Extracellular glutathione triggers innate immunity responses in the Arabidopsis leaf through a glutamate receptor3.3-dependent pathway homologous with that of the central nervous system.
Abstract
The tripeptide reduced glutathione (GSH; γ-glutamate [Glu]-cysteine [Cys]-glycine) is a major endogenous antioxidant in both animal and plant cells. It also functions as a neurotransmitter mediating communication among neurons in the central nervous system of animals through modulating specific ionotropic Glu receptors (GLRs) in the membrane. Little is known about such signaling roles in plant cells. Here, we report that transient rises in cytosolic calcium triggered by exogenous GSH in Arabidopsis (Arabidopsis thaliana) leaves were sensitive to GLR antagonists and abolished in loss-of-function atglr3.3 mutants. Like the GSH biosynthesis-defective mutant PHYTOALEXIN DEFICIENT2, atglr3.3 showed enhanced susceptibility to the bacterial pathogen Pseudomonas syringae pv tomato DC3000. Pathogen-induced defense marker gene expression was also decreased in atglr3.3 mutants. Twenty-seven percent of genes that were rapidly responsive to GSH treatment of seedlings were defense genes, most of which were dependent on functional AtGLR3.3, while GSH suppressed pathogen propagation through the AtGLR3.3-dependent pathway. Eight previously identified putative AtGLR3.3 ligands, GSH, oxidized glutathione, alanine, asparagine, Cys, Glu, glycine, and serine, all elicited the AtGLR3.3-dependent cytosolic calcium transients, but only GSH and Cys induced the defense response, with the Glu-induced AtGLR3.3-dependent transcription response being much less apparent than that triggered by GSH. Together, these observations suggest that AtGLR3.3 is required for several signaling effects mediated by extracellular GSH, even though these effects may not be causally related.
Reduced glutathione (GSH; γ-Glu-Cys-Gly) is the most abundant short-chain peptide in cells and is enzymatically synthesized from Glu, Cys, and Gly. It is present in both intra- and extracellular compartments, although at least 90% is found inside the cell. Depending on the cell type, glutathione levels have been estimated to range from 1 to 10 mm, 90% of which is in reduced form, thus representing the major pool of endogenous nonprotein thiols within organisms (Noctor et al., 2011). Its Cys sulfur group can perform reversible redox reactions to form disulfides, notably with another glutathione Cys residue to produce oxidized glutathione (GSSG), which is continuously recycled at high rates to GSH by glutathione reductase existing in the cytosol and other cellular organelles. Therefore, glutathione serves a central role in free radical scavenging, maintaining the thiol status of proteins, and functions as a general antioxidant protecting cell membranes against oxidative stress and DNA against radiation (for review, see Franco and Cidlowski, 2009; Schmidt and Dringen, 2012).
In plants, GSH typically accumulates in cells to millimolar concentration. Intracellular GSH is essential for development and reactive oxygen species scavenging as well as oxidative signaling during stress responses (Foyer and Noctor, 2011; Noctor et al., 2012). Arabidopsis (Arabidopsis thaliana) knockout mutants completely deficit in GSH biosynthesis show either embryo- or seedling-lethal phenotypes (Cairns et al., 2006; Pasternak et al., 2008), demonstrating its indispensable role in plant development. Other Arabidopsis mutants that have various degrees of reduction in endogenous GSH content show altered expression of stress-related genes (Ball et al., 2004; Mhamdi et al., 2010; Han et al., 2013a, 2013b), enhancing sensitivity to excessive zinc (Shanmugam et al., 2012) and cadmium (Jozefczak et al., 2012) as well as pathogens (Parisy et al., 2007; Dubreuil-Maurizi and Poinssot, 2012).
In animal cells, GSH not only functions as an essential antioxidant as in plant cells, but GSH in the extracellular space of neurons in the central nervous system can act like a neurotransmitter or neuromodulator through specific interactions with Glu receptors (Levy et al., 1991; Juurlink, 1999; Dringen, 2000; Oja et al., 2000; Shaw et al., 2001; Aoyama et al., 2008). Glu receptors are either ionotropic (iGLRs) or metabotropic. iGLRs form ionic cation channels permeable to Ca2+, Na+, and K+ and are named after their agonists N-methyl-d-Asp (NMDA), 2-amino-3-hydroxy-5-methyl-4-isoxazolepropionate, and kainite (Flores-Soto et al., 2012). Metabotropic Glu receptors are coupled to G proteins and functionally linked either to the formation of inositol phosphates and diacylglycerol or to the metabolism of cyclic nucleotides (Loane et al., 2012). The molecular structure of GSH tends to make it neuroactive, because all of its three individual amino acid residues can either function as neurotransmitters or interfere with glutamatergic neurotransmission, with Glu and Gly being excitatory and inhibitory neurotransmitters, respectively (Hnasko and Edwards, 2012). Furthermore, Cys is neurotoxic at high concentrations (Slivka and Cohen, 1993). GSH can stimulate NMDA receptor-dependent membrane depolarization and Ca2+ entry into rat neurons (Leslie et al., 1992; Janáky et al., 1993; Ogita et al., 1995; Varga et al., 1997), cultured hippocampal neurons (Chin et al., 2006), and pig cerebral cortical synaptic membranes (Janáky et al., 2000).
To the best of our knowledge, the first report of a potentially similar signaling role of extracellular GSH in plants to that found in the animal central nervous system was a study with tobacco (Nicotiana tabacum) expressing the Ca2+ reporter luminescence protein aequorin in the cytosol (Gomez et al., 2004). The authors reported that exogenous application of 5 mm GSH induces cytosolic Ca2+ ([Ca2+]cyt) transient elevation in the leaf (Gomez et al., 2004). In Arabidopsis roots, our previous study also demonstrated that 1 mm GSH can induce strong [Ca2+]cyt elevation (Qi et al., 2006). Furthermore, GSH can induce AtGLR3.3-dependent membrane depolarization (Qi et al., 2006). AtGLR3.3 is one of 20 Glu receptor genes in Arabidopsis (Lacombe et al., 2001). The predicted protein structure of AtGLRs includes conserved transmembrane and extracellular ligand binding domains of iGLRs and G protein-coupled receptors (GPCRs; Chiu et al., 1999; Turano et al., 2001). Different plant GLR members are suggested to play roles in various physiological processes, including mineral nutrient homeostasis (Kim et al., 2001; Dubos et al., 2003; Kang et al., 2006; Aouini et al., 2012), carbon/nitrogen balance (Kang and Turano, 2003), root development (Li et al., 2006), stomata movement (Cho et al., 2009), abscisic acid sensing (Kang et al., 2004), gravitropism sensing (Miller et al., 2010), and pollen tube elongation (Michard et al., 2011). Recently, pharmacological approaches produced evidence that AtGLRs could be involved in innate immunity response in Arabidopsis seedlings (Kwaaitaal et al., 2011, 2012). In light of the established roles of GSH in plant defense responses (Ferrari et al., 2003; Parisy et al., 2007; Schlaeppi et al., 2008) and the requirement of AtGLR3.3 for GSH-induced signaling events in the roots (Qi et al., 2006), we hypothesized that GSH could mediate innate immunity responses in the leaf through AtGLR3.3-dependent pathways. This study reports evidence that strongly supports this notion and gives rise to several intriguing new questions about this novel signaling cascade in the defense response.
RESULTS
Exogenous GSH Induces AtGLR3.3-Dependent Heterogeneous [Ca2+]cyt Rise
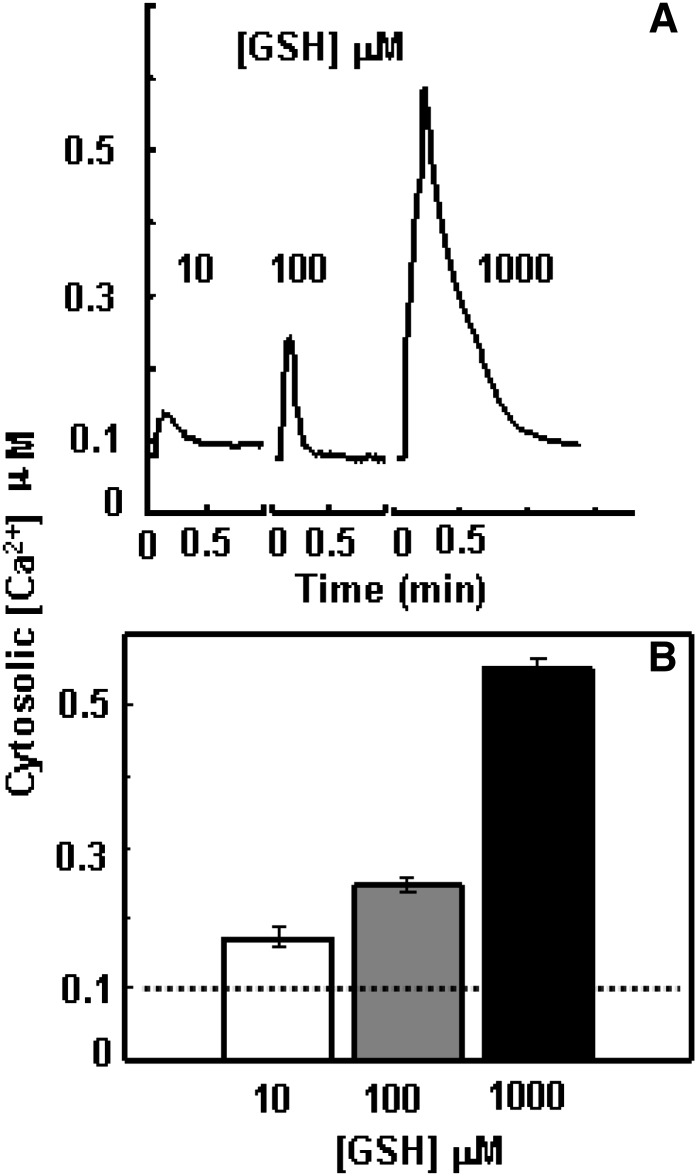
To investigate the signaling role of extracellular GSH in the leaf, we monitored how exogenous application of GSH ([GSH]ext) affects [Ca2+]cyt in detached Arabidopsis leaves, using cytosolic expression of the Ca2+ reporter protein aequorin. As observed previously (Qi et al., 2006), the background [Ca2+]cyt concentration was around 0.1 μm (Fig. 1). Immediately after the delivery of a 10 µm GSH solution into the detached leaf, a rise in [Ca2+]cyt was observed (Fig. 1). Increasing the [GSH]ext to 100 and 1,000 μm further promoted the response, which showed a strong concentration-dependent pattern (Fig. 1).
Figure 1.
[GSH]ext-induced concentration-dependent [Ca2+]cyt rise in the leaf. A, Representative recording curve of 10, 100, and 1,000 µm GSH-induced [Ca2+]cyt transient rise in a detached leaf. B, Averaged peak values with se of the responses (n = 5), which are significantly different to each other at P < 0.01. The dash line indicates the background [Ca2+]cyt.
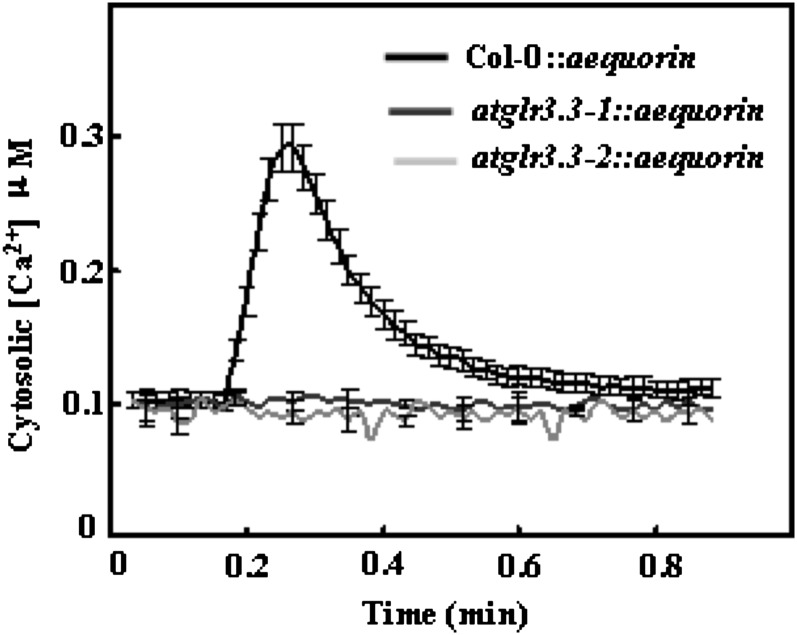
In our previous study with Arabidopsis roots, we demonstrated that 1 mm [GSH]ext induced membrane depolarization and that this effect largely depends on AtGLR3.3 (Qi et al., 2006), one of 20 Glu receptor-like genes in Arabidopsis. We hypothesized that the observed [GSH]ext-induced [Ca2+]cyt rise in this study was also mediated by AtGLR3.3. As an initial test of this hypothesis, we pretreated the leaf with the iGLRs antagonists (2R)-amino-5-phosphonopentanoate (AP5) and 6,7-dinitroquinoxaline-2,3-dione. Both of them, especially AP5, significantly suppressed the [GSH]ext-induced responses (Fig. 2). We next introduced aequorin gene into the atglr3.3-1 and atglr3.3-2 knockout mutant backgrounds through cross pollination and monitored its response to [GSH]ext. The GSH-induced [Ca2+]cyt rise observed in the wild type was completely absent in the mutants (Fig. 3).
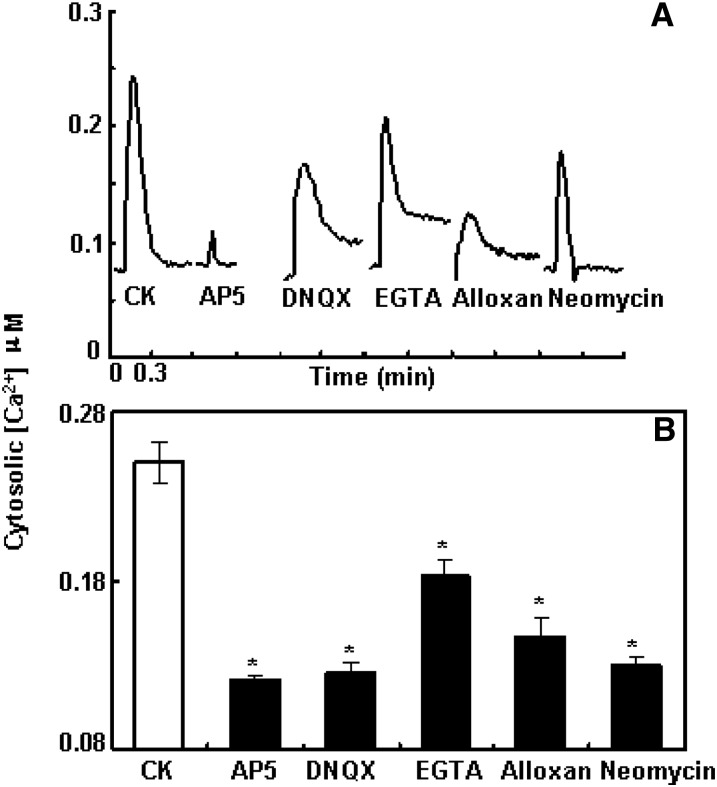
Figure 2.
Pharmacological study of the [GSH]ext-induced [Ca2+]cyt transient response in the leaf. A, Representative recording curve of 100 µm [GSH]ext-induced [Ca2+]cyt rise in the CK and pretreated with 1 mm various Ca2+ mobility pathway blockers as indicated. B, Averaged peak values of these responses with se (n = 4 for EGTA and n = 5 for the rest). The asterisk stands for the sd to the CK at P < 0.01. The average background [Ca2+]cyt was around 0.08 µm.
Figure 3.
[GSH]ext-induced [Ca2+]cyt response was absent in the atglr3.3 mutant. Averaged recording curves with se of 100 µm [GSH]ext-induced [Ca2+]cyt rise in the leaf cells of the ecotype Columbia (Col-0) and atglr3.3-1 and atglr3.3-2 mutants expressing aequorin (n = 10 for each).
We next used pharmacological approaches to investigate which Ca2+ mobility pathways contribute to the [GSH]ext-induced and AtGLR3.3-dependent [Ca2+]cyt rise. Removal of extracellular Ca2+ and including 1 mm Ca2+ chelator EGTA in the recording buffer significantly, but not completely, suppressed 100 μm GSH-induced [Ca2+]cyt rise (Fig. 2). This indicates that extracellular Ca2+ influx through certain plasma membrane Ca2+-permeable channels partially, but not fully, contributes to the observed response. cAMP-gated calcium channels have been identified in the plasma membrane of Arabidopsis leaf protoplasts (Ali et al., 2007). Alloxan can interrupt the cAMP-dependent Ca2+ influx pathway by inhibiting adenylate cyclase activity and so blocking cAMP production. Pretreatment for 1 h with 1 mm alloxan significantly inhibited the rise in [Ca2+]cyt (Fig. 2). Our previous study implicated Cyclic Nucleotide Gated Channel2 (CNGC2) in mediating the [Ca2+]cyt rise associated with innate immunity (Ma et al., 2009). However, in this study, we found a normal response of the cngc2 mutant expressing aequorin to the [GSH]ext (data not shown).
Chelating extracellular Ca2+ did not abolish the GSH-induced [Ca2+]cyt response (Fig. 2), implicating Ca2+ release from certain internal Ca2+ storage pools in the response. 1,4,5-trisphosphate (IP3)-mediated Ca2+ release from internal Ca2+ pools has been demonstrated in Arabidopsis guard cells (Tang et al., 2007). Neomycin can block the IP3-mediated Ca2+ mobility pathway by inhibiting the activity of phospholipase C, which catalyzes production of IP3 from phosphatidylinositol 4,5 bisphosphate. One-hour 1 mm neomycin pretreatment also significantly suppressed the [GSH]ext-induced Ca2+ response (Fig. 2). Taken together, these data demonstrated that the [GSH]ext-induced Ca2+ response fully depended on AtGLR3.3 but was caused by increased Ca2+ mobility through various pathways.
AtGLR3.3 Mediates Part of the Early Transcriptional Response of the Leaf to [GSH]ext
Ligand-induced [Ca2+]cyt rise is often linked to downstream transcriptional responses (Dodd et al., 2010). To focus on the early transcriptional responses to [GSH]ext and to establish the importance of AtGLR3.3 for the GSH-mediated signaling cascades, we treated the wild-type and atglr3.3-2 seedlings for 1 h with either the control (CK) recording buffer or the buffer containing 100 μm GSH. Two independent biological replicates were conducted for whole genome transcriptional profiling analysis with Agilent Arabidopsis microarrays. Under the CK condition, there were 35 genes showing at least 3-fold differential expression between the mutant and the wild type (Supplemental Table S1). As expected, AtGLR3.3 was the lowest expressed transcript in the atglr3.3 mutant compared with the wild type, with 47- and 50-fold changes in the two biological replicates, respectively (Supplemental Table S1).
GSH treatment of the wild type altered expression of 97 genes, with at least 3-fold transcript abundance change (Fig. 4A; Supplemental Tables S2 and S3). By comparing the genes modulated by GSH in the atglr3.3 mutant, we found that 70 (Supplemental Table S2) of the 97 genes showed no significant change in the mutant. The remaining 27 genes showed the same expression trends in response to GSH in both the wild type and the mutant (Fig. 4A; Supplemental Tables S3 and S4). Thus, this expression profiling analysis indicated that 72% (70/97) of the genes rapidly modulated by GSH in the wild type were dependent on the AtGLR3.3 pathway and 28% (27/97) independent of it.
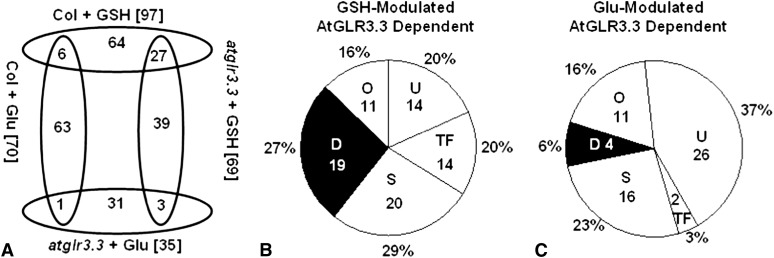
Figure 4.
Overview of the GSH- and Glu-mediated AtGLR3.3-dependent gene expression. A, Number of overlapping genes modulated by GSH and Glu in the ecotype Columbia (Col-0) and atglr3.3 mutant. B, Functional category of the genes modulated by GSH with AtGLR3.3 dependence. C, Functional category of the genes modulated by Glu with AtGLR3.3 dependence. The numbers in and the percentage symbol outside the pie chart indicate the number and percentage of the genes in the category, respectively. U, Unknown; TF, Transcription Factor; S, Signaling; O, Others; D, Defense.
To validate the transcriptional profiling data set, independent biological samples were produced, and eight genes selected from the list of genes regulated by GSH in the wild type were chosen for quantitative reverse transcript (qRT)-PCR analysis (Supplemental Tables S2 and S3). All eight genes showed significantly increased expression in response to GSH, an effect that was significantly attenuated in the atglr3.3-2 mutant background (Supplemental Fig. S1). The trends of gene expression in Supplemental Figure S1 were consistent with those revealed by the microarray (Supplemental Table S2). These observations provide further evidence that early transcriptional responses induced by GSH are at least partially acting through AtGLR3.3.
GSH-Regulated AtGLR3.3-Dependent Defense and Signaling-Related Genes
To explore which cellular pathways were regulated by GSH in an AtGLR3.3-dependent manner, we performed categorization of these genes by Gene Ontology annotation analysis combined with manual checking of available literature associated with each of the 70 genes in Supplemental Table S2 through searching The Arabidopsis Information Resource (http://www.arabidopsis.org). This analysis showed that 27%, 20%, 20%, 20%, and 16% of these genes were functionally categorized as Defense, Signaling, Unknown, Transcription Factor, and Others, respectively (Fig. 4B; Supplemental Table S2). There were eight genes that were common to the categories Defense and Transcription Factor. All of the genes in the Defense category have been demonstrated using corresponding mutants to play roles during defense responses or found to be induced by various pathogens at transcriptional level (Table I; Supplemental Table S2). These genes notably include the essential salicylic acid (SA) biosynthesis gene ISOCHORISMATE SYNTHASE1 (ICS1; Wildermuth et al., 2001) and its key regulator SYSTEMIC ACQUIRED RESISTANCE DEFICIENT1 (SARD1; Zhang et al., 2010) as well as the transcription factors CALMODULIN BINDING PROTEIN60-LIKEG (CBP60G) (Wang et al., 2009), WRKY33, and WRKY46 (Birkenbihl et al., 2012; Hu et al., 2012; Moreau et a., 2012), all of which are essential genes for mediating defense response of Arabidopsis.
Table I. Defense genes induced by GSH in ecotype Columbia but not glr3.3-2.
| Gene ID | FC01a | FC02a | Average | Annotation | Reference |
|---|---|---|---|---|---|
| Having confirmative role in defense | |||||
| AT5G13320 | 17.9 | 6.1 | 12 | GRETCHEN HAGEN 3.12; SA biosynthesis | Okrent et al., 2009 |
| AT2G46400 | 15.7 | 4.9 | 10.3 | WRKY46 transcription factor | Hu et al., 2012 |
| AT1G33960 | 12.5 | 3 | 7.75 | AVIRULENCE-INDUCED GENE1 | Reuber and Ausubel, 1996 |
| AT1G74710 | 5.8 | 4.8 | 5.3 | Isochorismate Synthase1; SA biosynthesis | Wildermuth et al., 2001 |
| AT1G72520 | 3.3 | 6.6 | 4.95 | Lipoxygenase4; jasmonic acid biosynthesis | Acosta and Farmer, 2010 |
| AT2G35980 | 5.1 | 3.9 | 4.5 | Yellow-leaf-specific gene9 | Po-Wen et al., 2013 |
| AT5G26920 | 4.4 | 3.4 | 3.9 | CBP60G | Wang et al., 2009 |
| AT1G73805 | 3.6 | 3.5 | 3.55 | SARD1; SA biosynthesis | Zhang et al., 2010 |
| AT2G38470 | 3.4 | 3.4 | 3.4 | WRKY33 transcription factor | Birkenbihl et al., 2012 |
| Having putative role in defense | |||||
| AT5G36970 | 14.8 | 23.1 | 18.95 | HAIRPIN-INDUCED1 LIKE25; induced by pathogens | Varet et al., 2002 |
| AT5G47850 | 7 | 13.2 | 10.1 | CRINKLY44 related4; protein kinase; response to insect | Little et al., 2007 |
| AT1G21240 | 7.5 | 3.3 | 5.4 | Wall associated kinase3; induced by SA | He et al., 1999 |
| AT2G18060 | 3.5 | 6.3 | 4.9 | NAC (NAM, ATAF1/2 and CUC2) domain containing protein37; induced by chitin | Libault et al., 2007 |
| AT3G50930 | 3.7 | 5.7 | 4.7 | ATPases associated with diverse cellular activities type; induced by bacterial pathogen | Navarro et al., 2004 |
| AT3G23230 | 3 | 6.3 | 4.65 | Ethylene response factor98; induced by pathogen and jasmonic acid | McGrath et al., 2005 |
| AT2G43000 | 3.7 | 5.1 | 4.4 | NAC domain containing protein42; camalexin biosynthesis | Saga et al., 2012 |
| AT1G14540 | 4 | 3.8 | 3.9 | Peroxidase4; induced by oxidative stress and fungus infection | Rasul et al., 2012 |
| AT1G18570 | 3.2 | 4.1 | 3.65 | MYB51 transcription factor; induced by pathogen | Maekawa et al., 2012 |
| AT4G33050 | 4 | 3.1 | 3.55 | Embryo sac development arrest39; response to chitin and virus | Libault et al., 2007 |
Fold changes of two replicates.
Genes in the Signaling category include a kinase (At1g67000), receptor-like proteins RLP7 and RLP20, calcium homeostasis-related proteins such as Glu receptors AtGLR1.2, AtGLR2.8, and AtGLR2.9, and CALMODULIN LIKE37. These proteins could be involved in relaying the GSH-induced AtGLR3.3-dependent rise in [Ca2+]cyt transient to downstream signaling cascades (Fig. 3). In addition, this category includes At3g28580, an AAA-ATPase, which was up-regulated 6-fold by GSH in an AtGLR3.3-dependent manner (Supplemental Table S2). This component has recently been reported to play a central role in mediating plant responses to reactive oxygen species in conditions of environmental stress (Simková et al., 2012). Taken together, this analysis demonstrated that the AtGLR3.3-dependent early transcriptional responses of Arabidopsis to [GSH]ext primarily involved cellular defense and signaling, functional categories that account for 47% of the modulated genes (Supplemental Table S2).
AtGLR3.3 Is Required for GSH-Mediated Innate Immunity Response in Arabidopsis Leaf
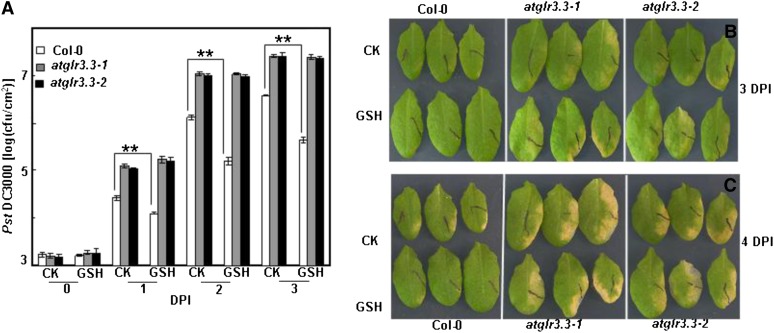
Arabidopsis mutants defective in GSH biosynthesis show altered defense responses to various pathogens (Glazebrook and Ausubel, 1994; Roetschi et al., 2001; van Wees et al., 2003; Bohman et al., 2004; Parisy et al., 2007; Dubreuil-Maurizi and Poinssot, 2012). Innate immunity responses involve complicated signaling networks. We hypothesized that the atglr3.3 mutants may have overlapping defense phenotypes with Arabidopsis mutants, such PHYTOALEXIN DEFICIENT2-1 (pad2-1), defective in GSH biosynthesis. To test its pathogen phenotype, we inoculated mature leaves of the wild type and two allelic knockout mutants (atglr3.3-1 and atglr3.3-2) with the hemibiotrophic model bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000). Pathogen growth was monitored at 0, 1, 2, and 3 d post inoculation (DPI). Figure 5 showed that the two atglr3.3 mutants had significantly higher susceptibility to the pathogen infection than the wild type, indicating that AtGLR3.3 has a role in innate immunity of Arabidopsis. To validate our experimental system, we reexamined the pathogen phenotype of pad2-1 along with the wild type. Supplemental Figure S3 showed that the pathogen population per unit in the mutant at 2 DPI was significantly higher than that of the wild type, which was consistent with the previous report (Glazebrook and Ausubel, 1994).
Figure 5.
Role of AtGLR3.3 in the innate immunity and GSH-triggered defense response in the leaf. A, Leaves of ecotype Columbia (Col-0) and atglr3.3-1 and atglr3.3-2 mutants were infiltrated with either CK solution or the solution containing 100 µm GSH 1 d before the inoculation of Pst DC3000 at 5 × 105 cfu mL–1. The pathogen proliferation was monitored at 0, 1, and 2 DPI. Values presented are average with se from at least four leaves from different plants. At 1, 2, and 3 DPI, there is sd between the Col-0 and two mutants at P < 0.01. Two asterisks indicated significance at P < 0.01. Five independent experiments were conducted and showed comparable results. B, Leaf symptoms of the Col-0, atglr3.3-1, and atglr3.3-2 plants preinfiltrated with CK solution or the solution containing 100 µm GSH 3 DPI with Pst DC3000. C, Same leaves in B were kept 1 d longer, e.g. 4 DPI, in a sealed agar plate.
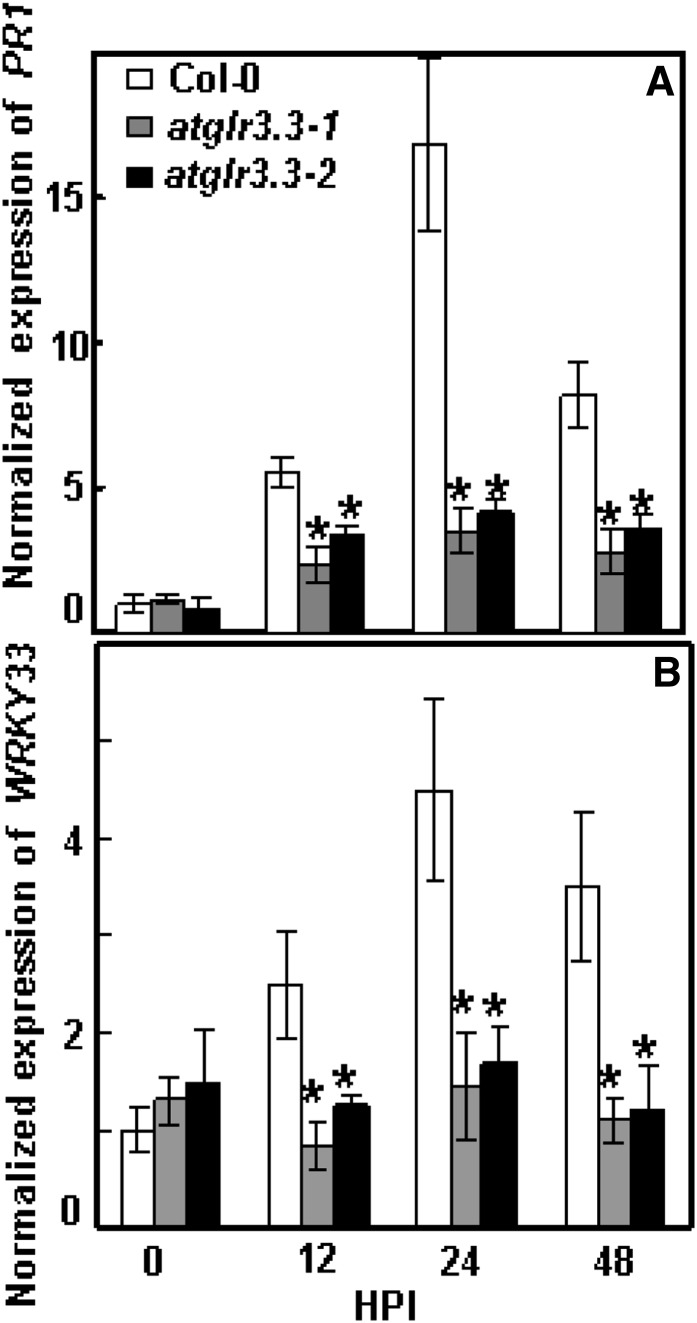
To investigate the role of AtGLR3.3 in the response, we monitored expression of two defense marker genes, PATHOGENESIS-RELATED GENE1 (PR1) and WRKY33, in response to pathogen infection in both the wild type and the mutants at 0, 12, 24, and 48 h post inoculation. The pathogen-induced expression of the defense marker genes was significantly attenuated in both allelic atglr3.3 loss-of-function mutants (Fig. 6). To investigate the physiological significance of the GSH-mediated AtGLR3.3-dependent defense gene expression (Fig. 4; Table I), we next examined the effect of [GSH]ext on pathogen propagation inside the leaves of the wild type and the mutants. One day prior to the pathogen infiltration, the leaves were infiltrated with the CK buffer or the CK buffer containing100 μm GSH. On the day after, all leaves were infiltrated with the pathogen suspension at 5 × 105 colony-forming units (cfu) mL–1. The GSH pretreatment significantly suppressed pathogen propagation in the wild-type leaf. However, suppression of propagation by [GSH]ext was not evident in the atglr3.3 mutants (Fig. 5). Thus, [GSH]ext promotes innate immunity responses in the leaf through AtGLR3.3-dependent pathways.
Figure 6.
Pst DC3000-induced defense market gene expression in ecotype Columbia (Col-0) and atglr3.3 mutants. Pst DC3000 at 5 × 105 cfu mL–1 was infiltrated into the leaves of Col-0, atglr3.3-1, and atglr3.3-2. The leaves were harvested at 0, 12, 24, and 48 h post infection (HPI) for PR1 (A) and WRKY33 (B) gene expression analysis with qRT-PCR. The transcript abundance was normalized with reference gene Tubulin2 in Col-0 at 0 h post infection. Values presented are average with se from three independent experiments. The asterisk indicated the sd between the mutants and Col-0 at same time point at P < 0.05.
Ligand Specificity of AtGLR3.3 in the Innate Immune Response
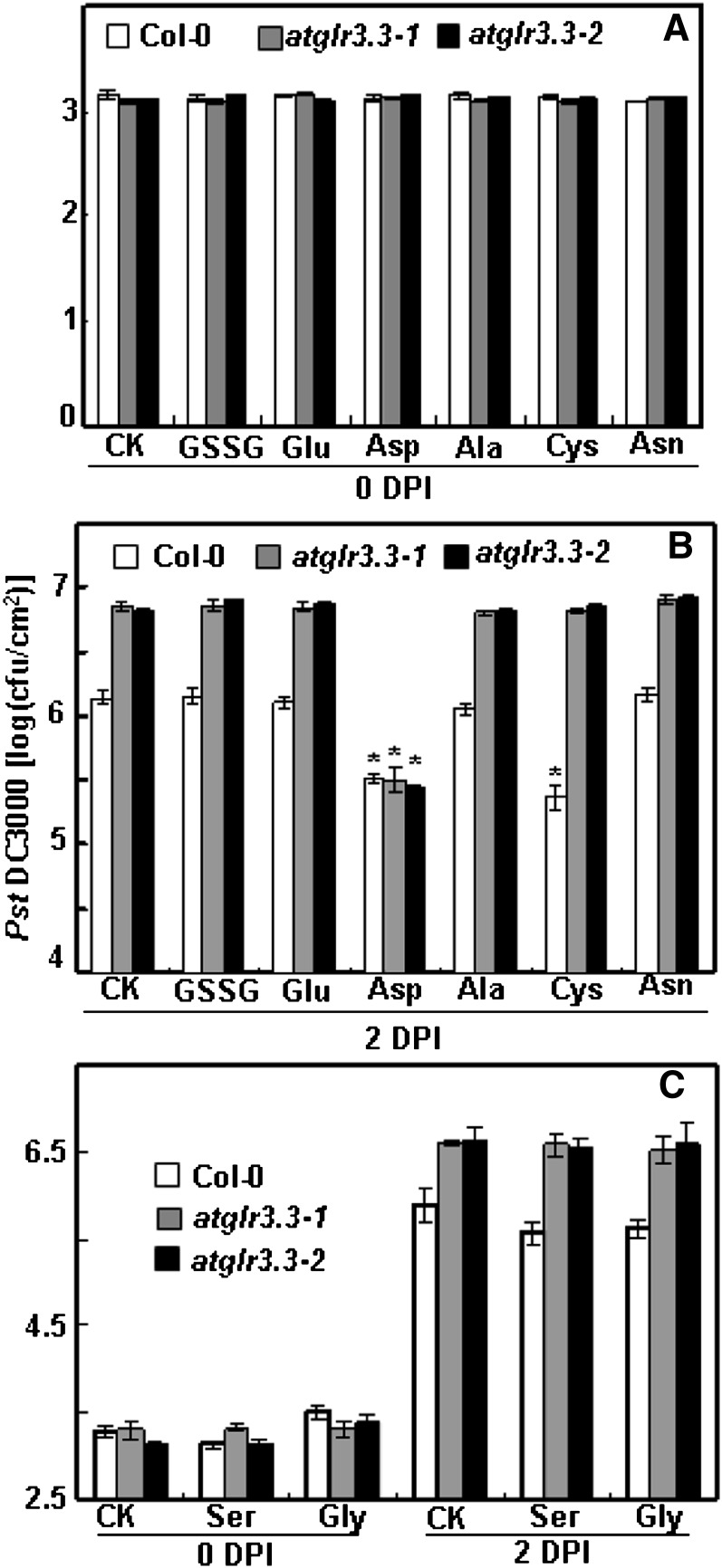
Our previous study with Arabidopsis root and hypocotyl cells suggests that there are up to eight putative ligands of AtGLR3.3: GSH, GSSG, Ala, Asn, Cys, Gly, Glu, and Ser (Qi et al., 2006; Stephens et al., 2008). Among these, Glu plays a dominant role (Stephens et al., 2008). Given the results shown in Figure 5, we hypothesized that if any of the eight putative ligands are true biological ligands of AtGLR3.3 in vivo, they should be able to evoke AtGLR3.3-dependent innate immunity response in the leaf by exogenous application, in a similar fashion to GSH (Figs. 3–5). To test this hypothesis, we infiltrated the leaf of the wild type and two alleles of mutants with CK or CK solution containing 100 μm GSSG, Ala, Asn, Cys, Gly, and Glu 1 d before the pathogen inoculation. We also included Asp, a nonexciting amino acid, as a control. At 2 DPI, consistent with the previous results (Fig. 5), the atglr3.3 mutants showed enhanced bacterial growth (Fig. 7). Among the compounds tested, only Cys and Asp were able to act similarly to GSH to significantly suppress pathogen propagation in the wild-type leaf (Fig. 7). The suppressing effects of Cys, but not Asp, were annulled in the atglr3.3 loss-of-function mutants (Fig. 7). Thus, of the two amino acids, only the suppressive effect of Cys acts through an AtGLR3.3-dependent pathway, similar to GSH.
Figure 7.
Pst DC3000 infection assay of ecotype Columbia (Col-0) and atglr3.3 with putative ligands and Asp. The experiment procedure and analysis methods are same as those in Figure 5. Pst DC3000 proliferation in these leaves was monitored at 0 (A) and 2 DPI (B) preinfiltrated with either CK solution or the solution containing 100 µm GSSG, Glu, Asp, Ala, Cys, and Asn. C, The same assay was conducted separately for Ser and Gly. At 2 DPI in B and C, except for Asp, the values of atglr3.3-1 and atglr3.3-2 were significantly higher than that of Col-0 at P < 0.01. The asterisk in B indicated there was sd between the treatment and the CK for the corresponding genotype at P < 0.01.
The Eight Putative Ligands, but Not Asp, Induce an AtGLR3.3-Dependent [Ca2+]cyt Rise
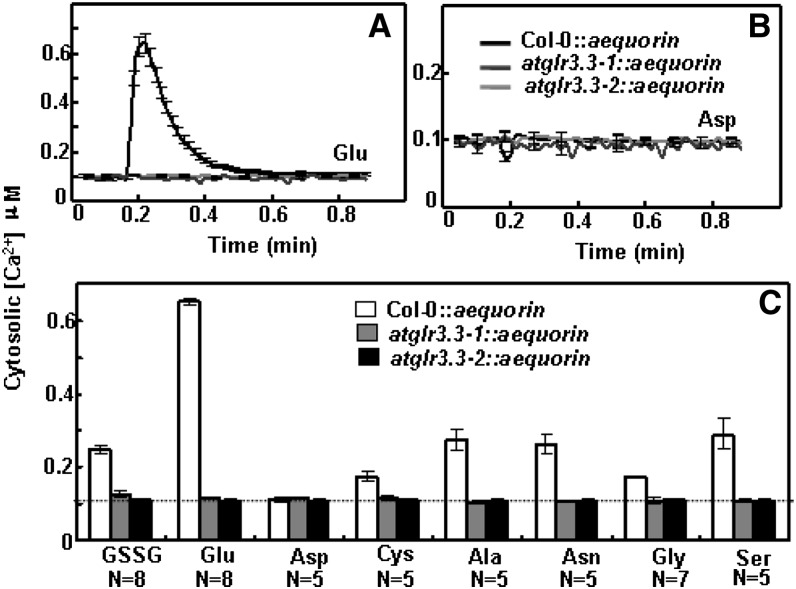
It was unexpected that the eight ligands and Asp would have distinct effects on the plant’s innate immunity responses (Figs. 5 and 7). We hypothesized that the difference could be related to (1) varying ability to initiate the transient rise in [Ca2+]cyt and (2) different dependence on AtGLR3.3 in the leaves. We therefore examined effects of seven ligands and Asp on the [Ca2+]cyt in the leaf cells of the wild type and atglr3.3 mutants. All the tested ligands, except Asp but including Cys, evoked significant [Ca2+]cyt responses (Fig. 8). These were of varying amplitudes, but all were dependent on AtGLR3.3 (Fig. 8). This result demonstrated that innate immunity responses triggered by the ligands GSH and Cys are not simply correlated with their ability to induce [Ca2+]cyt transients.
Figure 8.
Effects of the seven putative ligands of AtGLR3.3 and Asp on the [Ca2+]cyt in the leaf cells of ecotype Columbia (Col-0) and atglr3.3 expressing aequorin. Averaged recording curves with se of 100 µm Glu (A) and Asp (B) induced [Ca2+]cyt rise in the leaf cells of Col-0 and atglr3.3-1 and atglr3.3-2 mutants expressing aequorin (n = 8 for Glu and n = 5 for Asp). C, the average peak values with se of the response to the indicated ligands (n = 8 for GSSG and Glu, n = 5 for Asp, Cys, Ala, Asn, and Ser, and n = 7 for Gly). The asterisk indicated sd at P < 0.01 between the mutants and Col-0 for the same treatment. The dash line indicated the background [Ca2+]cyt.
The Glu-Mediated Early Transcriptional Response Is Distinct to That Induced by GSH
Glu is the principle exciting amino acid that activates GLRs in the central nervous system (Flores-Soto et al., 2012). It also evokes strong [Ca2+]cyt responses in the leaf (Fig. 8A), which are at least comparable with, if not stronger than, those induced by GSH (Figs. 2 and 3). However, Glu and GSH had essentially distinct effects on the plant defense responses. Most notably, Glu had no suppressive effect on the pathogen propagation (Figs. 5 and 7).
To further explore the influence of the two compounds, we compared Glu-elicited transcriptomic responses with those induced by GSH (Fig. 4B; Table I; Supplemental Table S2). In total, treatment of wild-type leaves with 100 μm Glu modulated expression of 70 genes, 69 of which showed no significant changes in response to Glu in the atglr3.3 mutant and can therefore be categorized as Glu-modulated AtGLR3.3-dependent genes (Fig. 4A; Supplemental Tables S5, S6, and S7). Among the 69 genes, only six genes were common to the GSH-modulated AtGLR3.3-dependent group (Fig. 4A; Supplemental Tables S2 and S5). Functional categorization of the 69 Glu-modulated AtGLR3.3-dependent genes showed that 37%, 16%, 23%, 3%, and 6% of these genes were classified as Unknown, Others, Signaling, Transcription Factor, and Defense, respectively (Fig. 4C). The Defense section only included four genes: PR1, FLAVIN-DEPENDENT MONOOXYGENASE1 (Mishina and Zeier, 2006), CELL WALL-ASSOCIATED KINASE1, and CELL WALL-ASSOCIATED KINASE3 (Brutus et al., 2010; De Lorenzo et al., 2011). Hence, it was strikingly different from the transcriptomic patterns elicited by GSH in an AtGLR3.3-dependent manner (Fig. 4B). Taken together, these results demonstrate that although GSH and Glu induce comparable AtGLR3.3-dependent [Ca2+]cyt responses, they induce distinct downstream early transcription responses. Crucially, only GSH was able to induce expression of a significant proportion of defense-related genes, which could explain the specific capacity of GSH to suppress pathogen propagation in the leaves (Fig. 5).
DISCUSSION
Since the discovery of AtGLRs in Arabidopsis genome (Lam et al., 1998), there has been considerable interest in deciphering their physiological functions (Kudla et al., 2010). Previously, in root cells, we demonstrated that AtGLR3.3 is required for the membrane depolarization induced by eight putative ligands, including GSH, GSSG, Ala, Asn, Cys, Glu, Gly, and Ser (Qi et al., 2006). It is possible that different AtGLRs expressed in different tissue and under various conditions show different ligand specificity. The first evidence to support this notion was the finding that d-Ser, but not Glu and Gly, activates AtGLR1.2-dependent Ca2+ conductance in the plasma membrane of the pollen tube (Michard et al., 2011). Among the eight putative ligands of AtGLR3.3, GSH has established physiological function as an essential antioxidant and signaling molecule involved in plant defense responses (Noctor et al., 2012). It led us to form hypotheses that AtGLR3.3 may play a role in innate immunity and in mediating GSH-induced defense responses. Results presented in this study support this hypothesis and open up several intriguing avenues of research for further exploration.
The GSH-Induced AtGLR3.3-Dependent [Ca2+]cyt Transient Involves Several Calcium Stores
It was previously observed that exogenous application of GSH at millimolar concentrations induced a [Ca2+]cyt rise in tobacco leaf and Arabidopsis root cells (Gomez et al., 2004; Qi et al., 2006). However, the identity of the proteins involved in this response was not known. In this study, we first demonstrated that as little as 10 μm GSH could evoke significant elevation of the [Ca2+]cyt (Fig. 1), suggesting that the cell has sensitive mechanisms to detect extracellular GSH at physiological concentrations. Furthermore, using a pharmacological approach and a transfer DNA (T-DNA) insertion knockout mutant expressing the Ca2+ reporter protein aequorin, we demonstrated that the response fully depended on AtGLR3.3 in Arabidopsis leaf cells (Fig. 3). The result is consistent with the previous observation that AtGLR3.3 is required for GSH-induced membrane depolarization in the roots (Qi et al., 2006). In animal cells, iGLRs form cation channels permeable to Ca2+, K+, and Na+ (Flores-Soto et al., 2012). Recently, AtGLR3.4, the closest homolog of AtGLR3.3, has been demonstrated to form an amino acid-gated Ca2+ channel when heterologously expressed in the plasma membrane of human embryonic kidney cells (Vincill et al., 2012).
It is possible that AtGLR3.3 forms similar Ca2+ channels to AtGLR3.4 in the plasma membrane of the leaf cells and is gated by extracellular GSH to conduct Ca2+ influx into the cytosol. However, chelating extracellular Ca2+ didn’t completely abolish the GSH-mediated [Ca2+]cyt response (Fig. 2). This implies that mobilization of internal Ca2+ pools also contributes to the response. AtGLRs are predicted to contain conserved domains of both iGLRs and GPCRs (Turano et al., 2001). It is possible that the AtGLR3.3, similar to GPCRs, is coupled to certain internal Ca2+ mobility-signaling pathways, such as IP3-mediated Ca2+ release from internal Ca2+ pools (Loane et al.,2012). Support for this notion comes from the observation that pharmacologically interfering with IP3 production significantly suppressed the GSH-mediated response (Fig. 2). In summary, the observed [Ca2+]cyt elevation could be explained by (1) direct influx of extracellular Ca2+ through the AtGLR3.3 channel into the cytosol and (2) indirect activation of internal Ca2+-releasing pathways through unknown intermediate signal transducers.
Role of AtGLR3.3 in Innate Immunity and GSH-Induced Defense Responses
Two alleles of atglr3.3 loss-of-function mutants showed consistently enhanced susceptibility to Pst DC3000 (Fig. 5) and decreased expression of defense-related marker genes in response to the pathogen (Fig. 6). This implicates AtGLR3.3 as a novel genetic component of the innate immunity system in the leaf. Theoretically, any of the previously identified eight ligands could be biologically significant ligands of the receptor. By testing the effects of the exogenous application of the ligands individually on pathogen propagation, we found that both GSH and Cys were able to significantly suppress pathogen growth in an AtGLR3.3-dependent manner (Fig. 7). The common feature that GSH and Cys share, and which is absent from the other potential ligands, is a thiol group. It is possible that it is these groups that interact with AtGLR3.3, which in turn evokes specific downstream signaling cascades that ultimately lead to the defense responses. It is well documented that cytosolic GSH can move into the extracellular space during cell death in animal cells (Hammond et al., 2004; Schmidt and Dringen, 2012). In plants, pathogen infection increases GSH production (Vanacker et al., 2000; Parisy et al., 2007). Possibly, cytosolic GSH can be exported to the extracellular space through oligopeptide transporters (Koh et al., 2002) during the interaction between the plant cells and pathogens.
GSH serves as reservoir for Cys, and the two flanking amino acids Glu and Gly protect the sulfhydryl group of Cys against oxidation (Meyer, 2008; Hell and Wirtz, 2011). It is known that externally supplied Cys, after being transported into the cytosol, is transiently formed into GSH (Hell and Wirtz, 2011). Therefore, it is possible that in addition to its direct binding to the AtGLR3.3, Cys can reinforce the GSH production and indirectly induce the defense response. Conversion of Cys to GSH may have contributed to the induced resistance to bacteria (preinfiltration 1 d prior to the pathogen inoculation), but the much shorter incubations used for the luminescence studies (minutes) mean that effects of Cys on [Ca2+]cyt are likely to be direct. It is a very intriguing question how perception of extracellular GSH and Cys by the membrane-anchored AtGLR3.3 relays into downstream signaling cascade.
The suppressing effect of Asp on the pathogen propagation through the AtGLR3.3-independent pathway was unexpected (Fig. 7). Asp is closely linked to several metabolites that play essential roles in primary metabolic pathways such as the tricarboxylic acid cycle that contributes to cellular energy metabolism (Kirma et al., 2012). It was possible that excess Asp could interrupt the energy production process and in turn decrease the energy in the plant cells available for the pathogen consumption. It is also possible that excess Asp drives production of certain defense compounds to either directly limit the pathogen propagation or trigger the plant cells’ innate immunity responses. However, Glu, another amino acid that is closely linked to the tricarboxylic acid cycle, did not suppress pathogen propagation.
Our study gives rise to questions about the physiological ligands of AtGLR3.3. In animal cells, [GSH]ext can induce release of other compounds, for example dopamine from mouse striatal slices (Janáky et al., 2007) and taurine from developing mouse hippocampus (Janáky et al., 2008). We cannot exclude the possibility that [GSH]ext induces release of other compounds from the plant cells to activate the AtGLR3.3-dependent innate immunity response.
GSH-Mediated AtGLR3.3-Dependent Early Transcription Response
Several studies report that [GSH]ext induces stress-associated gene expression, and partial and whole genome data have been presented for GSH- and glutathione reductase-deficient mutants (Ball et al., 2004; Mhamdi et al., 2010; Han et al., 2013b). In this study, we analyzed the early transcription responses of the plant cells to [GSH]ext with the aim of studying gene regulation effects of GSH acting as a signaling molecule rather than in its well-known role as an antioxidant. Because we only had two biological replicates, we set very strict standards as detailed in “Materials and Methods” to process the raw data. Its validity was supported by two findings: first, the AtGLR3.3 transcript was the most down-regulated in the atglr3.3 mutant compared with that in the wild type (Supplemental Table S1); second, real-time qRT-PCR with independent biological samples were largely consistent with the microarray data set (Supplemental Fig. S1). The GSH treatment modulated expression of 97 genes in the wild type, 70 of which were not significantly changed in atglr.3.3, accounting for 72% of the total (Fig. 4A; Supplemental Table S2). It clearly indicated that AtGLR3.3 plays an essential role in the early transcriptional responses of the leaf to GSH. The genetic mechanism of the GSH-mediated and AtGLR3.3-independent gene expression is unknown. In animal cells, in addition to the GLRs, GSH has been shown to have profound regulative effects on the ryanodine receptor (Zable et al., 1997), death receptor (Morito et al., 2003), and GPCRs (Wang et al., 2006). It is possible that certain unknown receptors in Arabidopsis mediate the AtGLR3.3-independent GSH signaling cascade.
For the GSH-modulated AtGLR3.3-dependent genes, we did standard Gene Ontology annotation analysis combined with manual literature checking. It revealed that 27% of the genes have either confirmative roles in defense or are induced by biotic stresses (Table I; Supplemental Table S2), which accounted for the largest subgroup in the early transcription response. It makes strong sense in context of the defense function of GSH observed in this study (Fig. 5). Particularly interesting is the finding that GSH induced expression of ICS1 and SARD1 in an AtGLR3.3-dependent manner, two essential genes for SA biosynthesis (Wildermuth et al., 2001; Zhang et al., 2010). A regulatory role for SA in GSH production in plants has been described (Srivastava and Dwivedi, 1998; Yoshida et al., 2009). Our finding that GSH can strongly induce ICS1 expression (Table I) implies that there is a positive AtGLR3.3-dependent feedback mechanism for GSH to enhance SA biosynthesis under stress conditions. Hydrogen peroxide induces production of SA in tobacco (Leon et al., 1995), while the GSH-deficient mutant pad2-1 lost pathogen Phytophthora brassicae-triggered expression of ICS1 (Dubreuil-Maurizi and Poinssot, 2012). Recently, it has been reported that hydrogen peroxide-triggered accumulation of SA in a catalase-deficient mutant can be largely blocked by blocking glutathione synthesis, and that is associated with the failure to up-regulate ICS1 (Han et al., 2013a). It will be very interesting to investigate the role of AtGLR3.3 in the interaction between SA and GSH homeostasis.
Correlation of the [Ca2+]cyt Transient Rise with the Defense Response
A large number of environmental and internal signals are able to evoke [Ca2+]cyt transients in plant cells (Kudla et al., 2010). In most of these cases, two fundamental questions remain: (1) the molecular component underlying the transient [Ca2+]cyt rise and (2) how the Ca2+ responses are linked to the downstream physiological function mediated by the signaling. In this study, we provided genetic evidence that the transient [Ca2+]cyt rise induced by the eight putative ligands were fully dependent on AtGLR3.3 (Figs. 3 and 8). However, the link between the GSH-induced transient [Ca2+]cyt rise and the defense functions of GSH still remain elusive.
It is possible that the two effects of GSH are not related. Evidence to support this argument is that Glu-induced AtGLR3.3-dependent [Ca2+]cyt transient rise was stronger than that of GSH (Fig. 8), even though unlike GSH, Glu did not induce significant amount of defense gene expression and defense responses (Figs. 4C and 7). Key points are the still unknown structure of the Glu receptor complex and the mechanism through which it specifically recognizes different ligands. In animal cells, functioning iGLRs are composed of different subunits encoded by individual iGLR genes and activated by different ligands. A typical case in this context is the NMDA iGLR, which is composed of four subunits, two from NMDA Receptor A (NRA), NRB, NRC, and NRD and two NR1s. NRA to NRD is activated by Glu and NR1 by Gly (Flores-Soto et al., 2012).
It is possible that the AtGLRs are also formed by different subunits that recognize different ligands. GSH and Cys could activate a unique subunit through their specific thiol group and induce additional signaling cascades that occur alongside, rather than downstream, of the transient [Ca2+]cyt rise. Our previous study with the cross-desensitization technique indirectly reveals that the AtGLRs are composed by AtGLR3.3 and other subunits specifically activated by different putative ligands (Stephens et al., 2008).
The situation could be even more complex. It has been proposed for some time that transient [Ca2+]cyt rises induced by different signals could be different in strength, frequency, and spatial distribution (Kudla et al., 2010). A recent study provides evidence in support of this and shows that transient [Ca2+]cyt rises with different strength and spatial distribution are linked to different transcription responses (Wheeler et al., 2012). Such concepts could explain why GSH and Glu induce comparable changes in [Ca2+]cyt but distinct gene expression responses. To completely elucidate this mechanism, studies will require more precise measurement and subcellular imaging of dynamic changes in [Ca2+]cyt. Whether the two GSH-mediated AtGLR3.3-dependent effects are related or not, our data suggest that the GSH-related functions of AtGLR3.3 are involved in signaling. The atglr3.3 mutant showed wild-type growth on cadmium (Supplemental Fig. S2), a condition in which GSH plays an important but nonsignaling role as a precursor molecule of phytochelatins that sequester cadmium (Cobbett et al., 1998; Jozefczak et al., 2012).
CONCLUSION
Glu receptors play vital roles in mediating information exchange among neurons in the central nervous system. Their complex functions are determined by their versatile structures, subunit composition, various ligand types, and binging sites as well as binding affinity in different tissues and under different physiological conditions (Flores-Soto et al., 2012; Popescu, 2012). In this study, we provided evidence to support a novel role of AtGLR3.3 in the innate immunity and GSH-mediated defense response in Arabidopsis leaf. Our findings promote understanding of the AtGLR gene family and the related signaling roles of extracellular GSH and raise intriguing questions for further study.
MATERIALS AND METHODS
Arabidopsis AtGLR3.3 Mutants Expressing Aequorin
The Arabidopsis (Arabidopsis thaliana) atglr3.3-1 and atglr3.3-2 (At1g42540) homozygous knockout mutants used in this study were described previously (Qi et al., 2006) and were SALK_040458 and SALK_066009, respectively (Alonso et al., 2003), obtained from The Arabidopsis Biological Resource Center. To introduce the aequorin gene into the mutant background, mutants were crossed with the aequorin transgenic Arabidopsis line (Knight et al., 1991). In the F2 segregation population, homozygous T-DNA insertions were detected with gene-specific primers (SALK_040458, 5′-TTGTTTGTTGCAGCATTGAG-3′, 5′-GCCAATCTTGAGTTCTTTCCC-3′ and SALK_066009, 5′-AAGCACCAGACATCTTACGC-3′, 5′-TCAGATTGGTCCAATTTTAGC-3′) and the T-DNA left border-specific primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′). The presence of active aequorin was validated by measuring luminescence intensity in the leaf, as described below.
[Ca2+]cyt Measurement in Arabidopsis Leaf Expressing Aequorin
To measure dynamic changes in [Ca2+]cyt in the leaf, Arabidopsis seeds were surface sterilized with 100% ethanol (v/v) for 15 min and dried on sterilized filter paper in a fume hood. Sterilized seeds were sown on 0.7% (w/v) agar medium containing one-half-strength Murashige and Skoog basic salts with 1% (w/v) Suc and 5 mm MES, pH 5.7. After 3-d treatment at 4°C, the plates were placed in a growth chamber at 22°C to 24°C , 12-h light/12-h dark, and a light intensity of 100 to 120 μmol E m–2 s–1. One leaf per 12-d-old seedlings was cut and placed in a 1.5-mL centrifuge tube with 100 μL control buffer (CK buffer) composed of 1 mm KCl, 1 mm CaCl2, and 5 mm MES, pH 5.7. We observed that leaves from precut seedlings had a very weak [Ca2+]cyt response. To ensure consistent results, therefore, care was taken to sample only one leaf per seedling. Under dim light, coelenterazine hcp (Promega) was added into the buffer to a final concentration of 1 μm. After 12- to 14-h incubation in the dark, the tube with the leaf was put into a single tube luminometer with a detection limit of 10–21 mol luciferase (Turner Biosystem, 20/20) in a room with dim light. The tube was left to stand for 3 to 5 min to allow the leaf to recover from potential disturbances of [Ca2+]cyt triggered by the handling. For the treatment, 100 μL control buffer with 2 times the final concentration of GSH was quickly delivered into the tube by positioning the pipette tip on the internal walls to enable the treatment solution to diffuse into the solution with the leaf. This procedure avoided directly dropping the treatment solution into the solution with the leaf and so aimed to minimize disturbance of the leaf [Ca2+]cyt background.
Luminescence intensity was measured at 1-Hz frequency. At the end of each recording, 500 μL ice-cold 1 m CaCl2 in 20% ethanol (v/v) was delivered into the tube to saturate the Ca2+ and aequorin protein-dependent luminescence. The absolute [Ca2+]cyt concentration was calculated based on the method described previously (Rentel and Knight, 2004). To explore the contribution of external Ca2+ to the observed [Ca2+]cyt rise, we incubated the leaf in Ca2+-free CK buffer with 1 mm EGTA, a Ca2+ chelator. The GSH treatment was also performed in the same buffer. To address effects of AP5, alloxan, neomycin, and 6,7-dinitroquinoxaline-2,3-dione on the [Ca2+]cyt responses, these chemicals were added into the recording buffer at the indicated concentration 1 h before the GSH treatments. All chemicals were purchased from Sigma.
Arabidopsis Seedling Treatment and RNA Extraction for Complementary DNA Microarray
Arabidopsis seedlings were cultured on 0.7% (w/v) agar medium containing one-half-strength Murashige and Skoog basic salt with 1% Suc (w/v) and 5 mm MES, pH 5.7. Ten-day-old seedlings were equilibrated in the CK buffer for 24 h. Then, these seedlings were either treated with the CK buffer or the CK buffer containing100 μm GSH and 100 μm Glu for 1 h, respectively. Total RNA was extracted from each sample using EasyPure Plant RNA kit (Transgene). Further RNA purification, probe preparation, hybridization, and microarray scanning were done at the National GeneChip Engineering Center (http://www.ebioserve.com). Labeled samples were hybridized with one Agilent Arabidopsis 4×44k array by following the manufacturer’s instructions. Four biologically independent sets of Arabidopsis seedlings were prepared, two for the microarray experiment and two for real-time qRT-PCR analysis.
The raw data were normalized by quantile algorithm using Gene Spring Software 11.0. The following criteria were used in the National GeneChip Engineering Center for defining significantly differentially expressed genes: (1) genes that are statistically significant at the level of P < 0.05 after false discovery rate correction and labeled “Present” call by the GeneChip Operation System output and (2) genes showing a change in the expression level between treatment and control at least 3-fold in the two replicates. The Gene Ontology annotation analysis was conducted using the online program at http://www.Arabidopsis.org/tools/bulk/go/index.jsp. The raw data of the microarray experiment was deposited at http://www.ebi.ac.uk/miamexpress/ with access number E–MEXP–3885.
qRT-PCR Analysis
Tissue preparation and RNA extraction were as for the microarray analyses. Total RNA was treated with RNAse-free DNAse to remove all DNA contamination (Takara). One microgram of RNA samples were used for complementary DNA synthesis with oligo(dT) primer and PrimeScript II reverse transcriptase (Takara) following a standard protocol. DNA contamination of the complementary DNA was further tested by PCR with a pair of primers spanning an intron. Before the qRT-PCR analysis, all primers were tested for their specificity and primer dimer formation by conducting semiquantitative reverse transcript PCR. qRT-PCR was conducted on a Biorad Chromo 4 real-time PCR system using SYBR Green Premix Ex Taq II kit (Takara). The Arabidopsis Tubulin2 gene (At5g62690) was used as a constitutive reference to normalize the gene expression. Relative expression to the wild type under CK condition was presented in Figure 6 and Supplemental Figure S1. Primer sequences for the PCR are listed in Supplemental Table S8.
Bacterial Growth Assay
To prepare Arabidopsis plants for the assay, we grew two wild-type and two mutant plants in the same pot to minimize side effects of growth environment difference on the pathogen propagation results. The pots were maintained in a growth chamber with 50 to 70% humidity at 22°C to 24°C, 12-h light/12-h dark, and a light intensity of 100 to 120 μmol E m–2 s–1. Four-week-old leaves were used in this study. For each plant, we chose two leaves for syringe-based infiltration. One day before the pathogen inoculation, we infiltrated the leaf with either the CK or CK containing100 μm GSH, GSSG, Ala, Asn, Cys, Gly, Glu, Ser, and Asp.
The bacterial strain used in this study was Pst DC3000. The –80°C stock of the pathogen was first streaked onto a low-salt Luria-Bertani plate (tryptone 10 g L–1, yeast [Saccharomyces cerevisiae] extract 5 g L–1, and NaCl 5 g L–1) with 50 μg mL–1 rifampin. Plates with bacteria were cultured at 28°C for 38 to 42 h, then subcultured in liquid Luria-Bertani solution growing up to an optical density at 600 nm of 0.8 to 1.0. After collection by centrifuging, the pathogen was resuspended in sterile water to an optical density at 600 nm of 0.001, corresponding to 5 × 105 cfu mL–1 bacteria, and infiltrated into the leaf with a needless 1-mL syringe. At 0, 1, 2, and 3 DPI, the inoculated leaf was taken and briefly rinsed in 75% ethanol (v/v) and sterile water. A leaf disc with 0.5-cm diameter was sampled from each of four to eight different leaves from separate plants for each treatments of each genotype and ground in sterile water. A standard colony-forming unit dilution assay was conducted (Katagiri et al., 2002). Five independent biological replicates were performed.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. qRT-PCR analysis of GSH-induced AtGLR3.3-dependent gene expression.
Supplemental Figure S2. Root elongation responses of ecotype Columbia and atglr3.3 to Cd2+.
Supplemental Figure S3. Pst DC3000 infection assay of Arabidopsis pad2-1 mutants.
Supplemental Table S1. Differential expressed gene in the atglr3.3-2 mutant compared with that in ecotype Columbia.
Supplemental Table S2. Genes modulated by GSH in ecotype Columbia but not atglr3.3-2 mutant.
Supplemental Table S3. Genes modulated by GSH in ecotype Columbia and atglr3.3-2 mutant.
Supplemental Table S4. Genes modulated by GSH in atglr3.3-2 but not ecotype Columbia.
Supplemental Table S5. Genes modulated by Glu in ecotype Columbia but not atglr3.3-2 mutant.
Supplemental Table S6. Genes modulated by Glu in ecotype Columbia and atglr3.3-2 mutant.
Supplemental Table S7. Genes modulated by Glu in atglr3.3-2 but not ecotype Columbia.
Supplemental Table S8. Primers used for the quantitative PCR.
Supplementary Material
Acknowledgments
We thank Shengyang He (Michigan State University) for the strain of bacterial pathogen Pst DC3000, Dr. Marc R. Knight (Durham University) for the Arabidopsis transgenic seeds expressing the aequorin gene, Felix Mauch (University of Fribourg) for the pad2-1 mutant seeds, Turner Biosystem USA for donation of the luminometer instrument, and Dr. Huifeng Zhu (Capital Normal University-Beijing) for initial technical assistance on the bacterial growth assay.
Glossary
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- iGLR
ionotropic Glu receptor
- NMDA
N-methyl-d-Asp
- [Ca2+]cyt
cytosolic Ca2+
- GPCR
G protein-coupled receptor
- [GSH]ext
exogenous glutathione
- AP5
(2R)-amino-5-phosphonopentanoate
- IP3
1,4,5-trisphosphate
- CK
control
- qRT
quantitative reverse transcript
- DPI
d post inoculation
- cfu
colony-forming unit
- T-DNA
transfer DNA
- Pst DC3000
Pseudomonas syringae pv tomato DC3000
- SA
salicylic acid
References
- Acosta IF, Farmer EE. (2010) Jasmonates. The Arabidopsis Book 8: e0129, doi/10.1199/tab.0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA. (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aouini A, Hernould M, Ariizumi T, Matsukura C, Ezura H, Asamizu E. (2012) Overexpression of the tomato glutamate receptor-like genes SlGLR1.1 and SlGLR3.5 hinders Ca2+ utilization and promotes hypersensitivity to Na+ and K+ stresses. Plant Biotechnol 29: 229–235 [Google Scholar]
- Aoyama K, Watabe M, Nakaki T. (2008) Regulation of neuronal glutathione synthesis. J Pharmacol Sci 108: 227–238 [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechtold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, et al. (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16: 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE. (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohman S, Staal J, Thomma BP, Wang M, Dixelius C. (2004) Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J 37: 9–20 [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ. (2006) Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin TY, Chueh SH, Tao PL. (2006) S-Nitrosoglutathione and glutathione act as NMDA receptor agonists in cultured hippocampal neurons. Acta Pharmacol Sin 27: 853–860 [DOI] [PubMed] [Google Scholar]
- Chiu J, DeSalle R, Lam HM, Meisel L, Coruzzi G. (1999) Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol 16: 826–838 [DOI] [PubMed] [Google Scholar]
- Cho D, Kim SA, Murata Y, Lee S, Jae SK, Nam HG, Kwak JM. (2009) De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J 58: 437–449 [DOI] [PubMed] [Google Scholar]
- Cobbett CS, May MJ, Howden R, Rolls B. (1998) The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in γ-glutamylcysteine synthetase. Plant J 16: 73–78 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, Brutus A, Savatin DV, Sicilia F, Cervone F. (2011) Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett 585: 1521–1528 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D, Merchant S, Briggs W, Ort D. (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Dringen R. (2000) Metabolism and functions of glutathione in brain. Prog Neurobiol 62: 649–671 [DOI] [PubMed] [Google Scholar]
- Dubos C, Huggins D, Grant GH, Knight MR, Campbell MM. (2003) A role for glycine in the gating of plant NMDA-like receptors. Plant J 35: 800–810 [DOI] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C, Poinssot B. (2012) Role of glutathione in plant signaling under biotic stress. Plant Signal Behav 7: 210–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Flores-Soto ME, Chaparro-Huerta V, Escoto-Delgadillo M, Vazquez-Valls E, González-Castañeda RE, Beas-Zarate C. (2012) Structure and function of NMDA-type glutamate receptor subunits. Neurologia 27: 301–310 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Cidlowski JA. (2009) Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ 16: 1303–1314 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM. (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez LD, Noctor G, Knight MR, Foyer CH. (2004) Regulation of calcium signalling and gene expression by glutathione. J Exp Bot 55: 1851–1859 [DOI] [PubMed] [Google Scholar]
- Hammond CL, Madejczyk MS, Ballatori N. (2004) Activation of plasma membrane reduced glutathione transport in death receptor apoptosis of HepG2 cells. Toxicol Appl Pharmacol 195: 12–22 [DOI] [PubMed] [Google Scholar]
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G. (2013a) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid Redox Signal 18: 2106–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Mhamdi A, Chaouch S, Noctor G. (2013b) Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ 36: 1135–1146 [DOI] [PubMed] [Google Scholar]
- He ZH, Cheeseman I, He D, Kohorn BD. (1999) A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol Biol 39: 1189–1196 [DOI] [PubMed] [Google Scholar]
- Hell R, Wirtz M. (2011) Molecular biology, biochemistry and cellular physiology of cysteine metabolism. The Arabidopsis Book 9: e0154, doi/10.1199/tab.0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko TS, Edwards RH. (2012) Neurotransmitter corelease: mechanism and physiological role. Annu Rev Physiol 74: 225–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Dong Q, Yu D. (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185-186: 288–297 [DOI] [PubMed] [Google Scholar]
- Janáky R, Dohovics R, Saransaari P, Oja SS. (2007) Modulation of [3H]dopamine release by glutathione in mouse striatal slices. Neurochem Res 32: 1357–1364 [DOI] [PubMed] [Google Scholar]
- Janáky R, Shaw CA, Oja SS, Saransaari P. (2008) Taurine release in developing mouse hippocampus is modulated by glutathione and glutathione derivatives. Amino Acids 34: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janáky R, Shaw CA, Varga V, Hermann A, Dohovics R, Saransaari P, Oja SS. (2000) Specific glutathione binding sites in pig cerebral cortical synaptic membranes. Neuroscience 95: 617–624 [DOI] [PubMed] [Google Scholar]
- Janáky R, Varga V, Saransaari P, Oja SS. (1993) Glutathione modulates the N-methyl-d-aspartate receptor-activated calcium influx into cultured rat cerebellar granule cells. Neurosci Lett 156: 153–157 [DOI] [PubMed] [Google Scholar]
- Jozefczak M, Remans T, Vangronsveld J, Cuypers A. (2012) Glutathione is a key player in metal-induced oxidative stress defenses. Int J Mol Sci 13: 3145–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juurlink BH. (1999) Management of oxidative stress in the CNS: the many roles of glutathione. Neurotox Res 1: 119–140 [DOI] [PubMed] [Google Scholar]
- Kang J, Mehta S, Turano FJ. (2004) The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol 45: 1380–1389 [DOI] [PubMed] [Google Scholar]
- Kang J, Turano FJ. (2003) The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6872–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Kim HB, Lee H, Choi JY, Heu S, Oh CJ, Kwon SI, An CS. (2006) Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fungal infection. Mol Cells 21: 418–427 [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY. (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: 0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Kwak JM, Jae SK, Wang MH, Nam HG. (2001) Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol 42: 74–84 [DOI] [PubMed] [Google Scholar]
- Kirma M, Araújo WL, Fernie AR, Galili G. (2012) The multifaceted role of aspartate-family amino acids in plant metabolism. J Exp Bot 63: 4995–5001 [DOI] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Koh S, Wiles AM, Sharp JS, Naider FR, Becker JM, Stacey G. (2002) An oligopeptide transporter gene family in Arabidopsis. Plant Physiol 128: 21–29 [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. (2011) Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem J 440: 355–365 [DOI] [PubMed] [Google Scholar]
- Kwaaitaal M, Maintz J, Cavdar M, Panstruga R. (2012) On the ligand binding profile and desensitization of plant ionotropic glutamate receptor (iGluR)-like channels functioning in MAMP-triggered Ca2+ influx. Plant Signal Behav 7: 1373–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe B, Becker D, Hedrich R, DeSalle R, Hollmann M, Kwak JM, Schroeder JI, Le Novère N, Nam HG, Spalding EP, et al. (2001) The identity of plant glutamate receptors. Science 292: 1486–1487 [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G. (1998) Glutamate-receptor genes in plants. Nature 396: 125–126 [DOI] [PubMed] [Google Scholar]
- Leon J, Lawton MA, Raskin I. (1995) Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol 108: 1673–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie SW, Brown LM, Trent RD, Lee YH, Morris JL, Jones TW, Randall PK, Lau SS, Monks TJ. (1992) Stimulation of N-methyl-d-aspartate receptor-mediated calcium entry into dissociated neurons by reduced and oxidized glutathione. Mol Pharmacol 41: 308–314 [PubMed] [Google Scholar]
- Levy DI, Sucher NJ, Lipton SA. (1991) Glutathione prevents N-methyl-d-aspartate receptor-mediated neurotoxicity. Neuroreport 2: 345–347 [DOI] [PubMed] [Google Scholar]
- Li J, Zhu S, Song X, Shen Y, Chen H, Yu J, Yi K, Liu Y, Karplus VJ, Wu P, et al. (2006) A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell 18: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Little D, Gouhier-Darimont C, Bruessow F, Reymond P. (2007) Oviposition by pierid butterflies triggers defense responses in Arabidopsis. Plant Physiol 143: 784–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loane DJ, Stoica BA, Faden AI. (2012) Metabotropic glutamate receptor-mediated signaling in neuroglia. Wiley Interdiscip Rev Membr Transp Signal 1: 136–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Qi Z, Smigel A, Walker RK, Verma R, Berkowitz GA. (2009) Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc Natl Acad Sci USA 106: 20995–21000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Sato T, Asada Y, Yasuda S, Yoshida M, Chiba Y, Yamaguchi J. (2012) The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol Biol 79: 217–227 [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ. (2008) The integration of glutathione homeostasis and redox signaling. J Plant Physiol 165: 1390–1403 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis-Bourguet E, Renou JP, et al. (2010) Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, Feijó JA. (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil d-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Miller ND, Durham Brooks TL, Assadi AH, Spalding EP. (2010) Detection of a gravitropism phenotype in glutamate receptor-like 3.3 mutants of Arabidopsis thaliana using machine vision and computation. Genetics 186: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Degrave A, Vedel R, Bitton F, Patrit O, Renou JP, Barny MA, Fagard M. (2012) EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora. Mol Plant Microbe Interact 25: 421–430 [DOI] [PubMed] [Google Scholar]
- Morito N, Yoh K, Itoh K, Hirayama A, Koyama A, Yamamoto M, Takahashi S. (2003) Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene 22: 9275–9281 [DOI] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH. (2012) Glutathione in plants: an integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH. (2011) Glutathione. The Arabidopsis Book 9: e0142, doi/10.1199/tab.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogita K, Enomoto R, Nakahara F, Ishitsubo N, Yoneda Y. (1995) A possible role of glutathione as an endogenous agonist at the N-methyl-d-aspartate recognition domain in rat brain. J Neurochem 64: 1088–1096 [DOI] [PubMed] [Google Scholar]
- Oja SS, Janáky R, Varga V, Saransaari P. (2000) Modulation of glutamate receptor functions by glutathione. Neurochem Int 37: 299–306 [DOI] [PubMed] [Google Scholar]
- Okrent RA, Brooks MD, Wildermuth MC. (2009) Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J Biol Chem 284: 9742–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F. (2007) Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J 49: 159–172 [DOI] [PubMed] [Google Scholar]
- Pasternak M, Lim B, Wirtz M, Hell R, Cobbett CS, Meyer AJ. (2008) Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J 53: 999–1012 [DOI] [PubMed] [Google Scholar]
- Popescu GK. (2012) Modes of glutamate receptor gating. J Physiol 590: 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Po-Wen C, Singh P, Zimmerli L. (2013) Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol Plant Pathol 14: 58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Stephens NR, Spalding EP. (2006) Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol 142: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasul S, Dubreuil-Maurizi C, Lamotte O, Koen E, Poinssot B, Alcaraz G, Wendehenne D, Jeandroz S. (2012) Nitric oxide production mediates oligogalacturonide-triggered immunity and resistance to Botrytis cinerea in Arabidopsis thaliana. Plant Cell Environ 35: 1483–1499 [DOI] [PubMed] [Google Scholar]
- Rentel MC, Knight MR. (2004) Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol 135: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber TL, Ausubel FM. (1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetschi A, Si-Ammour A, Belbahri L, Mauch F, Mauch-Mani B. (2001) Characterization of an Arabidopsis-Phytophthora pathosystem: resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J 28: 293–305 [DOI] [PubMed] [Google Scholar]
- Saga H, Ogawa T, Kai K, Suzuki H, Ogata Y, Sakurai N, Shibata D, Ohta D. (2012) Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol Plant Microbe Interact 25: 684–696 [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Bodenhausen N, Buchala A, Mauch F, Reymond P. (2008) The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J 55: 774–786 [DOI] [PubMed] [Google Scholar]
- Schmidt MM, Dringen R. (2012) GSH synthesis and metabolism. In Gruetter R, Choi I, eds, Advance in Neurobiology, Vol 4 Springer, New York, pp 1029–1050 [Google Scholar]
- Shanmugam V, Tsednee M, Yeh KC. (2012) ZINC TOLERANCE INDUCED BY IRON 1 reveals the importance of glutathione in the cross-homeostasis between zinc and iron in Arabidopsis thaliana. Plant J 69: 1006–1017 [DOI] [PubMed] [Google Scholar]
- Shaw C, Janaky R, Ogita K. (2001) Glutathione-mediated signal transduction in mammalian CNS. J Neurochem 78: 120. [DOI] [PubMed] [Google Scholar]
- Simková K, Moreau F, Pawlak P, Vriet C, Baruah A, Alexandre C, Hennig L, Apel K, Laloi C. (2012) Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 16360–16365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivka A, Cohen G. (1993) Brain ischemia markedly elevates levels of the neurotoxic amino acid, cysteine. Brain Res 608: 33–37 [DOI] [PubMed] [Google Scholar]
- Srivastava MK, Dwivedi UN. (1998) Salicylic acid modulates glutathione metabolism in pea seedlings. J Plant Physiol 153: 409–414 [Google Scholar]
- Stephens NR, Qi Z, Spalding EP. (2008) Glutamate receptor subtypes evidenced by differences in desensitization and dependence on the GLR3.3 and GLR3.4 genes. Plant Physiol 146: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang RH, Han S, Zheng H, Cook CW, Choi CS, Woerner TE, Jackson RB, Pei ZM. (2007) Coupling diurnal cytosolic Ca2+ oscillations to the CAS-IP3 pathway in Arabidopsis. Science 315: 1423–1426 [DOI] [PubMed] [Google Scholar]
- Turano FJ, Panta GR, Allard MW, van Berkum P. (2001) The putative glutamate receptors from plants are related to two superfamilies of animal neurotransmitter receptors via distinct evolutionary mechanisms. Mol Biol Evol 18: 1417–1420 [DOI] [PubMed] [Google Scholar]
- van Wees SC, Chang HS, Zhu T, Glazebrook J. (2003) Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol 132: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Carver TL, Foyer CH. (2000) Early H2O2 accumulation in mesophyll cells leads to induction of glutathione during the hyper-sensitive response in the barley-powdery mildew interaction. Plant Physiol 123: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet A, Parker J, Tornero P, Nass N, Nürnberger T, Dangl JL, Scheel D, Lee J. (2002) NHL25 and NHL3, two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol Plant Microbe Interact 15: 608–616 [DOI] [PubMed] [Google Scholar]
- Varga V, Jenei Z, Janáky R, Saransaari P, Oja SS. (1997) Glutathione is an endogenous ligand of rat brain N-methyl-d-aspartate (NMDA) and 2-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptors. Neurochem Res 22: 1165–1171 [DOI] [PubMed] [Google Scholar]
- Vincill ED, Bieck AM, Spalding EP. (2012) Ca2+ conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol 159: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yao Y, Kuang D, Hampson DR. (2006) Activation of family C G-protein-coupled receptors by the tripeptide glutathione. J Biol Chem 281: 8864–8870 [DOI] [PubMed] [Google Scholar]
- Wheeler DG, Groth RD, Ma H, Barrett CF, Owen SF, Safa P, Tsien RW. (2012) CaV1 and CaV2 channels engage distinct modes of Ca2+ signaling to control CREB-dependent gene expression. Cell 149: 1112–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Tamaoki M, Ioki M, Ogawa D, Sato Y, Aono M, Kubo A, Saji S, Saji H, Satoh S, et al. (2009) Ethylene and salicylic acid control glutathione biosynthesis in ozone-exposed Arabidopsis thaliana. Physiol Plant 136: 284–298 [DOI] [PubMed] [Google Scholar]
- Zable AC, Favero TG, Abramson JJ. (1997) Glutathione modulates ryanodine receptor from skeletal muscle sarcoplasmic reticulum. Evidence for redox regulation of the Ca2+ release mechanism. J Biol Chem 272: 7069–7077 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.