SUMMARY
Stalled replication forks are sources of genetic instability. Multiple fork remodeling enzymes are recruited to stalled forks, but how they work to promote fork restart is poorly understood. By combining ensemble biochemical assays and single molecule studies with magnetic tweezers, we show that SMARCAL1 branch migration and DNA annealing activities are directed by the single-stranded DNA binding protein RPA to selectively regress stalled replication forks caused by blockage to the leading-strand polymerase and to restore normal replication forks with a lagging-strand gap. We unveil the molecular mechanisms by which RPA enforces SMARCAL1 substrate preference. E. coli RecG acts similarly to SMARCAL1 in the presence of E. coli SSB, whereas the highly related human protein ZRANB3 has different substrate preferences. Our findings identify the important substrates of SMARCAL1 in fork repair, suggest that RecG and SMARCAL1 are functional orthologues, and provide a comprehensive model of fork repair by these DNA translocases.
INTRODUCTION
During S-phase, DNA replication forks encounter many obstacles that block the replicative DNA polymerase and induce fork stalling including unrepaired DNA damage, DNA-bound proteins, and DNA secondary structure. If left unrepaired, stalled forks can collapse, generate DNA double strand breaks (DSB), and be a source of the chromosome rearrangements frequently observed in cancer cells.
The DNA damage response (DDR) pathway works to prevent fork collapse by stabilizing the stalled fork, regulating DNA repair, and promoting replication restart (Branzei and Foiani, 2010). DDR proteins are recruited to stalled forks through multiple mechanisms including interactions with the single-stranded DNA (ssDNA) binding protein replication protein A (RPA). A small amount of RPA-ssDNA is present on the lagging-strand template during normal replication due to the discontinuous nature of lagging-strand synthesis. However, DNA damage can generate RPA-ssDNA on the leading-strand template due to uncoupling of the replicative DNA helicase and leading-strand polymerase (Byun et al., 2005). Fork stalling induces activation of the checkpoint kinase ATR which phosphorylates hundreds of proteins to control the replication stress response (Cimprich and Cortez, 2008).
One of these ATR substrates is SMARCAL1, otherwise known as HARP (Bansbach et al., 2009; Postow et al., 2009). SMARCAL1 travels with the replisome during an unperturbed S-phase (Betous et al., 2012), and is concentrated at stalled forks via a direct interaction with RPA (Bansbach et al., 2009; Ciccia et al., 2009; Yuan et al., 2009; Yusufzai et al., 2009). Cells lacking SMARCAL1 are hyper-sensitive to replication stress (Bansbach et al., 2009; Ciccia et al., 2009; Yuan et al., 2009; Yusufzai et al., 2009) and accumulate DSBs during DNA replication due to fork cleavage by the MUS81 endonuclease (Betous et al., 2012). Too much SMARCAL1 activity also causes fork-related damage indicating that SMARCAL1 must be regulated to prevent it from interfering with normal DNA replication (Bansbach et al., 2009). Inherited, biallelic, loss of function mutations in SMARCAL1 cause the disease Schimke immunoosseous dysplasia (SIOD) characterized by bone growth defects, renal failure, immune deficiencies and cancer predisposition (Baradaran-Heravi et al., 2012).
Biochemically, SMARCAL1 is a DNA-dependent ATPase in the SNF2 family. It binds a broad range of DNA substrates that have both single and double-stranded regions (Betous et al., 2012). SMARCAL1 has the ability to anneal two complementary DNA strands (Yusufzai and Kadonaga, 2008). Furthermore, SMARCAL1 binds and branch migrates synthetic Holliday junctions and model replication forks (Betous et al., 2012; Ciccia et al., 2012). This activity promotes fork regression into a “chicken foot” structure, which can be an intermediate in damaged replication fork repair.
Several other enzymes including FANCM, ZRANB3, WRN and BLM can catalyze similar reactions on synthetic DNA substrates (Ciccia et al., 2012; Gari et al., 2008a; Gari et al., 2008b; Machwe et al., 2006; Ralf et al., 2006). ZRANB3 is also a SNF2 family member with sequence similarity to SMARCAL1. ZRANB3 is recruited to stalled replication forks but through an interaction with PCNA instead of RPA (Ciccia et al., 2012; Yuan et al., 2012). SMARCAL1 and ZRANB3 do not act redundantly since knockdown of either causes replication-associated DNA damage. It is unclear how these enzymes convert the energy of ATP hydrolysis into fork remodeling activity. One model is that they act like E. coli RecG to translocate on double-stranded DNA and have a domain that inserts into the DNA branch point to facilitate fork regression (Singleton et al., 2001).
Here we investigated the mechanism by which SMARCAL1 repairs damaged replication forks. We find that SMARCAL1 can catalyze both fork regression and restoration. Its interaction with RPA directs these activities to provide the specificity needed to yield regression of damaged forks and restoration of normal fork structures. Single molecule studies reveal how RPA enforces this substrate preference, and comparisons to other enzymes indicate a functional relationship between SMARCAL1 and E. coli RecG. Together our results explain how SMARCAL1 is directed to remodel and repair stalled replication forks.
RESULTS
RPA directs SMARCAL1 to selectively remodel stalled replication forks with a leading-strand gap
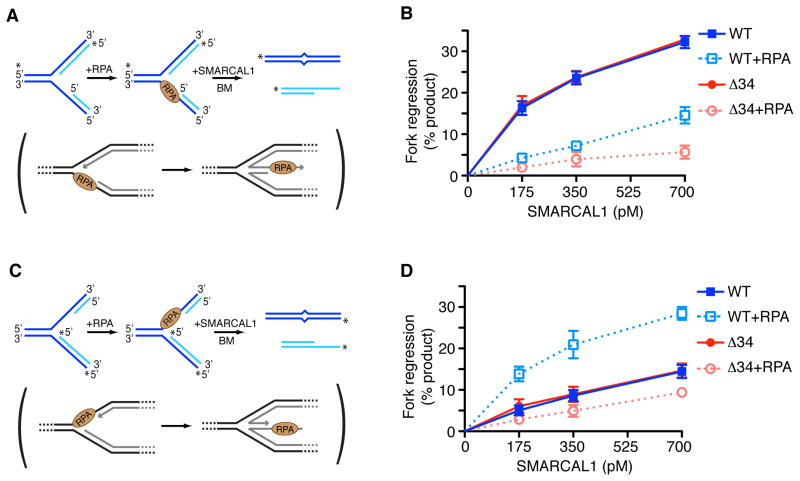
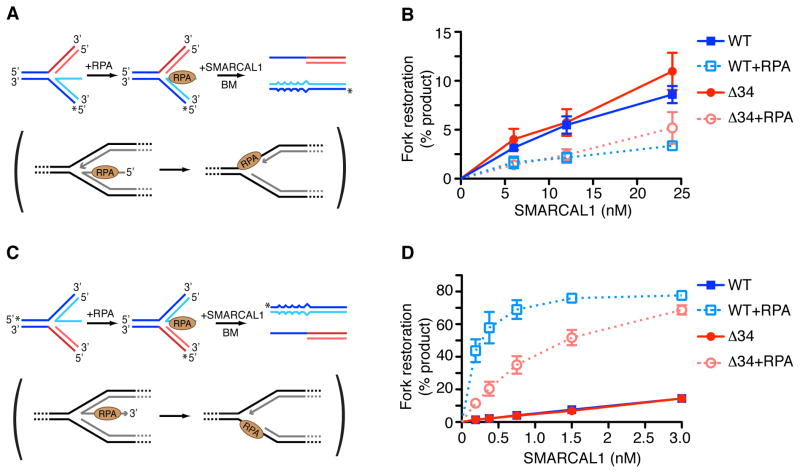
SMARCAL1 catalyzes fork regression of synthetic replication forks at least when no single-stranded DNA gap or RPA is present on the substrate (Betous et al., 2012; Ciccia et al., 2012). Fork regression provides a mechanism of repair but should not happen at actively elongating forks. Yet, SMARCAL1 is present even at elongating replication forks since its interaction with RPA is not regulated (Betous et al., 2012). Therefore, there must be mechanisms to direct its activity specifically to damaged forks and restrain its activity at normal forks. A ssDNA gap is present on the lagging strand at normal elongating forks. When a replisome stalls at a DNA lesion on the leading-strand template, uncoupling between the replicative polymerase and helicase creates a ssDNA gap on the leading-strand template. In both cases the ssDNA is bound by RPA. To test if ssDNA gaps and RPA influence SMARCAL1 function, we designed leading- and lagging-strand gap fork substrates containing a 32nt ssDNA gap that can accommodate one RPA molecule in its high affinity DNA bound conformation (Fig. 1A and C). In the absence of RPA, SMARCAL1 has higher activity on a lagging-strand gap replication fork than on a leading-strand gap replication fork (Fig. 1B, 1D, and Fig S1A, S1B).
Figure 1.
RPA directs SMARCAL1 specifically to a damaged replication fork substrate and inhibits its action at a normal fork. (A,C) Diagram of the lagging (A) or leading (C) gap replication fork regression assay. 32P-labeled strands are indicated with asterisks. A two base-pair mismatch is present on the parental (black) strands to prevent spontaneous branch migration. The physiological reaction mimicked by the experimental assay is shown in brackets. (B,D) Lagging (B) or leading (D) gap replication fork substrates were incubated 15min at room temperature in the presence or absence of RPA. Increasing amount of SMARCAL1 WT or Δ34 were added to the reaction and further incubated 20min at 30°C. DNA products were analyzed by native gel electrophoresis and phosphorimager quantitation of three experiments is shown (mean +/− SD). See also Fig. S1.
However, RPA inhibits SMARCAL1-catalyzed fork regression of a lagging gap replication fork and stimulates fork regression of a leading gap substrate (Fig. 1B, 1D, and Fig. S1A and S1B). In these experiments RPA was pre-bound to the gapped substrates such that all of the substrate molecules had one RPA molecule bound to the gap (Fig. S1C and S1D). This stimulation of SMARCAL1 on the leading gap substrate relies on a direct SMARCAL1-RPA interaction since RPA failed to stimulate a SMARCAL1-Δ34 mutant that lacks its RPA binding domain and cannot interact with RPA (Bansbach et al., 2009). Moreover, the bacterial single-strand binding protein SSB that does not interact with SMARCAL1 inhibits SMARCAL1 fork regression of both leading- and lagging-strand gap replication forks (Fig. S1E). Thus, RPA switches SMARCAL1 substrate preference for fork regression from a lagging to a leading gapped replication fork. This change in substrate preference enforced by RPA would help inhibit SMARCAL1 from working to regress an elongating, normal replication fork and stimulate it to regress a damaged fork.
SMARCAL1 binds the two strands of the DNA duplex asymmetrically
To understand how RPA modulates SMARCAL1 we asked if it affects SMARCAL1 ATPase activity. RPA minimally stimulates SMARCAL1 activity when it is bound both to the leading- and lagging-template strand (Fig. S2A and B). Thus, a change in ATPase activity cannot explain how RPA switches SMARCAL1 substrate preference. Likewise, the difference in substrate preference cannot be explained by a change in affinity to the substrate since SMARCAL1 binds similarly to both substrates with and without RPA (Fig. S2C and D).
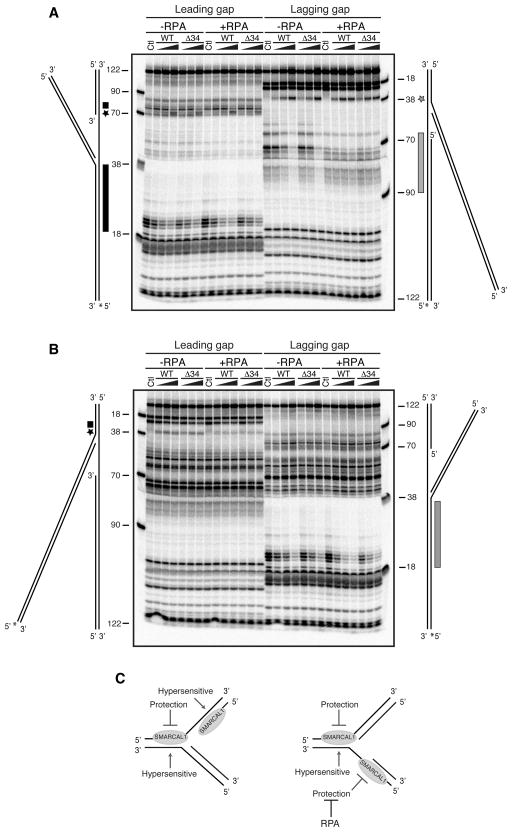
The DNA substrates used in the fork regression assay present two possible binding sites for SMARCAL1–either at the junction of parental strands forming the fork itself or on the other side of the gap at the junction of nascent/parental strands. To test whether the differences in substrate preference were due to changes in where SMARCAL1 binds when RPA is present we performed DNA footprinting studies. First we labeled either the leading or lagging template strand containing the ssDNA gap. SMARCAL1-WT or Δ34 protected about 20nt of the dsDNA portion of the parental strand duplex on the leading gap substrate (Fig. 2A, black rectangle). A DNase hypersensitive site is observed at the same position on the lagging gap substrate perhaps suggesting a bending of the DNA induced by SMARCAL1 (Fig. 2A, grey star). SMARCAL1 strongly protects the lagging-strand template duplexed with the lagging nascent strand (Fig. 2A, grey bar). In contrast, only high concentrations of SMARCAL1 induce a change in digestion pattern at this position on the leading gap substrate (Fig. 2A, black star and small rectangle). Labeling the other template DNA strand shows that SMARCAL1 protects this strand on the lagging gap substrate (Fig. 2B, grey bar), whereas it induces a hypersensitive site on the leading gap substrate (Fig. 2B, black star).
Figure 2.
SMARCAL1 binds asymmetrically to double-stranded DNA on the gapped fork substrates. (A,B) In each experiment, increasing amounts of SMARCAL1 WT or Δ34 was incubated with the labeled substrate for 15min prior to addition of nuclease. A diagram of each DNA substrate is placed next to the gel and stretched to correspond to the location of the size standards. The 32P-labelled DNA strand is indicated with an asterisk (*). Where indicated RPA was pre-bound to the substrate at a concentration sufficient to yield 100% binding. After nuclease digestion at levels titrated to yield single cleavage events per DNA substrate, the reaction products were heat denatured and separated on an 8% polyacrylamide denaturing sequencing gel. The control (Ctl) samples are the DNA substrates in the absence of SMARCAL1 protein. The stars and boxes next to the DNA substrate indicate nuclease hypersensitive and protected regions induced by SMARCAL1. (C) Summary of the modification of the digestion patterns observed in footprinting studies.
Most of the footprinting patterns were not affected by RPA. However, the binding of SMARCAL1 to the nascent/parental strand duplex on the lagging gap replication fork substrate was reduced (Fig. 2A, grey bar) suggesting that RPA prevents SMARCAL1 binding at this position of the DNA substrate (Fig. 2A). This effect was independent of the SMARCAL1-RPA interaction since both SMARCAL1 WT and Δ34 binding were affected.
The footprinting results are summarized in Fig. 2C. These data confirm that these substrates present two potential binding sites for SMARCAL1, although only binding at the fork junction is likely to allow fork regression. Both wild type and Δ34 SMARCAL1 binding to the parental and nascent strand duplex is reduced in the presence of RPA on the lagging strand gap substrate. The data also indicate that SMARCAL1 binds asymmetrically to the dsDNA. SMARCAL1 protects the parental leading-strand template at the fork regardless of the position of the gap or presence of RPA. SMARCAL1 binding to the fork also always induces a hypersensitive site on the lagging-strand template consistent with a conformational change of the DNA. Asymmetric binding to the dsDNA suggests SMARCAL1 may translocate with a specific polarity along a single strand of the DNA duplex similar to other dsDNA translocases (Singleton et al., 2007). The direct interaction of SMARCAL1 with RPA has no effect on these DNA binding patterns; therefore, they cannot explain why RPA stimulates the fork regression activity of the wild-type but not Δ34 SMARCAL1 protein on a leading-strand gap substrate.
SMARCAL1 catalyzes repetitive bursts of annealing activity
We next considered the possibility that RPA alters the ability of SMARCAL1 to convert ATP hydrolysis into movement. Fork regression requires concerted annealing of the parental strands, displacement of the nascent strands, and annealing of the nascent strands to one another. Since SMARCAL1 lacks helicase activity in standard helicase assays (Yusufzai and Kadonaga, 2008), strand displacement must be coupled to strand annealing. To understand if RPA changes the annealing or strand displacement ability of SMARCAL1, we used a single-molecule approach.
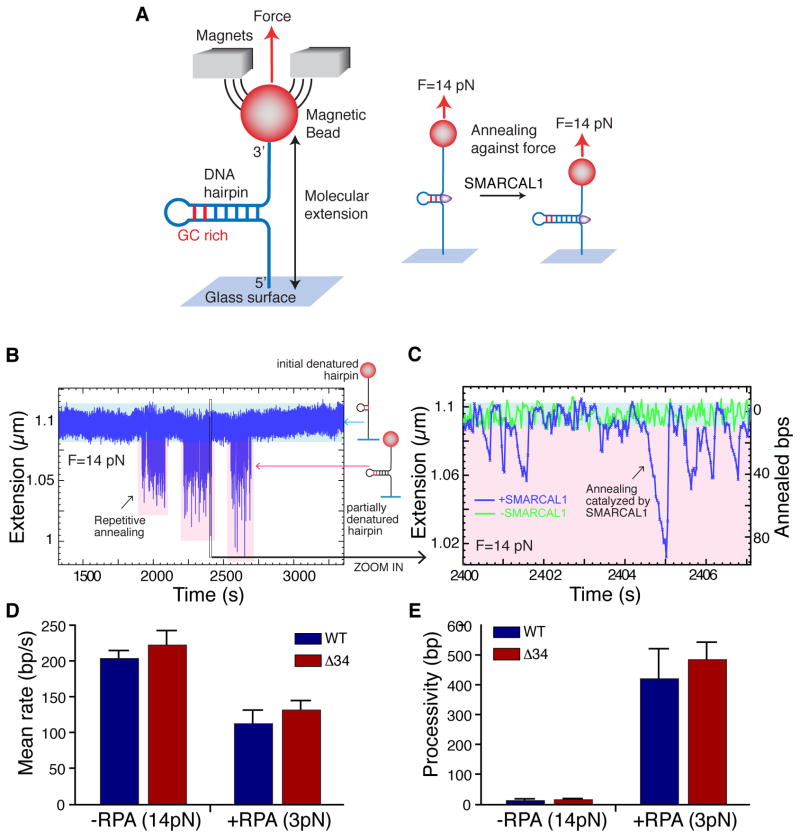
First, we used an assay to monitor strand-annealing (Manosas et al., 2012a) in which one end of a 1.2kbp DNA hairpin substrate is attached to a glass slide and the other end is attached to a magnetic bead. Application of a magnetic field gradient to generate a force unwinds the hairpin except for the last 20–30 nucleotides, which stay in a double stranded conformation due to high GC content. Annealing of the unwound strands is monitored after injection of enzyme and ATP by measuring the distance between the glass surface and magnetic bead (the molecular extension) (Fig. 3A).
Figure 3.
SMARCAL1 catalyzes repetitive rounds of DNA annealing. (A) Schematic of the magnetic tweezers single molecule annealing experiment. Details are described in supplemental methods. (B and C) Experimental traces corresponding to background fluctuation (green) or SMARCAL1 annealing activity (blue). Example of repetitive annealing events catalyzed by single SMARCAL1 molecules at 30pM enzyme concentration is shown. Panel C is a zoom in of a small portion of panel B. (D, E) Mean annealing rates (D) and processivity (E) of SMARCAL1 wild type (WT) or RPA-binding mutant (Δ34) in presence or absence of RPA calculated by fitting the distribution of rates and processivity to a Gaussian function and exponential function respectively. At least 75 traces were analyzed per condition. See also Fig. S3.
In the presence of ATP, SMARCAL1 catalyzes bursts of repetitive annealing of the DNA strands against the applied force as seen by a decrease in the extension length (Fig. 3B and C). At low enzyme concentration, these bursts of activity last for 150–250 seconds and are separated from each other by times of no activity (few hundreds of seconds), suggesting that a single molecule of SMARCAL1 performs each repetitive annealing burst.
Each burst corresponds to multiple cycles of ATP hydrolysis and protein movement. In this system the annealing of a single base-pair (bp) corresponds to an extension change of approximately 1nm (Manosas et al., 2012a). At 14pN the measured SMARCAL1 annealing rate is approximately 200 bp/second at physiological levels of ATP (Fig. S3A and B). The mean processivity, measured as the number of base pairs annealed prior to pausing, dissociation, or strand switching is 15 +/− 3 bp per annealing event at this force level (Fig. S3C).
To address whether RPA affects SMARCAL1 annealing activity, we added RPA to the single molecule substrate prior to introducing SMARCAL1. At high forces, RPA destabilizes the DNA helix resulting in melting/unwinding of dsDNA. Consequently, generation of the RPA-bound substrate and its analysis had to be completed at reduced force (3pN) (see supplemental methods). In these conditions the SMARCAL1 annealing rate is reduced modestly (40%) indicating the increased difficulty in reannealing the DNA strands when RPA must be displaced (Fig. 3D). However, SMARCAL1 processivity is greatly increased to approximately 400bp/annealing event in the presence of RPA (Fig. 3E). Wild-type and the Δ34 protein exhibit similar annealing rates and processivity at the same force in the absence and presence of RPA (Fig. 3D and E). Therefore, the SMARCAL1-RPA interaction does not influence SMARCAL1 annealing activity when RPA is bound to both ssDNA segments. The increase in SMARCAL1 processivity when RPA is present is likely due to the decrease in force needed to complete the experiment with RPA although we cannot rule out other effects of RPA in this context.
RPA influences SMARCAL1 fork regression activity by affecting the distance it moves
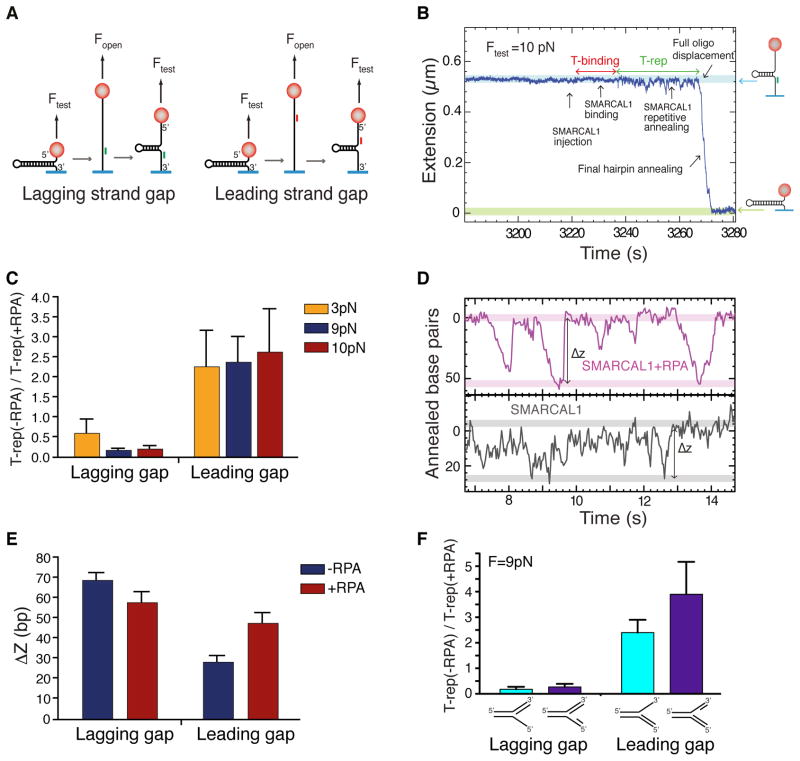
Next we asked whether RPA influences substrate specificity by altering SMARCAL1 DNA-strand displacement activity during the fork regression reaction. To generate the strand displacement substrates an oligonucleotide complementary to a central region of the hairpin is annealed to a fully denatured hairpin, and the force is decreased to allow the hairpin to spontaneously reanneal until the stem of the hairpin encounters the oligonucleotide. This creates a hairpin with long ssDNA tails and containing a duplex region at the junction between the hairpin stem and the 3′ or 5′ arm (Fig. 4A and Fig. S4). Hairpins with a duplex region on the 3′ or 5′ arms were respectively called lagging- and leading-strand gap substrates since they mimic replication forks with long lagging- and leading-strand gaps.
Figure 4.
RPA increases the distance SMARCAL1 travels per annealing event specifically on a DNA substrate that mimics a stalled replication fork with a leading strand gap. (A) Schematics of the strand displacement substrate construction with a gap on the lagging or leading strand. Details are described in Extended Experimental Procedures and Fig. S4. (B) Example of a typical experimental trace displaying characteristic features of the strand displacement reaction (SMARCAL1 binding, SMARCAL1 repetitive annealing, full oligonucleotide displacement and final hairpin annealing) at 200pM enzyme concentration. The molecular extensions corresponding to the initial strand displacement substrate and the fully formed hairpin are highlighted in blue and green respectively. (C) The ratio of T-rep measured in the absence or presence of RPA was measured at three different Ftest forces for both lagging and leading strand gap substrates. (n = 58–89 depending on condition). Error bars represent SD. The experiment at 10pN was done with a wash step after RPA binding to remove any free RPA molecules excluding the possibility that free RPA contributes to the differences. (D) Typical trace of the leading strand gap substrate showing repetitive annealing events in absence or presence of RPA. ΔZ represent the amplitude of the repetitive annealing events measured in number of annealed base pairs. (E) Mean and SD of ΔZ in the absence or presence of RPA for lagging or leading strand gap substrates. The difference in ΔZ measured in assays in absence and presence of RPA is significant (independent 2-group t-test: P=0.006 and P= 1.9e–07 for lagging and leading strand gap substrates). (F) The presence of a second oligonucleotide to generate a true 64 bp gap on the lagging or leading strand did not change the ability of RPA to inhibit or activate SMARCAL1 respectively. See also Fig. S4.
After SMARCAL1 injection there is a period of inactivity (T-binding) representing the time required for SMARCAL1 to bind the DNA substrate (Fig. 4B). Upon SMARCAL1 binding a period of sustained unproductive repetitive movement was observed (T-rep). Eventually, full oligonucleotide displacement allows rapid final hairpin annealing (Fig. 4B). Thus, T-binding measures any effect of oligonucleotide position or RPA on the ability of SMARCAL1 to load onto the substrate while T-rep measures the efficiency with which it displaces the oligonucleotide during the annealing reaction.
T-rep is influenced by the length of the duplexed region with longer oligonucleotides requiring more time to be fully displaced. In each case though, T-rep is considerably shorter for the lagging gap substrate compared to the leading gap substrate (Fig. S4C and D), and the ratio of the T-rep for the lagging vs. leading substrate remains the same (Fig. S4E). This is consistent with the observations from the ensemble studies where SMARCAL1 also showed a preference for the lagging strand gap substrate in the absence of RPA (Fig. 1).
We measured T-binding and T-rep at three different forces for both the leading and lagging strand gap substrates in the absence or presence of RPA. T-binding was equivalent in all conditions (data not shown) indicating that neither the oligonucleotide orientation nor RPA affects SMARCAL1 binding to the DNA substrates consistent with the ensemble results. Addition of RPA significantly increases the T-rep measured on the lagging gap substrate and decreases the T-rep measured for the leading gap substrate (Fig. 4C and Fig. S4). Again, these results are consistent with the RPA effects we observed on SMARCAL1 fork regression activity in the ensemble reactions although the stimulation of SMARCAL1 by RPA on the leading gap substrate and inhibition on the lagging gap is not sufficient to fully reverse the substrate preference in this experimental setup. Despite absolute differences in T-rep in the three force conditions the relative effects of RPA were similar suggesting that the inhibition and stimulation on the lagging and leading gap substrates respectively by RPA are not artifacts of the applied force.
The average distance that SMARCAL1 moves (ΔZ, see Fig. 4D) during each repetitive annealing event prior to full oligonucleotide dissociation and final hairpin annealing is different for each substrate. In the absence of RPA, ΔZ is greater for the lagging gap substrate than the leading gap substrate (Fig. 4E). RPA bound to the lagging gap substrate decreased ΔZ while RPA bound to the leading gap substrate increased ΔZ (Fig. 4E). Therefore, the values of T-rep and ΔZ are inversely correlated (Fig. S4I). RPA also had the same effects on SMARCAL1 activity toward single molecule substrates containing true gaps on either the leading or lagging strands, which were generated by annealing a second oligonucleotide to either the template leading or lagging strands respectively (Fig. 4F).
These results indicate that RPA influences SMARCAL1 fork regression activity by affecting the distance it translocates during each repetitive annealing and strand-displacement event. Specifically, RPA increases the distance SMARCAL1 moves on a leading gap substrate and decreases it on the lagging gap substrate.
RPA stimulates SMARCAL1 to catalyze normal replication fork restoration when the nascent leading strand is longer than the lagging strand
The regression of a fork into a chicken-foot structure could be an intermediate in fork repair or a pathological event caused by torsional stress or aberrant fork processing. In either case restarting replication requires restoration of a normal fork structure. In principle, SMARCAL1 could participate in the fork restoration reaction. Restoration would require branch migration of the regressed chicken-foot structure back towards the fork, and our previous studies indicated that SMARCAL1 can catalyze this reaction (Betous et al., 2012). Furthermore, using magnetic tweezers and a substrate that mimics a stalled fork without a gap, we have observed that SMARCAL1 regresses the fork forming a Holliday junction, migrates the Holliday junction, and frequently switches directions towards either fork regression or restoration (Fig. S5). However, in cells the leading and lagging nascent strands are likely to have different lengths. Therefore, regressed forks may have an extra ssDNA protrusion, which will be bound by RPA. Thus, either a nuclease activity to remove this ssDNA end or an activity to remove the RPA and anneal the nascent ssDNA to the template strand would be needed to fully restore the fork.
To investigate whether SMARCAL1 could catalyze this reaction we designed a replication fork restoration assay using DNA substrates mimicking partially regressed replication forks with a protruding 32nt ssDNA tail (Fig. 5A and C). The substrates contain mismatches to prevent spontaneous branch migration, contain heterologous arms to allow measurement of only fork restoration, bind one molecule of RPA in its high-affinity DNA bound state, and contain either a 3′ or 5′ “nascent” ssDNA tail. Branch migration of these substrates would restore replication forks with a ssDNA gap either on the lagging- or the leading-template strand depending on whether the nascent leading or lagging strand was longer respectively.
Figure 5.
SMARCAL1 preferentially catalyzes fork restoration that yields a normal lagging strand gap replication fork in the presence of RPA. (A,C) Schematic of the leading (A) or lagging (C) gap replication fork restoration assay. 32P-labeled strands are indicated with asterisks. Mismatches in the DNA strands were inserted between the longest nascent strand and the corresponding parental strand to prevent spontaneous fork restoration. In brackets is the physiological reaction mimicked by the experimental assay. (B,D) Leading (B) or lagging (D) gap replication fork restoration substrates were incubated 15min at room temperature in the presence or absence of RPA sufficient to bind 100% of the DNA substrate. Increasing amounts of SMARCAL1 WT or Δ34 were added to the reaction and further incubated 20min. DNA products were analyzed by native gel electrophoresis. Mean +/− SD from three independent experiments is depicted. See also Fig. S6 and Extended Experimental Results.
SMARCAL1 catalyzed fork restoration of both substrates but with very different efficiencies. In the absence of RPA, SMARCAL1 has a strong preference for restoring a lagging-strand gap since much higher SMARCAL1 concentrations were required to restore a replication fork with a leading-strand gap (Fig. 5B and Fig. S6). This difference in substrate preference was exacerbated when RPA was bound. RPA strongly stimulated SMARCAL1 activity to restore a normal fork configuration with a lagging-template strand gap while inhibiting its ability to restore the leading-strand gap fork (Fig. 5D and Fig. S6). The RPA stimulation partially depends on the direct interaction between SMARCAL1 and RPA since the SMARCAL1-Δ34 protein activity is also stimulated although not as strongly (Fig. 5B and Fig. S6).
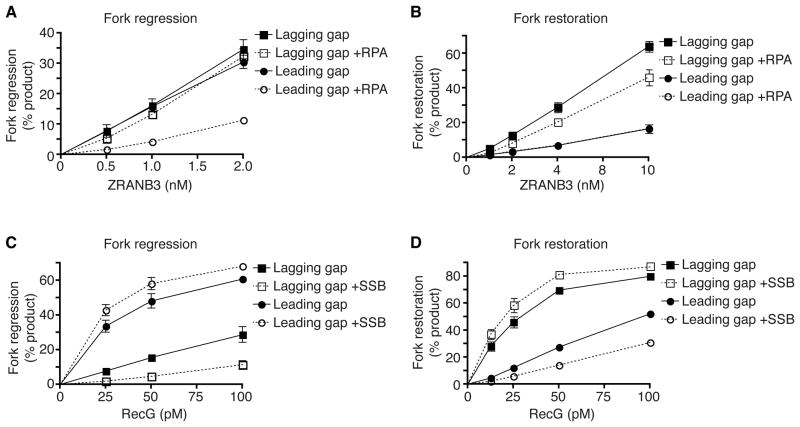
E. coli RecG, but not human ZRANB3, shares similar substrate preferences to SMARCAL1
To determine if the SMARCAL1 family member ZRANB3 shares a similar substrate preference we assayed its activity using substrates with ssDNA gaps. In contrast to SMARCAL1, ZRANB3 has no intrinsic preference for regressing either lagging- or leading-strand gap forks (Fig. 6A). Furthermore, unlike SMARCAL1, RPA inhibits ZRANB3 fork regression activity on leading-strand gap substrates. When tested in the fork restoration assay, ZRANB3 exhibits a preference to restoring a normal replication fork but the presence of RPA inhibits its activity (Fig. 6B). These results suggest that SMARCAL1 and ZRANB3 may have different substrates at stalled forks perhaps explaining why they work non-redundantly to promote fork repair and restart. Future studies to determine how PCNA affects ZRANB3 may be informative.
Figure 6.
E. coli RecG but not human ZRANB3 exhibits similar substrate preferences to SMARCAL1 in the presence of ssDNA binding proteins. (A,C) Leading or lagging strand gap fork regression substrates were incubated 15min in the presence or absence of RPA (A) or SSB (C). Increasing amounts of ZRANB3 (A) or RecG (C) were then added to the reaction and further incubated 20min. The DNA products were analyzed by native gel electrophoresis. (B,D) Fork restoration substrates were incubated with RPA or SSB and ZRANB3 or RecG as indicated. Reaction products were analyzed by native gel electrophoresis. Mean +/−SD from three experiments is shown in all graphs.
SMARCAL1 fork remodeling activities are reminiscent of the bacterial helicase RecG (McGlynn and Lloyd, 2000). Analogously to SMARCAL1, RecG directly interacts with E. coli single strand DNA binding protein (SSB) (Buss et al., 2008), SSB modestly enhances RecG activity (Robu et al., 2004), and RecG catalyzed DNA unwinding in the absence of SSB is greater on model substrates with ssDNA on the leading strand arms (modeling a leading strand gap) than when the ssDNA is on the lagging strand arms (McGlynn and Lloyd, 2001). To more directly compare RecG and SMARCAL1 we tested if RecG has any specificity for fork regression to normal (lagging strand gap) or damaged (leading strand gap) forks in the presence or absence of SSB. RecG prefers to regress a leading-strand gap substrate in the absence of SSB, and SSB further accentuates this difference in specificity by inhibiting RecG activity on the lagging-strand gap substrate and slightly stimulating its activity on the leading-strand gap substrate (Fig. 6C).
When tested using the fork restoration assay, RecG also exhibited a similar substrate preference as SMARCAL1. RecG prefers to work on a regressed fork with a longer nascent leading-strand yielding restoration of a normal fork with a lagging-strand gap (Fig. 6D). Again, SSB further accentuates this substrate preference. As might be expected for a prokaryotic enzyme, RecG has significantly higher activity at lower enzyme concentrations than SMARCAL1 (Fig. S6F). Nonetheless. in the presence of their ssDNA binding protein partners, both human SMARCAL1 and E. coli RecG prefer to regress stalled forks that would be generated by leading-strand template damage and restore normal replication forks with a lagging-strand gap.
DISCUSSION
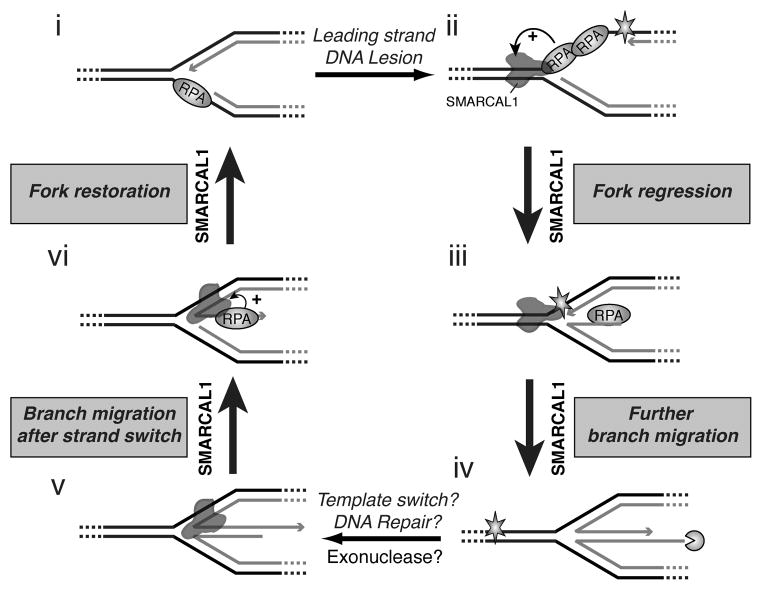
Our results support a comprehensive model of SMARCAL1 function in fork stabilization and repair (Fig. 7). First, the ssDNA binding protein RPA directs SMARCAL1 to catalyze fork regression on only a damaged replication fork that would be generated by stalling of the leading strand polymerase and prevents this activity at normal forks. Also, RPA inhibits SMARCAL1 binding to the lagging nascent/parental strands junction (Fig. 2), which may help prevent SMARCAL1 from interfering with Okazaki fragment processing. Other mechanisms, such as the coupling of helicase and polymerase are also likely to prevent SMARCAL1 from acting at actively elongating forks as they do in other systems (Manosas et al., 2012a). The overexpression of SMARCAL1 may cause fork damage (Bansbach et al., 2009) because higher concentrations of SMARCAL1 overcome these regulatory mechanisms.
Figure 7.
Model for damaged replication fork repair by SMARCAL1. (i–ii) RPA inhibits SMARCAL1 from regressing normal elongating forks with lagging-strand template gaps. DNA damage on the leading-strand template induces stalling of the replicative DNA polymerase and generation of a leading-strand ssDNA. (ii–iii) RPA stimulates SMARCAL1 fork regression activity on this stalled fork. (iii–iv) Continued branch migration yields a true chicken foot structure. (iv–v) Fork regression permits repair of the DNA lesion in the context of dsDNA or could allow template switching. The nascent lagging-strand of the regressed fork can be digested by a 5′ to 3 ′ exonuclease, which may form regressed fork with a longer nascent leading strand. (v–vi) Strand switching and branch migration would yield a partially regressed replication fork with a ssDNA strand 3 ′ tail corresponding to the nascent leading-strand. (vi–i) RPA stimulates SMARCAL1 to re-anneal the nascent leading ssDNA strand with the complementary parental strand to reform a normal DNA replication fork that can resume DNA replication. This model is similar to those described for E. coli RecG function (Gregg et al., 2002; McGlynn and Lloyd, 2000), although it also incorporates the substrate specificity dictated by single-stranded DNA binding proteins.
Second, SMARCAL1 catalyzes fork restoration after fork regression. Critically, RPA again enforces a strong specificity on SMARCAL1 that ensures it catalyzes fork restoration when the nascent leading-strand is longer than the nascent lagging-strand. Thus, SMARCAL1 will restore a normal fork configuration with a lagging template-strand gap but is inhibited from restoring a damaged fork configuration with a leading template-strand gap.
The combination of these RPA-directed substrate preferences combined with its Holliday junction migration activity allows SMARCAL1 to catalyze fork remodeling to deal with leading strand damage. Fork regression would place the DNA lesion back into the context of dsDNA where it can be repaired or template switching could bypass the lesion. Either template switching or limited 5′ to 3′ exonuclease activity may yield a blunt end in the middle toe of the chicken foot structure. Additional exonuclease degradation of the nascent lagging-strand would generate a longer 3′ tail. Branch migration and RPA-stimulated, SMARCAL1-dependent fork-restoration would then yield a normal fork configuration with a lagging-template strand gap (Fig. 7).
This model predicts that a 5′– 3′ exonuclease is involved prior to the fork restoration step. Indeed, in S. cerevisiae both Dna2 and Exo1 process regressed forks, and the Mre11 nuclease is present at damaged forks in vertebrates to promote fork restart (Costanzo et al., 2001; Cotta-Ramusino et al., 2005; Hu et al., 2012; Mirzoeva and Petrini, 2003; Trenz et al., 2006). A second prediction of the model is that in the absence of SMARCAL1, stalled forks with either a leading-strand gap or perhaps an intermediate in fork reversal should accumulate. These intermediates would be good substrates for the MUS81 structure-specific endonuclease (Osman and Whitby, 2007). Indeed, MUS81 catalyzes double-strand break formation in SMARCAL1-deficient cells (Betous et al., 2012).
Mechanism of SMARCAL1 fork remodeling
Our single-molecule studies reveal that SMARCAL1 acts to anneal DNA in a repetitive fashion. This repetitive activity is independent of the applied force or presence of RPA. Repetitive bursts of activity have also been observed for other DNA tracking enzymes like BLM and may be important for the enzyme’s function (Yodh et al., 2009). This repetitive action might correspond to repetitive cycles of catalysis followed by slippage of the enzyme allowing mechanical unwinding of the substrate due to the applied force. Slippage in the activity of several nucleic acid translocases has been observed (Manosas et al., 2012b; Myong et al., 2007; Sun et al., 2011). Alternatively, the repetitive activity might be generated through a strand switching mechanism in which the enzyme changes how it is bound to the substrate (Dessinges et al., 2004; Yodh et al., 2009). Strand switching would enable a switch from fork regression to restoration (Fig. S7).
The single-molecule studies also suggest that the mechanism for how RPA binding to SMARCAL1 enforces a substrate preference for fork regression is at least partially due to a change in distance of SMARCAL1 movement per round of activity before slipping or switching strands. When RPA is bound to a substrate mimicking a leading-strand gap, the distanceSMARCAL1 moves per enzymatic burst is larger than when RPA is absent. The direct interaction between SMARCAL1 and RPA may assist in the RPA displacement reaction by serving as an anchoring point for SMARCAL1 thereby allowing it to move further during each reaction cycle.
The RPA stimulation of SMARCAL1 is dependent on the DNA substrate. In fact, RPA inhibits SMARCAL1 when it is bound to a lagging strand gap or a 5′ DNA tail in the fork regression or fork restoration assays respectively. This difference may be explained by the asymmetric binding of RPA to ssDNA with four DNA binding domains (Fanning et al., 2006) SMARCAL1 prefers fork regression and fork restoration substrates in which the highest affinity DNA binding domains are positioned on the ssDNA closest to the fork and the weaker DNA binding domains are positioned further away (Fig. S7). The interaction surface of RPA for SMARCAL1 is attached to the weak DNA binding domain in RPA32. When all four DNA binding domains of RPA are bound, the ssDNA adopts a conformation that brings the RPA70A and RPA32D domains closer together (Fan and Pavletich, 2012). Nonetheless, the orientation specificity of RPA on the ssDNA necessarily controls the distance between the DNA and protein interaction surfaces for SMARCAL1 on the RPA-bound fork substrates. The combination of RPA DNA binding orientation and location of the SMARCAL1 binding surface provide a basis for the differences in SMARCAL1 movement and enzymatic activity.
Are RecG and SMARCAL1 functional orthologues?
Fork repair and restart through fork regression has been documented in prokaryotes and even bacteriophage (Gregg et al., 2002; Long and Kreuzer, 2009; Manosas et al., 2012a; McGlynn and Lloyd, 2000, 2001). The E. coli fork regression enzyme RecG shares some properties with SMARCAL1, and our model of SMARCAL1 function to repair damaged replication forks is similar to the models developed for RecG (Gregg et al., 2002; McGlynn and Lloyd, 2000). Our data further extend these models by demonstrating that ssDNA binding proteins help to enforce substrate specificity on these proteins.
RecG activity is critical to prevent lethality arising from 3′ ssDNA flap accumulation especially during DNA replication stress (Rudolph et al., 2010). RPA strongly stimulates SMARCAL1 activity to restore normal replication forks from partially regressed replication forks with 3′ ssDNA flaps. Therefore, we propose that like RecG, SMARCAL1 may prevent accumulation of these partially regressed 3′ ssDNA flap structures in cells, which would be efficiently cleaved by MUS81 yielding the replication associated DSBs observed in SMARCAL1-deficient cells (Betous et al., 2012).
Structural studies show that RecG catalyzes fork regression by acting as a dsDNA translocase with a wedge domain that binds at the fork junction to promote specific DNA remodeling activities (Singleton et al., 2001). SMARCAL1 also acts as a dsDNA translocase. Based on homology with the RAD54 SNF2 family member and our footprinting studies, the SMARCAL1 ATPase domains likely bind to the dsDNA (Durr et al., 2005). The SMARCAL1 HARP domains are candidates for wedge domain activity since they are essential for DNA binding and enzyme function (Betous et al., 2012; Ghosal et al., 2011).
There are also some differences between these enzymes. For example, RecG has a preference to regress DNA substrates with leading-strand gaps even without SSB, whereas, SMARCAL1 requires RPA to be present to have this preference. In addition, RecG has measurable helicase activity, which has not yet been observed for SMARCAL1. Nonetheless, the similarities in substrate preference indicate that they are likely functional orthologues with similar mechanism to maintain genome integrity during DNA replication.
SMARCAL1 has unique biochemical activities not shared by related fork processing enzymes
A close sequence paralog of SMARCAL1, ZRANB3, also is recruited to stalled replication forks to promote stabilization and restart (Ciccia et al., 2012; Yuan et al., 2012). Knockdown of ZRANB3 and SMARCAL1 yield similar phenotypes and ZRANB3 also has annealing helicase and fork regression activities (Ciccia et al., 2012; Yuan et al., 2012; Yusufzai and Kadonaga, 2010). So why are there two enzymes with similar functions in the cell? One difference is that ZRANB3 does not bind RPA. Instead it is recruited to damaged forks through an interaction with PCNA (Ciccia et al., 2012; Yuan et al., 2012). Our data indicate that this difference in interacting proteins also yields a difference in substrate preference. ZRANB3 is not stimulated by RPA. In fact, its abilities to regress a damaged replication fork or restore a normal fork are inhibited when RPA is present. Thus, ZRANB3 is likely to participate in a different reaction at stalled forks. Similarly, we also have compared SMARCAL1 to the WRN helicase and found that SMARCAL1 is much more active as a fork regression enzyme (R.B. and D.C., unpublished data). Comparisons with additional fork remodeling enzymes like FANCM and HLTF in the presence of RPA and other accessory proteins will be needed to fully understand the unique abilities of each of these enzymes. For example, the FANCM accessory protein FAAP24 has ssDNA binding properties and dramatically increases FANCM binding to splayed arm substrates (Ciccia et al., 2007).
Conclusions
Our discovery that SMARCAL1 is directed by RPA to have specific substrate preferences explains how it acts to promote damaged replication fork repair and restart. This activity appears to be functionally similar to the activity of E. coli RecG in fork repair. The ability of SMARCAL1 to restore a regressed fork into a normal fork structure may be important more generally outside the context of damaged fork repair. Fork regression can happen due to torsional stress when forks encounter tethered regions of chromosomes or during replication termination. SMARCAL1 may be needed in these circumstances if RPA stabilizes the reversed fork structure. In the absence of SMARCAL1, the regressed forks become substrates for endonucleases yielding DSBs increasing the chance of chromosomal rearrangements or cell death.
EXPERIMENTAL PROCEDURES
Protein purification and ensemble biochemical assays
Flag-SMARCAL1 was purified from baculovirus-infected insect cells as described previously (Betous et al., 2012). ZRANB3 was purified from HEK-293T cells transfected with His-Flag-ZRANB3 expression plasmid as described for SMARCAL1 human cell expression (Betous et al., 2012). Recombinant RecG was purified from E. coli using pGS772-RecG expression plasmid (Lloyd and Sharples, 1993) obtained from Piero Bianco (University of Buffalo, NY). Details of protein purification and substrate preparation are presented in extended experimental procedures. For fork regression and restoration assays, 3nM of DNA substrates were incubated with 6nM of RPA or SSB and increasing amounts of SMARCAL1, ZRANB3 or RecG in reaction buffer (40mM Tris (pH=7.5), 100mM KCl, 5mM MgCl2, 100μg/ml BSA, 2mM ATP and 2mM DTT) for 20min at 30°C. Fork restoration assays with SMARCAL1 and ZRANB3 were performed at 37°C for 20min. Samples were separated on 8% polyacrylamide gels. The DNA binding and ATPase assays were performed as described previously (Betous et al., 2012).
DNA footprinting
5nM of purified DNA substrate was incubated with or without RPA and increasing concentration of SMARCAL1 (2.5, 5 and 10nM) in footprinting buffer (20mM Hepes pH7.6, 0.1M KCl, 5mM MgCl2, 250ng/ml BSA, 1% glycerol, 0.1% IGEPAL CA-630 and 1mM DTT). Samples were cooled on ice for 3min and incubated for 2min in the presence of 40mU of Benzonase (Novagen). Reactions were stopped with 95% formamide and 20mM EDTA, denatured, and separated by electrophoresis.
Single molecule studies
Bead images were acquired at 30 Hz using a PicoTwist inverted microscope and the DNA extension was measured by tracking the bead position in real time (Gosse and Croquette, 2002). The force was estimated by using a force versus magnet vertical position calibration curve. The 1.2 Kbp hairpin with modified tails to attach to the magnetic bead and the glass surface was prepared as described elsewhere (Manosas et al., 2009). Experiments with SMARCAL1 were performed at 37°C in 10 mM Tris–Ac (pH 7.5), 40 mM KOAc, 1 mM Mg(OAc)2, 0.5 mM dithiothreitol and 1.5 mM ATP unless otherwise indicated. The protein concentration was 30–200pM SMARCAL1 and 3nM RPA.
Supplementary Material
HIGHLIGHTS.
RPA directs SMARCAL1 to regress stalled replication forks with a leading strand gap
RPA directs SMARCAL1 to restore normal forks with a lagging strand gap
Single molecule analyses indicate RPA controls bursts of SMARCAL1 translocation
E. coli RecG, but not ZRANB3, works similarly to SMARCAL1
Acknowledgments
This work was supported by NIH grant R01CA136933 to DC, a DOD BCRP fellowship (W81XWH-10-1-0581) to RB, ACS fellowship (PF-12-220-01) to ACM, and NIH fellowship (F31CA171586) to FBC. MM is supported by Juan de la Cierva Program MICINN-JDC. Recombinant protein expression was assisted by core facilities supported by P01CA092584. Michelle Spiering (The Pennsylvania State University, PA) prepared the 1.2 Kbp hairpin. Vincent Croquette (ENS Paris) built the magnetic tweezers instrument, contributed to the initial discussions of the research, and critically read the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Maria Manosas, Email: mmanosas@ub.edu.
David Cortez, Email: david.cortez@vanderbilt.edu.
References
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran-Heravi A, Raams A, Lubieniecka J, Cho KS, DeHaai KA, Basiratnia M, Mari PO, Xue Y, Rauth M, Olney AH, et al. SMARCAL1 deficiency predisposes to non-Hodgkin lymphoma and hypersensitivity to genotoxic agents in vivo. American journal of medical genetics Part A. 2012;158A:2204–2213. doi: 10.1002/ajmg.a.35532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Buss JA, Kimura Y, Bianco PR. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic acids research. 2008;36:7029–7042. doi: 10.1093/nar/gkn795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Molecular cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. Polyubiquitinated PCNA Recruits the ZRANB3 Translocase to Maintain Genomic Integrity after Replication Stress. Molecular cell. 2012 doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Molecular cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Molecular cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci U S A. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic acids research. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008a;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Molecular cell. 2008b;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Ghosal G, Yuan J, Chen J. The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1. EMBO reports. 2011;12:574–580. doi: 10.1038/embor.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosse C, Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophysical journal. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Molecular cell. 2002;9:241–251. doi: 10.1016/s1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W, et al. The intra-s phase checkpoint targets dna2 to prevent stalled replication forks from reversing. Cell. 2012;149:1221–1232. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Lloyd RG, Sharples GJ. Dissociation of synthetic Holliday junctions by E. coli RecG protein. The EMBO journal. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Kreuzer KN. Fork regression is an active helicase-driven pathway in bacteriophage T4. EMBO reports. 2009;10:394–399. doi: 10.1038/embor.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- Manosas M, Perumal SK, Croquette V, Benkovic SJ. Direct observation of stalled fork restart via fork regression in the T4 replication system. Science. 2012a;338:1217–1220. doi: 10.1126/science.1225437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Spiering MM, Ding F, Bensimon D, Allemand JF, Benkovic SJ, Croquette V. Mechanism of strand displacement synthesis by DNA replicative polymerases. Nucleic acids research. 2012b;40:6174–6186. doi: 10.1093/nar/gks253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nature chemical biology. 2009;5:904–912. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc Natl Acad Sci U S A. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res. 2003;1:207–218. [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem. 2009;284:35951–35961. doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- Robu ME, Inman RB, Cox MM. Situational repair of replication forks: roles of RecG and RecA proteins. J Biol Chem. 2004;279:10973–10981. doi: 10.1074/jbc.M312184200. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Mahdi AA, Upton AL, Lloyd RG. RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics. 2010;186:473–492. doi: 10.1534/genetics.110.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- Sun B, Johnson DS, Patel G, Smith BY, Pandey M, Patel SS, Wang MD. ATP-induced helicase slippage reveals highly coordinated subunits. Nature. 2011;478:132–135. doi: 10.1038/nature10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. The EMBO journal. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. The EMBO journal. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The HARP-like Domain-Containing Protein AH2/ZRANB3 Binds to PCNA and Participates in Cellular Response to Replication Stress. Molecular cell. 2012 doi: 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc Natl Acad Sci U S A. 2010;107:20970–20973. doi: 10.1073/pnas.1011196107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.