Abstract
The cyclic peptide phalloidin binds and stabilizes actin filaments. It is widely used in studies of actin filament assembly, including analysis of branch formation by Arp2/3 complex, but its influence on the branching reaction has not been considered. Here we show that rhodamine-phalloidin binds both Arp2/3 complex and the VCA domain of Arp2/3 complex activator, hWASp, with dissociation equilibrium constants of about 100 nM. Not only does phalloidin promote nucleation of pure actin monomers but it also dramatically stimulates branch formation by actin, Arp2/3 complex, and hWASp-VCA more than 10-fold and inhibits dissociation of branches. Therefore, the appearance of more branches in samples treated with rhodamine-phalloidin arises from multiple influences of the peptide on both the formation and dissociation of branches.
Many studies of actin filaments in cells and in biochemical experiments make use of fluorescent derivatives of the peptide phalloidin to label filaments. Fluorescent phalloidin stabilizes actin filaments and allows for dilution and the visualization of the products of the reactions. A useful feature of rhodamine-phalloidin is that its fluorescence is much higher when bound to actin filaments than when free (1).
One application of rhodamine-phalloidin has been to visualize actin filament branches formed by Arp2/31 complex and a nucleation promoting factor such as the VCA domain of human Wiskott–Aldrich syndrome protein (hWASp-VCA) (2). Further experiments showed that the number of branches preserved for observation declined exponentially with the interval of time between the onset of the polymerization and the addition of rhodamine-phalloidin (3, 4). This observation was interpreted to be due to stabilization of branches by phalloidin.
We revisited the interactions of phalloidin with actin in the presence of Arp2/3 complex and hWASp-VCA. We found that phalloidin binds not only actin filaments but also Arp2/3 complex and hWASp-VCA. Phalloidin dramatically increases nucleation of filaments by pure actin and by actin with Arp2/3 complex and hWASp-VCA. Arp2/3 complex forms many more new barbed ends in solution than observed by fluorescence microscopy of the same samples labeled with rhodamine-phalloidin, so many branches dissociate quickly in both the presence and absence of phalloidin. The number of branches observed microscopically declines over time but can be stabilized with rhodamine-phalloidin. Thus phalloidin has multiple influences on actin filament branch formation by Arp2/3 complex.
EXPERIMENTAL PROCEDURES
Proteins
We purified actin from a chicken skeletal muscle acetone powder using one round of polymerization and depolymerization followed by gel filtration on Sephacryl S300 (5). Part of the actin was labeled on Cys374 with pyrenyliodoacetamide (6). Fresh actin was stored in buffer G (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT) at 4 °C and used within 3 weeks of preparation. Immediately before use we dialyzed the actin for 24 h vs buffer G and centrifuged at 100000g for 2 h.
Arp2/3 complex was prepared from bovine thymus and stored frozen in KMEI buffer (50 mM KCl, 1 mM EGTA, 1 mM MgCl2, and 10 mM imidazole, pH 7.0) containing an additional 0.2 mM ATP and 20% glycerol (7).
We expressed hWASp-VCA and hWASp-CA fused to glutathione S-transferase (GST) in Escherichia coli BL21 cells and purified the fusion proteins by affinity chromatography on glutathione–Sepharose with elution by excess glutathione (7). Alternatively, we cleaved VCA or CA from the GST with thrombin while the fusion protein was bound to the affinity column. Both products were purified further by ion-exchange chromatography on Source Q-Sepharose. Fusion proteins were stored at −80 °C in 10 mM imidazole, pH 7.0. The C domain fragment (463–484) was synthesized, purified by HPLC (SYNPEP, Inc., SynPep Corp., Dublin, CA), stored dry, and resuspended in 10 mM imidazole, pH 7.0, prior to use.
Protein concentrations were determined by absorbance using the following extinction coefficients: actin (∊290 = 25974 M−1 cm−1), Arp2/3 complex (∊290 = 139000 M−1 cm−1), GST-hWASp-VCA (∊280 = 46200 M−1 cm−1), hWASp-VCA (∊280 = 5600 M−1 cm−1), and hWASp-CA (∊280 = 5600 M−1 cm−1).
Microscopy Assays
We used rhodamine-phalloidin (phalloidin labeled with tetramethylrhodamine B isothiocyanate; Fluka) to visualize actin filaments by fluorescence microscopy. Rhodamine-phalloidin was prepared in methanol according to the manufacturer’s specifications (∊552 = 85000 M−1 cm−1) and stored at −20 °C. We mixed actin, 10 nM Arp2/3 complex, and 300 nM GST-hWASp-VCA in KMEI buffer at time zero and added rhodamine-phalloidin equimolar to actin either together with the proteins or separately at a later time point in the reaction. After 30 min 10 μL of the reaction mixture was transferred carefully with a blunt pipet tip into 100 μL of fluorescence buffer [50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10 mM imidazole, pH 7.0, 100 mM DTT, 20 μg/mL catalase (Sigma C-40 from bovine liver), 100 μg/mL glucose oxidase (Sigma G-6125 type II from Aspergillus niger), 3 mg/mL glucose, and 0.5% methylcellulose (2000 cps at 2%)] (8). The sample was diluted further by gently adding another 900 μL of fluorescence buffer to the existing 110 μL for a total dilution of 100-fold. Mixing was accomplished by gently rotating the tube. Two microliters of the diluted sample was placed in the center of a glass slide, and a glass coverslip was lowered slowly onto the drop so that it spread smoothly out from the center to the edge. Coverslips were sealed around the edges with nail polish, and images were taken with a Hamamatsu ER ORCA CCD camera on an Olympus IX70 inverted microscope with epifluorescence illumination. Branches were stable for several hours.
To quantify the branch density, we flattened the average image intensity across the field and used a threshold of fluorescence to form a binary image of the filaments. Using ImageJ, we recorded the area (A) and perimeter (p) of each fluorescent structure and calculated the length (l) using the formula:
To obtain the total length of filament in each image, we added the calculated lengths of all the fluorescent structures that satisfied the following constraint: p2 > 16A. Branches in the image were counted by hand and recorded separately. Combined with the total length, we determined the branches per unit length for each condition.
Nucleation Assays
Nucleation experiments were performed in Corning 96-well flat-bottom plastic half-area plates (no. 3694, Corning Inc., Corning, NY) on a Molecular Devices Spectromax fluorescence plate reader (excitation, 362 nm; emission, 407 nm). Experiments for a range of actin concentrations (30% pyrenyl-actin) were initiated simultaneously by the addition of concentrated KMEI buffer and in some experiments addition of 5 or 10 nM Arp2/3 complex and 300 nM GST-VCA. Unlabeled phalloidin to match the actin concentration was introduced simultaneously with concentrated KMEI buffer, complex, and activator in the relevant experiments to mimic conditions used in phalloidin microscopy assays. The total volume in each well was 150 μL, and the time between starting a reaction and beginning readings was timed (average 15 s).
To obtain the polymer concentration from the fluorescence change, we subtracted the baseline fluorescence from the total fluorescence signal obtained with the same concentration of actin in G-buffer. We then normalized the maximum fluorescence signal with the final predicted concentration of actin polymer. In the absence of phalloidin, this quantity was the total monomer concentration minus the critical concentration (0.17 μM). In the presence of phalloidin we assumed a critical concentration of zero.
Concentrations of ends ([ends]) were calculated from the slopes of the polymerization curves (slope), actin elongation rate constants k+ of 7.1 μM−1 s−1 with phalloidin (9) or 12.9 μM−1 s−1 without phalloidin (10), dissociation rate constants k− of 0 with phalloidin or 1.3 s−1 without phalloidin, and instantaneous actin concentration ([G]):
Initial portions of plots of [ends] vs time were fit to a straight line as a measure of nucleation rate.
Binding Assays by Fluorescence Change
All equilibrium binding reactions were prepared in glass tubes at room temperature and incubated in the dark for 30 min before the fluorescence was measured with a PTI Alphascan fluorometer (excitation, 535 nm; emission, 575 nm). Reactions included 10 nM rhodamine-phalloidin in KMEI buffer with a range of concentrations of the target molecule (actin filaments, Arp2/3 complex, or hWASp-VCA). Competition experiments began with samples containing a high concentration of target molecule and 10 nM rhodamine-phalloidin in KMEI buffer. We added to these samples small aliquots of a concentrated solution of unlabeled phalloidin (Sigma p-2141) with 10 nM rhodamine-phalloidin and the same concentration of the target molecule as the test sample and reread the fluorescence after 30 min.
Binding Assays by Fluorescence Anisotropy
We titrated Arp2/3 complex into a sample containing 25 nM hWASp-VCA labeled with rhodamine-maleimide on a cysteine added to the N-terminus (11). We measured fluorescence anisotropy with a PTI Alphascan spectrofluorometer (excitation, 535 nm; emission, 570 nm) equipped with film polarizers in a T-configuration. Anisotropy was calculated using PTI Felix software.
Calculation of Dissociation Equilibrium Constants
To obtain the dissociation equilibrium constants for each interaction, we first subtracted from each data point the signal with no target molecule. Experimental values were inverted for competition assays where the fluorescence signal drops as unlabeled ligands saturate the targets. The data were then fit to the equation:
The dissociation equilibrium constant (Kd) and fluorescence conversion factor (cf) are fitting parameters while the concentrations of target molecule (x) and rhodamine-labeled partner ([R]) are known. The final fluorescence signal (fs) is either the increase in fluorescence seen on binding, decrease in fluorescence seen in competition experiments, or change in anisotropy observed in the reaction between rhodamine-labeled hWASp-VCA with Arp2/3 complex. The error values given are standard errors of the deviation of the data points from the curve fit, not the variation of multiple measurements of the Kd.
RESULTS
Rhodamine-Phalloidin Binding to Actin Filaments, Arp2/3 Complex, and VCA ActiVators
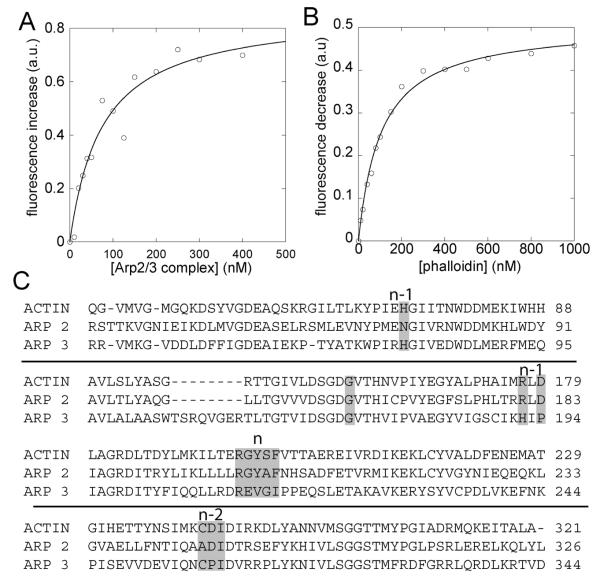
We used the fluorescence of rhodamine-phalloidin to study its interaction with Arp2/3 complex and VCA activators. The fluorescence of rhodamine-phalloidin increased nearly 2-fold with the concentration of Arp2/3 complex up to a plateau (Figure 1A). Fitting the binding isotherm gave a Kd of 67 ± 16 nM for a 1:1 bimolecular reaction of rhodamine-phalloidin with Arp2/3 complex. Unlabeled phalloidin competed rhodamine-phalloidin from Arp2/3 complex. The dependence on the concentration of unlabeled phalloidin gave a Kd of 25 ± 4 nM for unlabeled phalloidin binding Arp2/3 complex (Figure 1B).
Figure 1.
Binding of rhodamine-phalloidin and phalloidin to Arp2/3 complex measured by steady-state fluorescence. Experiments were performed in 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, and 10 mM imidazole, pH 7.0. (A) Dependence of the fluorescence of 10 nM rhodamine-phalloidin on the concentration of Arp2/3 complex. The data fit a bimolecular reaction curve with Kd = 67 ± 16 nM. (B) Titration of 10 nM rhodamine-phalloidin and 500 nM Arp2/3 complex with unlabeled phalloidin. This competition curve fits a Kd = 25 ± 4 nM for unlabeled phalloidin binding to Arp2/3 complex. (C) Comparison of partial amino acid sequences of actin, Arp2, and Arp3. Highlighted residues are in contact with or in close proximity to phalloidin bound to actin filaments.
hWASp-VCA also binds rhodamine-phalloidin. The fluorescence of rhodamine-phalloidin increased over 2-fold when titrated with GST-hWASp-VCA (Figure 2A) or hWASp-VCA (Figure 2B), giving Kd values of approximately 100 nM. Rhodamine-phalloidin also bound the CA region of VCA with a 6-fold lower affinity (Figure 2B) and the C region of VCA with a 12-fold lower affinity than VCA (Figure 2B). Purified GST had no effect on the fluorescence of rhodamine-phalloidin.
Figure 2.
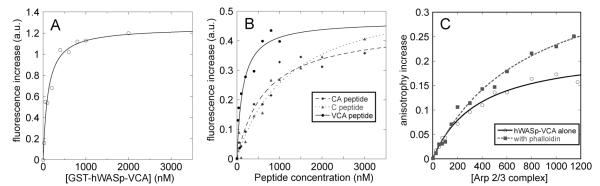
Interactions of phalloidin with human hWASp-VCA (residues 427–502) and of hWASp-VCA with Arp2/3 complex measured by fluorescence. Experiments were performed in 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, and 10 mM imidazole, pH 7.0. (A–C) Interaction of rhodamine-phalloidin with hWASp-VCA measured by steady-state fluorescence. (A) Titration of 10 nM rhodamine-phalloidin with GST-hWASp-VCA. The curve corresponds to a bimolecular reaction with Kd = 110 ± 24 nM. (B) Titration of 10 nM rhodamine-phalloidin with hWASp-VCA, hWASp-CA (residues 448–502), and hWASp-C (residues 463–485). The Kd’s are 134 ± 55 nM for VCA, 650 ± 175 nM for CA, and 1.22 ± 0.36 μM for C. (C) Interaction of rhodamine-hWASp-VCA with Arp2/3 complex measured by fluorescence anisotropy. Titration of 25 nM rhodamine-hWASp-VCA with Arp2/3 complex ± 2 μM unlabeled phalloidin. These data were fit to a bimolecular reaction curve in both the presence (Kd = 740 ± 110 nM) and absence of phalloidin (Kd = 350 ± 50 nM), although these are approximate values owing to the lack of saturation.
Effect of Phalloidin on Interaction of Arp2/3 Complex and VCA Activators
Because the phalloidin binds both the VCA activator and Arp2/3 complex, we tested whether the unlabeled phalloidin influences binding of hWASp-VCA to Arp2/3 complex (Figure 2C). We measured the anisotropy of rhodamine-labeled hWASp-VCA peptide as a function of the concentration of Arp2/3 complex. The presence of phalloidin increased the amplitude of the anisotropy change, but the anisotropy change did not plateau at 1.2 μM Arp2/3 complex, the highest concentration tested. We estimate that phalloidin reduced the affinity of hWASp-VCA for Arp2/3 complex by at least a factor of 2 and that a higher plateau value of the anisotropy would be reached in the presence of phalloidin.
Nucleation of Actin Filaments with and without Phalloidin
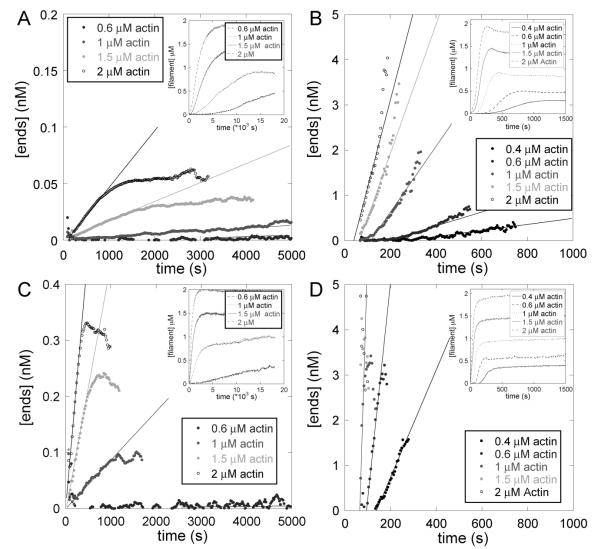
We used the fluorescence of pyrenyl-actin to measure the time course of polymerization of actin monomers alone (Figure 3A) or with Arp2/3 complex or with Arp2/3 complex and the hWASp-VCA nucleation-promoting factor (Figure 3B), each with and without phalloidin. At each time point we calculated the concentration of filament ends from the rate of polymerization, the rate constants for actin filament elongation, and the actin monomer concentration at that time point.
Figure 3.
Measurement of actin filament nucleation rates from the time course of polymerization of bulk samples of pyrenyl-actin. Conditions: 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, and 10 mM imidazole, pH 7.0, with various concentrations of 30% pyrenyl-actin monomers with or without 10 nM Arp2/3 complex and 300 nM GST-hWASp-VCA. The concentrations of ends were calculated from the slopes of the pyrenyl fluorescence vs time. The lines in the graphs of the concentration of ends vs time represent the initial average nucleation rate. (A) Time course of spontaneous polymerization (inset) and ends created for four concentrations of purified actin monomers in the absence of Arp2/3 complex and hWASp-VCA. (B) Time course of spontaneous polymerization (inset) and ends created for five concentrations of purified actin monomers in the presence of 10 nM Arp2/3 complex and 300 nM GST-hWASp-VCA. (C) Time course of spontaneous polymerization (inset) and ends created for four concentrations of purified actin monomers with equimolar concentrations of phalloidin. (D) Time course of spontaneous polymerization (inset) and ends created for five concentrations of purified actin monomers with equimolar phalloidin in the presence of 10 nM Arp2/3 complex and 300 nM GST-hWASp-VCA.
As actin alone polymerized over 10000 s (Figure 3A, inset), the concentration of ends increased initially but leveled off at less than 0.1 nM as monomers were consumed and the rate of nucleation fell to zero (Figure 3A). The presence of phalloidin equimolar to the actin increased the rate of pure actin polymerization and the final concentration of ends produced (Figure 3C). Addition of 10 nM Arp2/3 complexto actin had no effect on the time course of polymerization with or without phalloidin (data not shown), so a nucleation-promoting factor is required to activate Arp2/3 complex even in the presence of phalloidin. Actin polymerized much faster with 10 nM Arp2/3 complex and 300 nM GST-hWASp-VCA (Figure 3B inset), as new ends were created rapidly even at very low concentrations of actin monomer (Figure 3B). With Arp2/3 complex, GST-hWASp-VCA, and phalloidin, polymerization and the end creation rate were even faster than the same reaction without phalloidin (Figure 3D).
We used the slopes of plots of [ends] vs time to calculate the rates of end creation during the initial phase of polymerization (Figure 3). Unlabeled phalloidin increased the nucleation rate of actin alone, and the dependence of the nucleation rate on the concentration of actin was nonlinear (Figure 4, inset). Rhodamine-phalloidin also increased the rate of spontaneous nucleation, although the effect was only half as great as an equal concentration of unlabeled phalloidin, as expected from its lower affinity for actin filaments (12). In the presence of 10 nM Arp2/3 complex and 300 nM VCA the nucleation rate was 50 times greater than actin alone and was directly proportional to the actin concentration (above the monomer concentration of 0.4 μM) (Figure 4A). Unlabeled phalloidin increased nucleation by Arp2/3 complex and activator more that 10 times, also linear with the concentration of actin (Figure 4A). Rhodamine-phalloidin increased nucleation with Arp2/3 complex to the same extent as unlabeled phalloidin. Initial nucleation rates with Arp2/3 complex, VCA, and 100 nM actin filament seeds were similar whether or not the seeds were preincubated with unlabeled phalloidin (Figure 4B).
Figure 4.
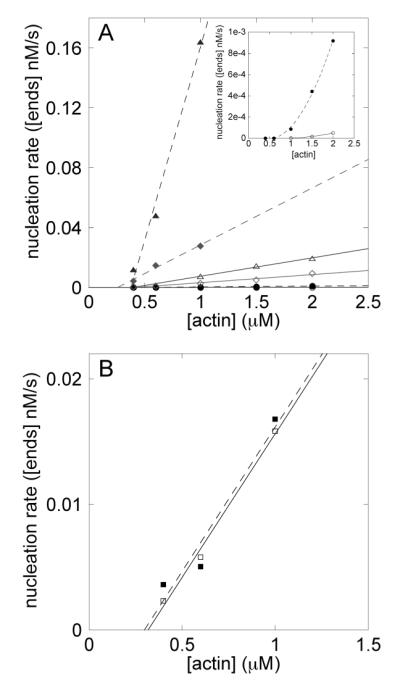
(A) Dependence of the average nucleation rate on the actin monomer concentration with (filled symbols) and without (open symbols) equimolar phalloidin. The inset shows the nucleation rate of actin alone. The main plot shows actin without Arp2/3 complex (circles) and with 300 nM GST-hWASp-VCA and 5 nM (diamonds) or 10 nM (triangles) Arp2/3 complex. Phalloidin increases the nucleation rate significantly when combined with Arp2/3 complex and activator. (B) Dependence of the initial nucleation rate on actin monomer concentration using 100 nM seeds with bound phalloidin (filled squares) or no phalloidin (open squares).
Visualizing Branches with Rhodamine-Phalloidin
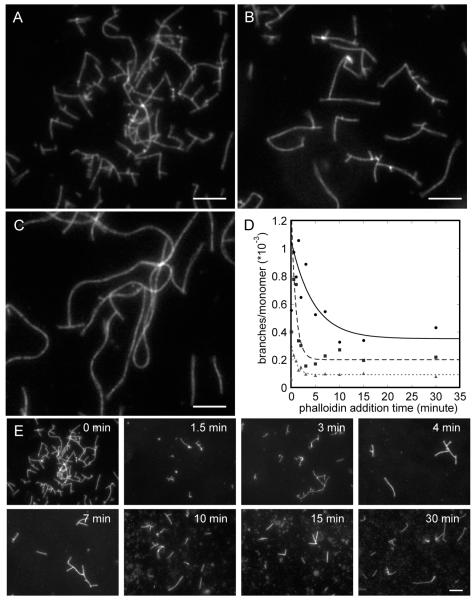
The density of branches observed microscopically in the final dilution of the products depended on both the concentration of actin monomer and the time between initiating a reaction and adding rhodamine-phalloidin. The density of branches was highest with 1 μM actin (Figure 5A) and progressively lower with 2 μM actin (Figure 5B) and 4 μM actin (Figure 5C). These reactions had run to completion, and the amount of active complex limited the number of branches formed. Higher concentrations of actin allow filaments to grow longer between branches. Figure 5E shows images from reactions with 1 μM actin with rhodamine-phalloidin added at increasing delay times during the reaction. The number of branches observed in the micrographs declined with the length of the delay before adding phalloidin (Figure 5D,E). Exponential fits to these data gave half-times of 4.6 ± 2.5 min (mean ± SD) with 1 μM actin monomers, 0.9 ± 0.3 min with 2 μM actin, and 1.1 ± 0.2 min with 4 μM actin.
Figure 5.
Fluorescence micrographs of actin filaments stained with rhodamine-phalloidin. Actin monomers at concentrations of 1, 2, or 4 μM were polymerized with 10 nM Arp2/3 complex and 300 nM GST-hWASp-VCA in buffer containing 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, and 10 mM imidazole, pH 7.0. (A–C) Reactions with equimolar rhodamine-phalloidin present during the polymerization reaction. Images are shown for (A) 1 μM actin, (B) 2 μM actin, and (C) 4 μM actin. (D, E) Rhodamine-phalloidin equimolar to actin was added at times ranging from 0 to 30 min after initiating the reaction. (D) Counts of the number of branches per subunit of filament in the micrograph. Exponential fits through the data (excluding the points at zero time) yield half-times of 4.6 ± 2.5 min with 1 μM actin (filled circles), 0.9 ± 0.3 min with 2 μM actin (filled squares), and 1.1 ± 0.2 min with 4 μM actin (triangles). (E) Micrographs from an experiment with phalloidin added from 0 to 30 min to a reaction with 1 μM actin, 10 nM Arp2/3 complex, and 300 nM GST-hWASp-VCA.
DISCUSSION
Our discovery that phalloidin binds Arp2/3 complex and hWASp-VCA requires a reexamination of the use of rhodamine-phalloidin to study actin filament branching by Arp2/3 complex. We consider each binding reaction and then how phalloidin influences branch formation and stability.
Rhodamine-Phalloidin Binding to Arp2/3 Complex
The increase in fluorescence of rhodamine-phalloidin when titrated with Arp2/3 complex and the reversal of this change upon titration with unlabeled phalloidin are consistent with a simple bimolecular binding reaction with a dissociation equilibrium constant in the range of 25–70 nM. The affinity of rhodamine-phalloidin for actin filaments is 2–4-fold higher (1, 13).
We looked for potential phalloidin binding sites on Arp2/3 complex by comparison with the phalloidin binding site on actin filaments (Figure 1C). Phalloidin binds filaments in a pocket at the interface of three subunits in the actin filament, which we will number n − 2, n − 1, and n from the pointed to the barbed end along the short-pitch helix (14). The pocket is lined with residues H73, R177, and D179 near the nucleotide binding cleft of subunit n − 1 and residues 196–201 in subdomain 4 of subunit n. Residues 285–287 in subdomain 3 of subunit n − 2 are more distant from the bound phalloidin in the structure (14). Mutation of residue G158, R177, or D179 compromises phalloidin binding (15, 16).
At a branch junction, Arp3 is proposed to occupy the n − 2 site, Arp2 the n − 1 site, and the first actin subunit the n site at the pointed end of the daughter filament (17). Arp2 has the residues corresponding to G158, R177, and D179 of actin (n − 1) in addition to residues similar to residues 196–201 of actin (n). The Arp3 subunit lacks these strong similarities to the actin residues proposed to interact with phalloidin, so Arp2 is a better candidate for the primary phalloidin binding site on Arp2/3 complex. When bound to the n − 1 site on Arp2, phalloidin is in an ideal position to stabilize the interaction of Arp2 with the first actin subunit (n) in the daughter filament helping to stabilize and activate the complex. Separation of the Arps in the inactive complex (17) and the weak n − 2 site on Arp3 provide an explanation for the inability of phalloidin to activate Arp2/3 complex on its own.
Rhodamine-Phalloidin Binding to hWASp-VCA
Rhodamine-phalloidin also binds hWASp-VCA with a Kd of about 100 nM. The C motif contributes to the binding site, but we know little else about this interaction. The fluorescence anisotropy of rhodamine-hWASp-VCA bound to Arp2/3 complex is greater in the presence of phalloidin. The rhodamine on this VCA construct was bound to a cysteine incorporated at the N-terminus of the V domain. Thus the N-terminal end of the VCA peptide was less mobile when bound to Arp2/3 complex in the presence of phalloidin.
Effects of Phalloidin on Actin Polymerization
Phalloidin speeds up the spontaneous polymerization of pure actin at actin concentrations greater than 0.6 μM, in spite of the fact that phalloidin slows elongation at the barbed end by almost half (9, 18). Spontaneous polymerization is faster, because phalloidin strongly promotes nucleation. Given that phalloidin inhibits subunit dissociation at both ends of actin filaments (18, 19), it may promote nucleation by inhibiting the very rapid dissociation of spontaneously formed actin dimers and trimers. At concentrations of actin <0.6 μM, the spontaneous formation of nuclei is too unfavorable to form filaments even with phalloidin.
Effects of Phalloidin on Actin Filament Nucleation by VCA and Arp2/3 Complex
Several pathways lead from mixtures of actin monomers, VCA, Arp2/3 complex, and filaments to an elongating branch (20). Under conditions of our experiments the main pathway is (i) A + V ⇆ AV, (ii) AV + Arp ⇆ AVArp (ternary complex with the inactive Arp2/3 complex), (iii) AVArp + Fil ⇆ AVArp-Fil, and (iv) AVArp-Fil + A ⇆ elongation, where A is actin monomer, V is VCA, Arp is Arp2/3 complex, and Fil is actin filament. The ternary complex of AVArp is inactive in nucleation until it binds the side of a filament. A rate-limiting activation step may be required between steps iii and iv (20) (21).
Under the conditions of our experiments phalloidin is prebound to Arp2/3 complex and VCA, allowing it to influence formation of the ternary complex as well as branch formation after the ternary complex binds a mother filament. Phalloidin binding to filaments might influence their interactions with ternary complexes, but this is unlikely, because phalloidin-labeled actin filaments and unlabeled actin filaments had the same effect on the initial nucleation rate (Figure 4B).
Phalloidin most likely stimulates branching nucleation by stabilizing interactions of Arp2/3 complex with the first or second actin subunit in the new filament either during the formation of the ternary complex (reaction ii) or during activation of Arp2/3 complex on the side of a filament. No information is available on the influence of phalloidin on reaction i. Phalloidin strongly stimulates branching nucleation in spite of reducing the affinity of VCA alone for Arp2/3 complex and inhibiting elongation (iv).
We can rule out effects of phalloidin on two side reactions. Phalloidin binding alone does not stabilize the Arp2-Arp3 dimer in a conformation that mimics the barbed end of a filament, because a nucleation-promoting factor (hWASp-VCA in our experiments) is required for phalloidin to promote nucleation by Arp2/3 complex. The mechanism is unlikely to involve Arp2/3 complex capturing preformed actin dimers, because phalloidin allows Arp2/3 complex and hWASp-VCA to form branches, even at low actin concentrations where spontaneous nucleation of actin is unfavorable.
Effect of Phalloidin on Branch Stability
We detected many more barbed ends by pyrene fluorescence in bulk samples than we observed by microscopy of samples polymerized with or treated with rhodamine-phalloidin (Table 1). In the absence of phalloidin, this mismatch arises from spontaneous dissociation of many branches before they grow long enough to be detected by light microscopy (7). Even in the presence of rhodamine-phalloidin many branches must dissociate before phalloidin has time to bind.
Table 1.
Comparison of Barbed Ends Produced in 30 min by Actin, 10 nM Arp2/3 Complex, and 300 nM hWASp-VCA in Bulk Samples and of Branches Observed by Fluorescence Microscopy of Parallel Samplesa
| [ends]/[polymerized actin subunits] × 10−3 in 30 min |
||||
|---|---|---|---|---|
| phalloidin throughout reaction |
phalloidin added at end of reaction |
|||
| [actin] | ||||
| (μM) | bulk sample | microscopy | bulk sample | microscopy |
| 1 | 3.4 ± 0.8 | 0.56 ± 0.13 | 2.3 ± 0.6 | 0.43 ± 0.1 |
| 2 | 1.9 ± 0.5 | 0.40 ± 0.10 | 1.7 ± 0.43 | 0.22 ± 0.05 |
| 4 | 0.91 ± 0.2 | 0.07 ± 0.02 | 0.91 ± 0.2 | 0.08 ± 0.02 |
The concentration of barbed ends in bulk samples was calculated from the time course of polymerization. Fluorescence microscopy was used to measure the number of branches per length of filament. Rhodamine-phalloidin equimolar to actin was present throughout the reaction or added at the end of the 30 min reaction.
Rhodamine-phalloidin stabilizes actin filament branches for much longer than their natural half-lives of less than 10 min (7). The number of branches preserved for observation declined with the interval of time between initiating a polymerization reaction and adding rhodamine-phalloidin (Figure 5) (3, 4). Two mechanisms may contribute to the ability of rhodamine-phalloidin to stabilize branches. One possibility is that phalloidin stabilizes branches directly, just as it stabilizes actin filaments, by binding between Arp2/3 complex and the first subunit of the daughter filament that is crucial for initiating the branch. Phalloidin also inhibits dissociation of γ-phosphate from ADP-Pi on actin filaments (3, 22) and may also influence γ-phosphate dissociation from Arp2. Branches dissociate faster after actin filaments (7) and Arp2/3 complex (23) hydrolyze their bound ATP and dissociate the γ-phosphate. A mutant of Arp2 that inhibits ATP hydrolysis prolongs the lifetime of branches (23). When combined with its ability to enhance nucleation of branches, these stabilizing effects increase the number of branches observed by fluorescence microscopy in the presence of phalloidin.
Our observations of phalloidin binding to Arp2/3 complex and VCA show that cellular components other than actin filaments bind rhodamine-phalloidin. Given that both new phalloidin receptors described here are present at high concentrations in cells, around 10 μM in leukocytes (24), some rhodamine-phalloidin is bound to sites other than actin filaments in cells stained with rhodamine-phalloidin. Rhodamine-phalloidin will also influence branching nucleation by Arp2/3 complex in permeabilized cells and in biochemical assays.
Footnotes
Abbreviations: Arp, actin-related protein; GST, glutathione S-transferase; hWASp, human Wiskott–Aldrich syndrome protein; hWASp-VCA, verprolin-connecting-acidic domain of hWASp.
This work was supported by Research Grant GM-026338 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Huang ZJ, Haugland RP, You WM, Haugland RP. Phallotoxin and actin binding assay by fluorescence enhancement. Anal. Biochem. 1992;200:199–204. doi: 10.1016/0003-2697(92)90299-m. [DOI] [PubMed] [Google Scholar]
- 2.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASp/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 3.Blanchoin L, Pollard TD, Mullins RD. Interaction of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 2000;10:1273–1282. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- 4.Weaver AM, Karginov AV, K. AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- 5.MacLean-Fletcher S, Pollard TD. Identification of a factor in conventional muscle actin preparation which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- 6.Pollard TD. Polymerization of ADP-actin. J. Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches by Arp2/3 complex. Biophys. J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection microscopy. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampath P, Pollard TD. Effects of cytochalasin, phalloidin, and pH on the elongation of actin-filaments. Biochemistry. 1991;30:1973–1980. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- 10.Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASp/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 12.De La Cruz EM, Pollard TD. Kinetics and thermodynamics of phalloidin binding to actin filaments from three divergent species. Biochemistry. 1996;35:14054–14061. doi: 10.1021/bi961047t. [DOI] [PubMed] [Google Scholar]
- 13.De La Cruz E, Pollard TD. Transient kinetic analysis of rhodamine phalloidin binding to actin filaments. Biochemistry. 1994;33:14387–14392. doi: 10.1021/bi00252a003. [DOI] [PubMed] [Google Scholar]
- 14.Oda T, Namba K, Maeda Y. Position and orientation of phalloidin in F-actin determined by X-ray fiber diffraction analysis. Biophys. J. 2005;88:2727–2736. doi: 10.1529/biophysj.104.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol. Biol. Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belmont LD, Patterson ML, Drubin DG. New actin mutants allow further characterization of the nucleotide binding cleft and drug binding sites. J. Cell Sci. 1999;112:1325–1336. doi: 10.1242/jcs.112.9.1325. [DOI] [PubMed] [Google Scholar]
- 17.Robinson RC, Turbedsky K, Kaiser D, Higgs H, Marchand JB, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1660–1661. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 18.Coluccio LM, Tilney LG. Phalloidin enhances actin assembly by preventing monomer dissociation. J. Cell Biol. 1984;99:529–535. doi: 10.1083/jcb.99.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dancker P, Low I, Hasselbach W, Wieland T. Interaction of actin with phalloidin: polymerization and stabilization of F-actin. Biochim. Biophys. Acta. 1975;400:407–414. doi: 10.1016/0005-2795(75)90196-8. [DOI] [PubMed] [Google Scholar]
- 20.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J. Biol. Chem. 2008;283:7135–7144. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- 21.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 22.Dancker P, Hess L. Phalloidin reduces the release of inorganic phosphate during actin polymerization. Biochim. Biophys. Acta. 1990;1035:197–200. doi: 10.1016/0304-4165(90)90116-e. [DOI] [PubMed] [Google Scholar]
- 23.Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch M, Drubin DG. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J. Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgs HN, Blanchoin L, Pollard TD. Influence of the Wiskott-Aldrich syndrome protein (WASp) C terminus and Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]