Abstract
We designed a series of mutations to separately destabilize two helical stems (designated S3 and S4) predicted by a covariation-based model of the coronavirus 3′UTR (Zust et al., 2008). Mouse hepatitis virus genomes containing three or four nucleotide mutations that destabilize either S3 or S4 were viable, whereas genomes carrying these mutations in both S3 and S4 were not viable. A genome carrying these mutations in S3 and S4 plus compensatory mutations restoring base-pairing yielded a virus with wild type phenotype. Larger mutations which completely disrupt S3 or S4 generated various phenotypes. Mutations opening up S3 were lethal. Disruptions of S4 generated both viable and lethal mutants. Genomes carrying the original mutations in S3 or S4 plus compensatory mutations restoring base pairing were viable and had robust growth phenotypes. These results support the Zust model for the coronavirus 3′UTR and suggest that the S3 stem is required for virus viability.
Keywords: Coronavirus, Cis-acting elements, 3′UTR, RNA secondary structure, MHV, Mutation, Reverse genetics
Highlights
-
•
Genetic studies of MHV 3′UTR RNA secondary structure support the Zust et al. model.
-
•
The 3′UTR S3 helical stem is required for virus viability.
-
•
The 3′UTR S4 helical stem is not required for virus viability.
Introduction
Coronaviruses are single-stranded positive-sense RNA viruses that contain a genomic RNA from 26–32 kb in length (Lai and Cavanagh, 1997). Coronaviruses have been divided into four genera based on antigenic and sequence comparisons (Siddell, 1995). Mouse hepatitis virus (MHV), one of the most extensively studied coronaviruses, is a 32 kb betacoronavirus (Britton, 2008). Cells infected with MHV contain genomic RNA and six to seven subgenomic mRNAs that make up a 3′ co-terminal nested set (Leibowitz et al., 1981, van der Born, 2008). Analysis of deletion mutants of MHV defective interfering (DI) RNAs indicated that DI replication requires cis-acting elements located within the 3′ terminal 436 nucleotides of the virus (Kim et al., 1993, Lin and Lai, 1993). This 436 nts region extends upstream of the 301 nts 3′ UTR into the nucleocapsid (N) protein gene. Subsequent studies demonstrated that in the context of the entire virus, the N protein gene can be moved to a different part of the genome with no effect on virus replication, implying that the 3′ UTR contains all the 3′ cis-acting sequences necessary for this process (de Haan et al., 2002, Goebel et al., 2004).
Multiple studies have been performed to determine the RNA secondary structures present in the 3′UTR and their functional role in viral replication (Goebel et al., 2004, Goebel et al., 2007, Hsue et al., 2000, Hsue and Masters, 1997, Johnson et al., 2005, Liu et al., 2001, Liu et al., 1997, Williams et al., 1999, Yu and Leibowitz, 1995a, Yu and Leibowitz, 1995b, Zust et al., 2008). Initial studies of the MHV 3′UTR RNA secondary structure predicted three stem-loop structures in the 3′-most 171 nts of the genome [excluding the poly(A) tail] (Yu and Leibowitz, 1995b). Subsequently, the Hsue and Masters also examined the secondary structure of the 3′UTR and predicted that the 5′ 68 nts of the 3′ UTR folded into a bulged stem-loop required for viral replication (Hsue and Masters, 1997). Biochemical studies supported this bulged stem-loop structure and suggested that it was necessary for RNA replication, although the terminal loop and the first internal loop near the base of the stem were found to be larger than originally thought (Hsue et al., 2000, Hsue and Masters, 1997). Williams et al. (Williams et al., 1999) examined the 3′UTR in bovine coronavirus and reported evidence for a phylogenetically conserved pseudoknot. This pseudoknot encompasses nts 238-185 (note that the 3′-most nucleotide upstream of the poly(A) tail is position 1 in this numbering scheme, with numbers ascending in the 5′ direction) in MHV, and its 5′ stem overlaps with the last segment of the upstream bulged stem-loop (positions 301–224 in MHV), making the two secondary structures in part mutually exclusive. This led to the hypothesis that these two structures may be alternate conformers of the same region of genomic RNA and constitute a molecular switch (Hsue et al., 2000). A genetic analysis using viral mutants isolated by targeted recombination supports this hypothesis (Goebel et al., 2004). A study examining the secondary structure of the last 166 nucleotides of the 3′UTR, a region downstream of the pseudoknot predicted a long multi-branch stem loop in this region of the genome, a model that was largely supported by enzymatic probing of RNA secondary structure (Liu et al., 2001). DI RNA replication assays supported a functional role for several stems in this structure. Subsequent re-examination of this data favored a somewhat different conformation for the last 42 nucleotides in the 3′UTR (Johnson et al., 2005). A series of DI RNA replication assays showed that mutations within this region (nts 42-1) that disrupted (ATW3′ and ATW5′) or restored (ATW) a predicted stem in this region prevented the accumulation of DI RNAs (Johnson et al., 2005). When these same mutations (ATW, ATW3′, and ATW5′) were introduced into the 3′UTR of the complete viral genome, only the ATW mutation was lethal; viruses containing the ATW3′ and ATW5′ mutations grew almost as well as wild type virus in a one-step growth curve experiment although they did produce significantly smaller plaques than wild type (Johnson et al., 2005). Goebel et al. (Goebel et al., 2007) demonstrated that a hyper variable bulged stem loop spanning nts 46–156 was not essential for MHV replication, even though this otherwise poorly conserved region contains a highly conserved octanucleotide sequence present in virtually all coronaviruses (Boursnell et al., 1985), and proposed a secondary structure model of the complete MHV 3′UTR (Goebel et al., 2004, Goebel et al., 2007). Based in part on sequence co-variation, Zust et al. (Zust et al., 2008) subsequently revised the 3′ region of this model (see Fig. 1) to reflect potential conserved base pairings present in viruses representing all three coronavirus subgroups. Although the phylogenetic conservation of the proposed secondary structural model is persuasive and suggests that the proposed secondary structures are functionally important, a genetic test of the functional importance of this structure was not performed.
Fig. 1.
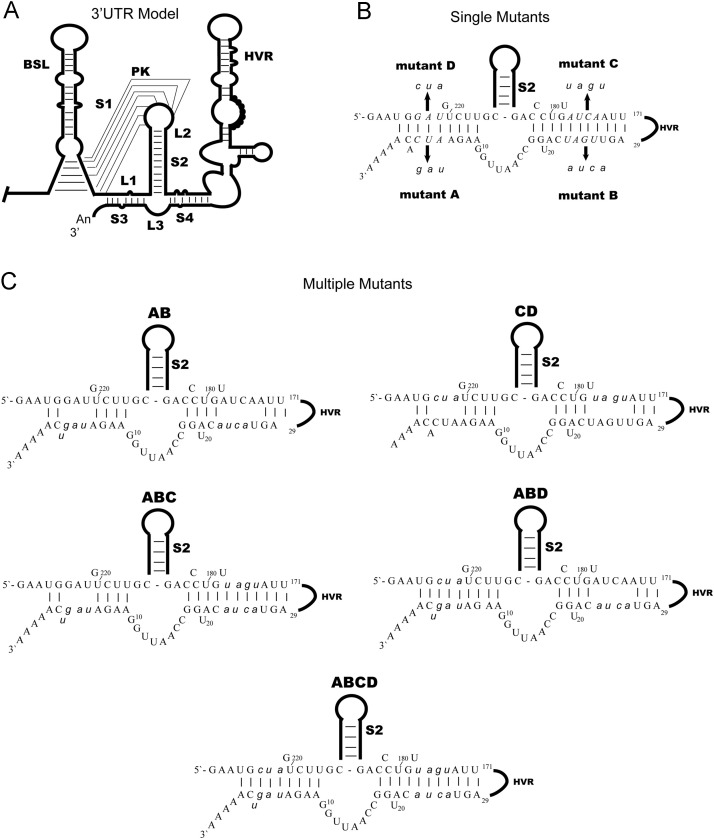
Proposed RNA secondary structures and mutations in the MHV 3′ UTR. (A) 3′ UTR model proposed by Zust et al. (2008), including the region downstream of S2 comprised of the HVR and the two helical stem structures (S3 and S4). (B) Mutations made to locally disrupt S3 and S4. The C and D mutations were made in this study and the A and B were previously made by Johnson et al. (2005). The wild type sequence is shown and the nucleotides that are altered by mutation italicized. Arrows indicate the sequence of each mutant. (C) Mutated nucleotides are indicated by italicized lower case letters.
We report here a functional analysis of the RNA secondary structure model proposed by Zust et al. (Zust et al., 2008). We utilized reverse genetic approaches to experimentally test this model through mutagenesis of the novel S3 and S4 stems (see Fig. 1) predicted by this model. Mutations opening up S3 were lethal, but large disruptions in S4 generated both viable and lethal mutants. Genomes carrying the original mutations in S3 or S4 plus compensatory mutations that restored base pairing in these stems were all viable and had robust growth phenotypes. Overall our results support the Zust model for the coronavirus 3′UTR and suggest that the S3 stem is required for virus viability, whereas at least some mutations that disrupt the S4 stem can be tolerated.
Results
Small disruptions of S3 or S4 have little effect on viral phenotype and negative-strand subgenomic RNA synthesis
In the Zust et al. model of the MHV 3′UTR (Zust et al., 2008), nucleotides 0 to 9 (note that the 5′-most A of the poly(A) tail is designated as position 0) are base paired with the L1 loop of the pseudoknot to form a helical stem (labeled S3 in Fig. 1A); nucleotides 18–29 are base-paired (S4 in Fig. 1A) with sequences downstream of the pseudoknot stem S2. In an earlier study of protein interacting sequences in the 3′UTR (Johnson et al., 2005), three mutations that mapped to S3 and/or S4 (see Fig. 1) in the Zust et al. model of the MHV 3′UTR (Zust et al., 2008) were examined for their effects on replication. These mutations, originally named ATW3′, ATW5′, and ATW (Johnson et al., 2005) have been renamed as A, B, and AB in the current study to increase clarity (see Fig. 1B and C). When these three mutations are introduced into the genomic RNA, genomes containing the A and B mutations lead to viable viruses, however, the genome containing the AB mutation is lethal (Johnson et al., 2005). Based on our earlier results with the A, B, and AB mutants we hypothesized that the presence of either of the two stems is sufficient for a viable virus, but disruption of both stems is lethal. To investigate this hypothesis and to perform a genetic test of the Zust secondary structure model of the 3′UTR, we made a series of additional mutations targeting S3 and S4. Mutations C and D, were designed to destabilize S3 and S4 respectively; mutation CD destabilizes both S3 and S4 (Fig. 1B). Mutations that affected both sides of S3 and S4 were designated ABD, ABC, and ABCD (Fig. 1C). When mutations C, D, ABC, and ABD, which destabilize base-pairing in either the S3 (D, ABC) or S4 (C, ABD) helices were introduced into the genomic RNA, they all produced viable virus with somewhat smaller plaque sizes compared to the wild type virus ( Fig. 2A), grew with near identical kinetics to wild type virus, and achieved peak titers that were at least 25% of those achieved by wild type virus (Fig. 2B). However, for genomes carrying mutation CD, a mutation which disrupts base-pairing in both S3 and S4, we were unable to recover virus in multiple trials, thus we concluded that this mutation was lethal, similar to the results we obtained with the A, B, and AB mutations in our previous work (Johnson et al., 2005). Furthermore, when mutant ABCD, which restores base-pairing in stems S3 and S4 but alters the sequences of these stems, was introduced into the genomic RNA it was viable and generated a virus that forms plaques almost identical in size to those formed by wild type virus, grew with nearly identical kinetics to wild type virus, and achieved a peak titer nearly identical to that of wild type virus (Fig. 2A and B). These results are entirely consistent with the Zust model (Zust et al., 2008) of the 3′UTR and our hypothesis that base-pairing in either S3 or S4 are necessary for MHV viability.
Fig. 2.
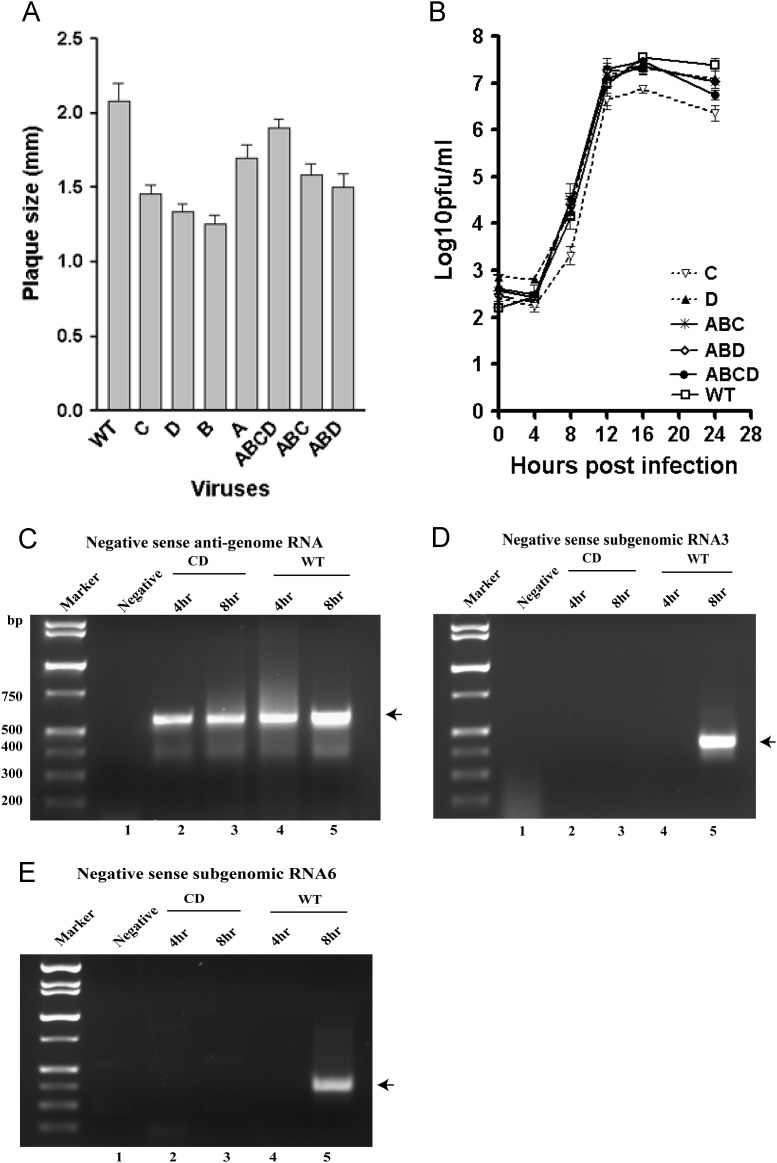
Growth phenotypes for viable mutant viruses and RNA phenotypes for lethal mutant. (A) Plaque size (mm) of viable viruses. (B) Growth curve of viable viruses. (C) RT-PCR assays for negative sense RNA corresponding to genomic RNA, (D) subgenomic RNA3, and (E) RNA6 for wild type MHV-A59 and lethal mutant CD. Lanes labeled Negative represent RNA extracted from cells that were mock electroporated with buffer rather than with in vitro transcribed RNA. The positions of the expected PCR products for each RNA species are indicated by arrows.
We had previously demonstrated that MHV genomes carrying the lethal AB mutation are unable to direct the synthesis of subgenomic RNA, although they are able to direct the synthesis of a minus sense complement of the genome (Johnson et al., 2005). To determine the RNA species that might have been generated in cells electroporated with genomes containing the CD mutation, we performed nested RT-PCR assays to detect negative-strand genomic RNA and negative-strand subgenomic RNA3 and mRNA6. These RNAs serve as templates for genomic and subgenomic mRNA synthesis (Pasternak et al., 2003, Sawicki and Sawicki, 1990, Sola et al., 2005, Zuniga et al., 2004). Nested RT-PCR results showed that negative-strand genomic RNAs were present in cells at 4 and 8 h after electroporation with in vitro transcribed genomes carrying the CD mutation, similar to what we observed after electroporation of WT genomes (Fig. 2C). In contrast, neither negative-strand subgenomic RNA3 nor RNA6 were detected in cells electroporated with in vitro transcribed genomes carrying the CD mutation, whereas cells electroporated with WT genomes contained negative-strand subgenomic RNA3 and RNA6 at 8 h incubation (Fig. 2D and E). For each sample, parallel RT-PCR reactions without an RT step were performed to ensure that residual DNA templates taken up by the cells during electroporation did not produce PCR signals (data not shown). These data show that genomes carrying the CD mutation destabilizing stems S3 and S4 are defective in directing subgenomic RNA synthesis, the identical phenotype detected with the AB mutations on the opposing sides of the stem in our previous work (Johnson et al., 2005).
Larger disruptions of S3 or S4 generate various viral phenotypes
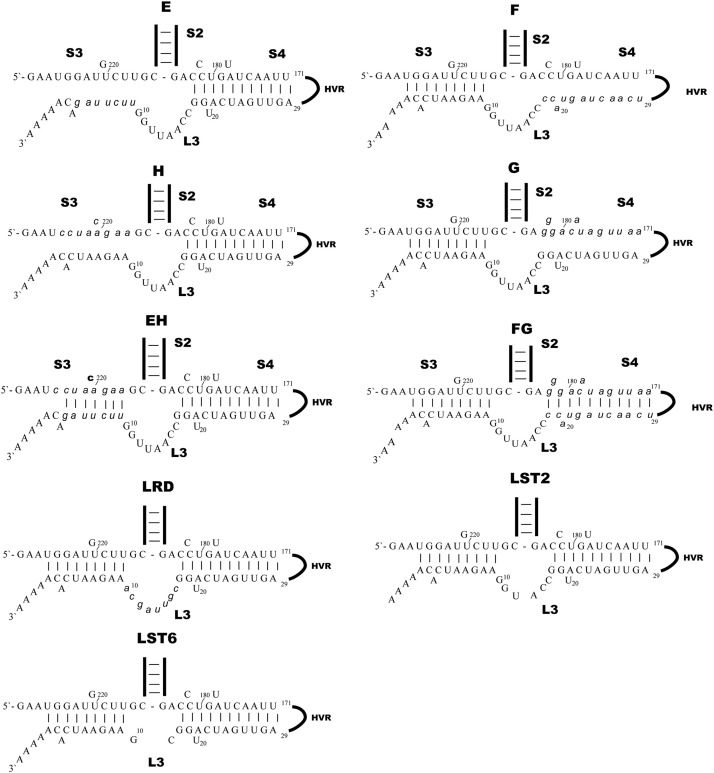
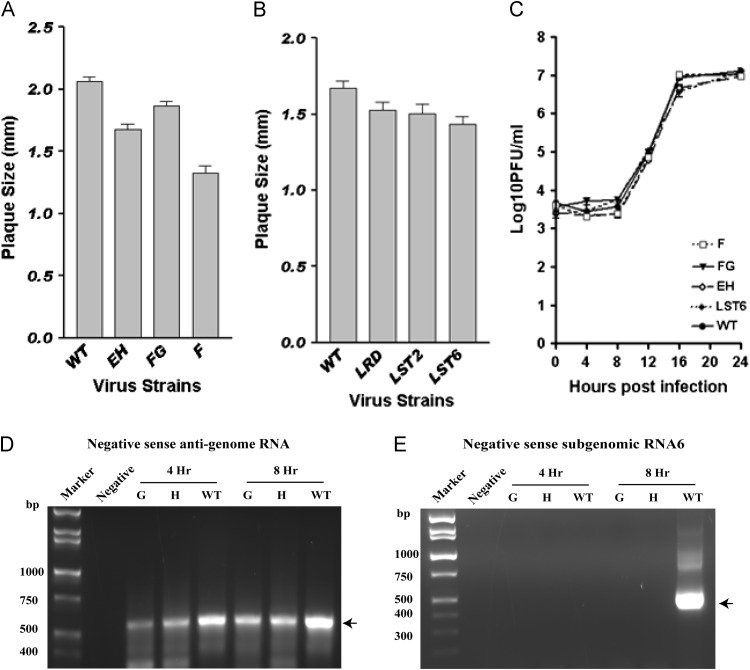
Based on the above results, we further tested if larger sequence disruptions in S3 or S4 affect virus viability. A series of mutants E, F, G, H, EH, and FG were made for this purpose ( Fig. 3). Mutations E and H, two mutants that completely disrupt S3, were lethal; a mutation in the 5′ side of S4 that completely disrupted this stem, mutation G, produced the same lethal phenotype. However, the mutation in the 3′ side of S4, mutation F, produced a viable virus. All plaque isolates of F mutant virus, from two independent electroporations, also contained a second site mutation, either A5/C5 or A6/C6. C5 or C6 can base-pair with G221 which is extruded in the WT S3 stem; this base-pairing increased the stability of S3 in viruses we recovered containing the F mutation. Sequencing of the nsp8 and nsp9 coding regions of these mutants failed to reveal additional second site mutations. Unsurprisingly, mutations that restored the S3 and S4 helices, mutations EH and FG, both generated viable mutants viruses EH and FG. These viable mutants have smaller plaque sizes, but essentially equivalent one-step growth curve compared to wild type virus ( Fig. 4A and C). RNA species present in cells electroporated with the lethal G and H mutants were analyzed by RT-PCR as described above for the AB and CD lethal mutants; negative sense genomic RNAs were detected when cells were electroporated with genomes containing the lethal mutants H in S3 and G in S4; however, subgenomic RNA synthesis is defective in these mutants (Fig. 4D and E). These results suggest that S3 is critical for virus viability, with complete disruption of S3 leading to a defect in subgenomic RNA synthesis and thus a lethal phenotype. The disparate effects of the F (viable), G (lethal), and FG (viable) mutations in S4 led us to model the possible effects of these three mutations on the overall folding of this portion of the 3′UTR in Mfold. The Mfold models suggest that the G mutant has the potential to fold into a very different structure than the wild type structure but with a similar thermodynamic stability, perhaps accounting for the G mutant's lethal phenotype. In contrast the F mutation is predicted to simply result in local unfolding of S4. This suggests that S4 is not essential for virus viability.
Fig. 3.
Mutations causing complete disruption of S3 or S4 helices or the L3 loop. Mutated nucleotides are indicated by italicized lower case letters.
Fig. 4.
Growth and RNA phenotypes of S3, S4, and L3 mutations. (A) Plaque size (mm) of S3 and S4 viable viruses. (B) Plaque size (mm) of L3 viable viruses. (C) Growth curve of S3, S4 and L3 viruses. (D) RT-PCR assay for negative sense RNA corresponding to genomic RNA for wild type MHV-A59 and lethal mutants G and H. (E) RT-PCR assays for negative sense RNA corresponding to subgenomic RNA6 for wild type MHV-A59 and lethal mutants G and H. Lanes labeled Negative represent RNA extracted from cells that were mock electroporated with buffer rather than with in vitro transcribed RNA. The positions of the expected PCR products are indicated by arrows.
Disruption of L3 has no effect on viral phenotypes
The 8 nts that make up loop 3 (L3, see Fig. 1A and Fig. 3) have a perfect palindromic sequence arrangement and led us to investigate the role of L3, if any, in viral replication. Three mutations, a loop randomized mutation (LRD), a mutation that shortened L3 by 2-nts (LST2), and a mutation that shortened L3 by 6-nts (LST6), were designed to test the functional role of L3 (Fig. 3). Interestingly, these mutations all resulted in viable viruses with only minor differences in plaque size and no differences in their replication kinetics or final titer compared to wild type virus (Fig. 4B and C). This result showed that L3 likely functions as a linker between S3 and S4 with few constraints on its sequence and length.
Discussion
Cis-acting sequences important for controlling viral replication are typically found in the 3′ and 5′ UTRs of RNA viruses. RNA secondary structural features are often important in the function of these cis-acting regions. In this study we have performed a functional analysis of the RNA secondary structural model of the MHV 3′UTR originally proposed by Zust et al. (Zust et al., 2008) utilizing a reverse genetics approach. The conservation amongst group 2 coronaviruses of the structural elements of the 3′UTR, namely the bulged stem-loop and overlapping pseudoknot and the conserved S3 and S4 helices connected by a variable length loop (see Fig. 1A) (Zust et al., 2008), and the ability of other group 2 coronavirus (SARS-CoV and BCoV) 3′UTRs with primary sequences that diverge significantly from MHV to functionally replace the MHV 3′UTR (Goebel et al., 2004, Hsue and Masters, 1997, Kang et al., 2006), suggests the RNA secondary structure of the 3′UTR plays a key functional role in coronavirus replication. The overlap of the most distal portion of the bulged stem-loop with the pseudoknot makes these two structures mutually exclusive. This led to the hypothesis that these two structures may be alternate conformations of the same region of genomic RNA and constitute a molecular switch (Hsue et al., 2000). Genetic studies demonstrated that both the pseudoknot and base-pairing of the lower region of the bulged-stem-loop were functionally important, supporting this hypothesis (Goebel et al., 2004). Biophysical studies (Stammler et al., 2011) demonstrate that the pseudoknotted conformation is much less stable than the double-hairpin conformation but suggest that stacking of the pseudoknot with the S3 helix can stabilize the pseudoknotted conformation allowing it to form. Mutations predicted to disrupt the S3 helix, (mutations E and H) were both lethal, whereas the double mutation EH predicted to restore the S3 helix, resulted in a viable virus, consistent with the idea that this stem may be necessary to allow the pseudoknotted conformation to form, or is necessary for viral replication in its own right.
Mutations in S4 predicted to disrupt this helix had differing effects. Mutation G, in the 5′ side of S4 and predicted to completely disrupt the S4 helix produced a lethal phenotype. This contrasted with the mutation in the 3′ side of the S4 helix, mutation F, which reproducibly produced a viable virus with a second site mutation in the S3 helix that stabilized this structure. The double mutant FG, predicted to restore the predicted structure, resulted in a viable virus, entirely consistent with the Zust (Zust et al., 2008) model. Modeling the possible effects of these three mutations on the overall folding of this portion of the 3′UTR in Mfold, suggests that the G mutant has the potential to cause the region containing the triple helix junction (S2, S3, and S4, see Fig. 1) to fold into a completely different two helix structure with a similar thermodynamic stability as the wild type structure. This structure, lacking the S2 stem-loop and both the S3 and S4 helices, would not be able to form the pseudoknotted structure or the double stem-loop structure (extended bulged stem-loop plus S2-L2 stem-loop), likely accounting for the G mutant's lethal phenotype. In contrast the F mutation is predicted to simply result in local unfolding of S4. All of the viruses we recovered with the F mutation in S4 also contained second site A5C or A6C mutations in S3 that resulted in a G–C base pair replacing an A–U base pair, thus increasing in the stability of S3 by a small amount. Our failure to recover viruses that contain the F mutation without any second site mutations, raises the possibility that S4 interacts (stacks) with the S3 helix, making a further necessary contribution to the stability of the 3′UTR, particularly in the pseudoknotted configuration.
Mutations with smaller sequence disruptions designed to separately destabilize the predicted S3 (mutations A, D, ABC) or S4 (mutations B, C, and ABD) helical stems were all viable, whereas mutants that destabilized both of these helices (mutations AB and CD) were lethal. A quadruple mutation (ABCD) that maintained both S3 and S4 while altering the sequences of these stems yielded a virus with an in vitro phenotype virtually identical to that of wild type virus. These results strongly support the Zust (Zust et al., 2008) model. Modeling the possible effects of the lethal AB and CD mutations on the overall folding of this portion of the 3′UTR in Mfold, suggests that the CD mutant has only one thermodynamically stable conformation in which the region containing the triple helix junction (S2, S3, and S4, see Fig. 1) folds into a completely different structure consisting of a long bulged stem-loop linked to a shorter bulged stem-loop by a seven nucleotide single-stranded linker. This structure, lacking the S2-L2 stem-loop and both the S3 and S4 helices, would not be able to form the pseudoknotted structure or the alternative two stem-loop structure that make up the putative molecular switch (Goebel et al., 2004), possibly accounting for the CD mutant's lethal phenotype. It is less clear from our modeling of the AB mutant why it is lethal. The three structures predicted by Mfold all maintain the S2-L2 stem-loop with the most stable predicted structure containing an additional bulge in S4 and a somewhat reconfigured and shorter S3 helix (see Fig. 1). These same altered S3 and S4 helices are predicted to be present singly in the A and B mutants, thus the predicted structure of the AB mutant does not provide a ready explanation for its lethal phenotype. It is also possible that the lethal AB and CD mutations perturb the equilibrium between the extended bulged stem-loop structure and the pseudoknotted structure to a degree that is incompatible with viability.
Mutations disrupting the L3 loop (LRD, LST2, and LST6) between S3 and S4 were all viable with no apparent phenotypic differences compared to wild type virus. This leads us to conclude that L3 might function as a linker between S3 and S4 with little sequence and length preference. Overall, the genetic studies of the 3′UTR strongly support the Zust model. Several alternative secondary structures for this region of the 3′UTR were proposed prior to the development of the current model. (Goebel et al., 2007, Johnson et al., 2005, Liu et al., 2001). Our results suggest that it is unlikely that these alternative RNA secondary structures have a functional role in replication and support the Zust model of the 3′UTR (Zust et al., 2008).
Zust et al. (Zust et al., 2008) introduced insertion mutations in the loop 1 (L1) sequence on the 5′ side of the S3 stem and recovered second site suppressor mutations in the nsp8 and nsp9 proteins in viruses recovered after limited passage. Based on these results these authors speculated that the L1 loop plus the S3 helix might serve as a binding site for a primase complex containing the nsp8 RNA dependent RNA polymerase (RdRp) activity (Imbert et al., 2006), and that this complex initiates minus sense RNA synthesis. In this model, once RNA synthesis had commenced from the 3′ end of the genome the base pairing in S3 would be disrupted, allowing formation of the upstream pseudoknot (see Fig. 1), enabling assembly of a complete replicase complex containing the nsp12 RNA RdRp and other nsps to subsequently elongate the RNA primer. Sequencing of the nsp8 and nsp9 coding sequences of viable mutant viruses A, D, and ABC, all containing mutations predicted to disrupt a small portion of the S3 helix, failed to demonstrate second site suppressor mutations in nsp8 and nsp9 (data not shown). Similarly, sequencing of this region in all mutant F isolates failed to demonstrate second site mutations. Additionally, the RNA phenotype of all of the lethal mutations that we examined (AB, CD, H, and G; Figs. 2C and 4E) indicated that genomes with these mutations were able to direct the synthesis of negative sense RNAs corresponding to the genome but were not able to synthesize negative sense templates for subgenomic RNA synthesis. This result is generally consistent with a model for subgenomic RNA synthesis (Sola et al., 2005, Zuniga et al., 2004) that hypothesized that genome circularization mediated by RNA–protein or RNA–RNA interactions plays an important role in leader-body sequence joining during transcription of minus sense subgenomic RNAs, providing a role for the 3′UTR in subgenomic RNA synthesis. It should be noted that the Zust (Zust et al., 2008) and Zuniga/Sola (Sola et al., 2005, Zuniga et al., 2004) models are not necessarily mutually exclusive and elements of both of them could explain the observed RNA phenotype of our mutants. Our data does not distinguish between these two models, but it does demonstrate that in either scenario the complex and very likely dynamic structure of the 3′UTR is necessary for discontinuous synthesis of subgenomic RNAs. Our studies do provide data supporting the idea that the 3′UTR likely is a single functional unit consisting of several interacting structures.
We also noticed that the MHV 5′UTR SL1 potentially forms base-pairing interactions with 3′UTR S3; the 5′UTR SL1 sequence 5′G8UGAUU133′ can be base paired to 3′UTR S3 3′C1ACUAA65′ and 5′UTR SL1 5′G14GCGUCCGUAC243′ can be base paired to 3′UTR S3 3′C218UGUAGGUA AG228 5′. We hypothesized that these interactions might be necessary for the genome circulization postulated by the Zuniga/Sola (Sola et al., 2005, Zuniga et al., 2004) model to promote the switch between transcription and replication. A series of mutations were designed to investigate these hypothesized interactions between the 5′ and 3′UTRs and we attempted to recover infectious virus harboring these mutations. These studies failed to support these hypothesized interactions (data not shown). Additional structural and biophysical studies to determine in detail the interactions of the various structures in the entire 3′UTR and their dynamics are required to more fully understand their roles in viral replication.
Materials and methods
Cells and viruses
Murine 17Cl-1, DBT, and L2 cells were grown in Dulbecco's modified Eagle medium (DMEM; Life Technologies) supplemented with 10% bovine calf serum (Hyclone), 4 mM glutamine, and penicillin and streptomycin, each at 50 μg/ml. BHK-R cells were maintained in DMEM supplemented as described above but containing 800 μg/ml G418. MHV-A59 1000 (Yount et al., 2002), was the virus used as our wild type strain. Wild type and mutant virus stocks were grown and tittered by plaque assay as described previously (Johnson et al., 2005).
Generation of recombinant viruses
The reverse genetic system described by Yount et al. (Yount et al., 2002) was used to recover wild-type MHV-A59 (MHV-A59 1000) and mutant viruses. Mutant viruses containing the A and B mutations had previously been isolated by Johnson et al. (Johnson et al., 2005). Mutations creating small sequence disruptions of S3 and S4 (C, D, CD, ABC, ABD, and ABCD) were generated in a G plasmid template by site-directed mutagenesis; mutations creating larger sequence disruptions in S3, S4, and L3 (E, F, G, H, EH, FG, LRD, SLT2, SLT6) were generated by overlapping PCR with G plasmid as template. The sequences of mutagenic oligonucleotides are available on request. All mutated target sites were sequence verified and subcloned back into the original G plasmid as a 285 nt NruI-PacI fragment to obviate the potential for undesired mutants being present outside the 3′UTR after site directed mutagenesis. The presence of the desired mutation was again verified by sequencing and these plasmids were subsequently used to regenerate infectious virus. Bacteria containing the seven plasmids (A–G) making up the MHV-A59 reverse genetic system were propagated at 30 °C and plasmid DNA purified using a plasmid Midi prep kit from OMEGA BIO-TEK according to the manufacturer's protocol. The MHV cDNAs were excised from mutant and wild type G plasmids, gel purified, and ligated to MHV-A59 cDNAs A–G to generate cDNAs corresponding to the entire MHV genome under the control of a T7 promoter. Full length cDNAs were in vitro transcribed using the Ambion mMessage mMachine T7 kit. Transcripts corresponding to the N gene coding sequence were also transcribed in vitro as described by Yount et al. (Yount et al., 2002). The N gene was mixed with the in vitro transcribed RNA and electroporated into BHK-R cells using a Biorad Gene Pulser. Electroporation was carried out three times at 0.85 kV. After electroporation, BHK-R cells were overlaid onto freshly seeded DBT cells in a T75 flask. Cultures were incubated for up to 3 days and monitored by phase microscopy for the development of cytopathic effect (CPE). Cultures that did not demonstrate CPE were blind passaged in DBT cells. A mutation was not considered to be lethal until at least 3 independent trials were performed, at least one of which was done at 34 °C and 39 °C to allow for recovery of temperature sensitive viruses. The cultures were frozen at −80 °C, sonicated, clarified by low speed centrifugation and virus plaque cloned on L2 cells. Individual plaques were then inoculated onto a confluent monolayer of DBT cells to generate small virus stocks. After maximal CPE developed the cell culture fluids were frozen at −80° and RNA was extracted for later analysis.
Characterization of recombinant viruses
RNA was extracted from infected monolayers of DBT cells using RNeasy RNA extraction kits (Qiagen). All recombinant viruses that were recovered had their 3′ UTRs sequenced. RT-PCR sequencing was carried out as described below. Reverse transcription reactions were carried out using 1 µg of total RNA, 1 µl of primer 20TG and the RNA-primer mixture was heated to 65 °C for 5 min. After heating, the reaction was put on ice for ∼1 min and then 1 μl 0.1 M DTT and 4 μl 5× First-Strand buffer was added along with 1 µl of Superscript III Reverse Transcriptase (Invitrogen Cat. no. 18080-044). The reaction was incubated at 42 °C for 60 min. The RT reaction was then heat inactivated at 70 °C for 15 min. The generated cDNA was PCR amplified using primers A59(+)30948 and Eco20TG ( Table 1). The conditions for the PCR reaction were as follows: 94 °C for 1 min followed by 30 cycles each consisting of 30 s at 94 °C, 1 min at 58 °C, and 1 min 68 °C. Amplified fragments were gel purified and sequenced with the primer A59(+)30948 (Table 1).
Table 1.
Sequencing, RT-PCR and PCR Primers used in this study+.
| Primer | Sequence |
|---|---|
| A59(+)30948-68 (PCR, sequencing) | TAA CCC CAG AGG ATA GAA GTC |
| 20TG (RT-PCR) | TTT TTT TTT TTT TTT TTT TTG |
| Eco20TG (PCR) | GAA TCC TTT TTT TTT TTT TTT TTT TTG |
| A59 (+)14369–14658 (RT-PCR) | GTG GAT ACA CAT CGT TAT CG |
| A59 (−)16577–16596 (RT-PCR) | TAC TGT GGT TTA TGG TCC TC |
| A59 (+)16038–16059 (RT-PCR) | ATG AAG TCT ACC TTC CAT ACC C |
| A59 (+)1–20 (RT-PCR) | TAT AAG AGT GAT TGG CGT CC |
| A59 (+)7–23 (RT-PCR) | AGT GAT TGG CGT CCG TA |
| A59 (+)26–47 (RT-PCR) | TAC CCT CTC AAC TCT AAA ACT C |
| A59 (−)24672-24654 (RT-PCR) | TGG GTT GCA GAT GAA AGG T |
| A59 (−)24284–24265 (RT-PCR) | GCA CCT GAT GGC GTA CTT GT |
| A59 (−)29593–29613 (RT-PCR) | TTG AGG GCA GTC GGT AAT TTC |
| A59 (−)29327–29345 (RT-PCR) | CAT AAG GTT GTT TGT TTC G |
All nucleotide positions are from Genbank Accession NC001846.
Viral plaque sizes were measured by projecting the individual well plates and an adjacent millimeter ruler with an overhead projector to achieve at least 10× magnification. Thirty plaques were measured for each virus in order to encompass the entire range of plaque phenotypes. Mean plaque sizes were calculated using the magnification factor determined from the millimeter ruler. Only clear plaques with a distinct circumference were measured.
To determine one-step growth kinetics, DBT cells were grown in 96-well plates and replicate cultures infected with either WT or mutant viruses at a MOI of 3. Samples were frozen at 0, 4, 8, 12, 16 and 24 h post infection. Triplicates samples were obtained at all time points and virus titers were determined by plaque assay.
Detection of negative-strand genomic and subgenomic RNAs for non-viable mutants
For the nonviable mutants CD, G, and H. RNA from electroporated cells was extracted at 4 and 8 h post-electroporation using the RNeasy Mini kits (Qiagen). Any residual DNA was removed from 10 µg of total RNA using DNase Treatment and Removal Reagents (AM1906, Applied Biosystems) according to the manufacturer's instructions. Negative-sense genomic and subgenomic RNAs in the cellular RNA was assayed by nested RT-PCR as previously described (Johnson et al., 2005, Liu et al., 2007, Yount et al., 2002). To detect negative-strand RNA complementary to the genome, oligo A59 (+) 14639–14658 (Table 1) was used as the RT primer for cDNA synthesis. Primers A59 (+)14639–14658 and A59 (−)16596–16577 were used in the first PCR reaction and primers A59 (+)16038–16059 and A59 (−)16596–16577 were used in the nested PCR reaction (Table 1). A no RT control was always run for each sample to be certain that any signal detected did not arise from residual plasmid DNA. To detect negative-strand RNA complementary to subgenomic mRNA 6, primer A59(+)1–20 was used as the RT primer, primers A59(+)7–23 and A59(−)29593–29613 were used in the first PCR reaction, and primers A59(+)26–47 and A59(−)29327–29345 were used in a nested PCR reaction. To detect negative-strand RNA complementary to subgenomic mRNA 3, primer A59(+)1–20 was used as the RT primer, primers A59(+)7–23 and A59(−)(24672–24654) were used in the first PCR reaction and primers A59(+)26–47 and A59(−)(24284–24265) were used in a nested PCR reaction. Amplified PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
Acknowledgments
The authors gratefully acknowledge Melissa Wilmarth for performing the initial site directed mutagenesis of the G plasmid and all of the members of the Leibowitz Lab for their support and encouragement. The authors gratefully acknowledge support from US National Institutes of Health grant AI067416.
Contributor Information
Pinghua Liu, Email: pliu@tmhs.org.
Julian L. Leibowitz, Email: leibowitz@medicine.tamhsc.edu.
References
- Boursnell M.E.G., Binss M.M., Foulds I.J., Brown T.K.K. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J. Gen. Virol. 1985;66:573–580. doi: 10.1099/0022-1317-66-3-573. [DOI] [PubMed] [Google Scholar]
- Britton P.a.D.C. Nidovirus genome organization and expression mechanism. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; Washington D.C: 2008. pp. 29–46. [Google Scholar]
- de Haan C.A., Volders H., Koetzner C.A., Masters P.S., Rottier P.J. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 2002;76:12491–12502. doi: 10.1128/JVI.76.24.12491-12502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78:669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Miller T.B., Bennett C.J., Bernard K.A., Masters P.S. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 2007;81:1274–1287. doi: 10.1128/JVI.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Hartshorne T., Masters P.S. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2000;74:6911–6921. doi: 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Masters P.S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.-C., Bourhis J.-M., Bussetta C., Coutard B., Egloff M.-P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS Coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.F., Feng M., Liu P., Millership J.J., Yount B., Baric R.S., Leibowitz J.L. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 2005;79:14570–14585. doi: 10.1128/JVI.79.23.14570-14585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Feng M., Schroeder M.E., Giedroc D.P., Leibowitz J.L. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 2006;80:10600–10614. doi: 10.1128/JVI.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jeong Y.S., Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J.L., Wilhelmsen K.C., Bond C.W. The virus-specific intracellular RNA Species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Lai M.M.C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J. Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li L., Millership J.J., Kang H., Leibowitz J.L., Giedroc D.P. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA. 2007;13:763–780. doi: 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Johnson R.F., Leibowitz J.L. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J. Virol. 2001;75:12105–12113. doi: 10.1128/JVI.75.24.12105-12113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yu W., Leibowitz J.L. A specific host cellular protein binding element near the 3′ end of mouse hepatitis virus genomic RNA. Virology. 1997;232:74–85. doi: 10.1006/viro.1997.8553. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J., Snijder E.J. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 2003;77:1175–1183. doi: 10.1128/JVI.77.2.1175-1183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.I. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S.G. Plenum Press; New York: 1995. The Coronaviridae: An Introduction. [Google Scholar]
- Sola I., Moreno J.L., Zuniga S., Alonso S., Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stammler S.N., Cao S., Chen S.-J., Giedroc D.P. A conserved RNA pseudoknot in a putative molecular switch domain of the 3′-untranslated region of coronaviruses is only marginally stable. RNA. 2011;17:1747–1759. doi: 10.1261/rna.2816711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Born E.a.E.J.S. RNA singnals regulating nidovirus RNA synthesis. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidovirus. ASM Press; Washington, D.C: 2008. pp. 115–131. [Google Scholar]
- Williams G.D., Chang R.Y., Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Denison M.R., Weiss S.R., Baric R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Leibowitz J.L. A conserved motif at the 3′ end of mouse hepatits virus genomic RNA required for host protein binding and viral RNA replication. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Leibowitz J.L. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J. Virol. 1995;69:2016–2023. doi: 10.1128/jvi.69.4.2016-2023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Miller T.B., Goebel S.J., Thiel V., Masters P.S. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2008;82:1214–1228. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]