Abstract
mRNA export is a critical step in gene expression. Export of transcripts can be modulated in response to cellular signaling or stress. Consistently, mRNA export is dysregulated in primary human specimens derived from many different forms of cancer. Aberrant expression of export factors can alter export of specific transcripts encoding proteins involved in proliferation, survival and oncogenesis. These specific factors, which are not used for bulk mRNA export, are obvious therapeutic targets. Indeed, given the emerging role of mRNA export in cancer, it is not surprising that efforts to target different aspects of this pathway have reached the clinical trial stage. Thus, like transcription and translation, mRNA export may also play a critical role in cancer genesis and maintenance.
Global overview of mRNA export and the nuclear pore
Dysregulation of transcription and translation have long been demonstrated to contribute to the genesis and maintenance of cancer. Given the critical role that mRNA export plays in gene expression, it is not surprising to find that is it also dysregulated in many malignancies [1–4]. Indeed, mRNA export factors and relevant components of the nuclear pore complex (NPC) contribute to preferential export of transcripts encoding proteins involved in proliferation, survival, metastases and invasion (Figure 1, Table 1). Trafficking of these transcripts profoundly affects their ultimate protein levels. Although for many years mRNA export was considered a default pathway with little to no regulation, evidence that these are both regulated by and regulators of signaling networks involved in oncogenesis is now emerging. In fact, modulation of levels of individual export factors and NPC components themselves can alter proliferation rates and response to extracellular stimuli. Given these roles, it is not surprising that novel therapeutic approaches are emerging to target this process.
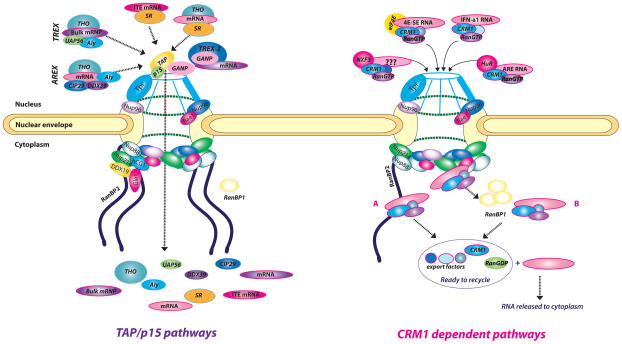
Figure 1. TAP1 and CRM-1 dependent mRNA export pathways are heterogenous demonstrating substantial plasticity.
As described in the text, factors associating with specific RNAs, often through USER codes, underlie formation of specific export mRNPs. The NPC dependent RNA export pathways are divided into two major subtypes, TAP and CRM1 dependent. Further subdivisions are indicated by the different composition of the nuclear mRNPs. For TAP and its cofactor NXT1/p15 dependent export, complexes that depend on Aly/REF (TREX and AREX), additionally on GANP (as a part of TREX-2 complex), and distinct complexes involving the SR proteins are shown (ITE mRNA represents intronless mRNAs). For the TAP pathways, release of cargoes is depicted using the DDX19/Gle1 model, which is hypothesized primarily from yeast data to be present in humans [9, 25]. Four subdivisions of the CRM1 pathway are similarly depicted by different nuclear mRNP complexes for HuR, eIF4E, IFN-α1 and NXF3. At the nuclear basket and cytoplasmic face, nucleoporins and co-factors described in the text are shown. The cytoplasmic side depicts only 4 fibrils, which is a simplification for presentation purposes (as there are known to be eight fibrils per NPC [8, 10]). For CRM1 dependent pathways, the RanGTP cycle is shown for cargo release, which can occur using RanBP2 (A) or RanBP1 (B). For the case of the eIF4E dependent mRNA export pathway, RanBP2 is depleted, RanBP1 is elevated and thus the RanBP1 release pathway is thought to predominate. Once cargoes are released, factors are recycled. Note the color difference of RanGTP (purple) and RanGDP (green). Many of these factors are dysregulated in cancer (see text).
Table 1. Nucleoporins and factors involved in mRNA export are dysregulated in cancer.
Most of Nups and export factors are multifunctional proteins and thus, only data with regard to their role in mRNA export are shown here. Note that these are a few examples. Further, not all phenotypes are observed amongst model systems, thus the relevant phenotypes from mammalian systems are shown.
| Nucleoporin or export factor | Examples of dysregulation in cancer | Biological effects of modulating expression | Reference |
|---|---|---|---|
| CRM1 | Overexpression in gliomas, cervical and pancreatic cancer | Reduction in CRM1 levels in cervical cancer cells leads to decreased proliferation and export generally | [45] |
| THOC1 | Elevated lung, ovarian and colon cancer | Reduction impairs proliferation and export | [41] |
| GANP | Elevated in mantle cell, diffuse large B cell and Hodgkins lymphomas | Knockdown represses proliferation and mRNA export | [47] |
| Aly/REF | Elevated in oral squamous cell carcinoma | [42] | |
| Rae1 | Breast cancer | [55] | |
| Nup88 | Elevated in prostate, ovarian, breast, mesotheliomas, hepatocellular, colon, some lymphomas and many forms of lung cancer | [51–53] | |
| RanBP2/Nup358 | Reduction leads to tumors in mice Reduction leads to increased eIF4E mediated mRNA export Overexpression suppresses eIF4E mediated transformation |
[24, 60] | |
| Nup96 | Reduction leads to increased proliferation Reduction leads to increased export of specific mRNAs |
[68] | |
| Nup214 | Chromosomal translocations associated with rare forms of acute myeloid and acute nonlymphoblastic leukemia | Overexpression leads to growth arrest and apoptosis Overexpression leads to mRNA export defects Knockout mice not viable |
[54] |
| eIF4E | AML, lymphoma, prostate, breast, head and neck, colon and many other cancers | Overexpression leads to increased proliferation, cell survival, transformation and increased export of specific subset of mRNAs | [83] |
When conceptualizing the impact of mRNA export on oncogenesis, it is useful to recall the RNA regulon model [5, 6]. This model provides a framework in which the production of proteins can be coordinated, both transcriptionally and post-transcriptionally. Different cis-acting elements in the relevant mRNAs recruit specific RNA binding proteins to act at distinct levels of post-transcriptional control including mRNA export, stability and translation. These cis-acting elements, usually located within the untranslated regions of RNAs, are referred to as USER (untranslated sequence elements for regulation) codes [5, 6]. Transcripts encoding proteins that act in similar processes will contain USER codes that coordinate their export, as well as other post-transcriptional stages of control. Multiple USER codes can have combinatorial and/or competitive effects depending on specific cellular context, which ultimately decides the fate of a particular mRNA. Therefore USER codes play an integral role in coordinating responses to cellular stimuli and stress and ultimately cellular processes such as proliferation and survival. The roles of specific USER codes in mRNA export are described in Box 1.
BOX 1. The roles of specific USER codes in mRNA export.
A theme that is relevant to all the described mRNA export pathways is the importance of cis-acting elements in the transcripts themselves, which select for the RNA pathway to be used [5, 6]. For instance, the ARE and 4E-SE elements select for HuR-CRM1 or eIF4E-CRM1 pathways respectively and as such, are USER codes for export [5, 36]. A USER code for the export of intronless H2a mRNAs is a 22 nucleotide transport element ITE (see SR section) [26]. Another element identified in several intronless mRNAs (HSPB3, IFNα1 and IFNβ1) is the cytoplasmic accumulation region (CAR) found in the coding region, which recruits the TREX complex [86]. USER codes are important to viral mRNA export as well. For instance, the constitutive transport element (CTE) in small retroviruses allows the direct binding of viral RNA to TAP and thus permits export independent of other host cell co-factors [87]. Similarly, the Rev response element in HIV allows recruitment of the viral protein Rev and association with CRM1, permitting export [88]. Importantly, as the RNA regulon would predict, the combination of elements in the RNAs themselves will set up combinatorial and competitive scenarios thereby selecting pathways for export depending on context driven features such as levels of appropriate RNA binding proteins etc.
With one exception, nucleo-cytoplasmic trafficking of mRNAs requires transit through the nuclear membrane using the nuclear pore complex (NPC). In this exception, large mRNPs (messenger ribonucleoprotein particles) exit the nucleus by budding at the nuclear membrane [7]. For the remaining cases, cargo-receptor mRNP export complexes interact with nuclear pore proteins, and together with other crucial export factors mediate active and directional transport [8–10]. The NPC is comprised of the nuclear basket, central membrane-traversing channel and cytoplasmic fibrils (Figure 1) [1, 4, 8, 10, 11] each component playing important roles in the overall export process. The NPC and its constituent nucleoporin proteins (Nups) are also implicated in non-transport functions [1, 2, 4, 10–14]. However, in this review we will focus on their mRNA export functions and evidence that these can be dysregulated in cancer.
The mRNA export superhighway
mRNA export is a multi-step process whereby transcripts must associate with the nuclear basket of the NPC, transit through the central channel and be released at the cytoplasmic fibrils[1, 9, 15]. mRNA export can be roughly divided into two forms: bulk and specific export. Bulk refers to the majority of poly-A transcripts. For the most part, mRNAs must be correctly processed to undergo efficient export. Such processing includes the addition of a methyl-7-guanosine (m7G) cap structure to their 5′ end, splicing and appropriate 3′ end formation, typically in the form of a poly-A tail [3, 9, 16]. For the export of the majority of transcripts, the addition of the m7G cap is particularly important as it recruits the cap-binding complex (CBC), which then recruits key export factors to the mRNP. Each processing step aids in the recruitment of factors, which allow the export mRNPs to bind the NPC and traverse the hydrophobic central channel. TAP/NXF1 is the major factor that bridges the interaction between the export mRNP and the NPC. CRM1 similarly binds the NPC and is used for specific subsets of mRNAs as well as other types of RNA (see Box 2)[9, 11, 16]. Upon arrival to the cytoplasm, mRNP cargoes must be released and export factors recycled to the nucleus for future rounds of export. Dysregulation can occur at any step leading to a variety of phenotypes from proliferation to growth arrest.
BOX 2. Different types of RNAs are exported in distinct manners.
Small nuclear RNAs (U snRNAs), which are involved in splicing also contain the m7G cap and bind the CBC, similarly to mRNAs. However, U snRNAs are exported via the CRM1 pathway whereby the NES containing co-factor PHAX, associates with both the U snRNA and CRM1 for export. This pathway is RanGTP dependent [89]. Ribosomal RNAs (rRNAs) are exported as the 40S and 60S ribosomal subunits. Interestingly, the 60S subunit utilizes both CRM1 and TAP [16]. The NES containing adaptor protein Nmd3 is recruited to the 60S subunit along with CRM1 and RanGTP. Additionally, TAP/p15 associate with a distinct part of the surface of the 60S subunit indicating that ribosomal subunit export uses both CRM1 and TAP. tRNAs are exported to the cytoplasm by direct interactions with either Exportin-t or Exportin-5; while miRNAs are exported via Exportin-5 [90]. Exportin-t and Exportin-5 require no additional factors to traverse the NPC.
Diversity of the TAP mediated export pathway
In humans, the transcription export (TREX) complex plays a major role in bulk mRNA export. TREX consists of UAP56, Aly/REF, CIP29 and the multi-subunit THO complex, which is comprised of THOC1/Hpr1, hTho2, THOC5, THOC6 THOC7 and Tex1 [9, 17–19]. Through interactions with Aly/REF the THO complex bridges the interaction of cargo mRNAs and the TAP/p15 receptor [9, 19]. Recent reports suggest more diversity with specific complexes perhaps playing a role in the export of subsets of transcripts. TREX2 and alternative TREX (AREX) export complexes, have been isolated and provide examples of this diversity [17, 18] (Figure 1). All of these complexes, through TAP, associate with the nuclear basket via the ribonucleic acid export protein Rae1 and Nup98 to permit passage through the central channel [20, 21]. As described below these proteins can be dysregulated in a variety of cancers (Table 1).
Once the cargo mRNPs have reached the cytoplasmic face, they typically associate with the cytoplasmic fibrils of the NPC where they undergo cargo release and export factor recycling. This is a highly regulated process that can potently impact export efficiency. The long fibrils at the cytoplasmic face, mainly comprised of Nup358 also known as RanBP2, contain binding sites for several proteins including TAP, RanGAP, Ran and others [10, 11]. RanBP2 associates withthe NPC via two nucleoporins: Nup88 and Nup214 [22]. It plays a very important role in the cargo release and recycling for both bulk and some specific forms of mRNA export. Hypomorph mice, defined as having genetically reduced levels of RanBP2, do not have bulk mRNA export defects (but there is an upregulation of the export of specific mRNAs, see below) whereas knockout cells have severely impaired mRNA export [23, 24]. Interestingly the hypomorph mice get spontaneous cancers (see below).
The majority of transcripts are then released from the cytoplasmic face via the ATP dependent DEAD box helicase DDX19 and its co-factor Gle1. Interestingly, this step relies on a potent signaling molecule Inositol-hexakisphosphate (InsP6) where the Gle1-InsP6 complex stimulates DDX19 binding to mRNA, which triggers ATP hydrolysis and cargo release [25] [9, 10]. This provides an excellent example of how intracellular signaling can impact mRNA export. Importantly, not all mRNAs will be transported via this pathway with equal efficiency in all contexts. Further, factors such as Nup214 and Nup88 are not exclusively used for this export pathway (see below).
The SR-TAP alternative
To date, the best-defined alternative TAP export pathway involves the serine and arginine rich SR proteins. The first reports on SR proteins involvement in mRNA export were focused on intronless H2a transcripts showing that two SR proteins SRp20 and 9G8 interact with a specific 22 nucleotide element in H2a mRNA (referred to as the intronless transport element, ITE), and recruit TAP to facilitate export (see Box 1). However, further studies indicated that these factors are essential for the export of some spliced transcripts as well [26, 27]. For spliced mRNAs, SR proteins, such as hyperphosphorylated 9G8, are recruited to pre-mRNA, and become hypophosphorylated after splicing, permitting preferentially binding to TAP in the nucleus. Once in the cytoplasm, SR proteins are rephosphorylated presumably enabling release of TAP and the mRNA cargo, and facilitating their recycling into the nucleus [26]. Thus, at least two classes of adaptors, Aly/REF and SR proteins, engage the TAP receptor and promote export of mRNAs. In this way, specific subsets of mRNAs can be differentially exported and regulated despite using the same nuclear transport receptor e.g. TAP [26]. Further, this is another example of how cellular signaling can regulate mRNA export.
There is always another way out: multiple exit strategies via CRM1
Although the majority of mRNAs use the TAP receptor to transit the NPC, subsets of transcripts are exported via the CRM1 pathway. CRM1 is the major protein export receptor in the nucleus [11, 28] and directly interacts with Nups in the central channel [11]. Additionally, through protein co-factors, it is involved in the export of specific types of mRNAs, small nuclear RNAs (U snRNAs) and ribosomal RNAs (see Box 2). CRM1 interacts with its cargoes via a leucine rich nuclear export signal (NES) found in many shuttling proteins [29]. To date, CRM1 does not bind RNA directly, but rather via NES containing adaptor proteins that bind RNA or other RNA binding proteins [11]. In the nucleus, CRM1 binds its cargo in the presence of the GTP-bound form of Ran [11]. Release in the cytoplasm requires association with the RanGTPase activating protein (RanGAP) and either RanBP1 or RanBP2 enabling GTPase hydrolysis for Ran. Once this step is completed, CRM1-cargo complexes dissociate permitting the RNA to enter the cytoplasm and to recycle export factors [11]. As in bulk mRNA export, Nup88, Nup214 and RanBP2 play critical roles in the recycling and release steps for CRM1 dependent export [30, 31].
Similar to TAP, CRM1 exports mRNA cargoes via multiple pathways based on adaptor proteins and USER codes within the mRNAs (Figure 1). This heterogeneity is demonstrated in humans, where some mRNAs that contain AU-rich elements (AREs) in their 3′ UTR are subject to CRM1 dependent export via HuR [32]. CRM1 dependence is demonstrated by the nuclear accumulation of some ARE containing mRNAs, but not bulk mRNA, upon treatment with the CRM1 inhibitor Leptomycin B. Importantly, some ARE containing mRNAs can be exported in a CRM1 dependent but HuR independent manner e.g. human Interferon-alpha-1 (IFNα1)[33] suggesting further functional diversity in these pathways. Interestingly one study reports that HuD, an HuR neuron specific family member, associates with mRNA and TAP suggesting more plasticity in terms of export options for HuR proteins [34]. In another pathway, CRM1 acts in the NXF3 mediated export of tissue specific mRNAs. Unlike TAP, NXF3 (a TAP family member) cannot bind Nups and thus uses CRM1 to transit through the NPC [35]. Presumably these transcripts have a specific USER code(s) that selects for the CRM1-NXF3 pathway, but this has yet to be identified.
CRM1 also plays a major role in eukaryotic translation initiation factor 4E (eIF4E) dependent mRNA export [36, 37]. eIF4E is best known for its functions in translation of mRNAs where it binds the m7G cap and through association with cofactors, recruits these transcripts to the ribosome. However, up to 70% of eIF4E is found in the nuclei of cells depending on the tissue type [38]. eIF4E associates with the m7G cap of a subset of mature nuclear mRNAs. In this case, the CBC associates with the pre-mRNA forms of these messages and is replaced by eIF4E in the nucleoplasm [37], in contrast to other described mRNA export-pathways where transcripts associate with eIF4E only upon arrival to the cytoplasm. In this pathway, eIF4E overexpression leads to enhanced mRNA export for a subset of mRNAs that encode proteins involved in proliferation, survival, metastases and invasion [38]. Sensitive mRNAs contain an ~50 nucleotide element in their 3′UTR (4E-sensitivity element, 4E-SE), which acts as a USER code for export [36]. Transcripts must be capped and contain the 4E-SE to be eIF4E export targets. eIF4E export mRNPs contain some factors shared with the bulk export pathway: UAP56, hnRNPA1 and DDX3 but not TAP, CBC or REF/Aly [37]. Importantly, endogenous 4E-SE mRNAs are targets of both bulk and eIF4E dependent processes, where 3′ UTRs can be 1000s of nucleotides long and contain many USER codes. Thus, eIF4E competes with the bulk mRNA export pathway to preferentially enhance the export of a specific subset of transcripts.
Taken together, diversity in export pathways underpins selection of specific subsets of transcripts. In this way, cell cycle, survival and stress responses can be coordinately modulated via mRNA export as predicted by the RNA regulon model. Importantly, dysregulation of mRNA export is observed at multiple levels (Table 1) as discussed below.
mRNA export: the good, the bad and the dysregulated
Given its critical and selective role in gene expression, it is not surprising that substantial data from primary patient specimens demonstrate dysregulation of the mRNA export machinery in cancer. Overall, dysregulation of export factors and Nups in different types of cancer are diverse and appear to depend on a very context specific landscape. For instance, THOC1 (component of the TREX complex), is highly elevated in the nucleus of primary lung, ovarian and colon cancer specimens but is downregulated in skin and testes cancer specimens [39, 40]. In breast cancer, elevation of THOC1 levels is positively correlated with tumor size and metastatic state [41]. Here, THOC1 reduction leads to impaired proliferation and inhibited mRNA export suggesting that it contributes to the oncogenic phenotype by increasing export of transcripts encoding proteins involved in proliferation and survival [41]. Aly/REF protein levels are elevated in oral squamous cell carcinoma patient material and cell lines suggesting that they can promote export of transcripts encoding oncogenes [42]. Similarly, CRM1 expression is elevated in many cancers including gliomas, cervical and pancreatic cancers [43–46]. Reduction in CRM1 levels in some cell types leads to decreased proliferation suggesting a causal link between its elevation and cancer [45]. The germinal centre associated protein (GANP), a constituent of the TREX-2 complex is highly elevated in mantle cell, diffuse large B cell and Hodgkin’ s lymphomas [47]. Although initial observations suggested a more general role for GANP in bulk mRNA export [48], a recent report suggests that depletion of GANP in human cells may inhibit export of specific mRNAs [49]. In either case, GANP elevation in tumors likely drives expression of a subset of transcripts by increasing recruitment of the corresponding cargo mRNPs to the nuclear basket thereby increasing export efficiency.
Nups are also dysregulated in cancer (reviewed in [2, 4, 50]). Nup88 is elevated in many cancers including prostate, ovarian, breast, mesotheliomas, hepatocellular, colon, some lymphomas and lung cancer, andits expression correlates with advanced tumor grade [51–53]. Although the stability of Nup88 is dependent on its ability to heterodimerize with Nup214 in normal cells, Nup214 is not elevated in the above cancers indicating that this interdependence can be uncoupled during oncogenesis [52]. This suggests that the export machinery may be rearranged in cancer cells. Nup88 may be more active not only because of increased levels but also because of reduced association with Nup214, which appears to have an inhibitory role on export. For instance, under certain conditions Nup214 overexpression in human cells led to poly(A) nuclear accumulation, cell cycle arrest and apoptosis [54]. Another example of dysregulation comes from the nuclear basket protein Rae1, which is amplified in breast cancer [55].
Chromosomal translocations have been identified for many Nups. Nup214 translocations are associated with rare forms of acute myeloid and acute non-lymphoblastic leukemias. Nup98, is involved in at least 14 translocations most of which are associated with hematological malignancies including AML, CML, and MDS [2, 50]. Tpr, a nuclear basket protein involved in mRNA export, is found in the TPR-Met translocation associated with gastric carcinomas [56, 57], while the Tpr-NTrk1 translocation associates with papillary thyroid carcinomas [58]. In these translocations, the role of the fusion partner is usually unrelated to mRNA export and substantially impacts the oncogenic potential of the fusion protein. For instance, Met is a potent receptor tyrosine kinase that controls cellular proliferation, survival, migration and morphogenesis. The Tpr-Met fusion protein is a constitutively activated kinase [4, 50]. In contrast, the fusion protein associated with the RanBP2-ALK translocation, found in inflammatory myofibroblastic tumors, associates with the NPC, potentially modifying nuclear pore functions [59]. For a detailed list of observed chromosomal translocations of Nups involved in human malignancies refer to [2, 50].
Recently it was shown that the cytoplasmic fibril protein Nup358/RanBP2 acts as a tumor suppressor impairing eIF4E-mediated transformation [60]. eIF4E is highly elevated in about 30% of cancers [38], and its overexpression leads to oncogenic transformation of cell lines and tumor formation in mouse models [61, 62]. Mutational studies indicate that eIF4E’s mRNA export function contributes to its oncogenic potential by enhancing expression of target mRNAs involved in proliferation and survival [60, 63, 64]. Interestingly, eIF4E overexpression also leads to downregulation of RanBP2 and relocalization of Nup214. RanBP2 reduction is sufficient to promote eIF4E dependent (but not bulk) mRNA export while overexpression of a RanBP2 fragment that binds CRM1 impairs this export. eIF4E overexpression leads to a loss of contact inhibition, one of the hallmarks of oncogenic transformation [61]. RanBP2 overexpression inhibits this eIF4E activity [60]. Consistent with these observations, RanBP2 hypomorph mice develop more spontaneous tumors than littermate controls [24]. Aside from defects in mRNA export, severe mitotic defects in these hypomorph animals likely also contribute to tumorogenesis [23, 24]. Thus, it seems that RanBP2 slows down the release and recycling of eIF4E dependent mRNA export cargoes. In order to maximize export, eIF4E downregulates RanBP2 to reduce sequestration and increases RanBP1 to enable efficient cargo release in the cytosol, this is likely less sterically hindered than on RanBP2 fibrils. In summary, these findings suggest that the NPC can be reprogrammed by oncogenes to promote the export of specific mRNAs as part of the transformation process.
Reprogramming of the NPC has been observed in other circumstances. For instance, vesicular stomatitis virus (VSV) infection promotes export of specific viral mRNAs whereby the VSV matrix M protein disrupts interaction of Nup98 with Rae1 inhibiting host cell mRNA export [65]. Reprogramming could also occur during oxidative and metabolic stress where the NPC is known to change composition [66, 67]. Future studies will tell whether these stress-mediated changes are associated with altered mRNA export and oncogenic potential as already observed in virus infection and eIF4E overexpression.
Cell cycle dysregulation, proliferation, and stress have all been shown to contribute to the cancer phenotype and therefore the role of mRNA export in controlling these aspects of cellular physiology is of importance. For instance, Nup96, an autoproteolytic fragment of Nup98 located in the nuclear basket [21], provides an excellent example of differential regulation of mRNA export modulating progression of the cell cycle [68]. Nup96+/− T cells cycle more quickly, whereas Nup96 overexpression leads to a delay in the G1/S transition. Nup96+/− T cells show enhanced export of cell cycle specific genes, cyclin D3, CDK6 and IkB mRNAs, while Eβ mRNAs are retained in the nucleus and GAPDH, ICAM and Tubulin mRNAs are unaffected. These results suggest that Nup96 selectively impairs expression of transcripts encoding proteins that regulate cell cycle progression.
Cellular responses to stress have important physiological effects and are often disrupted in cancer. Examples of preferential mRNA export are observed during heat shock or response to cell signaling. Genome wide studies indicated that in Drosophila, hsp70 and Hsp83 transcripts are differentially exported relative to bulk mRNA [69]. In yeast, phosphorylation of specific mRNA export factors via the MAPK pathway (Slt2 kinase) in response to heat shock, inhibits bulk mRNA while promoting hsp mRNA export [70]. Importantly, export of hsp mRNA does not require factors essential for bulk mRNA export, but involves new factors such as Nup42 or THOC5 [71–73]. Thus, subgroups of transcripts can use novel adaptors to provide specificity of mRNA export under specific cellular conditions. Although these findings have been reported in yeast and Drosophila, these proteins are highly conserved suggesting that similar effects will be observed in humans.
Several signaling pathways are known to modulate mRNA export under normal conditions, and thus when dysregulated as reported in many cancers, it is likely to have impact on these processes. Such pathways include PI3K, AKT and MAPK. Aside from involvement of MAPK as described above for Slt2 kinase [70], another example includesPI3K pathway, which targets Aly/REF [74]. Aly binds PI(3,4,5)P3 and PI(4,5)P2. PI3K regulates mRNA export through the direct association of Aly with PI(3,4,5)P3 and nuclear Akt. Aly mutants that do not bind PI(3,4,5)P3 have reduced mRNA export and proliferation activity. Importantly, Akt phosphorylation is required for the PI(3,4,5)P3 binding to Aly. Similarly, the cargo release of many mRNAs is also dependent on nuclear inositol signaling. Specifically, it relies on the interaction of InsP6 with the export factor Gle1 and its partner, the ATP helicase DDX19 (discussed above) [25]. Further, it was shown that production of InsP6 is required for efficient mRNA export in human cells [75]. Thus, release and recycling steps are regulated by inositol metabolism. Further, eIF4E acts both upstream and downstream of Akt [76]. Through its mRNA export activity, eIF4E leads to enhanced export of NBS1 mRNA. The NBS1 protein directly binds PI3K enhancing Akt activation [77]. Thus, the export pathways respond to cellular stress and modulate signaling pathways, acting as regulators of gene expression during signal transduction [2] These findings suggest they may play important roles in oncogenesis.
Therapeutic benefit to targeting mRNA export?
If mRNA export contributes to oncogenesis, the expectation is that targeting this activity should have a therapeutic benefit. Obviously, given the essential nature of the process, targeting has to be done in a well-considered manner focusing only on the export of transcripts involved in oncogenesis. Currently, there is substantial interest in targeting specific export factors for therapeutic benefit. One of the first targets was CRM1 [11]. Knocking down CRM1 using siRNA or specific inhibitors restored apoptotic activity and tumor sensitivity towards the chemotherapies doxorubicin, etoposide, cisplatinum and imatinib mesylate in cell lines [46]. The well-known CRM1 inhibitor, Leptomycin B, entered phase I clinical trials, but failed due to severe toxicity. Despite this, Leptomycin B remains a profoundly useful laboratory tool. Currently, drugs to target CRM1 activity in cancer cells are under development [46]. One such compound, KPT330 is currently in phase I clinical trials (www.karyopharm.com) [78]. Normal cells tolerate KPTs but not Leptomycin B potentially because KPTs bind CRM1 reversibly whereas Leptomycin B is not reversible. In terms of specificity for cancer cells, the precise molecular mechanisms underlying these observations are not well defined but are certainly an exciting emerging area. Given the requirement of CRM1 for the export of some mRNA (and other types of RNA), the anti-oncogenic properties of KPTs and Leptomycin B may act by impairing mRNA export. Most importantly, these observations provide strong evidence that these export factors can be targeted in patients.
In M4 and M5 acute myeloid leukemia (AML), eIF4E is highly elevated and localized primarily within the nucleus [79, 80] [64]. We previously identified a cap competitor, ribavirin, which disrupts eIF4E dependent mRNA export [79, 81–83]. In a multi-centre phase II clinical trial, refractory and relapsed patients were treated with ribavirin monotherapy. Ribavirin treatment led to reduced eIF4E dependent mRNA export and this correlated with clinical responses including remissions [79]. By way of comparison, 5/11 patients had objective clinical improvement using ribavirin monotherapy (remissions and blast responses) [79] whereas in a similar patient population rapamycin led to 0/22 responses [84]. Ribavirin also impairs eIF4E dependent translation (e.g. VEGF) [38, 85] and thus its affects likely arise from inhibiting multiple functions simultaneously. Nonetheless, these observations provide a direct correlation between impairing eIF4E dependent mRNA export and clinical responses. Thus, specific targeting of mRNA export pathways can lead to clinical benefit.
Concluding remarks
A plethora of studies demonstrate that the mRNA export machinery is dysregulated in a wide variety of human tumors. Further, mRNA export factors and associated Nups can modulate cellular processes that impact malignant phenotypes such as proliferation and survival, as well as oncogenic transformation. Importantly, mRNA export can both respond to and modulate major signaling pathways consistent with being positioned as a key integrator of gene expression and cell physiology. Also, targeting mRNA export in at least one clinical trial demonstrated therapeutic benefit. It is important to note that many of the factors discussed have additional cellular roles, and it is likely that their other activities also contribute to the oncogenic phenotype. In all, mRNA export should be considered a critical step in gene regulation that likely contributes to many human malignancies.
Highlights.
There is a causal link between mRNA export and oncogenesis
Changes to the nuclear pore alter mRNA export and cancer
Strategies are emerging to target mRNA export for therapeutic benefit
Acknowledgments
KLBB is supported by grants from the LLS and NIH. She is a Canada Research Chair.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 2.Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui N, Borden KL. mRNA export and cancer. Wiley Interdiscip Rev RNA. 2011 doi: 10.1002/wrna.101. [DOI] [PubMed] [Google Scholar]
- 4.Kohler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 5.Keene JD. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology. 2010;151:1391–1397. doi: 10.1210/en.2009-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansfield KD, Keene JD. The ribonome: a dominant force in coordinating gene expression. Biol Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walde S, Kehlenbach RH. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 2010;20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Chatel G, Fahrenkrog B. Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal. 2011;23:1555–1562. doi: 10.1016/j.cellsig.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Bermejo R, Kumar A, Foiani M. Preserving the genome by regulating chromatin association with the nuclear envelope. Trends Cell Biol. 2012;22:465–473. doi: 10.1016/j.tcb.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Hou C, Corces VG. Nups take leave of the nuclear envelope to regulate transcription. Cell. 2010;140:306–308. doi: 10.1016/j.cell.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddiqui N, Borden KL. mRNA export and cancer. Wiley Interdiscip Rev RNA. doi: 10.1002/wrna.101. [DOI] [PubMed] [Google Scholar]
- 16.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Navarro S, Hurt E. Linking gene regulation to mRNA production and export. Curr Opin Cell Biol. 2011;23:302–309. doi: 10.1016/j.ceb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Luna R, Rondon AG, Aguilera A. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim Biophys Acta. 2012;1819:514–520. doi: 10.1016/j.bbagrm.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Katahira J. mRNA export and the TREX complex. Biochim Biophys Acta. 2012;1819:507–513. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, Seo HS, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci U S A. 2010;107:10406–10411. doi: 10.1073/pnas.1005389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontoura BM, Dales S, Blobel G, Zhong H. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc Natl Acad Sci U S A. 2001;98:3208–3213. doi: 10.1073/pnas.061014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernad R, van der Velde H, Fornerod M, Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Walde S, Joseph J, Kehlenbach RH, van Deursen JM. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J Cell Biol. 2011;194:597–612. doi: 10.1083/jcb.201102018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2:540–548. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen KT, Holloway MP, Altura RA. The CRM1 nuclear export protein in normal development and disease. Int J Biochem Mol Biol. 3:137–151. [PMC free article] [PubMed] [Google Scholar]
- 29.Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutten S, Kehlenbach RH. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol Cell Biol. 2006;26:6772–6785. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura T, Hashimoto I, Nagase T, Fujisawa J. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-alpha1 mRNA. J Cell Sci. 2004;117:2259–2270. doi: 10.1242/jcs.01076. [DOI] [PubMed] [Google Scholar]
- 34.Saito K, Fujiwara T, Katahira J, Inoue K, Sakamoto H. TAP/NXF1, the primary mRNA export receptor, specifically interacts with a neuronal RNA-binding protein HuD. Biochem Biophys Res Commun. 2004;321:291–297. doi: 10.1016/j.bbrc.2004.06.140. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Bogerd HP, Wang PJ, Page DC, Cullen BR. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol Cell. 2001;8:397–406. doi: 10.1016/s1097-2765(01)00303-3. [DOI] [PubMed] [Google Scholar]
- 36.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–426. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Topisirovic I, Siddiqui N, Lapointe VL, Trost M, Thibault P, Bangeranye C, Pinol-Roma S, Borden KL. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009 doi: 10.1038/emboj.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Culjkovic B, Borden KL. Understanding and Targeting the Eukaryotic Translation Initiation Factor eIF4E in Head and Neck Cancer. J Oncol. 2009;2009:981679. doi: 10.1155/2009/981679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez-Sanchez MS, Saez C, Japon MA, Aguilera A, Luna R. Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer. 2011;11:77. doi: 10.1186/1471-2407-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Li Y, Khoury T, Alrawi S, Goodrich DW, Tan D. Relationships of hHpr1/p84/Thoc1 expression to clinicopathologic characteristics and prognosis in non-small cell lung cancer. Ann Clin Lab Sci. 2008;38:105–112. [PMC free article] [PubMed] [Google Scholar]
- 41.Guo S, Hakimi MA, Baillat D, Chen X, Farber MJ, Klein-Szanto AJ, Cooch NS, Godwin AK, Shiekhattar R. Linking transcriptional elongation and messenger RNA export to metastatic breast cancers. Cancer Res. 2005;65:3011–3016. doi: 10.1158/0008-5472.CAN-04-3624. [DOI] [PubMed] [Google Scholar]
- 42.Saito Y, Kasamatsu A, Yamamoto A, Shimizu T, Yokoe H, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. ALY as a potential contributor to metastasis in human oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2012 doi: 10.1007/s00432-012-1361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen A, Wang Y, Zhao Y, Zou L, Sun L, Cheng C. Expression of CRM1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery. 2009;65:153–159. doi: 10.1227/01.NEU.0000348550.47441.4B. discussion 159–160. [DOI] [PubMed] [Google Scholar]
- 44.Huang WY, Yue L, Qiu WS, Wang LW, Zhou XH, Sun YJ. Prognostic value of CRM1 in pancreas cancer. Clin Invest Med. 2009;32:E315. [PubMed] [Google Scholar]
- 45.van der Watt PJ, Maske CP, Hendricks DT, Parker MI, Denny L, Govender D, Birrer MJ, Leaner VD. The Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int J Cancer. 2009;124:1829–1840. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83:1021–1032. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujimura S, Xing Y, Takeya M, Yamashita Y, Ohshima K, Kuwahara K, Sakaguchi N. Increased expression of germinal center-associated nuclear protein RNA-primase is associated with lymphomagenesis. Cancer Res. 2005;65:5925–5934. doi: 10.1158/0008-5472.CAN-04-3259. [DOI] [PubMed] [Google Scholar]
- 48.Wickramasinghe VO, Stewart M, Laskey RA. GANP enhances the efficiency of mRNA nuclear export in mammalian cells. Nucleus. 2010;1:393–396. doi: 10.4161/nucl.1.5.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto N, Kuwahara K, Ohta K, Kitabatake M, Takagi K, Mizuta H, Kondo E, Sakaguchi N. Germinal center-associated nuclear protein (GANP) is involved in mRNA export of Shugoshin-1 required for centromere cohesion and in sister-chromatid exchange. Genes Cells. 2010;15:471–484. doi: 10.1111/j.1365-2443.2010.01396.x. [DOI] [PubMed] [Google Scholar]
- 50.Xu S, Powers MA. Nuclear pore proteins and cancer. Semin Cell Dev Biol. 2009;20:620–630. doi: 10.1016/j.semcdb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez N, Alonso A, Moragues MD, Ponton J, Schneider J. The nuclear pore complex protein Nup88 is overexpressed in tumor cells. Cancer Res. 1999;59:5408–5411. [PubMed] [Google Scholar]
- 52.Gould VE, Martinez N, Orucevic A, Schneider J, Alonso A. A novel, nuclear pore-associated, widely distributed molecule overexpressed in oncogenesis and development. Am J Pathol. 2000;157:1605–1613. doi: 10.1016/S0002-9440(10)64798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emterling A, Skoglund J, Arbman G, Schneider J, Evertsson S, Carstensen J, Zhang H, Sun XF. Clinicopathological significance of Nup88 expression in patients with colorectal cancer. Oncology. 2003;64:361–369. doi: 10.1159/000070294. [DOI] [PubMed] [Google Scholar]
- 54.Boer J, Bonten-Surtel J, Grosveld G. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol Cell Biol. 1998;18:1236–1247. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Soman NR, Correa P, Ruiz BA, Wogan GN. The TPR-MET oncogenic rearrangement is present and expressed in human gastric carcinoma and precursor lesions. Proc Natl Acad Sci U S A. 1991;88:4892–4896. doi: 10.1073/pnas.88.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu J, Miehlke S, Ebert MP, Hoffmann J, Breidert M, Alpen B, Starzynska T, Stolte Prof M, Malfertheiner P, Bayerdorffer E. Frequency of TPR-MET rearrangement in patients with gastric carcinoma and in first-degree relatives. Cancer. 2000;88:1801–1806. [PubMed] [Google Scholar]
- 58.Pierotti MA, Greco A. Oncogenic rearrangements of the NTRK1/NGF receptor. Cancer Lett. 2006;232:90–98. doi: 10.1016/j.canlet.2005.07.043. [DOI] [PubMed] [Google Scholar]
- 59.Ma Z, Hill DA, Collins MH, Morris SW, Sumegi J, Zhou M, Zuppan C, Bridge JA. Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;37:98–105. doi: 10.1002/gcc.10177. [DOI] [PubMed] [Google Scholar]
- 60.Culjkovic-Kraljacic B, Baguet A, Volpon L, Amri A, Borden KL. The oncogene eIF4E reprograms the nuclear pore complex to promote mRNA export and oncogenic transformation. Cell Rep. 2012;2:207–215. doi: 10.1016/j.celrep.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 62.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 63.Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. Embo J. 2001;20:4547–4559. doi: 10.1093/emboj/20.16.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Topisirovic I, Guzman ML, McConnell MJ, Licht JD, Culjkovic B, Neering SJ, Jordan CT, Borden KL. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol. 2003;23:8992–9002. doi: 10.1128/MCB.23.24.8992-9002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17:93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 66.Kodiha M, Tran D, Morogan A, Qian C, Stochaj U. Dissecting the signaling events that impact classical nuclear import and target nuclear transport factors. PLoS One. 2009;4:e8420. doi: 10.1371/journal.pone.0008420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crampton N, Kodiha M, Shrivastava S, Umar R, Stochaj U. Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol Biol Cell. 2009;20:5106–5116. doi: 10.1091/mbc.E09-05-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chakraborty P, Wang Y, Wei JH, vanDeursen J, Yu H, Malureanu L, Dasso M, Forbes DJ, Levy DE, Seemann J, et al. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev Cell. 2008;15:657–667. doi: 10.1016/j.devcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farny NG, Hurt JA, Silver PA. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 2008;22:66–78. doi: 10.1101/gad.1616008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carmody SR, Tran EJ, Apponi LH, Corbett AH, Wente SR. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol Cell Biol. 2010;30:5168–5179. doi: 10.1128/MCB.00735-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rollenhagen C, Hodge CA, Cole CN. Following temperature stress, export of heat shock mRNA occurs efficiently in cells with mutations in genes normally important for mRNA export. Eukaryot Cell. 2007;6:505–513. doi: 10.1128/EC.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vainberg IE, Dower K, Rosbash M. Nuclear export of heat shock and non-heat-shock mRNA occurs via similar pathways. Mol Cell Biol. 2000;20:3996–4005. doi: 10.1128/mcb.20.11.3996-4005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katahira J, Inoue H, Hurt E, Yoneda Y. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 2009;28:556–567. doi: 10.1038/emboj.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okada M, Jang SW, Ye K. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc Natl Acad Sci U S A. 2008;105:8649–8654. doi: 10.1073/pnas.0802533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Y, Wente SR, Majerus PW. Overexpression of the inositol phosphatase SopB in human 293 cells stimulates cellular chloride influx and inhibits nuclear mRNA export. Proc Natl Acad Sci U S A. 2001;98:875–879. doi: 10.1073/pnas.021558098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Culjkovic B, Tan K, Orolicki S, Amri A, Meloche S, Borden KL. The eIF4E RNA regulon promotes the Akt signaling pathway. J Cell Biol. 2008;181:51–63. doi: 10.1083/jcb.200707018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen YC, Su YN, Chou PC, Chiang WC, Chang MC, Wang LS, Teng SC, Wu KJ. Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J Biol Chem. 2005;280:32505–32511. doi: 10.1074/jbc.M501449200. [DOI] [PubMed] [Google Scholar]
- 78.Azmi AS, Aboukameel A, Bao B, Sarkar FH, Philip PA, Kauffman M, Shacham S, Mohammad RM. Selective Inhibitors of Nuclear Export Block Pancreatic Cancer Cell Proliferation and Reduce Tumor Growth in Mice. Gastroenterology. doi: 10.1053/j.gastro.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH, Jr, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114:257–260. doi: 10.1182/blood-2009-02-205153. [DOI] [PubMed] [Google Scholar]
- 80.Kraljacic BC, Arguello M, Amri A, Cormack G, Borden K. Inhibition of eIF4E with ribavirin cooperates with common chemotherapies in primary acute myeloid leukemia specimens. Leukemia. 2011;25:1197–1200. doi: 10.1038/leu.2011.57. [DOI] [PubMed] [Google Scholar]
- 81.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kentsis A, Volpon L, Topisirovic I, Soll CE, Culjkovic B, Shao L, Borden KL. Further evidence that ribavirin interacts with eIF4E. Rna. 2005;11:1762–1766. doi: 10.1261/rna.2238705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borden KL, Culjkovic-Kraljacic B. Ribavirin as an anti-cancer therapy: Acute Myeloid Leukemia and beyond? Leukemia and Lymphoma. 2010 doi: 10.3109/10428194.2010.496506. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizzieri DA, Feldman E, Dipersio JF, Gabrail N, Stock W, Strair R, Rivera VM, Albitar M, Bedrosian CL, Giles FJ. A phase 2 clinical trial of deforolimus (AP23573, MK-8669), a novel mammalian target of rapamycin inhibitor, in patients with relapsed or refractory hematologic malignancies. Clin Cancer Res. 2008;14:2756–2762. doi: 10.1158/1078-0432.CCR-07-1372. [DOI] [PubMed] [Google Scholar]
- 85.Graff JR, Zimmer SG. Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin Exp Metastasis. 2003;20:265–273. doi: 10.1023/a:1022943419011. [DOI] [PubMed] [Google Scholar]
- 86.Lei H, Dias AP, Reed R. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc Natl Acad Sci U S A. 2011;108:17985–17990. doi: 10.1073/pnas.1113076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 88.Yi R, Bogerd HP, Cullen BR. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J Virol. 2002;76:2036–2042. doi: 10.1128/jvi.76.5.2036-2042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 90.Leisegang MS, Martin R, Ramirez AS, Bohnsack MT. Exportin T and Exportin 5: tRNA and miRNA biogenesis - and beyond. Biol Chem. 2012;393:599–604. doi: 10.1515/hsz-2012-0146. [DOI] [PubMed] [Google Scholar]