Abstract
Non-apoptotic cell death mechanisms are largely uncharacterized despite their importance in physiology and disease (P. Kreuzaler and C.J. Watson, Nature Reviews Cancer). Here we sought to systematically identify non-apoptotic cell death pathways in mammalian cells. We screened 69,612 compounds for those that induce non-canonical cell death by counter screening in the presence of inhibitors of apoptosis and necrosis. We further selected compounds that require active protein synthesis for inducing cell death. Using this tiered approach, we identified NID-1 (Novel Inducer of Death-1), a small molecule that induces an active, energy-dependent cell death in diverse mammalian cell lines. NID-1-induced death required components of the autophagic machinery, including ATG5, and the lysosomal hydrolase cathepsin L, but was distinct from classical macroautophagy. Since macroautophagy can prevent cell death in several contexts, we tested and found that NID-1 suppressed cell death in a cell-based model of Huntington’s disease, suggesting that NID-1 activates a specific pathway. Thus the discovery of NID-1 identifies a previously unexplored cell death pathway, and modulating this pathway may have therapeutic applications. Furthermore, these findings provide a proof-of-principle for using chemical screening to identify novel cell death paradigms.
Keywords: cell death, autophagy, ATG5, cathepsin L, Huntington’s disease, vacuolization
Introduction
Cell death is central to normal development and the maintenance of tissue homeostasis. Deregulation of cell death is implicated in several diseases including cancers and neurodegeneration.. Historically, two cell death phenotypes, apoptosis and necrosis, have been well studied. However, these programs cannot account for all cell death mechanisms [2], as extensive developmental cell death has been observed in mice with defective apoptotic machinery [3]. Additional forms of cell death, such as necroptosis, ferroptosis, entosis and autophagy-like cell death have been described [4–7]. Such non-apoptotic cell death programs likely mediate distinct physiological and pathological processes. For example, necroptosis is a form of cell death that occurs when apoptosis is initiated but its execution is blocked by caspase inhibition. A suppressor of necroptosis, necrostatin (nec-1), is protective in a mouse model of ischemic-hypoxic neuronal death, indicating the biological relevance of this non-apoptotic pathway [8]. Thus, identifying novel cell death pathways and their regulatory mechanisms can enhance our understanding of the repertoire of cell death programs, and may provide new therapeutic targets.
Identification of experimental paradigms for dissecting non-canonical cell death programs and discovering tools to study them remains a major challenge. Much of our understanding of cell death processes comes from genetic screens performed in C. elegans [9]. While canonical death pathways unveiled in this model organism are evolutionarily conserved, additional cell death programs unique to mammalian cells are not accessible in simpler organisms. Large-scale RNAi screens can reveal the mechanisms of death in mammalian systems to some extent, however, several limitations remain: RNAi requires several days before knockdown is achieved for many proteins, some proteins with slow turnover are resistant to knockdown, certain cell types are not easily transfected, and RNAi reagents may produce off-target effects. Perturbations induced by small molecules complement genetic manipulations because they can be readily administered, used independently or in combinations, and the level of inhibition can be regulated by adjusting concentration [10]. Additionally, one can affect a single domain of a multifunctional protein or simultaneously inhibit paralogous proteins with small molecules [11].
Chemical screening thus offers a systematic approach to identify biological mechanisms and provides versatile tools with which to study them. We screened more than 69,000 compounds to identify a small molecule inducer of non-apoptotic cell death that we designated NID-1 (Novel Inducer of Death-1). We found that NID-1-induced cell death required new protein synthesis, was characterized by extensive cytosolic vacuolization, and involved components of the autophagic machinery, including ATG5 and the lysosomal protease cathepsin L, yet the death phenotype observed was distinct from classic macroautophagy. Furthermore, NID-1 was protective against mutant-huntingtin-(htt)-induced cell death, a model of polyglutamine neurodegeneration, suggesting the activation of a specific pathway. Recent studies have revealed other non-autophagic cellular processes involving vacuolization and cell engulfment that also utilize components of autophagic machinery [12, 13]. NID-1 will be a useful probe to study this ATG-5-and-cathepsin-L-dependent, non-apoptotic cell death pathway in mammalian cells and its potential role in neuroprotection.
Results
High-throughput screening identifies NID-1, an inducer of non-apoptotic cell death
We used a human fibroblast cell line (BJeLR) for this screen; this line is derived from human fibroblasts (BJeH) immortalized by expressing human telomerase and transformed by oncogenic Ras and SV40 large T antigen [14]. BJeLR cells are ideal for large-scale screening because they grow rapidly and have a small number of characterized genetic alterations, making it less likely that they contain mutations in regulators of unexplored cell death mechanisms that frequently occur in tumor cell lines. We screened 69,612 diverse small molecules in a cell viability assay based on measuring cellular reductive potential (see Experimental Methods). We identified 1,980 lethal compounds in a primary screen; these were further subjected to two criteria: first, we excluded compounds that induced death by canonical mechanisms involving apoptosis and necrosis by testing the ability of a collection of cell death inhibitors, including caspase, calpain and serine protease inhibitors, calcium channel blockers, and antioxidants, to block the lethality of each compound (Fig. 1A). Next, we selected lethal compounds that were suppressed by the protein synthesis inhibitor cycloheximide (CHX), in order to eliminate compounds that were killing cells via a non-active process or were non-specifically toxic (e.g. detergents). This tiered screen focused our analysis on non-canonical cell death inducers that required ongoing protein synthesis for their lethality. Three compounds met these criteria; the most potent of these (EC50 = 0.5 μg/ml), a nitrothiophenylpropenamide, was designated Novel Inducer of Death-1 (NID-1) and chosen for further investigation (Fig. 1A and 1B). NID-1’s structure was confirmed by NMR, and mass spectrometry (see Methods).
Fig. 1.
NID-1 induces cell death in diverse cell types. (A) Schematic of flow diagram used for identification of NID-1; structure of NID-1 (right panel). (B) BJeLR cells were treated with NID-1 (10 μg/ml) alone or co-treated with the protein synthesis inhibitor cycloheximide (CHX, 3 μM); cell viability was measured by a trypan blue dye-exclusion assay 8 h after treatment. Data represent mean ± S.D. of an experiment performed in duplicate, * (p< 0.05, left panel). The dose-response for cell death induced by NID-1 in BJeLR and HT-1080 cells was determined using the Alamar blue viability assay (right panel). (C) Time course of NID-1 (10 μg/ml)-induced cell death in BJeLR cells (left panel). Phase contrast micrographs of untreated BJeLR cells (middle) and after 8 h NID-1 (10 μg/ml) treatment (right panel). (D) BJeLR cells were treated with NID-1 and the supernatant (unattached fraction) was removed, and the cell viability for the attached fraction was determined by trypan blue exclusion. U2OS cells do not show cell detachment upon NID-1 treatment, and cell viability in these cells was assayed 30 h after NID-1 treatment, * (p< 0.05). (E) BJeLR or HT-1080 cells were seeded in 6-well plates, allowed to adhere overnight, and treated with NID-1 (5 μg/ml) for time points indicated. Cells were then trypsinized and re-plated into 0.3% agar-media mixture. Cells were stained with 0.005% crystal violet after culture for two weeks and the number of colonies per well was counted (left panel) and imaged (right panel), * (p<0.05). Data is representative of two independent experiments. (F) Time course of cell viability upon NID-1 treatment (10 μg/ml) in the indicated cell lines using trypan blue dye-exclusion assay. The experiments are representative of at least two independent experiments.
Structure activity relationship for NID-1
In order to identify the functional groups that are critical for NID-1’s activity, we performed a structure activity relationship (SAR) analysis for NID-1. Nine commercially available analogs of NID-1 were tested for their effects on cell viability. This analysis revealed that while some minor modifications to the phenyl ring were tolerated, others such as removal of the nitro group on the thiophene ring resulted in complete loss of activity (SI Fig. 1). The observation that small changes in the scaffold could lead to complete loss of activity, suggests that NID-1’s effects are likely to be mediated by a specific target. We also developed a three-step synthetic route to NID-1 and confirmed the activity of the re-synthesized NID-1 (SI Fig. 1). This SAR information and availability of a synthetic route will aid future studies with NID-1.
NID-1 induces cell death in diverse cell lines
We confirmed NID-1’s lethality by additional cell viability criteria—trypan blue dye-exclusion to assess membrane integrity, microscopic examination of cellular morphology, and colony formation assays to test long-term viability. NID-1 induced rapid cell death: death was initiated 3 h after treatment and most cells were dead by 10 h. NID-1-treated cells started to round up and lose adhesion by 4 h after treatment, and cell detachment correlated with cell death (Fig. 1C). However, NID-1 did not cause anoikis, a form of caspase-dependent death that is secondary to cell detachment [15]. We tested whether cell death preceded detachment, by evaluating death in both the attached and detached (supernatant) fraction after NID-1 treatment. Though cell death was greater in the detached fraction (74%), a substantial percentage of attached cells (34%) were dead by 8 h, indicating that cell death can occur prior to detachment (Fig. 1D). Furthermore, in the U2OS cell line, the cells remained adherent upon NID-1 treatment, yet 50% of attached cells were dead 30 hours after treatment (Fig. 1D). Thus, NID-1-induced cell death is not a consequence of detachment. Next, we confirmed that NID-1 induces cell death rather than senescence. In a long-term clonogenic assay, treatment with NID-1 for 3 h followed by washout of the compound abrogated colony formation in soft agar over the course of two weeks (Fig. 1E). Finally, we found that NID-1 induced cell death in several non-transformed human cells, including BJeH cells, and mouse embryo fibroblasts (MEF), as well as transformed human cell lines (DRD, RKO, H1299), and over 60 cancer cell lines representing diverse tissues of origin (Fig. 1F and SI Table1), indicating that NID-1 activates death across distinct cell and tissue types.
NID-1 induced death is independent of caspases, Bax and Bak
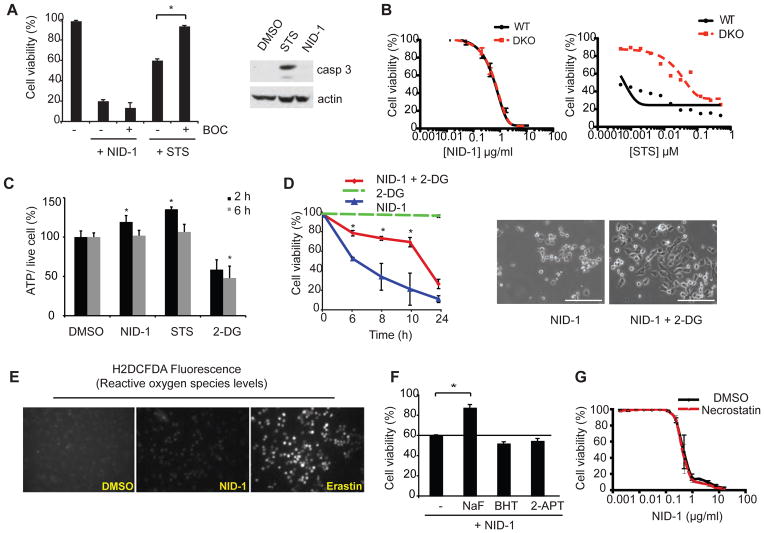
Since caspases are key regulators of apoptotic cell death, we had included broad-spectrum pan-caspase inhibitors in the panel of death inhibitors used to discover NID-1. We verified that NID-1-induced death was not suppressed by a broad-spectrum caspase inhibitor (BOC-D-Fmk) (Fig. 2A). In contrast, BOC-D-Fmk suppressed cell death caused by staurosporine (STS), a known inducer of apoptotic cell death. As expected, NID-1-induced death was not accompanied by cleavage of the effector caspase 3, whereas STS induced caspase 3 cleavage (Fig. 2A, right panel). Finally, we tested the role of two pro-apoptotic Bcl-2 family members, Bax and Bak, which are key regulators of mitochondrial outer membrane permeabilization, an initial step in the intrinsic apoptotic cell death pathway [16]. NID-1 was equally lethal in isogenic mouse embryonic fibroblasts (MEFs) derived from wild type (WT) and Bax/Bak double knockout (DKO) mice. In contrast, Bax/Bak DKO MEFs were resistant to STS induced apoptosis (Fig. 2B). Together, these data indicate that NID-1 induces a caspase-independent, non-apoptotic cell death.
Fig. 2.
NID-1 induces an active, non-apoptotic cell death. (A) BJeLR cells were treated with NID-1 (10 μg/ml) or staurosporine (STS, 0.5 μM) alone or in combination with BOC (50 μM). Cell viability was assayed after 8 h (NID-1) or 20 h (STS) using the trypan blue dye-exclusion assay (left panel), * (p<0.05). Western blot of cleaved caspase 3: cells were treated with vehicle (DMSO), STS (0.5 μM) or NID-1 (5 μg/ml) and harvested after 4 h, and subjected to western blot for cleaved caspase 3. Actin was used as a loading control (right panel). (B) Dose-response for NID-1 and STS in WT MEFs and in isogenic bax−/−, bak−/− double knockout (DKO) MEFs. Viability was measured by Alamar blue assay 48 h after treatment. (C) ATP levels were determined following 2 and 6 h treatments with NID-1 (2 μg/ml), staurosporine (STS, 0.1 μM) and 2-deoxyglucose (2-DG, 5mM) and compared to DMSO control; * (p<0.05) (D) BJeLR cells were either pretreated with 2-deoxyglucose (2-DG, 5 mM) for 2 h, or left untreated, and then treated with NID-1 (10 μg/ml). Cell viability was determined using the trypan blue dye-exclusion assay, * (p<0.05, left panel). Data is representative of two independent experiments. Phase contrast images of NID-1 and NID-1 + 2-DG (5 mM) treated cells after 8 h. Scale bar, 200μm (right panel). (E) Reactive oxygen species were detected using H2DCFDA after 6 h of treatment with NID-1 (10 μg/ml), or erastin (10 μM), and compared to DMSO-treated control cells. (F) Cells were left untreated or pretreated with the various antioxidants (butylated hydroxytoluene (BHT, 150 μM) or 2-acetylphenothiazine (2-APT, 10 μM)) for 2 h before NID-1 (10 μg/ml) treatment. Sodium flouride (5 mM) was a positive control. Cell viability was determined using the trypan blue dye-exclusion assay, * (p<0.05). (G) BJeLR cells were treated with dose dilution of NID-1 alone or in combination with necrostatin-1 (5 μg/ml) and viability was determined by an Alamar Blue assay. Experiments are representative of at least 2 independent experiments.
NID-1-induced death requires ATP and is distinct from necrosis
Necrosis is a form of cell death triggered in response to cell trauma, toxins, and hypoxia. Bio-energetic failure is a hallmark of necrosis, and cellular ATP levels decrease before necrotic cell death occurs [17]. NID-1 treatment did not decrease ATP levels, suggesting that the death was not necrotic (Fig. 2C). We further tested whether decreasing ATP levels could enhance NID-1-induced death. Cells were treated with NID-1 in the presence and absence of a glycolysis inhibitor, 2-deoxyglucose (2-DG), in order to decrease cellular ATP levels (Fig. 2C). We found that 2-DG as well as sodium fluoride (NaF), another glycolytic inhibitor, in fact attenuated NID-1-induced cell death (Fig. 2D and 2F), suggesting a requirement for ATP in NID-1 induced death.
Necrosis is also associated with increased reactive oxygen species (ROS) generation. However, NID-1 treatment did not increase ROS levels, whereas erastin, a previously described inducer of oxidative cell death in BJeLR cells [18], increased fluorescence of the ROS sensitive dye H2DCFDA (Fig. 2E). Additionally, cell death was not suppressed by antioxidants such as butylated hydroxytoluene (BHT) or 2-acetylphenothiazine (2-APT) but was suppressed by NaF, a glycolytic inhibitor (Fig. 2F). Antioxidants were tested at the same concentration demonstrated to protect against erastin-induced death in the same cell line [18]. Finally, necrostatin-1 a small molecule inhibitor of programmed necrotic cell death [4] did not prevent NID-1 induced cell death at concentrations up to 10-fold times EC50 needed to inhibit necroptosis (Fig. 2G).
NID-1 does not induce DNA damage, and death is independent of cell proliferation
We tested whether NID-1 induced either nuclear condensation or DNA damage, prominent features of apoptotic cells. In cells stained with the nuclear dye Hoechst, the nuclei of NID-1 treated cells were diffuse and unfragmented, in contrast to cells treated with STS that undergo apoptosis (SI Fig. 2A). We monitored two additional indicators of DNA damage: p53 phosphorylation at serine 15, and nuclear speckling of phospho-H2AX [19]. While the DNA-damaging agent doxorubicin (Dox) increased p53 phosphorylation and phospho-H2AX nuclear specking, both these parameters were unaffected by NID-1 (SI Fig. 2B). Since several chemotherapeutic agents preferentially kill rapidly dividing cells, we tested whether NID-1 specifically targeted proliferating cells. Cells that were arrested in G1/S using hydroxyurea (HU) or aphidocolin (APH) before NID-1 treatment remained at least as sensitive to NID-1 as proliferating cells (SI Fig. 2C). Together, these data indicate that NID-1-induced cell death is executed actively, is non-necrotic, non-oxidative, and proceeds independently of DNA damage or cell proliferation.
NID-1-induced death is characterized by extensive cytosolic vacuolization
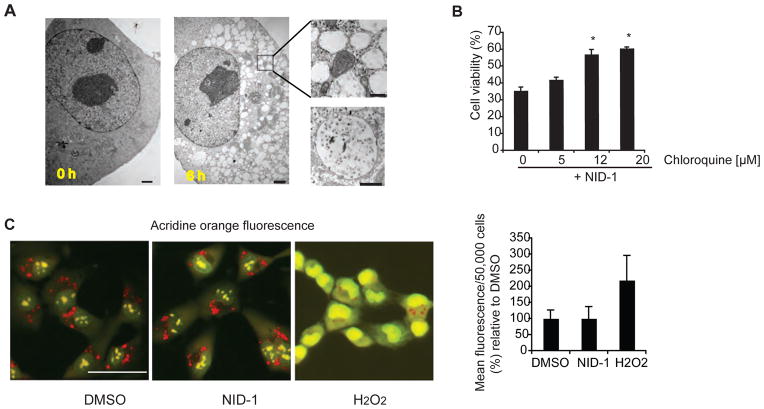
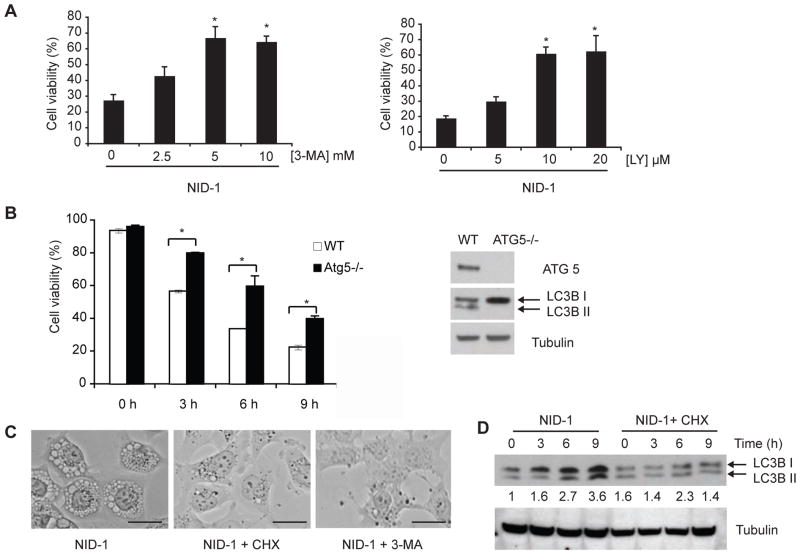
NID-1-treated cells showed prominent cytosolic vacuoles by light microscopy. Vacuolization did not appear to be a general stress response to lethal stimuli in this cellular system as none of the other lethal compounds that induce diverse death pathways, including staurosporine (apoptosis), doxorubicin (DNA damage), H2O2 (oxidative death), erastin (ferroptosis) among others, shared this morphology [7]. Time-course analysis using transmission electron microscopy demonstrated cytosolic vacuoles within 3 to 6 h after NID-1 treatment (Fig. 3A). Multiple vacuoles ranging from 0.5 to 1.0 microns in diameter were observed at the onset of cell death. These vacuoles were lined by single membranes and contained cellular debris, in sharp contrast to classic macroautophagy where double membrane-bound vacuoles are characteristic [20]; the vacuoles in NID-1 treated cells were bound by single membranes even as early as one hour following NID-1 treatment when vacuoles begin to appear (Fig. 3A, lower right panel).
Fig. 3.
NID-1 induced death requires lysosomal activity and is accompanied by cytosolic vacuolization (A) Electron micrographs of BJeLR cells treated with DMSO or NID-1 (2 μg/ml) for 6 h; scale bar = 2μm. Inset: higher magnification of vacuoles at 6 h; scale bar = 500 nm. Lower right panel: EM image of a single-membrane bound vacuole in a NID-1-treated cell at 1 h after NID-1 treatment; scale bar = 500 nm. (B) Cells were pretreated with vehicle, or indicated concentration of chloroquine for 12 h and then treated with NID-1 (10 μg/ml). Cell viability was determined 8 h post-treatment by the trypan blue dye-exclusion assay. Data represent the mean ± S.D. of an experiment performed in duplicate and is representative of two independent experiments, * (p< 0.05). (C) BJeLR cells were treated with DMSO, NID-1 (10 μg/ml) or H2O2 (1 mM) for 4 h and lysosomal accumulation of Acridine Orange (red) was determined. Representative fluorescence micrographs are shown (left three panels, scale bar 50 μm). BJeLR cells were treated with DMSO or NID-1 (10 μg/ml) or H2O2 (0.5 mM) for 2 h and the release of acridine orange (AO) into the cytosol was determined using flow cytometry. The results represent the mean ± S.D. of an experiment with 50,000 cells counted per sample (right panel).
NID-1 induced death involves lysosomal hydrolytic enzymes
Lysosomes are single membrane bound organelles containing hydrolytic enzymes that are active at the low pH within lysosomes and are involved in digesting cellular contents [21]. Because of the extensive vacuolization observed upon NID-1 treatment, we tested whether lysosomal activity was required for death induced by NID-1. Chloroquine is a strong base that selectively accumulates in lysosomes and inhibits lysosomal acidification and thus prevents activation of hydrolytic enzymes [22]. Chloroquine attenuated NID-1-induced cell death (Fig. 3B), suggesting that lysosomal hydrolytic activity is required for NID-1 induced death.
Lysosomes are implicated in cell death through multiple mechanisms. One well-documented mechanism of lysosome-mediated cell death involves aberrant release of lysosomal enzymes into the cytosol by lysosomal membrane permeabilization (LMP) [23]. The pH-sensitive fluorescent dye acridine orange (AO) localizes to lysosomes and emits a red fluorescent signal in the highly acidic environment of the lysosome. Upon LMP, AO is released into the cytosol and emits green fluorescence in the neutral pH of the cytosol [24]. We found that hydrogen peroxide (H2O2) a known inducer of LMP [25], decreased lysosomal (red) and increased diffuse cytosolic (green) fluorescence (Fig. 3C). In contrast, NID-1-treated cells showed no change in lysosomal localization of AO. Flow cytometric quantification of fluorescence in the green channel confirmed that H2O2 increased AO fluorescence by 120% (Fig. 3C, right panel). However, fluorescence in NID-1-treated cells was comparable to DMSO-treated cells. The results average data for 50,000 cells per treatment and a time course analysis at 2, 4 and 6 h after treatment did not show LMP in NID-1 treated cells (data shown for 2 h). Since LMP triggers death via apoptosis or necrosis depending upon the extent of LMP [26], the lack of LMP upon NID-1 treatment further supports that the selection criteria identified a compound that induces death that is neither apoptotic nor necrotic.
NID-1 induced death involves cathepsin L
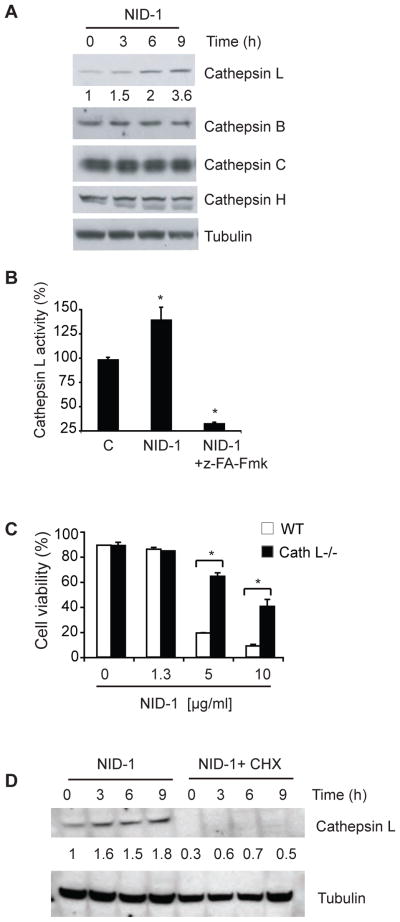
Since LMP was not involved in NID-1 induced death, yet cell death was inhibited by chloroquine, we tested whether NID-1-induced death was dependent upon lysosomal activity. First, we tested whether NID-1 altered the expression of lysosomal enzymes. Cathepsins comprise a large family of lysosomal proteases, of which cathepsin L and B are involved in lysosomal degradation [27]. We observed a specific increase in cathepsin L levels upon NID-1 treatment, whereas other detectable cathepsins (B, C, and H) were unchanged (Fig. 4A). [28] The increase in cathepsin L protein was accompanied by enhanced (25–40%) cathepsin L enzymatic activity that was suppressed by a cathepsin inhibitor (Fig. 4B). Additionally, mouse embryo fibroblasts derived from cathepsin L knock-out mice [29] were resistant to NID-1-induced cell death (Fig. 4C). Finally, cycloheximide, the protein synthesis inhibitor that suppressed NID-1 induced death also suppressed cathepsin L induction upon NID-1 treatment (Fig. 4D), suggesting that cathepsin L contributes to NID-1-induced cell death. We attempted to determine if NID-1 altered cathepsin L localization. However, the cathepsin L immunofluorescence staining was very weak (possibly due to sub-threshold abundance) preventing determination of Cathepsin L localization. However, the localization of Cathepsin C, another lysosomal cathepsin was unaltered by NID-1 treatment (SI Fig. 3C). Similarly, lysotracker staining of cells did not reveal major alteration in abundance or localization of lysosomes upon NID-1 treatment (data not shown). Together with the acridine orange data (Fig. 3C) these results suggests that NID-1 did not induce LMP or substantial leakage of cathepsins into cytosol, though more subtle leakage of cathepsins cannot be excluded.
Fig. 4.
NID-1-induces Cathepsin-L-dependent cell death. (A) NID-1 (5 μg/ml) treated BJeLR cells were analyzed by western blotting for the indicated cathepsins. Tubulin was the loading control. The levels of cathepsin L were quantitated, normalized to tubulin and indicated below the cathepsin L blot. (B) Cells were treated with vehicle control, NID-1 (5 μg/ml) alone or with the cathepsin inhibitor z-FA-Fmk (50 μM) and harvested after 3 h. Cathepsin L activity was measured by monitoring cleavage of AFC-conjugated substrate. Data are displayed relative to control (set as 100%), and are representative of two independent experiments where mean ± S.D. of experiments performed in duplicate, * (p< 0.05). (C) Embryonic fibroblasts from wild type and cathepsin L knock-out mice were treated with NID-1. Cell viability was determined 8 h post-treatment by the trypan blue dye-exclusion assay. Data represent the mean ± S.D. of an experiment performed in duplicate and are representative of three independent experiments, * (p< 0.05). (D) BJeLR cells were untreated or pretreated with CHX (3 μM) for 12 h and then with NID-1 (5 μg/ml) and the levels of indicated proteins were analyzed by western blotting. The levels of cathepsin L were quantitated, normalized to tubulin and indicated below the cathepsin L blot.
NID-1 engages the autophagic machinery to activate a process distinct from autophagy
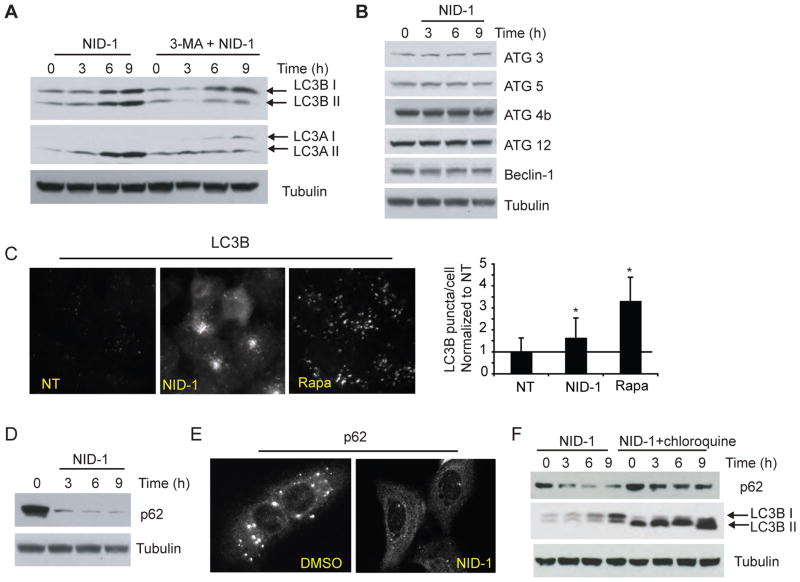
The vacuoles induced by NID-1 treatment were clearly distinct from characteristic double-membrane autophagic vacuoles. However, recent studies have uncovered a role for autophagic machinery in generating single membrane-bound vacuoles that are involved in phagocytosis and entosis [12, 13]. Therefore we assessed whether components of autophagic machinery were involved in NID-1 induced vacuolization and cell death. We evaluated the levels of two isoforms (A and B) of microtubule-associated light chain 3 (LC3) protein. A hallmark of autophagy, as well of single membrane vacuoles in entosis and phagocytosis is the conversion of cytosolic LC3-I to autophagic vacuole-bound LC3-II [30]. [12, 13]; membrane-bound LC3-II has increased electrophoretic mobility, allowing these two forms to be distinguished by western blot [30]. NID-1 treatment increased levels of both LC3A and of LC3B, and the conversion of both to their autophagic vacuole bound forms (II) (Fig. 5A). Transcriptional increase in the levels of LC3, is associated with increased autophagy in several mammalian systems [31, 32]. While levels of LC3 were increased, expression of other ATG proteins remained unchanged (Fig. 5B). The recruitment of LC3 from the cytosol to entotic vacuoles/phagosomes, is also detectable by immunofluorescence as increased formation of LC3B puncta [12, 32]. NID-1 treatment increased LC3B puncta formation by 60% (Fig. 5C). Rapamycin, an inducer of classical macroautophagy, caused an even greater increase in puncta formation. However, the pattern of LC3B puncta induced by NID-1 was distinct from that induced by rapamycin: NID-1 induced large coalescent clusters of LC3B puncta; in contrast, rapamycin treatment resulted in an evenly distributed increase in discrete puncta (Fig. 5C). Next, we measured the levels of p62 (sequestosome-1) protein. p62 targets substrates for degradation and is itself degraded by autophagy [33]. p62 levels rapidly decreased upon NID-1 treatment (Fig. 5D). p62 exists prominently as cytosolic puncta, and these puncta are sites of cargo recruitment for degradation and are themselves subsequently degraded [34]. Immunofluorescence revealed that p62 puncta were rapidly depleted within 3–6 hours of NID-1 treatment (Fig. 5E). We also found that chloroquine, an inhibitor of lysosomal degradation [35], attenuated NID-1-induced p62 degradation (Fig. 5F). Since LC3B-II is also recycled and degraded by lysosomal enzymes, chloroquine causes accumulation of LC3B-II. As expected, chloroquine increased LC3B-II levels, however it could not prevent the further increase in LC3-I caused by NID-1 (Fig. 5E), suggesting that NID-1 increases vacuole formation rather than inhibiting the processing of these vacuoles. Together, these data suggest that NID-1 activates a cellular degradative process that is distinct from macroautophagy but involves autophagic-lysosomal machinery.
Fig. 5.
NID-1 activates autophagic machinery. (A) Cells were untreated or pretreated with 3-methyladenine (3-MA, 10 mM) for 12 h and then with NID-1 (5 μg/ml) and harvested for western blot analysis, and analyzed for LC3 A and B protein levels. Tubulin was used as a loading control. (B) NID-1 (5μg/ml) treated BJeLR cell lysates were subjected to western blotting for several proteins involved in autophagy. (C) U2OS cells were untreated, or treated with NID-1 (5 μg/ml) or rapamycin (10 nM) for 9 h and then fixed and stained for LC3B immunofluorescence. The puncta in the images were quantified using Image J software (NIH), n=10, * (p<0.05, student’s t-test). (D) NID-1 (5 μg/ml)-treated BJeLR cells were analyzed for p62 by western blotting. (E) U2OS cells were treated with vehicle (DMSO) or NID-1 (10 μg/ml) and the cells were fixed after 6 h treatment. Cells were stained for p62 by immunofluorescence as described in materials and methods. Representative micrographs are shown. (F) BJeLR cells were pretreated with chloroquine (12 μM) for 10 h, or untreated, and then treated with NID-1. Cell lysates were subjected to western blotting for indicated proteins.
NID-1-induced death is PI3K and ATG5 dependent
The initial step in the formation of single membrane vacuoles reported in entosis and phagocytosis is a phosphoinositol 3-kinase (PI3K) dependent step [12]. We found that structurally diverse inhibitors of PI3K, 3-methyl adenine (3-MA) and LY294002, that inhibit biogenesis of single-membrane vacuoles in entosis [12, 36], both suppressed NID-1-induced cell death (Fig. 6A). 3-MA treatment also attenuated the conversion of LC3-I to LC3-II (Fig. 5A). Furthermore, these inhibitors suppressed NID-1-induced death in several cell lines (SI Fig. 3). A critical autophagy gene, ATG5 is involved in the formation of autophagosomes and of single-membraned vacuoles in entosis [12, 37, 38]. As autophagy can act as a pro-survival or death-inducing mechanism in response to various stimuli, deletion of ATG5 also has differing effects depending on the nature of cellular insult [39]. While ATG5 loss was not protective against staurosporine or ER stress-induced death [39], we observed that genetic deletion of ATG5 attenuated NID-1 induced death in MEFs derived from ATG5 knockout mice compared to wild-type MEFs (Fig. 6B). We verified that ATG5 knockout MEFs lack ATG5 expression and therefore do not convert LC3B-I to II (Figure 6B). (Fig. 6B), indicating the lack of LC3 recruitment to vacuoles. Finally, we found that both protein synthesis inhibition with cycloheximide, and PI3K inhibition with 3-MA blocked vacuole formation induced by NID-1 (Fig. 6C), and cycloheximide applied at the same concentration that was protective against cell death also prevented the induction of LC3B upon NID-1 treatment (Fig. 6D). These data indicate that NID-1 activates a cytosolic vacuolization process distinct from classical autophagy, yet cell death is dependent, at least in part, on components of autophagic machinery.
Fig. 6.
NID-1 induced cell death is PI3K-and-ATG5-dependent. (A) BJeLR cells were pretreated with vehicle, 3-MA or LY 294002 for 12 h and then with NID-1 (10 μg/ml). Cell viability was measured by trypan blue dye-exclusion assay 8 h after treatment. Data represent the mean ± S.D. of an experiment performed in duplicate and is representative of three independent experiments, * (p< 0.05). (B) Mouse embryonic fibroblasts derived from WT and ATG5 knockout mice were treated with NID-1 (5 μg/ml) and cell viability was determined at indicated times using trypan blue dye-exclusion. The data is representative of 2 independent experiments, * (p< 0.05). Levels of ATG5 and LC3B II conversion in WT and ATG5−/− MEFs were analyzed by western blotting (right panel). (C) BJeLR cells were untreated, treated with CHX (3 μM) or 3-MA (10 mM) for 12 h and then with NID-1 (5 μg/ml) for 9 h. Light photomicrographs of cells demonstrating vacuoles (scale bar, 25 μm). (D) BJeLR cells were untreated or pretreated with CHX (3 μM) for 12 h and then with NID-1 (5 μg/ml) and the levels of indicated proteins were analyzed by western blotting. The levels of LC3B were quantitated, normalized to tubulin and are indicated below the blot.
NID-1 is protective in a Huntington’s Disease model
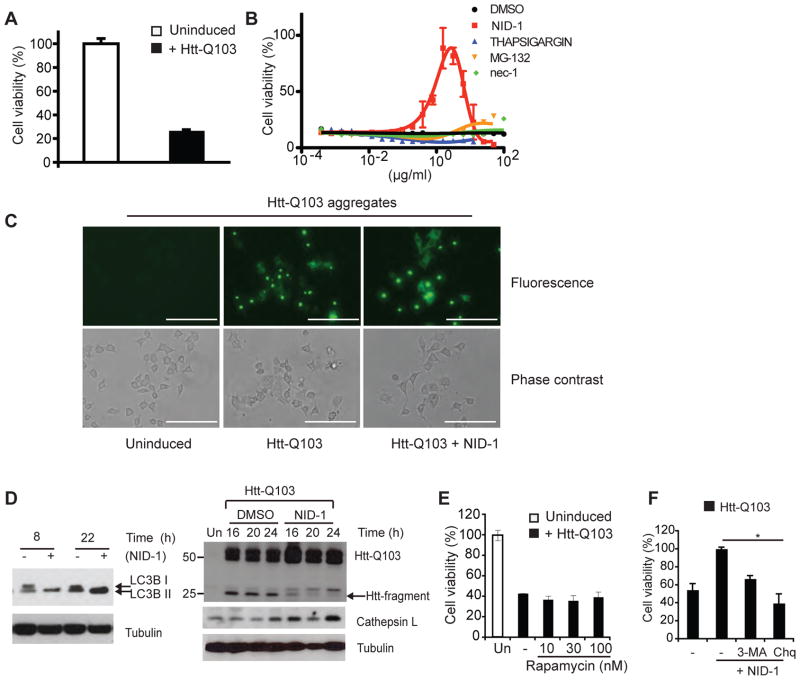
Modulation of classical and non-canonical autophagic activity has been shown to be protective in several neurodegenerative disease models [40]. We tested whether NID-1 may have a protective effect in one such disease context. Huntington’s Disease is an autosomal dominant neurodegenerative disease caused by mutant huntingtin allele that encodes the huntingtin (htt) protein with a polyglutamine repeat expansion (> 36 glutamine). We used a previously described PC12 cell culture model of Huntington’s disease [41] wherein inducible expression of mutant htt containing 103 glutamine repeats (htt-Q103) causes cell death within 48 hours (Fig. 7A). Htt-Q103 is expressed as a fusion protein with GFP (Green Fluorescence Protein) to enable monitoring of protein aggregates in live cells [41]. NID-1 reproducibly rescued mutant-htt-induced cell death, whereas modulators of several other pathways including necroptosis (nec-1), ER stress (thapsigargin), and proteosomal degradation (MG132) were ineffective. We noted that higher concentrations of NID-1 were toxic, suggesting a window of concentration range for the protective effect. NID-1 did not alter mutant htt expression or aggregate formation, indicating that NID-1 did not simply turn-off expression or enhance degradation of mutant protein (Fig. 7C and 7D). Furthermore, rapamycin was unable to rescue cell death in this model, indicating that induction of classical macroautophagy was not involved in NID-1’s rescue (Fig. 7E). However, NID-1 at the protective concentration (1 μg/ml) induced LC3B and cathepsin L in PC12 cells (Fig. 7D). Additionally, both 3-MA and chloroquine partially inhibited NID-1’s protective effect (Fig. 7F) suggesting that NID-1 affects the same pathway in PC12 cells as in BJeLR cells. We also observed that the levels of a 25kD fragment of mutant htt were lower in NID-1 treated cells compared to DMSO treated cells suggesting differential processing of mutant htt in NID-1 treated cells (Fig. 7D, right panel). Since mutant htt is processed to smaller fragments in vivo by several proteases including caspases and cathepsins; and these processed fragments are linked to toxicity, these results suggest that NID-1 modulates a pathway distinct from classical autophagy that protects against mutant htt-toxicity, possibly by altered processing of mutant htt.
Fig. 7.
NID-1 suppresses mutant-huntingtin-induced cell death. (A) PC12 cells were un-induced or induced to express htt-Q103. Cell viability was determined 48 h after induction of mutant htt expression using the Alamar blue assay. Data represent the mean ± S.D. of an experiment performed in triplicate is representative of three independent experiments. (B) PC12 cells were induced to express htt-Q103 and treated with a dose-dilution of indicated compounds. Cell viability was determined 48 h later as in (A). Data represent the mean ± S.D. of an experiment performed in triplicate. The rescue by NID-1 was confirmed in three independent experiments. (C) PC12 cells were un-induced or induced to express mutant htt-GFP fusion protein and then treated with NID-1 (1 μg/ml) or DMSO. Htt-Q103 aggregates were visualized using fluorescence microscopy after 15 hr of treatment. Scale bar = 200 μm. (D) PC12 cells expressing mutant htt-Q103 were incubated with control (DMSO) or NID-1 (1 μg/ml), harvested at the times indicated, and levels of indicated proteins were analyzed by western blotting. Uninduced (Un) cells were controls for lack of htt-Q103 expression. (E) PC12 cells were uninduced or induced to express htt-Q103 and treated with a dose-dilution of rapamycin. Cell viability was determined 30 h later. The results represent the mean ± S.D. of an experiment performed in duplicate and is representative of 2 independent experiments. (F) PC12 cells were untreated or pretreated with 3-MA (5mM) or chloroquine (Chq, 15 μM) for 15 h. Cells were then treated with NID-1 (1μg/ml) and htt-Q103 was induced. Cell viability was determined 30 h after induction and viability of NID-1 treated cells was arbitrarily set as 100% (*, p< 0.05, student’s t-test). The results represent the mean ± S.D. of an experiment performed in duplicate and is representative of 2 independent experiments.
Discussion
Most paradigms for identifying cell death programs have used genetic screens in eukaryotes such as C. elegans, overlooking death programs that are unique to mammals. Large-scale RNAi screens address this challenge to some extent, but have limitations [42, 43]. As an alternative, we used a tiered small molecule screening approach to identify uncharacterized cell death programs in mammalian cells. Having initially identified 1,980 lethal compounds in a primary screen of 69,612 compounds, we selected compounds that maintained their lethality in the presence of inhibitors of necrosis and apoptosis, and further selected those requiring new protein synthesis to induce cell death. Applying these criteria, we identified NID-1, a potent inducer of non-canonical cell death in diverse cell types.
Cell death induced by NID-1 was distinct from apoptosis or necrosis. Though NID-1-induced death shares some features of autophagy, such as suppression by PI3K inhibitors, ATG5 dependence, degradation of an autophagy substrate, p62 (sequestosome-1) and increased LC3 puncta, however, NID-1’s effects are clearly distinct from classical macroautophagy. For example, the cytosolic vacuoles induced upon NID-1 treatment are bound by single membranes and appear to be either empty or contain some cellular debris (Fig. 3A). This is in contrast to double-membrane-bound vacuoles containing electron dense organelles reported in macroautophagy [44], as well as in other specialized forms of autophagy [45–48]. Although NID-1 may induce rapid formation of single membrane-bound autophagolysosomes by the fusion of autophagosomes to lysosomes, we did not observe double membrane vacuoles even as early as 1 h after NID-1 treatment when vacuoles were just beginning to form (Fig. 3A, lower right panel). Additionally, NID-1-induced LC3II puncta were distinct from those induced by an activator of macroautophagy, rapamycin, (Fig. 5C). Recent studies have demonstrated the involvement of autophagic machinery in the generation of single membrane-bound vacuoles that are employed in phagocytosis and entosis, a form of cellular engulfment that results in the degradation of neighboring cells [12, 13]. In these cases, LC3 is recruited to phagosomes and entotic vacuoles in a PI3K-dependent and ATG-dependent manner, phagosomes then fuse with lysosomes, and vacuolar contents are digested. Such overlap in the use of cellular machinery is well documented in other cellular processes; for example, electron transport and apoptosis both utilize cytochrome c [49]. Our results suggest that NID-1-induced cell death employs a similar overlapping use of the autophagic machinery.
We also found a role for cathepsin L in NID-1 induced cell death. NID-1 induced cathepsin L protein and enzymatic activity, and NID-1 induced death was suppressed in MEFs derived from cathepsin L knock-out mice (Fig. 4A–D). A related question was the role of LMP and sub-cellular site of cathepsin L action in NID-1 induced death. NID-1 did not induce LMP (Fig. 4C). Though we were unable to detect cathepsin L by immunofluorescence, however, NID-1 treatment did not alter the cellular localization of cathepsin C, a related lysosomal cathepsin, (SI Fig. 3C). Together these data suggest that NID-1 is unlikely to cause major lysosomal leakage, though we cannot exclude a scenario where cathepsin L slowly leaks into the cytosol and contributes to cell death by degrading cytosolic substrates. The site of cathepsin L action and its relevant substrates in NID-1 induced death remains to be determined.
An interesting result of our study was the protection conferred by NID-1 in a Huntington’s Disease model. Alterations in autophagy are implicated in numerous neurodegenerative diseases, and modulation of both canonical and non-canonical autophagy pathways are being pursued as therapeutic avenues [50]. Our results suggest that induction of autophagic machinery was not responsible for the rescue by NID-1, since rapamycin, an inducer of classical autophagy was not protective in PC12-Q103 cells (Fig. 7E) and NID-1 did not decrease mutant htt levels. However, NID-1 treatment increased cathepsin L and decreased the levels of a 25 kD mutant htt fragment. Cathepsin L is implicated in processing mutant htt and polyglutamine containing protein fragments within lysosomes [51, 52] and increased expression of cathepsins can suppress mutant htt toxicity in primary neurons [53]. Furthermore, cathepsin L and cathepsin B double knockout mice show massive neurodegeneration [54] that is reversed by re-expressing cathepsin L [55]. Thus altered processing of mutant htt by increased cathepsin L is a potential mechanism for the rescue. Alternatively, other substrates of cathepsin L or lysosomal enzymes that are involved in mutant htt toxicity may mediate the rescue. We observed robust protection against mutant htt toxicity by lower concentrations of NID-1 (up to 2 μg/ml), while higher concentrations were toxic (Fig. 7B), suggesting that modest activation of the NID-1-induced pathway may be protective. It would also be interesting to test whether NID-1 is protective in models of other neurodegenerative disorders, such as Parkinson’s and Alzheimer’s disease, where altered autophagy is implicated in pathogenesis [56].
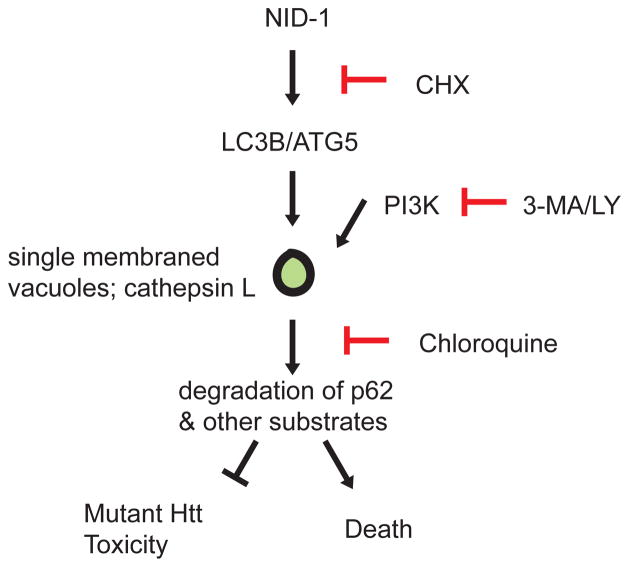
We propose a model of NID-1-induced death: NID-1 induces formation of single membrane-bound vacuoles in a PI3K-dependent and ATG5-dependent manner. Cellular substrates essential for survival are recruited by p62 to these vacuoles and digested by lysosomal proteases such as cathepsin L. Depending on the cellular context, a loss of these cellular components causes cell death, or in the case of mutant htt induced toxicity, survival (Fig. 8). In support of this model, perturbations such as serum-deprivation-induced cell death involve transient intralysosomal cathepsin L activation [57], and prostaglandin J2-induced cell death that is characterized by a non-autophagic cytosolic vacuolization, but requires the autophagic machinery [58]. Examples of physiological non-apoptotic cell death involving vacuolization include developmental linker-cell death in C. elegans [59], and developmental spinal neuronal death in caspase deficient mice [3]. Prominent cellular vacuolization and cell death is also induced by pathogenic organisms such as H. pylori [60] and M. pneumoniae [61] via specific cytotoxins. The mechanism(s) of cell death in these processes remain unclear, but underscore the relevance of alternative cell death paradigms involving cellular vacuolization to normal physiology and diseases (reviewed in [62]).
Fig. 8.
Model of NID-1’s effect on cell death. NID-1 induces LC3A, LC3B and formation of single-membrane bound vacuoles in a protein synthesis, PI3K/ATG5 dependent manner. These vacuoles are likely sites of degradation of key cellular substrates whose degradation causes cell death. However in the context of mutant huntingtin toxicity, the substrates degraded may be involved in toxicity and thus their degradation confers protection.
Conclusions
Non-canonical cell death pathways are important in physiological and pathological processes but remain poorly understood. Using a small molecule screening approach, we have identified an inducer of a non-apoptotic ATG5-Cathepsin L cell death that involves the autophagic machinery, yet is clearly distinct from autophagy. Modest activation of this pathway also has a protective role in a model of polyglutamine toxicity. Further dissection of the cellular pathway involved in this novel cell death may reveal the link(s) between autophagic machinery and cell death and its relevance to physiological and pathological processes including neurodegeneration.
Materials and Methods
High throughput Cell Viability Screen
Cells were seeded in 384-well black, clear-bottom plates using the Biomek FX dual arm with multi-channel-L (Beckman Coulter). The Biomek was also used to add compound treatments in an arrayed format. Library compounds are stored as DMSO stocks and diluted 1:75 in media to create 10X daughter plates. Final treatments were added 1:10 to cells in media for a final concentration of 5μg per ml on assay plates. The total assay volume was 40 μl per well. Screening was performed in triplicate. Alamar Blue (AB) was added after 24 h of compound treatment. Cells were then incubated for an additional 12 h in the presence of the dye, and fluorescence intensity was detected by a Victor3 plate reader (Perkin Elmer).
Chemicals and Antibodies
All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. Cathepsin (Z-FA-fmk) and Cathepsin L inhibitor I (Catalog no. 219421) were from EMD biosciences. Antibodies were obtained from Cell Signaling Technology and Santa Cruz Biotechnology except secondary antibodies: IRDye 800CW Goat-anti-Rabbit, IRDye 680CW Goat-anti-Mouse (LI-COR); Alexa Fluor® 594 goat anti-mouse IgG, Alexa Fluor® 488 goat anti-rabbit IgG (Invitrogen). Primary antibodies for immunofluorescence: rabbit anti-LC3B (Cell Signaling, cat no. 2775), phospho-H2AX (Cell Signaling, cat no. 9718).
Compound Libraries
Chemical libraries were composed of a combination of synthetic and natural product-like compounds. Small molecules were obtained from different providers including TimTec, IBS, Chembridge, Asinex, and Life chemicals. Also screened were compounds from the Annotated Compound Library (ACL) of bioactive compounds selected for their chemical diversity [63] and the Blood-Brain Barrier (BBB) set of compounds selected for chemical properties suitable for penetrating the blood-brain barrier. All compounds were prepared as 4 mg/ml solutions in DMSO in 384-well polypropylene plates (Greiner, cat. #781280) and stored at −80°C.
Chemical Characterization of NID-1
The structure of NID-1 was confirmed by 1H-NMR, 13C-NMR and HRMS:
1H-NMR (400MHz, MeOD): δ 8.00 (1H, J = 4.3 Hz, d), 7.77 (1H, J = 15Hz, d), 7.47-s7.45 (1H, m), 7.40 (1H, J = 4.2 Hz, d), 7.35-7.23 (3H, m), 6.98 (1H, J = 16 Hz, d), 2.70 (2H, J = 7.6 Hz, q), 1.23 (3H, J = 7.6 Hz, t); 13C-NMR (100MHz, MeOD): δ 164.83, 152.92, 146.70, 139.01, 134.88, 132.68, 129.62, 129.36, 129.01, 126.97, 126.33, 124.93, 24.36, 13.81; High Resolution Mass Spectrum (m/z): (MH)+ calculated for C15H14N2O3S, 303.0725; found 303.0803.
Trypan Blue dye-exclusion assay
Samples of 1 ml of cells in culture media were prepared for analysis of cell viability. The ViCell (Beckman Coulter) performs trypan blue exclusion and images 100 frames per sample to count the total number of cells per sample and the percentage of live, trypan blue excluding, cells. Trypan blue is a viability dye, which is excluded by the intact membrane of live cells, and stains positive in dying cells whose membranes have been permeabilized. Error bars represent standard deviation from mean intensity based on duplicate treatments. For determining cell viability in attached cells, after treatment, the supernatant was collected and the cells were washed twice with 2 mL of PBS and also collected to capture all detached cells. Adherent cells were trypsinized, harvested and collected separately.
Cathepsin L activity assay
Cathespsin L activity assay kit (Catalog no. K142-100) was purchased from Biovision Inc. (Mountainview, CA) and the assay performed based on manufacturer instructions.
Alamar Blue assay
Alamar Blue (Invitrogen) is a non-toxic, cell viability indicator that contains a cell permeable nonfluorescent molecule, resazurin, which is converted to the fluorescent molecule resorufin upon reduction by living cells. 50% Alamar Blue solution was prepared in cell culture media and added 1:4 to assay plates for a final concentration of 10%. Error bars represent standard deviation from mean intensity based on treatments applied in triplicate. Graphs were created using Prism software and best-fit curves displayed were calculated using a nonlinear regression of the log(agonist) vs. response with a variable slope.
Soft Agar Colony Formation Assay
BJeLR or HT-1080 cells were seeded at 1×105 in 6-well plates, allowed to adhere overnight, and treated with 5 μg/mL NID-1 for time points indicated. Cells were then washed with fresh media, trypsinized and counted, and 2×104 cells were plated into 2 mL of 0.3% agar-media mixture on top of a 0.6% agar-media layer. Cells were cultured for two weeks, then stained with 0.005% crystal violet and imaged.
Western Blotting
For western blots, cells were collected, and washed with cold PBS. Cell lysates were prepared, denatured, separated by SDS-polyacrylamide gel electrophoresis on 4–20% gradient Tris-HCl gels (Invitrogen), transferred to a polyvinylidene difluoride membrane, probed with the indicated primary antibodies overnight at 4°C, and HRP conjugated secondary antibodies for 1 h at room temperature. Blots were developed using ECL (Pierce) and exposed to photographic film. The bands on scanned films were quantitated using Image J software (NIH).
Immunofluorescence
Cells were seeded at 50,000 cells per well onto coverslips in 6-well plates. After treatment, cells were fixed in −20°C methanol for 7 m on ice and rinsed three times in TBS. Slips were incubated in primary antibody diluted 1:100 in 1% goat serum in TBS for 2 h at RT, rinsed for 10 min in TBS, incubated in secondary antibody diluted 1:400 in 1% goat serum in TBS for 1 h at RT and rinsed again for 10 m in TBS. Slips were mounted in Fluoromount containing 2 μg/ml Hoechst and sealed. Samples were imaged using a 63X objective.
Reactive oxygen species detection
Cells were treated in 6-well plates. Medium was removed and 5-(6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Invitrogen) was added to cells and incubated at 37°C for 30 minutes. The dye is non-fluorescent until the acetate groups are cleaved away by intracellular esterases and the molecule is oxidized by reactive oxygen species in the cell. Plates were imaged to detect fluorescein intensity using an inverted fluorescent microscope.
Hoechst Staining
Hoechst nucleic acid stain (Invitrogen) was diluted 1:1500 in PBS for a final concentration of 10 μM and added to cells on cover slips for 10 min at room temperature. Cells were washed with PBS to decrease background signal.
Electron Microscopy
Cells were fixed with 2.5% glutaraldehyde in 0.1M Sorenson’s buffer (PH 7.2) for at least one hour. Cells were then post-fixed with 1% OsO4 in Sorenson’s buffer for 1 h. En bloc staining was performed using 1% tannic acid. After dehydration, cells were embedded in a mixture of Lx-112 (Ladd Research Industries, Inc.) and Embed-812 (EMS, Fort Washington, PA). Thin sections were cut on the MT-7000 ultramicrotome. The sections were stained with uranyl acetate and lead citrate to enhance contrast between various cellular structures and examined under a JEOL JEM-1200 EXII electron microscope. Pictures were taken on an ORCA-HR digital camera (Hamamatsu) and recorded with an AMT Image Capture Engine.
LC3B Puncta Quantification
LC3B puncta per image were quantified with ImageJ Analyze Particles function, lower threshold set at 80, and higher threshold set at 255. Cells per image were counted visually in order to determine puncta/cell. At least ten images were analyzed per condition. Significance was determined using a two-tailed, two sample equal variance t-test.
Atg5−/− and cathepsin L−/− MEF Viability Analysis
Atg5−/−, cathepsin L−/− MEFs and their corresponding littermate WT MEFs that had been immortalized with SV40 large T antigen were kindly provided by N. Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) and C. Borner (Albert Ludwigs University, Freiburg, Germany) and have been previously described [29, 38]. Cells were maintained in DMEM with 10% FBS and 5% CO2 at 37°C. For viability analysis, cells were seeded at 5×104 cells/well of a 6-well plate and allowed to adhere overnight. Cells were then treated with 10 μg/mL NID-1 for 8h, and viability was analyzed by trypan blue exclusion method with quantification performed by Vi-Cell (Beckman Coulter). Differential interference contrast (DIC) images at the indicated time points were acquired using a 20X objective.
PC12 viability assay
PC12 cells that conditionally express a mutant huntingtin protein containing a 103 polyglutamine expansion fused to enhanced GFP (mHtt-GFP) upon tebufenozide (Teb) induction were used to test for NID-1 rescue against mutant huntingtin toxicity. Teb-induced and uninduced cells were seeded in 384-well plates and treated with 2-fold dose-response series of the indicated compounds at the time of induction. Survival following 48 h of induction and compound treatment was measured using Alamar Blue. Percent survival was determined with respect to viability of uninduced cells, and Prism software was used to fit curves.
Lysosomal membrane permeabilization assay
BJeLR cells were seeded in six well dishes at a density of 200,000 cells/well and incubated overnight. Cells were incubated in media containing 0.1 μg/mL acridine orange for 15 min, washed once with regular media, followed by incubation with regular media for an additional 15 min. Drug treatments of vehicle (DMSO), NID-1 (10 μg/mL) or H2O2 (500 μM) were performed for 2, 4 or 6 hours. After drug treatment, cells were washed once with 1 mL of PBS, trypsinized, harvested and pelleted (3 min at 1000rpm). The cell pellet was resuspended in 1 mL of PBS again and re-pelleted. Cells were resuspended in 500 μL of HBSS and transferred to a micro-centrifuge tube prior to analysis on an Accuri C6 flow cytometer. Fluorescence in 50,000 cells were analyzed per condition and time point.
Synthesis of NID-1
Methyl 3-(5-nitrothiophen-2-yl) prop-2-enoate (2) procedure was modified from [64]. Trimethyl phosphonoacetate (4.7 mmol) was added to a solution of LiCl (3.7 mmol) in 10 mL CH3CN and allowed to stir for 5 minutes. Et3N (3.7 mmol) was added and the solution was stirred for 10 minutes, at which point a solution of 5-nitro-2-thiophene-carboxaldehyde (1, 3.1 mmol) in 5 ml CH3CN was added. A yellow precipitate formed. The solution was stirred overnight. 20 ml Et2O and 30 ml 2M NH4Cl were added and then separated. The aqueous phase was extracted 3 times with 20 ml Et2O. The combined organic layers were washed with brine and dried over Na2SO4. TLC showed two spots in 25% EtOAc in hexanes, one with RF ≈ 0.05 corresponding to the starting aldehyde and the other at RF ≈ 0.15. Column chromatography (25% EtOAc in hexanes) separated the spots and gave 1.9 mmol (61%) of the expected product. 1H NMR (400 MHz, CDCl3) δ 7.85 (d, 1H), 7.67 (d, 1H), 7.18 (d, 1H), 6.42 (d, 1H), 3.83 (s, 3H). 3-(5-nitrothiophen-2-yl)prop-2-enoic acid (3). The procedure was modified from [65]. 2 (0.7 mmol) was dissolved in 6 ml of THF:H2O (3:1) and the solution was cooled in ice water. LiOH (2.1 mmol) was added and the solution was stirred for 30 minutes in the ice bath. The solution turned from yellow to dark red. The ice bath was removed, allowing the reaction to warm to room temperature, and stirred for 20 hours. 1% HCl was added to pH 3. The solution was extracted 3 times with EtOAc, washed with brine, and dried over NaSO4. The solvent was evaporated to give 0.64 mmol (92%) of the expected product. 1H NMR (400 MHz, CD3OD) δ 7.95 (d, 1H), 7.72 (d, 1H), 7.39 (d, 1H), 6.50 (d, 1H).
Supplementary Material
Acknowledgments
We thank Kristy Brown for assistance in preparing electron microscopy samples, Craig Thompson for providing the Bax−/− Bak−/− MEFs and Mark Czaja and Noboru Mizushima for providing the Atg5−/− MEFs. This research was supported by grants from the US National Institutes of Health R01CA097061, R01GM085081, R01CA161061, the Arnold and Mabel Beckman Foundation and NYSTAR. B.R.S. is an Early Career Scientist of the Howard Hughes Medical Institute.
Abbreviations used
- APH
aphidocolin
- ATG
Autophagy gene
- DKO
double knock out
- 3-MA
3-methyl adenine
- HU
Hydroxyurea
- MEF
mouse embryo fibroblasts
- NID
Novel inducer of death
- PI3K
phosphotidyl inositol 3-kinase
- RIP-1
Receptor Interacting Protein -1
- STS
staurosporine
References
- 1.Kreuzaler P, Watson CJ. Killing a cancer: what are the alternatives? Nat Rev Cancer. 12:411–424. doi: 10.1038/nrc3264. [DOI] [PubMed] [Google Scholar]
- 2.Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev. 2010;24:2592–2602. doi: 10.1101/gad.1984410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oppenheim RW, Flavell RA, Vinsant S, Prevette D, Kuan CY, Rakic P. Programmed cell death of developing mammalian neurons after genetic deletion of caspases. J Neurosci. 2001;21:4752–4760. doi: 10.1523/JNEUROSCI.21-13-04752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 5.Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–979. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 6.Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 7.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B, 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 10.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 11.Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 14.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 15.Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 16.Antignani A, Youle RJ. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol. 2006;18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Skulachev VP. Bioenergetic aspects of apoptosis, necrosis and mitoptosis. Apoptosis. 2006;11:473–485. doi: 10.1007/s10495-006-5881-9. [DOI] [PubMed] [Google Scholar]
- 18.Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, Smith R, Lessnick SL, Sahasrabudhe S, Stockwell BR. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalil A, Morgan RN, Adams BR, Golding SE, Dever SM, Rosenberg E, Povirk LF, Valerie K. ATM-dependent ERK signaling via AKT in response to DNA double-strand breaks. Cell Cycle. 2011;10:481–491. doi: 10.4161/cc.10.3.14713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, Lynch JP, Uehara T, Sepulveda AR, Davis LE, Winkler JD, Amaravadi RK. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson AC, Appelqvist H, Nilsson C, Kagedal K, Roberg K, Ollinger K. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis. 2010;15:527–540. doi: 10.1007/s10495-009-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusuzaki K, Murata H, Takeshita H, Hashiguchi S, Nozaki T, Emoto K, Ashihara T, Hirasawa Y. Intracellular binding sites of acridine orange in living osteosarcoma cells. Anticancer Res. 2000;20:971–975. [PubMed] [Google Scholar]
- 25.Zdolsek J, Zhang H, Roberg K, Brunk U. H2O2-mediated damage to lysosomal membranes of J-774 cells. Free Radic Res Commun. 1993;18:71–85. doi: 10.3109/10715769309147344. [DOI] [PubMed] [Google Scholar]
- 26.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 27.Dennemarker J, Lohmuller T, Muller S, Aguilar SV, Tobin DJ, Peters C, Reinheckel T. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391:913–922. doi: 10.1515/BC.2010.097. [DOI] [PubMed] [Google Scholar]
- 28.Ravanko K, Jarvinen K, Helin J, Kalkkinen N, Holtta E. Cysteine cathepsins are central contributors of invasion by cultured adenosylmethionine decarboxylase-transformed rodent fibroblasts. Cancer Res. 2004;64:8831–8838. doi: 10.1158/0008-5472.CAN-03-2993. [DOI] [PubMed] [Google Scholar]
- 29.Oberle C, Huai J, Reinheckel T, Tacke M, Rassner M, Ekert PG, Buellesbach J, Borner C. Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death Differ. 2010;17:1167–1178. doi: 10.1038/cdd.2009.214. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y, Pei Y, Wan B, Guo J, Yu L. Molecular cloning and characterization of rat LC3A and LC3B--two novel markers of autophagosome. Biochem Biophys Res Commun. 2006;339:437–442. doi: 10.1016/j.bbrc.2005.10.211. [DOI] [PubMed] [Google Scholar]
- 31.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 32.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zois CE, Giatromanolaki A, Sivridis E, Papaiakovou M, Kainulainen H, Koukourakis MI. “Autophagic flux” in normal mouse tissues: focus on endogenous LC3A processing. Autophagy. 2011;7:1371–1378. doi: 10.4161/auto.7.11.16664. [DOI] [PubMed] [Google Scholar]
- 36.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2011;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Singh R, Massey AC, Kane SS, Kaushik S, Grant T, Xiang Y, Cuervo AM, Czaja MJ. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J Biol Chem. 2008;283:4766–4777. doi: 10.1074/jbc.M706666200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Burgoyne RD. Identification of common genetic modifiers of neurodegenerative diseases from an integrative analysis of diverse genetic screens in model organisms. BMC Genomics. 2012;13:71. doi: 10.1186/1471-2164-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2010;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haney SA. Increasing the robustness and validity of RNAi screens. Pharmacogenomics. 2007;8:1037–1049. doi: 10.2217/14622416.8.8.1037. [DOI] [PubMed] [Google Scholar]
- 43.Luesch H, Chanda SK, Raya RM, DeJesus PD, Orth AP, Walker JR, Izpisua Belmonte JC, Schultz PG. A functional genomics approach to the mode of action of apratoxin A. Nat Chem Biol. 2006;2:158–167. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 46.van der Vaart A, Mari M, Reggiori F. A picky eater: exploring the mechanisms of selective autophagy in human pathologies. Traffic. 2008;9:281–289. doi: 10.1111/j.1600-0854.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Yang Q, Mao Z. Chaperone-mediated autophagy: machinery, regulation and biological consequences. Cell Mol Life Sci. 2011;68:749–763. doi: 10.1007/s00018-010-0565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 49.Blackstone NW, Green DR. The evolution of a mechanism of cell suicide. Bioessays. 1999;21:84–88. doi: 10.1002/(SICI)1521-1878(199901)21:1<84::AID-BIES11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 50.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhutani N, Piccirillo R, Hourez R, Venkatraman P, Goldberg AL. Cathepsins L and Z are critical in degrading polyglutamine-containing proteins within lysosomes. J Biol Chem. 2012;287:17471–17482. doi: 10.1074/jbc.M112.352781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim YJ, Sapp E, Cuiffo BG, Sobin L, Yoder J, Kegel KB, Qin ZH, Detloff P, Aronin N, DiFiglia M. Lysosomal proteases are involved in generation of N-terminal huntingtin fragments. Neurobiol Dis. 2006;22:346–356. doi: 10.1016/j.nbd.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 53.Liang Q, Ouyang X, Schneider L, Zhang J. Reduction of mutant huntingtin accumulation and toxicity by lysosomal cathepsins D and B in neurons. Mol Neurodegener. 2011;6:37. doi: 10.1186/1750-1326-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Felbor U, Kessler B, Mothes W, Goebel HH, Ploegh HL, Bronson RT, Olsen BR. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci U S A. 2002;99:7883–7888. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sevenich L, Pennacchio LA, Peters C, Reinheckel T. Human cathepsin L rescues the neurodegeneration and lethality in cathepsin B/L double-deficient mice. Biol Chem. 2006;387:885–891. doi: 10.1515/BC.2006.112. [DOI] [PubMed] [Google Scholar]
- 56.Lee JA. Autophagy in neurodegeneration: two sides of the same coin. BMB Rep. 2009;42:324–330. doi: 10.5483/bmbrep.2009.42.6.324. [DOI] [PubMed] [Google Scholar]
- 57.Kaasik A, Rikk T, Piirsoo A, Zharkovsky T, Zharkovsky A. Up-regulation of lysosomal cathepsin L and autophagy during neuronal death induced by reduced serum and potassium. Eur J Neurosci. 2005;22:1023–1031. doi: 10.1111/j.1460-9568.2005.04279.x. [DOI] [PubMed] [Google Scholar]
- 58.Kar R, Singha PK, Venkatachalam MA, Saikumar P. A novel role for MAP1 LC3 in nonautophagic cytoplasmic vacuolation death of cancer cells. Oncogene. 2009;28:2556–2568. doi: 10.1038/onc.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abraham MC, Lu Y, Shaham S. A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev Cell. 2007;12:73–86. doi: 10.1016/j.devcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 60.Catrenich CE, Chestnut MH. Character and origin of vacuoles induced in mammalian cells by the cytotoxin of Helicobacter pylori. J Med Microbiol. 1992;37:389–395. doi: 10.1099/00222615-37-6-389. [DOI] [PubMed] [Google Scholar]
- 61.Kannan TR, Baseman JB. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc Natl Acad Sci U S A. 2006;103:6724–6729. doi: 10.1073/pnas.0510644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aki T, Nara A, Uemura K. Cytoplasmic vacuolization during exposure to drugs and other substances. Cell Biol Toxicol. 2012;28:125–131. doi: 10.1007/s10565-012-9212-3. [DOI] [PubMed] [Google Scholar]
- 63.Root DE, Flaherty SP, Kelley BP, Stockwell BR. Biological mechanism profiling using an annotated compound library. Chem Biol. 2003;10:881–892. doi: 10.1016/j.chembiol.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 64.Dineen TA, Roush WR. Total Synthesis of Cochleamycin A. Organic Letters. 2004;6:2043–2046. doi: 10.1021/ol049331x. [DOI] [PubMed] [Google Scholar]
- 65.Jia Y, Bois-Choussy M, Zhu J. Synthesis of DEFG Ring of Complestatin and Chloropeptin I: Highly Atropdiastereoselective Macrocyclization by Intramolecular Suzuki-Miyaura Reaction. Organic Letters. 2007;9:2401–2404. doi: 10.1021/ol070889p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.