Abstract
Vaccination is the most effective way to reduce the impact of epidemic as well as pandemic influenza. However, the licensed inactivated influenza vaccine induces strain-specific immunity and must be updated annually. When novel viruses appear, matched vaccines are not likely to be available in time for the first wave of a pandemic. Yet, the enormous diversity of influenza A viruses in nature makes it impossible to predict which subtype or strain will cause the next pandemic. Several recent scientific advances have generated renewed enthusiasm and hope for universal vaccines that will induce broad protection from a range of influenza viruses.
Keywords: Influenza virus, Universal vaccine
Influenza virus diversity and vaccines
Influenza viruses belong to the family Orthomyxoviridae and are classified into three types, A, B, and C; type A influenza viruses are further divided into subtypes based on the antigenicity of the hemagglutinin (HA) and neuraminidase (NA) surface glycoproteins. Currently 16 HA and 9 NA subtypes are recognized, all of which exist in aquatic birds that are their natural reservoirs. Influenza B viruses only infect humans but two antigenically and phylogenetically distinct lineages co-circulate [1]. Seasonal influenza epidemics caused by influenza A and B viruses are responsible for up to five million cases of severe illness and between 250,000 and 500,000 deaths each year, worldwide (see: http://www.who.int/mediacentre/factsheets/fs211/en/). In addition to seasonal epidemics, sporadic influenza pandemics, which are global outbreaks caused by newly emerged influenza A viruses, result in similar or greater morbidity and mortality (see: http://www.who.int/mediacentre/factsheets/fs211/en/). Vaccination is the most effective way to reduce the impact of epidemic as well as pandemic influenza though available vaccines are not uniformly efficacious across age groups. Recent meta-analyses of randomized controlled studies over several influenza seasons showed 59% pooled efficacy in adults aged 18-65 years with trivalent inactivated vaccine (TIV) [2] and high efficacy in children with live attenuated influenza vaccine (LAIV) [2, 3].
A truly universal vaccine should cover all subtypes of influenza A viruses and both lineages of influenza B viruses. However, due to the significant genetic and antigenic differences between influenza A and B viruses, a single vaccine that protects against both types may not be realistic. Thus, for the purposes of this review, ‘universal vaccines’ focus on influenza A viruses. Although the ideal universal vaccine would provide protection against several or all subtypes of influenza A viruses, the first step may be a vaccine that is more broadly cross-protective than the currently licensed influenza vaccines and one that would not need to be administered or updated almost every year.
The case for a universal influenza vaccine
Antibody mediated protection directed against the influenza HA protein is generally strain-specific because the dominant epitopes on the globular head of the HA that are the target of the antibody response are under immune pressure to drift. As a result, currently licensed influenza vaccines, which induce a protective immune response directed mainly at the viral HA, have to be updated annually. The selection of virus strains to be included in seasonal influenza vaccines is based on global virologic surveillance coordinated by the World Health Organization (WHO) and the strains are selected several months in advance of the next influenza season because the manufacture, release and distribution of the egg-based trivalent influenza vaccine is a long process; it takes approximately 6 months from the time the vaccine composition is decided to distribution. A seasonal influenza vaccine would require less frequent updating if it induced broader cross-protection and longer lasting immunity; this is one of the key arguments in favor of a universal influenza vaccine.
The enormous diversity of influenza A viruses in nature makes it impossible to predict which subtype or strain will cause the next pandemic. The recent experience with the 2009 H1N1 influenza pandemic has driven a surge of interest in a universal influenza vaccine. Decisions to produce and to deploy a pandemic H1N1 (pH1N1) vaccine were made by the WHO and various national authorities shortly after the virus began to spread around the world in April 2009. Yet, vaccine was not widely available to the public in the Northern hemisphere until after the peak of the second wave of the pandemic [4]. This delay in the availability of vaccine has led to a re-evaluation of the existing strategy of manufacturing a matched vaccine after the pandemic strain emerges. In the event of a pandemic, prior immunization with a vaccine that induces broad but less robust protection against a range of viruses would have some advantage over a potentially more robust, strain-specific vaccine that is manufactured after the pandemic is declared.
Another reason for renewed interest in the development of a universal influenza vaccine is the recent (re-)discovery by several investigators of broadly cross-reactive neutralizing antibodies (Abs) directed against an epitope in the highly conserved stem or stalk of the influenza HA. These broadly cross-reactive HA stem Abs had not been previously identified in human sera because they are not easily detected; the dominant antibody response is directed at epitopes on the globular head of the HA. In addition to its less frequent presence compared to Abs against globular head region, the Abs against stem region cannot be detected by the hemagglutination inhibition (HAI) assay that is widely used for detection of influenza-neutralizing Abs. The application of newer technologies including immortalization of memory B cells, isolation of plasmablasts and combinatorial libraries made it possible to generate HA stem Abs from the peripheral blood of vaccinated or naturally infected people [5-8]. Also, stem Abs were detected much more frequently following the emergence of the novel 2009 pH1N1 virus [9, 10]. It has been proposed that exposure to seasonal influenza viruses or vaccines selects for expansion of influenza-specific memory B cells that recognize shared epitopes on the globular head of the HA that are subject to antigenic drift [11]. However, there is a preferential expansion of memory B cells specific for conserved epitopes in the HA stem upon infection or vaccination with an antigenically distinct virus such as the 2009 pH1N1 virus that does not share epitopes on the globular head [11].
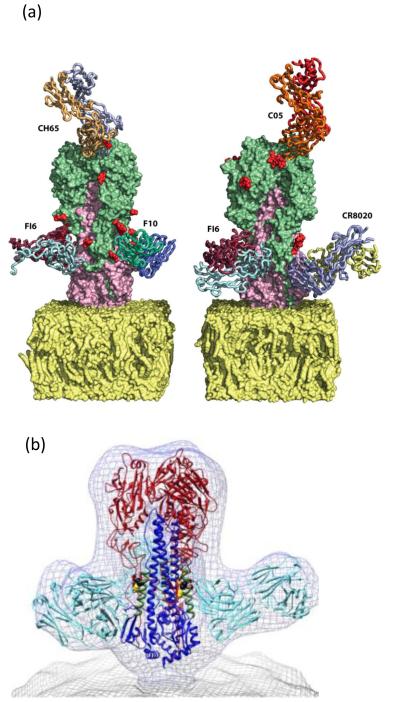
The 16 subtypes of influenza A HAs fall into two distinct phylogenetic groups: group 1 includes H1, H2, H5, H6, H8, H9, H11, H12, H13 and H16 and group 2 includes H3, H4, H7, H10, H14 and H15. Broadly cross-reactive HA stem Abs can neutralize the infectivity of a range of influenza A viruses in vitro and are effective in prophylaxis and treatment of influenza virus infection in animal models [12]. Some of the Abs cross-neutralize antigenically and genetically distinct viruses within the same subtype [6], some cross-neutralize viruses within a phylogenetically related group of HA subtypes [7, 8, 13], while rare Abs are able to neutralize a wide range of subtypes across both groups of HAs [14, 15]. The binding sites of the Abs have been determined by x-ray crystallography [7, 13, 14, 16] as well as by cryo-electron microscopy [17] (Figure 1). One broadly neutralizing Ab C05 binds to a conserved element of the receptor binding site on the HA head of both group 1 and 2 viruses [16]. The molecular characterization of these Abs and their target epitopes has generated optimism for the prospect of eliciting cross-protective Abs by rationally designed vaccines.
Figure 1.
Binding sites of selected broadly neutralizing antibodies on the HA. (a) Specific binding sites of various mAbs specific for phylogenetically distinct influenza A viruses are shown; F10 (group 1), F16 (Group 1 and 2), CR8020 (Group 2), C05 (Group 1 and 2) and CH65 (H1N1). The figure was adapted with permission from [12]. (b) Wire-mesh representation of front view of the density map of the complex formed by the broadly neutralizing murine mAb C179 (cyan) with trimeric HA, showing that intact virions are available to bind the anti-stem antibody. This figure was adapted from [17].
The immunologic basis for universal vaccine strategies
Antibodies directed at the viral HA and NA mediate protection from infection while cellular immune responses directed at internal proteins of the virus are necessary for viral clearance. The role of cytotoxic T lymphocytes (CTLs) in mediating heterosubtypic immunity in influenza was recognized many years ago in mice [18, 19]. In a study of human volunteers infected with a 1979 H1N1 virus, in the absence of H1N1-specific HAI Abs, CTL responses were associated with reduced viral shedding [20]. Although the CTL response does not prevent virus infection, it plays an important role in viral clearance and in heterosubtypic protection [21-23]. Most neutralizing Abs are directed against the globular head of the HA and both HA and NA glycoproteins are highly variable and drift under immune pressure. In contrast, the stem region of the HA and the internal viral proteins including the nucleoprotein (NP), polymerase acidic (PA) protein, matrix (M) and membrane ion channel (M2) proteins are highly conserved across influenza subtypes and are thus viewed as desirable targets for a universal vaccine. Table 1 lists possible viral targets of universal vaccines.
Table1.
Viral targets of universal vaccines
| Protein | Targeted site | Function of target | Proposed mechanism (s) of protection |
|---|---|---|---|
| Hemagglutinin (HA) | Stem | Fusion activity | Inhibition of fusion, maturation of the HA, viral egress and ADCC |
| Matrix 2 (M2) | Ectodomain of M2 (M2e) |
Ion channel | Alveolar macrophages and (M2e) Fc ADCC Antibody dependent NK cell activity Complement mediated lysis |
| Nucleoprotein (NP) | T-cell and antibody epitopes |
T-cell stimulation and non- neutralizing antibody |
Cell lysis by CD8+c ytotoxic T-lymphocytes (CTL) CD4+T-lymphocyte mediated cytolysis and B- cell stimulation |
| Matrix 1(M1) | T-cell epitopes | T-cell stimulation | Cell lysis by CD8+c ytotoxic T-lymphocytes (CTL) CD4+T- lymphocyte mediated cytolysis and B- cell stimulation |
| Neuraminidase (NA) | Conserved sialidase active site |
Sialidase | Inhibition of viral spread |
Target antigens for universal vaccines
Among the eleven viral proteins encoded by influenza A viruses, several proteins including HA, M (M1 and M2e), NP and NA proteins have been evaluated as promising candidate antigens for universal vaccines. The nature of the antigens and delivery methods as well as the type of immune responses induced by each antigen will be discussed below.
HA protein
HA is synthesized as a precursor (HA0) that is cleaved into HA1 and HA2 domains. The cleavage site of HA with the fusion peptide and N-terminal portion of HA2 is the most conserved sequence among influenza A viruses. In general terms, two approaches have been undertaken to induce broadly cross-reactive immunity to HA. One focuses on full length HA constructs and the other on the conserved HA2 domain.
For the full length HA, ‘centralized’ HA genes representing a putative ancestor were synthesized from consensus sequences within subtype (H1 or H5) or among different subtypes (H1, H2, H3, H4 and H5) and delivered to mice using a replication defective adenovirus vector [24], virus like particles (VLPs) derived from mammalian cells [25] or as a DNA vaccine [26]. Protection conferred by these vaccines measured as survival from lethal challenge was limited to viruses within the same subtype [24].
Although it is clear that infection or seasonal influenza vaccines can elicit rare broadly neutralizing monoclonal antibodies (mAb) directed at the HA stem in mice [27, 28] and humans [12], it has been a challenge to develop vaccines designed to induce HA stem-specific responses exclusively. The frequency of anti-stem Abs is considerably lower than that of anti-globular head Abs [6]. In fact, immune reactivity to peptides corresponding to a consensus sequence of the cleavage peptide consisting of 8 amino acids from the C-terminus of HA1 and 11 amino acids from the N-terminus of HA2 of H1 or H3 viruses was absent in sera from 100 randomly selected individuals with prior laboratory confirmed exposure to influenza A viruses [29]. The HA stem is thought to be less immunogenic than the globular head, (i) because the immunodominant head physically masks the stem region on the influenza virion and (ii) because of the close proximity of the stem epitope(s) to the viral membrane [30]. The challenge is to elicit an effective immune response to a less immunogenic HA stem; success will require innovative methods for delivery of the peptide. If all Abs directed against the HA stem do not share the same characteristics, a combination of peptides covering various epitopes may be needed.
Peptide constructs corresponding to the HA cleavage site from influenza A and B viruses elicited neutralizing antibody responses and confered protection against morbidity and mortality associated with lethal challenge in mice [29, 31]. A synthetic peptide corresponding to the highly conserved long α-helix of HA2 of an H3 virus elicited different levels of heterosubtypic protective immunity against H5 and H1 viruses in mice [32]. Vaccination provided protection against weight loss caused by both viruses and 60% of mice survived lethal challenge with H5 virus but none survived challenge with an H1 virus. The peptide was designed based on the binding properties of a murine mAb (12D1) that recognizes the HA2 domain and neutralized a range of H3 subtype viruses [33]. It would be interesting to know whether this linear epitope vaccine binds and/or neutralizes other HA subtypes from groups 1 and 2. An HA2-based construct containing part of the HA1 and mutations to stabilize the low pH conformation of HA2, also designed based on the 12D1 epitope, was expressed in Escherichia coli [34]. This immunogen protected mice from death but not weight loss following challenge with the homologous H3 viruses and from neither with a heterologous H1 virus.
Several different adjuvants have been used with peptide-based vaccines including the outer membrane protein complex of Neisseria meningitides [29], incomplete Freund’s adjuvant [31], complete Freund’s adjuvant [32] and CpG7909 [34].
Another approach to deliver HA stem peptides is the use of VLPs. The 20-residue α-helix of the HA2 that forms the major component of the epitope of mAb CR6261, a heterotypic neutralizing human mAb that binds several group 1 HAs [8], was expressed on VLPs derived from Flock House virus [35]. Although Abs induced by this VLP in mice had binding characteristics similar to CR6261, they did not exhibit neutralizing activity in vitro or in vivo. Thus, further research is needed for the design of functional and protective peptide-based vaccines.
Two novel approaches have been undertaken to focus the immune responses against the HA stem while eliminating the response to the immunodominant globular head. First, a headless HA was designed by introducing a deletion in the HA1 [36]. Mice immunized with cells expressing a headless HA derived from an H2N2 virus were fully protected from homologous virus challenge but were only partially protected from heterologous H1N1 virus challenge [36]. More recently, Steel et al. prepared similar headless HA constructs based on H1N1 and H3N2 viruses and immunized mice with a combination of plasmid DNA encoding the headless HA gene and VLPs co-expressing the headless HA and human immunodeficiency virus Gag protein [37]. Although the H1N1 headless HA vaccine strategy elicited ELISA Abs against heterologous group 1 viruses, efficacy of protection against heterologous viruses was not assessed [37].
The second approach used to overcome the subdominance of stem epitopes is to boost anti-stem Abs by immunization with antigenically distant viruses. Various platforms were used for sequential immunization with divergent influenza viruses, including sequential DNA vaccines [33], priming with a DNA vaccine followed by a boost with an inactivated vaccine or with an adenovirus vectored vaccine expressing the HA [38, 39] or sequential infection with influenza viruses [40]. Priming with a DNA vaccine expressing a 1999 H1 HA followed by a boost with seasonal TIV from the same year or matched adenovirus-encoded HA stimulated the production of cross-reactive HA stem Abs in mice, ferrets and non-human primates [38]. Later, these investigators demonstrated that the prime-boost immunization strategy elicited cross-reactive anti-HA stem Abs in mice and ferrets regardless of previous influenza exposure, although prior infection with more divergent strains seemed to direct a stronger anti-stem response [39]. Support for the role of infection or immunization with divergent strains was observed in humans, where robust immune responses against the HA stem were detected in individuals after 2009 pH1N1 virus infection or vaccination [9-11, 41]. In another recent example, human subjects primed with an H5 HA DNA vaccine that received an inactivated H5 vaccine boost developed anti-HA stem Abs that cross-neutralized group 1 viruses [42]. In a short period of time, there has been tremendous progress in this area and there is hope that one or more of these approaches will be successful.
M2e protein
M2 is a small integral membrane protein that functions as a pH-dependent proton channel, essential for proper maturation of the HA and release of the viral genome into the cytoplasm [43, 44]. Interest in the M2 protein for a universal vaccine is based on two observations: (i) passive transfer of an anti-M2 mAb that lacked neutralizing activity reduced the level of replication of a challenge virus in the lungs of mice [45] and (ii) the sequence of the extracellular domain of M2 (M2e) is conserved among human influenza A viruses. However, the frequency and levels of Abs against M2 or M2e are very low in influenza-infected animals as well as in humans [46, 47]. Therefore, several strategies were tried to overcome the poor immunogenicity of M2e, including fusing the peptide to immunogenic carrier proteins or delivery of VLPs generated from proteins expressed in baculovirus or mammalian cells or as icosahedral particles [48-53]. Antibody-dependent cell cytotoxicity (ADCC), antibody-dependent NK cell activity and complement mediated lysis have been proposed as mechanisms of M2e-mediated protection [54, 55]. In a recent study, Fc receptors were shown to be essential for anti-M2e IgG-mediated immune protection in mice, indicating that ADCC and Ab-dependent cell-mediated phagocytosis are the main mechanisms involved [56]. Additionally, in combination with pre-existing anti-M2e Abs, alveolar macrophages that are resident in the lung suppress influenza virus infection and spread by eliminating infected cells more effectively and rapidly than in naive animals [56].
In early studies, immunization of mice with M2e peptide genetically fused to the N-terminus of the hepatitis B virus core antigen (HBc) elicited a humoral immune response and enhanced pulmonary clearance of challenge virus [57]. Although vaccination with M2e-HBc induced cross-reactive humoral immune responses against several influenza A viruses and reduced morbidity following challenge, it did not confer complete protection against infection. Strategies that have been used to enhance the immunogenicity of M2e-HBc include the incorporation of M2e into the immunodominant loop of HBc, the insertion of multiple copies of M2e peptides at the N-terminus of HBc, and intranasal immunization [58, 59]. The M2e peptide has also been chemically fused to HBc [60]. When immunogenicity of HBc VLPs and a conjugated vaccine containing the same M2e peptide sequence were compared in mice and non-human primates, the immunogenicity of the two vaccines in the two species was different; both vaccines were equally immunogenic in mice, whereas the HBc VLP vaccine was inferior in rhesus monkeys [61]. Thus, rodents may not be the appropriate model for preclinical evaluation of these vaccines.
Another promising delivery method for M2e is a baculovirus-based VLP, where M2 protein is expressed as a native membrane-anchored protein on the VLP surface, without HA and NA proteins which mask the exposure of M2e on virions [62]. M2 VLPs were immunogenic without adjuvant and conferred cross protection against lethal challenge with different subtypes in mice although some morbidity (weight loss) was reported. Cross-protective immunity was enhanced when mice were immunized with inactivated vaccine supplemented with M2 VLPs [63] or with engineered M2 protein containing four repetitive M2e regions fused with bacterial flagellin incorporated into an influenza virus M1-based VLP [64]. The sequence of one region (amino acid 10-20) on M2e is consistent with host-range restriction [65]. In order to improve heterosubtypic cross-protection against viruses isolated from different species, Kim et al. [66] produced VLPs expressing tandem repeats of M2e peptides containing two human, two avian and one swine origin M2e peptide sequence fused with HA transmembrane and cytoplasmic domains. Interestingly, M2e proteins were incorporated into these VLPs to levels 100-fold higher than on influenza virions and sera from mice immunized with the VLPs reacted with a range of influenza viruses including H1N1 (1934 and 2009 strains), H3N2 (1982 strain) and H5N1 (2004 strain) viruses.
T7 bacteriophage nanoparticles are another attractive delivery system that has been studied for the delivery of viral, bacterial and cancer vaccines [67]. M2e protein induced immune responses that are comparable to those seen with an adjuvanted-M2e peptide vaccine in mice [51].
NP and M1 proteins
The amino acid sequences of the NP and M1 proteins are well conserved among influenza A viruses. Since these proteins are not exposed on the surface of virions or infected cells, they mainly induce cellular immune responses, particularly CTL responses against processed peptides [68, 69]. In addition to CD8+ CTLs, NP Abs are involved in viral clearance through ADCC in mice [70]. However, in humans the levels of anti-NP Abs vary and are rarely boosted by trivalent inactivated vaccines. Thus the importance of NP Ab-mediated ADCC in heterosubtypic protection in humans is not clear. Mice can also protected from lethal challenge by immunization with conserved CD4+ T cell epitopes [71]. Influenza-specific memory CD4+ T cells contribute to protective immune responses via multiple pathways [72].
Vector-based and linear peptide-based antigen delivery are two approaches that have been undertaken recently for the induction of effective CTL responses in humans. Phase 1 and 2 clinical studies using modified vaccinia virus Ankara (MVA) expressing NP and M1 proteins demonstrated that the MVA-NP+M1 vaccine was generally safe, though the higher dose was associated with a significant increase in malaise, nausea/vomiting and rigors [73]. The investigators reported a 60% reduction of laboratory-confirmed influenza infection following experimental challenge in subjects vaccinated with a single intramuscular dose of vaccine [73, 74]. However, it is difficult to assess the true efficacy of this vaccine because only a small number of subjects were studied (11 vaccinees and 11 controls). Another interesting approach that was recently tested in humans was recombinant protein containing multiple linear epitopes from different influenza proteins (HA, NP and M1) derived from both influenza A and B viruses [75]. This vaccine was well tolerated despite large doses (2 doses of 250 μ g [M250] or 500 μg [M500]) of vaccine administered with the adjuvant Montanide, and both humoral and cellular immunity were elicited in the group that received M500 with adjuvant. It is interesting that the Abs elicited by this vaccine demonstrated ADCC activity, although the contribution of such humoral responses in influenza infection is not clear. Humoral responses to peptide antigens were also reported when mice were immunized with multiple epitopes from HA, NP and M1 proteins inserted into bacterial flagellin [76].
NA protein
As in the case of the HA, the nine subtypes of NA fall into two phylogenetic groups [77]. Although there are some efforts to develop NA based vaccines, it is generally accepted that immunization with NA alone would not prevent infection. However, the immune response against NA limits viral spread and reduces the severity of disease [78]. Baculovirus expressed VLPs containing N1 NA protein induced heterosubtypic NA antibodies in mice, protected them from lethal challenge with homologous (H1N1) and heterologous (H3N2) viruses and reduced viral replication in the lungs [79]. Two highly conserved sequences near the NA enzymatic site could be valuable for quantification of NA in vaccines [80].
One component versus a multicomponent vaccine
Although the first steps in identifying critical components of a broadly cross-protective, universal influenza vaccine focus on single peptides or proteins as immunogens, a sound argument can be made to combine several components into a single vaccine because vaccines targeting NA, NP or M1 proteins that mediated CTLs or ADCC responses alone would not provide protection from infection. Viral immunogens could be combined, e.g. NP + M or HA + M2e or components that stimulate different arms of the immune system could be combined e.g. non-neutralizing anti-NP antibody and CD8+ T cells. A combination approach was demonstrated by Goodman et al. [81] who used T-cell epitopes from multiple internal proteins as well as conserved epitopes of HA and NA as immunogens. Alternatively, vaccines that are anticipated to ameliorate disease rather than prevent infection, but that induce broader immunity, could be used to complement existing vaccine strategies directed at HA. While such a strategy may not obviate the need for annual vaccination, it could provide some protection against antigenic drift variants and could blunt the effect of the first wave of a pandemic.
Challenges
Despite recent scientific advances in vaccinology, until 2012 all influenza vaccines licensed in the US were produced by methods that were established more than 30 years ago. The approaches discussed above suggest that a new generation of influenza vaccines is feasible but many technical, regulatory and logistical challenges remain (Box 1).
Box 1. Challenges in the development of universal vaccines.
- Technical
- Eliciting a reproducible T cell response in outbred species
- Maintenance or rapid recall of memory T cells to the site of infection
- Delivery of the immunogen
- Regulatory
- Identification of an immune correlate of protection
- Assays to measure immune correlates of protection
- Path to licensure
- Measuring efficacy in humans: challenge studies or large field studies
- Demonstrating non-inferiority vs. licensed vaccine
- Conducting placebo controlled trials in populations for whom licensed influenza vaccine is recommended
- Cost
- Implementation
- Who should be vaccinated, when and how often?
- Public education regarding the goals and expectations of universal vaccine
Immunization with T-cell vaccines could provide resistance to disease caused by a wide range of influenza viruses. The repertoire of CTLs in response to influenza A viruses in humans is much broader than that observed in mice [82] and the usage of some CTL epitopes is more dominant than others [83]. In addition, the human leukocyte antigen (HLA) background has a major influence on the magnitude and specificity of CTL responses [83-85]. These factors must be taken into consideration in the design of CTL-based influenza virus vaccines. Recent advances in immunoinformatics may improve the selection of epitopes for T-cell based vaccines [86]. One example of such an application is a peptide-based vaccine that included selected CD8+ T-cell epitopes that were conserved among different influenza virus subtypes and that were recognized by HLA types that are commonly seen in humans. Reduced viral replication and protection from mortality following infection with a heterosubtypic virus were reported in HLA-A2 transgenic HHD mice immunized with this vaccine [87].
Other technical challenges that must be considered in the design of T-cell based vaccines for influenza are the kinetics of influenza virus replication and of the cell-mediated response; influenza viruses replicate very rapidly in the host. Peak viral titers correlate with disease and are achieved before a cell-mediated immune response that restricts viral replication can be generated from memory. Therefore, a T-cell based vaccine must induce T cells and maintain them in a highly functional state without causing immunopathology. It would be essential to maintain this state of immunity through the duration of a typical first wave of a pandemic [88].
In general, peptides or fusion proteins are poor immunogens and require the use of adjuvants. However, few adjuvants are licensed for use in humans and new adjuvanted vaccines will require extensive safety studies in humans [89, 90]. A technical challenge that applies to all new immunogens is the choice of a suitable delivery platform. Several approaches have been applied in this field of research (Box 2).
Box 2.
Delivery of immunogens
Universal influenza vaccines require innovative delivery systems since most antigens that might provide broad heterosubtypic protection are poorly immunogenic. Modification of antigens, addition of immune stimulating components (adjuvants) and/or rationally designed vaccination regimens must be applied. The most promising systems currently sought as universal vaccine delivery methods are summarized.
-
Peptides with adjuvants
Peptide antigens are simple to produce. They can be designed to deliver multiple epitopes in single vaccine. However, almost all peptide-based vaccines require a large dose of antigen, addition of adjuvants and/or multiple doses of vaccine to elicit a robust immune response. In addition, regulatory approval for a new adjuvant will require extensive safety studies in humans.
-
Peptides fused to carrier proteins
Fusion protein antigens are used for effective presentation of antigens as well as for enhancement of immunogenicity. Hepatitis B core antigen (HBc) has been used for delivery of influenza virus antigens including M2e peptides. M2e fused with bacterial flagellin has been used in conjunction with a VLP delivery system.
-
Peptides or proteins expressed in viral vectors
Presentation of antigens delivered by viral vectors mimics viral infection and can induce both humoral and cellular immunity. However, preexisting immunity or a response to the vector can hinder the immune response to the target antigen. Modified vaccinia virus based vaccines with influenza NP and M1 proteins have been tested recently in phase 1 and 2 clinical studies.
-
VLPs
VLP vaccines can present membrane-anchored antigens such as HA, NA and M2 in their native forms. In addition to the epitopes for which native structure is critical, VLPs can present multimeric peptide antigens as has been explored for the M2e domain. However, in some cases, incorporation of proteins into VLPs is poor or not possible (headless HA).
-
Phage based nanoparticles
The phage delivery system has some attractive advantages: (i) phage vaccines can be produced easily at large scale and low cost, (ii) phages are highly stable, (iii) phage particles are naturally immunogenic and (iv) phages can be used as particles as well as DNA vaccines. Potential disadvantages are that only small peptides can be expressed in high copy number. Although the application of this system to influenza universal vaccines has been limited to an M2e protein vaccine studied in mice, we can expect to see further studies using this approach for universal influenza vaccines.
-
Prime boost regimes with DNA prime and recombinant adenovirus or protein vaccine boost
The effectiveness of this approach to induce immune responses against the group-specific HA stem region has been proven in several animal models as well as in humans. Further fine-tuning of selection of antigens that will cover both phylogenetic groups of HA and the design of effective vaccine regimens will be the next steps.
The key regulatory challenges that can be anticipated are the need for qualitative and quantitative methods to determine and define the potency of the vaccine, the need to identify immune correlates of protection and the development of validated assays to measure them. The path to licensure of novel influenza vaccines is likely to be long. Regulatory authorities are likely to require demonstration of efficacy in humans. Clinical trials of vaccines that do not prevent infection but ameliorate disease will require large numbers of subjects and will be very costly. It is not clear whether it will be necessary to demonstrate non-inferiority compared to the licensed vaccine and it may be ethically challenging to conduct placebo-controlled clinical trials in populations for whom licensed influenza vaccine is recommended.
When a novel, broadly protective influenza vaccine becomes available, public health authorities will have to determine how truly universal it is. Important decisions will include whom to vaccinate, whether and when they should be revaccinated and how often the universal vaccine may require updating. Both health care professionals and the public will need to be educated about the goals and expectations of a universal vaccine.
Concluding remarks
Although the concept of a universal influenza vaccine is not new, a confluence of recent developments in molecular virology, immunology and vaccine delivery suggest that a new generation of broadly cross-protective influenza vaccines is on the horizon. Depending on the design, these vaccines may ameliorate disease rather than prevent infection. It is likely that we will achieve the goal of a truly universal influenza vaccine in a step-wise fashion. However, the intermediate milestones will be of great value if they complement and broaden the protection conferred by existing vaccines.
*. Highlights.
A new generation of broadly cross-protective influenza vaccines is on the horizon.
These vaccines may ameliorate disease rather than prevent infection.
The HA stem, M2, NP and M proteins of the virus are the preferred targets.
Innovative methods to enhance vaccine immunogenicity and delivery will be needed.
Acknowledgements
The authors work is supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rota PA, et al. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- 2.Osterholm MT, et al. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 2012;12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 3.Ambrose CS, et al. The efficacy of intranasal live attenuated influenza vaccine in children 2 through 17 years of age: a meta-analysis of 8 randomized controlled studies. Vaccine. 2012;30:886–892. doi: 10.1016/j.vaccine.2011.11.104. [DOI] [PubMed] [Google Scholar]
- 4.Borse RH, et al. Effects of vaccine program against pandemic influenza A (H1N1) virus, United States, 2009–2010. Emerg. Infect. Dis. 2013;19:439–448. doi: 10.3201/eid1903.120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomson CA, et al. Pandemic H1N1 influenza infection and vaccination in humans induces cross-protective antibodies that target the hemagglutinin stem. Front Immunol. 2012;3:87. doi: 10.3389/fimmu.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li GM, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–42. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 13.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 15.Clementi N, et al. A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes. PLoS ONE. 2011;6:e28001. doi: 10.1371/journal.pone.0028001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris AK, et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Effros RB, et al. Generation of both cross-reactive and virus-specific T-cell populations after immunization with serologically distinct influenza A viruses. J. Exp. Med. 1977;145:557–568. doi: 10.1084/jem.145.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap KL, et al. Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature. 1978;273:238–239. doi: 10.1038/273238a0. [DOI] [PubMed] [Google Scholar]
- 20.McMichael AJ, et al. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 1983;309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 21.Epstein SL, et al. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine. 2005;23:5404–5410. doi: 10.1016/j.vaccine.2005.04.047. [DOI] [PubMed] [Google Scholar]
- 22.Okuda K, et al. Protective immunity against influenza A virus induced by immunization with DNA plasmid containing influenza M gene. Vaccine. 2001;19:3681–3691. doi: 10.1016/s0264-410x(01)00078-0. [DOI] [PubMed] [Google Scholar]
- 23.Roy S, et al. Partial protection against H5N1 influenza in mice with a single dose of a chimpanzee adenovirus vector expressing nucleoprotein. Vaccine. 2007;25:6845–6851. doi: 10.1016/j.vaccine.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver EA, et al. Protection against divergent influenza H1N1 virus by a centralized influenza hemagglutinin. PLoS ONE. 2011;6:e18314. doi: 10.1371/journal.pone.0018314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giles BM, et al. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin. Vaccine Immunol. 2012;19:128–139. doi: 10.1128/CVI.05533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen MW, et al. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13538–13543. doi: 10.1073/pnas.0806901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuno Y, et al. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okuno Y, et al. Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J. Virol. 1994;68:517–520. doi: 10.1128/jvi.68.1.517-520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianchi E, et al. Universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J. Virol. 2005;79:7380–7388. doi: 10.1128/JVI.79.12.7380-7388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong PD, Wilson IA. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 2009;10:573–578. doi: 10.1038/ni.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvath A, et al. A hemagglutinin-based multipeptide construct elicits enhanced protective immune response in mice against influenza A virus infection. Immunol. Lett. 1998;60:127–136. doi: 10.1016/s0165-2478(97)00137-5. [DOI] [PubMed] [Google Scholar]
- 32.Wang TT, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang TT, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bommakanti G, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13701–13706. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneemann A, et al. A virus-like particle that elicits cross-reactive antibodies to the conserved stem of influenza virus hemagglutinin. J. Virol. 2012;86:11686–11697. doi: 10.1128/JVI.01694-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sagawa H, et al. The immunological activity of a deletion mutant of influenza virus haemagglutinin lacking the globular region. J. Gen. Virol. 1996;77(Pt 7):1483–1487. doi: 10.1099/0022-1317-77-7-1483. [DOI] [PubMed] [Google Scholar]
- 37.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1:e00018–00010. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 39.Wei CJ, et al. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci. Transl. Med. 2012;4:147ra114. doi: 10.1126/scitranslmed.3004273. [DOI] [PubMed] [Google Scholar]
- 40.Krammer F, et al. Hemagglutinin stalk-reactive antibodies are boosted following sequential infection with seasonal and pandemic H1N1 influenza virus in mice. J. Virol. 2012;86:10302–10307. doi: 10.1128/JVI.01336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He XS, et al. Heterovariant cross-reactive B-cell responses induced by the 2009 pandemic influenza virus A subtype H1N1 vaccine. J. Infect. Dis. 2013;207:288–296. doi: 10.1093/infdis/jis664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ledgerwood JE, et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect. Dis. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamb RA, et al. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 44.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treanor JJ, et al. Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J. Virol. 1990;64:1375–1377. doi: 10.1128/jvi.64.3.1375-1377.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Black RA, et al. Antibody response to the M2 protein of influenza A virus expressed in insect cells. J. Gen. Virol. 1993;74(Pt 1):143–146. doi: 10.1099/0022-1317-74-1-143. [DOI] [PubMed] [Google Scholar]
- 47.Feng J, et al. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 2006;3:102. doi: 10.1186/1743-422X-3-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarez P, et al. Tandem repeats of the extracellular domain of Matrix 2 influenza protein exposed in Brucella lumazine synthase decameric carrier molecule induce protection in mice. Vaccine. 2013;31:806–812. doi: 10.1016/j.vaccine.2012.11.072. [DOI] [PubMed] [Google Scholar]
- 49.Bessa J, et al. Efficient induction of mucosal and systemic immune responses by virus-like particles administered intranasally: implications for vaccine design. Eur. J. Immunol. 2008;38:114–126. doi: 10.1002/eji.200636959. [DOI] [PubMed] [Google Scholar]
- 50.Denis J, et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26:3395–3403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 51.Hashemi H, et al. Immunization with M2e-displaying T7 bacteriophage nanoparticles protects against influenza A virus challenge. PLoS ONE. 2012;7:e45765. doi: 10.1371/journal.pone.0045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matic S, et al. Efficient production of chimeric human papillomavirus 16 L1 protein bearing the M2e influenza epitope in Nicotiana benthamiana plants. BMC Biotechnol. 2011;11:106. doi: 10.1186/1472-6750-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turley CB, et al. Safety and immunogenicity of a recombinant M2e-flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine. 2011;29:5145–5152. doi: 10.1016/j.vaccine.2011.05.041. [DOI] [PubMed] [Google Scholar]
- 54.Jegerlehner A, et al. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 2004;172:5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 55.Tompkins SM, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg. Infect. Dis. 2007;13:426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.El Bakkouri K, et al. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J. Immunol. 2011;186:1022–1031. doi: 10.4049/jimmunol.0902147. [DOI] [PubMed] [Google Scholar]
- 57.Neirynck S, et al. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999;5:1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 58.De Filette M, et al. Universal influenza A vaccine: optimization of M2-based constructs. Virology. 2005;337:149–161. doi: 10.1016/j.virol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 59.De Filette M, et al. The universal influenza vaccine M2e-HBc administered intranasally in combination with the adjuvant CTA1-DD provides complete protection. Vaccine. 2006;24:544–551. doi: 10.1016/j.vaccine.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 60.Blokhina EA, et al. A molecular assembly system for presentation of antigens on the surface of HBc virus-like particles. Virology. 2013;435:293–300. doi: 10.1016/j.virol.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 61.Fu TM, et al. Comparative immunogenicity evaluations of influenza A virus M2 peptide as recombinant virus like particle or conjugate vaccines in mice and monkeys. Vaccine. 2009;27:1440–1447. doi: 10.1016/j.vaccine.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 62.Song JM, et al. Influenza virus-like particles containing M2 induce broadly cross protective immunity. PLoS ONE. 2011;6:e14538. doi: 10.1371/journal.pone.0014538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song JM, et al. Vaccination inducing broad and improved cross protection against multiple subtypes of influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:757–761. doi: 10.1073/pnas.1012199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang BZ, et al. Enhanced influenza virus-like particle vaccines containing the extracellular domain of matrix protein 2 and a Toll-like receptor ligand. Clin. Vaccine Immunol. 2012;19:1119–1125. doi: 10.1128/CVI.00153-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu W, et al. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect. 2005;7:171–177. doi: 10.1016/j.micinf.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Kim MC, et al. Virus-like particles containing multiple M2 extracellular domains confer improved cross-protection against various subtypes of influenza virus. Mol. Ther. 2013;21:485–492. doi: 10.1038/mt.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao J, et al. Phage display and its application in vaccine design. Ann Microbiol. 2010;60:13–19. [Google Scholar]
- 68.Hillaire ML, et al. Induction of virus-specific cytotoxic T lymphocytes as a basis for the development of broadly protective influenza vaccines. J Biomed Biotechnol. 2011;2011:939860. doi: 10.1155/2011/939860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan PT, et al. Highly conserved influenza A sequences as T cell epitopes-based vaccine targets to address the viral variability. Hum. Vaccin. 2011;7:402–409. doi: 10.4161/hv.7.4.13845. [DOI] [PubMed] [Google Scholar]
- 70.Lamere MW, et al. Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J. Virol. 2011;85:5027–5035. doi: 10.1128/JVI.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexander J, et al. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine. 2010;28:664–672. doi: 10.1016/j.vaccine.2009.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKinstry KK, et al. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J. Clin. Invest. 2012;122:2847–2856. doi: 10.1172/JCI63689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berthoud TK, et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin. Infect. Dis. 2011;52:1–7. doi: 10.1093/cid/ciq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lillie PJ, et al. Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP+M1, in humans. Clin. Infect. Dis. 2012;55:19–25. doi: 10.1093/cid/cis327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atsmon J, et al. Safety and immunogenicity of multimeric-001--a novel universal influenza vaccine. J. Clin. Immunol. 2012;32:595–603. doi: 10.1007/s10875-011-9632-5. [DOI] [PubMed] [Google Scholar]
- 76.Adar Y, et al. A universal epitope-based influenza vaccine and its efficacy against H5N1. Vaccine. 2009;27:2099–2107. doi: 10.1016/j.vaccine.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 77.Russell RJ, et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 78.Johansson BE, Cox MM. Influenza viral neuraminidase: the forgotten antigen. Expert Rev. Vaccines. 2011;10:1683–1695. doi: 10.1586/erv.11.130. [DOI] [PubMed] [Google Scholar]
- 79.Quan FS, et al. Influenza M1 VLPs containing neuraminidase induce heterosubtypic cross-protection. Virology. 2012;430:127–135. doi: 10.1016/j.virol.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gravel C, et al. Qualitative and quantitative analyses of virtually all subtypes of influenza A and B viral neuraminidases using antibodies targeting the universally conserved sequences. Vaccine. 2010;28:5774–5784. doi: 10.1016/j.vaccine.2010.06.075. [DOI] [PubMed] [Google Scholar]
- 81.Goodman AG, et al. A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus. PLoS ONE. 2011;6:e25938. doi: 10.1371/journal.pone.0025938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jameson J, et al. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 1998;72:8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akram A, Inman RD. Immunodominance: a pivotal principle in host response to viral infections. Clin. Immunol. 2012;143:99–115. doi: 10.1016/j.clim.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 84.Boon AC, et al. Preferential HLA usage in the influenza virus-specific CTL response. J. Immunol. 2004;172:4435–4443. doi: 10.4049/jimmunol.172.7.4435. [DOI] [PubMed] [Google Scholar]
- 85.Boon AC, et al. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 2002;76:582–590. doi: 10.1128/JVI.76.2.582-590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salimi N, et al. Design and utilization of epitope-based databases and predictive tools. Immunogenetics. 2010;62:185–196. doi: 10.1007/s00251-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan AC, et al. The design and proof of concept for a CD8(+) T cell-based vaccine inducing cross-subtype protection against influenza A virus. Immunol. Cell Biol. 2013;91:96–104. doi: 10.1038/icb.2012.54. [DOI] [PubMed] [Google Scholar]
- 88.Subbarao K, et al. Development of effective vaccines against pandemic influenza. Immunity. 2006;24:5–9. doi: 10.1016/j.immuni.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 89.Nohynek H, et al. AS03 adjuvanted AH1N1 vaccine associated with an abrupt increase in the incidence of childhood narcolepsy in Finland. PLoS ONE. 2012;7:e33536. doi: 10.1371/journal.pone.0033536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mutsch M, et al. Use of the inactivated intranasal influenza vaccine and the risk of Bell’s palsy in Switzerland. N. Engl. J. Med. 2004;350:896–903. doi: 10.1056/NEJMoa030595. [DOI] [PubMed] [Google Scholar]