Abstract
Transcriptome analysis of polar bears (Ursus maritimus) yielded sequences with highest similarity to the human endogenous retrovirus group HERV-K(HML-2). Further analysis of the polar bear draft genome identified an endogenous betaretrovirus group comprising 26 proviral copies and 231 solo LTRs. Molecular dating indicates the group originated before the divergence of bears from a common ancestor but is not present in all carnivores. Closely related sequences were identified in the giant panda (Ailuropoda melanoleuca) and characterized from its genome. We have designated the polar bear and giant panda sequences Ursus maritimus endogenous retrovirus (UmaERV) and Ailuropoda melanoleuca endogenous retrovirus (AmeERV), respectively. Phylogenetic analysis demonstrated that the bear virus group is nested within the HERV-K supergroup among bovine and bat endogenous retroviruses suggesting a complex evolutionary history within the HERV-K group. All individual remnants of proviral sequences contain numerous frameshifts and stop codons and thus, the virus is likely non-infectious.
Keywords: Polar bear, giant panda, endogenous retrovirus, genomics
Endogenous retroviruses are a complex and large (up to 10%) part of the genome of vertebrates. They represent the successful colonization of the genome by exogenous retroviruses upon infection of the germline or hybridization with a species or population in which endogenization has occurred (Gifford and Tristem, 2003). The classification of retroviruses as endogenous or exogenous is not always clearly delineated as some may exist in both states and thus spread by both Mendelian transmission and by infection. For example, the mouse mammary tumor viruses (MMTV) are both transmitted to offspring as Mendelian traits and by infection from maternal breast milk. Exogenous and endogenous betaretroviruses are associated with mammary tumors in mice. Though definitive proof is not available, ERVs have been associated with various diseases such as cancer, neurodegenerative diseases and autoimmune diseases (Denner et al., 1995; Greenwood et al., 2011a; Sugimoto et al., 2001). Betaretroviruses, in particular HERV-K (HML-2), several loci of which encode functional proteins, have been implicated in various human tumour diseases (Ruprecht et al., 2008). A betaretrovirus in sheep, endogenous Jaagsiekte sheep retrovirus (enJSRV), the exogenous counterpart of which is strongly supported as the causative agent of a transmissible lung cancer in sheep, protects against exJSRV infection and is required for sheep placental development (Varela et al., 2009). The diversity of tumor types associated with betaretroviruses contrasts somewhat with gammaretroviruses, another retroviral group specifically associated with oncogenesis. Gammaretroviruses are typically associated with leukemia such as murine leukemia viruses (MLV) or koala retrovirus (KoRV) (Avila-Arcos et al., 2012; Tarlinton et al., 2005).
Most exogenous retrovirus groups identified to date have endogenous counterparts. However, not all groups have endogenous counterparts in all species, for example, endogenous retroviruses closely related to lentiviruses have only been identified in lemurs, rabbits, weasels and ferrets to date (Cui and Holmes, 2012; Gilbert et al., 2009; Han and Worobey, 2012; Katzourakis et al., 2007). Endogenous counterparts of delta retroviruses and HIV/SIV have not been identified to date. Gammaretroviruses, foamy retroviruses, and betaretroviruses have been discovered in a greater number of species. For example, it was recently proposed that betaretroviruses have been evolving within the genomes of murid rodents for at least the last 20 million years and were occasionally transmitted to non-rodent species in the course of the global spread of murids (Baillie et al., 2004). However, knowledge about distribution and diversity of ERVs is limited by lack of characterization of genomes as opposed to their absence or lack of diversity.
As more genomes become available, the opportunity to characterize novel retroviruses is increasing. Both the polar bear (Ursus maritimus) and the giant panda (Ailuropoda melanoleuca) have recently been sequenced to the draft genome level (Li, 2011; Li et al., 2010). Endogenous retroviruses have not been described in bears. As part of a study to identify novel viral and bacterial microbes from two polar bears, brain and liver cDNA were deep sequenced to generate transcriptomes and microbial sequences were characterized from the sequence reads. While the majority of sequences identified by shotgun sequencing were of polar bear origin, a subset of transcribed viral sequences identified were most similar to HERV-K(HML-2) as determined by genetic database searches. A number of corresponding endogenous retroviral loci were found in various scaffold sequences of a recently generated polar bear draft genome sequence (Li, 2011). We characterized this newly discovered endogenous betaretrovirus group regarding species distribution, evolutionary age and phylogenetic relationship with other retroviruses, and established a limited tissue transcription profile. We document here the full-length consensus polar bear ERV that we designated Ursus maritimus endogenous retrovirus (UmaERV) and its close relative in giant pandas, Ailuropoda melanoleuca endogenous retrovirus (AmeERV).
Results
Identification of UmaERV from polar tissues
RNA was extracted from brain and liver from two polar bears (Knut of the Berlin Zoological Garden and Jerka of the Wuppertal Zoological Garden) both of whom died as a result of viral encephalitis. Approximately 260 million 100 nt sequences were generated by Illumina shotgun sequencing of ribosome-subtracted libraries (74, 63, 58, and 65 million each from liver and brain from Knut and Jerka, respectively). These datasets were searched for possible pathogen-derived sequences, and the results of these searches will be described elsewhere. The searches also revealed the presence of apparent endogenous retrovirus-like sequences, including HERV-K(HML-2) gag and pol sequences. Primers were designed in both gag and pol to amplify a larger portion of the genome from the bear cDNAs and a PCR product was amplified from all four polar bear tissues from which the sequence reads were derived. Direct sequencing of the products and blastn searches again revealed highest similarity to HERV-K(HML-2).
Identification of UmaERV integration sites in polar bear and in panda bear genomes
PCR product sequences identified a subregion within the polar bear draft genome scaffold000030 sequence. A “seed” UmaERV (Ursus maritimus endogenous retrovirus) locus was identified in that scaffold subregion using RetroTector (Sperber et al., 2009; Sperber et al., 2007) and Repeatmasker (Tempel, 2012). A BLASTn search of all the 72,214 polar bear scaffold sequences, using the proviral body sequence of the seed UmaERV as probe, identified 26 UmaERV loci in the polar bear draft genome. Another BLASTn search with the seed UmaERV LTR sequence as probe identified 261 UmaERV locus-associated and solitary LTRs. Multiple alignments of identified proviral and LTR sequences were generated, and majority rule-based consensus sequences were generated. Characteristics of the UmaERV consensus provirus are shown in Figure 1 (and Figure S1-S2 in the supplementary data). Further sequence analysis of consensus protein sequences employing RetroTector and NCBI CD Search identified typical retroviral motifs and also a dUTPase domain within the protease coding sequence. The UmaERV LTR was most similar to an LTR sequence annotated in the giant panda as LTR1_AMe, and UmaERV like sequences were found in the giant panda by PCR. The giant panda genome draft assembly (BGI-Shenzhen AilMel 1.0 Dec. 2009), as provided by the UCSC Genome Browser, was therefore BLAT-searched with UmaERV LTR and body consensus sequences as probe. We detected ca. 20 loci similar to the UmaERV body sequence and about 145 loci similar to the UmaERV LTR sequence in the giant panda draft assembly. We propose to name the UmaERV-similar sequences in the panda Ailuropoda melanoleuca Endogenous Retrovirus (AmeERV). Characteristics of the AmeERV sequence can be found in Figure S3. Characteristics of UmaERV and AmeERV sequences as they are found in the respective draft genome sequences are provided as supplementary data (Tables S1-S6) and the relative similarity of the UmaERV and AmeERV consensus sequences is shown in Figure 2. The respective consensus sequences are also provided in a supplementary text file.
Figure 1. Consensus sequence of UmaERV provirus.
The consensus provirus sequence of UmaERV is displayed in A. ORFs for proviral gag, pro, pol and env genes, resulting protein sequences and protein domains, the latter as predicted by NCBI Conserved Domain Search (Marchler-Bauer et al., 2013) and Retrotector (Sperber et al., 2009)(Sperber et al., 2007), are indicated. Starts and ends of ORF are further highlighted by colored arrows and lines ending in diamonds respectively. The generated consensus sequence did not result in complete ORFs for pol and env gene regions, and frameshifts are indicated. Proviral 5′ and 3′ LTRs are highlighted in light green. Note that the PBS predicted by Retrotector (Sperber et al., 2007) overlaps with the 5′LTR 3′ end by 5 nt. The Pustell matrix diagram in Figure 2 and the comparative alignment in Figure S1 demonstrates the near identity of the consensus sequences of UmaERV and AmeERV.
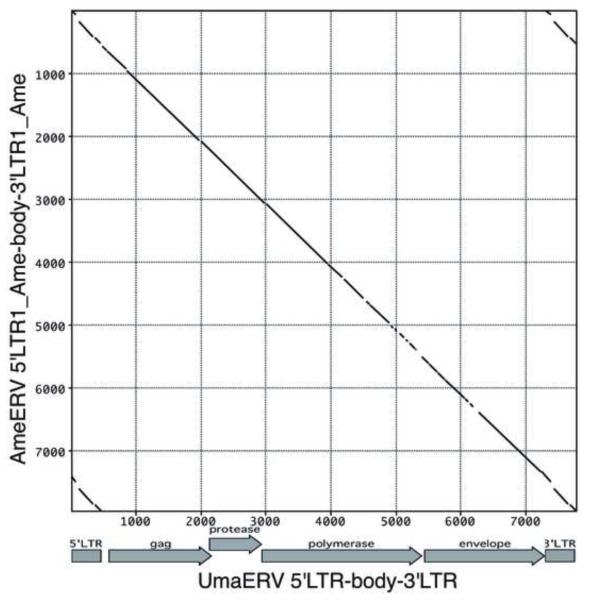
Figure 2. High sequence similarity between UmaERV and AmeERV proviral sequences.
Shown is a Pustell matrix comparison of UmaERV and AmeERV proviral consensus sequences (window size=30; min. % score=90; jump=1). Note that the LTR1_Ame sequence, as provided by Repbase v17.08, in the AmeERV provirus sequence displays some sequence differences to the actual majority rule consensus sequence for AmeERV-associated LTRs, the latter of which is very similar to the consensus sequence of UmaERV-associated LTRs. A pairwise sequence comparison of both proviral sequences is shown in Figure S1.
Most UmaERV loci were severely mutated and 5′ or 3′ or internal proviral regions were often missing (Figure S2). Similar results were obtained for AmeERV (Figures 2 and S3). Although retroviral gag, pro, pol or env gene regions were often present within the proviruses, none of them appeared capable of encoding retroviral proteins of significant length. Thus, it is unlikely that any single UmaERV locus could produce retroviral proteins, let alone infectious virus. The state of the UmaERV loci in the polar bear genome thus suggests that UmaERV is exclusively endogenous. A comparison of the consensus sequence of UmaERV and AmeERV demonstrate their overall high similarity (Figure S1).
Age estimates of UmaERV and distribution in bears
As the data suggested UmaERV is an ERV, the age of the ERV group was estimated using two different approaches. First, UmaERV LTR sequences identified by BLASTn searches were multiply aligned using MAFFT, the alignment was manually optimized and Kimura-2-parameter distances of LTR sequences to a majority-rule consensus sequence were calculated for three LTR subregions and excluding CpG dinucleotide positions because they are prone to higher mutations rates due to 5-methyl cytosine spontaneous deamination (Katoh et al., 2005; Kimura, 1980). Using a previously published bear-specific mutation rate of 0.0015/nt/year (Hailer et al., 2012a), UmaERV sequences were estimated to be approximately 48.28 (±42.24) million years old.
The second dating method employed was based on sequence divergence of the proviral 5′ and 3′ LTRs. Upon provirus formation, the 5′ and 3′ LTRs are identical in sequence due to the strategy by which pre-proviral dsDNA is reverse transcribed from a retroviral RNA genome. For ERVs, accumulation of sequence differences between proviral 5′ and 3′ LTRs can be used to estimate the age of a given provirus (Dangel et al., 1995). For UmaERV loci where age determination based on LTR-LTR divergence could be applied the ages were similar to those obtained based on the phylogenetic based dating of the UmaERV LTRs (Table S7). Thus, we conclude that the UmaERV group is approximately 45 million years old.
Bears are estimated to have separated from seals and their relatives (pinnipeds) 35 million years ago (Krause et al., 2008). The age estimates for UmaERV suggested that this viral group should thus be present in all bears. To further test the so far estimated age of UmaERV sequences, genomic DNA was extracted from brown bear (Ursus arctos), black bear (Ursus americanus), spectacled bear (Tremarctos ornatus) and giant panda (Ailuropoda melanoleuca). Genomic DNAs were screened with primers that yielded an approximately 1 kb fragment in all bears tested. Direct sequencing of the products demonstrated that the virus obtained was similar to UmaERV in each bear species tested (Figure S4). This supports the age estimates for UmaERV as the giant panda and other bears diverged from a common ancestor ca. 20 million years ago (Krause et al., 2008). Suitable pinniped tissue was not available for testing. However, searching the dog and cat genomes for UmaERV sequences using BLAT at the UCSC Genome Browser (Karolchik et al., 2004) yielded no positive identification. Thus, UmaERV is a bear virus, likely in pinnipeds but is not present in all carnivores.
Phylogeny of UmaERV and AmeERV
Consensus proviral protein sequences were generated for UmaERV and AmeERV as described in Material and Methods (Figure 1). Both UmaERV and AmeERV consensus sequences contained GAG, PRO, POL and ENV coding sequences. The N-terminal portion of the PRO coding sequence had a betaretrovirus typical dUTPase domain. The resulting amino acid sequences were aligned to representative murine, cervid, bovine, bat and human betaretroviruses, particularly the HERV-K(HML) supergroup, and other retroviruses. Lentiviral sequences were used as an outgroup in Bayesian analysis of GAG, PRO, POL and ENV sequences. UmaERV and AmeERV were sister taxa in all analyses for each protein (Figure 3). The trees were largely consistent with murine, ovine and rabbit betaretrovirus forming a distinct clade and UmaERV and AmeERV belonging to a clade including HERV-K(HMLs), a bovine ERV and notably closer relationship with some recently described bat ERVs (Hayward et al., 2013). The UmaERV-AmeERV containing clade was generally structured such that HERV-K(HML-5) and HML-6 were basal to a clade containing the bear ERVs, bovine ERV and the remaining HML groups. An exception was PRO in which the UmaERV-AmeERV clade is located between HERV-K(HML-5) and HML-6. Thus, the here described UmaERV clade of Ursus maritimus and AmeERV of Ailuropoda melanoleuca is nested within the HERV-K(HML) clade of betaretroviruses. Phylogenetic analysis based on nucleotide sequences, where alignable, yielded consistent results with the protein results with respect to UmaERV and AmeERV’s placement within the HERV-K group (Figure S5).
Figure 3. Phylogenetic relationship of UmaERV and AmeERV within the Retroviridae.
Bayesian phylogenetic trees are shown for the GAG, PRO, POL and ENV proteins. Posterior probabilities >50% are shown. All sequences from taxa represented in the trees are described in the Materials and Methods. The overall topology with respect to UmaERV and AMeERV was consistent regardless of the protein analyzed except for PRO where HML-6 was not basal to UmaERV and AMeERV.
A dUTPase domain is not uniformly distributed among retroviruses. For example, it is found primarily in betaretroviruses and in two lentiviral groups. Even among HERV-K(HML) groups, it is notably absent from HML-7 and HERV-KC4 though this may be the result of mutations occurring post endogenization that subsequently spread by retrotransposition (Mayer and Meese, 2003). As the evolution of this viral activity apparently differs from the virus as a whole, the dUTPase was analyzed separately phylogenetically for indication of inconsistent tree placement relative to the rest of the viral proteins. Despite its apparent dispensability, the phylogenetic placement of UmaERV and AmeERV dUTPase was consistent with all other protein sequences examined and did not alter the phylogenic placement of the group based on PRO which contains the dUTPase domain (not shown and Figure 3).
A phylogenetic analysis of the LTRs of UmaERV and AmeERV demonstrated that each clade contained representative LTRs from both polar bear and panda (Figure 4). Thus UmaERV and AmeERV LTRs are largely homologous further supporting a common ancient origin of the viral group prior to the diversification of the bear lineage.
Figure 4. Phylogenetic relationship of UmaERV and AmeERV LTR sequences.
A neighbor joining tree is shown for the 261 UmaERV and 145 AmeERV proviral and solitary LTR sequences identified in the polar bear and panda draft genomes. Green circles represent UmaERV LTRs and blue circles AmeERV.
Expression of UmaERV
Although unlikely to produce complete proteins or infectious virus, UmaERV could be transcribed as has been shown for many betaretrovirus related HERVs. To examine the transcription profile of the virus, polar bear brain, liver, kidney, spleen and lung RNA was extracted and RT-PCR was performed (see Materials and Methods). The primers were designed to amplify any of the identified UmaERVs but still be able to distinguish the source virus using the sequences between the primers. Only brain was positive for UmaERV transcripts. The expressed sequence detected was identical to the scaffold000030 identified by RNA-Seq by direct sequencing (Figure S4).
Discussion
The surreptitious discovery of UmaERV and AmeERV by screening of polar bear transcriptomes for microbial sequences represents the first endogenous retroviruses described in detail from bears based on available genome sequence data. In a dual approach both individual sequence reads and de novo assembled contigs showed considerable similarities to different regions of Retroviridae genomes. As short (100 nt) sequences distinct viral genes (gag and pol) were obtained they could be used as anchor sequences to a fragment of sufficient length for characterization of the full-length proviruses from the polar bear draft genome using various strategies including Repeatmasker and RetroTector.
Age estimates based on a molecular clock of the full complement of LTRs and estimates based on 5′ and 3′ proviral LTR divergence indicated that UmaERV endogenization predated the separation of the bear and seal lineages from a common ancestor. However, genomic invasion occurred subsequent to the separation of seals and bears from other eutherian carnivores such as dogs and cats as closely related sequences were not found in the cat or dog genomes. Even the UmaERV provirus with the lowest age estimate predates the radiation of bears from a common ancestor though age estimates based on sequence comparisons must be regarded as relatively rough estimates for individual loci. We were furthermore able to retrieve a related ERV, named AmeERV, from the giant panda from the draft genome sequence of that species. PCR experiments indicate that related viruses are present in several additional bear species (Figure S4). A rodent source for all betaretroviruses within the last 20 million years has been hypothesized previously (Baillie et al., 2004). However, the age estimates for the divergence of UmaERVs and AmeERVs do not support a cross species transfer that recently although it remains possible that rodents were the source of betaretroviruses. Similarly, HERV-K(HML-6) has been proposed to be ca. 20 million years old although older ages (30 million years) have also been estimated (Yin et al., 1999). However, in almost all phylogenetic analyses performed, this group was basal to bear ERVs estimated to be much older than 20 million years. Also, divergences of HERV-K(HML-6) sequences from a consensus sequence indicate an age similar or even greater to that of HERV-K(HML-5) which was previously estimated approximately 55 million years (Lavie et al., 2004) (see also below). The wide distribution of similar ERVs in giant pandas and polar bears is consistent with an older origin of the UmaERV viral group as giant pandas and polar bears diverged from a common ancestor ca. 20 million years ago. The common occurrence of this ERV group in all bear genomic DNA tested suggests invasion occurred well before divergence of the bear lineages.
Phylogenetic analysis yielded a consistent placement of the UmaERV/AmeERV clade nested within the HERV-K supergroup. Interestingly, HERV-K(HML-5) and HML-6 were basal to the bear viruses and a cattle ERV identified in Bos Taurus and bats. Based on phylogenetic analysis, HML-6 and HML-5 are evolutionarily old HERV-K supergroup members that likely predate the radiation of strepsirrhine and catarrhine primates 55 Mya (Jern, Sperber, and Blomberg, 2005; Medstrand et al., 1997) (Lavie et al., 2004). The ancestral nature of HML-6 and HML-5 suggests that exogenous counterparts of these ERV groups were transspecies viruses that were able to invade the genomes of distantly related mammalian groups ranging from primates to bears. There are likely additional intermediate species that served to bridge transfer of basal HML-6 and HML-5 groups among taxa that remain to be identified provided they are not extinct. Noteworthy in this context are closer relationships of bear ERVs and HERV-K(HMLs) with certain betaretroviral ERV groups recently identified in bats (Hayward et al., 2013). Interestingly, subsequent to the genome invasions, it seems further propagation of these betaretroviral groups was highly species specific. For example, the remaining HERV-K(HML) groups are restricted to Old World monkeys and hominoid primates, as far as we know. Similarly, while the overall betaretroviral group examined suggests cross-species viral transfer, the phylogenetic relationships of UmaERV and AmeERV indicates virus-host co-evolution and does not reflect subsequent transspecies transmission events. All LTR clades found in polar bears are found in pandas consistent with viral endogenization and proliferation prior to bear speciation. LTR lineage differences among the bears likely reflect within species duplication of specific ERVs (Figure 4).
The dUTPase protein is not universally present among betaretroviruses including among the HERV-K supergroup. Whether this reflects selection against this activity is unclear. However, it is apparently a dispensable function. If selected against, it could be hypothesized that the phylogenetic placement might be inconsistent with other genes such as the HML-6 Envelope protein which switched from a basal position relative to the bear ERVs to a derived position (Figure 1). However, the UmaERV and AmeERV dUTPase domain’s phylogenetic grouping was consistent with all other proteins examined.
Consistent with the great age of the retrovirus group, expression was very limited. In the different tissue types available for study from polar bears, expression could be detected only in the brain and only from the most complete UmaERV present in the genome. Brain expression is consistent with ERV expression in many other species where transcription of ERVs can be detected (Greenwood et al., 2011b; Stengel et al., 2006). However, given that the results are derived from post mortem tissues, it cannot be ruled out that RNA quality may have failed to detect low level transcription in additional tissues. The viral protein in all identified UmaERVs contained premature stops and deletions that coupled with the lack of detection of widespread transcriptional activity suggest this ERV group has not been recently active. This contrasts with other betaretroviral groups such as HERV-K related elements in humans and non-human primates for which both young and old elements show transcriptional activity (Seifarth et al., 2005; Stengel et al., 2006). Whether this reflects differences in suppression of ERVs in different species or a biological difference in bears such as lower concentration of ERV relevant transcription factors remains to be determined.
The identification of novel ERV sequences is useful for resolving the phylogeny of retroviruses given that ERVs in wildlife often reflect unknown and no longer exogenously circulating retrovirus variants from many millions of years ago (Han and Worobey, 2012; Lamere et al., 2009). Given the number of species’ genomes sequenced currently or in the future, there will be a huge amount of sequence data including endogenous retroviral sequences providing a source of information for further resolving the evolution of retroviruses. Bioinformatic tools for detection of retroviral sequences in genome sequence, such as Retrotector employed in our study, are proving themselves to be highly useful, efficient and accurate. UmaERV and AmeERV, although containing remnants of all viral genes, likely only exist as genomic fossils of viruses that no longer have exogenous counterparts. However, these fossils demonstrate that viruses in taxa with no recent common ancestry such as bears, primates and cattle share viral sequences with a common ancestry more recent than their hosts. The further screening of genomic data of wildlife will continue to elucidate the relationships and genetic history of both endogenous and exogenous retroviruses.
Materials and Methods
Samples
Polar bear samples included brain, liver, and kidney from Knut (male) were kindly provided by the Zoological Garden Berlin ( Bernhard Blaszkiewitz, Andre Schüle and Heiner Klös). The Zoological Garden Berlin also provided muscle from Bao Bao, a male giant Panda (Ailuropoda melanoleuca). Brain and liver samples from Jerka (female) were kindly provided by the Zoological Garden Wuppertal by Arne Lawrenz. Spectacled (Tremarctos ornatus), black (Ursus americanus), and brown (Ursus arctos) bear samples were provided by Tierpark Berlin, and Allwetterzoo Münster, respectively. Samples were stored frozen at −80°C.
Nucleic acid preparation and next generation sequencing
Approximately 25 mg of each tissue was used for DNA or RNA extraction using QIAmp DNA mini and RNeasy Lipid tissue kits according to manufacturer instruction. For transcriptome sequencing, ribosomal RNA was selectively degraded to increase the complexity of the obtained sequence reads that would otherwise be dominated by such highly abundant transcripts. rRNA-depleted RNA was selected by using the Ribo-ZeroTM rRNA removal kit following manufacturer’s protocol (EpiCentre) and quantified using a Nanodrop 7500 spectrophotometer.
100 ng of rRNA-depleted RNA was fragmented and RNA-seq library preparation was carried out as described previously (Adamidi et al., 2011). RNA-seq was performed on a HiSeq2000 sequencing platform with 1 × 100 cycles of single read single-plex sequencing, in accordance with manufacturer’s instructions (Illumina).
PCR and expression analysis
For PCR and reverse transcription PCR (RT-PCR), DNA or cDNA was diluted to include100 ng for each reaction. For RT-PCR, RNA was DNAse treated and aliquots of non-reverse transcribed RNA tested for amplification of a portion of the retrovirus using primers UmaERV F1, UmaERVR1, UmaERV F3 ,UmaERV R3, UmaERV F4, and UmaERV R4 to ensure that DNA was removed prior to reverse transcription. cDNA was prepared with Invitrogen Superscript III and random primers according to manufacture instructions. PCR primers used for expression analysis-PCR included UmaERV F1 (5′-TTTCCCTAGTCTTTGTTCCCG-3′), UmaERV R1 (5′-CGTAACCCATTTCCCTGTAGAG-3′), UmaERV F3 (5′-TGCTGCATTAACCGCTCTTA-3′), UmaERV R3 (5′-TAAGTAAAGGCCATCTTCCA-3′), UmaERV F4 (5′-ATTTCCCTAGTCTYTGTCCC-3′), and UmaERV R4 (5′-GYGGCATGTAACAAATCTAAAATTG-3′). cDNA PCR was performed in 25 l reactions containing 0.5U of My Taq HS polymerase mix (Bioline), 200 nM primers, and 130 ng of template. Thermocycling conditions were 95 C denaturing for 5 min followed by 33 cycles of 95 C for 15 sec, 55 C for 20 sec, 72 C for 13 sec, with a final extension of 72 C for 13 sec. Genomic DNA from all the bears was amplified using primers KTRV F1 (5′-TGGTAC TGCTCTACAGGGAA-3′) and KTRV R1 (5′-GTGCCACTCTAAAGTTCACG-3′). DNA PCR was performed in 25 l reactions containing 0.5U of My Taq HS polymerase mix (Bioline), 200 nM primers, and 100 ng of template. Thermocycling conditions were 95 C denaturing for 3 min followed by 32 cycles of 95 C for 15 sec, 55 C for 20 sec, 72 C for 35 sec, with a final extension of 72 C for 35 sec. PCR products were visualized on a 1.5% [w/v] agarose gel using GelRed Nucleic acid gel stain (Biotium). Positive PCR amplification products were purified using the Qiaquick PCR clean up kit (Qiagen), and Sanger sequenced using forward and reverse primers (StarSeq GmbH).
Bioinformatic analysis
The Illumina generated shotgun reads were analyzed in two different ways. First they were filtered in several steps for polar bear sequences by subtracting matches to the polar bear genome. The remaining reads were blastx searched against all virus sequences from the NCBI protein database. Blast matches with e-values <0.001 were used to assign each read to a virus. The resulting dataset was then analysed to find species that had multiple non-overlapping hits to different parts of its genome. To estimate an overall probability measure for each occurring species, the p-values of non-overlapping hits were considered independent and thus multiplied. For each group of overlapping hits the smallest p-value was used, as overlapping hits cannot be considered independent and the minimum p-value gives an upper bound for the combined p-value of the group.
In the second approach reads were assembled into contigs. The assembly was carried out with Velvet (version 1.2.03) (Zerbino and Birney, 2008) using standard parameters and hash length 23. No expected coverage was entered as DNA from different microbial species (and thus with different coverage) was expected in the sample. Contigs longer than 200 bp were selected and a blastn search against the NCBI Nucleotide database was performed.
Identification of UmaERV sequences in the polar bear genome sequence
PCR primers (UmaERV F1 and R1) were designed based on the identified retrovirus-like sequences to amplify an approximately 1 kb fragment and the resulting PCR product was sequenced. The sequence was then used as a probe for identifying closely related sequences in the 72,214 scaffolds of the polar bear genome sequence. Genome scaffolds were initially retrieved from the polar bear draft genome and were indexed for mapping with BWA (version 0.5.9-r26-dev) using bwa index. Fasta sequences of the PCR fragments were converted into fastq format giving each base the highest possible quality (i.e. 41). Sequences were then mapped against the polar bear genome scaffolds using bwa bwasw, which is optimized for long queries, with default parameters. Both PCR products mapped to the same scaffold (Scaffold000030) in close proximity.
Based on the significant hits of the two PCR product sequences in Scaffold00030, a sufficiently large surrounding sequence portion of that scaffold was examined for retroviral sequences using Repeatmasker and RetroTector (Sperber et al., 2009; Sperber et al., 2007; Tempel, 2012). For Repeatmasker analysis, when using the abblast search engine, default speed/sensitivity, and mammal as DNA source, UmaERV LTRs were annotated as LTR1_AMe, an LTR identified in the panda (Ailuropoda melanoleuca), and proviral body sequences as ERV2-2-EC_I-int, an ERV group identified in the horse (Equus caballus). LTR and proviral body regions were defined based on Repeatmasker and RetroTector output. Those sequences then served as probes for identifying UmaERV proviral body and LTR sequences in the other scaffolds using BLAST as implemented in Geneious v5.6. We identified in the various scaffolds 27 UmaERV proviral bodies and 267 UmaERV LTRs, or remnants thereof (described in Tables S1-S3). For the identified proviral body sequences, we extracted for each the matching region plus 5 kb of upstream and downstream flanking sequence and delineated UmaERV content and boundaries based on Repeatmasker output.
Generation of UmaERV and AmeERV LTR and proviral consensus sequences
We generated multiple alignments of UmaERV proviral amino acid and LTR sequences employing MAFFT (Katoh et al., 2005). Multiple alignments were manually optimized and majority rule consensus sequences were generated from each alignment. Employing RetroTector, protein (putein) sequences of retroviral Gag, Protease, Polymerase and Envelope were generated based on the proviral consensus sequence. Retroviral sequence motifs within those protein sequences were also identified by RetroTector and NCBI CD Search. RetroTector also served to predict retroviral protein sequences for HERV-K(HML) groups for which there were no good protein sequences reported before. HERV-K(HML) and other HERV-K reference (consensus) sequences, as included in Repbase, then served as template for RetroTector analysis.
We identified UmaERV-like sequences in the panda genome ailMel1 draft assembly by BLAT searches at the UCSC Genome Browser (Karolchik et al., 2004) with the UmaERV body and LTR consensus sequences as probe. We retrieved genomic sequences corresponding to coordinates of matching regions in the panda genome draft sequence using the UCSC Table Browser (Karolchik et al., 2004). We multiply aligned retrieved LTR and body sequences separately using MAFFT. Majority rule consensus sequences and retroviral protein (putein) sequences were generated using RetroTector as described above.
Phylogenetic analysis
Multiple alignments of amino acid sequences were generated using MAFFT (Katoh et al., 2005). The divergence among UmaERVs and between the UmaERV consensus and other retroviruses made nucleotide sequence alignments unreliable. Preliminary searches of UmaERV Protein (putein) sequences where performed on the NCBI BLASTX database (Pirooznia, Perkins, and Deng, 2008; Pirooznia, Perkins, and Deng, 2007). Gag, Polymerase, Protease, and Envelope consensus protein (putein) sequences reported here were compared with HERV-K(HML) protein sequences already known or likewise generated by RetroTector and other betaretroviruses were obtained from Genbank and Ensembl: MMPV (accession numbers: NP 056893.1, NP 056892.1, NP 056891.1), exogenous MMTV (AAF31472.1, AAF31468.1, AAF31464.1, AAF31470.1), MMTV (NP 056881.1, BAA03766.1, NP 056880.1, AAF31465.1), enJRSV (ABV71123.1, ABV71078.1, ABV71120.1, ABV71098.1), JSRV (NP041184.1, AAD45225.1, CAA77121.1, ENTV2 (NP_862831.2, ADI50272.1, ADI50273.1, NP862834.2), Ovine ENTV (ACX93967.1, ACX93968.1, ADK26418.1, ACX93970.1), Mtv1 (AF228550.2, AAF31465.1, AF228550.1), SRV-1 (P04024.1, P04027.1), SRV-2 (AAA47561.1, AF126467_2, AF126467.3, P51515.1), SRV-4 ( YP_003864100.1, ADC52788.1, YP_003864103.1, YP_003864101.1), SRV-Y (BAM71049.1), SRV-D (BAD89356.1), SERV (AAC97563.1, AAC97566.1, AAC97567.1), SMRV (NP_041259, NP_041260.2, NP_041261.1, NP_041262.1), PyERV (AAN77283.1), MlERV-A (Scaffold_GL429780:11816573-11826438), MlERV-B(Scaffold_GL429905:2902336-2910456), MlERV-C (Scaffold_AAPE02058399:20007-28108), PvERV-A (Scaffold_22753:8224-518), Cavia porcelus (XP_003460945.1), Monodelphis domestica (XP_003342276.1), Bos Taurus endogenous retrovirus (ABM73644.1, DAA22938.1, ABM73646.1 ABM73647.1), Oryctolagus cuniculus retrovirus (XP 002714483.1, XP 002710662.1, XP 002712621.1), Sarcophilus harrisii (XP_003775259.1), GaLV (U60065), KoRV (AF151794.2), Bovine Leukemia Virus (AAA427784.1, NC_001414.1, AAA42786.1), Avian Leukemia virus (YP_004222726.1, NC 001408.1,YP_004222727.1), Equine Infectious Anemia Virus (AAA43011.1, AFV61764.1, AAC58658), Caprine Athritis Encephalities Virus (ACA81610.2, NP_040942.1, NP_040938.1). Human Immunodeficiency Virus-2 (AAB0737.1, CAA00302.1, CAA43572.1) and Human Immunodeficiency Virus-1 (ACT76482, AER12461.1, AAF35356.1) was used as an outgroup. The evolutionary model for the phylogenetic analysis was selected using ProtTest 2.4 (Abascal, Zardoya, and Posada, 2005) with “Wheland and Goldman model applied with invariable sites and gamma distribution (WAG+I+G)” determined as the optimal model for Protease, Envelope, and Gag. “Rtrev model with invariable sites and gamma distribution (rtrev+I+G)” was the determined optimal model for Polymerase. Bayesian Inference analysis was performed with MrBayes 3.2.1 (Huelsenbeck and Ronquist, 2001) for each gene’s protein/putein alignment using the above-determined model. Default number of Markov Chain Monte Carlo (MCMC) runs of 1 million generations, sampling trees every 200 generations, generated majority consensus trees after a burn in of 1250 generations.
Age estimation of UmaERV proviral sequences
We calculated the age of UmaERV sequences by two different methods both employing a molecular clock. First, using PAUP* (Swofford, 2003), we determined sequence divergence of UmaERV LTR sequences from the UmaERV LTR consensus sequence based on a method previously described for Alu subfamilies (Kapitonov and Jurka, 1996). Hypermutable CpG sites were excluded from the analysis. Sequence divergence was corrected according to the Kimura-2-parameter (K2P) model (Kimura, 1980). Calculated sequence divergences from the consensus were used to estimate evolutionary ages of UmaERV LTR sequences assuming a molecular clock. A reported polar bear mutation rate of 0.0015/nt/myr (Hailer et al., 2012b) was used. Second, we determined sequence divergence between proviral 5′ and 3′ LTRs that were identical at the time of provirus formation and accumulated mutations independently since then, employing T = D/2•0.0015, where D is the K2P-corrected sequence divergence between a proviral 5′ and 3′ LTRs (Dangel et al., 1995). Mean and standard deviations were calculated from values obtained from each method.
Supplementary Material
Highlights.
The first bear endogenous retroviruses described in detail
A novel betaretroviruses related to human HERV-K
Present in all bears but not all carnivores
Acknowledgements
The authors wish to thank Claudia Szentiks for providing tissues and information about specific bears. The authors thank Arne Lawrenz and Katrin Griess of the Zoological Garden Wuppertal and Bernhard Blaszkiewitz, Andre Schüle and Heiner Klös of the Zoological Garden Berlin for providing bear samples for this study. The authors also thank Joe DeRisi for valuable assistance during the early phases of this project. The described research on polar bear and panda post mortem tissue was approved by the Internal Ethics Committee of the Leibniz-Institute for Zoo and Wildlife Research (IZW), Approval No. 2012-05-01. The project described was supported by Grant Number R01GM092706 from the National Institute of General Medical Sciences (NIGMS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the National Institutes of Health. The research of Jens Mayer is supported by grants from Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–5. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adamidi C, Wang Y, Gruen D, Mastrobuoni G, You X, Tolle D, Dodt M, Mackowiak SD, Gogol-Doering A, Oenal P, Rybak A, Ross E, Sanchez Alvarado A, Kempa S, Dieterich C, Rajewsky N, Chen W. De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 2011;21(7):1193–200. doi: 10.1101/gr.113779.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Arcos MC, Ho SY, Ishida Y, Nikolaidis N, Tsangaras K, Honig K, Medina R, Rasmussen M, Fordyce SL, Calvignac-Spencer S, Willerslev E, Gilbert MT, Helgen KM, Roca AL, Greenwood AD. 120 years of koala retrovirus evolution determined from museum skins. Mol Biol Evol. 2012 doi: 10.1093/molbev/mss223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GJ, van de Lagemaat LN, Baust C, Mager DL. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J Virol. 2004;78(11):5784–98. doi: 10.1128/JVI.78.11.5784-5798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Holmes EC. Endogenous lentiviruses in the ferret genome. J Virol. 2012;86(6):3383–5. doi: 10.1128/JVI.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangel AW, Baker BJ, Mendoza AR, Yu CY. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K(C4) are a molecular clock of evolution. Immunogenetics. 1995;42(1):41–52. doi: 10.1007/BF00164986. [DOI] [PubMed] [Google Scholar]
- Denner J, Phelps RC, Lower J, Lower R, Kurth R. Expression of the Human Endogenous Retrovirus Herv-K in Tumor and Normal-Tissues and Antibody-Response of Pregnant-Women, Tumor and Aids Patients against Herv-K Gag and Env Peptides. Aids Research and Human Retroviruses. 1995;11(S103):S103. [Google Scholar]
- Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26(3):291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Maxfield DG, Goodman SM, Feschotte C. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 2009;5(3):e1000425. doi: 10.1371/journal.pgen.1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood AD, Vincendeau M, Schmadicke AC, Montag J, Seifarth W, Motzkus D. Bovine spongiform encephalopathy infection alters endogenous retrovirus expression in distinct brain regions of cynomolgus macaques (Macaca fascicularis) Molecular Neurodegeneration. 2011a:6. doi: 10.1186/1750-1326-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood AD, Vincendeau M, Schmadicke AC, Montag J, Seifarth W, Motzkus D. Bovine spongiform encephalopathy infection alters endogenous retrovirus expression in distinct brain regions of cynomolgus macaques (Macaca fascicularis) Mol Neurodegener. 2011b;6(1):44. doi: 10.1186/1750-1326-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailer F, Kutschera VE, Hallstrom BM, Klassert D, Fain SR, Leonard JA, Arnason U, Janke A. Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science. 2012a;336(6079):344–7. doi: 10.1126/science.1216424. [DOI] [PubMed] [Google Scholar]
- Hailer F, Kutschera VE, Hallstrom BM, Klassert D, Fain SR, Leonard JA, Arnason U, Janke A. Nuclear Genomic Sequences Reveal that Polar Bears Are an Old and Distinct Bear Lineage. Science. 2012b;336(6079):344–347. doi: 10.1126/science.1216424. [DOI] [PubMed] [Google Scholar]
- Han GZ, Worobey M. Endogenous lentiviral elements in the weasel family (mustelidae) Mol Biol Evol. 2012;29(10):2905–8. doi: 10.1093/molbev/mss126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JA, Tachedjian M, Cui J, Field H, Holmes EC, Wang LF, Tachedjian G. Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology. 2013;10(35) doi: 10.1186/1742-4690-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jern P, Sperber GO, Blomberg J. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology. 2005;2:50. doi: 10.1186/1742-4690-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42(1):59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–6. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–8. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci U S A. 2007;104(15):6261–5. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Krause J, Unger T, Nocon A, Malaspinas AS, Kolokotronis SO, Stiller M, Soibelzon L, Spriggs H, Dear PH, Briggs AW, Bray SC, O’Brien SJ, Rabeder G, Matheus P, Cooper A, Slatkin M, Paabo S, Hofreiter M. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol Biol. 2008;8:220. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamere SA, St Leger JA, Schrenzel MD, Anthony SJ, Rideout BA, Salomon DR. Molecular characterization of a novel gammaretrovirus in killer whales (Orcinus orca) J Virol. 2009;83(24):12956–67. doi: 10.1128/JVI.01354-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie L, Medstrand P, Schempp W, Meese E, Mayer J. Human endogenous retrovirus family HERV-K(HML-5): status, evolution, and reconstruction of an ancient betaretrovirus in the human genome. J Virol. 2004;78(16):8788–98. doi: 10.1128/JVI.78.16.8788-8798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zhang G, Willerslev E, Wang J, Wang J. Genomic data from the polar bear (Ursus maritimus) GigaScience. 2011 [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, Zhang Z, Zhang Y, Wang W, Li J, Wei F, Li H, Jian M, Nielsen R, Li D, Gu W, Yang Z, Xuan Z, Ryder OA, Leung FC, Zhou Y, Cao J, Sun X, Fu Y, Fang X, Guo X, Wang B, Hou R, Shen F, Mu B, Ni P, Lin R, Qian W, Wang G, Yu C, Nie W, Wang J, Wu Z, Liang H, Min J, Wu Q, Cheng S, Ruan J, Wang M, Shi Z, Wen M, Liu B, Ren X, Zheng H, Dong D, Cook K, Shan G, Zhang H, Kosiol C, Xie X, Lu Z, Li Y, Steiner CC, Lam TT, Lin S, Zhang Q, Li G, Tian J, Gong T, Liu H, Zhang D, Fang L, Ye C, Zhang J, Hu W, Xu A, Ren Y, Zhang G, Bruford MW, Li Q, Ma L, Guo Y, An N, Hu Y, Zheng Y, Shi Y, Li Z, Liu Q, Chen Y, Zhao J, Qu N, Zhao S, Tian F, Wang X, Wang H, Xu L, Liu X, Vinar T, Wang Y, Lam TW, Yiu SM, Liu S, Huang Y, Yang G, Jiang Z, Qin N, Li L, Bolund L, Kristiansen K, Wong GK, Olson M, Zhang X, Li S, Yang H. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–7. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41(D1):D348–52. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Meese EU. Presence of dUTPase in the various human endogenous retrovirus K (HERV-K) families. J Mol Evol. 2003;57(6):642–9. doi: 10.1007/s00239-003-2514-6. [DOI] [PubMed] [Google Scholar]
- Medstrand P, Mager DL, Yin H, Dietrich U, Blomberg J. Structure and genomic organization of a novel human endogenous retrovirus family: HERV-K (HML-6) Journal of General Virology. 1997;78(Pt 7):1731–44. doi: 10.1099/0022-1317-78-7-1731. [DOI] [PubMed] [Google Scholar]
- Pirooznia M, Perkins EJ, Deng Y. Batch Blast Extractor: an automated blastx parser application. BMC Genomics. 2008;9(Suppl 2):S10. doi: 10.1186/1471-2164-9-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Perkins EJ, Deng YP. Batch Blast Extractor: an automated blastx parser application. BMC Genomics. 2007:9. doi: 10.1186/1471-2164-9-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell Mol Life Sci. 2008;65(21):3366–82. doi: 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol. 2005;79(1):341–52. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber G, Lovgren A, Eriksson NE, Benachenhou F, Blomberg J. RetroTector online, a rational tool for analysis of retroviral elements in small and medium size vertebrate genomic sequences. BMC Bioinformatics. 2009;10(Suppl 6):S4. doi: 10.1186/1471-2105-10-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber GO, Airola T, Jern P, Blomberg J. Automated recognition of retroviral sequences in genomic data--RetroTector. Nucleic Acids Res. 2007;35(15):4964–76. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A, Roos C, Hunsmann G, Seifarth W, Leib-Mosch C, Greenwood AD. Expression profiles of endogenous retroviruses in Old World monkeys. J Virol. 2006;80(9):4415–21. doi: 10.1128/JVI.80.9.4415-4421.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto J, Matsuura N, Oda T, Jinno Y. Novel HERV-K genes (ERV3 and ERV4) were mapped to autoimmune disease loci on chromosome 3. American Journal of Human Genetics. 2001;69(4):371–371. [Google Scholar]
- PAUP* Phylogenetic Analysis Using Parsimony (* and Other Methods) Sinauer Associates; Sunderland Massachusetts: p. 4. [Google Scholar]
- Tarlinton R, Meers J, Hanger J, Young P. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. Journal of General Virology. 2005;86:783–787. doi: 10.1099/vir.0.80547-0. [DOI] [PubMed] [Google Scholar]
- Tempel S. Using and understanding RepeatMasker. Methods Mol Biol. 2012;859:29–51. doi: 10.1007/978-1-61779-603-6_2. [DOI] [PubMed] [Google Scholar]
- Varela M, Spencer TE, Palmarini M, Arnaud F. Friendly viruses: the special relationship between endogenous retroviruses and their host. Ann N Y Acad Sci. 2009;1178:157–72. doi: 10.1111/j.1749-6632.2009.05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Medstrand P, Kristofferson A, Dietrich U, Aman P, Blomberg J. Characterization of human MMTV-like (HML) elements similar to a sequence that was highly expressed in a human breast cancer: further definition of the HML-6 group. Virology. 1999;256(1):22–35. doi: 10.1006/viro.1998.9587. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.