Abstract
Background
Maternal asthma and child’s sex are among the most significant and reproducible risk factors for the development of asthma. Although the mechanisms for these effects are unknown, they likely involve non-classical genetic mechanisms. One such mechanism could involve the transfer and persistence of maternal cells to her offspring, a common occurrence known as maternal microchimerism (MMc). MMc has been associated with many autoimmune diseases, but has not been investigated for a role in asthma or allergic disease.
Objective
We hypothesized that some of the observed risks for asthma may be due to different rates of transmission or persistence of maternal cells to children of mothers with asthma compared to children of mothers without asthma, or to sons compared to daughters. We further hypothesized that rates of MMc differ between children with and without asthma.
Methods
We tested these hypotheses in 317 subjects from three independent cohorts using a real-time quantitative PCR assay to detect a non-inherited HLA allele in the child.
Results
MMc was detected in 20.5% of subjects (range 16.8% – 27.1% in the three cohorts). We observed lower rates of asthma among MMc positive subjects compared to MMc negative subjects (odds ratio [OR] 0.38, 95% CI 0.19, 0.79; P=0.029). Neither maternal asthma nor sex of the child was a significant predictor of MMc in the child (P = 0.81 and 0.15, respectively).
Conclusions
Our results suggest for the first time that MMc may protect against the development of asthma.
Keywords: Microchimerism, maternal, asthma
INTRODUCTION
Asthma is a common disease with a multifactorial etiology that involves both genetic and environmental risk factors.1–5 Epidemiologic and birth cohort studies have revealed intriguing sex-specific effects on risk that are poorly understood.6–11 For example, maternal asthma and male sex are among the most significant and reproducible risk factors for the development of childhood onset asthma, whereas paternal asthma and female sex are more significant risk factors for asthma with onset at older ages. 6, 7, 11–13 Moreover, the risks associated with a family of maternal or paternal asthma may vary in sons compared to daughters.14 These effects are unlikely to be due to classical genetic mechanisms, and have been variously attributed to X-linked15 or mitochondrial16, 17 inheritance, imprinting,18, 19 and hormonal factors.20, 21 Yet none have provided satisfactory explanations for these sex-specific patterns, suggesting that other as yet unidentified mechanisms play a role.
Microchimerism (Mc) refers to the presence of small numbers of non-self cells in an individual. The normal exchange of cells between mother and fetus during pregnancy is the most common source of Mc. The presence of maternal cells in her offspring is referred to as maternal microchimerism (MMc), while the presence of fetal cells in the mother is referred to as fetal microchimerism (FMc). MMc is detectable in approximately 17–55% of healthy children,22, 23 and can persist in the offspring well into adulthood (at least to age 69).24 FMc is detectable in 22–31% of healthy women with at least one previous pregnancy,25–27 and has been detected up to 40 years after childbirth.25 The roles that these foreign cells play in human health and disease are controversial, with evidence supporting both beneficial and harmful effects to the host.28, 29 In addition, although the precise mechanisms through which foreign cells influence either health or disease are unknown, a study of 31 healthy women characterized peripheral blood MMc cells as T cells in 25% of women, B cells in 14% of women, monocytes/macrophages in 16% of women, and NK cells in 28% of women.30. The investigators suggested that persistent chimeric cells may be stem cell-like and able to differentiate into cell types that perform diverse immunological, and possibly other, functions.
Both fetal28, 29, 31, 32 and maternal23, 29 microchimerism have been associated with many autoimmune diseases and, more recently, with non-autoimmune diseases, such as cancer28, 33. However, to date, a role for MMc in asthma or any allergic disease has not been investigated. We hypothesized that rates of MMc differ between children of asthmatic and non-asthmatic mothers, between sons and daughters, and between children with asthma and children without asthma. To test these hypotheses, we studied 91 asthmatic and 220 non-asthmatic subjects from three cohorts. Our study suggests that MMc protects against the development of asthma in childhood, but does not support a role for MMc in the increased risks associated with maternal asthma or for the observed sex ratio skewing in asthma.
METHODS
Subjects
This study was performed in subjects from three independent cohorts, described below.
Childhood Origins of ASThma (COAST) Cohort
The COAST study is a prospective birth cohort study in Madison, Wisconsin. The cohort included 289 children with at least one parent with respiratory allergies and/or a history of physician-diagnosed asthma who were enrolled between 1998 and 2000, as previously described 36. DNA, isolated from whole blood, was available from 107 mother-child pairs with an informative non-inherited maternal allele (NIMA; for description see Detection of Microchimerism); all children were sampled between and one and seven years of age.
Asthma in the child was diagnosed at age 6 and at age 8 based on the presence of at least one of the following in the previous year: (1) physician-diagnosed wheezing, (2) use of albuterol for coughing or wheezing as prescribed by a physician, (3) use of daily asthma medication, (4) step-up plan including use of albuterol or short-term use of inhaled corticosteroids during illness, or (5) use of prednisone for asthma exacerbation. Subjects were classified as positive for atopy by skin prick testing when the mean diameter of the wheal (half the sum of the largest diameter and its perpendicular measurement) was ≥3mm in response to any of the following allergens: eastern tree mix, grass mix, weed mix, ragweed, Alternaria alternata, Cladosporium herbarum, Aspergillus fumigatus, Dermatophagoides pteronyssinus, Dermatophagoides farinae, dog epithelium, cat hair, and American/German cockroach. EDTA-anticoagulated whole blood was collected during yearly visits and DNA was isolated using Gentra Puregene DNA extraction reagents from the whole blood (QIAGEN Inc., 158389). One DNA sample, collected from each child between the ages of 1 and 7 years, was selected for the studies. The study was approved by the University of Wisconsin Human Subjects Committee and the University of Chicago Institutional Review Board.
HLA genotypes in the COAST children and their mothers were determined using reverse strip blots.37 The 107 mother-child pairs in this study included 24 pairs in which only the mother had asthma, 25 pairs in which only the child had asthma, 17 pairs in which both the mother and child had asthma, and 41 pairs in which neither mother nor child had asthma. This sample is 86% European American, 8% African American, 4% Latino, and 2% Native American.
Tucson Infant Immune Study (IIS)
The Tucson IIS is a prospective birth cohort study of 482 children who were enrolled between 1996 and 2004, without regard to asthma or allergy status in the parents.38–40 DNA, isolated from peripheral blood mononuclear cells (PBMCs), was available from 107 mother-child pairs with an informative NIMA; all children were sampled between one and eight years of age.
Asthma in the child was based on a doctor diagnosis at 5 years of age in addition to either asthma symptoms or use of medication since the age of 4 years. Asthma was physician-diagnosed in the mothers. A diagnosis of atopy in the child was based on skin prick test reactivity to 18 allergens. Wheal sizes were measured using the longest diameter plus the perpendicular diameter and the two diameters were summed. Wheal sizes ≥3mm after subtraction of the control wheal size were considered positive. Heparinized blood samples were collected during doctor’s visits between 1 and 8 years of age; DNA was isolated from ficoll-separated PBMCs using QiaAMP DNA Blood Mini Kit (QIAGEN Inc.; 51106). The Tucson IIS was approved by the University of Arizona Institutional Review Board.
The Tucson IIS children and mothers were genotyped for the 10 HLA alleles that had validated qPCR assays for microchimerism studies, as described above, using allele-specific primers.35 The 107 mother-child pairs in this study included 13 pairs in which only the mother had asthma, nine pairs in which only the child had asthma, two pairs in which both the mother and child had asthma, and 83 pairs in which neither mother nor child had asthma. This sample was 71% European American, 22% Hispanic, 2% African American, and 5% other ethnicities.
Hutterites
The Hutterites are a religious isolate who originated in Europe in the 16th century and now live in the northern plains states (U.S.) and western provinces (Canada).41, 42 Our asthma studies in approximately 1,200 South Dakota Hutterites are population-based, and include all individuals age 6 years and older who were home during our visits to their communal farms in South Dakota and were able to perform lung function and undergo methacholine challenge testing, as described.43, 44 To best match the Hutterite sample to the COAST and Tucson IIS samples and to maximize the number of children with asthma in our study, we first selected all Hutterite children who were ≤12 years of age at the time of our studies (N=66) and then selected from that sample those with both a diagnosis of asthma and an informative NIMA (N=18). We further included all of their siblings with an informative NIMA, except for married sisters who were excluded to avoid the possibility of FMc. This yielded a sample of 63 individuals from 18 families, only three of which had a mother with asthma. Therefore, to increase the number of families with an asthmatic mother, we included 12 additional families (40 children) with a mother with asthma and an informative NIMA, also excluding married daughters. The final sample was comprised of 103 offspring from 30 families (range of 1–10 children per family), and included 15 mothers and 38 children (range of 1–7 per family) with asthma. DNA for these individuals was isolated from whole blood.
Phenotyping studies were conducted in South Dakota between 1996–1997 and again between 2006–2009 44, 45; the criteria for a diagnosis of asthma were (1) the presence of at least two symptoms (wheezing, cough, or shortness of breath), (2) a >20% fall in baseline FEV1 at ≤ 25 mg/mL or a 15% improvement in baseline FEV1 following inhalation of albuterol, and (3) a doctor’s diagnosis of asthma. A diagnosis of atopy was based on a positive skin prick test for reactivity to at least one of the 14 tested allergens. A reaction was considered positive if the mean diameter of the wheal size was ≥3mm larger than the mean diameter of the negative control reaction. EDTA-anticoagulated whole blood was collected and DNA was isolated using an isopropanol precipitation-based protocol using an Autogen AGF3000 DNA Extractor (Autogen, Inc.). These studies were approved by The University of Chicago’s Institutional Review Board.
Asthma status was unavailable for six offspring in the Hutterite families; these individuals were therefore included only in analyses of variables that could influence MMc rates but not in analyses of asthma. HLA typing in the Hutterites was performed using a combination of serological and molecular methods, as previously described.46
Detection of MMc
MMc detection was based on the presence of the non-inherited maternal (HLA) allele, referred to as the NIMA, as described previously.34 We used validated assays for eight common HLA alleles (DRB1*01, DRB1*15/16, DRB1*04, DRB1*07, DRB1*08, DRB1*14, DQA1*01, DQA1*05),35 and considered the NIMA to be informative if it differed from both alleles present in the child. All NIMAs were confirmed to be present in the mother and absent in the child by allele-specific PCR in mother-child pairs prior to inclusion in studies of microchimerism.
Real-time quantitative PCR was performed using a 7900HT Fast Real Time PCR System to detect the presence and quantity of maternal cells in the child, following established protocols (see 35 for details). Briefly, all reactions were performed in a final volume of 20 µL using TaqMan Universal Master Mix (Applied Biosystems) and 300 µM of each amplification primer and 100 µM of the allele-specific probe. Each DNA sample was tested for the presence of the NIMA in six aliquots of approximately 20,000 genome equivalents (GE) each; the sensitivity of the assay is 1 cell/20,000 GE as determined by testing dilutions of 0.5, 1, 5, and 10 GEs of DNA that is homozygous for a specific allele allele, each in a background of 10,000, 15,000, 20,000, 25,000 and 30,000 GEs of DNA that is negative for that allele. Using a conversion factor of 6.6 pg of DNA per cell, 1 cell in a background of 20,000 GE could be detected. The number of cell/genome equivalents surveyed is calculated based on a standard curve using DNA from source DNA that is homozygous for the HLA allele (NIMA) being tested and amplified with control (β-globin) primers. GEs of NIMA per million host cells are determined using standard curves generated with the same control DNA amplified with the HLA-specific primer. We required that a sample test positive in at least one out of six wells to be considered positive for microchimerism. The range of microchimerism detected is expressed as the number of maternal GE within a sample containing one million host (child’s) cells. All studies were performed in one laboratory and by a single individual (E.E.T.) who was blind to affection status, sex and maternal asthma status of the samples at the time of the testing.
Statistical Analyses
All analyses were performed using mixed logistic models (SAS PROC GLMMIX), which allowed us to take into account the relatedness between Hutterite subjects. A random genetic effect for each Hutterite subject was modeled with a mean of zero and a covariance structure that is dependent upon the relationships between pedigree members (using the kinship coefficient between them). The restricted pseudo-likelihood estimation method was applied to estimate the variance components. The best linear unbiased estimators of fixed effects were obtained with the generalized least square method.
Because the extent to which a number of potential variables influence rates of MMc is unknown, we first evaluated the effects of age of child, birth order, NIMA, sample (COAST, Tucson IIS, Hutterite), and DNA source (whole blood or PBMCs) on rates of MMc in the offspring using a mixed logistic model. We then tested our hypotheses that prevalences of MMc differ between (i) children of asthmatic mothers and children of non-asthmatic mothers, ii) sons and daughters, or iii) children with asthma and children without asthma using logistic regression, and included in these analyses the significant covariate (DNA source) identified in the first analyses described above.
Power Analyses
We used 100,000 permutations to assess the power of logistic regression to detect a significant result given odds ratios (OR) of 1.5, 2.0, and 2.5, an average rate of MMc of 26.5% 22, 23, 35, 47, 48, and prevalences of 0.2 for maternal asthma, 0.15 for asthma and 0.5 for female/male sex.
RESULTS
The composition of the three study samples is described in Table 1 and Supplemental Table 1. In the sample of 317 mother-child pairs, we had power of 0.25, 0.22, and 0.33 to detect an OR of 1.5 for maternal asthma, asthma, and sex after 100,000 permutations; 0.60, 0.56, and 0.78 to detect an OR of 2.0, and 0.85, 0.81, and 0.96, to detect an OR of 2.5, respectively. Overall, we detected MMc in 65 of 317 (20.5%) subjects, with rates of 16.8% in COAST subjects (GE range: 4.1 to 925.2), 27.1% in Tucson IIS subjects (GE range: 0.2 to 312.0), and 17.5% in Hutterite subjects (GE range: 1.5 to 374.7) (Table 2 and Supplemental Table 2). These rates of MMc are within the range of those previously reported.23, 35, 49
Table 1.
Characteristics of the study samples.
| Sample Size (# Mother- Child Pairs) |
Male:Female (% male) |
Number with Asthma (%) |
Number with Maternal Asthma (%) |
Mean Age, years (SD) [Range] |
Mean Birth Order (SD) [Range] |
Samples of European descent (%) |
DNA Source |
|
|---|---|---|---|---|---|---|---|---|
| COAST | 107 | 60:47 (56.1) | 42 (39.2) | 41 (38.3) | 5.1 (1.6) [1–7] | 2.0 (1.1) [1–6] | 92 (85.9) | WB |
| Tucson IIS | 107 | 50:57 (46.7) | 11 (10.3) | 15 (14.2) | 4.2 (1.5) [1–8] | 1.6 (0.92) [1–6] | 76 (71.0) | PBMC |
| Hutterites | 103 | 55:48 (53.4) | 36 (37.1) | 48 (46.6) | 13.9 (5.8) [6–35] | 3.8 (2.3) [1–10] | 103 (100) | WB |
| Combined Sample | 317 | 165:152 (52.1) | 91 (29.2) | 104 (32.8) | 7.7 (5.6) [1–35] | 2.4 (1.8) [1–10] | 271 (85.5) | - |
WB, whole blood; PBMC, peripheral blood mononuclear cell.
Table 2.
Number of MMc positive (+) individuals and range of genome equivalents (GE) in each cohort and the combined sample.
| Sample Size | MMc+ (%) | Range of GE | |
|---|---|---|---|
| COAST | 107 | 18 (16.8) | 4.1–925.2 |
| Tucson IIS | 107 | 29 (27.1) | 0.2–312.0 |
| Hutterites | 103 | 18 (17.5) | 1.5–374.7 |
| Combined | 317 | 65 (20.8) | 0.2–925.2 |
Before testing our main hypotheses, we first examined six variables that could potentially influence rates of MMc using a multivariate logistic model (Table 3). Rates of MMc did not significantly differ with respect to birth order (P=0.42; Supplemental Figure 1A), age (P=0.95; Supplemental Figure 1B), ethnicity (European American, African American, Hispanic, other; P=0.62), or HLA type of the NIMA (P=0.30; Supplemental Table 3). Both cohort (COAST, Tucson, Hutterite) and source of DNA (whole blood, PMBCs) were each significant predictors of MMc rates (P=0.039 and 0.036, respectively), which were higher in the Tucson IIS cohort and in DNA derived from PBMCs. Because the only DNA derived from PBMCs was from the Tucson cohort, these two covariates were completed confounded. As a result, we could not determine if the higher rate of MMc in the Tucson IIS cohort was due to the source of DNA (PMBCs) or to other features of this sample that differ from the other two, and we included DNA source (as a surrogate for Tucson) as a covariate in all subsequent analyses in the 317 mother-child pairs,
Table 3.
Results of multivariate logistic regression analysis of potential covariates.
| Variable | β (SE) | P value |
|---|---|---|
| Variables with potential effects on MMc rate | ||
| Birth order | 0.077(0.096) | 0.42 |
| Age | −0.003(0.047) | 0.95 |
| Ethnicity (European American as reference) | ||
| African American | 0.274(0.748) | 0.71 |
| Hispanic | −0.275(0.537) | 0.61 |
| Other | −1.339(1.104) | 0.23 |
| DNA source (whole blood as reference)b | 0.785(0.374) | 0.036 |
| HLA type of NIMA (DRB1*07 as reference) | ||
| DRB1*01 | −0.357(0.696) | 0.61 |
| DRB1*15/16 | 0.671(0.543) | 0.22 |
| DRB1*04 | 0.22(0.528) | 0.68 |
| DQA1*05 | 0.798(0.506) | 0.12 |
| Othera | 0.617(0.593) | 0.29 |
HLA alleles that occur in less than 10% of the sample were pooled (DRB1*01, DRB1*08, DRB1*14).
NA source (PBMC) is included as a surrogate for the Tucson IIS sample, which has the same effect on MMc rates (see text for details).
We first tested the null hypothesis that maternal asthma status was not a significant predictor of MMc in her offspring. We could not reject the null hypothesis because rates of MMc did not differ significantly between offspring of mothers with and without asthma (19.2% and 21.1%, respectively; P=0.81; Table 4A). We next tested the null hypothesis that sex of the offspring was not a significant predictor of MMc. MMc rates were higher in daughters (24.3%) compared to sons (16.9%), but these differences were not significant (OR for MMc in daughters compared to sons: 1.57 (95% CI 0.91, 2.73; P=0.15; Table 4B) and we could not reject the null hypothesis.
Table 4.
Results of multivariate logistic analyses on maternal asthma and sex of child as predictors of MMc.
| Sample Size |
MMc Rates |
Odds Ratio (95% CI) |
DNA source |
Main effect |
|||
|---|---|---|---|---|---|---|---|
| β (SE) |
P value |
β (SE) |
P value |
||||
| A. Model 1: Maternal asthma (MA) as a predictor of MMc | |||||||
| MA - No | 213 | 21.1% | 1.12 (0.62, 2.03) | 0.57 (0.29) | 0.06 | 0.08 (0.32) | 0.81 |
| MA - Yes | 104 | 19.2% | |||||
| B. Model 2: Male sex as a predictor of MMc | |||||||
| Sons | 165 | 16.9% | 1.57 (0.91, 2.73) | 0.52 (0.29) | 0.07 | 0.41 (0.28) | 0.15 |
| Daughters | 152 | 24.3% | |||||
CI = confidence interval
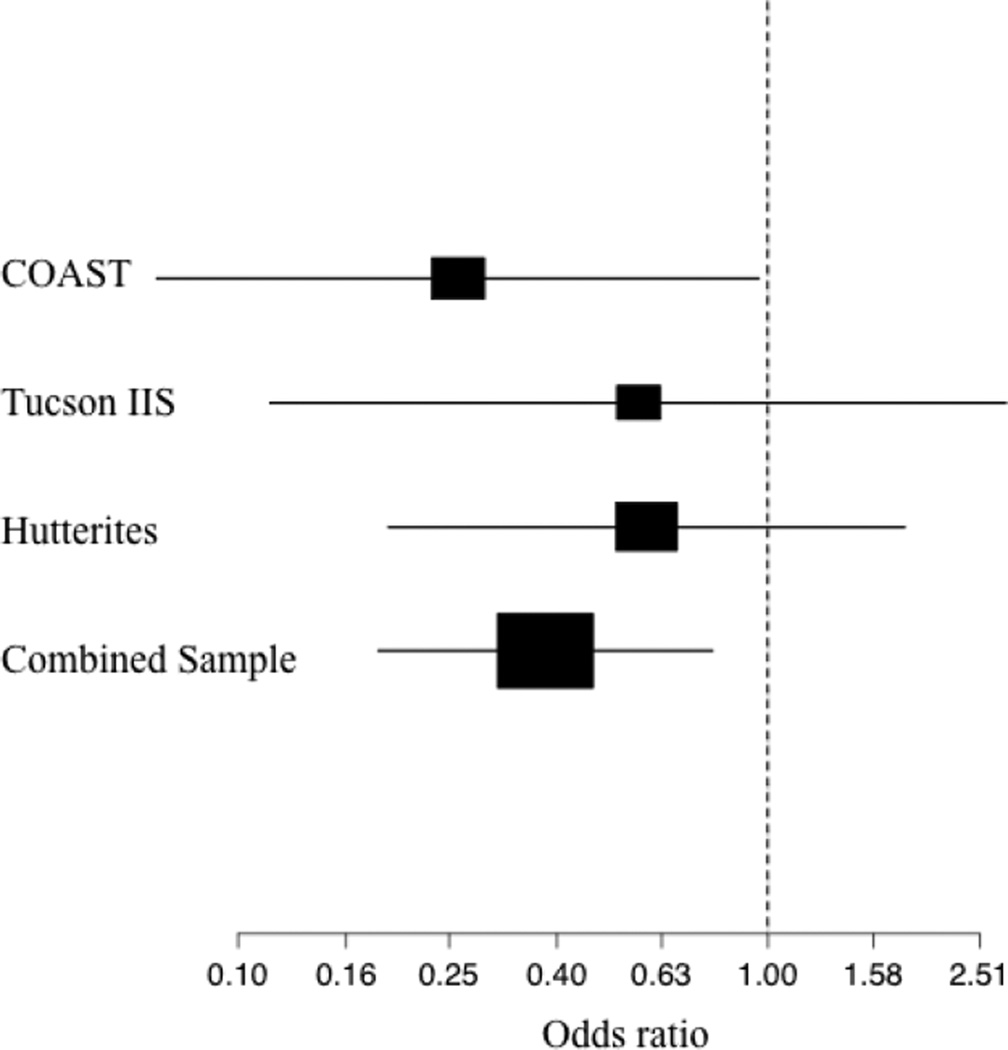
Lastly, we tested the null hypothesis that MMc was not a significant predictor of asthma, including DNA source as a covariate. The rate of asthma was significantly lower among MMc positive subjects compared to MMc negative subjects (15.4% and 32.1%, respectively; OR for asthma among MMc positive compared to MMc negative individuals: OR 0.38, 95% CI 0.19, 0.79; P=0.029; Table 5 and Figure 1). DNA source was also a significant predictor of asthma in this model (OR 0.18, 95% CI 0.09, 0.37; P=4.4×10−7), reflecting the lower prevalence of asthma in the Tucson IIS cohort in which the DNA source was PBMCs compared to the COAST and Hutterite cohorts in which the DNA source was whole blood. The different prevalence rates in the three cohorts reflect the different ascertainment schemes (see Methods), and there was no interaction between MMc and DNA source on asthma risk (P=0.23). The proportion of atopic subjects did not differ between MMc positive and MMc negative individuals, or between MMc positive and MMc negative individuals with asthma (P=0.17 and P=0.73, respectively; Supplementary Table 1). Among the offspring with detectable MMc, the mean GEs were not significantly different between MMc positive children with and without asthma (41.8 and 105.9, respectively; P=0.43) or between MMc positive sons and daughters (88.2 and 102.0, respectively; P=0.62).
Table 5.
Results of multivariate logistic analysis of MMc as a predictor of asthma.
| Model 3: MMc as a predictor of asthma | |||||||
|---|---|---|---|---|---|---|---|
| Sample Size |
Asthma Rate |
Odds Ratio (95% CI) |
DNA Source | Main Effect | |||
| β (SE) | P value | β (SE) | P value | ||||
| MMc - No | 246 | 32.1% | 0.38 (0.19, 0.79) | 0.27 (0.052) | 4.4×10–7 | 0.13 (0.06) | 0.029 |
| MMc - Yes | 65 | 15.4% | |||||
CI = confidence interval
Figure 1.
Forest plot showing the ORs and 95% confidence intervals for asthma risk among MMc positive subjects. The box sizes are proportional to the sample sizes.
DISCUSSION
The long-term survival of maternal cells in healthy individuals is a common occurrence,22, 35 but the immunologic consequences of harboring maternal cells well into adulthood are still largely unknown. Our study implicates maternal microchimerism for the first time in risk for asthma, suggesting a protective role for persisting maternal cells in her offspring.
Although both asthma and autoimmune diseases are immune-mediated conditions, their underlying immune aberrations may be complementary, or even opposite with respect to T cell polarization and subsequent disease risk. For example, a meta-analysis of studies in more than 1,000 European school children reported significantly reduced risk for the development of type 1 diabetes (T1D) by age 15 years among children with asthma (OR 0.70, 95% CI 0.54–0.91); interestingly, the protective effect was stronger with respect to asthma compared to other allergic conditions (atopy, atopic dermatitis).50 The converse association was reported in study of 181 British school children with T1D, which showed a significantly reduced risk for self-reported ever wheezing compared to control children from the same study (OR 0.36, 95% CI 0.25–0.52).51 In the latter study, more frequent or more severe wheezing attacks were associated with lower risk of T1D (trend P <0.01). A recent meta-analysis of studies in almost 4,000 children (including the two studies discussed above) reported a significantly reduced risk for asthma by age 18 among children with T1D (OR 0.82, 95% CI 0.68–0.99).52 Moreover, while variants in many of the same genes are associated with both asthma and autoimmune diseases,53 the effect is often in opposite directions so that the allele associated with risk for autoimmune disease is associated with protection from asthma (or allergic diseases), and vice versa.54–56 Those observations, combined with the results of our study, would be consistent with a model of opposite immune dysregulation in autoimmune disease and asthma, in which persistent MMc in the children predisposes to autoimmune diseases and protects from asthma and allergic diseases.
Further studies are warranted to both validate the results presented here and to elucidate the mechanism(s) by which persistent maternal cells in her offspring modulate risk for asthma and other immune-mediated diseases. Moreover, although the higher rates of MMc in daughters compared to sons (24.3% vs. 16.9%) was not significant in this study, it is a potentially intriguing observation because it could suggest a mechanism for the higher prevalence of asthma in boys during childhood10, 21 and the higher rates of autoimmune diseases in females throughout life.57 Larger studies would be needed to further evaluate this observation, although studies of MMc and disease are challenging for a number of reasons. First, the availability of samples suitable for these studies is limiting because DNA must be available from mother-child pairs and informative markers with highly specific assays are required. Although these studies could be performed using other informative genetic markers, HLA offers a highly informative and robust system for detecting Mc. However, HLA typing is not only expensive but requires specialized assays that are not available in all laboratories. Moreover, validated assays for Mc studies are currently available for a limited number of HLA alleles. As a result, only approximately half of the mother-child pairs in the cohorts in our study had an informative NIMA. Thus, although this is one of the largest studies of MMc to date, it is still relatively small. Second, the presence of microchimerism can vary in DNA derived from different sources (i.e. peripheral blood vs. tissues),35, 48, 58 and possibly by DNA isolation methods. As a result, it is essential that DNA samples from cases and controls within any one study are collected from the same blood components or tissues and processed using identical protocols. The higher rate of MMc in the Tucson IIS cohort in this study could be due to DNA source (PBMC vs whole blood), although comparative studies of Mc rates in paired DNA samples from PBMCs and whole blood from the same individual are ambiguous, suggesting that relative rates may differ depending on disease status or possibly other variables. 59
Lastly, while MMc in the periphery may have direct effects on immune development, it is likely that the presence and/or abundance of maternal cells in tissues, i.e., the lung in this case, may differ between individuals with and without asthma, as has been observed in pancreatic beta cells in T1D 23 and muscle tissue in juvenile dermatomyositis.47 However, performing MMc studies directly in lung tissues from asthmatic and non-asthmatic individuals has not been possible because maternal DNA (or HLA information) is rarely, if ever, available for adults who participate in studies of lung derived cells. Nonetheless, future efforts to study MMc in lung-derived tissues from individuals with and without asthma may provide valuable insights into the mechanisms by which persistent maternal chimeric cells in her children protect them against the development of childhood asthma, and possibly suggest novel therapeutic interventions.
Supplementary Material
Clinical Implications.
The transfer and persistence of maternal cells to her offspring during pregnancy may protect against the development of asthma, suggesting a previously unexplored mechanism that influences asthma risk.
ACKNOWLEDGEMENTS
This work was supported by P01 HL070831 (COAST), AI 42268 (Tucson IIS), R01 HL085197 (Hutterites). E.E.T was supported by K12 HL090003.
The authors thank J. Lee Nelson for invaluable comments and discussion, Dan Nicolae for comments on statistical analyses, and Kevin Ross for technical assistance.
Abbreviations Used
- COAST
Childhood Origins of ASThma
- FMc
Fetal microchimerism
- GE
Genome equivalents
- HLA
Human leukocyte antigen
- IIS
Infant Immune Study
- MMc
Maternal microchimerms
- NIMA
Non-inherited maternal allele
- OR
Odds ratio
- PBMC
Peripheral blood mononuclear cell
- WB
Whole blood
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27:107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 4.Cookson W, Moffatt M, Strachan DP. Genetic risks and childhood-onset asthma. J Allergy Clin Immunol. 2011;128:266–270. doi: 10.1016/j.jaci.2011.06.026. quiz 71–2. [DOI] [PubMed] [Google Scholar]
- 5.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leynaert B, Sunyer J, Garcia-Esteban R, Svanes C, Jarvis D, Cerveri I, et al. Gender differences in prevalence, diagnosis and incidence of allergic and non-allergic asthma: a population-based cohort. Thorax. 2012 doi: 10.1136/thoraxjnl-2011-201249. [DOI] [PubMed] [Google Scholar]
- 7.Litonjua AA, Carey VJ, Burge HA, Weiss ST, Gold DR. Parental history and the risk for childhood asthma. Does mother confer more risk than father? Am J Respir Crit Care Med. 1998;158:176–181. doi: 10.1164/ajrccm.158.1.9710014. [DOI] [PubMed] [Google Scholar]
- 8.Wright AL, Holberg CJ, Taussig LM, Martinez FD. Factors influencing the relation of infant feeding to asthma and recurrent wheeze in childhood. Thorax. 2001;56:192–197. doi: 10.1136/thorax.56.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loisel DA, Tan Z, Tisler CJ, Evans MD, Gangnon RE, Jackson DJ, et al. IFNG genotype and sex interact to influence the risk of childhood asthma. J Allergy Clin Immunol. 2011;128:524–531. doi: 10.1016/j.jaci.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almqvist C, Worm M, Leynaert B. Impact of gender on asthma in childhood and adolescence: a GA2LEN review. Allergy. 2008;63:47–57. doi: 10.1111/j.1398-9995.2007.01524.x. [DOI] [PubMed] [Google Scholar]
- 11.Postma DS. Gender differences in asthma development and progression. Gend Med. 2007;4(Suppl B):S133–S146. doi: 10.1016/s1550-8579(07)80054-4. [DOI] [PubMed] [Google Scholar]
- 12.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995;332:133–138. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 13.Wright AL, Stern DA, Kauffmann F, Martinez FD. Factors influencing gender differences in the diagnosis and treatment of asthma in childhood: the Tucson Children's Respiratory Study. Pediatr Pulmonol. 2006;41:318–325. doi: 10.1002/ppul.20373. [DOI] [PubMed] [Google Scholar]
- 14.Mandhane PJ, Greene JM, Cowan JO, Taylor DR, Sears MR. Sex differences in factors associated with childhood- and adolescent-onset wheeze. Am J Respir Crit Care Med. 2005;172:45–54. doi: 10.1164/rccm.200412-1738OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haagerup A, Bjerke T, Schiotz PO, Binderup HG, Dahl R, Kruse TA. Asthma and atopy - a total genome scan for susceptibility genes. Allergy. 2002;57:680–686. doi: 10.1034/j.1398-9995.2002.23523.x. [DOI] [PubMed] [Google Scholar]
- 16.Raby BA, Klanderman B, Murphy A, Mazza S, Camargo CA, Jr, Silverman EK, et al. A common mitochondrial haplogroup is associated with elevated total serum IgE levels. J Allergy Clin Immunol. 2007;120:351–358. doi: 10.1016/j.jaci.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 17.Zifa E, Daniil Z, Skoumi E, Stavrou M, Papadimitriou K, Terzenidou M, et al. Mitochondrial genetic background plays a role in increasing risk to asthma. Mol Biol Rep. 2011 doi: 10.1007/s11033-011-1262-8. [DOI] [PubMed] [Google Scholar]
- 18.Cookson WO, Young RP, Sandford AJ, Moffatt MF, Shirakawa T, Sharp PA, et al. Maternal inheritance of atopic IgE responsiveness on chromosome 11q. Lancet. 1992;340:381–384. doi: 10.1016/0140-6736(92)91468-n. [DOI] [PubMed] [Google Scholar]
- 19.Durham AL, Wiegman C, Adcock IM. Epigenetics of asthma. Biochim Biophys Acta. 2011;1810:1103–1109. doi: 10.1016/j.bbagen.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Townsend EA, Miller VM, Prakash YS. Sex differences and sex steroids in lung health and disease. Endocr Rev. 2012;33:1–47. doi: 10.1210/er.2010-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloney S, Smith A, Furst DE, Myerson D, Rupert K, Evans PC, et al. Microchimerism of maternal origin persists into adult life. J Clin Invest. 1999;104:41–47. doi: 10.1172/JCI6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson JL, Gillespie KM, Lambert NC, Stevens AM, Loubiere LS, Rutledge JC, et al. Maternal microchimerism in peripheral blood in type 1 diabetes and pancreatic islet beta cell microchimerism. Proc Natl Acad Sci U S A. 2007;104:1637–1642. doi: 10.1073/pnas.0606169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan WF, Atkins CJ, Naysmith D, van der Westhuizen N, Woo J, Nelson JL. Microchimerism in the rheumatoid nodules of patients with rheumatoid arthritis. Arthritis Rheum. 2012;64:380–388. doi: 10.1002/art.33358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans PC, Lambert N, Maloney S, Furst DE, Moore JM, Nelson JL. Long-term fetal microchimerism in peripheral blood mononuclear cell subsets in healthy women and women with scleroderma. Blood. 1999;93:2033–2037. [PubMed] [Google Scholar]
- 26.Gannage M, Amoura Z, Lantz O, Piette JC, Caillat-Zucman S. Feto-maternal microchimerism in connective tissue diseases. Eur J Immunol. 2002;32:3405–3413. doi: 10.1002/1521-4141(200212)32:12<3405::AID-IMMU3405>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 27.Nelson JL, Furst DE, Maloney S, Gooley T, Evans PC, Smith A, et al. Microchimerism and HLA-compatible relationships of pregnancy in scleroderma. Lancet. 1998;351:559–562. doi: 10.1016/S0140-6736(97)08357-8. [DOI] [PubMed] [Google Scholar]
- 28.Fugazzola L, Cirello V, Beck-Peccoz P. Fetal microchimerism as an explanation of disease. Nat Rev Endocrinol. 2011;7:89–97. doi: 10.1038/nrendo.2010.216. [DOI] [PubMed] [Google Scholar]
- 29.Gammill HS, Nelson JL. Naturally acquired microchimerism. Int J Dev Biol. 2010;54:531–543. doi: 10.1387/ijdb.082767hg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loubiere LS, Lambert NC, Flinn LJ, Erickson TD, Yan Z, Guthrie KA, et al. Maternal microchimerism in healthy adults in lymphocytes, monocyte/macrophages and NK cells. Lab Invest. 2006;86:1185–1192. doi: 10.1038/labinvest.3700471. [DOI] [PubMed] [Google Scholar]
- 31.Ando T, Imaizumi M, Graves PN, Unger P, Davies TF. Intrathyroidal fetal microchimerism in Graves' disease. J Clin Endocrinol Metab. 2002;87:3315–3320. doi: 10.1210/jcem.87.7.8656. [DOI] [PubMed] [Google Scholar]
- 32.Klintschar M, Schwaiger P, Mannweiler S, Regauer S, Kleiber M. Evidence of fetal microchimerism in Hashimoto's thyroiditis. J Clin Endocrinol Metab. 2001;86:2494–2498. doi: 10.1210/jcem.86.6.7540. [DOI] [PubMed] [Google Scholar]
- 33.Kallenbach LR, Johnson KL, Bianchi DW. Fetal cell microchimerism and cancer: a nexus of reproduction, immunology, and tumor biology. Cancer Res. 2011;71:8–12. doi: 10.1158/0008-5472.CAN-10-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gammill HS, Adams Waldorf KM, Aydelotte TM, Lucas J, Leisenring WM, Lambert NC, et al. Pregnancy, microchimerism, and the maternal grandmother. PLoS One. 2011;6:e24101. doi: 10.1371/journal.pone.0024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert NC, Erickson TD, Yan Z, Pang JM, Guthrie KA, Furst DE, et al. Quantification of maternal microchimerism by HLA-specific real-time polymerase chain reaction: studies of healthy women and women with scleroderma. Arthritis Rheum. 2004;50:906–914. doi: 10.1002/art.20200. [DOI] [PubMed] [Google Scholar]
- 36.Lemanske RF., Jr The childhood origins of asthma (COAST) study. Pediatr Allergy Immunol. 2002;13(Suppl 15):38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 37.Bugawan TL, Apple R, Erlich HA. A method for typing polymorphism at the HLA-A locus using PCR amplification and immobilized oligonucleotide probes. Tissue Antigens. 1994;44:137–147. doi: 10.1111/j.1399-0039.1994.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 38.Rothers J, Stern DA, Spangenberg A, Lohman IC, Halonen M, Wright AL. Influence of early day-care exposure on total IgE levels through age 3 years. J Allergy Clin Immunol. 2007;120:1201–1207. doi: 10.1016/j.jaci.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 39.Stern DA, Guerra S, Halonen M, Wright AL, Martinez FD. Low IFN-gamma production in the first year of life as a predictor of wheeze during childhood. J Allergy Clin Immunol. 2007;120:835–841. doi: 10.1016/j.jaci.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 40.Su Y, Rothers J, Stern DA, Halonen M, Wright AL. Relation of early antibiotic use to childhood asthma: confounding by indication? Clin Exp Allergy. 2010;40:1222–1229. doi: 10.1111/j.1365-2222.2010.03539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hostetler J. Hutterite Society. Baltimore: John Hopkins University Press; 1974. [Google Scholar]
- 42.Steinberg A. Genetic studies in an inbred human isolate. Baltimore: John Hopkins University Press; 1967. [Google Scholar]
- 43.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67:1154–1162. doi: 10.1016/s0002-9297(07)62946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cusanovich DA, Billstrand C, Zhou X, Chavarria C, De Leon S, Michelini K, et al. The combination of a genome-wide association study of lymphocyte count and analysis of gene expression data reveals novel asthma candidate genes. Hum Mol Genet. 2012;21:2111–2123. doi: 10.1093/hmg/dds021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ober C, Aldrich CL, Chervoneva I, Billstrand C, Rahimov F, Gray HL, et al. Variation in the HLA-G promoter region influences miscarriage rates. Am J Hum Genet. 2003;72:1425–1435. doi: 10.1086/375501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed AM, Picornell YJ, Harwood A, Kredich DW. Chimerism in children with juvenile dermatomyositis. Lancet. 2000;356:2156–2157. doi: 10.1016/S0140-6736(00)03500-5. [DOI] [PubMed] [Google Scholar]
- 48.Suskind DL, Kong D, Stevens A, Wahbeh G, Christie D, Baxter-Lowe LA, et al. Maternal microchimerism in pediatric inflammatory bowel disease. Chimerism. 2011;2:50–54. doi: 10.4161/chim.2.2.16556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cox LA, Ramos RC, Dennis TN, Jimenez SA, Smith JB, Artlett CM. Detection of microchimeric cells in the peripheral blood of nonpregnant women is enhanced by magnetic cell sorting before PCR. Clin Chem. 2003;49:309–312. doi: 10.1373/49.2.309. [DOI] [PubMed] [Google Scholar]
- 50.Decreased prevalence of atopic diseases in children with diabetes. The EURODIAB Substudy 2 Study Group. J Pediatr. 2000;137:470–474. doi: 10.1067/mpd.2000.109109. [DOI] [PubMed] [Google Scholar]
- 51.Douek IF, Leech NJ, Gillmor HA, Bingley PJ, Gale EA. Children with type-1 diabetes and their unaffected siblings have fewer symptoms of asthma. Lancet. 1999;353:1850. doi: 10.1016/S0140-6736(99)00988-5. [DOI] [PubMed] [Google Scholar]
- 52.Cardwell CR, Shields MD, Carson DJ, Patterson CC. A meta-analysis of the association between childhood type 1 diabetes and atopic disease. Diabetes Care. 2003;26:2568–2574. doi: 10.2337/diacare.26.9.2568. [DOI] [PubMed] [Google Scholar]
- 53.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 54.Munthe-Kaas MC, Carlsen KH, Helms PJ, Gerritsen J, Whyte M, Feijen M, et al. CTLA-4 polymorphisms in allergy and asthma and the TH1/TH2 paradigm. J Allergy Clin Immunol. 2004;114:280–287. doi: 10.1016/j.jaci.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 55.Li X, Ampleford EJ, Howard TD, Moore WC, Torgerson DG, Li H, et al. Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Rose-Zerilli MJ, Koppelman GH, Sandling JK, Holloway JW, Postma DS, et al. Evidence of association between interferon regulatory factor 5 gene polymorphisms and asthma. Gene. 2012;504:220–225. doi: 10.1016/j.gene.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 57.Jacobson DL, Gange SJ, Rose NR, Graham NM. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin Immunol Immunopathol. 1997;84:223–243. doi: 10.1006/clin.1997.4412. [DOI] [PubMed] [Google Scholar]
- 58.Cirello V, Perrino M, Colombo C, Muzza M, Filopanti M, Vicentini L, et al. Fetal cell microchimerism in papillary thyroid cancer: studies in peripheral blood and tissues. Int J Cancer. 2010;126:2874–2878. doi: 10.1002/ijc.24993. [DOI] [PubMed] [Google Scholar]
- 59.Rak JM, Pagni PP, Tiev K, Allanore Y, Farge D, Harle JR, et al. Male microchimerism and HLA compatibility in French women with sclerodema: a different profile in limited and diffuse subset. Rheumatology (Oxford) 2009;48:363–366. doi: 10.1093/rheumatology/ken505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.