Abstract
We assess the progress in biomolecular modeling and simulation, focusing on structure prediction and dynamics, by presenting the field’s history, metrics for its rise in popularity, early expressed expectations, and current significant applications. The increases in computational power combined with improvements in algorithms and force fields have led to considerable success, especially in protein folding, specificity of ligand/biomolecule interactions, and interpretation of complex experimental phenomena (e.g. NMR relaxation, protein-folding kinetics and multiple conformational states) through the generation of structural hypotheses and pathway mechanisms. Although far from a general automated tool, structure prediction is notable for proteins and RNA that preceded the experiment, especially by knowledge-based approaches. Thus, despite early unrealistic expectations and the realization that computer technology alone will not quickly bridge the gap between experimental and theoretical time frames, ongoing improvements to enhance the accuracy and scope of modeling and simulation are propelling the field onto a productive trajectory to become full partner with experiment and a field on its own right.
1. Introduction
1.1 Motivation for field assessment
During a recent play in New York’s Under the Radar Festival named Invisible Atom, the protagonist Atom begins to question his own existence in the light of recent personal and world troubles. It appears to him that through attempts for growth and discovery, humankind has brought destruction. The theory of the atom – the never-ending quest by scientists to reduce the atom to smaller and smaller parts – serves in the play as an allegory to excess and greed, which also led to the economic crisis that personally affects Atom, and to the dark consequences of ‘progress ’.
Like Atom’s questioning and quest for truth, the biological community has continually inspected its progress. Recent NIH and DOE workshops have explored the impact of modeling on biology and the potential utilization of next-generation computing technology by the biological community (DOE, 2009; IMAG, 2009). Recognizing the immense challenges in biology in the 21st century, the NAS has urged for biology’s integration with other fields as well as biology’s consolidation of its many sub-disciplines (NAS, 2009). Furthermore, the general realization that many problems in biology have turned out to be much more difficult than originally perceived (Hayden, 2010) also affects the impact that modeling can have on solving them. Finally, suggestions have been made of a growing gap between academe and industry as far as molecular modeling is concerned (Nicholls, 2010) and a cultural disparity between academic and industrial sectors, including basic versus applied research (Mowery et al. 2004; Washburn, 2005).
Taken together, it is now valuable to consider progress in the field of biomolecular modeling and simulation, which appears in a critical ‘coming-of-age ’ time, roughly 40 years after it started. How has modeling fulfilled initial expectations ? Has it become a field on its own right ? Are the technological advances paying off? How successful are expansions of the scope of the simulation subjects – molecular size, complexity and simulation time? How is modeling perceived in the experimental community, by scientists outside of academe, and by the general public ? What are future prospects for the field ?
Through specific examples, some of which were provided in response to a questionnaire (Appendix A), we sketch the field’s progress over the past few decades ; see Schlick et al. (1999) for an early assessment of algorithmic challenges in computational molecular biophysics, and recent perspectives regarding protein-folding advances (Dill et al. 2008), protein modeling’s impact on biomedical research (Schwede et al. 2009) and the usage of dynamics simulations to interpret single-molecule pulling experiments (Lee et al. 2009), for example.
Our examination of computing trends and specific examples of success suggest that biomolecular modeling and simulation is a vibrant field with demonstrated successes and immense potential to impact biology, medicine, and technology. Thus, despite unrealistic high expectations expressed in the late 1980s, technical advances in computer technology and algorithms have transitioned the field well onto a productive and exciting trajectory of being not only full partner with experiment but eventually a field on its own right.
In the next section, we provide a brief historical perspective of the field. Section 2 presents metrics of the field’s rise in popularity, as reflected by publication records and computer power progress. Section 3 collects statements concerning the field’s expectations, and Section 4 presents a proposed expectation curve for the field. Sections 5 and 6 discuss in turn examples of success and failure. Our summary in Section 7 is followed by general recommendations to accelerate the field’s progress. The appendix presents our community questionnaire, used to sample some opinions rather than represent the field exhaustively, and mentions some observations shared by respondents.
1.2 What is molecular modeling ?
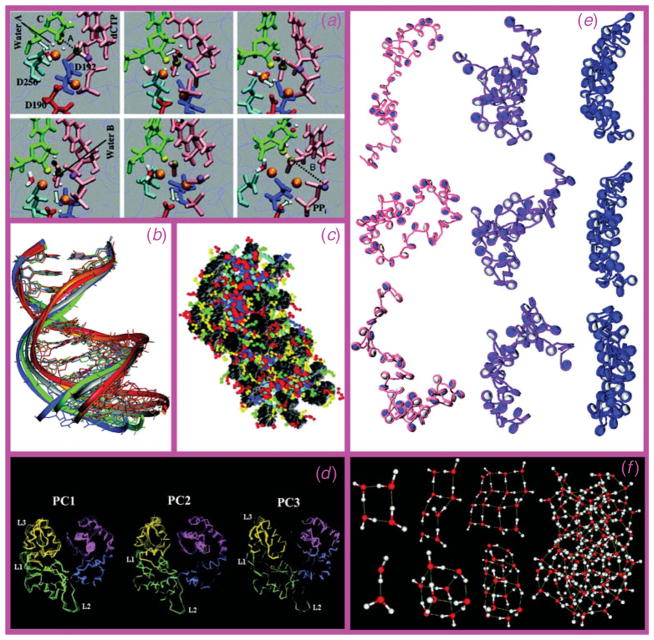
Molecular modeling is the science and art of studying molecular structure and function through model building and computation. The model building can be as simple as plastic templates or metal rods, or as sophisticated as interactive, animated color stereo-graphics and laser-made wooden sculptures. The computations encompass ab initio and semi-empirical quantum mechanics, empirical (molecular) mechanics, visualization, homology modeling, docking, molecular dynamics (MD), Monte Carlo, free energy and solvation methods, enhanced sampling and pathway methods, principal component analysis (PCA), structure/activity relationships (SAR), chemical/biochemical information and databases and many other established procedures and methods (see Fig. 1 for examples).
Fig. 1.
Examples of various modeling methods and applications. (a) QM/MM pathway (from top left to bottom right) of DNA repair enzyme polymerase β nucleotide incorporation (Radhakrishnan & Schlick, 2006); (b) superimposed MD configurations from a solvated dodecamer simulation ; (c) mesoscale oligonucleosome model with nucleosome cores in grey and tails and linker histones in color (see Arya & Schlick, 2009; Schlick, 2009a for details) ; (d) top three principal component motions in the thumb and finger subdomains of DNA polymerase β (Arora & Schlick, 2004); (e) representative Monte Carlo snapshots in the sampling of 48-unit oligonucleosomes at three different salt environments using the model shown in (c) ; and (f) minimized water clusters.
The questions being addressed by computational approaches today are as intriguing and as complex as the biological systems themselves. They range from understanding the equilibrium structure of a small biopolymer subunit to the energetics of hydrogen-bond formation in proteins and nucleic acids, binding affinities of ligands/drugs to their target, complex kinetics of protein folding and the complex functioning of a supramolecular aggregate. Indeed, given many experimental triumphs, modeling approaches are needed to pursue many fundamental questions concerning the biological motions and functions of complex systems like ion channels, signaling receptors, membrane transporters, ribosomes, nucleosomes and non-coding RNAs. Modeling can provide a way to systematically explore structural/dynamical/thermodynamic patterns, and test and develop hypotheses to help understand and extend basic laws that govern molecular structure, flexibility and function.
With improved modeling algorithms, better force fields, faster computers and experimental advances (e.g. single-molecule techniques and high-speed X-rays), modeling and simulation have expanded in both quality and scope. Problems and approaches that were insurmountable a few years ago are now possible.
Yet, a field’s maturity also implies that some current users may not be familiar with caveats and inherent approximations that field pioneers clearly recognized. Indeed, the readily available programs for simulation and visualization make usage much more facile but possibly also easier to abuse. Moreover, advanced programs and simulation methods like MD and folding are far from generally applicable automated procedures ; their application relies on user expertise as well as biological intuition.
1.3 Field roots
The field of biomolecular modeling is relatively young, having started in the 1960s, with pioneers like Lifson, Scheraga, Allinger, Levitt, Warshel and others. Building upon the simulation technique described in 1959 by Alder and Wainwright and applied to hard spheres (Alder & Wainwright, 1959), Rahman and Stillinger described in the early 1970s the first MD simulation of a polar molecule, liquid water (Rahman & Stillinger, 1971, 1974).
These developments led quickly to using molecular mechanics force fields with energy minimization as a tool for refinement of crystal structures in the late 1970s (Jack & Levitt, 1978; Konnert & Hendrickson, 1980), enzyme energetics (Warshel & Levitt, 1976) and later to MD approaches (McCammon et al. 1977) (see below) that excited practitioners.
It took a few more years, however, for the field to gain legitimacy, because the work of these computational chemistry pioneers could not be easily attributed to a specific discipline ; moreover, the notion of transferability of force field parameters was criticized by spectroscopists1.
When the first-generation biomolecular force fields were established, the rising power of supercomputers widened the possibilities for application. Most programs and force fields today, for both small and large molecules, are based on the works of the pioneers mentioned above and their co-workers, with the addition of water force fields developed in the late 1970s and early 1980s by Berendsen and co-workers (e.g. Ryckaert et al. 1977) and by Jorgensen and co-workers (Jorgensen et al. 1983) (SPC and TIP3P/TIP4P, respectively), concepts in protein electrostatics and enzyme/substrate complexes in solution by Warshel and colleagues (Warshel & Levitt, 1976; Warshel & Russell, 1984), and pioneering pharmaceutical applications by the late Peter Kollman (Wang et al. 2001).
1.4 Current simulation trends
Modern versions of these and other molecular simulation packages have led to competition for ‘ better, bigger, and faster ’ program design. Programs like NAMD (Phillips et al. 2005), Desmond (Bowers et al. 2006), or GROMACS (Berendsen et al. 1995; Lindahl et al. 2001) have focused on efficient multi-processor implementations. In addition, a specialized computer hard wired for long MD simulations, called Anton, has been launched with the explicit goal of generating long simulations (approaching the millisecond and beyond) (Shaw et al. 2007, 2009, 2010).
For a historical perspective (see details in Schlick, 2010), the first MD simulation of a biological process was for the small protein BPTI (bovine pancreatic trypsin inhibitor) in vacuum (McCammon et al. 1977). In 1998, a 1 μs simulation of a villin-headpiece (using periodic boundary conditions) made headlines (Duan & Kollman, 1998; Duan et al. 1998) as its duration was longer by three orders of magnitude than all prior simulations. Three years later, the longest simulation of 38 μs reflected aggregate dynamics by the ingenious folding@home distributed computing approach (Shirts & Pande, 2000) – many short trajectories performed on contributed processors worldwide to simulate the microsecond timescale – for the C-terminal β-hairpin from protein G (16 residues) (Zagrovic et al. 2001) ; see later works in (Ensign et al. 2007; Snow et al. 2002) extending the work up to hundreds of 1 μs simulations.
Modelers have continued to simulate larger biomolecular systems (e.g. entire satellite mosaic virus with one million atoms (Freddolino et al. 2006)) as well as longer time frames (e.g. B-DNA dodecamer (Pérez et al. 2007), ubiquitin (Maragakis et al. 2008), and β2 AR protein receptor (Dror et al. 2009) for over 1 μs, and small proteins for one millisecond (Shaw et al. 2010)) with specialized MD programs and dedicated supercomputers. In fact, these achievements suggest by simple extrapolation that 1-second simulations will be attainable by 2015 (Fig. 2f) ! At the same time, coarse-grained models and combinations of enhanced sampling methods are emerging as viable alternatives for simulating complex biomolecular systems (recently reviewed in (Earl & Deem, 2008; Klein & Shinoda, 2008; Lei & Duan, 2007; Liwo et al. 2008; Maisuradze et al. 2010; Schlick, 2009b, c)).
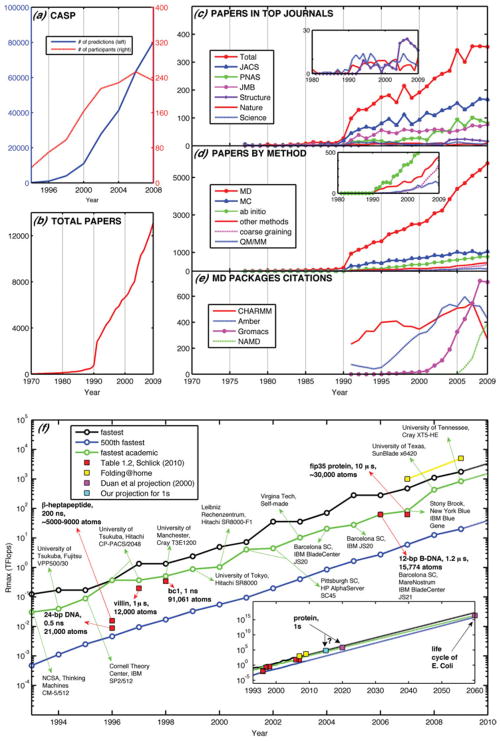
Fig. 2.
Metrics of the field’s rise in popularity and the evolution of computational performance. (a) CASP predictions (left-axis) and participants (right-axis) ; (b) modeling papers in peer-reviewed journals as found in the ISI Web of Science using the query words molecular dynamics, biomolecular simulation, molecular modeling, molecular simulation and/or biomolecular modeling; (c) papers from (b) appearing in high-impact-factor journals ; (d) papers from (b) decomposed by method; (e) citations to the original papers of four popular MD packages ; (f) computational systems ranked first, 500th, and highest- ranked academic facility assembled in the Top500 supercomputer lists (http://www.top500.org). The total speed for folding@home is also shown for 2007 and 2009. Some biomolecular modeling milestones are also connected to a specific computing system, assuming that the computations were performed about a year prior to publication, except for the two 1998 publications which we associate with computations started in 1996, for : 24-bp DNA system using NCSA SGI machines (Young & Beveridge, 1998), β-heptapeptide using SGI Power Challenge (Daura et al. 1998), villin using the Cray T3E900 (Duan & Kollman, 1998), bc1 membrane complex using the Cray T3E900 (Izrailev et al. 1999), B-DNA dodecamer using MareNostrum, Barcelona (Pérez et al. 2007) and fip35 protein ran on NCSA Dell clusters (Freddolino et al. 2008), with full details presented in Schlick (2010) ; also shown are future predictions (Duan et al. 2000) (see text).
Long simulations, however, open interesting questions regarding the adequacy of force fields as well as long-time stability and error propagation of the simulation algorithms. Although for some proteins, folding simulations can be very successful (e.g. Bowman et al. 2009; Day et al. 2010; Dror et al. 2009; Freddolino & Schulten, 2009; Kelley et al. 2009; Mittal & Best, 2010; Noé et al. 2009; Pitera et al. 2008; Voelz et al. 2010), long simulations can reveal inadequacies in force fields and therefore help advance force field development. For example, a 10 μs simulation of the β-protein Fip35 (Freddolino et al. 2008) did not provide the anticipated folded conformation nor the folding trajectory from the extended state, as expected from experimental measurements; it was determined in a subsequent work that force field inaccuracies for β-protein interactions affected the results and not incorrect sampling (Freddolino et al. 2009).
Overall, the sampling problem in MD remains a fundamental limitation of the technique’s general utility, including its usefulness in drug design applications.
2. Metrics of the field’s rise in popularity
As mentioned above, protein folding has attracted many to biomolecular modeling and simulation, as indicated by the rise of CASP (critical assessment of techniques for protein structure prediction) participants in Fig. 2a. To measure the rise in popularity of the field of biomolecular modeling, more generally, Fig. 2b shows the rise in the number of scientific publications since 1970 (see caption for key words searched). A notable increase can be seen with sharper linear increase since 2005.
The quality and relevance of the publications, however, speaks more for a field’s stature than sheer number alone. As an estimate of the change in relevance and quality of the publications in the field of biomolecular modeling, we quantify the number of papers in several high-impact journals in Fig. 2c. Again, we note a sharp rise in the number of biomolecular modeling papers around 1990. Since the selected journals are not specialized in modeling, this trend also implies that the field is gaining more attention from a wider community of scientists.
A decomposition of the papers measured in Fig. 2b according to the simulation technique can be seen in Fig. 2d. Clearly, MD overwhelms all other techniques, and it is followed by Monte Carlo simulations, ab initio quantum mechanics (QM), coarse graining, and quantum-mechanics/molecular-mechanics (QM/MM) methods. Indeed, several sophisticated MD programs and force field packages are now available ; Fig. 2e shows trends in citations to the original papers of popular MD programs (Amber (Pearlman et al. 1995; Wang et al. 2004), CHARMM (Brooks et al. 1983), GROMACS (Berendsen et al. 1995; Hess et al. 2008; Lindahl et al. 2001) and NAMD (Phillips et al. 2005)) ; the low number of 2009 citations likely represents a lag in the update of manuscript databases rather than a decrease in program usage. While the number of citations for Amber and CHARMM has remained steady in recent years, the number of citations for GROMACS and NAMD continues to rise, likely because the latter are especially suitable to parallelized computer architectures and present open-source environments.
Has the biology community utilized computer power well ? A dramatic increase in computational power over the past 20 years has certainly helped fuel the field, but how do the computers used compare with the fastest computer system available ?
Figure 2f shows that since 1993 the speed of the fastest computer system in the world has increased exponentially according to the Top500 list (http://www.top500.org), a biannual list of the 500 fastest non-distributed computers ranked by their performance on the LINPACK benchmark. From the Top500 list, the speed of the fastest computer systems used in academia is shown in Fig. 2f and listed in Table 1. We have also added the current total speed achieved by the folding@home project which has now passed the Petaflop mark. These trends of computer power follow an exponential growth. The figure also shows that many milestones in the field of biomolecular simulation –including the villin (Duan & Kollman, 1998) and bc1 membrane complex (Izrailev et al. 1999) on the Cray T3E900, B-DNA dodecamer on MareNostrum (Pérez et al. 2007), and fip35 protein on NCSA Dell clusters (Freddolino et al. 2008) – have been obtained using one of the fastest Top500 computers available.
Table 1.
Fastest academic computer systems as ranked in the November’s Top500 lists, http://www.top500.org. Rank represents the position of the fastest academic system in the Top500 list per year. The systems are ranked in terms of Rmax, which is an estimate of the maximum performance of a computer measured using a freely available implementation of the High Performance Computing LINPACK Benchmark
| Year | Rank | Institution | Computer | Rmax (GFlops/s) |
|---|---|---|---|---|
| 1993 | 4 | NCSA | Thinking Machines Corporation, CM-5/512 | 30·4 |
| 1994 | 6 | University of Tsukuba | Fujitsu, VPP500/30 | 39·8 |
| 1995 | 6 | Cornell Theory Center | IBM, SP2/512 | 88·4 |
| 1996 | 1 | Center for Computational Science University of Tsukuba | Hitachi, CP-PACS/2048 | 368·2 |
| 1997 | 4 | Center for Computational Science University of Tsukuba | Hitachi CP-PACS/2048 | 368·2 |
| 1998 | 7 | CSAR at the University of Manchester | Cray Inc., T3E1200 | 509 |
| 1999 | 5 | University of Tokyo | Hitachi, SR8000/128 | 873 |
| 2000 | 7 | Leibniz Rechenzentrum | Hitachi, SR8000-F1/112 | 1035 |
| 2001 | 2 | Pittsburgh Supercomputing Center | Hewlett-Packard, AlphaServer SC45 1 GHz | 4059 |
| 2002 | 6 | Pittsburgh Supercomputing Center | Hewlett-Packard AlphaServer SC45, 1 GHz | 4463 |
| 2003 | 3 | Virginia Tech | Self-made 1100 Dual 2·0 GHz Apple G5/Mellanox Infiniband 4X/Cisco GigE | 10 280 |
| 2004 | 4 | Barcelona Supercomputing Center | IBM BladeCenter JS20 (PowerPC970 2·2 GHz) | 20 530 |
| 2005 | 8 | Barcelona Supercomputing Center | IBM JS20 Cluster, PPC 970, 2·2 GHz, Myrinet | 27 910 |
| 2006 | 5 | Barcelona Supercomputing Center | IBM BladeCenter JS21 Cluster, PPC 970, 2·3 GHz, Myrinet | 62 630 |
| 2007 | 10 | Stony Brook/BNL, New York Center for Computational Sciences | IBM eServer Blue Gene Solution | 82 161 |
| 2008 | 6 | Texas Advanced Computing Center University of Texas | Sun Microsystems SunBlade r6420, Opteron QC 233 GHz, Infiniband | 433 200 |
| 2009 | 3 | National Institute for Computational Sciences, University of Tennessee | Cray Inc. Cray XT5-HE Opteron Six Core 2·6 GHz | 831 700 |
More generally, this publication volume and computation perspective is optimistic. The trends suggest that biomolecular researchers have utilized well and will continue to have access to ever more powerful machines that in turn push the frontiers of the field.
3. Field expectations
As the field of biomolecular modeling gained momentum in the late 1980s with the increasing availability of high-speed computers, introduction of standardized programs, and significant technological innovations, the notable application and algorithmic (for minimization, integration, and data analysis) successes that followed stirred a new enthusiasm in the molecular biophysics community and beyond. Biomolecular modeling and simulation, in addition to high-throughput structure technology and other experimental information, was expected to revolutionize many applications through systematic structure-to-function links.
As early as 1986, the computational quantum chemistry pioneer Henry Schafer wrote (Schaefer, 1986) :
It is clear that theoretical chemistry has entered a new stage … with the goal of being no less than full partner with experiment.
Commenting in 1988 on two exciting MD applications to materials and liquids, the late Sir John Maddox, former editor of Nature, wrote (Maddox, 1989):
Molecular dynamics is clearly well on the way to being a universal tool, as if it were the differential calculus. So much is plain from the range of problems now being tackled, and sometimes at least partially solved, by the application of this technique. Gone, it seems, are the days when people’s ambitions in the field were restricted to the calculation of the properties of smallish molecules, with only distant hopes of being able to tackle the problems of, say, protein molecules. Now, molecular dynamics is being used to tackle problems which are quite general.
A decade later, as technological advances in high-throughput technology provided biological data as never before, it was believed that computers would allow us to quickly sift through all these data and transform it into relevant biological information. The New York Times reporter Andrew Pollack wrote in 1998 (Pollack, 1998):
In a marriage of biotech and high tech, computers are beginning to transform the way drugs are developed, from the earliest stage of drug discovery to the late stage of testing the drugs in people.
Similarly, Stu Borman, reporter for Chemical & Engineering News, wrote in connection to drug design using combinatorial chemistry approaches in 1998 (Borman, 1998):
Recent advances in solid-phase synthesis, informatics, and high-throughput screening suggest combinatorial chemistry is coming of age.
Commenting on the two 1998 Chemistry Nobel Prize awardees in quantum chemistry – Walter Kohn of the University of California, Santa Barbara and John Pople of Northwestern University who ‘have developed computational methods to crunch the numbers involved in quantum theory in ways that allow chemists to make sense of the behavior of atoms as they bond together to form molecules ’ – a reporter in The Economist went as far to suggest that experimentalists will soon face competition (Economist, 1998):
In the real world, this could eventually mean that most chemical experiments are conducted inside the silicon of chips instead of in the glassware of laboratories. Turn off that Bunsen burner; it will not be wanted these ten years.
Have these high expectations been met? While it remains to be seen how effective in-silico biology will be over the long run, it appears that those early expressed hopes for quick break-throughs have not been realized consistently. Or, perhaps, expectations were simply too high for this young field.
In response to our questionnaire question regarding fulfillment of initial expectations (Appendix A), computer scientist Jacques Cohen writes:
These are very difficult modeling problems and nature has to be considered as the ultimate referee.
Structural biologist and crystallographer Stephen Neidle put it best:
My expectations have been conservative, and the reality has thus not disappointed me.
For the subfield of computer-aided drug design (CADD), a 2007 perspective (Drie, 2007) suggests that only since 2000 have structure-based design and virtual screening technologies begun to pay off in a realistic way; still, the future is considered bright, with new avenues for CADD – like new drug target classes, novel drug mechanisms, multiple drug targets and automated technologies – expected to bear fruit.
In their 2005 review of MD and protein function, Karplus and Kuriyan state that, at the very least, simulations have reached the stage that they can be critically evaluated by experimentalists (Karplus & Kuriyan, 2005):
The combination of increased computer power and improved potential functions has resulted in an ability to generate simulations that approach the point at which they can survive critical examination by the experimentalists who determine the structures of the proteins being simulated.
They continue to write that statistically meaningful folding simulations of proteins with all-atom models are not yet possible but that it is likely that within the next decade (i.e. by around 2015) folding mechanisms and folded structures might be simulated directly from sequence by MD. Indeed, recent dynamics simulations of protein folding have already been impressive using innovative conformational sampling approaches like Markov state models (Noé et al. 2007, 2009; Voelz et al. 2010), network models (Berezhkovskii et al. 2009), or replica-exchange MD (Day et al. 2010; Garcia & Pascheck, 2008; Kelley et al. 2009; Mittal & Best, 2010; Ozkan et al. 2007; Pitera et al. 2008).
In 2000, Duan et al. projected that, in 20 years, simulating a second in the lifetime of medium-sized proteins will be possible, as well as in 50–60 years, the entire life cycle of an Escherichia coli cell ; these projections assume a computational power increase of one order of magnitude every 3–4 years (Duan et al. 2000). From Fig. 2, this computational increase has been more than met, but the issues of force fields and sampling have not been taken into account in setting those goals.
Folding simulations have of course received substantial attention. Folding proteins by 2005 was the stated goal of the IBM Blue Gene over a decade ago. When a new computer game called Foldit based on Rosetta@home software was launched for the general public, a reporter in the local newspaper ScienceDaily enticed readers with the headline (ScienceDaily, 2008):
Computer Game’s High Score Could Earn The Nobel Prize in Medicine.
Foldit has since showed great promise, because some folding tasks are better handled visually than automatically (Cooper et al. 2010). More generally, a rise in the community’s participation in CASP predictions shows the rising interest in protein folding (Fig. 2a) : since CASP1 in 1994 where 35 research groups participated to predict 33 target proteins, CASP8 in 2008 has attracted 233 participant groups to predict 121 target proteins.
Medical applications remain the most prominent driving force for modeling. Current NIH Director Francis S. Collins recently wrote (Collins, 2010):
The power of the molecular approach to health and disease has steadily gained momentum over the past several decades and is now poised to catalyze a revolution in medicine.
Modeling and simulation is certainly playing an important role in this new era.
4. Field evolution assessed via an expectation curve
In reference to the unanticipated challenges that we face today concerning unveiling the genetic basis for disease, NIH Director Francis S. Collins also stated (Borman, 2010):
It’s been said that a truly transformational technology will always have its immediate consequences overestimated and its long-term consequences underestimated.
Based on the data collected here, this certainly appears to be true for the field of biomolecular modeling and simulation.
In 1993, computer scientist James Bezdek introduced the general notion of a technology expectation (or hype) curve in his first editorial of the new journal he founded on fuzzy models, a branch of mathematics that attempts to formulate the notion of vagueness (Bezdek, 1993). Bezdek’s technology development framework was later popularized by the Gartner group. This expectation curve describes the evolution of a new technology or field by several common stages in terms of expectation levels : (1) a technology trigger that serves as the starting point for the field ; (2) an early high peak corresponding to initial euphoria and inflated expectations when a frenzy of publicity envelopes some isolated successes ; (3) a trough of disillusionment or depth of cynicism, when failures become apparent, obstacles appear, and general support and interest wane; (4) recovery phase, where practitioners continue to develop and refine the technique or technology ; and (5) steady progress of productivity when realistic expectations and true benefits emerge. Not all new technologies reach the final stage ; some new approaches fade away after the cynicism stage.
Our version of the expectation curve for biomolecular modeling and simulation is presented in Fig. 3. Based on the quotes above and general information discussed here, the starting point for the field is the 1970s, when force field pioneers began reporting their applications. Rising expectations continued (especially from the late 1980s) until the late 1990s, where the height of naive euphoria for biomolecular modeling occurred (stage 2). Structure-based rational drug design was then well recognized and expected to replace older, less efficient and laborious ways of screening. Some disappointments became apparent as this 21st century began: pharmaceutical industries have expended huge resources on drug design modeling initiatives that only led to modest successes at best (Munos, 2009), and the revolutionary transformations expected following completion of the Human Genome Project in our understanding of ancestry and human disease diagnosis and treatment did not bear fruit right away as our appreciation of the enormous complexity of biology set in (Hayden, 2010). While the new technologies helped generate voluminous amounts of data, the complexity of information storage involved (in genes, regulation, etc.) has also escalated into a picture of intertwined complex layers of regulatory networks. This problem is also manifested in the recent declining production of new drug entities (Munos, 2009) (further discussed in Schlick, 2010).
Fig. 3.
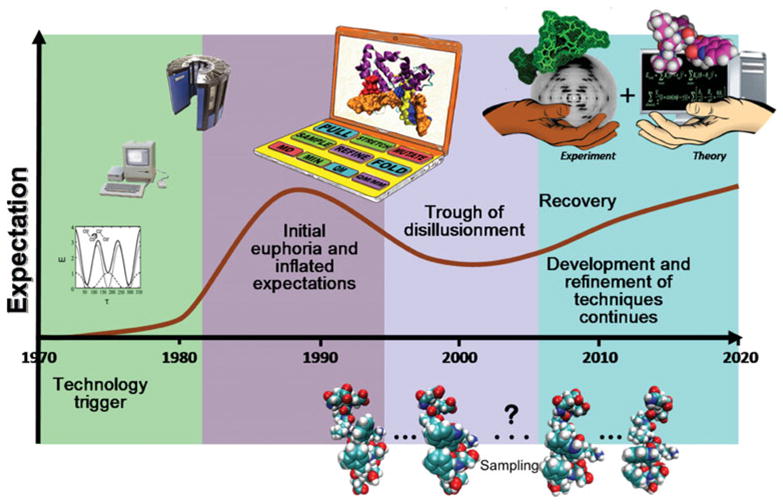
Proposed expectation curve for the field of biomolecular modeling and simulation, with approximate timeline. The field started when comprehensive molecular mechanics efforts started, and it took off with the increasing availability of fast workstations and later supercomputers. Following unrealistically high expectations and disappointments, the field is expected to make more realistic progress so that eventually theory and experiment will be hand-in-hand partners.
Although it is difficult to pinpoint exactly where the trough in the expectation curve (stage 3) lies for the field, we suggest that it may have occurred roughly one decade ago. Our optimism also suggests that the productivity phase is well underway, following the appreciation for the field’s great potential to help understand biological systems, despite the apparently complex intertwined regulatory networks and the realization that force fields and sampling algorithms require improvement.
5. Modeling and simulation successes
Many notable successes in various areas – from experimental interpretation to structure prediction to drug design – have been reported for biomolecular modeling, and it is instructive to examine them. Our examples include MD theory and applications, algorithms for biomolecular simulation, structure prediction, protein-folding theory, modeling-aided drug design, and interpretation of complex experimental phenomena.
5.1 MD as a technique to study atomic motions
A spectacular example of success reflecting a modern marriage among modeling theory, technology and biology involves the theory and practice of MD. Essentially, MD is statistical mechanics by numbers, or Laplace’s vision of Newtonian physics (de Laplace, 1820) on modern supercomputers. The impressive progress in the development of biomolecular force fields, coupled to spectacular computer technology advances, has now made it possible to transform this vision into a reality, by overcoming the difficulty noted by Dirac of solving the equations of motion for multi-body systems (Dirac, 1929).
Since successful applications were reported in the 1970s in protein dynamics (McCammon et al. 1977), MD – with variations and extensions – has now become a popular (Fig. 2d) and universal tool, ‘as if it were the differential calculus ’ (Maddox, 1989). MD is in fact one of the few tools available, by both experiment and theory, to probe molecular motion on the atomic scale. By following the equations of motion as dictated by a classic molecular mechanics force field, complex relationships among biomolecular structure, flexibility, and function can be investigated.
Today’s sophisticated dynamics programs, like NAMD or GROMACS, adapted to parallel and massively parallel computer architectures, and specialized hardware have made simulations of biomolecular systems in the microseconds feasible in several weeks of computing. Anton’s hardware/software co-design is pushing the envelope to the millisecond timeframe (Shaw et al. 2009, 2010). Although the well-recognized limitations of sampling in atomistic dynamics, as well as in the governing force fields, have led to many innovative sampling alternatives to enhance coverage of the thermally accessible conformational space (as recently reviewed in Earl & Deem, 2008; Klein & Shinoda, 2008; Lei & Duan, 2007; Liwo et al. 2008; Schlick, 2009b, c), many still rely on MD for local sampling.
Overall, MD simulations have been used for numerous applications, including to refine experimental structures, extending most recently to low-resolution crystal data (Schröder et al. 2010), interpret various experimental data such as single-molecule force-extension curves (e.g. Lee et al. 2009) or NMR spin-relaxation in proteins (e.g. Case, 2002; Henzler-Wildman et al. 2007; Tsui et al. 2000), improve structure-based function predictions, for example, by predicting calcium-binding sites (Altman et al. 2009), link static experimental structures to implied pathways (e.g. Golosov et al. 2010; Radhakrishnan & Schlick, 2004), estimate the importance of quantum effects in lowering free-energy barriers of biomolecular reactions (Hu et al. 2003), make structural predictions, deduce reaction mechanisms, propose free-energy pathways and associated mechanisms (e.g. Faraldo-Gomez & Roux, 2007; Radhakrishnan & Schlick, 2005), resolve or shed light on experimental ambiguities (see Subsection 5.6), and design new folds and compounds, including drugs and enzymes (e.g. Baker et al. 2003; Hornak & Simmerling, 2007; Jiang et al. 2008; Neidle et al. 2001). Challenging applications to complex systems like membranes, to probe associated structures, motions and interactions (e.g. Grossfield et al. 2008; Khelashvili et al. 2009; Vasquez et al. 2008) are also noteworthy. Specific success stories from such MD simulations are discussed separately in the subsections below.
5.2 Establishment of reliable algorithms
Rigorous algorithm development and analysis provide a clear framework by which to define the important notions of accuracy, stability and error control. This understanding in turn provides practical guides to parameter selection and trajectory analysis. Three notable areas of algorithmic contributions in recent years include methods for MD integration, evaluation of non-bonded electrostatic interactions and enhanced sampling methods for biomolecules.
The establishment of symplectic integrators such as leap frog, velocity Verlet, and constrained dynamics formulations (e.g. Leimkuhler & Reich, 2004; Schlick, 2010) has allowed researchers to correctly generate MD trajectories and analyze the stability of a simulation in terms of energy conservation, and the robustness of the simulation with respect to the timestep size.
Highlighting resonance artifacts in MD simulations (Mandziuk & Schlick, 1995; Schlick et al. 1998), predicting resonant timesteps, and establishing stochastic solution to resonances (Barth & Schlick, 1998; Schlick et al. 1997; Sweet et al. 2008) have all led to an improved understanding and quality of MD simulations (Morrone et al. 2010).
The advent of efficient and straightforward-to-program particle mesh Ewald (PME) (Darden et al. 1993; Essmann et al. 1995; York & Yang, 1994) and related methods (Duan & Krasny, 2000; Greengard & Rokhlin, 1987; Saito, 1992; Saito, 1997; Sandak, 2001; Skeel et al. 2002) for evaluation of the long-range electrostatic interactions, which constitute the most time-consuming part of a biomolecular simulation, has made possible more realistic MD simulations without non-bonded cutoffs. A problem that in part remains unsolved in their implementation involves the optimal integration of PME methods with multiple-timestep methods and parallelization of PME implementations. The presence of fast terms in the reciprocal Ewald component limits the outer timestep and hence the speedup (Morrone et al. 2010; Procacci et al. 1998; Qian & Schlick, 2002; Stuart et al. 1996; Zhou et al. 2001). Moreover, memory requirements create a bottleneck in typical PME implementations in MD simulations longer than a microsecond. This is because the contribution of the long-range electrostatic forces imposes a global data dependency on all the system charges ; in practice, this implies communication problems (Fitch et al. 2006). Thus, much work goes into optimizing associated mesh sizes, precision, and sizes of the real and inverse spaces to delay the communication bottleneck as possible (e.g. Shaw et al. 2009), but overall errors in long simulations are far from trivial (Morrone et al. 2010; Snir, 2004).
In addition to MD integration and electrostatic calculations, sampling the vast configurational space has also triggered many innovative approaches to capture ‘ rare events ’. The many innovative enhanced sampling methods are either independent of MD or based on MD. In the former class, as recently surveyed (Earl & Deem, 2008; Liwo et al. 2008; Schlick, 2009b), are various Monte Carlo approaches, harmonic approximations, and coarse-grained models. These can yield valuable conformational insights into biomolecular structure and flexibility, despite altered kinetics. Although Monte Carlo methods are not always satisfactory for large systems on their own right, they form essential components of more sophisticated methods (e.g. transition path sampling (Dellago & Bolhuis, 2007) or Markov chain Monte Carlo sampling (Pan & Roux, 2008)), as recently surveyed (Schlick, 2009c).
More generally, MD-based methods for enhanced sampling of biomolecules can involve modification of the potential (like accelerated MD or AMD (Grant et al. 2010; Hamelberg et al. 2004), simulation protocol (like replica-exchange MD or REMD (Sugita & Okamoto, 1999)), or algorithm; as well as global reformulations, such as transition path sampling (Bolhuis et al. 2002; Dellago & Bolhuis, 2007), forward flux simulation (Borrero & Escobedo, 2008), and Markov state models (Noé & Fischer, 2008). Simple modifications like AMD can aid in the interpretation of residual dipolar coupling and chemical shift measurements in NMR (Cervantes et al. 2009; Markwick et al. 2009, 2010). Global formulations, however, are needed more generally not only to generate more configurations or to suggest mechanistic pathways but also to compute free-energy profiles for the reaction and detailed kinetics including reaction rates.
Divide and conquer methods for sampling and for piecing together reaction rate information are especially suitable for readily available computer cluster networks. REMD is a popular method, with many extensions (e.g. Roitberg et al. 2007), but implementations for large systems are problematic due the need for many replicas and the non-uniform sampling. There are many successful reports of using tailored enhanced sampling methods for biomolecular applications (e.g. Abrams & Vanden-Eijnden, 2010; Berezhkovskii et al. 2009; Chennamsetty et al. 2009; Noé et al. 2007, 2009; Ozkan et al. 2007; Radhakrishnan & Schlick, 2004; Voelz et al. 2010), but applications at large to biomolecules and/or in the absence of experimental data remain a challenge.
Among the numerous other examples of algorithms, PCA, clustering techniques and simple consistency tests have helped to analyze complex groups of experimental data. For example, clustering graphs of RNA secondary structures led to a proposal of novel RNA motifs that are RNA-like (i.e. physical) and non-physical (Kim et al. 2004a, b) ; some in the former class have been designed computationally and also later experimentally confirmed (Kim et al. 2010; Schlick, 2010). Interestingly, a simple tool to analyze PDB structures in terms of quantifying the interior protein volume called RosettaHoles (Sheffler & Baker, 2008) detected a group of significant outliers, some of which were traced to a single author ; this eventually led to the retraction of structures (Borrell, 2009).
Thus, algorithms are playing an increasing role in a wide range of problems dealing with biomolecular structure, thermodynamics, and dynamics, as well as design.
5.3 Structure predictions that preceded experiment
Although the general problem of structure prediction is far from solved, predicting a peptide structure has been more amenable to present modeling, and there have also been notable success stories for larger systems, as early as the late 1980s, for human immunodeficiency virus (HIV) protease (PR), and for complex RNAs, by sequence alignment. The successes for RNA are especially noteworthy since far fewer structures of RNAs are available at atomic resolution and the field is younger than protein prediction. Some interesting early prediction cases are collected below, following general trends for peptides and proteins.
5.3.1 HIV-1 PR
HIV-1 PR is an aspartic PR essential for the replication of the HIV. Already in 1987, Pearl & Taylor attempted to predict the HIV-1 PR tertiary structure by computational modeling alone (Fig. 4a) (Pearl & Taylor, 1987). Their approach involved multiple sequence alignment to predict a secondary structure followed by a knowledge-based method that matches fragments from large related structures to produce a tertiary structure model.
Fig. 4.
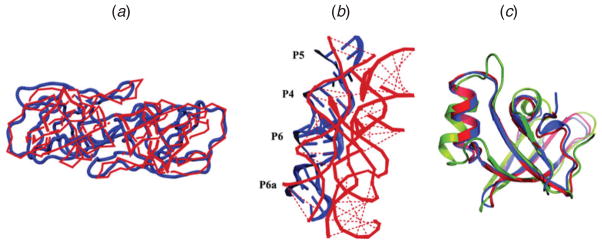
Examples of folding predictions that preceded experiment. (a) HIV PR: tertiary structure of the homodimer (red), predicted by homology modeling (Pearl & Taylor, 1987), superimposed with the crystal structure 3HVP (blue), 198 residues shown (Wlodawer et al. 1989); (b) group-I intron : P4–P6 domain (red), predicted by a comparative modeling (Michel & Westhof, 1990), aligned with crystal structure 1GID (blue), 38 residues (Cate et al. 1996); (c) CASP protein prediction by the Baker group for target T0492 (Raman et al. 2009), kindly provided by Srivatsan Raman and David Baker; crystal structure, 73 residues (blue) of this all-β protein (SH3-like barrel fold) is shown against the best detectable template found by comparative modeling (green) and the predicted structure (red) built using Rosetta using the best template found.
Two years later, another theoretical model was produced (Weber et al. 1989) using related information from the Rous sarcoma virus (RSV) determined at 3 Å resolution (Miller et al. 1989). In the same year, the HIV-1 PR structure was resolved by X-ray crystallography at 3 Å resolution (Navia et al. 1989). Interestingly, the authors noted a local structural discrepancy with respect to the predicted models (Pearl & Taylor, 1987; Weber et al. 1989) in the topology of the N- and C-terminal regions and the helix near the C-terminal ; some of these residues at the N-terminal were not observed in the electron density map, and so inherent disorder and flexibility were presented as possible explanations for these differences. However, when another structure of HIV-1 PR at 2·8 Å resolution was solved soon after, residues not captured in the first crystal were explicitly resolved (Wlodawer et al. 1989), and subsequent analysis showed excellent agreement with the modeling predictions rather than the previous crystal. The studies of HIV-1 PR initiated by the theoretical models and confirmed later by the crystallography eventually led to development, largely due to molecular modeling, of the first PR inhibitor, saquinavir, which was approved by FDA in 1995, and nine other PR inhibitors approved by 2006.
5.3.2 Group I intron
The group I intron is a self-splicing ribozyme present in a wide range of pathogenic organisms. The conserved core of a tertiary structure, where two parallel coaxial stacking of helices were juxtaposed to form an active site, was first modeled in 1990 by Michel & Westhof using comparative sequence analysis (Michel & Westhof, 1990). Specifically, they aligned the 87 available sequences of group I introns and derived some tertiary contacts from a distance co-variation analysis. Using stereochemical modeling, a tertiary structure of the conserved core of group I introns was then predicted. It took six additional years until the predicted model was confirmed by crystallography of the catalytic core structure (P4–P6 domain) at 2·8 Å resolution (Cate et al. 1996) (Fig. 4b). Since the self-splicing function of group I introns from ribosomal RNA genes is essential for maturation of ribosome, inhibition of self-splicing is a therapeutic goal. Indeed, various subsequent RNA–ligand interaction studies have led to the design of inhibitors for RNA function (Labuda et al. 2009).
5.3.3 Peptide structure prediction
Unlike many proteins, peptides can adopt a variety of conformations in solution depending on the conditions and presence of ligands, and thus there has been great interest in design of stable peptides, such as the synthetic β-peptides composed of β amino acids. Already in 1998, reversible β-peptide folding in solution in atomic detail was demonstrated (Daura et al. 1998), in agreement with NMR data (Daura et al. 1999). Other interesting examples include an engineered 20-residue Trp-cage peptide with a novel fold and fast folding kinetics (Neidigh et al. 2002), Amber-based folding of the Trp-cage peptide at 1 Å resolution (Simmerling et al. 2002) and mini-protein simulations using folding@home whose predicted folding rates agree well with experimental measurements of mean folding times and equilibrium constants by circular dichroisms and fluorescence (Snow et al. 2002). Some successful programs for peptide prediction include PEPstr which uses clever initial modeling followed by Amber-based energy minimization (Kaur et al. 2007), PepLook based on a Monte-Carlo refinement procedure in the peptide conformational space sampled systematically (Thomas et al. 2009) and Robetta (a descendant of Rosetta) based on homology modeling and knowledge-based potentials (Kim et al. 2004a).
5.3.4 Protein structure prediction: lessons from CASP
The high-profile community-wide exercise for protein structure prediction, CASP, held biannually since 1994, has sharpened our understanding of what works and what fails in current protein structure prediction. The exercise consists of released protein sequence targets, soon to be resolved by X-ray crystallography or NMR spectroscopy, serving as prediction targets in various categories : template-based or template-free modeling for tertiary structure prediction, substructure prediction for high-resolution models, disordered protein-region identification, function prediction, and more; predictions are then collected and assessed by independent experts who meet together with the predictor teams to discuss the work. The growth of participants in CASP (Fig. 2a) has also led to significant computational advances in structure determination and has identified select groups whose methods were consistently found to score high. For example, Baker’s group has been singled out as a good performer using template-based predictions (assembly of short fragments of known proteins using a Monte Carlo sampling procedure followed by all-atom refinement) with the Rosetta program; in CASP8, prediction of accuracy up to 1–2 Å resolution were demonstrated (Raman et al. 2009) (e.g. Fig. 4c).
More generally, comparative modeling (template based) and knowledge-based approaches (using statistical potentials based on analysis of known protein structures) have been found to produce reasonably good structural models, with notably more accurate predictions becoming possible. Predicting regions that are substantially different from the target appear far more difficult.
This is somewhat disappointing to devotees of ab initio folding, relying on physics-based force fields, a technique that has shown some, but more modest, progress (see next section). Still, template-free approaches based on statistical instead of molecular mechanics potentials have also emerged as competitive. Nonetheless, force-field-based approaches are essential for interpreting protein interactions and energies, such as required for drug design, and also for later stages of refinement, following template and homology modeling.
Ultimately, modeling work in this field is invaluable because it teaches us to ask, and seek answers to, systematic questions about sequence/structure/function relationships and about the underlying forces that stabilize biomolecular structure.
5.4 Protein-folding theory
Besides the folded protein structures discussed above, other parts of the protein-folding puzzle that have intrigued experimentalists and theoreticians alike are understanding the associated protein-folding code (thermodynamics) and pathways (kinetics) (Dill et al. 2008). A statistical– mechanics framework based on the density of states has been pioneered by Frauenfelder, Wolynes, Dill, Onuchic, Thirumalai and others for answering these questions (e.g. Dill et al. 2008; Frauenfelder et al. 1991; Wolynes, 2005) by analyzing energy landscapes of proteins on the basis of various experimental studies of proteins (fluorescence spectroscopy, NMR, single-molecule experiments, fast-laser temperature jumps, mutational studies, etc.). Essentially, the density of states arranged in the funnel landscape describes the conformational heterogeneity of the protein. Such free-energy landscapes can help interpret conformational substates and folding mechanisms and thereby relate protein dynamics, folding kinetics, and function. For example, fast protein folders must navigate through mostly local contacts in smoother free-energy landscapes, while slow-folding kinetics can be attributed to rugged energy landscapes, with more global contacts and more conformational traps and barriers. Standard chemical kinetics and transition state theory in combination with simplified models have also been important in analyzing folding events (e.g. Berezhkovskii et al. 2009; Best & Hummer, 2006; Ozkan et al. 2007; Yang & Grueble, 2003).
By relating folding kinetics to mutation profiles, mutation experiments have also helped further validate the utility of such theoretical frameworks. For example, recent folding kinetics experiments on mutants of α-spectrin, a protein of the intracellular matrix of red blood cells important for membrane elasticity (Wensley et al. 2010), have confirmed the notion that slow kinetics is correlated to rugged landscapes. By swapping domains through chimeric constructs for protein domains of α-spectrin, the reasons for disparate folding rates (by three orders of magnitude) of the protein relative to two homologues were traced to internal friction resulting from residue-specific interactions that can lead to misdocking of helices. For this membrane protein, slow unfolding kinetics can be advantageous because fewer rearrangements during the cell’s lifetime, and hence fewer possible degradations, result.
That theory contributes significantly to experiment was also echoed by Fersht and colleagues in a separate work on a three-helix bundle protein and various mutants which showed via simulation that folding intermediates are on the folding pathway (Mayor et al. 2003):
… combining molecular dynamics and experiment has now allowed us to characterize all of the necessary structures along the pathway. … Simulation could well be the answer in general for solving such problems in mechanism.
Numerous other general areas of theory, such as protein electrostatics and enzymatic reactions (Kamerlin et al. 2009; Warshel et al. 2006), conformational transitions in DNA polymerases in relation to fidelity mechanisms (Bebenek et al. 2008; Foley & Schlick, 2009; Golosov et al. 2010; Radhakrishnan et al. 2006), and interpretation of NMR signals (Case, 2002; Henzler-Wildman et al. 2007; Markwick et al. 2010; Tsui et al. 2000) have also benefited from simplified theoretical frameworks and models developed to interpret complex biological phenomena.
5.5 Modeling-aided drug discovery and design
Modeling molecular structures and dynamics has already helped to define molecular specificity and clarify functional aspects that are important for drug development (see also Schwede et al. 2009). Although modeling suggestions are not always verified by experiment, they help make important suggestions and offer general leads.
Clearly, as biological structures and functions are being described, natural disease targets that affect the course of disease can be proposed. Examples include specific regions of HIV enzymes or misfolded protein fibrils known as amyloids (for related misfolding disorders like Alzheimer’s and Creutzfeltdt–Jakob disease). Modeling work is important for pinpointing specific targets in these regions from structural, stability, and flexibility considerations and also testing resulting interactions (as reviewed in Ghosh et al. 2006; Zhou, 2008). For example, the sequence-specific triggers for amyloid formation, as recently uncovered (Goldschmidt et al. 2010) – self-complementary segments that can stick together – suggest capping alterations to slow down their formation or small molecules to act as folding chaperones (Schnabel, 2010). Supplementing these structure–function drug design paradigms is a systems-biology approach that attempts to modify response of genes, proteins and metabolites by integrating organ- and system-level modeling (Csermely et al. 2005; Kitano, 2007).
The solved HIV enzymes (PR, integrase, and reverse transcriptase) led to the development of many inhibitors that are now in clinical usage, some of which were aided by computer simulations. For example, besides the structural determination mentioned above for HIV PR, MD simulations using various HIV PR models have suggested that a fully open conformation of the PR ‘flaps’ may be favorable for drug access to the active site (Collins et al. 1995; Hamelberg & McCammon, 2005; Hornak et al. 2006; Tozzini & McCammon, 2005). Recent simulations have also led to design proposals (Scott & Schiffer, 2000) and other insights into PR/drug interactions (Hornak & Simmerling, 2007).
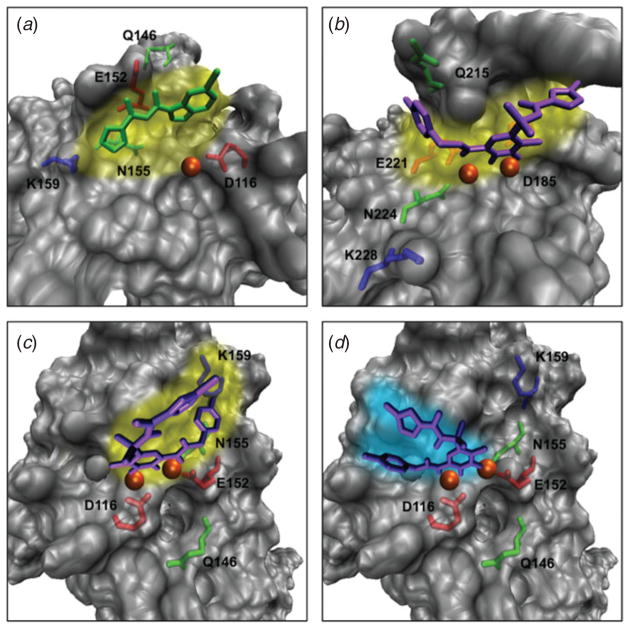
The solved complex of HIV integrase with an inhibitor in 1999 (Goldgur et al. 1999) attracted considerable attention since it provided a general platform for the drug design of another class of HIV inhibitors besides PR inhibitors. MD simulations have suggested that inhibitors could bind in more than one orientation to HIV-1 integrase, preferred and flipped (Perryman et al. 2010; Schames et al. 2004), as shown in Fig. 5, which also shows an X-ray structure of an inhibitor bound to an evolutionary similar virus (Hare et al. 2010). Simulations also showed that binding modes can be selected to exploit stronger interactions in specific regions and orientations (Hazuda et al. 2004; Lin et al. 2002; Schames et al. 2004) and that different divalent-ion arrangements are associated with these binding sites and fluctuations (Perryman et al. 2010). Thus, modeling can help outline new possibilities that stimulate further modeling and experimentation. More generally, emerging experimental findings combined with modeling are expected to aid the development of antiretroviral drugs.
Fig. 5.
Drug/inhibitor-binding pockets to HIV integrase as solved experimentally and as predicted by modeling. (a) Preferred (or primary) binding (yellow shading) of the inhibitor 5-CITEP (green) to the active site of the HIV-1 integrase core domain as determined by crystallography (Goldgur et al. 1999); (b) preferred binding (yellow shading) of the anti-HIV drug Raltegravir (purple) to the active site of the prototype foamy virus (PFV) integrase, a structural homologue of HIV-1 integrase, as determined by crystallography (Hare et al. 2010); (c) predicted preferred binding (yellow shading) for Raltegravir to the HIV-1 integrase catalytic core domain as determined by modeling (Perryman et al. 2010); (d) alternative, or flipped binding (turquoise shading) for Raltegravir to the HIV-1 integrase catalytic core domain as determined by modeling (Perryman et al. 2010). C and D are based on PDB entry 1QS4, with missing residues in panel (A) reconstructed by homology. The magnesium ions are shown as orange spheres.
Other examples of drug successes based on molecular modeling include the design of potent thrombin inhibitors to treat a variety of blood coagulation and clotting-related diseases by tailoring ligands to the enzyme pocket (Brady et al. 1998), the SARS virus inhibitor (Dooley et al. 2006), glutamate nanosensors to monitor neurologic functions (Okumoto et al. 2005), agonists to treat anxiety and depression (Becker et al. 2006), and a migraine treatment (Boyd, 1998).
While both the monetary cost and development time required for each successful drug remains very high (Amir-Aslani, 2008; Munos, 2009), and great successes are now few and far between, it is clear that molecular modeling will be important for the future of drug discovery and for many other design applications in biomedicine and nanotechnology. What is not obvious, however, is what type of modeling tool will prove to be most useful. This is because the usage of MD simulations by pharmaceutical scientists appears to be minimal (Nicholls, 2010), compared to tools from cheminformatics, ligand similarity searches and visualization, which are heavily used, and quantitative structure-activity relationships (QSAR), docking, homology modeling, bioinformatics and quantum mechanics, to a lesser extent. Limitations of force fields and sampling are most often quoted as reasons for the curbed application of dynamics to real-life problems.
The emergence of another class of successful drugs distinct from the traditional small-molecule drugs, namely biologics – biomolecules derived from living cells – might also increase the role of modeling in pharmaceutical discovery. This is because understanding the complex biomolecular function is important to this alternative approach, which also relies on a systems biology viewpoint. Overall, it is becoming clear that integrated advances on several fronts – small-molecule drug design, biologics, high-throughput approaches, pharmacogenomics and other innovations – are required to develop new paradigms to manage the complex scientific, technological and economic factors involved in drug design.
5.6 Interpretation of experimental structures and related data disparities
In a recent review of their group’s applications of biomolecular dynamics for interpretation of atomic force microscopy studies and electron microscopy structures, Schulten and colleagues write (Lee et al. 2009) that :
… the power of molecular dynamics (MD) simulations to make relevant discoveries on their own, i.e., without corroborating experimental evidence, is a hotly debated topic today.
They continue to argue that :
… many important research problems in modern cell biology require a combined experimental-computational approach and computing has to be a reliable partner. Indeed, in many cases, experimental researchers must team up with MD experts who complement and resolve ambiguity in experimental data.
Other examples were provided above in Subsection 5.1, on using MD simulations and other modeling techniques (spectral decompositions, enhanced sampling methods, QM/MM techniques, etc.) to gain insights into biological structure and function. Many of these predictions can ultimately be tested by further experimentation.
Importantly, modeling can help sift through conflicting experimental information. Two such examples where different experimental techniques have produced different structural perspectives involve the organization of G quadruplexes and of chromatin fibers.
5.6.1 G-quadruplexes
G-quadruplexes are organized topologies of G-rich sequences in telomeric DNA (chromosome ends) held by Hoogsteen base pairing. Such sequences are of great interest because of their role in preserving genomic stability ; a better understanding of their structural properties could help interpret processes related to human aging caused by chromosome degradation.
Crystal structures of short telomeric sequences are available, as are several NMR solution structures. The topologies from these studies, however, differ in the orientation of the loops connecting the G-tetrad planes (Fig. 6) and in the overall stacking arrangement of the complexes. Namely, whereas the crystal topologies suggest parallel stranded constructs, the NMR structures suggest anti-parallel backbone arrangements with stabilizing ions (Burge et al. 2006).
Fig. 6.
G quadruplex architecture and drug binding. (a) Antiparallel and (b) parallel folds of human telomeric G-quadruplexes; (c) biological unit of the complex between the drug candidate molecule BRACO-19 and two bimolecular human telomeric quadruplexes (Campbell et al. 2008). Figure kindly provided by Stephen Neidle.
By modeling guanine-rich quadruplexes using MD, free-energy simulations, and PCA, Neidle and co-workers determined stable structures of these complexes in parallel orientations and predicted higher-order organizations (Haider & Neidle, 2009; Haider et al. 2008). These structures help propose plausible models on longer systems that could not be solved experimentally as well as build higher-order repeat models. Such detailed models can examine the relative stability of parallel and anti-parallel arrangements and suggest conditions under which each form will be stable. Ultimately, atomic models could be used for ligand-binding drug design, already an active area to fight cancer (Neidle & Parkinson, 2002). Figure 6c shows a crystal structure of an anti-cancer drug, at the interface of two parallel-folded quadruplexes and stabilized by a water network, bound between two human telomeric quadruplexes (Campbell et al. 2008). The structure confirms and extends modeling predictions and underscores that, while theory provides generally correct ideas, experimental data clarify and refine the modeling suggestions.
5.6.2 Chromatin organization
Chromatin structure represents another higher-order structure of great interest. Understanding chromatin organization is important because the chromatin structure affects genome accessibility to the protein synthesis machinery. Hence, transcription regulation is ultimately a structural problem.
Although several levels of folding are recognized for the chromatin fiber, only the basic unit of the chromatin fiber – the nucleosome, or the protein octamer core around which about 200 bp of DNA are wound (Davey et al. 2002; Kornberg & Thomas, 1974; Luger et al. 1997) – and the associated low-level packaging are well characterized (Felsenfeld & Groudine, 2003).
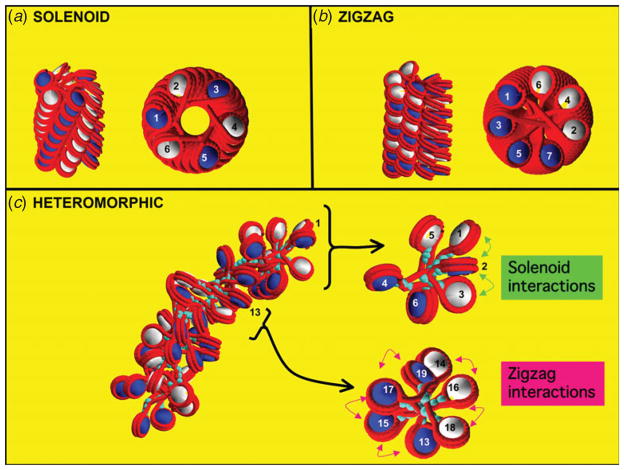
At physiological ionic strengths, it is believed that the chromatin forms a ‘30 nm’ chromatin filament observed by electron microscopy, but several models for this polynucleosomal level of folding have been suggested based on X-ray crystallography and electron microscopy imaging, with yet no consensus (Tremethick, 2007; van Holde & Zlatanova, 2007). For example, the two-start zigzag model, where the nucleosomes crisscross one another around the helical axis with straight linker DNA (Dorigo et al. 2004) is evident in the recent crystallographic structure of the tetranucleosome (Schalch et al. 2005), while the solenoid topology (Finch et al. 1977), in which the nucleosomes arrange helically around the fiber axis with bent linker DNA, is supported by electron micrographs (Fig. 7).
Fig. 7.
Chromatin organization : ideal models and simulation-generated model. (a) Ideal solenoid model for the chromatin fiber (side, top, and upper top layer views), in which DNA linkers are bent and neighboring nucleosomes (i±1) are in closest contact ; (b) ideal zigzag model for the chromatin fiber (side, top, and upper top-layer views), in which DNA linkers are straight and next-nearest neighbors (i±2) are in closest contact; (c) heteromorphic architecture obtained by modeling (Grigoryev et al. 2009) at divalent ion environments with the chromatin model shown in Fig. 1c in which mostly zigzag forms with straight linker DNAs are interspersed with bent DNA linkers. In all views, linker and wrapped DNA are colored red ; odd and even nucleosomes are white and blue, respectively ; and linker histones are turquoise.
Only recently have researchers begun to dissect the influence of key internal and external factors such as length of the connecting linker DNA segments between nucleosomes (which can vary from 10 to 70 bp), the binding of linker histones, and the presence of various concentrations of monovalent and divalent ions on chromatin structure (e.g. Kruithof et al. 2009; Robinson et al. 2006). Modeling has played an important role (e.g. Arya & Schlick, 2006; Grigoryev et al. 2009; Schlick & Perisic, 2009; Stehr et al. 2008; Sun et al. 2005; Wong et al. 2007) in examining different models, dissecting the dependence of fiber width on the linker DNA length, the effect of linker histones of fiber structure and the condensation roles of divalent ions on chromatin organization. For example, our Monte Carlo simulations of a coarse-grained oligonucleosome model have suggested a compact zigzag organization for the chromatin fiber at typical linker DNA lengths with linker histones and a more heteromorphic architecture of zigzag forms with straight linker DNA interspersed with bent DNA linkers at divalent ion environments (Fig. 7) (Grigoryev et al. 2009); this view thus merges the two ideal models and provides further explanations to the prevalence of both structures by experiments. While studies are ongoing, modeling has underscored the notion that the chromatin structure is heteromorphic (Grigoryev et al. 2009; Wong et al. 2007) rather than a single dominant structure.
Interestingly, recent folding simulations of proteins have indicated that conformational space may be more heterogeneous than originally believed (Ensign & Pande, 2009; Freddolino & Schulten, 2009).
6. Some modeling and simulation failures
Although publications of failures are not common, several general areas of failure can be noted in biomolecular modeling.
6.1 CASP failures in structure prediction
As mentioned above, prediction of successes for small proteins can be documented by the CASP experiments, especially from template-based modeling; indeed, proteins with >35% sequence similarity tend to be folded alike. However, exceptions are well known (Gan et al. 2002). In a recent CASP8 experiment, two protein targets (56 residues each) for template-based modeling were released containing>90% sequence identity ; the first protein falls in the α fold class, while the second has an α/β topology. Surprisingly, all 159 participant teams except four failed to recognize the two different folds. Instead, most predictors attributed the α/β fold to both targets ; the difficulty was traced to steric clashes occurring at the side chain of the non-identical residues (Alexander et al. 2007).
More common are failures in de novo or ab initio folding predictions. In CASP8, several groups submitted predictions for up to 10 targets. Except for one group that performed well, the performance of the others was inconsistent, i.e. some bad and some good predictions (Kryshtafovych et al. 2009). One difficult target, which contained five α-helices and four antiparallel β-strands, received no correct predictions (Ben-David et al. 2009). It has also been established that high-resolution modeling (<2 Å) for new folds is far beyond the current capacity of even the best of programs (Zhang, 2008). Moreover, protein structure refinement appears to be quite a difficult task, as evident from performance in CASP8 for this new refinement category (MacCallum et al. 2009).
6.2 Failures in configurational sampling
Sampling the high-dimensional rugged function of biomolecules to locate all the thermally accessible configurations is a well-recognized bottleneck in MD. Moreover, despite all the clever enhanced sampling methods that have been designed, the problem is far from solved. The sampling problem is also exacerbated by the parameter approximations in force fields and algorithms, and this rigorously requires stochastic frameworks for modeling as well as data analysis. In practice, this means that most MD simulations sample local states that are initial configuration, model, and parameter dependent, as shown for the narrow sampled ranges in dihedral angle distributions in peptides (Sugita & Okamoto, 1999), or the limited configurational space sampled for the ring peptide cyclosporin A, which could not be found in the all trans conformation except at very high temperatures (O’Donohue et al. 1995). Thus, convergence of structures in the global sense can almost never be proven in practice, even with simplified coarse-grained models and enhanced sampling methods.
6.3 Force field biases
Force field problems are well recognized, for example, in predicting β proteins (Ensign & Pande, 2009; Freddolino & Schulten, 2009; Maupetit et al. 2009; Mittal & Best, 2010) and the related over-stabilization of helices (Best et al. 2008; Freddolino et al. 2009; Hornak et al. 2006; Tanizaki et al. 2008), possibly due to systematic errors in protein backbone potentials and the approximate treatment of hydrogen bonds. Noted also in implicit solvent models are structural biases (e.g. towards α helices) compared to explicit-water systems (Roe et al. 2007) and limitations in electrostatic treatments due to simple atom-centered charged models (Beachy et al. 1997; Chen & Brooks, 2008; Halgren, 1999); polarizable force fields thus represent a more accurate description of molecular properties (Ponder et al. 2010).
Although modern force fields perform comparably in MD simulations (Price & Brooks, 2002), the general parameterization problem is difficult for several reasons. These include the enormity of the parameter space, the difficulty in validating models against the varied experimental data, the desire to simplify models, while at the same time extending the biological scope of the modeling, and the practical issue that force field development may not be as scientifically rewarding as are applications. Thus, no force field can accurately reproduce all the complex interactions and properties of real systems, but force fields need not be perfect to be meaningful ! Their overall utility is in generating qualitative and quantitative insights into structural, energetic, and dynamic properties of complex systems through systematic studies, especially when trends are compared for a related group of biological systems (like single base-pair variants of DNA or protein mutants). Ultimately, predictions or interpretations can be tested experimentally.
7. Summary
From the information and examples provided here, it appears that biomolecular modeling and simulation is a vibrant field with many avid proponents and numerous exciting areas of exciting activity. Like many new promising technologies, however, initial expectations might have been set too high. Indeed, the general appreciation for a greater biological complexity than originally believed in the wake of the Human Genome Project and beyond, together with the realization that more work on algorithms is essential to bridge the gap between experimental and theoretical time frames, has transitioned the field from a trough of sorts to a more realistic and productive phase of activity (Fig. 3). Clearly, a recognized strength of researchers in the field is their ability to synthesize information from various sources and develop innovative tools that build upon knowledge in different disciplines. The unprecedented opportunities in 21st century biology, in general, and biomolecular modeling, in particular, argue for a focused effort by the community to define key scientific challenges that build upon existing knowledge and technical capabilities to advance modeling to new levels of accuracy and reliability ; concerted advances can undoubtedly help our understanding of biological systems and lead to many beneficial applications in health, environment, and technology (NAS, 2009).
8. Recommendations
To help propel the field onto its productive trajectory, the following general recommendations and areas that require further progress can be identified.
8.1 Bridging Scales
Given the enormous range and complexity of biological temporal and spatial dimensions, work should continue on all levels of biomolecular modeling, from all-atom representations to coarse-grained models on both the classical and quantum-classical levels, despite imperfections and approximations. This is needed to study processes ranging from macromolecular folding (proteins, RNA, and other complexes) to biochemical binding and reaction mechanisms (enzyme catalysis or protein/ligand interactions) to macromolecular pathways (DNA replication and repair or chromatin organization) and up to supramolecular cellular processes like protein-signaling networks. Because for biology the ‘devil is in the details’, simulation efforts are required on all levels of biological complexity rather than focusing on very accurate studies on the finest level. This means that many approaches in addition to all-atom modeling are important to continue, such as coarse-grained modeling and mesoscale models. New methodological developments are needed in all levels (see the next item) so that, ultimately, an integration of all these tools could allow a kind of ‘telescoping’ from one level of resolution to the other to focus on specific aspects of a variety of biological processes.
8.2 Methodology advances
Studying biological systems and problems on the above levels successfully and reliably requires new methods and models to be systematically developed, including force fields, hybrid quantum/molecular mechanics models, enhanced sampling techniques, rigorous coarse graining of multiscale models, and tool integration. As mentioned for force fields (next item), developing models on other scales requires consolidated efforts of concept development, parameterization, and code sharing.
8.3 Force fields
Improved force fields are needed to address the above biological problems, with parameterization and validation conducted systematically and rigorously on both all-atom and coarse-grained models. The ideas needed to improve force fields appear to be known, like polarizable force fields (Ponder et al. 2010), but efforts need to be consolidated, prioritized, and organized. Special funding for such consolidated efforts could help attract researchers to work on these tasks.
8.4 Open-source codes
Better methods for data sharing are needed, including for force fields, algorithms, and many computer programs associated with research papers. For example, a requirement to deposit computer codes when submitting methodology papers, as done in some computer science journals, could help reviewers test the programs and ultimately allow users in the community to apply the algorithms. This will go a long way toward validating algorithms and identifying the most effective techniques. For enhanced sampling methods, which tend to be home grown, this program sharing could help practitioners apply clever ideas to important systems.
8.5 Better interdisciplinary education
One origin for a poor perception of the field is that many applications of molecular modeling packages as black boxes are not reliable, although the images may be impressive. This problem could be alleviated with better multidisciplinary education, including in modeling, to teach the basics of modeling studies and analysis, to stimulate modeling experimentation, and to cultivate sound scientific practices (such as parameter testing, statistical interpretations, and sensitivity analysis). For example, programs in computational biology or bioinformatics could offer under-graduate and graduate-level courses like bio/molecular modeling, computational chemistry, or modeling tools for experimentalists. Courses should introduce the fundamentals in the fields as well as provide practical experience through simple programming and modeling assignments to introduce students to common caveats and encourage critical thinking and healthy skepticism (e.g. as in Schlick, 2010).
8.6 Research collaborations
Although funding agencies have led to many teams working together, there remain many duplicate research efforts. A better deployment of truly interdisciplinary teams working together to advance specific areas of methodology and application will be beneficial. This could be achieved through large community efforts on several key fronts of algorithms (like MD, QM/MM and sampling), force fields, and specific applications.
8.7 Community assessment efforts
The CASP exercise has been an exemplary success in defining the state-of-the-art in protein folding and helping propel a field forward in an organized, community-wide manner. Similar efforts are needed for nucleic acids, force field development, MD practices, enhanced sampling methods, quantum-classical mechanics simulations, coarse-grained models, implicit solvation, and others.
8.8 Faster processors
Computer technology has certainly impacted the field tremendously, and will continue to do so. Efforts must continue to make available large central computing resources like supercomputing centers to biomolecular modelers. The sharing of an Anton supercomputer with the community is a commendable effort. In addition, because data management becomes an issue for biomolecular data, and queue times can be long when computations are conducted remotely, algorithms and programs that utilize distributed computing platforms and local networks of fast processors are also important.
8.9 Shared resources
Systematic advances in algorithms, force fields, and modeling approaches require establishment of better software repositories and shared resources. Infrastructural support for generating and analyzing such voluminous molecular data requires the development of efficient simulation management tools such as clustering, archiving, comparison, debugging, visualization, and communication protocols.
8.10 Better communication between academe and industry
Greater efforts are needed to bring open dialog and constructive research efforts concerning tools and applications used in academia and those in the pharmaceutical industry. For example, cross-community interactions could help lead to better efforts in establishing needed databases, such as for protein/ligand structures, or developing better approaches for binding free energies. Overall, better modes are needed for transforming basic or translational research efforts in academia to more applied problems in industry and technology. Economic incentives for such collaborations would be needed, and these could come from a combination of industrial, academic and government agencies.
8.11 Reports of failure
Progress in the field could be expedited by a venue that would allow scientists to publish modeling failures. Understanding the problems or failures could go a long way in helping finding the right solutions. Perhaps journal sections dedicated to such publications could inspire scientists to share such information.
Of course, these wide suggestive strokes require many of the details to be filled in. We invite readers to share comments and suggestions. Such concerted efforts can help move the field away from a local trough of disappointment (or a shattering of inflated expectations) toward a more realistic productive phase, where modeling and experiment can be hand-in-hand partners (Fig. 3).
Acknowledgments
Support from NSF, NIH, ACS and Philip-Morris International are gratefully acknowledged. The authors thank questionnaire respondents for their time and opinions: Russ B. Altman, Armando Bravo Garcia, Axel Brunger, Monica Canalizo, James Canary, Jianlin Cheng, Jacques Cohen, Gemma Comellas, Xavier Daura, Nanjie Deng, Carmen Domene-Nunez, Kevin Drew, Eric Drexler, Meredith Foley, Simone Furini, Hin Hark Gan, Jan Hermans, Kenneth L. Ho, Jonathan Ipsaro, Yuansheng Jiang, Nessim Kichik, John Klepeis, Axel Kohlmeyer, Elmar Krieger, Jonathan Lai, Christian Laing, Eric H. Lee, Ben Leimkuhler, Francis Lemon, Yunlang Li, Jing Ma, Aviv Madar, Tom Markland, John Markley, Andy McCammon, Steve McKeever, Jules Moskowitz, Veronica Murphy, Stephen Neidle, Mauricio Esguerra Neira, Chris Oostenbrink, Vijay Pande, Alex Pearlman, Ognjen Perisic, Giulio Quarta, David Rooklin, Harold A. Scheraga, Bohdan Schneider, Ned Seeman, David Shaw, Bob Skeel, Stephen Smith, Andrew Sundstrom, Bill Taylor, Alexander Vologodskii, Marcus Weck, John Zhang, and Nengjie Zhou.
10. Appendix A
The following questions were sent to a small sample of about 125 computational and biological scientists from students to experts across the globe.
Your background: Describe your field of research and professional position.
Overall assessment (a): Do you believe that bimolecular modeling has fulfilled the expectations for the field during the past three decades (e.g. in drug design, protein folding pathway prediction, etc.) ? Why or why not?
-
Overall assessment (b): Do you consider molecular modeling to be a complement to experiment or a field in its own right ?
If you are an experimentalist, do you use molecular simulation results/techniques to interpret experimental data ?
–OR–
If you are a theoretician, does your work help interpret experimental work? Please elaborate as possible.
Success story: Can you describe a striking example of a successful biomolecular modeling application, from your lab and/or one from another lab, i.e. cases in which molecular simulations have led to innovative scientific contributions or have correctly helped interpret experimental results ?
Failure: Can you describe a striking example of failure of biomolecular modeling, i.e. an example in which excessive/incorrect claims regarding the validity of molecular simulations have lead to misinterpretations of experimental data or misleading conclusions ?
Future (a): In the long term, what applications do you think will benefit most from the field of biomolecular modeling (e.g. medicine, research, technology, etc.) ?
Future (b): What specific advances do you believe the field of biomolecular modeling would benefit the most from (e.g. improved force fields, faster processors, increased funding, etc.).
Fifty-eight responses were obtained, most from academic scientists. Although the sample is small, it was striking to see the difference in opinions regarding the field’s progress as assessed by mostly algorithm developers (mathematicians, computer scientists, etc.) on one hand versus computational chemists and biologists on the other. The former group was far more reserved, and perhaps disappointed, at the relatively modest impact of algorithmic advances. Perhaps technology advances have overshadowed algorithmic work. Of course, without the formal guidance that rigorous algorithms provide (see Subsection 5.2), many of the simulations and applications would not have been possible. Regarding the independence of the field, opinions were divided in that almost all respondents believed that modeling is a complement to experiment. About half of the respondents also added that the field has become a discipline on its own right.
The majority of respondents believe that initial field expectations were not met, but agree that these might have been set too high.
Although no straight-forward approach to rational drug design has succeeded due to the underlying complexity of the systems and associated energy landscapes, modeling and simulation, specifically MD, has provided a way to probe molecular function. Many admit that modeling helps suggest new experiments and generate hypotheses.
Thus, respondents agree that modeling has provided a variety of creative tools for investigating biomolecular interactions, structures and energies, and that all these tools have evolved substantially. Researchers also admit that, to outsiders, these advances may not be as apparent, convincing, or useful. We suggest here that this is because tools and programs are not yet automated to the point that they could be used as ‘black box’ tools for specific applications. Still, a general optimism for the future is shared by many respondents, especially in biomedical applications.
Force field improvement was by far the most important advance that respondents recognized as essential for future progress of the field; this is followed by algorithm development. Some believe that better sampling, due to theoretical and computational advances, will impact drug modeling and design applications in particular.
Footnotes
This comment was shared by force-field pioneer Norman L. Allinger.
References
- Abrams CF, Vanden-Eijnden E. Large-scale conformational sampling of proteins using temperature-accelerated molecular dynamics. Proceedings of the National Academy of Sciences, USA. 2010;107:4961–4966. doi: 10.1073/pnas.0914540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder BJ, Wainwright TE. Studies in molecular dynamics. I. General method. Journal of Chemical Physics. 1959;31:459–466. [Google Scholar]
- Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proceedings of the National Academy of Sciences, USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman R, Radmer R, Glazer D. Improving structure-based function prediction using molecular dynamics. Structure. 2009;17:919–929. doi: 10.1016/j.str.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir-Aslani A. Toxicogenomic predictive modeling : emerging opportunities for more efficient drug discovery and development. Tech Forecast Soc Change. 2008;75:905–932. [Google Scholar]
- Arora K, Schlick T. In Silico evidence for DNA polymerase β’s substrate-induced conformational change. Biophysics Journal. 2004;87:3088–3099. doi: 10.1529/biophysj.104.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proceedings of the National Academy of Sciences, USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya G, Schlick T. A tale of tails : how histone tails mediate chromatin compaction in different salt and linker histone environments. Journal of Physical Chemistry A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D, Kuhlman B, Dantas G, Ireton G, Varani G, Stoddard B. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Barth E, Schlick T. Overcoming stability limitations in biomolecular dynamics: I. combining force splitting via extrapolation with Langevin dynamics in ln. Journal of Chemical Physics. 1998;109:1617–1632. [Google Scholar]
- Beachy MD, Chasman D, Murphy RB, Halgren TA, Friesner RA. Accurate ab initio quantum chemical determination of the relative energetics of peptide conformations and assessment of empirical force fields. Journal of the American Chemical Society. 1997;119:5908–5920. [Google Scholar]
- Bebenek K, Garcia-Diaz M, Foley MC, Pedersen LC, Schlick T, Kunkel TA. Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase μ. EMBO Reports. 2008;9:459– 464. doi: 10.1038/embor.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker OM, Dhanoa DS, Marantz Y, Chen D, Shacham S, Cheruku S, Heifetz A, Mohanty P, Fichman M, Sharadendu A. An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. Journal of Medical Chemistry. 2006;49:3116–3135. doi: 10.1021/jm0508641. [DOI] [PubMed] [Google Scholar]
- Ben-David M, Noivirt-Brik P, Paz A, Prilusky J, Sussman JL, Levy Y. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;9:50–65. doi: 10.1002/prot.22591. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communication. 1995;91:43–56. [Google Scholar]
- Berezhkovskii A, Hummer G, Szabo A. Reactive flux and folding pathways in network models of coarse-grained protein dynamics. Journal of Chemical Physics. 2009;130:205102. doi: 10.1063/1.3139063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Buchete NV, Hummer G. Are current molecular dynamics force fields too helical ? Biophysics Journal. 2008;95:L07–L09. doi: 10.1529/biophysj.108.132696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Hummer G. Diffusive model of protein folding dynamics with Kramers turnover in rate. Phys Rev Lett. 2006;96:228104. doi: 10.1103/PhysRevLett.96.228104. [DOI] [PubMed] [Google Scholar]
- Bezdek J. Fuzzy models – what are they, and why? IEEE Transactions on Fuzzy Systems. 1993;1:1–5. [Google Scholar]
- Bolhuis PG, Chandler D, Dellago C, Geissler PL. Transition path sampling: throwing ropes over rough mountain passes, in the dark. Annual Review of Physical Chemistry. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- Borman S. Reducing time to drug discovery. Chemical and Engineering News. 1998;77:33–48. [Google Scholar]
- Borman S. Human genome sequence milestone. Chemical and Engineering News. 2010;88:30–32. [Google Scholar]
- Borrell B. Fraud rocks protein community. Nature. 2009;462:970. doi: 10.1038/462970a. [DOI] [PubMed] [Google Scholar]
- Borrero EE, Escobedo FA. Optimizing the sampling and staging for simulations of rare events via forward flux sampling schemes. Journal of Chemical Physics. 2008;129:024115. doi: 10.1063/1.2953325. [DOI] [PubMed] [Google Scholar]
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (SC06); Tampa. New York: Florida ACM Press; 2006. [Google Scholar]
- Bowman GR, Beauchamp KA, Boxer G, Pande VS. Progress and challenges in the automated construction of Markov state models for full protein systems. Journal of Chemical Physics. 2009;131:124101. doi: 10.1063/1.3216567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DB. Rational drug design: controlling the size of the Haystack. Modern Drug Discovery. 1998;1:41–47. [Google Scholar]
- Brady SF, Stauffer KJ, Lumma WC, Smith GM, Ramjit HG, Lewis SD, Lucas BJ, Gardell SJ, Lyle EA, Appleby SD, Cook JJ, Holahan MA, Stranieri MT, Lynch JJ, Jr, Lin JH, Chen IW, Vastag K, Naylor-Olsen AM, Vacca JP. Discovery and development of the novel potent orally active thrombin inhibitor N-(9-hydroxy-9-fluorenecarboxy)prolyl trans-4-amino-cyclohexylmethyl amide (L-372,460): co-application of structure-based design and rapid multiple analogue synthesis on solid support. Journal of Medical Chemistry. 1998;41:401–406. doi: 10.1021/jm9705014. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. Journal of Computational Chemistry. 1983;4:187–217. [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Research. 2006;34:5402– 5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell H, Parkinson GN, Reszka AP, Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. Journal of the American Chemical Society. 2008;130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- Case DA. Molecular dynamics and NMR spin relaxation in proteins. Accounts of Chemical Research. 2002;35:325–331. doi: 10.1021/ar010020l. [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1785. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Cervantes CF, Markwick PR, Sue SC, McCammon JA, Dyson HJ, Komives EA. Functional dynamics of the folded ankyrin repeats of IκBα revealed by nuclear magnetic resonance. Biochemistry. 2009;48:8023–8031. doi: 10.1021/bi900712r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Brooks CL., III Implicit modeling of nonpolar solvation for simulating protein folding and conformational transitions. Physical Chemistry Chemical Physics. 2008;10:471–481. doi: 10.1039/b714141f. [DOI] [PubMed] [Google Scholar]
- Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proceedings of the National Academy of Sciences, USA. 2009;106:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS. Opportunities for research and NIH. Science. 2010;327:36–37. doi: 10.1126/science.1185055. [DOI] [PubMed] [Google Scholar]
- Collins JR, Burt SK, Erickson JW. Flap opening in HIV-1 protease simulated by activated’ molecular dynamics. Nature Structural and Molecular Biology. 1995;2:334–338. doi: 10.1038/nsb0495-334. [DOI] [PubMed] [Google Scholar]
- Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, Leaver-Fay A, Baker D, Popović Z, Players F. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Agoston V, Pongor S. The efficiency of multi-target drugs: The network approach might help drug design. Trends in Pharmacological Sciences. 2005;26:178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. Journal of Chemical Physics. 1993;98:10089– 10092. [Google Scholar]
- Daura X, Gademann K, Jaun B, Seebach D, Gunsteren WFV, Mark A. Peptide folding: when simulation meets experiment. Angewandte Chemie (International edition in English) 1999;38:236–240. [Google Scholar]
- Daura X, Jaun B, Seebach D, Gunsteren WFV, Mark A. Reversible peptide folding in solution by molecular dynamics simulation. Journal of Molecular Biology. 1998;280:925–932. doi: 10.1006/jmbi.1998.1885. [DOI] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Mäeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. Journal of Molecular Biology. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- Day R, Paschek D, Garcia AE. Microsecond simulations of the folding/unfolding thermodynamics of the Trp-cage miniprotein. Proteins. 2010;78:1889–1899. doi: 10.1002/prot.22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laplace PS. Thé orie Analytique des Probabilités. 3. VII. Paris, France: Gauthier-Villars; 1820. Oeuvres complètes de Laplace. [Google Scholar]
- Dellago C, Bolhuis PG. Transition path sampling simulations of biological systems. Topics in Current Chemistry. 2007;268:291–317. [Google Scholar]
- Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annual Reviews of Biophysics. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac PAM. Quantum mechanics of many-electron systems. Proceedings of the Royal Society of London A. 1929;123:714–733. [Google Scholar]
- DOE. Opportunities in Biology at the Extreme Scale of Computing. 2009 http://www.er.doe.gov/ascr/ProgramDocuments/Docs/BiologyReport.pdf.
- Dooley AJ, Shindo N, Taggart B, Park JG, Pang YP. From genome to drug lead: Identification of a small-molecule inhibitor of the SARS virus. Bioorganic and Medicinal Chemistry Letters. 2006;16:830–833. doi: 10.1016/j.bmcl.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- Drie JHV. Computer-aided drug design: The next 20 years. Journal of Computer-Aided Molecular Design. 2007;21:591–601. doi: 10.1007/s10822-007-9142-y. [DOI] [PubMed] [Google Scholar]
- Dror RO, Arlow DH, Borhani DW, Jensen MØ, Piana S, Shaw DE. Identification of two distinct inactive conformations of the 2-adrenergic receptor reconciles structural and biochemical observations. Proceedings of the National Academy of Sciences, USA. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Kollman PA. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- Duan Y, Kollman PA, Harvey SC. Protein folding and beyond. In: Keinam E, Schechter I, editors. Chemistry for the 21st Century. Weinheim, Germany: Wiley-VCH; 2000. [Google Scholar]
- Duan Y, Wang L, Kollman PA. The early stage of folding of Villin headpiece subdomain observed in a 200-nanosecond fully solvate molecular dynamics simulation. Proceedings of the National Academy of Sciences, USA. 1998;95:9897–9902. doi: 10.1073/pnas.95.17.9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan ZH, Krasny R. An Ewald summation based multipole method. Journal of Chemical Physics. 2000;113:3492–3495. [Google Scholar]
- Earl DJ, Deem MW. Monte Carlo simulations. Methods in Molecular Biology. 2008;443:25–36. doi: 10.1007/978-1-59745-177-2_2. [DOI] [PubMed] [Google Scholar]
- Economist. The 1998 Nobel Prizes. 1998. Oct 15, p. 97. [Google Scholar]
- Ensign DL, Kasson PM, Pande VS. Heterogeneity even at the speed limit of folding: large-scale molecular dynamics study of a fast-folding variant of the Villin headpiece. Journal of Molecular Biology. 2007;374:806–816. doi: 10.1016/j.jmb.2007.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensign DL, Pande VS. The Fip35 WW domain folds with structural and mechanistic heterogeneity in molecular dynamics simulations. Biophysics Journal. 2009;96:L53–L55. doi: 10.1016/j.bpj.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- Faraldo-Gomez J, Roux B. On the importance of a funneled energy landscape for the assembly and regulation of multidomain Src tyrosine kinases. Proceedings of the National Academy of Sciences, USA. 2007;104:13643–13648. doi: 10.1073/pnas.0704041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Finch JT, Lutter LC, Rhodes D, Brown AS, Rushton B, Levitt M, Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Fitch BG, Rayshubskiy A, Eleftheriou M, Ward TJC, Giampapa M, Pitman MC, Germain RS. Blue matter: approaching the limits of concurrency for classical molecular dynamics. Super-computing, 2006 SC’06; Proceedings of the ACM/IEEE SC 2006 Conference; 2006. p. 44. [Google Scholar]
- Foley M, Schlick T. The relationship between conformational changes in Pol μ’s active site upon binding incorrect nucleotides and mismatch incorporation rates. Journal of Physical Chemistry B. 2009;113:13035–13047. doi: 10.1021/jp903172x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- Freddolino PL, Arkhipov AS, Larson SB, McPherson A, Schulten K. Molecular dynamics simulations of the complete satellite tobacco mosaic virus. Structure. 2006;14:437–449. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Freddolino PL, Liu F, Gruebele M, Schulten K. Ten-microsecond molecular dynamics simulation of a fast-folding WW domain. Biophysics Journal. 2008;94:L75–L77. doi: 10.1529/biophysj.108.131565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freddolino PL, Park S, Roux B, Schulten K. Force field bias in protein folding simulations. Biophysics Journal. 2009;96:3772–3780. doi: 10.1016/j.bpj.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freddolino PL, Schulten K. Common structural transitions in explicit-solvent simulations of villin headpiece folding. Biophysics Journal. 2009;97:2338–2347. doi: 10.1016/j.bpj.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan HH, Perlow RA, Roy S, Ko J, Wu M, Huang J, Yan S, Nicoletta A, Vafai J, Sun D, Wang L, Noah JE, Pasquali S, Schlick T. Analysis of protein sequence/structure similarity relationships. Biophysics Journal. 2002;83:2781–2791. doi: 10.1016/s0006-3495(02)75287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AE, Pascheck D. Simulation of the pressure and temperature folding/unfolding equilibrium of a small RNA hairpin. Journal of the American Chemical Society. 2008;130:815–817. doi: 10.1021/ja074191i. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Nie A, An J, Huang Z. Structure-based virtual screening of chemical libraries for drug discovery. Current Opinion in Chemical Biology. 2006;10:194–202. doi: 10.1016/j.cbpa.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H, Davies DR. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: A platform for antiviral drug design. Proceedings of the National Academy of Sciences, USA. 1999;96:13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proceedings of the National Academy of Sciences, USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golosov AA, Warren JJ, Beese LS, Karplus M. The mechanism of the translocation step in DNA replication by DNA polymerase I : a computer simulation. Structure. 2010;18:83–93. doi: 10.1016/j.str.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BJ, Gorfe AA, McCammon JA. Large conformational changes in proteins: signaling and other functions. Current Opinion in Structural Biology. 2010;20:142–147. doi: 10.1016/j.sbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greengard L, Rokhlin V. A fast algorithm for particle simulation. Journal of Computational Physics. 1987;73:325–348. [Google Scholar]
- Greengard L, Rokhlin V. A new version of the fast multipole method for the Laplace equation in three dimensions. Acta Numerica. 1997;6:229–269. [Google Scholar]
- Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proceedings of the National Academy of Sciences, USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. Journal of Molecular Biology. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Parkinson GN, Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophysics Journal. 2008;95:296–311. doi: 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider SM, Neidle S. A molecular model for drug binding to tandem repeats of telomeric G-quadruplexes. Biochemical Society Transactions. 2009;37:583–588. doi: 10.1042/BST0370583. [DOI] [PubMed] [Google Scholar]
- Halgren TA. MMFF VII. Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for interaction energies and geometries. Journal of Computational Chemistry. 1999;20:730–748. doi: 10.1002/(SICI)1096-987X(199905)20:7<730::AID-JCC8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Hamelberg D, McCammon JA. Fast peptidyl cis–trans isomerization within the flexible gly-rich flaps of HIV-1 protease. Journal of the American Chemical Society. 2005;127:13778–13779. doi: 10.1021/ja054338a. [DOI] [PubMed] [Google Scholar]
- Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. Journal of Chemical Physics. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EC. Life is complicated. Nature. 2010;464:664–667. doi: 10.1038/464664a. [DOI] [PubMed] [Google Scholar]
- Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Zhuang L, Fisher TE, Embrey M, Guare JP, Jr, Egbertson MS, Vacca JP, Huff JR, Felock PJ, Witmer MV, Stillmock KA, Danovich R, Grobler J, Miller MD, Espeseth AS, Jin L, Chen IW, Lin JH, Kassahun K, Ellis JD, Wong BK, Xu W, Pearson PG, Schleif WA, Cortese R, Emini E, Summa V, Holloway MK, Young SD. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proceedings of the National Academy of Sciences, USA. 2004;101:11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hornak V, Okur A, Rizzo RC, Simmerling C. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proceedings of the National Academy of Sciences, USA. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V, Simmerling C. Targeting structural flexibility in HIV-1 protease inhibitor binding. Drug Discovery Today. 2007;12:132–138. doi: 10.1016/j.drudis.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Elstner M, Hermans J. Comparison of a QM/MM force field and molecular mechanics force fields in simulations of alanine and glycine ‘Dipeptides’ (Ace-Ala-Nme and Ace-Gly-Nme) in water in relation to the problem of modeling the unfolded peptide backbone in solution. Proteins : Structure, Function, and Genetics. 2003;50:451–463. doi: 10.1002/prot.10279. [DOI] [PubMed] [Google Scholar]
- IMAG. IMAG Futures Meeting: The Impact of Modeling on Biomedical Research. 2009 http://www.imagwiki.org/mediawiki/index.php?title=IFM_Agenda.
- Izrailev S, Crofts AR, Berry EA, Schulten K. Steered molecular dynamics simulation of the Rieske subunit motion in the cytochrome bc1 complex. Biophysics Journal. 1999;77:1753–1768. doi: 10.1016/S0006-3495(99)77022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A, Levitt M. Refinement of large structures by simultaneous minimization of energy and R factor. Acta Crystallographica A. 1978;34:931–935. [Google Scholar]
- Jiang L, Althoff EA, Clemente FR, Doyle L, Röthlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, III, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekar J, Madura J, Impey R, Klein M. Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- Kamerlin SCL, Haranczyk M, Warshel A. Progress in ab initio QM/MM free-energy simulations of electrostatic energies in proteins: accelerated QM/MM studies of pK, redox reactions and solvation free energies. Journal of Physical Chemistry B. 2009;113:1253–1272. doi: 10.1021/jp8071712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proceedings of the National Academy of Sciences, USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Garg A, Raghava GPS. PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein and Peptide Letters. 2007;14:626–631. doi: 10.2174/092986607781483859. [DOI] [PubMed] [Google Scholar]
- Kelley NW, Huang X, Tam S, Spiess C, Frydman J, Pande VS. The predicted structure of the headpiece of the Huntingtin protein and its implications on Huntingtin aggregation. Journal of Molecular Biology. 2009;388:919–927. doi: 10.1016/j.jmb.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelashvili G, Grossfield A, Feller SE, Pitman MC, Weinstein H. Structural and dynamic effects of cholesterol at preferred sites of interaction with Rhodopsin identified from microsecond length molecular dynamics simulations. Proteins. 2009;76:403–417. doi: 10.1002/prot.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Research. 2004a;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Izzo JA, Schlick T. RNAs with Novel Technologies: Predictions and Confirmations. 2010. Submitted. [Google Scholar]
- Kim N, Shiffeldrim N, Gan H, Schlick T. Candidates for novel RNA topologies. Journal of Molecular Biology. 2004b;341:1129–1144. doi: 10.1016/j.jmb.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Kitano H. A robustness-based approach to systems-oriented drug design. Nature Reviews Drug Discovery. 2007;6:202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- Klein ML, Shinoda W. Large-scale molecular dynamics simulations of self-assembling systems. Science. 2008;321:798–800. doi: 10.1126/science.1157834. [DOI] [PubMed] [Google Scholar]
- Konnert JH, Hendrickson WA. A restrained-parameter thermal-factor refinement procedure. Acta Crystallographica A. 1980;36:344–350. [Google Scholar]
- Kornberg R, Thomas JO. Chromatin structure : oligomers of histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Kruithof M, Chen FT, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force microscopy reveals a highly compliant Helical folding for the 30-nm chromatin fiber. Nature Structural and Molecular Biology. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- Kryshtafovych A, Fidelis K, Moult J. CASP8 results in context of previous experiments. Proteins : Structure, Function, and Genetics Supplement. 2009;9:217–228. doi: 10.1002/prot.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda LP, Pushechnikov A, Disney MD. Small molecule microarrays of RNA-focused peptoids help identify inhibitors of a pathogenic group I intron. ACS Chemical Biology. 2009;4:299–307. doi: 10.1021/cb800313m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EH, Hsin J, Sotomayor M, Comellas G, Schulten K. Discovery through the computational microscope. Structure. 2009;17:1295–1306. doi: 10.1016/j.str.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Duan Y. Improved sampling methods for molecular simulation. Current Opinion in Structural Biology. 2007;17:187–191. doi: 10.1016/j.sbi.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Leimkuhler B, Reich S. Simulating Hamiltonian dynamics (Cambridge Monographs on Applied and Computational Mathematics) Cambridge, UK: Cambridge University Press; 2004. [Google Scholar]
- Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility : the relaxed complex scheme. Journal of the American Chemical Society. 2002;124:5632–5633. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel D. GROMACS 3.0 : A package for molecular simulation and trajectory analysis. Journal of Molecular Modeling. 2001;7:306–317. [Google Scholar]
- Liwo A, Czaplewski C, Oldziej S, Scheraga HA. Computational techniques for efficient conformational sampling of proteins. Current Opinion in Structural Biology. 2008;18:134–139. doi: 10.1016/j.sbi.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2·8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- MacCallum JL, Hua L, Schnieders MJ, Pande VS, Jacobson MP, Dill KA. Assessment of the protein-structure refinement category in CASP8. Proteins. 2009;77 (Suppl 9):66–80. doi: 10.1002/prot.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox J. Statistical mechanics by numbers. Nature. 1989;334:561. [Google Scholar]
- Maisuradze GG, Senet P, Czaplewski C, Liwo A, Scheraga HA. Investigation of protein folding by coarse-grained molecular dynamics with the UNRES force field. Journal of Physical Chemistry A. 2010;114:4471–4485. doi: 10.1021/jp9117776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandziuk M, Schlick T. Resonance in the dynamics of chemical systems simulated by the implicit-midpoint scheme. Chemical Physics Letters. 1995;237:525–535. [Google Scholar]
- Maragakis P, Lindorff-Larsen K, Eastwood MP, Dror RO, Klepeis JL, Arkin IT, Jensen MØ, Xu H, Trbovic N, Friesner RA, Palmer AG, III, Shaw DE. Microsecond molecular dynamics simulation shows effect of slow loop dynamics on backbone amide order parameter of proteins. Journal of Physical Chemistry B. 2008;112:6155–6158. doi: 10.1021/jp077018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick PR, Bouvignies G, Salmon L, McCammon JA, Nilges M, Blackledge M. Toward a unified representation of protein structural dynamics in solution. Journal of the American Chemical Society. 2009;131:16968–16975. doi: 10.1021/ja907476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick PR, Cervantes CF, Abel BL, Komives EA, Blackledge M, McCammon JA. Enhanced conformational space sampling improves the prediction of chemical shifts in proteins. Journal of the American Chemical Society. 2010;132:1220–1221. doi: 10.1021/ja9093692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit J, Derreumaux P, Tufféry P. A fast method for large-scale de novo peptide and miniprotein structure prediction. Journal of Computational Chemistry. 2009;31:726–738. doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- Mayor U, Guydosh NR, Johnson CM, Grossmann JG, Sato S, Jas GS, Freund SM, Alonso DO, Daggett V, Fersht AR. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. Journal of Molecular Biology. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Miller M, Jaskolski M, Rao JK, Leis J, Wlodawer A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989;337:576–579. doi: 10.1038/337576a0. [DOI] [PubMed] [Google Scholar]
- Mittal J, Best RB. Tackling force-field bias in protein folding simulations: folding of Villin HP35 and Pin WW domains in explicit water. Biophysics Journal. 2010;99:L26–L28. doi: 10.1016/j.bpj.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone JA, Zhou R, Berne BJ. Molecular dynamics with multiple time scales : How to avoid pitfalls. Journal of Chemical Theory and Computation. 2010;6:1798–1804. doi: 10.1021/ct100054k. [DOI] [PubMed] [Google Scholar]
- Mowery DC, Nelson RR, Sampat BN, Ziedonis AA. Ivory Tower and Industrial Innovation : University-Industry Technology Transfer Before and After the Bayh–Dole Act. Stanford, CA: Stanford University Press; 2004. [Google Scholar]
- Munos B. Lessons from 60 years of pharmaceutical innovation. Nature Rev. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- NAS. A new biology for the 21st century: Ensuring the United States leads the coming biology revolution. 2009 http://www.nap.edu/catalog/12764.html. [PubMed]
- Navia MA, Fitzgerald PM, McKeever BM, Leu CT, Heimbach JC, Herber WK, Sigal IS, Darke PL, Springer JP. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989;337:615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- Neidigh JW, Fesinmeyer RM, Andersen NH. Designing a 20-residue protein. Nature Structural and Molecular Biology. 2002;9:425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nature. 2002;1:383–392. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- Neidle S, Read M, Harrison J, Romagnoli B, Tanious F, Gowan S, Reszka A, Wilson D, Kelland L. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proceedings of the National Academy of Sciences, USA. 2001;98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A. Computational Biology and Bioinformatics Seminar. Columbia University Medical Center; 2010. [Google Scholar]
- Noé F, Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Current Opinion in Structural Biology. 2008;8:154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Noé F, Horenko I, Schütte C, Smith JC. Hierarchical analysis of conformational dynamics in biomolecules : Transition networks of metastable states. Journal of Chemical Physics. 2007;126:155102. doi: 10.1063/1.2714539. [DOI] [PubMed] [Google Scholar]
- Noé F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proceedings of the National Academy of Sciences, USA. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donohue MF, Burgess AW, Treutlein HR, Walkinshaw MD. Modeling conformational changes in cyclosporin A. Protein Science. 1995;4:2191–2202. doi: 10.1002/pro.5560041025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ, Frommer WB. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proceedings of the National Academy of Sciences, USA. 2005;102:8740–8745. doi: 10.1073/pnas.0503274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan SB, Wu GA, Chodera JD, Dill KA. Protein folding by zipping and assembly. Proceedings of the National Academy of Sciences, USA. 2007;104:11987–11992. doi: 10.1073/pnas.0703700104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan AC, Roux B. Building Markov state models along pathways to determine free energies and rates of transitions. Journal of Chemical Physics. 2008;129:064107. doi: 10.1063/1.2959573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Taylor WR. A structural model for the retroviral proteases. Nature. 1987;329:351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, III, Debolt S, Ferguson D, David S, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Computational Physics Communications. 1995;91:1–41. [Google Scholar]
- Pérez A, Luque J, Orozco M. Dynamics of B-DNA on the Microsecond time scale. Journal of the American Chemical Society. 2007;129:14739–14745. doi: 10.1021/ja0753546. [DOI] [PubMed] [Google Scholar]
- Perryman AL, Forli S, Morris GM, Burt C, Cheng Y, Palmer MJ, Whitby K, McCammon JA, Phillips C, Olson AJ. A dynamic model of HIV integrase inhibition and drug resistance. Journal of Molecular Biology. 2010;397:600–615. doi: 10.1016/j.jmb.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitera JW, Swope WC, Abraham FF. Observation of noncooperative folding thermo-dynamics in simulations of 1BBL. Biophysics Journal. 2008;94:4837–4846. doi: 10.1529/biophysj.107.123265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack A. The New York Times. 1998. Drug testers turn to ‘ Virtual Patients’ as Guinea Pigs. [Google Scholar]
- Ponder JW, Wu C, Ren P, Pande VS, Chodera JD, Schnieders MJ, Haque I, Mobley DL, Lambrecht DS, DiStasio RA, Jr, Head-Gordon M, Clark GNI, Johnson ME, Head-Gordon T. Current status of the AMOEBA polarizable force field. Journal of Physical Chemistry B. 2010;114:2540–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DJ, Brooks CL., III Modern protein force fields behave comparably in molecular dynamics simulations. Journal of Computational Chemistry. 2002;23:1045– 1057. doi: 10.1002/jcc.10083. [DOI] [PubMed] [Google Scholar]
- Procacci P, Marchi M, Martyna GJ. Electrostatic calculations and multiple time scales in molecular dynamics simulation of flexible molecular systems. Journal of Chemical Physics. 1998;108:8799–8803. [Google Scholar]
- Qian X, Schlick T. Efficient multiple-timestep integrators with distance-based force splitting for particle-mesh-Ewald molecular dynamics simulations. Journal of Chemical Physics. 2002;116:5971–5983. [Google Scholar]
- Radhakrishnan R, Arora K, Wang Y, Beard WA, Wilson SH, Schlick T. Regulation of DNA repair fidelity by molecular checkpoints: ‘Gates’ in DNA polymerase β’s substrate selection. Biochemistry. 2006;45:15142–15156. doi: 10.1021/bi061353z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R, Schlick T. Orchestration of cooperative events in DNA synthesis and repair mechanism unraveled by transition path sampling of DNA polymerase β’s closing. Proceedings of the National Academy of Sciences, USA. 2004;101:5970–5975. doi: 10.1073/pnas.0308585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R, Schlick T. Fidelity discrimination in DNA polymerase β: differing closing profiles for a mismatched G:A versus matched G:C base pair. Journal of the American Chemical Society. 2005;127:13245–13252. doi: 10.1021/ja052623o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R, Schlick T. Correct and incorrect nucleotide incorporation pathways in DNA polymerase β’s. Biochemical and Biophysics Research Communication. 2006;350:521–529. doi: 10.1016/j.bbrc.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Stillinger FH. Molecular dynamics study of liquid water. Journal of Chemical Physics. 1971;55:3336–3359. [Google Scholar]
- Rahman A, Stillinger FH. Improved simulation of liquid water by molecular dynamics. Journal of Chemical Physics. 1974;60:1545–1557. [Google Scholar]
- Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D. Structure prediction for CASP8 with all-atom refinement using rosetta. Proteins Supplement. 2009;9:89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the ‘30-nm’ chromatin fiber: Evidence for a compact, interdigitated structure. Proceedings of the National Academy of Sciences, USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe DR, Okur A, Wickstrom L, Hornak V, Simmerling C. Secondary structure bias in generalized born solvent models: comparison of conformational ensembles and free energy of solvent polarization from explicit and implicit solvation. Journal of Physical Chemistry B. 2007;111:1846–1857. doi: 10.1021/jp066831u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitberg AE, Okur A, Simmerling C. Coupling of replica exchange simulations to a non-Boltzmann structure reservoir. Journal of Physical Chemistry B. 2007;111:2415–2418. doi: 10.1021/jp068335b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- Saito M. Molecular dynamics simulations of proteins in water without the truncation of long-range Coulomb interactions. Molecular Simulation. 1992;8:321–333. [Google Scholar]
- Sandak B. Multiscale fast summation of long-range charge and dipolar interactions. Journal of Computational Chemistry. 2001;22:717–731. [Google Scholar]
- Schaefer HF. Methylene: A paradigm for computational quantum chemistry. Science. 1986;231:1100–1107. doi: 10.1126/science.231.4742.1100. [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- Schames JR, Henchman RH, Siegel JS, Sotriffer CA, Ni H, McCammon JA. Discovery of a novel binding trench in HIV integrase. Journal of Medical Chemistry. 2004;47:1879–1881. doi: 10.1021/jm0341913. [DOI] [PubMed] [Google Scholar]
- Schlick T. From macroscopic to mesoscopic models of chromatin folding. In: Fish J, editor. Bridging The Scales in Science in Engineering. New York: Oxford University Press; 2009a. pp. 514–535. [Google Scholar]
- Schlick T. Monte Carlo, harmonic approximation, and coarse-graining approaches for enhanced sampling of biomolecular structure. F1000 Biology Reports. 2009b;1:48. doi: 10.3410/B1-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T. Molecular-dynamics based approaches for enhanced sampling of long-time, large-scale conformational changes in biomolecules. F1000 Biology Reports. 2009c;1:51. doi: 10.3410/B1-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T. Molecular Odeling : An Interdisciplinary Guide. 2. New York: Springer-Verlag; 2010. [Google Scholar]
- Schlick T, Barth E, Mandziuk M. Biomolecular dynamics at long timesteps: bridging the timescale gap between simulation and experimentation. Annual Review of Biophysics and Biomolecular Structure. 1997;26:179–220. doi: 10.1146/annurev.biophys.26.1.181. [DOI] [PubMed] [Google Scholar]
- Schlick T, Mandziuk M, Skeel R, Srinivas K. Nonlinear resonance artifacts in molecular dynamics simulations. Journal of Computational Physics. 1998;139:1–29. [Google Scholar]
- Schlick T, Perisic O. Mesoscale simulations of two nucleosome-repeat length oligonucleosomes. Physical Chemistry Chemical Physics. 2009;11:10729–10737. doi: 10.1039/b918629h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T, Skeel RD, Brünger AT, Kalé LV, Board JA, Jr, Hermans J, Schulten K. Algorithmic challenges in computational molecular biophysics. Journal of Computational Physics. 1999;151:9–48. [Google Scholar]
- Schnabel J. The dark side of proteins. Nature. 2010;464:828–829. doi: 10.1038/464828a. [DOI] [PubMed] [Google Scholar]
- Schröder G, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Sali A, Honig B, Levitt M, Berman HM, Jones D, Brenner SE, Burley SK, Das R, Dokholyan NV, Dunbrack RL, Jr, Fidelis K, Fiser A, Godzik A, Huang YJ, Humblet C, Jacobson MP, Joachimiak A, Krystek SR, Jr, Kortemme T, Kryshtafovych A, Montelione GT, Moult J, Murray D, Sanchez R, Sosnick TR, Standley DM, Stouch T, Vajda S, Vasquez M, Westbrook JD, Wilson IA. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17:151–159. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ScienceDaily. Computer Game’s High Score Could Earn The Nobel Prize In Medicine. 2008. May 9, [Google Scholar]
- Scott WR, Schiffer CA. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Structure. 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]
- Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, Eastwood MP, Gagliardo J, Grossman J, Ho CR, Ierardi DJ, Kolossváry I, Klepeis JL, Layman T, McLeavey C, Moraes MA, Mueller R, Priest EC, Shan Y, Spengler J, Theobald M, Towles B, Wang SC. Anton: a special-purpose machine for molecular dynamics simulation. Proceedings of the 34th annual international symposium on Computer architecture; ACM, San Diego, CA. 2007. pp. 1–12. [Google Scholar]
- Shaw DE, Dror RO, Salmon JK, Grossman JP, Mackenzie KM, Bank JA, Young C, Deneroff MM, Batson B, Bowers KJ, Chow E, Eastwood MP, Ierardi DJ, Klepeis JL, Kuskin JS, Larson RH, Lindorff-Larsen K, Maragakis P, Moraes MA, Piana S, Shan Y, Towles B. Millisecond-scale molecular dynamics simulations on anton. SC ‘09: Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis; ACM, San Diego, CA. 2009. pp. 1–11. [Google Scholar]
- Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- Sheffler W, Baker D. RosettaHoles : rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Science. 2008;18:229–239. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirts M, Pande V. Screen savers of the World Unite! Science. 2000;290:1903–1904. doi: 10.1126/science.290.5498.1903. [DOI] [PubMed] [Google Scholar]
- Simmerling C, Strockbine B, Roitberg AE. All-atom structure prediction and folding simulations of a stable protein. Journal of the American Chemical Society. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- Skeel RD, Tezcan I, Hardy DJ. Multiple grid methods for classical molecular dynamics. Journal of Computational Chemistry. 2002;23:673–684. doi: 10.1002/jcc.10072. [DOI] [PubMed] [Google Scholar]
- Snir M. A note on N-body computations with cutoffs. Theory of Computing Systems. 2004;37:295–318. [Google Scholar]
- Snow CD, Nguyen H, Pande VS, Gruebele M. Absolute comparison of simulated and experimental protein folding dynamics. Nature. 2002;420:102–106. doi: 10.1038/nature01160. [DOI] [PubMed] [Google Scholar]
- Stehr R, Kepper N, Rippe K, Wedemann G. The effect of the internucleosomal interaction potential on the folding of the chromatin fiber. Biophysics Journal. 2008;95:3677–3691. doi: 10.1529/biophysj.107.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart SJ, Zhou R, Berne BJ. Molecular dynamics with multiple time scales : the selection of efficient reference system propagators. Journal of Chemical Physics. 1996;105:1426–1436. [Google Scholar]
- Sugita Y, Okamoto Y. Replica-exchange molecular dynamics methods for protein folding. Chemical Physics Letters. 1999;314:141–151. [Google Scholar]
- Sun J, Zhang Q, Schlick T. Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proceedings of the National Academy of Sciences, USA. 2005;102:8180–8185. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet CR, Petrine P, Pande VS, Izaguirre JA. Normal mode partitioning of Langevin dynamics for biomolecules. Journal of Chemical Physics. 2008;128:145101. doi: 10.1063/1.2883966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizaki S, Clifford J, Connelly BD, Feig M. Conformational sampling of peptides in cellular environments. Biophysics Journal. 2008;94:747–759. doi: 10.1529/biophysj.107.116236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Deshayes S, Decaffmeyer M, Eyck MV, Charloteaux B, Brasseur R. PepLook: An Innovative in Silico Tool for Determination of Structure, Polymorphism and Stability of Peptides. New York: Springer- Verlag; 2009. [DOI] [PubMed] [Google Scholar]
- Tozzini V, McCammon JA. A coarse grained model for the dynamics of flap opening in HIV-1 protease. Chemical Physics Letters. 2005;413:123–128. [Google Scholar]
- Tremethick DJ. Higher-order structures of chromatin: The elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Tsui V, Radhakrishnan I, Wright PE, Case DA. NMR and molecular dynamics studies of the hydration of a zinc finger-DNA complex. Journal of Molecular Biology. 2000;302:1101–1117. doi: 10.1006/jmbi.2000.4108. [DOI] [PubMed] [Google Scholar]
- van Holde K, Zlatanova J. Chromatin fiber structure, where is the problem now? Seminars in Cell and Developmental Biology. 2007;18:651–658. doi: 10.1016/j.semcdb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Vasquez V, Sotomayor M, Cordero-Morales J, Schulten K, Perozo E. A structural mechanism for MscS gating in lipid bilayers. Science. 2008;321:1210–1214. doi: 10.1126/science.1159674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz VA, Bowman GR, Beauchamp K, Pande VS. Molecular simulation of Ab initio protein folding for a millisecond folder NTL9(1–39) Journal of the American Chemical Society. 2010;132:1526–1528. doi: 10.1021/ja9090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general AMBER force field. Journal of Computational Chemistry. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Wang W, Donini O, Reyes CM, Kollman PA. Biomolecular simulations: recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid non-covalent interactions. Annual Review of Biophysics and Biomolecular Structure. 2001;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- Warshel A, Levitt M. Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of carbonium ion in the reaction of lysozyme. Journal of Molecular Biology. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- Warshel A, Russell ST. Calculations of electrostatic interactions in biological systems and in solutions. Quarterly Review of Biophysics. 1984;17:283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olson MHM. Electrostatic basis for enzyme catalysis. Chemical Reviews. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- Washburn J. University Inc : The Corporate Corruption of Higher Education. New York: Basic Books; 2005. [Google Scholar]
- Weber IT, Miller M, Jaskolski M, Leis J, Skalka AM, Wlodawer A. Molecular modeling of the HIV-1 protease and its substrate binding site. Science. 1989;243:928–931. doi: 10.1126/science.2537531. [DOI] [PubMed] [Google Scholar]
- Wensley BG, Batey S, Bone FA, Chan ZM, Tumelty NR, Steward A, Kwa LG, Borgia A, Clarke J. Experimental evidence for a frustrated energy landscape in a three-helix-bundle protein family. Nature. 2010;463:685–688. doi: 10.1038/nature08743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Miller M, Jaskolski M, Sathyanarayana BK, Baldwin E, Weber IT, Selk LM, Clawson L, Schneider J, Kent SB. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- Wolynes PG. Recent successes of the energy landscape theory of protein folding and function. Quarterly Review of Biophysics. 2005;38:405–410. doi: 10.1017/S0033583505004075. [DOI] [PubMed] [Google Scholar]
- Wong H, Victor JM, Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS ONE. 2007;2:e877. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, Grueble M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- York D, Yang W. The fast Fourier Poisson method for calculating Ewald sums. Journal of Chemical Physics. 1994;101:3298–3300. [Google Scholar]
- Young MA, Beveridge DL. Molecular dynamics simulations of an Oligonucleotide duplex with adenine tracts phased by a full helix turn. Journal of Molecular Biology. 1998;281:675–687. doi: 10.1006/jmbi.1998.1962. [DOI] [PubMed] [Google Scholar]
- Zagrovic B, Sorin EJ, Pande V. β-hairpin folding simulations in atomistic detail using an implicit solvent model. Journal of Molecular Biology. 2001;313:151–169. doi: 10.1006/jmbi.2001.5033. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Progress and challenges in protein structure prediction. Current Opinion in Structural Biology. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JZ. Structure-directed combinatorial library design. Current Opinion in Chemical Biology. 2008;12:379–385. doi: 10.1016/j.cbpa.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Zhou R, Harder E, Xu H, Berne BJ. Efficient multiple time step method for use with Ewald and particle mesh Ewald for large biomolecular systems. Journal of Chemical Physics. 2001;115:2348–2358. [Google Scholar]