Summary
To accommodate the large cells following zygote formation, early blastomeres employ modified cell divisions. Karyomeres are one such modification, a mitotic intermediate wherein individual chromatin masses are surrounded by nuclear envelope, which then fuse to form a single mononucleus. We identified brambleberry, a maternal-effect zebrafish mutant that disrupts karyomere fusion resulting in formation of multiple micronuclei. brambleberry is a previously unannotated gene homologous to Kar5p, which participates in nuclear fusion in yeast. We demonstrate that Brambleberry is required for pronuclear fusion following fertilization in zebrafish. As karyomeres form, Brambleberry localizes to the nuclear envelope with prominent puncta evident near karyomere-karyomere interfaces corresponding to membrane fusion sites. Our studies identify the first factor acting in karyomere fusion and suggest that specialized proteins are necessary for proper nuclear division in large dividing blastomeres.
Keywords: membrane fusion, nuclear assembly, maternal-effect, karyomere, brambleberry, karyokinesis, cleavage stage of development, blastomeres, nuclear envelope, fertilization, pronuclear fusion
INTRODUCTION
The nuclear envelope forms a physical barrier between the cytoplasm and the nucleoplasm. It consists of an outer and inner membrane and is perforated by nuclear pore complexes, which regulate the import and export of macromolecules critical for cell function (Hetzer, 2010). In vertebrates, the nuclear envelope breaks down at the onset of mitosis and then reassembles. Our current understanding of nuclear envelope breakdown and assembly during cell cycle progression is based on a combination of cell free nuclear assembly assays (Anderson and Hetzer, 2007; Burke and Gerace, 1986; Hetzer et al., 2001; Larijani and Poccia, 2009; Newport, 1987; Newport and Dunphy, 1992; Rotem et al., 2009) and cell culture studies (Anderson and Hetzer, 2008; Ellenberg et al., 1997; Lu et al., 2011). These studies have provided insight into the basic mechanisms of nuclear dynamics during the cell cycle. However, in animals, cells come in many shapes and sizes, with large cells presenting a particular challenge for cytokinesis and karyokinesis during cell division.
The large cells of early vertebrate embryos exhibit altered cell division dynamics during development. Eggs are endowed with large maternal stores of RNA, protein and organelles that are required for the earliest stages of development prior to the onset of zygotic transcription (Cummins, 2002; Telford et al., 1990). Thus, the eggs and one-cell stage embryos of many species are very large. For example human one-cell stage embryos have diameters of 120 um, and fertilized zebrafish and Xenopus eggs have diameters of 500 um and 1000um, respectively (Bownds et al., 2010; Griffin et al., 2006). Division mechanics of these early blastomeres in frogs and zebrafish are modified to accommodate their large size. For example, in smaller somatic cells microtubules of the mitotic spindle can scale relative to the cell’s size; however, intrinsic properties of microtubules limit their ultimate growth to 60um in length (Wuhr et al., 2008). Therefore adjustments to spindle and astral microtubule organization and mechanics are required to achieve proper cell division in these large blastomeres (Wuhr et al., 2010). Although the mechanism mediating these adjustments is not clear, recent studies have shown that astral microtubules interact with Dynein, anchored in uncharacterized “interaction zones” to provide local counteracting forces (Wuhr et al., 2010). In addition, at least during the earliest divisions, the centrosomes are significantly detached from the spindle poles during mitosis, potentially expanding the spindle reach in these large cells (Gard et al., 1995; Wuhr et al., 2008).
Specialized mechanics for early cell division are also evident in mammals. In mice, maternal loss of chromokinesin Kid reveals a requirement for this motor protein specifically during the cleavage stage of development for preventing multi-micronucleation of blastomeres (Ohsugi et al., 2008). Through its DNA binding and kinesin motor functions Kid is proposed to promote axial compaction of chromosomes during anaphase. Maximum axial compaction of separating chromosomes normally occurs early in anaphase (Mora-Bermudez et al., 2007). Due to a failure of chromosomes to form compact masses in mutant embryos, it is suggested that the nuclear envelope can form around individual chromosomes ultimately resulting in blastomeres with multiple, micronuclei (Ohsugi et al., 2008). Thus, the anaphase chromosomes and/or cytoplasm of the large early developing mouse cells have a propensity to assemble nuclear envelopes around separate chromatin bodies. Therefore specialized mechanisms and molecules counteract this tendency and promote the formation of a mononucleus (Ohsugi et al., 2008).
Similarly, human embryos have an inherent propensity to form multiple, micronuclei following in vitro fertilization. Clinical studies have demonstrated that prescreening embryos to eliminate ones with multi-micronuclei following in vitro fertilization increases the rate of successful pregnancies (Balakier and Cadesky, 1997). Other reports show that knockdown of the DNA polymerase polD4 subunit in human cells results in the formation of multiple micronuclei (Huang et al., 2010).
Karyomeres are intermediate nuclear structures that normally form in cleavage stage blastomeres near the end of mitosis due to individual or groups of chromosomes becoming entirely enclosed by nuclear envelope. These nuclear bodies ultimately fuse to form a mononucleus. Such nuclear bodies do not appear to be present in later somatic cells and are likely required to accommodate the unique features of cleavage-stage embryos. The molecules required for regulating the activities of karyomere formation and fusion are entirely unknown. Karyomeres have been described in a number of organisms and in some instances, such as in sea urchins, karyomere assembly and fusion have been described ultrastructurally (Longo, 1972). More recently karyomeres were described in fish (Schoft et al., 2003), leech (Fernandez and Olea, 1995), and frog (Montag et al., 1988), where it was shown that DNA replication can initiate within karyomeres (Lemaitre et al., 1998). Karyomere formation and fusion have been described in the early rabbit embryo as well (Gulyas, 1972). It is not clear to what extent karyomeres play a role in nuclear assembly in other mammalian embryos.
Employing a molecular genetics approach, we identified Brambleberry (Bmb) as a protein required for karyomere fusion in the developing zebrafish embryo. In bmb maternal-effect mutant embryos, karyomeres persist during interphase resulting in multi-micronuclei formation. We positionally cloned the bmb mutant gene and identified it as an unannotated gene found in vertebrates and invertebrates, bearing 27% similarity to the yeast nuclear membrane fusion protein, Kar5p. We show that Bmb protein localization is dynamic. During metaphase Bmb is localized near the mitotic spindle region and its localization shifts to the chromosomes as they reach the end of the spindle. During karyomere fusion Bmb is found in prominent puncta, mainly at karyomere-karyomere interfaces corresponding to putative fusion sites. We also demonstrate that bmb is required for pronuclear fusion in zygote formation. Our results support the hypothesis that specialized proteins are necessary for proper nuclear division in large dividing blastomeres.
RESULTS
brambleberry is required for early development and normal nuclear morphology
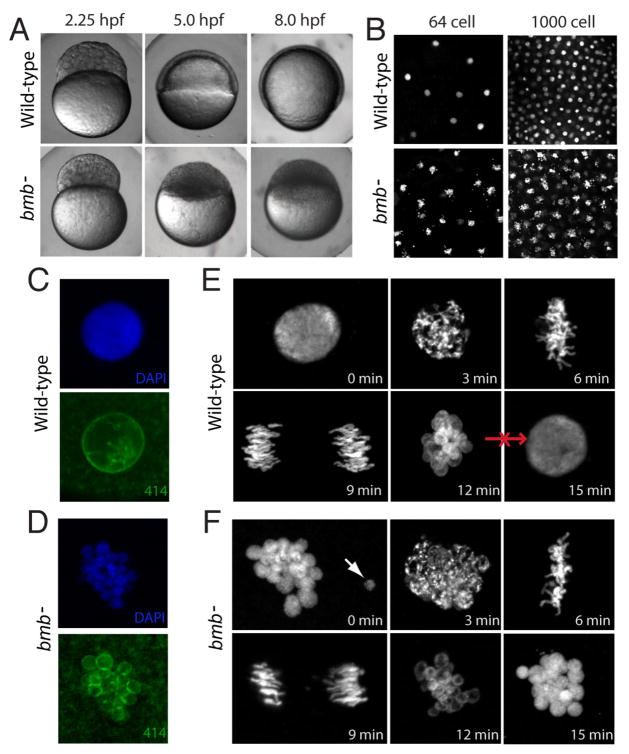
We performed a chemically-induced mutagenesis screen to identify maternal-effect mutants that specifically affect the cleavage stage in early zebrafish development (to be published elsewhere). Embryos derived from bmb mutant mothers (henceforth referred to as bmb embryos for simplicity) arrest development shortly after the mid-blastula transition (MBT; Figure 1A), a period corresponding to an important shift from maternal to zygotic control (Newport and Kirschner, 1982). We found that the cell cycle rate of bmb mutants is similar to that of WT prior to the MBT (data not shown). DAPI staining of fixed embryos selected for interphase at three time points during cleavage (2-, 64-, and 1000-cell stage embryos) revealed that all blastomeres of bmb embryos throughout this period contained morphologically abnormal nuclei that appeared fragmented (Figure 1B; data not shown). High resolution imaging of individual nuclei stained with DAPI in combination with the nuclear envelope marker, mab414, demonstrated that the abnormal nuclear morphology of bmb mutants is due to chromatin bodies that are separated from each other and are each associated with a nuclear envelope (compare Figure 1C with 1D). This result demonstrates that bmb nuclei are multi-micronucleated. Based on the resemblance of the nuclear morphology to the brambleberry or blackberry, we named this mutant gene brambleberry (bmb). The bmb nuclear morphology defect is 100% penetrant, strictly recessive-maternal and is uniform during the cleavage period.
Figure 1. bmb is required for early development and proper nuclear morphology.
(A) Prior to the MBT (2.25 hpf), embryos from bmb mutant females are morphologically similar to WT embryos. After the MBT at 5.0 and 8.0 hpf, bmb mutants fail to undergo cell movements associated with epiboly and gastrulation. (B) DAPI staining of interphase nuclei during cleavage in WT (top) and bmb− (bottom) reveals multiple chromatin bodies associated with each nucleus. (C) WT and (D) bmb− interphase nuclei stained with mab414 (green) indicate that bmb− nuclei are multi-micronucleated compared to WT. (E) WT and (F) bmb− cell cycle time course at the 2 to 4 cell transition (images are projections of multiple confocal Z-slices). Top left –interphase; Top middle- prophase, Top right-metaphase, bottom left -anaphase, bottom middle- telophase, bottom right-interphase. In E and F for each time point, n=3. In this and subsequent figures ‘n’ refers to number of embryos examined (unless otherwise noted). In each case multiple nuclei or cells of each embryo were also examined.
Next we compared distinct cell cycle transition points in bmb and WT embryos at the 2- to 4-cell stage to further investigate the nuclear defect in bmb mutant embryos. Embryos from synchronously developing clutches were fixed at three-minute intervals over a complete cell cycle and stained with DAPI to visualize the chromatin. Figures 1E and 1F show the basic stages of mitosis in WT and bmb respectively, beginning (0 min) and ending at interphase (15 min). Despite the altered nuclear morphology, bmb chromatin appeared to condense normally (compare Figure 1E, 3min with 1F, 3min), and progressed to metaphase (6 min) and anaphase (9 min). Occasionally, in bmb mutants individual chromatin bodies were found separated from the group (Figure 1F, 0 min., arrow) or chromosomes were misaligned at the metaphase plate or during anaphase (data not shown). Strikingly, the WT chromatin arrangement during telophase (Figure 1E, 12 min) resembles that of the bmb interphase arrangement (compare to Figure 1F, 15 min), suggesting that the telophase to interphase transition in bmb mutants may be disrupted, as an intact mononucleus never forms.
bmb is required for karyomere fusion
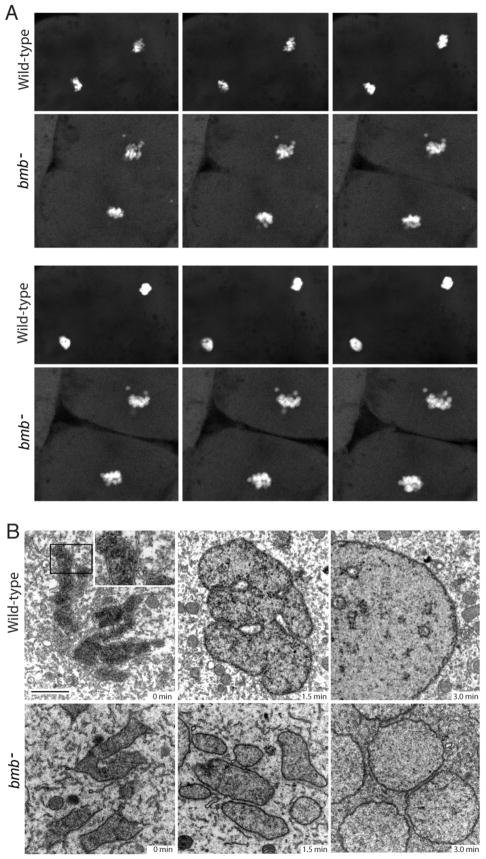
We hypothesized that the bmb phenotype is a defect in karyomere fusion. Karyomeres are intermediate cleavage stage structures of individual or groups of chromosomes enclosed by nuclear envelope, which fuse to form a mononucleus. To begin to examine karyomere dynamics in both WT and bmb embryos in real time, we performed time-lapse confocal microscopy. The WT experiment initiates during mitosis when distinct chromatin bodies are evident in an arrangement resembling the bmb phenotype (Figure 2A, Movie S1). As mitosis concludes in WT, chromatin bodies coalesced to form an intact mononucleus (Figure 2A, Movie S1). The chromosomes in the bmb mutant transitioned through the chromatin arrangement observed in WT but failed to ultimately coalesce and form an intact mononucleus as in WT (Figure 2A, Movie S2).
Figure 2. Nuclear membrane fusion is disrupted in bmb mutants.
(A) Frames from time-lapse experiments at the telophase-interphase transition demonstrate that chromatin bodies normally coalesce in WT (top) but fail to do so in bmb− (bottom). Note: 6 frames were selected (from Movie S1 and S2 to best align the sequence of events between WT and bmb− at the telophase to interphase transition. (B) Electron microscopy of WT versus bmb− at the telophase-interphase transition. Embryos at the 128-cell stage were fixed at 90-second intervals for TEM. Black bar = 2 microns. The inset in WT (0 min) is enlarged 2X to show the double membrane nuclear envelope. For each time point n=2 embryos. Multiple cells from each embryo were examined in both WT and bmb−.
Next we examined the telophase to interphase transition in WT and bmb mutants at the ultrastructural level. At a time point designated as 0 minutes, condensed WT chromosomes are each encased by a characteristic double membrane nuclear envelope, as karyomeres (Figure 2B top left; inset). After 1.5 minutes, the karyomeres in WT appear to have fused to form a lobular nuclear structure (Figure 2B top center). At the 3.0-minute time point, a round mononucleus has formed with some internal membranes present (Figure 2B top right). At the 0-minute time point in bmb mutants, chromosomes are enclosed in membrane similar to WT (Figure 2B bottom left). After 1.5 minutes separated chromosomes enclosed in nuclear envelope persist in bmb mutants (Figure 2B bottom center). At the 3.0-minute time point multiple micronuclei are present. Multiple micronuclei with no fusion intermediates or connectors of any type were found in all bmb− interphase cells examined (n=12). These data suggest that multiple micronuclei form in bmb− due to a failure in karyomere fusion.
bmb encodes a protein similar to yeast Kar5p
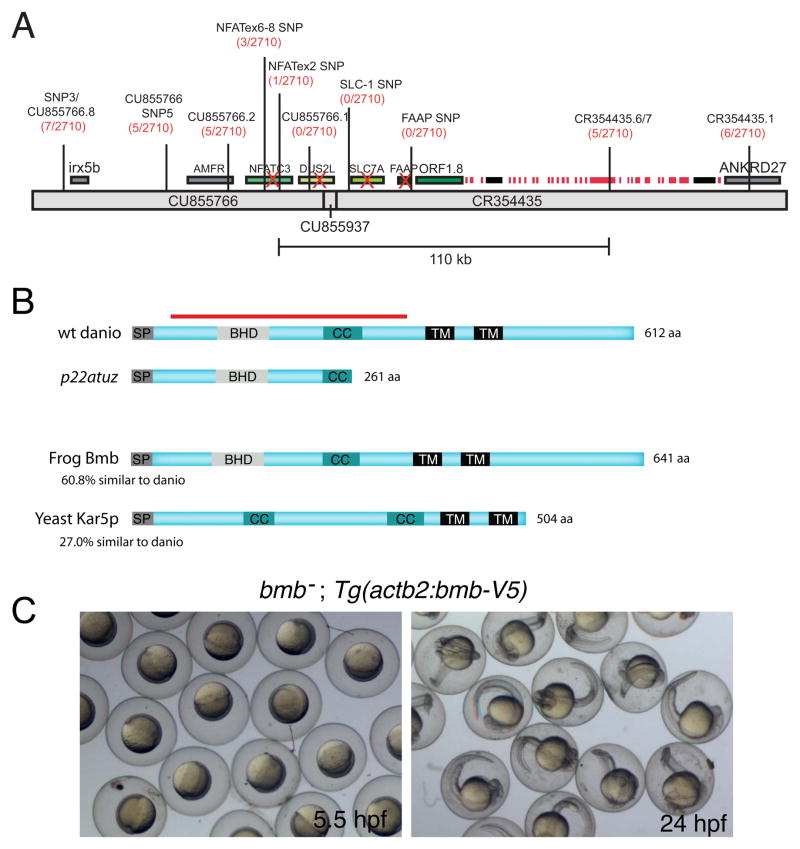
To identify the bmb gene, we mapped the bmb mutation to chromosome 25 through bulk segregation analysis (Knapik et al., 1996). We then narrowed the physical interval containing bmb by meiotic recombination to a 110 kb region (Figure 3A). All of the unique annotated genes within this interval were completely sequenced and none contained mutations within their open reading frames (ORFs).
Figure 3. bmb encodes a conserved novel predicted transmembrane protein.
(A) Recombinants over total meiotic events are indicated parenthetically in red below the marker name. Candidate genes eliminated based on their WT cDNA sequence are indicated with a red X. A histone gene cluster is indicated as red boxes. Two predicted transposable elements are indicated by black boxes. (B) ORF1.8 encodes a novel 612 residue protein with a predicted N-terminal signal peptide sequence (SP) followed by a Bmb Homology Domain (BHD), a coiled-coil (CC) and two C-terminal transmembrane domains (TM) initiating at amino acid residues 363 and 415 (TMpred program) (Hofmann and Stoffel, 1993). Yeast Kar5p is 27% similar (excluding the C-terminal region) to the D. rerio homolog. The red bar corresponds to the recombinant protein used to generate Bmb antibodies. (C) Rescued bmb− embryos at 50% epiboly (5.5 hpf) and 24 hpf. See also Figure S1.
Next we searched for unannotated genes in the interval and identified a 1839 bp ORF (ORF1.8) contained in 12 predicted exons (Figure S1). ORF1.8 overlaps with an EST (wu:fi04f09), containing both 5′ and 3′ UTR sequence, which along with ORF1.8 predict a 2.1 kb novel gene. We identified within this gene a single mutation in the splice acceptor site of exon 6 (Figure S1A), altering it from AG to TG. As a consequence an inappropriate AG 10 nucleotides downstream is used that introduces a premature stop codon 10 bases further downstream within the ORF (Figure S1A).
To determine if ORF1.8 corresponds to the bmb gene, we generated a transgene carrying ORF1.8 with a V5 epitope fused to its C-terminus (Tg(actb2:bmb-V5)). The transgene rescued the bmb multi-micronuclear mutant phenotype (Figure S3A, B), and fully rescued embryo development to 24 hpf (Figure 3C). Thus, ORF1.8 is the bmb gene. WT bmb encodes a 612 residue novel protein with a predicted N-terminal signal peptide and two downstream transmembrane domains (Figure 3B). In addition, a domain search program (COILS) (Lupas et al., 1991) predicts a coiled-coil domain spanning residues 247 through 274 (Figure 3B). The corresponding bmbp22atuz cDNA encodes a predicted 261 residue truncated protein lacking both transmembrane domains and part of the coiled-coil domain (Figure 3B). An NCBI BLAST search revealed predicted Bmb homologs encoded in the genomes of other vertebrate and invertebrate species (data not shown). Alignment of the D. rerio and X. tropicalis bmb genes reveals 61% similarity at the amino acid level (Figure 3B). Interestingly, there is strong conservation within a stretch of 69 residues in the N-terminal region of the Bmb protein among a diverse set of species (Figure S1B). We refer to this domain as the Bmb Homology Domain (BHD), as it is not found in other characterized proteins to date.
A Position Specific Iterated BLAST (PSI-BLAST) search, which can identify more distantly related proteins, revealed Saccharomyces cerevisiae Kar5p, a protein with 27% similarity to bmb. Interestingly, Kar5p is required for nuclear fusion during yeast mating (Beh et al., 1997). Similar to Kar5p, frog and fish Bmb contain an N-terminal coiled-coil domain, two C-terminal transmembrane domains (Figure 3C) and are highly basic at the C-terminus (data not shown). Kar5p has an additional coiled-coil domain (Figure 3B). Kar5p also lacks the BHD and compared to Bmb lacks a significant portion of the C-terminus beyond the second transmembrane domain (Figure 3B), raising the possibility that these domains have a karyomere-specific function.
Bmb is dynamically localized during cell cycle progression at the cleavage stage
To begin to understand Bmb’s role in nuclear envelope dynamics during embryonic development, we investigated its subcellular localization. In WT interphase nuclei, Bmb localized to the nuclear periphery and internal membranes (Figure S1C, D, 5H). In contrast, we failed to detect any signal in bmb mutant embryos (Figure S1E, F). We examined Bmb localization during mitosis at the 32- to 64-cell transition of the cleavage stage. At prophase Bmb is detected at the nuclear periphery (Figure S2A, Bmb, arrowheads). During prometaphase Bmb becomes highly concentrated in the mitotic spindle region near the chromosomes (Figure S2B–D). At metaphase Bmb localizes in the vicinity of the mitotic spindle marked with anti-alpha tubulin antibody and is reduced at the metaphase plate (Figure 4A, 0 minutes; Figure S2E). One minute later, now in anaphase, Bmb protein continues to be enriched in the region of the spindle, with Bmb foci intermingled with, although not yet assembled on, the separating chromosomes (Figure 4B, inset). Bmb foci then assemble along the separating chromosomes (Figure 4C), and by the 2-minute time point, Bmb localization shifts mainly onto the separated chromosomes (Figure 4D). We conclude that Bmb is sequestered near the mitotic spindle during metaphase and is rapidly localized to the chromosomes during their separation (Figure 4E). These early-formed (elongated) karyomeres continue to travel a significant distance to reach their ultimate central cellular positions (Figure 4D, E; 5B), indicating that membrane-enclosed chromosomes can be transported to their final destination.
Figure 4. Bmb protein dynamics during mitosis.
(A) Bmb protein localizes as distinct foci to the mitotic spindle region during metaphase (n=4). (B) During anaphase Bmb is interspersed between the separating chromosomes (inset; n=3) and in the region of the mitotic spindle. (C) Later in anaphase, Bmb foci are decorating the chromosomes (inset; n=3). (D) At the 2 min time point Bmb surrounds the individual chromosomes, which are still separating (n=5). (E) Schematic summarizing Bmb localization during metaphase to anaphase/telophase. Arrows in the right panel indicate that the karyomeres will continue to move to their final central position in the cell. 0 min corresponds to metaphase at the 32-to-64 cell transition. Bmb (green), microtubules (DM1a, red), chromosomes (DAPI, white and blue in merge). Scale bar = 5 μm and insets show 2X enlargements. See also Figure S2.
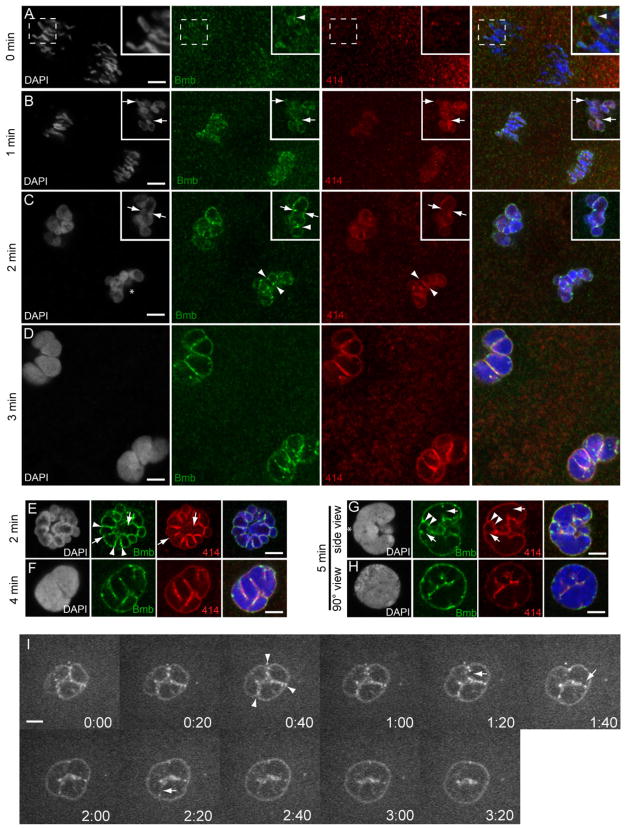
Figure 5. Bmb foci mark sites corresponding to pre-, post- and active karyomere fusion.
(A) At time point designated 0 min (mid-anaphase) at the 32- to 64-cell transition, Bmb protein (green) is first seen accumulating at the leading edge of the separating chromosomes (inset, arrowhead) prior to the assembly of nucleoporins (marked by mab414, red). (B) As the chromosomes continue to separate, nuclear membrane (marked by mab414) and Bmb surround individual chromosomes forming karyomeres. Insets in B show a more advanced example from the same time point (1 min). (C) Karyomeres become increasingly more spherical and prominent Bmb foci are more apparent at karyomere-karyomere interfaces (arrowheads). In another example, Bmb foci span a former interface, which now lacks nuclear membrane (inset, arrowheads) and a prominent Bmb aggregate interfaces adjacent karyomeres (inset, arrowhead). (D) At the 3 min time point larger secondary karyomeres are present. (E) Another 2 min example at a 90° view compared to C (an asterisk in C marks reference point). Bmb foci at karyomere-karyomere interfaces, where membrane is still present (arrowheads) or partially/completely absent (arrows). (F) Secondary karyomere formation at a 45° view. ( G) Bmb foci (Bmb, arrowheads) flank presumptive karyomere pre-fusion site (membrane still present- 414, arrowheads) and Bmb foci can be found at presumptive karyomere post-fusion sites (lack of membrane- 414, arrows). Note G–H shows mononucleus formation (5 min) from a lateral side view (G) and another sample from a 90° vantage point (reference point marked with asterisk in G) (H). All panels correspond to individual confocal Z-slices. Scale bar = 5 μm and insets show 2X enlargements. For each time point n ≥ 3. (I) Time-lapse imaging of Bmb-venus. Bmb foci are seen at the karyomere-karyomere interfaces (arrowheads) and are ultimately eliminated (arrows) as the mononucleus is formed. See also Movie S3 and Figure S3 for details.
Bmb protein localizes to pre-, post- and active karyomere fusion sites
Since Bmb is implicated in nuclear membrane fusion, we examined Bmb protein localization during karyomere fusion. During anaphase prior to karyomere formation (t= 0 minutes), Bmb protein is found in puncta (possibly vesicles) in close proximity with the separating sister chromatids (Figure 5A, Bmb). In some instances, Bmb puncta begin to associate with chromosomes at the leading edge (Figure 5A, merge inset, arrowhead). At this point, mab414 positive nucleoporins of the nuclear envelope are not yet assembled on chromosomes (Figure 5A, merge inset). At the next time point (1 min) Bmb is strongly associated with the separating chromosomes, with numerous Bmb foci evident (Figure 5B, Bmb). Nucleoporin positive chromosomal staining is now also detected (Figure 5B, 414). In a few embryos from the same time point, more advanced karyomere development can be detected (Figure 5B, Bmb, 414, inset): chromosomes are fully contained in nuclear envelope, significantly less elongated, and more oval in shape. In many instances tear-shaped karyomeres (Figure 5B, inset, arrows) are seen, which may represent a transitional state between elongated chromosomes to a more spherical shaped body.
Progressive stages of karyomere fusion can be found at the 2- and 3-minute time points. At both the 2-minute (Figure 5C, E) and 3-minute (Figure 5D) time points potential pre-karyomere and post-karyomere fusion events are represented. In some instances loosely associating karyomeres have a prominent Bmb punctum at their interface (Figure 5C, Bmb, inset, arrowhead). More tightly associated (pre-fusion) neighboring karyomeres that still have membrane between them (Figure 5C, 414, arrowheads) are frequently flanked by prominent Bmb foci (Figure 5C, Bmb, arrowheads). A view perpendicular to the division axis reveals multiple Bmb positive puncta along the karyomere-karyomere interface (Figure 5E, Bmb arrowheads). Adjacent karyomeres that appear to have most of the common membrane missing (likely fusion intermediates) also have prominent Bmb foci in the immediate vicinity (Figure 5C, inset arrows, Bmb and 414; 5E, Bmb and 414, arrows). At the 3-minute time point larger secondary karyomeres, also described in sea urchin (Longo, 1972), are detected, which presumably result from fusion of smaller primary karyomeres (Figure 5D, note that 5D is the same magnification as C). In embryo samples from the 4-minute time point, individual karyomeres are less recognizable (Figure 5F).
At the 5-minute time point a mostly contiguous round nucleus has formed (Figure 5H). Samples imaged perpendicular to the division plane (n=8) indicates a distinct polarity within the resulting nucleus along the dividing axis (Figure 5G). Prominent Bmb foci persist at putative post-fusion sites (Figure 5G, Bmb, arrows). Each mononucleus is formed with a significant amount of internal membrane, the so-called nuclear reticulum (Malhas et al., 2011), which stains with both mab414 and anti-Bmb (Figure 5G–H). These data indicate that Bmb foci correlate with points of pre-, post- and active karyomere membrane fusion during the cleavage stage of development.
We confirmed Bmb subcellular localization by staining transgenic Bmb-V5 embryos at distinct points of the cell cycle with both anti-V5 and anti-Bmb. Interphase nuclei from bmb−; Tg(actb2:bmb-V5) were indistinguishable from bmb−/+; Tg(actb2:bmb-V5) nuclei (Figure S3, compare A and B). Anti-V5 (green) and anti-Bmb (red) colocalize during metaphase and anaphase (Figure S3C, D). During telophase (karyomere stage), anti-Bmb foci colocalized with anti-V5 foci (Figure S3C, D, arrowheads).
To examine Bmb protein dynamics during karyomere fusion in real time, we injected one-cell stage bmb− embryos with bmb-venus mRNA. As early as the 64-to-128 cell transition, mutant embryos formed robust mononuclei, demonstrating that the Bmb-venus fusion protein can promote membrane fusion (Figure 5I; data not shown). Similar to fixed WT embryos (Figure 5C–F), Bmb positive foci were detected at karyomere-karyomere interfaces (Figure 5I, arrowheads), and more uniform staining was found at non-interface regions (Figure 5I; Movie S3). As in fixed samples karyomere-karyomere interfaces were more easily viewed 90° to the division axis. Time-lapse imaging at 20-second intervals demonstrates that Bmb foci begin to disappear near the mid-point of the karyomere-karyomere interface (Figure 5I, arrows). Loss of Bmb foci continues bidirectionally as karyomeres merge to form a mononucleus. Thus, live imaging demonstrates that Bmb foci are enriched at karyomere-karyomere interfaces during fusion and are also evident flanking post fusion sites.
Nuclear envelope assembles on elongated condensed chromosomes during cleavage
We next compared the dynamics of nuclear envelope assembly between cleavage and a mid-gastrula stage (8.0 hpf), which lacks karyomere intermediates. At mid-gastrulation, we found that the nuclear envelope is first evident surrounding a single chromatin mass (Figure S4D, arrows) and then later with the interphase nuclear periphery (Figure S4E). In contrast, during cleavage the nuclear envelope begins to assemble when the separating chromosomes are still elongated and appear condensed (Figure S4A, 5B). Furthermore during anaphase progression at 8.0 hpf, inter-chromosomal spacing is relatively absent (Figure S4B, C) compared to the cleavage stage (Figure S4A, 5A, B, 4B–D). Together, the data demonstrate that during the cleavage period, the nuclear envelope assembles on chromosomes that are in a more elongated, condensed state compared with later stages of embryonic development where membrane is assembled on a single, contiguous chromatin mass.
Bmb is required for pronuclear fusion
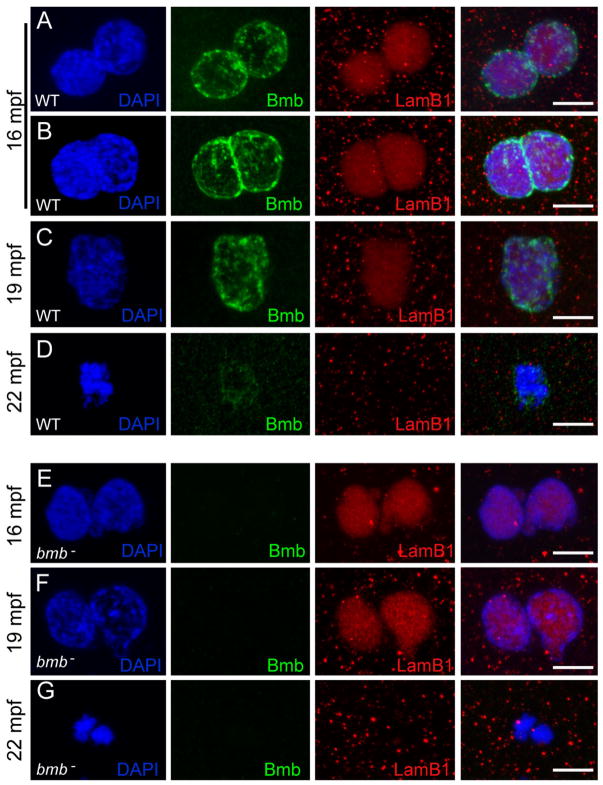
Since Bmb is homologous to Kar5p, we investigated its potential role in pronuclear fusion. In zebrafish, pronuclei normally fuse between 14–20 minutes post fertilization (mpf) (Dekens et al., 2003). Therefore, we performed a time course experiment of WT and bmb mutants spanning 10–25 mpf. In addition, we stained for Lamin B1 to follow the progression of nuclear fusion. As pronuclei congress in WT embryos, Bmb protein was detected in a punctate pattern throughout the nuclear periphery (Figure 7A; 16 mpf). As pronuclei begin to fuse, strong Bmb staining can be detected at the interface (Figure 7B; 16 mpf). Bmb protein is again seen throughout the nuclear periphery once fusion is complete (Figure 7C; 19mpf) and begins to disassociate from the chromatin during the first prometaphase (Figure 7D; 22 mpf). In bmb mutants pronuclei appeared to congress normally and were juxtaposed to one another at 16 mpf (Figure 7E). This configuration persists between 16–22 mpf (Figure 7F; data not shown). At 22 mpf the two DNA masses are found condensed separately (Figure 7F) and then ultimately unify on the metaphase plate at 25 mpf (data not shown).
Figure 7. Bmb is required for pronuclear fusion.
(A) At 16 mpf WT pronuclei are fully congressed. (B) Some WT pronuclei begin to fuse at 16 mpf. (C) At 19 mpf WT pronuclear fusion is complete. (D) At 22 mpf in WT embryos a single condensed DNA mass is detected at the first prometaphase. (E) At 16 mpf bmb− pronuclei are fully congressed. (F) At 19 mpf bmb− pronuclei fail to fuse as chromatin begins to condense. (G) At 22 min two separate condensed DNA masses are detected in bmb− embryos at the first prometaphase. Scale bars = 10 μm. At each time point n ≥ 4. Images correspond to Z-projections of individual confocal Z-planes. WT TL males were crossed to female bmb− mutants in E–G.
DISCUSSION
In this work we identify a novel nuclear membrane associated protein that is necessary for nuclear membrane fusion specifically during early embryonic development in zebrafish. The identification of Bmb and its role in nuclear membrane fusion illustrates the importance of examining nuclear assembly in the context of the developing animal at specific stages. Moreover, the bmb mutant exemplifies the power of forward genetic approaches in vertebrates to identify new factors acting in this process. Our results reveal the first molecular component required for this process, which appears specific for the nuclei of early, large dividing blastomeres of the embryo. bmb mutant males appear normal and are fertile. Thus, Bmb has a very specific function in facilitating membrane fusion, which is only required maternally during cleavage development.
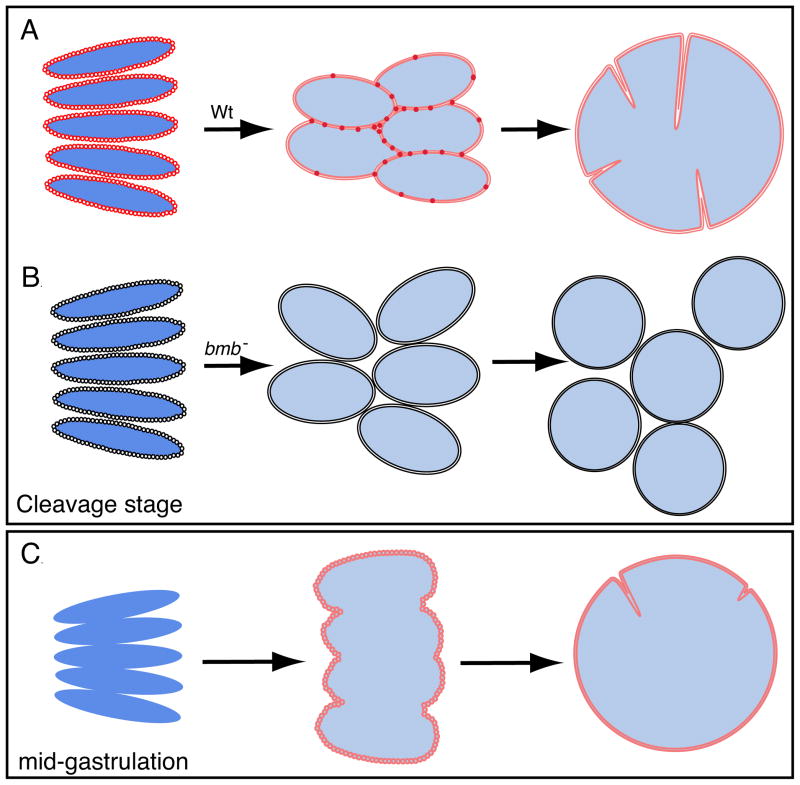
Figure 6 schematizes the WT and bmb telophase-to-interphase transition during the cleavage stage and later embryogenesis. At cleavage, Bmb puncta first localize to individual condensed (dark blue) chromosomes during mid to late anaphase. As the chromatin decondenses (light blue), Bmb foci accumulate at karyomere-karyomere interfaces and fusion ensues. In bmb mutants, karyomeres still form but cannot fuse, remain separated, and subsequently form multiple micronuclei. By mid-gastrulation, metaphase and anaphase chromosomes associate more closely and the nuclear envelope assembles around the entire decondensed chromatin mass instead of individual condensed chromosomes. Consequently, this results in the formation of a mononucleus directly.
Figure 6. Model of nuclear division during embryonic development.
(A) During cleavage, the nuclear envelope and Bmb assemble on individual chromosomes as they continue to separate in mitosis resulting in karyomeres forming. As elongated karyomeres transition into more spherical shaped bodies, Bmb puncta becomes more prevalent at the karyomere-karyomere interfaces and fusion ensues to form a mononucleus. (B) In bmb mutants karyomeres still form, however, the absence of Bmb leads to a failure in membrane fusion resulting in multiple micronuclei forming. (C) At mid-gastrulation Bmb protein and the nuclear envelope do not assemble on individual chromosomes, instead, they assemble on a single chromatin mass and karyomere fusion is not necessary.
We show that in the early embryo the nuclear envelope assembles around individual condensed, elongated chromosomes (Figure 2B and S4A). Intriguingly recent findings molecularly link chromatin decondensation to nuclear envelope assembly both in vitro and in vivo through the regulation of Aurora B kinase (Ramadan et al., 2007). Thus, it appears that factors during cleavage are present that allow nuclear envelope to assemble around condensed chromatin. Whether Aurora B is involved in this context and the exact relationship between nuclear envelope assembly and the level of chromosome condensation during cleavage remains to be determined. Nevertheless, the altered mode of nuclear assembly during cleavage could certainly influence karyomere formation by physically partitioning the chromosomes prior to forming a single chromatin mass.
Bmb encodes a highly dynamic putative nuclear envelope protein that is required for karyomere fusion
Bmb is predicted to be a transmembrane protein. Our immunofluorescence studies indicate that it associates with the nuclear envelope of karyomeres and the ensuing mononucleus. During most of mitosis, Bmb cytoplasmic puncta localize in the region of the mitotic spindle, suggesting that Bmb is a component of a nuclear membrane vesicle population that is either a component of, or independent of the ER. Interestingly, a novel inner nuclear membrane protein Samp1 from human cells, localizes to the mitotic spindle, thus defining a membrane domain associated with the spindle (Buch et al., 2009). It is not clear if Bmb is in contact with the spindle (directly or indirectly), which could serve to deliver Bmb-associated vesicles via the microtubules as the separated chromosomes reach the end of the spindle. Once karyomere structures are established, Bmb foci on the nuclear envelope correlate with pre-, post- and active fusion at karyomere-karyomere interfaces.
In bmb mutants multiple micronuclei continue to form uniformly at 3 hpf (Figure 1B). By 4 hpf, bmb mutant cells appear more mononuclear (unpublished), suggesting a possible reduced Bmb requirement at post cleavage stages. In WT at 4 hpf intermediate karyomere-like structures form with limited although not complete nuclear membrane partitioning of the chromosomes (unpublished). Low levels of Bmb are still present at mid-gastrulation (8 hpf) when karyomeres do not form. Interestingly, a ubiquitously expressed Bmb transgene does not affect viability or appear to affect normal nuclear dynamics throughout the life span of a WT fish.
Bmb is 27% homologous to and has a similar domain organization to yeast Kar5p (Figure 3B). Kar5p’s requirement for nuclear fusion during mating (Beh et al., 1997) is consistent with Bmb’s role in pronuclear and karyomere fusion. Interestingly, previous studies indicate that Kar5p is located largely in the periplasmic space of the nuclear membrane (Beh et al., 1997). Therefore Kar5p does not likely interact directly with apposing protein(s) on the distinct nuclei to promote nuclear fusion (Beh et al., 1997). Instead Kar5p may either influence the structure of the nuclear membrane or interact with local neighboring protein(s) to facilitate membrane fusion. Interestingly, small “lipid bridges” can be detected between unfused nuclei in kar5 zygotes in EM micrographs (Beh et al., 1997; Kurihara et al., 1994). This suggests that Kar5 may not be required for the early steps in nuclear fusion, but instead facilitates downstream events, such as expanding the fusion pore (Melloy et al., 2009). We did not detect membrane fusion intermediates at the EM level in bmb mutants, suggesting that Bmb may be necessary for earlier fusion events. Since Bmb has a C-terminal extension beyond the second presumptive transmembrane domain (108 residues in Danio and 137 residues in Tropicalis) not found in yeast, this portion of the protein may reflect a mechanistic distinction of Bmb’s versus Kar5p’s role in nuclear membrane fusion. Future experiments will be required to elucidate if Bmb’s role in membrane fusion is direct, mediated through protein-protein interactions, or more similar to Kar5p’s presumptive indirect role.
The requirement of Bmb for pronuclear fusion in zebrafish suggests functional conservation of the related protein over about 1.5 billion years. In yeast, Kar5p is required in both nuclear membranes for their fusion (Beh et al., 1997; Kurihara et al., 1994). Whether Bmb is similarly required in both pronuclei for fusion, or if Bmb in a single membrane is sufficient will be challenging to decipher. Examining a cross between a bmb−/− mutant female and a WT male (and vice versa) cannot test this mechanism, as shown in Figure 7. Here both pronuclei are void of the Bmb protein despite that the male pronucleus is WT. In the converse cross, WT female to bmb−/− male, pronuclear fusion is normal and Bmb is detected on both pronuclei (unpublished) despite the mutant male pronucleus. Although little is known about nuclear membrane biochemistry during fertilization in mammals, in sea urchins the male nuclear envelope breaks down almost completely and then re-forms with nuclear membrane components derived from the egg (Larijani and Poccia, 2009). This may also be the case in zebrafish. Thus, karyomere fusion and pronuclear fusion would be considered homotypic fusion events, as apposing membranes are of the same composition (Rothman and Warren, 1994).
The unilateral or bilateral role of Bmb in membrane fusion may be resolved through a combination of genetics and biochemistry. Karyomere formation has been demonstrated in vitro using early Xenopus embryonic extracts generated from G2 synchronized nuclei (Lemaitre et al., 1998). Developing such a system in zebrafish could allow the use of transgenic nuclear membrane markers in combination with bmb mutant and WT derived extracts. Such studies could ultimately test the question of whether Bmb is required in one or both apposing membranes during nuclear membrane fusion.
Membrane fusion during nuclear assembly
SNARES are important factors in various membrane fusion events throughout the cell (Jahn and Scheller, 2006). Recently it has been demonstrated that N-ethylmaleimide-sensitive factor (NSF) and SNARE molecules are required for nuclear envelope assembly in cell-free assays (Baur et al., 2007). In addition, inner and outer nuclear membrane fusion is important for nuclear pore complex formation (Fichtman et al., 2010). However, the in vivo role of membrane fusion in nuclear envelope assembly remains a topic of discussion within the field and the degree to which fusion of vesicles and/or membrane derived sheets play a role is not yet clear (Anderson and Hetzer, 2007; Larijani and Poccia, 2009). Also, differences among tissues and cell types in intact animals may complicate the issue further. Nevertheless, the identification of the bmb mutant reveals a case where nuclear membrane fusion is unequivocally essential. Future structural and biochemical studies of Bmb protein will shed light on the mechanism underlying double membrane fusion of the nuclear envelope where molecular mechanisms are greatly lacking.
Karyomere fusion in insects and mammals
Bmb is conserved among both vertebrate and invertebrate species (e.g., Figure S1B). A Bmb homolog exists in the tick genome where karyomere intermediates are described (Jasik, 2006). Although we found homologs in a variety of additional insects (e.g., ants, beetles, mosquitoes, bees), we did not identify a homolog in Drosophila despite its high level of genome annotation. Interestingly, nuclear envelopes do not breakdown completely during early development in Drosophila (Harel et al., 1989), and therefore, Bmb may not be necessary for this mode of nuclear division.
Identification of bmb in mammalian species has been elusive. Corresponding mammalian sequences may not be available in some of the current assemblies. It is possible that karyomere intermediates are relevant to species with extended cleavage stages under maternal control and therefore are not required in certain mammalian systems. This could explain the absence of bmb in mice, where the maternal-zygotic-transition (MZT) occurs at the 2-cell stage and karyomere intermediates appear to be absent. In some mammalian systems, however, the MZT occurs as late as the 16-cell stage. Indeed, this is the case in rabbit and, interestingly, karyomeres form and fuse to form a mononucleus during early development in the rabbit (Gulyas, 1972). Thus, Bmb or Bmb-like molecules may be relevant in certain mammals.
We demonstrated that bmb is also required for pronuclear fusion (Figure 7) analogous to the function of the Kar5p in yeast. In mice pronuclei do not fuse, paralleling an apparent lack of karyomere biology. Instead, the maternal and paternal pronuclear envelope breaks down prior to union of the respective chromosomes (Zamboni et al., 1972), similar to what occurs in bmb mutant embryos. Interestingly, pronuclei partially fuse in rabbit (Gondos et al., 1972), a mammalian system where karyomeres do form during the cleavage stage.
In human and mouse embryos the early large blastomeres also have a propensity to form micronuclei. Multiple micronuclei have been detected in in vitro fertilized embryos and correlate negatively with pregnancy success (Balakier and Cadesky, 1997). In mouse chromokinesin Kid is required to counter the tendency to form multiple micronuclei. Kid is a microtubule motor protein with a DNA binding domain (Tokai et al., 1996). Kid protein interacts with the metaphase chromosomes and its dynamic interactions are important to ensure that the dividing chromosomes undergo maximal axial compaction (Ohsugi et al., 2008). In Kid maternal-effect mutants, micronuclei form due to a mechanism that appears similar to karyomere formation (Ohsugi et al., 2008). Thus, dividing chromosomes in the early mouse embryo have the propensity to form karyomere-like structures, but this is prevented through Kid function.
We propose that some mammals, such as mice, use a different mechanism to form an intact mononucleus in a cellular environment that favors micronuclei formation. In mice, Kid ensures that chromosomes are maintained in a compact association during early anaphase to promote mononucleus formation (Ohsugi et al., 2008). This mechanism functions only during early cleavage, when cells are very large. In frogs and fish, an alternate mechanism is used to prevent micronuclei formation, which involves the formation of karyomere intermediates that require specialized molecules like Bmb to promote fusion into a single mononucleus.
EXPERIMENTAL PROCEDURES
Fish stocks
The TL strain was used for most WT experiments (except when bmb heterozygotes were used). The bmbp22atuz mutant allele was generated in an ENU-induced mutagenesis screen (unpublished), using a strategy similar to Dosch et al., 2004).
Chromosomal Mapping and Cloning of bmb
bmb was mapped through bulk segregation analysis (Pelegri and Mullins, 2004) to a 2.2 centiMorgen interval flanked by Simple Sequence Length Polymorphic (SSLP) markers z30279 and z1772 near the telomere of chromosome 25 (AB strain). cDNA from hetero- and homozygous ovaries was used as template for PCR to sequence candidate genes in the interval. A bmb cDNA including 5′ and 3′ UTRs was fully sequenced (GenBank accession number XXXXX). Tg(actb2:bmb-V5) was generated to confirm the identity of bmb by rescue. Further details on mapping, cloning and transgenesis are described in Extended Experimental Procedures.
Transmission Electron Microscopy
Embryos from controlled spawnings were collected at 26°C, manually dechorionated, aged to the 128-cell stage and fixed at 1.5 minute intervals over 21 minutes (14 time points) in 2.5% glutaraldehyde, 2.0% paraformaldehyde (PFA), 0.1 M sodium cacodylate overnight at 4°C. Fo ur embryos from each time point were removed and stained with DAPI to select for the telophase-interphase portion of the cell cycle. Two embryos from corresponding time points were processed for EM using standard methods at the UPenn EM core facility and imaged on a JEOL JEM 1010 electron microscope.
Immunofluorescence
Embryos were collected from controlled spawnings at the appropriate time interval (typically 1-minute for 32-to-64 cell transition and 3-minutes beginning at 10 mpf for the fertilization experiments) at 26°C a nd fixed overnight in 4% PFA in PBS at 4°C. Embryos were washed in PBS 0.1% triton X-100 (PBT), manually dechorionated, washed three additional times in PBT, transferred to 100% methanol and stored for at least 12 hours at −20°C. Embryos were immunostained as described (Dekens et al., 2003). with the primary antibodies: Lamin B1 (1:400; Abcam, Cambridge, MA), DM1α (1:500; Sigma), mab414 (1:500; Abcam, Cambridge, MA), α-Bmb (1:1000), V5 (1:500; Invitrogen). Appropriate secondary antibodies were used (Alexa 488 or 594; Molecular Probes, Carlesbad, Ca) at 1:500. Embryos were mounted in Vecta Shield with or without DAPI (Vector Laboratories, Burlingame, Ca) and imaged on a Zeiss 510 or 710 LSM. Typically, 7 to 20 Z-slices were captured depending on the stage of the cell cycle and the orientation of the division plane at 0.75μm or 0.50μm intervals. Confocal data was analyzed and processed using Image J software (Abramoff et al., 2004). Details of the Bmb antibody production are described in Extended Experimental Procedures.
Live Imaging
One-cell embryos were injected with 1 ng of Alexa 488 conjugate histone H1 (Sigma, St. Louis, Mo) and manually dechorionated. Embryos at the 16- to 32-cell stage were mounted in 5% methylcellulose and images taken at one-minute intervals on a Zeiss 510 Laser Scanning confocal Microscope (LSM) in an environmentally controlled chamber (26°C). The clon ing, expression details, and imaging of Bmb-venus are described in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Brambleberry (Bmb) functions in nuclear envelope fusion of karyomeres
Bmb localizes in prominent foci at pre-, post- and active karyomere fusion sites
Loss of maternal Bmb causes multiple micronuclei to form in cleavage stage cells
Bmb acts in pronuclear fusion during zygote formation following fertilization
Acknowledgments
We thank V. Lemon for technical assistance, P. Klein, E. Bi, S. DiNardo and P. Tran for critical reading of the manuscript, the Mullins Lab for thoughtful discussions and the fish facility staff for maintaining the fish stocks. This work was supported by grants R21HD062952 and R01HD065600 to M.C. Mullins, Training Grant T32-HD007516 to E. Abrams and NRSA postdoctoral fellowships 5F32GM77835 to E. Abrams, 1F32GM080926 to L. Kapp and DRG1826-04 to F. Marlow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramoff MD, Magalhaes PJ, JRS . Image Processing with ImageJ. Biophotonics International; 2004. [Google Scholar]
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol. 2007;9:1160–1166. doi: 10.1038/ncb1636. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakier H, Cadesky K. The frequency and developmental capability of human embryos containing multinucleated blastomeres. Hum Reprod. 1997;12:800–804. doi: 10.1093/humrep/12.4.800. [DOI] [PubMed] [Google Scholar]
- Baur T, Ramadan K, Schlundt A, Kartenbeck J, Meyer HH. NSF- and SNARE-mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J Cell Sci. 2007;120:2895–2903. doi: 10.1242/jcs.010181. [DOI] [PubMed] [Google Scholar]
- Beh CT, Brizzio V, Rose MD. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J Cell Biol. 1997;139:1063–1076. doi: 10.1083/jcb.139.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownds C, Wilson R, Marshall DJ. Why do colder mothers produce larger eggs? An optimality approach. J Exp Biol. 2010;213:3796–3801. doi: 10.1242/jeb.043356. [DOI] [PubMed] [Google Scholar]
- Buch C, Lindberg R, Figueroa R, Gudise S, Onischenko E, Hallberg E. An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J Cell Sci. 2009;122:2100–2107. doi: 10.1242/jcs.047373. [DOI] [PubMed] [Google Scholar]
- Burke B, Gerace L. A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell. 1986;44:639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- Cummins JM. The role of maternal mitochondria during oogenesis, fertilization and embryogenesis. Reprod Biomed Online. 2002;4:176–182. doi: 10.1016/s1472-6483(10)61937-2. [DOI] [PubMed] [Google Scholar]
- Dekens MP, Pelegri FJ, Maischein HM, Nusslein-Volhard C. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development. 2003;130:3907–3916. doi: 10.1242/dev.00606. [DOI] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell. 2004;6:771–780. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Olea N. Formation of the female pronucleus and reorganization and disassembly of the first interphase cytoskeleton in the egg of the glossiphoniid leech Theromyzon rude. Dev Biol. 1995;171:541–553. doi: 10.1006/dbio.1995.1303. [DOI] [PubMed] [Google Scholar]
- Fichtman B, Ramos C, Rasala B, Harel A, Forbes DJ. Inner/Outer nuclear membrane fusion in nuclear pore assembly: biochemical demonstration and molecular analysis. Mol Biol Cell. 2010;21:4197–4211. doi: 10.1091/mbc.E10-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Cha BJ, Schroeder MM. Confocal immunofluorescence microscopy of microtubules, microtubule-associated proteins, and microtubule-organizing centers during amphibian oogenesis and early development. Curr Top Dev Biol. 1995;31:383–431. doi: 10.1016/s0070-2153(08)60234-3. [DOI] [PubMed] [Google Scholar]
- Gondos B, Bhiraleus P, Conner LA. Pronuclear membrane alterations during approximation of pronuclei and initiation of cleavage in the rabbit. J Cell Sci. 1972;10:61–78. doi: 10.1242/jcs.10.1.61. [DOI] [PubMed] [Google Scholar]
- Griffin J, Emery BR, Huang I, Peterson CM, Carrell DT. Comparative analysis of follicle morphology and oocyte diameter in four mammalian species (mouse, hamster, pig, and human) J Exp Clin Assist Reprod. 2006;3:2. doi: 10.1186/1743-1050-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyas BJ. The rabbit zygote. 3. Formation of the blastomere nucleus. J Cell Biol. 1972;55:533–541. doi: 10.1083/jcb.55.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Zlotkin E, Nainudel-Epszteyn S, Feinstein N, Fisher PA, Gruenbaum Y. Persistence of major nuclear envelope antigens in an envelope-like structure during mitosis in Drosophila melanogaster embryos. J Cell Sci. 1989;94(Pt 3):463–470. doi: 10.1242/jcs.94.3.463. [DOI] [PubMed] [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 1993 [Google Scholar]
- Huang QM, Akashi T, Masuda Y, Kamiya K, Takahashi T, Suzuki M. Roles of POLD4, smallest subunit of DNA polymerase delta, in nuclear structures and genomic stability of human cells. Biochem Biophys Res Commun. 2010;391:542–546. doi: 10.1016/j.bbrc.2009.11.094. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jasik K. Cleavage in Ixodes ricinus (L.) (Acari: Ixodidae) Wiad Parazytol. 2006;52:291–297. [PubMed] [Google Scholar]
- Knapik EW, Goodman A, Atkinson OS, Roberts CT, Shiozawa M, Sim CU, Weksler-Zangen S, Trolliet MR, Futrell C, Innes BA, et al. A reference cross DNA panel for zebrafish (Danio rerio) anchored with simple sequence length polymorphisms. Development. 1996;123:451–460. doi: 10.1242/dev.123.1.451. [DOI] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larijani B, Poccia DL. Nuclear envelope formation: mind the gaps. Annu Rev Biophys. 2009;38:107–124. doi: 10.1146/annurev.biophys.050708.133625. [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Geraud G, Mechali M. Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication. J Cell Biol. 1998;142:1159–1166. doi: 10.1083/jcb.142.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo FJ. An ultrastructural analysis of mitosis and cytokinesis in the zygote of the sea urchin, Arbacia punctulata. J Morphol. 1972;138:207–238. doi: 10.1002/jmor.1051380206. [DOI] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol. 2011;194:425–440. doi: 10.1083/jcb.201012063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Malhas A, Goulbourne C, Vaux DJ. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Melloy P, Shen S, White E, Rose MD. Distinct roles for key karyogamy proteins during yeast nuclear fusion. Mol Biol Cell. 2009;20:3773–3782. doi: 10.1091/mbc.E09-02-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag M, Spring H, Trendelenburg MF. Structural analysis of the mitotic cycle in pre-gastrula Xenopus embryos. Chromosoma. 1988;96:187–196. doi: 10.1007/BF00302357. [DOI] [PubMed] [Google Scholar]
- Mora-Bermudez F, Gerlich D, Ellenberg J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat Cell Biol. 2007;9:822–831. doi: 10.1038/ncb1606. [DOI] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Newport J, Dunphy W. Characterization of the membrane binding and fusion events during nuclear envelope assembly using purified components. J Cell Biol. 1992;116:295–306. doi: 10.1083/jcb.116.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Ohsugi M, Adachi K, Horai R, Kakuta S, Sudo K, Kotaki H, Tokai-Nishizumi N, Sagara H, Iwakura Y, Yamamoto T. Kid-mediated chromosome compaction ensures proper nuclear envelope formation. Cell. 2008;132:771–782. doi: 10.1016/j.cell.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Pelegri F, Mullins MC. Genetic screens for maternal-effect mutations. Methods Cell Biol. 2004;77:21–51. doi: 10.1016/s0091-679x(04)77002-8. [DOI] [PubMed] [Google Scholar]
- Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell. 2009;20:4031–4042. doi: 10.1091/mbc.E09-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Schoft VK, Beauvais AJ, Lang C, Gajewski A, Prufert K, Winkler C, Akimenko MA, Paulin-Levasseur M, Krohne G. The lamina-associated polypeptide 2 (LAP2) isoforms beta, gamma and omega of zebrafish: developmental expression and behavior during the cell cycle. J Cell Sci. 2003;116:2505–2517. doi: 10.1242/jcs.00450. [DOI] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamota T. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 1996;15:457–467. [PMC free article] [PubMed] [Google Scholar]
- Wuhr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. Evidence for an upper limit to mitotic spindle length. Curr Biol. 2008;18:1256–1261. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhr M, Tan ES, Parker SK, Detrich HW, 3rd, Mitchison TJ. A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol. 2010;20:2040–2045. doi: 10.1016/j.cub.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni L, Chakraborty J, Smith DM. First cleavage division of the mouse zygot. An ultrastructural study. Biol Reprod. 1972;7:170–193. doi: 10.1093/biolreprod/7.2.170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.