turesky RJ. Chem Res Toxicol. 2010;23:1234. doi: 10.1021/tx100098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hang B, Chenna A, Guliaev AB, Singer B. Mutat Res. 2003;531:191. doi: 10.1016/j.mrfmmm.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 102.Bransfield LA, Rennie A, Visvanathan K, Odwin SA, Kensler TW, Yager JD, Friesen MD, Groopman JD. Chem Res Toxicol. 2008;21:1622. doi: 10.1021/tx800145w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Egner PA, Groopman JD, Wang JS, Kensler TW, Friesen MD. Chem Res Toxicol. 2006;19:1191. doi: 10.1021/tx060108d. [DOI] [PubMed] [Google Scholar]
- 104.Dudley E, Lemiere F, Van Dongen W, Esmans E, El Sharkawi AM, Games DE, Brenton AG, Newton RP. Nucleosides Nucleotides Nucleic Acids. 2003;22:987. doi: 10.1081/NCN-120022719. [DOI] [PubMed] [Google Scholar]
- 105.Chao MR, Wang CJ, Chang LW, Hu CW. Carcinogenesis. 2006;27:146. doi: 10.1093/carcin/bgi177. [DOI] [PubMed] [Google Scholar]
- 106.Chen HJ, Chang CM. Chem Res Toxicol. 2004;17:963. doi: 10.1021/tx0341963. [DOI] [PubMed] [Google Scholar]
- 107.Chen HJ, Chiu WL. Chem Res Toxicol. 2005;18:1593. doi: 10.1021/tx050145p. [DOI] [PubMed] [Google Scholar]
- 108.Goggin M, Loeber R, Park S, Walker V, Wickliffe J, Tretyakova N. Chem Res Toxicol. 2007;20:839. doi: 10.1021/tx700020q. [DOI] [PubMed] [Google Scholar]
- 109.Park S, Seetharaman M, Ogdie A, Ferguson D, Tretyakova N. Nucleic Acids Res. 2003;31:1984. doi: 10.1093/nar/gkg299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goggin M, Anderson C, Park S, Swenberg J, Walker V, Tretyakova N. Chem Res Toxicol. 2008;21:1163. doi: 10.1021/tx800051y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cadet J, Douki T, Frelon S, Sauvaigo S, Pouget JP, Ravanat JL. Free Radic Biol Med. 2002;33:441. doi: 10.1016/s0891-5849(02)00820-1. [DOI] [PubMed] [Google Scholar]
- 112.Ravanat JL, Cadet J, Douki T. Curr Mol Med. 2012;12:655. doi: 10.2174/156652412800792651. [DOI] [PubMed] [Google Scholar]
- 113.Ravanat JL, Douki T, Duez P, Gremaud E, Herbert K, Hofer T, Lasserre L, Saint-Pierre C, Favier A, Cadet J. Carcinogenesis. 2002;23:1911. doi: 10.1093/carcin/23.11.1911. [DOI] [PubMed] [Google Scholar]
- 114.Boysen G, Collins LB, Liao S, Luke AM, Pachkowski BF, Watters JL, Swenberg JA. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:375. doi: 10.1016/j.jchromb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dong M, Dedon PC. Chem Res Toxicol. 2006;19:50. doi: 10.1021/tx050252j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dong M, Vongchampa V, Gingipalli L, Cloutier JF, Kow YW, O'Connor T, Dedon PC. Mutat Res. 2006;594:120. doi: 10.1016/j.mrfmmm.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 117.Pang B, Zhou X, Yu H, Dong M, Taghizadeh K, Wishnok JS, Tannenbaum SR, Dedon PC. Carcinogenesis. 2007;28:1807. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 118.Ramesh A, Archibong AE, Niaz MS. J Toxicol Environ Health-Part A. 2010;73:1611. doi: 10.1080/15287394.2010.514225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen L, Wang M, Villalta PW, Hecht SS. Chem Res Toxicol. 2007;20:1498. doi: 10.1021/tx700147f. [DOI] [PubMed] [Google Scholar]
- 120.Kotandeniya D, Murphy D, Seneviratne U, Guza R, Pegg A, Kanugula S, Tretyakova N. Chem Res Toxicol. 2011;24:1966. doi: 10.1021/tx2002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ham AJ, Engelward BP, Koc H, Sangaiah R, Meira LB, Samson LD, Swenberg JA. DNA Repair (Amst) 2004;3:257. doi: 10.1016/j.dnarep.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 122.Giese RW. J Chromatogr A. 2000;892:329. doi: 10.1016/s0021-9673(00)00364-2. [DOI] [PubMed] [Google Scholar]
- 123.Kind T, Fiehn O. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dayringer HE, McLafferty FW. Organic Mass Spectrometry. 1977;12:53. [Google Scholar]
- 125.McLafferty FW, Turecek F. Interpretation of mass spectra. University Science Books; Mill Valley, CA: 1993. [Google Scholar]
- 126.Kind T, Fiehn O. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang J, Håkansson K. Tandem Mass Spectrometry in Mass Spectrometry of Nucleosides and Nucleic Acids. CRC Press; Boca Raton, FL: 2010. Mass Spectrometry of Nucleosides and Nucleic Acids; pp. 105–126. [Google Scholar]
- 128.Josephs JL, Sanders M. Rapid Commun Mass Spectrom. 2004;18:743. doi: 10.1002/rcm.1402. [DOI] [PubMed] [Google Scholar]
- 129.Dudley E. Analysis of Urinary Modified Nucleosides by Mass Spectrometry. CRC Press; Boca Raton, FL: 2010. Mass Spectrometry of Nucleosides and Nucleic Acids; pp. 163–194. [Google Scholar]
- 130.Gregson JM, McCloskey JA. Int J Mass Spectrom. 1997;165:475. [Google Scholar]
- 131.Inagaki S, Hirashima H, Esaka Y, Higashi T, Min JZ, Touo-Oka T. Chromatographia. 2010;72:1043. [Google Scholar]
- 132.Tuytten R, Lemiere F, Van Dongen W, Esmans EL, Witters E, Herrebout W, Van DV, Dudley E, Newton RP. J Am Soc Mass Spectrom. 2005;16:1291. doi: 10.1016/j.jasms.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 133.Jensen SS, Ariza X, Nielsen P, Vilarrasa J, Kirpekar F. J Mass Spectrom. 2007;42:49. doi: 10.1002/jms.1136. [DOI] [PubMed] [Google Scholar]
- 134.Alexander AJ, Kebarle P, Fuciarelli AF, Raleigh JA. Anal Chem. 1987;59:2484. doi: 10.1021/ac00147a010. [DOI] [PubMed] [Google Scholar]
- 135.Frycak P, Huskova R, Adam T, Lemr K. J Mass Spectrom. 2002;37:1242. doi: 10.1002/jms.389. [DOI] [PubMed] [Google Scholar]
- 136.Nelson CC, McCloskey JA. J Am Chem Soc. 1992;114:3661. [Google Scholar]
- 137.Tarun M, Rusling JF. Anal Chem. 2005;77:2056. doi: 10.1021/ac048283r. [DOI] [PubMed] [Google Scholar]
- 138.Seneviratne U, Antsypovich S, Goggin M, Quirk Dorr D, Guza R, Moser A, Thompson C, York DM, Tretyakova N. Chem Res Toxicol. 2010;23:118. doi: 10.1021/tx900312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. Proc Natl Acad Sci USA. 2007;104:12129. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rojas-Cherto M, Peironcely JE, Kasper PT, van der Hooft JJ, de Vos RC, Vreeken R, Hankemeier T, Reijmers T. Anal Chem. 2012;84:5524. doi: 10.1021/ac2034216. [DOI] [PubMed] [Google Scholar]
- 141.Sheldon MT, Mistrik R, Croley TR. J Am Soc Mass Spectrom. 2009;20:370. doi: 10.1016/j.jasms.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 142.Rasche F, Scheubert K, Hufsky F, Zichner T, Kai M, Svatos A, Bocker S. Anal Chem. 2012;84:3417. doi: 10.1021/ac300304u. [DOI] [PubMed] [Google Scholar]
- 143.Ridder L, van der Hooft JJ, Verhoeven S, de Vos RCH, van Schaik R, Vervoort J. Rapid Commun Mass Spectrom. 2012;26:2461. doi: 10.1002/rcm.6364. [DOI] [PubMed] [Google Scholar]
- 144.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. J Mass Spectrom. 2010;45:703. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 145.Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, Hirai MY, Sakurai T, Kikuchi J, Saito K. Silico Biol. 2008;8:339. [PubMed] [Google Scholar]
- 146.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. Anal Chem. 2006;78:779. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 147.Wolf SM, Vouros P. Chem Res Toxicol. 1994;7:82. doi: 10.1021/tx00037a013. [DOI] [PubMed] [Google Scholar]
- 148.Li L, Chiarelli MP, Branco PS, Antunes AM, Marques MM, Goncalves LL, Beland FA. J Am Soc Mass Spectrom. 2003;14:1488. doi: 10.1016/j.jasms.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 149.Turesky RJ, Vouros P. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:155. doi: 10.1016/j.jchromb.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 150.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Toxicol Lett. 2012;213:83. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Van den Driessche B, Van Dongen W, Lemiere F, Esmans EL. Rapid Commun Mass Spectrom. 2004;18:2001. doi: 10.1002/rcm.1578. [DOI] [PubMed] [Google Scholar]
- 152.Spilsberg B, Rundberget T, Johannessen LE, Kristoffersen AB, Holst-Jensen A, Berdal KG. J Agric Food Chem. 2010;58:6370. doi: 10.1021/jf903065a. [DOI] [PubMed] [Google Scholar]
- 153.Kato K, Yamamura E, Kawanishi M, Yagi T, Matsuda T, Sugiyama A, Uno Y. Mutat Res. 2011;721:21. doi: 10.1016/j.mrgentox.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 154.Kanaly RA, Hanaoka T, Sugimura H, Toda H, Matsui S, Matsuda T. Antioxid Redox Signal. 2006;8:993. doi: 10.1089/ars.2006.8.993. [DOI] [PubMed] [Google Scholar]
- 155.Kanaly RA, Matsui S, Hanaoka T, Matsuda T. Mutat Res. 2007;625:83. doi: 10.1016/j.mrfmmm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 156.Gangl ET, Turesky RJ, Vouros P. Chem Res Toxicol. 1999;12:1019. doi: 10.1021/tx990060m. [DOI] [PubMed] [Google Scholar]
- 157.Gangl ET, Turesky RJ, Vouros P. Anal Chem. 2001;73:2397. doi: 10.1021/ac0100401. [DOI] [PubMed] [Google Scholar]
- 158.Compagnone D, Curini R, D'Ascenzo G, Del Carlo M, Montesano C, Napoletano S, Sergi M. Anal Bioanal Chem. 2011;401:1983. doi: 10.1007/s00216-011-5261-3. [DOI] [PubMed] [Google Scholar]
- 159.Chou PH, Kageyama S, Matsuda S, Kanemoto K, Sasada Y, Oka M, Shinmura K, Mori H, Kawai K, Kasai H, Sugimura H, Matsuda T. Chem Res Toxicol. 2010;23:1442. doi: 10.1021/tx100047d. [DOI] [PubMed] [Google Scholar]
- 160.Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, Turesky RJ. Anal Chem. 2009;81:809. doi: 10.1021/ac802096p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Giese RW. Chem Res Toxicol. 1997;10:255. doi: 10.1021/tx9601263. [DOI] [PubMed] [Google Scholar]
- 162.Koivisto P, Peltonen K. Anal Bioanal Chem. 2010;398:2563. doi: 10.1007/s00216-010-4217-3. [DOI] [PubMed] [Google Scholar]
- 163.Dizdaroglu M, Jaruga P, Rodriguez H. Nucleic Acids Res. 2001;29:E12. doi: 10.1093/nar/29.3.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moser A, Range K, York DM. J Phys Chem B. 2010;114:13911. doi: 10.1021/jp107450n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Iwasaki Y, Nakano Y, Mochizuki K, Nomoto M, Takahashi Y, Ito R, Saito K, Nakazawa H. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:1159. doi: 10.1016/j.jchromb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 166.Yun BH, Rosenquist TA, Sidorenko V, Iden CR, Chen CH, Pu YS, Bonala R, Johnson F, Dickman KG, Grollman AP, Turesky RJ. Chem Res Toxicol. 2012;25:1119. doi: 10.1021/tx3000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kao CY, Giese RW. Chem Res Toxicol. 2005;18:70. doi: 10.1021/tx049854d. [DOI] [PubMed] [Google Scholar]
- 168.Saha M, Abushamaa A, Giese RW. J Chromatogr A. 1995;712:345. doi: 10.1016/0021-9673(95)00545-x. [DOI] [PubMed] [Google Scholar]
- 169.Yong LC, Schulte PA, Kao CY, Giese RW, Boeniger MF, Strauss GH, Petersen MR, Wiencke JK. Am J Ind Med. 2007;50:293. doi: 10.1002/ajim.20443. [DOI] [PubMed] [Google Scholar]
- 170.Rogstad DK, Darwanto A, Herring JL, Rogstad KN, Burdzy A, Hadley SR, Neidigh JW, Sowers LC. Chem Res Toxicol. 2007;20:1787. doi: 10.1021/tx700221x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen HJ, Lin TC, Hong CL, Chiang LC. Chem Res Toxicol. 2001;14:1612. doi: 10.1021/tx015551x. [DOI] [PubMed] [Google Scholar]
- 172.Chen HJ, Zhang L, Cox J, Cunningham JA, Chung FL. Chem Res Toxicol. 1998;11:1474. doi: 10.1021/tx980107o. [DOI] [PubMed] [Google Scholar]
- 173.Chen HJ, Chiang LC, Tseng MC, Zhang LL, Ni J, Chung FL. Chem Res Toxicol. 1999;12:1119. doi: 10.1021/tx990074s. [DOI] [PubMed] [Google Scholar]
- 174.Chen HJ, Chiu WL. Chem Res Toxicol. 2003;16:1099. doi: 10.1021/tx034057l. [DOI] [PubMed] [Google Scholar]
- 175.Morinello EJ, Ham AJ, Ranasinghe A, Sangaiah R, Swenberg JA. Chem Res Toxicol. 2001;14:327. doi: 10.1021/tx0002076. [DOI] [PubMed] [Google Scholar]
- 176.United States Surgeon General. Reducing the health consequences of smoking: 25 years of progress. U.S. Gov. Print Off; Washington, DC: 1989. [Google Scholar]
- 177.Wilm MW, Mann M. Int J Mass Spectrom Ion Proc. 1994;136:167. [Google Scholar]
- 178.El Faramawy A, Siu KW, Thomson BA. J Am Soc Mass Spectrom. 2005;16:1702. doi: 10.1016/j.jasms.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 179.Gale DC, Smith RD. Rapid Commun Mass Spectrom. 1993;7:1017. doi: 10.1002/rcm.1290090805. [DOI] [PubMed] [Google Scholar]
- 180.Beland FA, Churchwell MI, Hewer A, Phillips DH, da Costa GG, Marques MM. Biochem Biophys Res Commun. 2004;320:297. doi: 10.1016/j.bbrc.2004.05.168. [DOI] [PubMed] [Google Scholar]
- 181.Chao MR, Wang CJ, Yen CC, Yang HH, Lu YC, Chang LW, Hu CW. Biochem J. 2007;402:483. doi: 10.1042/BJ20061447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chao MR, Yen CC, Hu CW. Free Radic Biol Med. 2008;44:464. doi: 10.1016/j.freeradbiomed.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 183.Doerge DR, Churchwell MI, Fang JL, Beland FA. Chem Res Toxicol. 2000;13:1259. doi: 10.1021/tx0001575. [DOI] [PubMed] [Google Scholar]
- 184.Singh R, Teichert F, Verschoyle RD, Kaur B, Vives M, Sharma RA, Steward WP, Gescher AJ, Farmer PB. Rapid Commun Mass Spectrom. 2009;23:151. doi: 10.1002/rcm.3866. [DOI] [PubMed] [Google Scholar]
- 185.Wickremsinhe ER, Ackermann BL, Chaudhary AK. Rapid Commun Mass Spectrom. 2005;19:47. doi: 10.1002/rcm.1747. [DOI] [PubMed] [Google Scholar]
- 186.Liu J, Chen CF, Tsao CW, Chang CC, Chu CC, DeVoe DL. Anal Chem. 2009;81:2545. doi: 10.1021/ac802359e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Glick J, Zarbl H, Vouros P. Abstract presented at the 234th ACS National Meeting; Boston, MA. August 19-23, 2007; 2007. [Google Scholar]
- 188.Randall K, Argoti D, Paonessa J, Ding Y, Oaks Z, Zhang Y, Vouros P. J Chromatogr A. 2010;1217:4135. doi: 10.1016/j.chroma.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gu D, Turesky RJ, Tao Y, Langouet SA, Nauwelaers GC, Yuan JM, Yee D, Yu MC. Carcinogenesis. 2012;33:124. doi: 10.1093/carcin/bgr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jelakovic B, Karanovic S, Vukovic-Lela I, Miller F, Edwards KL, Nikolic J, Tomic K, Slade N, Brdar B, Turesky RJ, Stipancic Z, Dittrich D, Grollman AP, Dickman KG. Kidney Int. 2012;81:559. doi: 10.1038/ki.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen CH, Dickman KG, Moriya M, Zavadil J, Sidorenko V, Edwards KG, Gnatenko D, Wu L, Turesky RJ, Pu YS, Grollman AP. Proc Natl Acad Sci USA. 2012;109:8241. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Delaney JC, Essigmann JM. Chem Biol. 1999;6:743. doi: 10.1016/s1074-5521(00)80021-6. [DOI] [PubMed] [Google Scholar]
- 193.Loechler EL, Benasutti M, Basu AK, Green CL, Essigmann JM. Prog Clin Biol Res. 1990;340A:51. [PubMed] [Google Scholar]
- 194.Delaney JC, Essigmann JM. Biochemistry. 2001;40:14968. doi: 10.1021/bi015578f. [DOI] [PubMed] [Google Scholar]
- 195.Denissenko MF, Pao A, Tang M, Pfeifer GP. Science. 1996;274:430. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 196.Yoon JH, Lee CS, Pfeifer GP. Carcinogenesis. 2003;24:113. doi: 10.1093/carcin/24.1.113. [DOI] [PubMed] [Google Scholar]
- 197.Margolin Y, Shafirovich V, Geacintov N, Dedon PC. Chem Res Toxicol. 2004;17:1763. [Google Scholar]
- 198.Margolin Y, Cloutier JF, Shafirovich V, Geacintov NE, Dedon PC. Nat Chem Biol. 2006;2:365. doi: 10.1038/nchembio796. [DOI] [PubMed] [Google Scholar]
- 199.Mathur P, Xu J, Dedon PC. Biochemistry. 1997;36:14868. doi: 10.1021/bi971533w. [DOI] [PubMed] [Google Scholar]
- 200.Momparler RL, Bovenzi V. J Cell Physiol. 2000;183:145. doi: 10.1002/(SICI)1097-4652(200005)183:2<145::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 201.Matter B, Wang G, Jones R, Tretyakova N. Chem Res Toxicol. 2004;17:731. doi: 10.1021/tx049974l. [DOI] [PubMed] [Google Scholar]
- 202.Denissenko MF, Chen JX, Tang MS, Pfeifer GP. Proc Natl Acad Sci USA. 1997;94:3893. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dannenberg JJ, Tomasz M. J Am Chem Soc. 2000;122:2062. [Google Scholar]
- 204.Burrows CJ, Muller JG. Chem Rev. 1998;98:1109. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 205.Grotjahn L, Frank R, Blocker H. Nucleic Acids Res. 1982;10:4671. doi: 10.1093/nar/10.15.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Grotjahn L, Blocker H, Frank R. Biol Mass Spectrom. 1985;12:514. [Google Scholar]
- 207.McNeal CJ, Narang SA, Macfarlane RD, Hsiung HM, Brousseau R. Proc Natl Acad Sci USA. 1980;77:735. doi: 10.1073/pnas.77.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.McNeal CJ, Ogilvie KK, Theriault NY, Nemer MJ. J Am Chem Soc. 1982;104:972. [Google Scholar]
- 209.Cerny RL, Gross ML, Grotjahn L. Anal Biochem. 1986;156:424. doi: 10.1016/0003-2697(86)90276-9. [DOI] [PubMed] [Google Scholar]
- 210.Cerny RL, Tomer KB, Gross ML, Grotjahn L. Anal Biochem. 1987;165:175. doi: 10.1016/0003-2697(87)90217-x. [DOI] [PubMed] [Google Scholar]
- 211.Wang Z, Wan KX, Ramanathan R, Taylor JS, Gross ML. J Am Soc Mass Spectrom. 1998;9:683. doi: 10.1016/S1044-0305(98)00178-0. [DOI] [PubMed] [Google Scholar]
- 212.Nordhoff E, Ingendoh A, Cramer R, Overberg A, Stahl B, Karas M, Hillenkamp F, Crain PF. Rapid Commun Mass Spectrom. 1992;6:771. doi: 10.1002/rcm.1290061212. [DOI] [PubMed] [Google Scholar]
- 213.Nordhoff E, Karas M, Cramer R, Hahner S, Hillenkamp F, Kirpekar F, Lezius A, Muth J, Meier C, Engels JW. J Mass Spectrom. 1995;30:99. [Google Scholar]
- 214.Stemmler EA, Buchanan MV, Hurst GB, Hettich RL. Anal Chem. 1995;67:2924. doi: 10.1021/ac00113a029. [DOI] [PubMed] [Google Scholar]
- 215.Hettich R, Buchanan M. J Am Soc Mass Spectrom. 1991;2:402. doi: 10.1016/1044-0305(91)85006-R. [DOI] [PubMed] [Google Scholar]
- 216.Juhasz P, Roskey MT, Smirnov IP, Haff LA, Vestal ML, Martin SA. Anal Chem. 1996;68:941. doi: 10.1021/ac9510503. [DOI] [PubMed] [Google Scholar]
- 217.Talbo G, Mann M. Rapid Commun Mass Spectrom. 1996;10:100. doi: 10.1002/(SICI)1097-0231(19960115)10:1<100::AID-RCM402>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 218.Spengler B. J Mass Spectrom. 1997;32:1019. [Google Scholar]
- 219.Hagan NA, Smith CA, Antoine MD, Lin JS, Feldman AB, Demirev PA. J Am Soc Mass Spectrom. 2012;23:773. doi: 10.1007/s13361-011-0333-3. [DOI] [PubMed] [Google Scholar]
- 220.McLuckey SA, Van Berkel GJ, Glish GL. J Am Soc Mass Spectrom. 1992;3:60. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 221.Jacobson KB, Arlinghaus HF, Buchanan MV, Chen CH, Glish GL, Hettich RL, McLuckey SA. Genet Anal Tech Appl. 1991;8:223. doi: 10.1016/1050-3862(91)90032-m. [DOI] [PubMed] [Google Scholar]
- 222.Little DP, Chorush RA, Speir JP, Senko MW, Kelleher NL, McLafferty FW. J Am Chem Soc. 1994;116:4893. [Google Scholar]
- 223.Ni J, Pomerantz C, Rozenski J, Zhang Y, McCloskey JA. Anal Chem. 1996;68:1989. doi: 10.1021/ac960270t. [DOI] [PubMed] [Google Scholar]
- 224.Owens DR, Bothner B, Phung Q, Harris K, Siuzdak G. Bioorg Med Chem. 1998;6:1547. doi: 10.1016/s0968-0896(98)00098-4. [DOI] [PubMed] [Google Scholar]
- 225.Barry JP, Vouros P, Van Schepdael A, Law SJ. J Mass Spectrom. 1995;30:993. [Google Scholar]
- 226.Schurch S, Tromp JM, Monn ST. Nucleosides Nucleotides Nucleic Acids. 2007;26:1629. doi: 10.1080/15257770701549053. [DOI] [PubMed] [Google Scholar]
- 227.Murray KK. J Mass Spectrom. 1996;31:1203. doi: 10.1002/(SICI)1096-9888(199611)31:11<1203::AID-JMS445>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 228.Smith M. Rapid Commun Mass Spectrom. 2011;25:511. doi: 10.1002/rcm.4886. [DOI] [PubMed] [Google Scholar]
- 229.Iannitti P, Sheil MM, Wickham G. J Am Chem Soc. 1997;119:1490. [Google Scholar]
- 230.Marzilli LA, Wang D, Kobertz WR, Essigmann JM, Vouros P. J Am Soc Mass Spectrom. 1998;9:676. doi: 10.1016/S1044-0305(98)00039-7. [DOI] [PubMed] [Google Scholar]
- 231.Glover RP, Lamb JH, Farmer PB. Rapid Commun Mass Spectrom. 1998;12:368. [Google Scholar]
- 232.Iannitti-Tito P, Weimann A, Wickham G, Sheil MM. Analyst. 2000;125:627. doi: 10.1039/a908920i. [DOI] [PubMed] [Google Scholar]
- 233.Harsch A, Sayer JM, Jerina DM, Vouros P. Chem Res Toxicol. 2000;13:1342. doi: 10.1021/tx000140m. [DOI] [PubMed] [Google Scholar]
- 234.Colgrave MP, Iannitti-Tito P, Wickham G, Sheil MM. Aust J Chem. 2003;56:401. [Google Scholar]
- 235.Wang Y, Zhang Q, Wang Y. J Am Soc Mass Spectrom. 2004;15:1565. doi: 10.1016/j.jasms.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 236.Xiong W, Glick J, Lin Y, Vouros P. Anal Chem. 2007;79:5312. doi: 10.1021/ac0701435. [DOI] [PubMed] [Google Scholar]
- 237.Chan W, Yue H, Wong RN, Cai Z. Rapid Commun Mass Spectrom. 2008;22:3735. doi: 10.1002/rcm.3791. [DOI] [PubMed] [Google Scholar]
- 238.Chowdhury G, Guengerich FP. Angew Chem Int Ed Engl. 2008;47:381. doi: 10.1002/anie.200703942. [DOI] [PubMed] [Google Scholar]
- 239.Jamin EL, Arquier D, Tulliez J, Debrauwer L. Rapid Commun Mass Spectrom. 2008;22:3100. doi: 10.1002/rcm.3707. [DOI] [PubMed] [Google Scholar]
- 240.Anichina J, Zhao Y, Hrudey SE, Schreiber A, Li XF. Anal Chem. 2011;83:8145. doi: 10.1021/ac201646z. [DOI] [PubMed] [Google Scholar]
- 241.Sharma VK, Glick J, Vouros P. J Chromatogr A. 2012;1245:65. doi: 10.1016/j.chroma.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Schrader W, Linscheid M. J Chromatogr A. 1995;717:117. doi: 10.1016/0021-9673(95)00558-9. [DOI] [PubMed] [Google Scholar]
- 243.Schrader W, Linscheid M. Arch Toxicol. 1997;71:588. doi: 10.1007/s002040050431. [DOI] [PubMed] [Google Scholar]
- 244.Wang Y, Wang Y. Anal Chem. 2003;75:6306. doi: 10.1021/ac034683n. [DOI] [PubMed] [Google Scholar]
- 245.Gao L, Zhang L, Cho BP, Chiarelli MP. J Am Soc Mass Spectrom. 2008;19:1147. doi: 10.1016/j.jasms.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rozenski J, Crain PF, McCloskey J. A Nucleic Acids Res. 1999;27:196. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rozenski J, McCloskey J. A J Am Soc Mass Spectrom. 2002;13:200. doi: 10.1016/S1044-0305(01)00354-3. [DOI] [PubMed] [Google Scholar]
- 248.Oberacher H, Wellenzohn B, Huber CG. Anal Chem. 2002;74:211. doi: 10.1021/ac015595a. [DOI] [PubMed] [Google Scholar]
- 249.Oberacher H, Mayr BM, Huber CG. J Am Soc Mass Spectrom. 2004;15:32. doi: 10.1016/j.jasms.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 250.Oberacher H, Pitterl F. Int J Mass Spectrom. 2011;304:124. [Google Scholar]
- 251.Liao Q, Shen C, Vouros P. J Mass Spectrom. 2009;44:549. doi: 10.1002/jms.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sharma VK, Glick J, Liao Q, Shen C, Vouros P. J Mass Spectrom. 2012;47:490. doi: 10.1002/jms.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tretyakova N, Matter B, Ogdie A, Wishnok JS, Tannenbaum SR. Chem Res Toxicol. 2001;14:1058. doi: 10.1021/tx010062i. [DOI] [PubMed] [Google Scholar]
- 254.Pieles U, Zurcher W, Schar M, Moser HE. Nucleic Acids Res. 1993;21:3191. doi: 10.1093/nar/21.14.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mao B, Li B, Amin S, Cosman M, Geacintov NE. Biochemistry. 1993;32:11785. doi: 10.1021/bi00095a006. [DOI] [PubMed] [Google Scholar]
- 256.Tretyakova NY, Matter B, Jones R, Shallop A. Biochemistry. 2002;41:9535. doi: 10.1021/bi025540i. [DOI] [PubMed] [Google Scholar]
- 257.Rajesh M, Wang G, Jones R, Tretyakova N. Biochemistry. 2005;44:2197. doi: 10.1021/bi0480032. [DOI] [PubMed] [Google Scholar]
- 258.Guza R, Kotandeniya D, Murphy K, Dissanayake T, Lin C, Giambasu GM, Lad RR, Wojciechowski F, Amin S, Sturla SJ, Hudson RH, York DM, Jankowiak R, Jones R, Tretyakova NY. Nucleic Acids Res. 2011;39:3988. doi: 10.1093/nar/gkq1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Cooper DN, Youssoufian H. Hum Genet. 1988;78:151. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 260.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 261.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Science. 2011;333:1300. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tabish AM, Poels K, Hoet P, Godderis L. PLoS One. 2012;7:e34674. doi: 10.1371/journal.pone.0034674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yang I, Fortin MC, Richardson JR, Buckley B. Anal Biochem. 2011;409:138. doi: 10.1016/j.ab.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hu J, Zhang W, Ma H, Cai Y, Sheng G, Fu J. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:2765. doi: 10.1016/j.jchromb.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 266.Liu Z, Wu J, Xie Z, Liu S, Fan-Havard P, Huang TH, Plass C, Marcucci G, Chan KK. Anal Biochem. 2009;391:106. doi: 10.1016/j.ab.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hu CW, Liu HH, Li YJ, Chao MR. Chem Res Toxicol. 2012;25:462. doi: 10.1021/tx2004954. [DOI] [PubMed] [Google Scholar]
- 268.Song L, James SR, Kazim L, Karpf AR. Anal Chem. 2005;77:504. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- 269.Friso S, Choi SW, Dolnikowski GG, Selhub J. Anal Chem. 2002;74:4526. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 270.Yamagata Y, Szabo P, Szuts D, Bacquet C, Aranyi T, Paldi A. Epigenetics. 2012;7:141. doi: 10.4161/epi.7.2.18906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Crain PF, McCloskey JA. Anal Biochem. 1983;132:124. doi: 10.1016/0003-2697(83)90434-7. [DOI] [PubMed] [Google Scholar]
- 272.Le T, Kim KP, Fan G, Faull KF. Anal Biochem. 2011;412:203. doi: 10.1016/j.ab.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Zambonin CG, Palmisano F. Rapid Commun Mass Spectrom. 1999;13:2160. doi: 10.1002/(SICI)1097-0231(19991115)13:21<2160::AID-RCM768>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 274.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genes Dev. 2011;25:679. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. Nature. 2011;473:343. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hobartner C. Angew Chem Int Ed Engl. 2011;50:4268. doi: 10.1002/anie.201100350. [DOI] [PubMed] [Google Scholar]
- 277.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Nat Biotechnol. 2011;29:68. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhang L, Zhang L, Zhou K, Ye X, Zhang J, Xie A, Chen L, Kang JX, Cai C. J Biomol Screen. 2012;17:877. doi: 10.1177/1087057112447946. [DOI] [PubMed] [Google Scholar]
- 280.Prakash S, Johnson RE, Prakash L. Annu Rev Biochem. 2005;74:317. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 281.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. DNA Repair Amst. 2007;6:891. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 282.Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. J Biol Chem. 2001;276:43487. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 283.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. Mol Cell. 2001;8:7. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 284.Woodgate R. Genes Dev. 1999;13:2191. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 285.Yang W. FEBS Lett. 2005;579:868. doi: 10.1016/j.febslet.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 286.Yang W, Woodgate R. Proc Natl Acad Sci USA. 2007;104:15591. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Goodman MF. Annu Rev Biochem. 2002;71:17. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 288.Choi JY, Zang H, Angel KC, Kozekov ID, Goodenough AK, Rizzo CJ, Guengerich FP. Chem Res Toxicol. 2006;19:879. doi: 10.1021/tx060051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Christov PP, Petrova KV, Shanmugam G, Kozekov ID, Kozekova A, Guengerich FP, Stone MP, Rizzo CJ. Chem Res Toxicol. 2010;23:1330. doi: 10.1021/tx100082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Maddukuri L, Eoff RL, Choi JY, Rizzo CJ, Guengerich FP, Marnett LJ. Biochemistry. 2010;49:8415. doi: 10.1021/bi1009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Stover JS, Chowdhury G, Zang H, Guengerich FP, Rizzo CJ. Chem Res Toxicol. 2006;19:1506. doi: 10.1021/tx0601455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. J Biol Chem. 2005;280:29750. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 293.Levine RL, Miller H, Grollman A, Ohashi E, Ohmori H, Masutani C, Hanaoka F, Moriya M. J Biol Chem. 2001;276:18717. doi: 10.1074/jbc.M102158200. [DOI] [PubMed] [Google Scholar]
- 294.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Biochemistry. 2000;39:4575. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 295.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. J Biol Chem. 2002;277:37604. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 296.Stafford JB, Eoff RL, Kozekova A, Rizzo CJ, Guengerich FP, Marnett LJ. Biochemistry. 2009;48:471. doi: 10.1021/bi801591a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. J Biol Chem. 2006;281:2358. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 298.Eoff RL, Irimia A, Angel KC, Egli M, Guengerich FP. J Biol Chem. 2007;282:19831. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 299.Eoff RL, Angel KC, Egli M, Guengerich FP. J Biol Chem. 2007;282:13573. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- 300.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. J Biol Chem. 2006;281:38244. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 301.Eoff RL, Irimia A, Egli M, Guengerich FP. J Biol Chem. 2007;282:1456. doi: 10.1074/jbc.M609661200. [DOI] [PubMed] [Google Scholar]
- 302.Zhang H, Eoff RL, Kozekov ID, Rizzo CJ, Egli M, Guengerich FP. J Biol Chem. 2009;284:3563. doi: 10.1074/jbc.M807778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Zang H, Chowdhury G, Angel KC, Harris TM, Guengerich FP. Chem Res Toxicol. 2006;19:859. doi: 10.1021/tx060056s. [DOI] [PubMed] [Google Scholar]
- 304.Christov PP, Angel KC, Guengerich FP, Rizzo C. J Chem Res Toxicol. 2009;22:1086. doi: 10.1021/tx900047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kotapati S, Maddukri L, Wickramaratne S, Seneviratne U, Goggin M, Pence MG, Villalta PW, Guengerich FP, Marnett L, Tretyakova N. J Biol Chem. 2012;287:38800. doi: 10.1074/jbc.M112.396788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Yuan B, You C, Andersen N, Jiang Y, Moriya M, O'Connor TR, Wang Y. J Biol Chem. 2011;286:17503. doi: 10.1074/jbc.M111.232835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ellison KS, Dogliotti E, Connors TD, Basu AK, Essigmann JM. Proc Natl Acad Sci USA. 1989;86:8620. doi: 10.1073/pnas.86.22.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Tretyakova NY, Wishnok JS, Tannenbaum SR. Chem Res Toxicol. 2000;13:658. doi: 10.1021/tx000083x. [DOI] [PubMed] [Google Scholar]
- 309.Guza R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Chem Res Toxicol. 2006;19:531. doi: 10.1021/tx050348d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Malayappan B, Johnson L, Nie B, Panchal D, Matter B, Jacobson P, Tretyakova N. Anal Chem. 2010;82:3650. doi: 10.1021/ac902923s. [DOI] [PubMed] [Google Scholar]
- 311.Johnson LA, Malayappan B, Tretyakova N, Campbell C, Macmillan ML, Wagner JE, Jacobson PA. Pediatr Blood Cancer. 2012;58:708. doi: 10.1002/pbc.23254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Van den Driessche B, Esmans EL, Van der Linden A, Van Dongen W, Schaerlaken E, Lemiere F, Witters E, Berneman Z. Rapid Commun Mass Spectrom. 2005;19:1999. doi: 10.1002/rcm.2018. [DOI] [PubMed] [Google Scholar]
- 313.Churchwell MI, Beland FA, Doerge DR. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;844:60. doi: 10.1016/j.jchromb.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 314.Fu PP, Chou MW, Churchwell M, Wang Y, Zhao Y, Xia Q, Gamboa dC, Marques MM, Beland FA, Doerge DR. Chem Res Toxicol. 2010;23:637. doi: 10.1021/tx900402x. [DOI] [PubMed] [Google Scholar]
- 315.Soglia JR, Turesky RJ, Paehler A, Vouros P. Anal Chem. 2001;73:2819. doi: 10.1021/ac010218j. [DOI] [PubMed] [Google Scholar]
- 316.Ricicki EM, Soglia JR, Teitel C, Kane R, Kadlubar F, Vouros P. Chem Res Toxicol. 2005;18:692. doi: 10.1021/tx049692l. [DOI] [PubMed] [Google Scholar]
- 317.Doerge DR, Churchwell MI, Marques MM, Beland FA. Carcinogenesis. 1999;20:1055. doi: 10.1093/carcin/20.6.1055. [DOI] [PubMed] [Google Scholar]
- 318.Paehler A, Richoz J, Soglia J, Vouros P, Turesky RJ. Chem Res Toxicol. 2002;15:551. doi: 10.1021/tx010178e. [DOI] [PubMed] [Google Scholar]
- 319.Means JC, Olsen PD, Schoffers E. J Am Soc Mass Spectrom. 2003;14:1057. doi: 10.1016/S1044-0305(03)00409-4. [DOI] [PubMed] [Google Scholar]
- 320.Stout MD, Jeong YC, Boysen G, Li Y, Sangaiah R, Ball LM, Gold A, Swenberg JA. Chem Res Toxicol. 2006;19:563. doi: 10.1021/tx050346t. [DOI] [PubMed] [Google Scholar]
- 321.Matsuda T, Yabushita H, Kanaly RA, Shibutani S, Yokoyama A. Chem Res Toxicol. 2006;19:1374. doi: 10.1021/tx060113h. [DOI] [PubMed] [Google Scholar]
- 322.Loureiro AP, Marques SA, Garcia CC, Di MP, Medeiros MH. Chem Res Toxicol. 2002;15:1302. doi: 10.1021/tx025554p. [DOI] [PubMed] [Google Scholar]
- 323.Nakao LS, Fonseca E, Augusto O. Chem Res Toxicol. 2002;15:1248. doi: 10.1021/tx0255166. [DOI] [PubMed] [Google Scholar]
- 324.Monien BH, Muller C, Engst W, Frank H, Seidel A, Glatt H. Chem Res Toxicol. 2008;21:2017. doi: 10.1021/tx800217d. [DOI] [PubMed] [Google Scholar]
- 325.Arlt VM, Stiborova M, Henderson CJ, Thiemann M, Frei E, Aimova D, Singh R, Gamboa da Costa G, Schmitz OJ, Farmer PB, Wolf CR, Phillips DH. Carcinogenesis. 2008;29:656. doi: 10.1093/carcin/bgn002. [DOI] [PubMed] [Google Scholar]
- 326.Singh R, Gaskell M, Le Pla RC, Kaur B, zim-Araghi A, Roach J, Koukouves G, Souliotis VL, Kyrtopoulos SA, Farmer PB. Chem Res Toxicol. 2006;19:868. doi: 10.1021/tx060011r. [DOI] [PubMed] [Google Scholar]
- 327.Beland FA, Churchwell MI, Von Tungeln LS, Chen S, Fu PP, Culp SJ, Schoket B, Gyorffy E, Minarovits J, Poirier MC, Bowman ED, Weston A, Doerge DR. Chem Res Toxicol. 2005;18:1306. doi: 10.1021/tx050068y. [DOI] [PubMed] [Google Scholar]
- 328.Chen YL, Wang CJ, Wu KY. Rapid Commun Mass Spectrom. 2005;19:893. doi: 10.1002/rcm.1868. [DOI] [PubMed] [Google Scholar]
- 329.Zhang SM, Chen KM, Aliaga C, Sun YW, Lin JM, Sharma AK, Amin S, El Bayoumy K. Chem Res Toxicol. 2011;24:1297. doi: 10.1021/tx200188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Powley MW, Li Y, Upton PB, Walker VE, Swenberg JA. Carcinogenesis. 2005;26:1573. doi: 10.1093/carcin/bgi119. [DOI] [PubMed] [Google Scholar]
- 331.Powley MW, Walker VE, Li Y, Upton PB, Swenberg JA. Chem Biol Interact. 2007;166:182. doi: 10.1016/j.cbi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 332.Gamboa da Costa G, Churchwell MI, Hamilton LP, Von Tungeln LS, Beland FA, Marques MM, Doerge DR. Chem Res Toxicol. 2003;16:1328. doi: 10.1021/tx034108e. [DOI] [PubMed] [Google Scholar]
- 333.Huang CC, Shih WC, Wu CF, Chen MF, Chen YL, Lin YH, Wu KY. Rapid Commun Mass Spectrom. 2008;22:706. doi: 10.1002/rcm.3414. [DOI] [PubMed] [Google Scholar]
- 334.Marsden DA, Jones DJ, Lamb JH, Tompkins EM, Farmer PB, Brown K. Chem Res Toxicol. 2007;20:290. doi: 10.1021/tx600264t. [DOI] [PubMed] [Google Scholar]
- 335.Tompkins EM, Jones DJ, Lamb JH, Marsden DA, Farmer PB, Brown K. Rapid Commun Mass Spectrom. 2008;22:19. doi: 10.1002/rcm.3328. [DOI] [PubMed] [Google Scholar]
- 336.Pathak S, Singh R, Verschoyle RD, Greaves P, Farmer PB, Steward WP, Mellon JK, Gescher AJ, Sharma RA. Cancer Lett. 2008;261:74. doi: 10.1016/j.canlet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 337.Evans MD, Singh R, Mistry V, Sandhu K, Farmer PB, Cooke MS. Free Radic Res. 2008;42:831. doi: 10.1080/10715760802506323. [DOI] [PubMed] [Google Scholar]
- 338.Li CS, Wu KY, Chang-Chien GP, Chou CC. Environ Sci Technol. 2005;39:2455. doi: 10.1021/es0487123. [DOI] [PubMed] [Google Scholar]
- 339.Synold T, Xi B, Wuenschell GE, Tamae D, Figarola JL, Rahbar S, Termini J. Chem Res Toxicol. 2008;21:2148. doi: 10.1021/tx800224y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Monien BH, Engst W, Barknowitz G, Seidel A, Glatt H. Chem Res Toxicol. 2012;25:1484. doi: 10.1021/tx300150n. [DOI] [PubMed] [Google Scholar]
- 341.Ishii Y, Inoue K, Takasu S, Jin M, Matsushita K, Kuroda K, Fukuhara K, Nishikawa A, Umemura T. Chem Res Toxicol. 2012;25:1112. doi: 10.1021/tx300084p. [DOI] [PubMed] [Google Scholar]
- 342.Vanden Bussche J, Moore SA, Pasmans F, Kuhnle GG, Vanhaecke L. J Chromatogr A. 2012;1257:25. doi: 10.1016/j.chroma.2012.07.040. [DOI] [PubMed] [Google Scholar]
- 343.Shen LC, Chiang SY, Lin MH, Chung WS, Wu KY. Toxicol Lett. 2012;213:309. doi: 10.1016/j.toxlet.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 344.Mutlu E, Jeong YC, Collins LB, Ham AJ, Upton PB, Hatch G, Winsett D, Evansky P, Swenberg JA. Chem Res Toxicol. 2012;25:391. doi: 10.1021/tx200447w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Dennehy MK, Loeppky RN. Chem Res Toxicol. 2005;18:556. doi: 10.1021/tx049802o. [DOI] [PubMed] [Google Scholar]
- 346.Thomson NM, Mijal RS, Ziegel R, Fleischer NL, Pegg AE, Tretyakova NY, Peterson LA. Chem Res Toxicol. 2004;17:1600. doi: 10.1021/tx0498298. [DOI] [PubMed] [Google Scholar]
- 347.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Chem Res Toxicol. 2006;19:674. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Stepanov I, Hecht SS. Chem Res Toxicol. 2009;22:406. doi: 10.1021/tx800398x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Williams MV, Lee SH, Pollack M, Blair IA. J Biol Chem. 2006;281:10127. doi: 10.1074/jbc.M600178200. [DOI] [PubMed] [Google Scholar]
- 350.Zhang S, Villalta PW, Wang M, Hecht SS. Chem Res Toxicol. 2007;20:565. doi: 10.1021/tx700023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Zhang S, Villalta PW, Wang M, Hecht SS. Chem Res Toxicol. 2006;19:1386. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wang M, Yu N, Chen L, Villalta PW, Hochalter JB, Hecht SS. Chem Res Toxicol. 2006;19:319. doi: 10.1021/tx0502948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Chen L, Wang M, Villalta PW, Luo X, Feuer R, Jensen J, Hatsukami DK, Hecht SS. Chem Res Toxicol. 2007;20:108. doi: 10.1021/tx060232x. [DOI] [PMC free article] [PubMed] [Google Scholar]