Abstract
Quantitative measurement of specific protein phosphorylation sites is a primary interest of biologists, as site-specific phosphorylation information provides insights into cell signaling networks and cellular dynamics at a system level. Over the last decade, selective phosphopeptide enrichment methods including IMAC and metal oxides (TiO2 and ZrO2) have been developed and greatly facilitate large scale phosphoproteome analysis of various cells, tissues and living organisms, in combination with modern mass spectrometers featuring high mass accuracy and high mass resolution. Various quantification strategies have been applied to detecting relative changes in expression of proteins, peptides, and specific modifications between samples. The combination of mass spectrometry-based phosphoproteome analysis with quantification strategies provides a straightforward and unbiased method to identify and quantify site-specific phosphorylation. We describe common strategies for mass spectrometric analysis of stable isotope labeled samples, as well as two widely applied phosphopeptide enrichment methods based on IMAC(NTA-Fe3+) and metal oxide (ZrO2). Instrumental configurations for on-line LC-tandem mass spectrometric analysis and parameters of conventional bioinformatic analysis of large data sets are also considered for confident identification, localization, and reliable quantification of site-specific phosphorylation.
Introduction
Protein phosphorylation is one of the most widespread post translational modifications (PTMs) occurring on amino acid side chains of Ser, Thr, Tyr, His, Cys, Glu, Asp, Arg, and other residues in both eukaryotic and prokaryotic cells (1). The acid stable phosphomonoesters of Ser, Thr, and the relatively stable Tyr are easily amenable to proteomic analysis while the low stability of phosphohistidine and other phosphorylated residues makes their analysis problematic by conventional proteomic methods (2). Reversible phosphorylation and dephosphorylation are controlled by the dynamic interplay between protein kinases and phosphatases on their target proteins. Reversible phosphorylation, as a key mechanism of regulating protein activities, participates in diverse cellular processes including metabolism, cell communication, cell growth and development. Abnormal control of phosphorylation events due to an imbalance of kinase and phosphatase activities has been linked to a large number of disease states, including cancer (3–6), diabetes (7, 8), and Alzheimer’s disease (9–11). Phosphorylation is a central process in signal transduction to activate and propagate signals through phosphorylation cascades in response to specific stimuli such as hormones, growth factors, and drugs (1, 12, 13). In addition, phosphorylation has a broad range of effects on target proteins, including control of subcellular localization, protein structure, protein interactions, and enzymatic activity. These direct effects of phosphorylation on protein function make quantitative phosphoproteome analysis useful for identifying and characterizing changes in protein function and for mapping control pathways. The use of quantitative phosphoproteome analysis in conjunction with time course studies (1, 12, 14) and genetic interventions (1, 15), are particularly useful in deciphering signal transduction pathways and provide direct insights into many biological processes (16–26).
Modern mass spectrometers featuring high mass accuracy and resolution combined with the utility of various tandem mass spectrometry (MS/MS or MS2) techniques are capable of accurately and rapidly identifying, in a single experiment, thousands of peptides in a complex mixture derived from a whole cell digest. However, significant challenges remain in phosphopeptide analysis by mass spectrometry. In most cases, phosphorylation is a transient event (27–29), typically occurring at low stoichiometry, so detection of phosphorylated peptides in the presence of a large excess of non-phosphorylated peptides can still be a challenging task. The intrinsic negative charge of phosphomonoesters lowers phosphopeptide ionization efficiency in positive mode relative to non-phosphorylated peptides, while rendering enhanced ionization for phosphopeptides in negative ion mode relative to unphosphorylated cognates. Their low levels in complex mixtures also complicate the detection of phosphopepitdes due to ion suppression, limited sampling efficiency and detection thresholds of current mass spectrometers. These issues can be overcome in part by selective isolation methods to capture and concentrate phosphopeptides from complex mixtures before MS analysis.
Over the last decade, many phosphopeptide enrichment methods have been developed and applied to phosphorylation analysis by mass spectrometry. The most widely applied phosphopeptide enrichment methods are affinity chromatography using metal oxide supports (TiO2 or ZrO2) (30, 31), immobilized metal ions (IMAC) (32–34), and anti-phosphotyrosine immunoaffinity techniques (35–37). Metal oxides and IMAC have strong affinities toward p-Ser, p-Thr, and p-Tyr under slightly acidic conditions over other competing amino acid residues. This selectivity allows highly efficient phosphopeptide enrichment methods. Some selectivity of these materials towards highly acidic peptides (rich in Asp and Glu residues) can interfere and methods have been fine tuned to reduce background adsorption of acidic peptides. Methyl esterification of Asp and Glu residues in peptides prior to affinity enrichment do reduce background levels of acidic, non-phosphorylated peptides but generally do not warrant the additional effort (38, 39). Although each of these methods shares similar properties of ion affinity towards phosphate groups, they exhibit different isolation specificities with their own advantages and disadvantages. IMAC has been reported to have higher affinity for multiply phosphorylated peptides over singly phosphorylated peptides under typical conditions, while metal oxides appear to exhibit moderate or less preference for multiply phosphorylated peptides (31, 40). The efficiency and selectivity of affinity chromatography for phosphopeptide enrichment are greatly influenced by various factors, such as buffer concentration, pH, the ratio of sample to affinity support, and the levels of non-phosphorylated peptides and other ions present in the final sample. Several studies have shown that combining multiple enrichment methods into one analytical pipeline provides more comprehensive coverage of phosphopeptides in complex samples (12, 36, 41, 42). The affinity based enrichment methods have been modified and applied for enrichment of intact phosphorylated proteins. However in the subsequent MS analysis, the enriched phosphoproteins generate mixtures of phosphorylated and non-phosphorylated peptides due to low stoichiometries of phosphorylation at individual sites, making it difficult to detect and characterize the phosphorylated peptides. Most enrichment techniques have focused on enriching phosphorylated peptides from enzymatic digests which provides more highly enriched phosphopeptides.
Due to the low levels of Tyr-phosphorylated peptides in general(1), Tyr-phosphopeptides represent a very low fraction of phosphopeptides isolated using IMAC and metal oxide based enrichment relative to Ser/Thr-phosphopeptides. Anti-phosphotyrosyl (pY)-antibodies are frequently used to selectively enrich Tyr-phosphorylated proteins and peptides which enhances their detection (4, 12, 35–37, 43). Immunopurification of Tyr-phosphopeptides using anti-pY-antibodies has been successfully employed for quantitative studies of Tyr-directed phosphorylations, including receptor tyrosine kinase (RTK) signaling pathway analysis (35, 44–47). This approach can also be used prior to the general phospho-affinity media mentioned above to provide separate pools of Tyr-phosphorylated and Ser/Thr-phosphorylated peptides. Several anti-phosphoserine/threonine antibodies are commercially available, which recognize phosphorylated Ser/Thr residues in specific sequence motifs (48). These phospho-motif-antibodies that can be used in immune-blotting and immunoprecipitaion are recently gaining increased attention as a useful tool to identify novel kinase substrates and to investigate kinase activities but care must be taken to validate specificity(48–52). Other enrichment methods based on chemical modification (53, 54) are rarely used for complex mixtures in part because the high degree of sample handling, side reactions, yields, and variability in stoichiometry often leads to significant sample loss, false positives, and increases in sample heterogeneity.
In conjunction with phosphopeptide enrichment, peptide fractionation techniques such as SCX (strong cation exchange chromatography) (55, 56), HILIC (hydrophilic interaction chromatography) (57–59), and gel electrophoresis (60–62) have been applied to improve the efficiency of phosphopeptide enrichment in large scale phosphoproteomic analysis. SCX separates peptides based on peptide charges under acidic conditions (e.g., pH 2.7) and is a most efficient and widely used as a prefractionation method prior to phosphopeptide enrichment either by IMAC or metal oxides.
Although the ‘bottom-up’ approach, in which proteins are digested and the resulting peptides are separated by chromatography and analyzed by MS, is routinely applied due to the advantages of convenient peptide separation, ionization efficiencies, and efficient peptide fragmentation often yielding enough sequence information for protein identification, it is not optimal in global proteomics to detect combinatorial effects at multiple phosphorylation sites in a single protein. This is particularly the case when sequence coverage is low. ‘Top-down’ (intact) protein analysis can provide highly valuable information on protein variant forms and information for all PTMs present, as well as the combinatorial properties of multiple PTMs in the same protein (63–65). However fragmentation of intact protein ions is more challenging to generate sequence informative fragments and generally less efficient than fragmentation of peptide ions (66–68). As the increase of ion internal energy induces fragmentation in MS, large ions redistributing extra energy amongst many vibrational modes (degrees of freedom) requires higher internal energy to induce dissociation. Especially in low-energy collision-induced dissociation (CID) adapted in most of the modern hybrid type instruments, the amount of kinetic energy converted into internal energy by the collisions with target gases is limited to fragment large ions at observable time (66–68).
When phosphoproteome enrichment and MSMS approaches are combined with quantification methodologies, the relative abundances of up to a few thousand specific phosphorylation sites can be obtained. It is important to note that careful statistical analysis is required for quantitative phosphoproteomics. In addition to probabilities for confident peptide identification, it is necessary to establish probabilities for precise localization of phosphorylation sites and for reliable measurement of the quantitative changes in phosphorylation at those sites. In most cases investigators are interested primarily in those peptides that change their levels of phosphorylation and p-values based on frequency distribution are particularly useful in this regard. In situations where the absence of change is significant, p-values are less useful and coefficients of variation or other indications of variance are more appropriate. Manual verification of important phosphorylation sites/events identified from a proteomic experiment is highly advisable to verify the site assignment. Confirmation might be possible by simple inspection or may require de novo analysis. In rare cases, synthesis of the phosphopeptide to confirm the fragmentation pattern may be necessary. The previous identification of a site of phosphorylation in a phosphopeptide database is not sufficient precedence to confirm your site of phosphorylation, particularly if the same software packages are used for the reference dataset. High quality spectral data with sufficient sequence information to meet the stringent requirements for confident peptide identification is best achieved using high resolution, high mass accuracy instruments to minimize the impact of false positives. This is particularly important for phosphoproteome analysis where each identification is essentially a single peptide hit.
Profiling changes in protein phosphorylation in response to stimuli is particularly useful for elucidation of cell signaling pathways mediated by dynamic phosphorylation. Several methods for protein or peptide quantification are compatible with phosphoproteome analysis. Stable isotope labeling methods incorporating 2H, 13C, 15N, or 18O atoms into proteins or peptides via metabolic (69, 70), chemical (71–73), or enzymatic processes (74, 75) provide the most precise quantitative comparisons of multiple samples. The higher precision of isotope enrichment methods generally allow more sensitive detection of changes in phosphorylation levels but the isotopic impurity does limit the dynamic range to some extent. Stable isotope labeling in amino acids during cell culture (SILAC) (69) is widely applied for in vitro cell culture to acquire highly precise relative quantification of protein expression and phosphorylation levels in multiplex (routinely up to three conditions). Chemically identical isotope pairs/triplets in the downstream sampling processes can be detected and quantified simultaneously by measuring the relative intensities of isotope-labeled peptides with specific mass shifts in the mass spectrometer. Alternatively, iTRAQ (11, 76, 77), covalent chemical labeling of peptides with isobaric mass tags, and related methods have become feasible for quantitative phosphoproteomics using newer mass spectrometers with improved collision chambers and ion optics. The isobaric tags have the advantages of high multiplexing capacity and broad application to samples not amenable to SILAC.
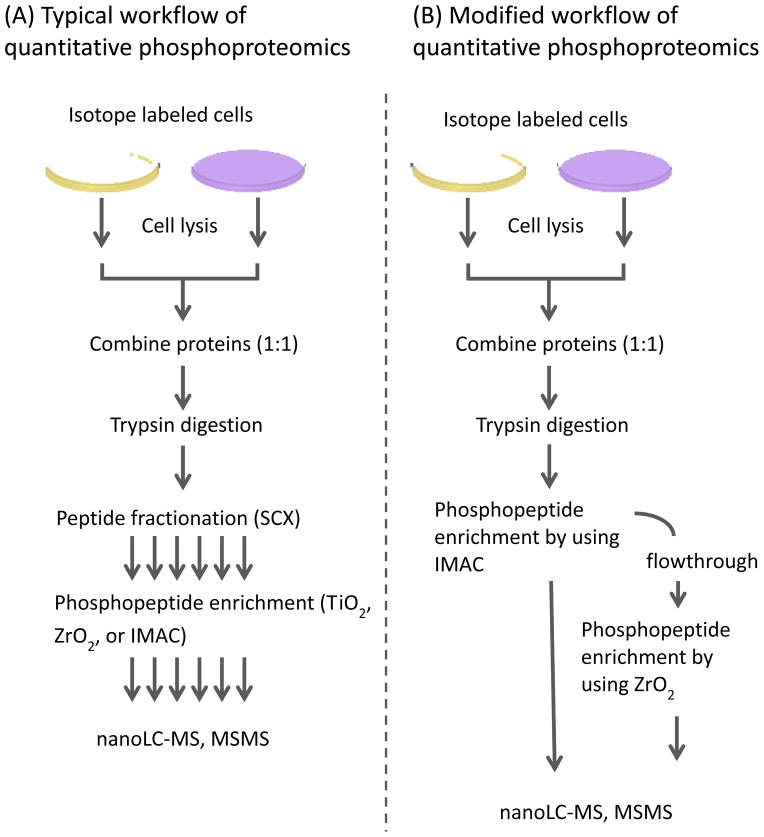
The label free quantification method provides a useful alternative approach, in which quantitative information is acquired by spectral counting (55, 78, 79) or from extracted ion intensities (80) of peptides from separate LC-MS runs for each sample. This approach provides comparisons of large numbers of samples and great flexibility for different isolation methods and sample types. However, the phosphopeptide enrichment step performed individually in phosphoproteomic analysis can introduce potentially large variance in the quantification unless considerable attention is paid to reproducibility and significant numbers of replicates are run. Phosphopeptides having low signal intensities or spectral counts may be difficult to quantify, particularly with the missing data problem of current proteomics technologies (low overlap of peptide IDs in replicate runs). Label-free approaches have been applied to large scale phosphoproteomics with the above caveats. In this article, we focus primarily on providing practical methods for large scale quantitative phosphoproteomic analysis based on the highly precise and sensitive SILAC labeling method, comprised of dual affinity chromatography-based phosphopeptide enrichment (IMAC, and ZrO2), and reversed-phase LC-MSMS with CID in an LTQ-Orbitrap. We also discuss certain aspects of data analysis pertinent to phosphoproteomics and briefly discuss alternative workflows. Here, we demonstrate dual phosphopeptide enrichment techniques combining IMAC and ZrO2 for phosphopeptide analysis of sub-milligram mouse macrophage cells; IMAC enrichment on a whole cell tryptic digest followed by ZrO2 enrichment from IMAC flowthrough. Figure 1 schematically outlines workflows for identification and quantification of a large scale phosphoproteomics study using SILAC. In the particular example shown, more unique phosphopeptides were identified from sub-milligram quantities of mouse macrophage protein extract using the second approach (single IMAC enrichment on whole cell lysate and ZrO2 enrichment of IMAC flow through). This approach reduces sampling steps, which reduces potential sample variance, yet provides similar or improved phosphoproteome coverage relative to the first example. A large number of alternative workflows for phosphoproteome analysis have been published that can be applied to a broad range of sample types and experimental aims (42, 81–84) and phosphoproteome analysis continues to evolve rapidly.
Figure 1.
(A) Schematic of a typical workflow of MS-based quantitative phosphoproteomics of metabolically labeled cells, and (B) a modified workflow employing two enrichment steps using IMAC and ZrO2 successively for a peptide mixture from the labeled whole cell lysates.
SILAC quantification is usually based on the ratios of integrated signal intensities for labeled (heavy) peptides in a chromatogram (XIC, extracted ion chromatography) relative to the signal intensities of the corresponding unlabeled (light) peptides for duplex analysis (and light to medium to heavy isotope ratios for triplex studies). For the examples shown, we used Mascot (ver. 2.2), MaxQuant (ver. 1.0.13.13) (85), and the PTM-score embedded in MaxQuant to identify peptides, determine phosphorylation sites, and quantify phosphorylations from SILAC proteome data acquired with an LTQ-Orbitrap instrument.
Methods
2.1 Establishing optimized phosphopeptide enrichment methods
SILAC labeling has been widely discussed elsewhere (19, 69, 86) and so is not discussed in detail here except to indicate that isotope incorporation proceeds effectively for most cells with medium to fast doubling times. Heavy isotope labeled Arg can be converted into heavy labeled Pro in some slow growing cells which complicates interpretation (86–90). Incorporation efficiency can be determined from a simple 1D LC-MSMS analysis of trypsinized extracts of cells grown on heavy isotope labeled Arg and Lys amino acids after an appropriate number of doublings. Major ions will be observed for the most abundant and highly ionizable peptides which are used to determine the efficiency of heavy isotope incorporation.
Reproducibility in phosphoproteomic analysis can be achieved through optimizing and standardizing enrichment methods. Especially for label-free quantification in which all samples are individually prepared and compared, each sample handling step needs to be performed carefully to reduce sample variability. Also minimizing sampling steps prior to MS analysis is critical.
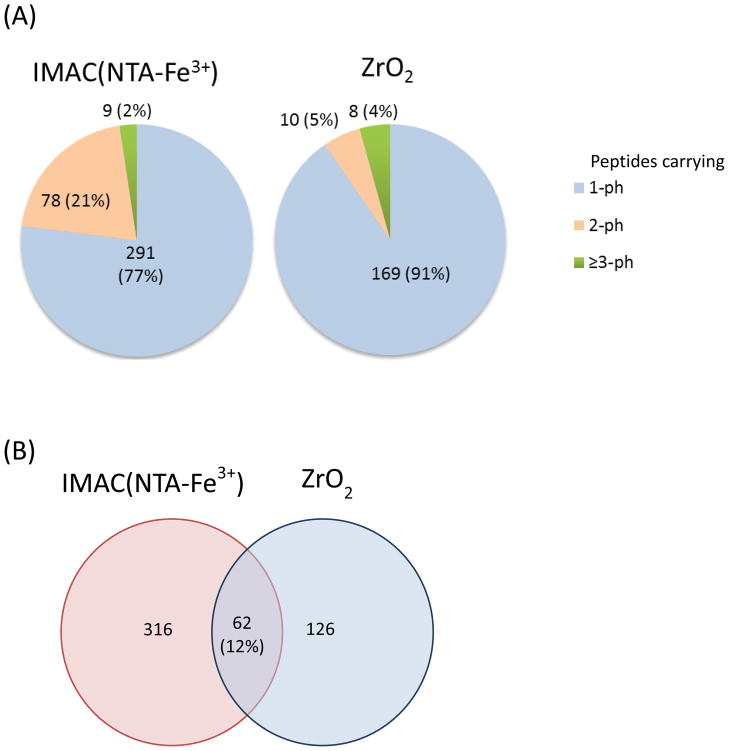
Optimization of conditions for phosphopeptide enrichment should consider the binding specificities of sorbents, ratio of sample to sorbent, binding capacities of sorbents, buffer conditions, ambient temperature, and compatibility with subsequent sample preparation and analysis steps. In large-scale phosphoproteomic analysis, metal oxides (TiO2, ZrO2) and IMAC are typically used owing to their high recovery and selectivity for phosphopeptides. As mentioned above, each affinity material exhibits different specificities; the specificity for multiply phosphorylated peptides is generally IMAC > TiO2 > ZrO2 (42, 83). Consistent with previous applications of multiple affinity enrichment media, combining two complementary enrichment steps, IMAC(NTA-Fe3+) and ZrO2, allows a more comprehensive analysis of phosphopeptides from sub-milligram quantities of mouse macrophages in the basal state. Mouse bone-marrow derived macrophages were SILAC encoded with normal and heavy Arg(6C13, 4N15) and Lys(6C13) during cell culture. Cells were lysed using a micro-sonic probe with protease inhibitors (complete, EDTA free, Roche) and phosphatase inhibitors (Pierce). Protein concentrations in cell lysates were determined by Bradford assay. Cell extracts were combined in a 1:1 ratio of protein amount (420 μg each, total 840 μg), treated with DTT and IAA to reduce and alkylate disulfide bonds, and digested with trypsin (TPCK-modified porcine trypsin) overnight. Phosphopeptides were enriched by IMAC(NTA-Fe3+) and ZrO2 successively, as in Figure 1B, by following procedures in section 2.1.1 below.. We evaluated the performance of phosphopeptide enrichment methods by comparing the ratio of the number of identified phosphopeptides versus the number of all peptides identified. Phospho-enriched fractions of IMAC and ZrO2 were analyzed by nanoLC-MSMS with an LTQ-Orbitrap XL (Thermo) interfaced with a 2D nanoLC (Eksigent). The analytes were separated over a 120 min gradient at a flow rate of 200 nL/min on a reversed-phase capillary column, packed in-house (75 μm ID × 15 cm) with 3 μm C18 resin (Sepax HP-C18). Separation method was used as follows: 0–5min, 10% B; 5–100, 10–40% B;100–110 min, 40–100% B; 110–120 min, 100% B, where solvent A (0.1% formic acid) and solvent B (90% acetonitrile with 0.1% formic acid). Peptides were identified and quantified by MaxQaunt(ver1.0.13.13) by applying p<0.01 for peptide and protein identification and modification site localization. Results of two LC-MSMS replicates of each fraction were combined for evaluation of one enrichment experiment. From the two LC-MSMS analyses of the IMAC(NTA-Fe3+) eluent, 378 unique phosphorylated peptides and only 17 non-phosphorylated peptides (acidic) were identified, providing 96% enrichment efficiency. Phosphorylated peptides identified in the IMAC fraction were composed of 77% singly phosphorylated, 21% doubly phosphorylated, and 2% triply or more highly phosphorylated peptides. In the fraction of the ZrO2 enrichment from IMAC flowthrough, 187 unique phosphopeptides and 525 unique non-phosphorylated peptides were identified, showing 26% enrichment efficiency. Of the 187 phosphopeptides eluted from the ZrO2 resin, 91% were singly phosphorylated peptides (Figure 2A.). The overlap of the identified phosphopeptides between IMAC and the consecutive ZrO2 enrichment was 12% (Figure 2B.). Overall, we identified 504 unique phosphopeptides containing 683 unique phosphorylation sites in 294 proteins in basal state sub-milligram mouse macrophage samples after four LC-MSMS runs.
Figure 2.
(A) Pie charts depicting the distribution of unique peptides carrying different numbers of phosphorylation groups identified by the NTA-Fe3+ and subsequent ZrO2 enrichment. (B) Venn diagram showing the number of unique phosphopeptides identified from fractions of IMAC enrichment and successive ZrO2 enrichment from IMAC flowthrough (by the modified workflow in Figure 1B.).
As mentioned above, the ratio of affinity resin to sample quantity is also an important factor for the selectivity of IMAC and metal oxides for phosphopeptides (31). Use of high ratios of affinity materials to peptides can lead to the increased binding of non-phosphorylated peptides competing with phosphopeptides. In practice, it is recommended to use lower ratios of resin to sample, considering the very low stoichiometry of phosphorylation and restricted quantities of biological samples. Also note that excess amounts of samples over the binding capacity of the resin can lead to sample loss and reduction in total phosphopeptide identifications. At more moderate ratios, high sample to resin ratios can skew the enrichment towards multiply phosphorylated peptides. As large amounts of proteins, typically more than 2 milligrams up to several tens of milligrams, can be used for large scale phosphoproteomic analysis, sample pre-fractionation is widely employed before the phosphopeptide enrichment step. SCX is a highly effective sample fractionation technique for peptides that is compatible with affinity based enrichment steps. However, C18 desalting of SCX fractions before phosphopeptide enrichment may lead to the loss of short or highly hydrophilic phosphopeptides. HILIC chromatography which requires no salt for elution is becoming more widely used for phosphopeptide separation because it avoids the desalting step (57, 58, 84, 91). The modified protocol in Figure 1B, for serial IMAC and ZrO2 enrichment, can reduce sample losses and variability that are exacerbated in smaller samples (sub-milligram).
2.1.1 NTA-Fe3+ enrichment for phosphopeptides, with minor modifications from reference (83)
Materials and reagents
microspin column “F” with screw cap and small 10 μm filter (Boca scientific), NTA(nitrilotriacetate) uncharged resin (Qiagen), 200 mM HOAc (acetic acid), 50 mM FeCl3 (Aldrich, anhydrous, ≥99.99%) in 100 mM HOAc, 20% formic acid, water (HPLC grade), C18 tip (Millipore, ZipTip), binding and washing solution:: 60% acetonitrile with 100 mM acetic acid; elution solution: 4% NH4OH
Activation of IMAC resin with Fe3+
-
1)
Transfer 0.5 mL of NTA resin suspension (approx. 50% resin slurry suspension) into a 2mL microtube.
-
2)
Wash the resin with 3 volumes of water in slow end-to-end motion for 10 min, and aspirate supernatant after the resin has settled.
-
3)
Wash the resin with 3 volumes of 200 mM acetic acid for 10 min.
-
4)
Activate with 3 volumes of 50 mM FeCl3 in 100 mM acetic acid for 1 hour in slow end-to-end motion.
-
5)
Wash with 3 volumes of 200 mM HOAc for 10 min, twice.
⋇ Activated bulk NTA-Fe3+) can be stored in 200 mM HOAc at 4 °C for several weeks.
Phosphopeptide enrichment with NTA-Fe3+
-
6)
Desalt sample peptide mixture with C18 Zip-tip, following the vendor’s protocol, and dry down in an eppendorf tube.
-
7)
Transfer an aliquot of activated NTA-Fe3+ resin suspension (50% resin slurry) into the tube with dried sample. (10 uL of NTA-Fe3+ resin for approximately 1mg of protein digest)
-
8)
Add binding solution (60% acetonitrile with 100 mM HOAc) to adjust pH value to 3 and a total volume of 50 μL.
-
9)
Incubate the sample with resin on a slow shaker for 1 hour at room temperature.
-
10)
Transfer sample mix with resin into a micro spin column with filter pre-washed with 100 mM HOAc.
-
11)
Spin out the binding solution in a microcentrifuge at 500 RCF for 30 sec.
-
12)
Save the flowthrough volume.
-
13)
Wash the resin with 50 μL of washing solution (60% acetonitrile with 100 mM HOAc) twice. Collect flowthrough and combine with the flowthrough from step (12).
-
14)
Elute bound phosphopeptides with elution solution (50 μL of 4% NH4OH).
-
15)
Immediately add 12 L of 20% formic acid to adjust the pH to approximately 3. Check pH by aliquoting a small volume to a pH indicator strip.
-
16)
3+ eluate and the combined flowthrough. NTA-Fe3+ eluate will be Dry down NTA-Fe reconstituted with 0.1% TFA before LC-MS analysis.
-
17)
Spin down for 5 min at 10,000 RGF, and load supernatant for LC injection.
2.1.2 ZrO2 based enrichment for phosphopeptides
Materials and reagents: ZrO2 resin (Glygen, 20–30 μm), microspin column “F” with screw cap and small 10 μm filter (Boca scientific), binding solution: 30% acetonitrile with 25 mg/mL 2,5-dihydroxybenzoic acid (DHB, Aldrich) with 200 mM formic acid; washing solution: 50% acetonitrile with 200 mM formic acid; elution solution: 4% NH4OH, 20% formic acid.
Weigh ZrO2 resin in a microcentrifuge tube. (2–5 mg of resin for 1 mg of peptide sample)
Wash ZrO2 resin with 100 μL of 100% acetonitrile and aspirate supernatant after resin has settled.
Activate ZrO2 resin with 100 μL of binding solution at least for 1 min and aspirate.
Add 50 μL of binding solution to the ZrO2 tube.
Transfer ZrO2 suspension with binding solution into the centrifuge tube with the dried flowthrough from NTA-Fe3+ enrichment above or other peptide samples, desalted with C18 and dried.
Incubate with slow shaking for 1 hour at room temperature.
Transfer the ZrO2 suspension (now with peptides bound) into a spin column.
Spin out the binding solution in a microcentrifuge at 500 RCF for 30 sec.
Wash the ZrO2 resin with 50 μL of washing solution, twice.
Elute bound peptides with elution solution (50 μL of 4% NH4OH).
Immediately add 12 μL of 20% formic acid to eluted peptides to adjust the pH to approximately 3. Check pH by spotting 0.5 μL onto pH indicator strips.
Dry down ZrO2 eluent on a SpeedVac with no heat, and reconstitute with 0.1% TFA solution before LC-MS.
-
Spin down for 5 min at 10,000 RGF, and load supernatant for LC injection.
Note 1. Peptide samples must be cleaned with C18 or graphite carbon before IMAC and ZrO2 enrichment.
Note 2. Elongated binding time at higher temperatures may lead to poor recovery of phosphopeptides due to the irreducible covalent bonds formed between phosphate group and resin (92, 93).
Note 3. When phosphopeptide enrichment efficiency (spectral counts of all phosphopeptides/total spectral counts of all identified peptides) is low, reduce the amount of resin used. A second enrichment of the IMAC eluate using a reduced amount of resin can improve recovery of phosphopeptides.
Note 4. ZrO2 resin in the protocol 2.1.2 above can be replaced with TiO2 resin (Glygen, 20–30 μm) without further modification.
2.2. Reversed-phase nanoLC separation for quantitative phosphoproteome analysis
Multidimensional chromatographic separation of biological samples combined with high resolution and high mass accuracy MSMS analysis is a core technology enabling high throughput detection of thousands of components within a single experiment. New stationary phases, different separation modes, and advanced instrumentation such as UPLC (ultra performance liquid chromatography) and chip-based chromatography have emerged in the field of proteomics, yet reversed-phase (C18) nano-scale HPLC performed with flow rates in the range of 100–500 nL/min on 50–100 μm ID columns is still prevalent as the front end separation of peptides for tandem MS instruments. Although reversed-phase LC columns retain very hydrophilic peptides such as short chain phosphopeptides poorly, reversed-phase LC is still widely and efficiently applied for phosphoproteomic analysis because of its high peak capacity, high reproducibility, and easy interface with MS instruments.
Efficient analysis of phosphopeptides in complex mixtures, requires optimal nanoLC resolution and stability. Long capillary columns (50–75 μm ID) packed with small size particles (3μm) and shallow gradients of acetonitrile at low pH are typically used for separation of extremely complex phosphopeptide mixtures. Long gradient times from 100 min up to 600 min (94) are often used for phosphoproteome analysis to increase the number of phosphopeptide identifications. The long gradient runs partially compensate for the limited duty cycles of current high resolution MS instruments. Alternatively, technical replicates of the same samples are acquired to serve the same purpose (95). As instrument duty cycles improve or more effective fragmentation schema (96, 97) are developed, good depth of phosphoproteome coverage should become more practical without excessive instrument time requirements.
2.3 Mass spectrometry for identification and quantification of isotope-labeled phosphopeptides
Localization of phosphorylation sites is a more difficult task than detection and identification of phosphopeptides. This is because a relatively complete series of fragment ions are required to unambiguously identify the specific amino acid residue(s) phosphorylated. The low abundance and low ionization efficiency of phosphopeptides contribute to low signal to noise ratios which impair their selection for MSMS and yield low fragment ion counts. During CID fragmentation of peptides, predominant neutral loss of phosphoric acid (H3PO4, 98 Da.) is often (but not always) observed from Ser-/Thr-phosphopeptides, resulting in limited fragmentation and thus little information on peptide sequence and location of phosphorylation. Neutral loss dependent scans (MS3) selectively triggered on the neutral loss fragment followed by MS/MS scan of phosphopeptides can improve the success rates and certainty of peptide sequencing, but adding the MS3 scan cycle to the MS acquisition scheme increases the duty cycle for peptide identification (98–100). Multistage activation (MSA) or pseudo-MS3 is designed to apply activation energy at the m/z (−98, −49, −32.7) of the corresponding neutral loss ions of the precursor phosphopeptides, simultaneous with CID fragmentation of the original precursor peptides. The resulting MSA spectra containing fragment ions from both MS2 and MS3 provide more sequence information without increasing the duty cycle for phosphopeptide identification. However CID-MS/MS itself showed similar performance for phosphopeptide identification to MS3 and MSA in several large-scale phosphoproteomic studies(101, 102), identifying large numbers of redundant phosphopeptides by all of these methods and small numbers of unique phosphopeptides from each approach. Other fragmentation techniques, such as electron capture dissociation (ECD)(103, 104) and electron transfer dissociation (ETD)(105, 106), have gained extensive attention as alternative fragmentation techniques that are capable of providing sequence information and exact localization for phosphopeptides.
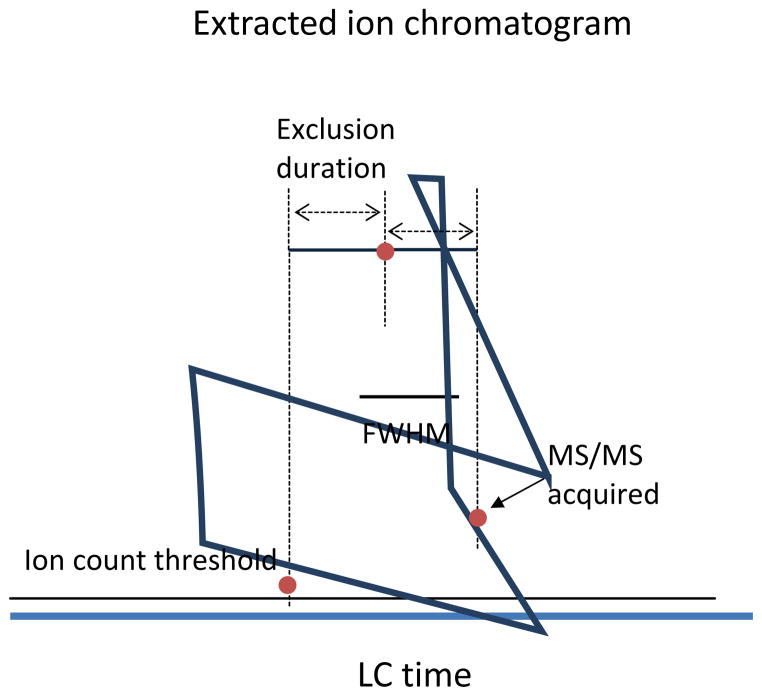
However the lower fragmentation efficiency and longer duty cycle of these techniques currently hamper their routine application to large scale phosphoproteome analysis on many instruments. Selective phosphopeptide enrichment and peptide isolation for LC-MS analysis is essential for highly reproducible and highly sensitive quantitative analysis of phosphoproteomics, but optimizing instrumental parameters for MS and MS/MS is also critical for successful proteomic analysis. Utilizing data-dependent MS/MS acquisition and dynamic exclusion of recurring ions can significantly increase the number of peptides/proteins identified. The hybrid LTQ-Orbitrap system consisting of a fast scanning ion trap mass reactor (LTQ) and a high resolution mass analyzer (Orbitrap) is one robust platform for high-throughput quantitative analysis of phosphoproteomes and instrument optimization is discussed below in the context of this instrument. Many of the same considerations are valid for other hybrid type high resolution, high mass accuracy instruments such as quadrupole time-of-flight (Q-TOF)(107–109). In a typical CID-MS/MS experiment, precursor ions are automatically selected based on ion intensities from the ions in the MS survey scans. Survey MS scans acquired with sub-ppm mass accuracy and high mass resolution of above 30,000 increases the confidence of peptide identification by filtering out large numbers of false positive matches. High mass accuracy can be achieved by internal lock mass calibration with polydimethyl cyclosiloxane ((Si(CH3)2O)6H+ at m/z 445.120025) as internal calibrant (110), and by routine external mass calibration. In addition to high mass resolution and high mass accuracy of the MS spectra, high quality MSMS spectra are also critical to successful identification. The quantity and quality of spectral data acquired on a hybrid LTQ-Orbitrap are determined by auto-gain control (AGC) target values of MS and MS/MS and ion-count threshold for data-dependent acquisition. A high ion-count threshold above noise level generally reduces the number of spectra acquired but increases the quality of data. Ion-count settings that are too high above noise levels leads to under-sampling which reduces the sensitivity of quantification based on XIC and spectral counts. Dynamic exclusion is used to limit identical precursor ions repeatedly selected for CID fragmentation within the width of a chromatographic window (111, 112). It increases the number of unique peptide fragmentation spectra acquired because instrument duty cycle time is not wasted collecting redundant spectra. Long exclusion duration times exclude abundant ions eluting in broad chromatographic bands from precursor selection subsequent to the first MS/MS acquisition of the ions over the period. Exclusion duration (113), a period of time for selected ions to be excluded from MSn acquisition, influences the number of spectra acquired as well as the quality of the spectra. Ideally, the highest quality MS/MS spectrum of an ion can be obtained at the chromatographic peak with the highest signal intensity of the target ion. To enhance the identification rates, the dynamic exclusion duration is optimized at the average chromatographic peak width at the half maximum intensity (FWHM) of the identified peptides. Ions picked for sequencing at the beginning of the chromatographic bands near the baseline can be selected for MS/MS sequencing near the chromatographic peak at the second acquisition. (Figure 3.) Instrument settings for the LTQ-Orbitrap XL (Thermo) typically used in phosphoproteomics are shown in Table 1.
Figure 3.
Schematic diagram depicting a precursor ion selected for MS/MS with dynamic exclusion applied. As an ion elutes from LC the first MS/MS scan of the ion is acquired near the ion count threshold, which likely yields poor quality spectrum. Exclusion duration set to the LC peak width allows the next MS/MS acquired near peak apex with high ion counts that provides high quality spectrum in general.
Table 1.
Experimental settings of LTQ-Orbitrap XL used in phosphoproteomic analysis.
| Parameters of LTQ- Orbitrap XL | Experimental settings* | Allowed range | Notes |
|---|---|---|---|
| Resolution | 60,000 | 7500– 100,000 |
|
| AGC target value of Orbitrap | 800,000 (Orbitrap) 30,000 (LTQ) |
|
|
| Minimum signal threshold(counts) | 15,000 | 0–10E(9) |
|
| Maximal ion accumulation time | 500 ms (Orbitrap) 250 ms (LTQ) |
|
|
| Duration of dynamic exclusion | 30–60 sec |
|
|
| Normalized collision energy (CID) | 30% | 0–100% |
|
| Activation time (CID) | 30 ms |
|
Values typically used for phosphoproteome analysis.
2.4 MS data processing and bioinformatics analysis for confident identification, site–localization and quantification of isotope-labeled phosphopeptides
Efficient phospho-specific enrichment methods integrated with advanced MS instrumentation allow generation of high quality phosphoproteomic datasets. Highly accurate precursor ion masses and sequence specific fragment ions in MS/MS spectra facilitate confident peptide and protein identification via database search engines, such as Mascot (114), Sequest (115), OMSSA (116), and X!Tandem (117, 118). The reliability of search results is statistically measured by false discovery rates (FDR) in the target-decoy search method, which provides simple and convenient filtering criteria for confident identification. A stringent filtering cutoff of FDR < 1% is typically applied for confident peptide identification.
Identification and quantification of phosphorylated peptides are two key goals in phosphoproteomics that can be accomplished readily. Identification of the actual site of phosphorylation is also very important to understand signaling mechanisms and it is necessary to establish probabilities for the precise localization of phosphorylation sites in addition to probabilities for reliable measurement of the quantitative changes in phosphorylation at those sites. However exact localization of phosphorylation sites is still challenging. Especially in tandem experiments by CID fragmentation where the abundant neutral loss of phosphorylation from precursor ions during CID often limits sequence coverage. Confident localization of phosphorylation sites on peptides requires diagnostic fragment ions bracketing the sites to be present in the MS/MS spectra. Probability based scoring algorithms efficiently extract structural information from each MS/MS spectrum in large proteomic datasets to provide high throughput analysis of phosphorylation sites. The ‘Ascore’ (119) algorithm measures the probability of correct phosphorylation site localization based on the presence and intensity of site-determining ions in the MS/MS spectra. Post-translational modification (PTM) scoring used in MaxQuant (ver 1. 0.13.13) (85, 120) for instance, extracts localization information based on an algorithm that makes use of the four most intense fragment ions per 100 m/z units in an MS2 or MS3 spectrum. The PTM-score tests for all possible combinations of peptide backbone fragments and phosphorylation sites and reports the combinations that matched to the spectra with the highest scores. While probability based scoring tools can process large proteomic datasets to extract sequence and localization information, peptides with multiple Ser, Thr, and Tyr residues may lead to ambiguous assignments of phosphorylation sites. Ambiguity in phosphorylation site-localization is further complicated by intramolecular transfer of phosphate group during CID fragmentation, which has been reported with ion-trap mass spectrometers using relatively long ion trapping times (121). Cofragmentation of coeluting isophosphomers (differing by location of the phosphorylation sites on the same peptides) is also a problem. Although they have been improved considerably in recent years, some level of miss-assignment of phosphorylation sites may still be expected at low but significant levels in all current versions of search tools. Therefore manual verification of crucial phosphopeptides is highly advisable. Confirmation may be possible by manual inspection, de novo analysis, or most stringently, by synthesis of the phosphopeptide to confirm the fragmentation pattern.
Quantification of isotopically labeled samples, such SILAC, is determined based on the ratios of integrated signal intensities for labeled (heavy) peptides in a chromatogram (XIC, extracted ion chromatogram) relative to the signal intensities of the corresponding unlabeled (light) peptides. Several software packages are available to effectively analyze SILAC data, including MaxQuant (84, 85), Mascot Distiller (122), Xpress (123), ASAPratio (124), and others (125, 126). The open source MaxQuant package was developed for SILAC data analysis, especially for MS data acquired in a hybrid tandem mass spectrometer featuring high mass resolution and high mass accuracy such as the LTQ-Orbitrap or LTQ-FTICR. For the results summarized in Figure 2, we used Mascot (ver. 2.2), MaxQuant (ver. 1.0.13.13), and PTM-score embedded in MaxQuant to identify peptides, determine phosphorylation sites, and quantify phosphorylations from SILAC proteome data acquired with an LTQ-Orbitrap instrument.
Probability values (p-values), reflecting statistical significance of measured changes in a frequency distribution, provides a good guide for investigators to decide which phosphorylation sites are differentially phosphorylated between different cell conditions. Changes below a threshold p < 0.05, or more stringently, p < 0.01, are typically considered statistically significant in large scale quantitative phosphoproteome analysis. With various bioinformatics analyses performed on searched data, extensive information collected from quantitative phosphoproteomic analysis can be integrated and expanded by combining with data extracted from curated databases to provide insights into phosphorylation-mediated signaling networks.
Concluding remarks
Protein phosphorylation is involved in a broad range of biological activities and processes. Characterizing distinct changes of site-specific phosphorylations may provide significant insights into specific cellular activities including cell signaling. In a typical approach for large-scale phosphoproteome analysis by mass spectrometry, selective phosphopeptide enrichment from non-phosphorylated peptides before MS analysis is the most effective strategy to improve detection sensitivity of low abundance phosphorylated peptides. While metal oxides (TiO2, ZrO2) and IMAC have been widely adapted for phosphorylation studies due to their highly efficient separation of phosphopeptides, careful optimization and control of separation conditions are necessary to achieve high specificity and reproducibility. Separation and mass spectrometry technologies are still evolving and multiple workflows for phosphoproteome have been established, including the use of multiple enrichment methods. While various protein and peptide labeling methods as well as label-free methods are available for relative or absolute quantification of proteins, peptides, and PTMs, stable isotope labeling methods such as SILAC and iTRAQ can provide the most sensitive and unbiased determinations of differential phosphorylation. The importance of instrument configuration, operating conditions, and bioinformatic analysis of large data sets cannot be underestimated for confident identification, localization, and quantification of phosphorylations.
Acknowledgments
This study was supported by NIH grant P41-RR18627. The authors thank Dr. Subramaniam Pennathur and Dr. Lixia Zeng for the mouse macrophage samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 2.Ciesla J, Fraczyk T, Rode W. Acta Biochim Pol. 2011;58:137–48. [PubMed] [Google Scholar]
- 3.Chong PK, Lee H, Kong JW, Loh MC, Wong CH, Lim YP. Proteomics. 2008;8:4370–82. doi: 10.1002/pmic.200800051. [DOI] [PubMed] [Google Scholar]
- 4.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 5.Harsha HC, Pandey A. Mol Oncol. 2010;4:482–95. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hers I, Vincent EE, Tavare JM. Cell Signal. 2011;23:1515–27. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Diabetes. 2006;55:2171–9. doi: 10.2337/db06-0148. [DOI] [PubMed] [Google Scholar]
- 8.Iwai LK, Benoist C, Mathis D, White FM. J Proteome Res. 2010;9:3135–45. doi: 10.1021/pr100035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudrabhatla P, Grant P, Jaffe H, Strong MJ, Pant HC. Faseb J. 2010;24:4396–407. doi: 10.1096/fj.10-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudrabhatla P, Jaffe H, Pant HC. Faseb J. 2010;25:3896–905. doi: 10.1096/fj.11-181297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe L, D’Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. Proteomics. 2007;7:3651–60. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray BN, Kweon HK, Argetsinger LS, Fingar DC, Andrews PC, Carter-Su C. Mol Endocrinol. 2012;26:1056–73. doi: 10.1210/me.2011-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan C, Olsen JV, Daub H, Mann M. Mol Cell Proteomics. 2009;8:2796–808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oyama M, Kozuka-Hata H, Tasaki S, Semba K, Hattori S, Sugano S, Inoue J, Yamamoto T. Mol Cell Proteomics. 2009;8:226–31. doi: 10.1074/mcp.M800186-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R, von Mering C, Aebersold R. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Proc Natl Acad Sci U S A. 2007;104:1488–93. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Proc Natl Acad Sci U S A. 2005;102:3948–53. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. Nature. 2005;438:679–84. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 19.Mann M. Nat Rev Mol Cell Biol. 2006;7:952–8. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 20.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Nature. 2002;415:180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 21.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Mol Cell Proteomics. 2005;4:310–27. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Everley PA, Krijgsveld J, Zetter BR, Gygi SP. Mol Cell Proteomics. 2004;3:729–35. doi: 10.1074/mcp.M400021-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Bonenfant D, Schmelzle T, Jacinto E, Crespo JL, Mini T, Hall MN, Jenoe P. Proc Natl Acad Sci U S A. 2003;100:880–5. doi: 10.1073/pnas.232735599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. J Proteome Res. 2005;4:1661–71. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- 25.Emmott E, Rodgers MA, Macdonald A, McCrory S, Ajuh P, Hiscox JA. Mol Cell Proteomics. 2010;9:1920–36. doi: 10.1074/mcp.M900345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collier TS, Randall SM, Sarkar P, Rao BM, Dean RA, Muddiman DC. Rapid Commun Mass Spectrom. 2011;25:2524–32. doi: 10.1002/rcm.5151. [DOI] [PubMed] [Google Scholar]
- 27.Stadtman ER, Chock PB. Proc Natl Acad Sci U S A. 1977;74:2761–5. doi: 10.1073/pnas.74.7.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shacter E, Chock PB, Stadtman ER. J Biol Chem. 1984;259:12252–9. [PubMed] [Google Scholar]
- 29.Karin M, Hunter T. Curr Biol. 1995;5:747–57. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 30.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Anal Chem. 2004;76:3935–43. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 31.Kweon HK, Hakansson K. Anal Chem. 2006;78:1743–49. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 32.Andersson L, Porath J. Anal Biochem. 1986;154:250–4. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 33.Posewitz MC, Tempst P. Anal Chem. 1999;71:2883–92. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 34.Hart SR, Waterfield MD, Burlingame AL, Cramer R. J Am Soc Mass Spectrom. 2002;13:1042–51. doi: 10.1016/S1044-0305(02)00432-4. [DOI] [PubMed] [Google Scholar]
- 35.Heibeck TH, Ding SJ, Opresko LK, Zhao R, Schepmoes AA, Yang F, Tolmachev AV, Monroe ME, Camp DG, 2nd, Smith RD, Wiley HS, Qian WJ. J Proteome Res. 2009;8:3852–61. doi: 10.1021/pr900044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boersema PJ, Foong LY, Ding VM, Lemeer S, van Breukelen B, Philp R, Boekhorst J, Snel B, den Hertog J, Choo AB, Heck AJ. Mol Cell Proteomics. 2010;9:84–99. doi: 10.1074/mcp.M900291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, Fang B, Liu RZ, Lin H, Kinose F, Bai Y, Oguz U, Remily-Wood ER, Li J, Altiok S, Eschrich S, Koomen JM, Haura EB. J Proteome Res. 10:305–19. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Nat Biotechnol. 2002;20:301–5. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 39.Simon ES, Young M, Chan A, Bao ZQ, Andrews PC. Anal Biochem. 2008;377:234–42. doi: 10.1016/j.ab.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunn JD, Reid GE, Bruening ML. Mass Spectrom Rev. 29:29–54. doi: 10.1002/mas.20219. [DOI] [PubMed] [Google Scholar]
- 41.Jensen SS, Larsen MR. Rapid Commun Mass Spectrom. 2007;21 doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- 42.Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. Mol Cell Proteomics. 2008;7:661–71. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang G, Neubert TA. J Proteome Res. 2011;10:5454–62. doi: 10.1021/pr200697x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Signorello MG, Giacobbe E, Leoncini G. J Cell Biochem. 2011;112:2794–802. doi: 10.1002/jcb.23194. [DOI] [PubMed] [Google Scholar]
- 45.Chen CY, Chi LM, Chi HC, Tsai MM, Tsai CY, Tseng YH, Lin YH, Chen WJ, Huang YH, Lin KH. Mol Cell Proteomics. 2011;11:M111.011270. doi: 10.1074/mcp.M111.011270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding VM, Boersema PJ, Foong LY, Preisinger C, Koh G, Natarajan S, Lee DY, Boekhorst J, Snel B, Lemeer S, Heck AJ, Choo A. PLoS One. 2011;6:e17538. doi: 10.1371/journal.pone.0017538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Science. 2011;332:1322–6. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White CD, Toker A. Curr Protoc Mol Biol. 2013;Chapter 18(Unit 18–20) doi: 10.1002/0471142727.mb1820s101. [DOI] [PubMed] [Google Scholar]
- 49.Doppler H, Storz P, Li J, Comb MJ, Toker A. J Biol Chem. 2005;280:15013–9. doi: 10.1074/jbc.C400575200. [DOI] [PubMed] [Google Scholar]
- 50.Gronborg M, Kristiansen TZ, Stensballe A, Andersen JS, Ohara O, Mann M, Jensen ON, Pandey A. Mol Cell Proteomics. 2002;1:517–27. doi: 10.1074/mcp.m200010-mcp200. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. J Biol Chem. 2002;277:39379–87. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 52.Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. J Biol Chem. 2000;275:36108–15. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 53.Zhou H, Watts JD, Aebersold R. Nat Biotechnol. 2001;19:375–8. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 54.Oda Y, Nagasu T, Chait BT. Nat Biotechnol. 2001;19:379–82. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 55.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 56.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Nat Biotechnol. 1999;17:676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 57.McNulty DE, Annan RS. Mol Cell Proteomics. 2008;7:971–80. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Boersema PJ, Mohammed S, Heck AJ. Anal Bioanal Chem. 2008;391:151–9. doi: 10.1007/s00216-008-1865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alpert AJ. Anal Chem. 2007;80:62–72. doi: 10.1021/ac070997p. [DOI] [PubMed] [Google Scholar]
- 60.Cargile BJ, Sevinsky JR, Essader AS, Stephenson JL, Jr, Bundy JL. J Biomol Tech. 2005;16:181–9. [PMC free article] [PubMed] [Google Scholar]
- 61.Cargile BJ, Talley DL, Stephenson JL., Jr Electrophoresis. 2004;25:936–45. doi: 10.1002/elps.200305722. [DOI] [PubMed] [Google Scholar]
- 62.Cargile BJ, Bundy JL, Freeman TW, Stephenson JL., Jr J Proteome Res. 2004;3:112–9. doi: 10.1021/pr0340431. [DOI] [PubMed] [Google Scholar]
- 63.Chait BT. Science. 2006;314:65–6. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 64.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Nature. 2011;480:254–8. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H, Ge Y. Cir Cardiovasc Genet. 2011;4:711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vekey K, Somogyi A, Wysocki VH. Rapid Commun Mass Spectrom. 1996;10:911–8. doi: 10.1002/(SICI)1097-0231(19960610)10:8<911::AID-RCM593>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 67.Galhena AS, Dagan S, Jones CM, Beardsley RL, Wysocki VH. Anal Chem. 2008;80:1425–36. doi: 10.1021/ac701782q. [DOI] [PubMed] [Google Scholar]
- 68.Wells JM, McLuckey SA. Methods Enzymol. 2005;402:148–85. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 69.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 70.Hernandez M, Simon O, Bergner H. Arch Tierernahr. 1981;31:651–60. doi: 10.1080/17450398109426872. [DOI] [PubMed] [Google Scholar]
- 71.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotechnol. 1999;17:994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 72.Hsu JL, Huang SY, Chow NH, Chen SH. Anal Chem. 2003;75:6843–52. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 73.Ji C, Guo N, Li L. J Proteome Res. 2005;4:2099–108. doi: 10.1021/pr050215d. [DOI] [PubMed] [Google Scholar]
- 74.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal Chem. 2001;73:2836–42. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 75.Reynolds KJ, Yao X, Fenselau C. J Proteome Res. 2002;1:27–33. doi: 10.1021/pr0100016. [DOI] [PubMed] [Google Scholar]
- 76.Ow SY, Cardona T, Taton A, Magnuson A, Lindblad P, Stensjo K, Wright PC. J Proteome Res. 2008;7:1615–28. doi: 10.1021/pr700604v. [DOI] [PubMed] [Google Scholar]
- 77.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Mol Cell Proteomics. 2010;9:1885–97. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 79.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ. Cell. 2006;127:1265–81. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 80.Bondarenko PV, Chelius D, Shaler TA. Anal Chem. 2002;74:4741–9. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 81.Imami K, Sugiyama N, Kyono Y, Tomita M, Ishihama Y. Anal Sci. 2008;24:161–6. doi: 10.2116/analsci.24.161. [DOI] [PubMed] [Google Scholar]
- 82.Pinkse MW, Mohammed S, Gouw JW, van Breukelen B, Vos HR, Heck AJ. J Proteome Res. 2008;7:687–97. doi: 10.1021/pr700605z. [DOI] [PubMed] [Google Scholar]
- 83.Ye J, Zhang X, Young C, Zhao X, Hao Q, Cheng L, Jensen ON. J Proteome Res. 2010;9:3561–73. doi: 10.1021/pr100075x. [DOI] [PubMed] [Google Scholar]
- 84.Engholm-Keller K, Hansen TA, Palmisano G, Larsen MR. J Proteome Res. 2011;10:5383–97. doi: 10.1021/pr200641x. [DOI] [PubMed] [Google Scholar]
- 85.Cox J, Mann M. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 86.Ong SE, Mann M. Nat Protoc. 2006;1:2650–60. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 87.Hwang SI, Lundgren DH, Mayya V, Rezaul K, Cowan AE, Eng JK, Han DK. Mol Cell Proteomics. 2006;5:1131–45. doi: 10.1074/mcp.M500162-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Ong SE, Kratchmarova I, Mann M. J Proteome Res. 2003;2:173–81. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 89.Schmidt F, Strozynski M, Salus SS, Nilsen H, Thiede B. Rapid Commun Mass Spectrom. 2007;21:3919–26. doi: 10.1002/rcm.3290. [DOI] [PubMed] [Google Scholar]
- 90.Van Hoof D, Pinkse MW, Oostwaard DW, Mummery CL, Heck AJ, Krijgsveld J. Nat Methods. 2007;4:677–8. doi: 10.1038/nmeth0907-677. [DOI] [PubMed] [Google Scholar]
- 91.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. Mol Cell Proteomics. 2008;7:1389–96. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Viornery C, Chevolot Y, Léonard D, Aronsson BO, Péchy P, Mathieu HJ, Descouts P, Grätzel M. Langmuir. 2002;18:2582–89. [Google Scholar]
- 93.Adden N, Gamble LJ, Castner DG, Hoffmann A, Gross G, Menzel H. Langmuir. 2006;22:8197–204. doi: 10.1021/la060754c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han G, Ye M, Liu H, Song C, Sun D, Wu Y, Jiang X, Chen R, Wang C, Wang L, Zou H. Electrophoresis. 31:1080–9. doi: 10.1002/elps.200900493. [DOI] [PubMed] [Google Scholar]
- 95.Ham BM, Yang F, Jayachandran H, Jaitly N, Monroe ME, Gritsenko MA, Livesay EA, Zhao R, Purvine SO, Orton D, Adkins JN, Camp DG, 2nd, Rossie S, Smith RD. J Proteome Res. 2008;7:2215–21. doi: 10.1021/pr700575m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Savitski MM, Sweetman G, Askenazi M, Marto JA, Lang M, Zinn N, Bantscheff M. Anal Chem. 83:8959–67. doi: 10.1021/ac201760x. [DOI] [PubMed] [Google Scholar]
- 97.Tolmachev AV, Monroe ME, Purvine SO, Moore RJ, Jaitly N, Adkins JN, Anderson GA, Smith RD. Anal Chem. 2008;80:8514–25. doi: 10.1021/ac801376g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ulintz PJ, Bodenmiller B, Andrews PC, Aebersold R, Nesvizhskii AI. Mol Cell Proteomics. 2008;7:71–87. doi: 10.1074/mcp.M700128-MCP200. [DOI] [PubMed] [Google Scholar]
- 99.Boersema PJ, Mohammed S, Heck AJ. J Mass Spectrom. 2009;44:861–78. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 100.Palumbo AM, Smith SA, Kalcic CL, Dantus M, Stemmer PM, Reid GE. Mass Spectrom Rev. 2011;30:600–25. doi: 10.1002/mas.20310. [DOI] [PubMed] [Google Scholar]
- 101.Villen J, Beausoleil SA, Gygi SP. Proteomics. 2008;8:4444–52. doi: 10.1002/pmic.200800283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ulintz PJ, Yocum AK, Bodenmiller B, Aebersold R, Andrews PC, Nesvizhskii AI. J Proteome Res. 2009;8:887–99. doi: 10.1021/pr800535h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Anal Chem. 2000;72:563–73. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 104.Cooper HJ, Hakansson K, Marshall AG. Mass Spectrom Rev. 2005;24:201–22. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 105.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci U S A. 2004;101:9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Proc Natl Acad Sci U S A. 2007;104:2193–8. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Campbell JM, Collings BA, Douglas DJ. Rapid Commun Mass Spectrom. 1998;12:1463–74. doi: 10.1002/(SICI)1097-0231(19980815)12:15<1003::AID-RCM275>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 108.Steen H, Kuster B, Mann M. J Mass Spectrom. 2001;36:782–90. doi: 10.1002/jms.174. [DOI] [PubMed] [Google Scholar]
- 109.Kristensen DB, Imamura K, Miyamoto Y, Yoshizato K. Electrophoresis. 2000;21:430–9. doi: 10.1002/(SICI)1522-2683(20000101)21:2<430::AID-ELPS430>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 110.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Mol Cell Proteomics. 2005;4:2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 111.Wang N, Li L. Anal Chem. 2008;80:4696–710. doi: 10.1021/ac800260w. [DOI] [PubMed] [Google Scholar]
- 112.Chen HS, Rejtar T, Andreev V, Moskovets E, Karger BL. Anal Chem. 2005;77:7816–25. doi: 10.1021/ac050956y. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y, Wen Z, Washburn MP, Florens L. Anal Chem. 2009;81:6317–26. doi: 10.1021/ac9004887. [DOI] [PubMed] [Google Scholar]
- 114.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 115.Eng JK, McCormack AL, Yates JRI. J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 116.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. J Proteome Res. 2004;3:958–64. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 117.Craig R, Beavis RC. Rapid Commun Mass Spectrom. 2003;17:2310–6. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 118.Pappin DJ, Hojrup P, Bleasby AJ. Curr Biol. 1993;3:327–32. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 119.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. Nat Biotechnol. 2006;24:1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 120.Olsen JV, Mann M. Proc Natl Acad Sci U S A. 2004;101:13417–22. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Palumbo AM, Reid GE. Anal Chem. 2008;80:9735–47. doi: 10.1021/ac801768s. [DOI] [PubMed] [Google Scholar]
- 122.Mueller LN, Brusniak MY, Mani DR, Aebersold R. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 123.Han DK, Eng J, Zhou H, Aebersold R. Nat Biotechnol. 2001;19:946–51. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li XJ, Zhang H, Ranish JA, Aebersold R. Anal Chem. 2003;75:6648–57. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 125.Huang X, Tolmachev AV, Shen Y, Liu M, Huang L, Zhang Z, Anderson GA, Smith RD, Chan WC, Hinrichs SH, Fu K, Ding SJ. J Proteome Res. 2011;10:1228–37. doi: 10.1021/pr1010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bakalarski CE, Elias JE, Villen J, Haas W, Gerber SA, Everley PA, Gygi SP. J Proteome Res. 2008;7:4756–65. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]