Abstract
Diagnostic and therapeutic radiation technology has developed dramatically in recent years, and its use has increased significantly, bringing clinical benefit. The use of diagnostic radiology has become widespread in modern society, particularly in paediatrics where the clinical benefit needs to be balanced with the risk of leukaemia and brain cancer increasing after exposure to low doses of radiation. With improving long-term survival rates of radiotherapy patients and the ever-increasing use of diagnostic and interventional radiology procedures, concern has risen over the long-term risks and side effects from such treatments. Biomarker development in radiology and radiotherapy has progressed significantly in recent years to investigate the effects of such use and optimise treatment. Recent biomarker development has focused on improving the limitations of established techniques by the use of automation, increasing sensitivity and developing novel biomarkers capable of quicker results. The effect of low-dose exposure (0–100 mGy) used in radiology, which is increasingly linked to cancer incidences, is being investigated, as some recent research challenges the linear-no-threshold model. Radiotherapy biomarkers are focused on identifying radiosensitive patients, determining the treatment-associated risk and allowing for a tailored and more successful treatment of cancer patients. For biomarkers in any of these areas to be successfully developed, stringent criteria must be applied in techniques and analysis of data to reduce variation among reports and allow data sets to be accurately compared. Newly developed biomarkers can then be used in combination with the established techniques to better understand and quantify the individual biological response to exposures associated with radiology tests and to personalise treatment plans for patients.
Research into the identification of biomarkers of radiation exposure is an emerging and developing area with multiple possible benefits for patients, doctors and the general public. A radiation biomarker is a biological entity that changes after exposure to radiation, allowing exposed individuals to be identified and, with some biomarkers, a dose to be estimated. There are different types of biomarkers, including chromosome aberrations, protein expression or gene expression. Some can measure accurately the dose received, while others can only indicate if a dose was received. Biomarkers can help clinicians manage treatment for a patient exposed accidentally to the wrong radiation dose or on purpose through radiotherapy. They may be able to predict the treatment response of a tumour and estimate the risk of acute or late effects in normal tissues. Biomarkers can also identify the dose received by the patient in a full or partial body exposure. Such information can help inform the necessary medical treatment plan for the patient, and it may also identify patients with a high likelihood of developing cancer in the future so that regular monitoring can be set up.
Biomarkers of ionising radiation
Ionising radiation (IR) is well known for the production of a range of deoxyribonucleic acid (DNA) damages, critical of which are DNA double-strand breaks (DSBs). Misrepair of these breaks can produce many different types of chromosomal aberrations, such as terminal deletions, translocations, ring chromosomes and, what is considered the most specific aberration for radiation exposure, dicentric chromosomes. As peripheral blood is a rapidly and easily accessible source of sample, most studies use whole blood isolated lymphocytes or lymphocyte subsets in assessing radiation response. Given that lymphocytes circulate throughout the body, partial exposure can still be determined using this cell type. Although a number of the techniques described can be used for other cell types, such as skin fibroblasts and buccal epithelial cells, the use of lymphocytes is what we will focus on in this review.
Cytogenetic techniques are well established for biodosimetry purposes and can be used for analysis of recent unstable aberrations (dicentric, micronucleus assay) or stable aberrations of past radiation exposure [translocation analysis using fluorescence in situ hybridisation (FISH)]. Owing to their time-consuming scoring requirements, developments in automating the established techniques are under way, and faster techniques, such as measuring protein and gene expression, are being developed. This is mainly for the rapid triage of patients and management of a mass casualty situation. With the increased use of radiological equipment for medical procedures, such as CT scanning, positron emission tomography and nuclear medicine, biomarkers are also being used to assess their long-term health risks.
Cytogenetic techniques
Dicentric assay
Dicentric aberrations occur from the misrepair of DSB ends on two different chromosomes, resulting in a chromosome with two centromeres and an acentric fragment (Figure 1). The dicentric assay measures the presence of these aberrations in lymphocyte metaphases. Dicentrics decrease over time post exposure, as lymphocytes are renewed with a half-life of approximately 3 years. Figure 2 illustrates the persistence of this and other biomarkers over time. This technique is therefore suited for recent exposures, with blood samples ideally taken within 4 weeks of exposure. It involves a cell culture time of 48 h and addition of colcemid to block mitosis, allowing visualisation of the dicentrics during metaphase. The dicentric assay is considered to be the gold standard of biodosimetry techniques, supported by the International Organization for Standardization [1,2]. The success of the dicentric assay as a biomarker is because of the fact that it is radiation specific and has a low spontaneous frequency. This allows accurate estimation of doses down to 0.1 Gy [3]. Although factors such as age and gender have been shown not to affect the baseline or radiation-induced yields of dicentrics, it is important to establish individual dose–response calibration curves for each radiation quality [4]. The assay can also be used to assess partial body exposure where a part of the body may be exposed to a high dose of radiation, leaving other parts unexposed. This results in an overdispersed or a non-Poisson distribution of dicentrics in the scored metaphases. Two calculations, Dolphin’s contaminated Poisson or Sasaki’s Quantity of Dicentrics and Rings, take the non-Poission distribution of dicentrics into account and therefore can provide a partial body fraction size and dose estimate [3]. A waiting period of 24 h must be used until blood is taken to allow complete mixing of the irradiated with the non-irradiated cells in the lymphoid system. Its limitation is that it takes more than 51 h for sample preparation alone, and scoring of 500–1000 metaphase spreads must be carried out by a trained cytogeneticist, which is time consuming. In case of a mass casualty situation, the number of metaphase spreads could be reduced down to 50. Although this means a reduced sensitivity of 0.5 Gy, it has been shown to provide enough information for the triage of mass casualties [5]. Also, recent work has shown that an automatic dicentric detection system produces a three-fold higher throughput than manual scoring, with comparable dose estimates for both whole and partial body exposure [6].
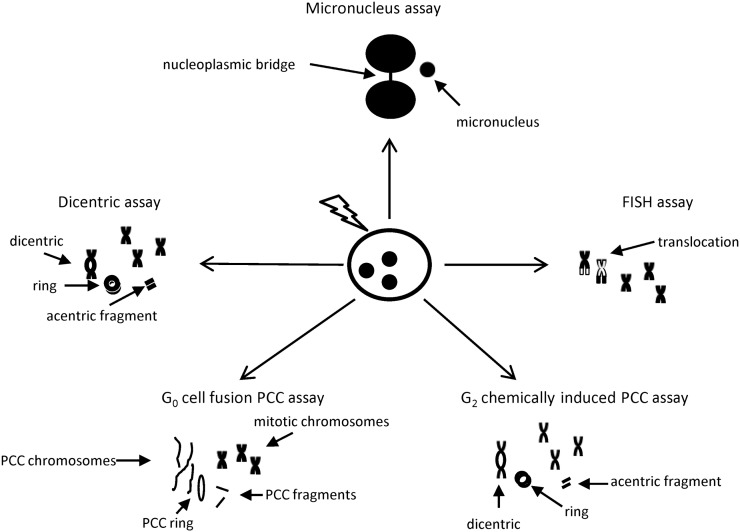
Figure 1.
Cytogenetic techniques covered in this review. After irradiation, cytogenetic assays are used to visualise different types of chromosome damage, dicentric, nucleoplasmic bridge, micronucleus, ring chromosome, acentric fragment and translocation. FISH, fluorescence in situ hybridisation; G0. resting phase; G2, interphase gap 2; PCC, premature chromosome condensation.
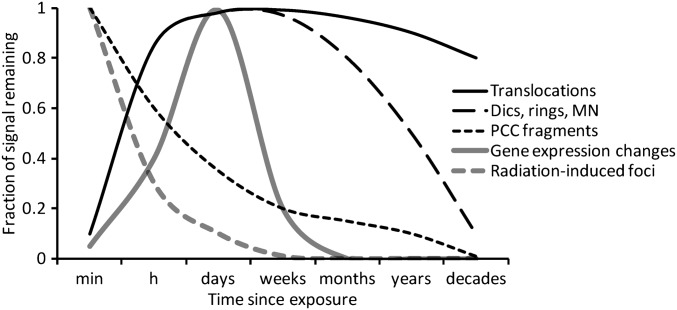
Figure 2.
Induction and persistence of different deoxyribonucleic acid damage-associated biomarkers in peripheral blood lymphocytes following ionising radiation exposure. dics, dicentrics; MN, micronucleus; PCC, premature chromosome condensation.
The micronucleus assay
Micronuclei are extranuclear spheres composed of acentric chromosome fragments or whole chromosomes that are left behind in the cytoplasm after mitosis. The cytokinesis-blocked micronucleus (CBMN) assay technique was developed in 1985 when Fenech and Morley introduced cytochalasin-B to lymphocyte cultures, effectively blocking cytokinesis and allowing identification of micronuclei and binucleated cells. Micronuclei are not radiation specific. They can occur spontaneously, generally containing whole chromosomes. Contributing factors include age, gender, diet and chemicals, which produce a variable background level and a higher threshold of detection for low doses ranging from 0.2 Gy to 0.3 Gy [3]. Recent work has improved on this limitation by utilising the fact that spontaneous micronuclei are generally composed of whole chromosomes, in contrast to radiation-induced micronuclei that develop from acentric chromosome fragments. Use of the FISH pan-centromere probe has allowed separation of micronuclei into centromere-positive spontaneous micronuclei and centromere-negative IR-induced micronuclei. It has been shown that an increase of micronuclei with dose is dependent on centromere-negative micronuclei [7,8]. This inclusion of centromere staining into the assay produced a lower detection limit of 0.1 Gy and, therefore, a more sensitive assay. Another measurement included in the assay is the nucleoplasmic bridges (NPBs) that are thought to be linked to dicentrics. The inclusion of NPB scoring in the CBMN assay is useful because it has low background frequency in contrast to the high background level of micronuclei, and therefore produces a more sensitive technique [9]. When compared with the dicentric assay in the accident in Istanbul, Turkey, where 10 workers were exposed to a Co-60 source, the micronucleus assay produced similar dose estimates [10]. A recent study involving 22 patients with rectal cancer showed no significant difference between the dicentric assay and the micronucleus assay after radiochemotherapy [11]. In terms of a diagnostic technique, scoring for the CBMN technique is quick, simple and can be automated for use in a mass casualty situation [12,13]: a distinct advantage over the dicentric assay, which is a time-consuming process requiring expert personnel. Automation, however, does not include centromere-negative micronuclei or NPB in its scoring: features that increased its sensitivity in biodosimetry [14]. Although it is suggested as an alternative for the dicentric assay, one major limitation with the CBMN assay in its use as a diagnostic tool is that it can only estimate the average total body dose and cannot be used for partial body exposure.
Premature chromosome condensation
The premature chromosome condensation (PCC) technique was first established by Johnson and Rao [15] and later developed for biodosimetry by Pantelias and Maille [16] when they fused interphase lymphocytes with mitotic cells, causing the chromosomes in the lymphocytes to condense prematurely. Chromosome condensation can be achieved by either fusion of interphase cells with mitotic cells, known as fusion-based PCC, or drugs such as okadaic acid or calyculin A, allowing aberrations such as chromosome breaks, resulting in excess PCC fragments, to be visualised quickly after exposure to radiation. Since cell culture is not required, the slide preparation process is much quicker than with the dicentric or micronucleus techniques at just 3 h for fusion-based PCC. Technical difficulties still remain with the fusion-based technique, and therefore it is not commonly used. Fusion can be achieved with either a virus or a fusing agent, such as polyethylene glycol (PEG), with associated problems such as monitoring of PEG concentrations, molecular weight and exposure time and the need for a virus handling approved laboratory [17].
For drug-induced PCC, lymphocytes need to be stimulated and cultured for 2 days to induce the expression of mitosis promoting factor, without which the drug cannot induce chromosome condensation. Scoring is still time consuming; however, automation systems are being developed. The use of FISH probes has been incorporated into the PCC assay, allowing the scoring of dicentrics [18] and translocations and producing a much more versatile assay. The PCC technique differs from the other cytogenetic techniques in that it does not require cells to enter mitosis, and therefore cells damaged from high doses are not subject to apoptosis or, more importantly, cell cycle arrest, which would prevent metaphase formation of the most severely damaged cells and result in the underestimation of the total dose. This allows large doses (>5 Gy) to be estimated in comparison with the dicentric and micronucleus assays that work best at doses below 5 Gy owing to the saturation effect from high doses. PCC can score chromosomal rings in cases of high exposure, such as victims of the Tokaimura accident exposed to lethal doses [19], where it has been shown to be more effective than dicentric scoring [20]. Thanks to the avoidance of cell cycle delays and higher yields of scorable aberrations, this technique also has advantages over the dicentric assay in partial body exposure dosimetry [21] and in elderly or immune-compromised patients, and it can produce results quicker than the dicentric assay [17].
Fluorescence in situ hybridisation
The techniques discussed so far are best used for determining dose estimates within weeks to months after radiation exposure, as the aberrations scored are unstable and disappear with cell turnover (lymphocytes have an average half-life of 2–4 years). FISH, however, detects stable aberrations such as translocations that are present in circulating lymphocytes years after exposure, as they can be passed on from irradiated stem cells to descendant lymphocytes [22,23]. Translocations involve the exchange of chromosomal segments between two different chromosomes. FISH paints one or more chromosomes using a chromosome-specific library of DNA probes with a fluorochrome attached, allowing the translocations to be visualised by fluorescence microscopy. Usually, three of the large chromosomes (chromosomes 1–12) are painted with one colour, with the rest only counter-stained with a fluorescent DNA dye. Using this approach, around 30% of all translocations are detected as bicoloured chromosomes [3]. Pancentromeric probes can also be incorporated into the technique to distinguish translocations from dicentrics [24]. A formula developed by Lucas et al [25] allows this detection of translocations to be scaled up to represent the full genome with near identical results in comparison to G-banding. This technique is used mainly on epidemiological study groups such as the A-bomb survivors, where the dicentric assay can no longer be useful, and radiation workers to analyse lifetime exposure, or as a follow-up on exposed individuals when assessing cancer risk. To ensure that the aberration will be persistent, only stable cells should be scored; these are cells without dicentrics, acentrics or centric rings [26]. Two factors that contribute significantly to background levels are age and smoking [27], with the baseline frequency of translocations and minimum detectable dose increasing linearly with age [28]. The detection limit of several hundred milligrays for individual cases can be lowered to <50 mGy by studying cohorts such as that used by Bhatti et al [29] by combining three studies on 362 radiologists and airline pilots. The technique is the most time-consuming biodosimetry technique by far, taking more than a week to produce results, and typically involves the scoring of about 3000 cells [3]. However, since it looks at retrospective exposures, the need for quick results is not vital.
Protein techniques
IR causes different types of DNA damage, such as base damage, single-strand breaks and DSBs, which are considered the most lethal and more specific for IR than most other lesions. Different techniques have been developed to measure DSB formation, such as neutral elution, pulsed field electrophoresis and comet assays; however, none is as sensitive as the γ-H2AX assay. On formation of a DSB, a variety of repair proteins accumulate at the break site, such as ATM, Rad51, 53BP1 and the MRN complex, which consists of Mre11, Rad50 and NBS1. The histone H2AX is phosphorylated within seconds to form γ-H2AX, reaching a peak at around 30 min [30]. The foci that form can be used to quantify DNA damage through antibody staining and immunofluorescence microscopy within a few hours after receiving a blood sample. The H2AX foci assay is the most sensitive method for detecting DSBs and can reach a lower detection limit of 1 mGy [31]. This is only achieved when pre-irradiation H2AX levels are known, as there is variation between individuals and cell types. Therefore, this method is suited more for planned exposures such as diagnostic radiology [32] or radiotherapy [33]. As a biodosimetry technique, it is useful immediately after exposure, as foci levels peak at <1 h but then decline within days [34]. Developments have been made to increase speed and throughput of results by measuring γ-H2AX intensity instead of scoring the number of foci [35], but this, as with most automated techniques, results in lower sensitivity. As this technique is mainly useful within hours of exposure, a recent study has aimed at developing a portable fluorescence spectrometer for use in triage of exposed individuals [36]. Other protein biomarkers not associated with DNA damage induction and signalling include C-reactive protein and serum amylase; however, they have limitations, such as interindividual variation, lack of specificity, and they cannot discriminate between partial and total body exposure [37]. They are used in conjunction with other techniques in clinical monitoring rather than as a principal biodosimetry technique; and for this reason, they will not be discussed in detail in this review.
Gene expression techniques
Gene expression has been investigated as an alternative technique that could produce results within hours. Gene expression analysis is considered mainly for rapid triage of individuals in a mass casualty situation where the priority is not to give an exact dose estimate but to separate the exposed from the “worried well” as quickly as possible. Following irradiation, the DNA damage response is activated with the expression of numerous signalling pathways. Genome studies have shown that the genes affected by radiation play a role in many processes, such as DNA repair, cell cycle checkpoints and/or apoptosis [38,39]. The use of microarray analysis of gene expression following IR exposure has produced hundreds of possible genes of interest. Gene expression varies depending on the dose, dose rate, cell type, type of radiation exposure and time after exposure. It was shown that the genes up-regulated at different times of 6 h, 24 h and 7 days have <20% overlap [40]. A long-term study of lymphocytes at 2, 7 and 55 days showed up to only five genes in common from the acute- to late-stage responses [41]. Different genes have also been shown to be up-regulated at low doses in comparison with high doses [42]. At low doses, a gender effect is also seen to influence the gene expression [40]. It is therefore likely that any tailor-made array will need to consist of a panel of dose–response and time–response genes. Gene expression studies so far have been able to identify irradiated subjects from non-irradiated subjects at doses down to 50 cGy and with an accuracy of 100% for mice and 90% for humans [43]. This dramatically dropped to 61% in patients who previously received chemotherapy, a problematic factor with this approach. There are a few consistently up-regulated genes, FDXR, CDKN1A, PUMA, PHPT1, SESN1 and GADD45, identified in microarray studies that have been followed up extensively by the more sensitive and reproducible quantitative polymerase chain reaction (QPCR) technique, which can produce data within hours [38,44–47]. A linear dose–reponse has been observed by reverse transcriptase-QPCR from doses 1–3 Gy [47] and 0.5–4 Gy [48]. A recent study by Manning et al [49] has illustrated the potential of gene expression to be used in the triage of patients by using dose–response curves to identify blood samples exposed to a low (5–100 mGy) or high (0.5–4 Gy) dose of irradiation. Although these results are encouraging, larger population studies must be carried out to assess the variation in response for different genes.
A quantitative nuclease protection assay has been developed that distinguishes exposed from non-exposed individuals from as little as 30 μl of blood, which would be collected from a fingerstick in a mass casualty situation [50]. The nuclease protection assay uses nucleic acid hybridisation and nuclease digestion of non-hybridised RNA to selectively detect and quantify individual RNA molecules, without the need for PCR amplification. The small sample volume and turnaround time of <12 h are attractive features of this assay that would be used for the triage of patients rather than accurate dose estimation. Successful detection of partial body exposure has been reported with an accuracy of 79–100%, a vital property of a diagnostic technique in accidental cases [51]. High throughput can be achieved through multiwell arrays that are widely available commercially. Robotics introduces its own source of variability from dispensing very low volumes in these high throughput systems. Although the accuracy is not 100%, it is consistently high in comparison with technical day-to-day variability when done by hand. Multiplexing in real-time QPCR reactions using four dyes or more has increased the throughput of samples, as well as reducing cost and labour, paving the way for the possible use of this technique in a clinical setting [47]. A widespread issue that must be addressed is the variability among experiments using QPCR. Huge variability can be introduced at multiple steps in the QPCR process, such as cell subpopulation differences [52,53], storage, RNA isolation, reverse transcription, PCR conditions and data analysis [54]. A need to clarify these variations by publishing detailed methods was set out in the MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments [55]. This has also been advised for microarrays [56].
Biomarkers for diagnostic and interventional radiology
X-rays are an established diagnostic tool in medicine, being used for more than a 100 years. Owing to the low doses being used, these techniques are considered acceptable when clinical benefit is clear and their use is widespread. The use of X-rays and diagnostic equipment has developed enormously in the last decade, with the biological and individual effect of irradiation still being investigated. There is now mounting epidemiological evidence that links low doses to cancer incidences [57].
CT has become an increasingly widespread diagnostic tool in the last decade. It is quick, accurate and very user-friendly. Even though it is often the method of choice, the associated organ doses are much larger than those obtained for conventional radiography, such as mammography or chest radiography [58]. One of the largest areas of increased use of CT has been seen for paediatric diagnosis. CT has established itself as the main diagnostic tool for appendicitis, replacing ultrasonography [59]. Children are inherently more sensitive than adults to radiation, and they have more years of life in which cancer may develop [58]. Its increased use is therefore of great concern, and many studies have linked cancer occurrence to radiation exposure in children [60,61]. To study the effects of low-dose exposure, γ-H2AX scoring is widely used, as it is sensitive to doses as low as 1 mGy. A dose-dependent linear increase in γ-H2AX foci was seen in peripheral blood mononuclear cells of adult patients undergoing a CT scan [32]. Interventional radiology, such as paediatric cardiac catheterisation, has also recently caused concern as a study by Beels et al [62] has shown an increase in γ-H2AX foci in peripheral blood lymphocytes after in vivo cardiac catheterisation. The in vivo dose–response curve appeared steeper at low doses in contrast to the current linear-no-threshold model, highlighting the need for further research into this effect. Studies on the low-dose effects of CT scans often use dose-enhancing contrast agents or involve blood from patients diagnosed with cancer [63]. A recent study by Jost et al [64] examined the dicentric and γ-H2AX dose–response in lymphocytes from healthy donors ex vivo with and without the use of iodine as a contrast agent, giving comparable results at low radiation doses for 5 mg iodine per millilitre of blood but showing a clear biological dose-enhancing effect for a contrast dosage well outside the diagnostic CT range.
Nationwide screening programmes have been introduced to monitor people at an age when cancers largely develop. Although they are vital for recognising the early forms of cancer, recent work has questioned the benefit of repeatedly screening susceptible individuals. Mammography screening uses low-energy X-rays that cause concern, as low-energy photons are generally more efficient in damaging cellular DNA. This property, known as the relative biological effectiveness (RBE) of the radiation, compares the end points of radiation exposure from one type of radiation quality relative to another and increases with very low-energy X-rays. Studies have reported different RBE values, with early work by Frankenberg et al [65] controversially reporting high RBE values of about 3.2 for 29 kV X-rays vs the conventional 220 kV X-rays and 8 for 29 kV X-rays vs Co-60 gamma rays. This work was later re-evaluated by Göggelmann et al [66] and the RBE found to be lower, on the order of 2. A study by Andrieu et al [67] suggested that BRCA carriers had a 1.54× higher risk of developing breast cancer after exposure to chest X-rays in comparison with non-exposed BRCA carriers. Individuals with a high risk of breast cancer from BRCA1 or BRCA2 mutations were also shown to produce higher levels of DSBs, identified by γ-H2AX scoring, after mammographic ex vivo irradiation of epithelial cells [68]. There are many factors that contribute to the organ dose received, among the most important being the number of scans performed. Breast cancer is a common cancer in the UK, with 41 259 cases per 100 000 females reported in 2010 [69]. High incidences of breast cancer were observed in females exposed to multiple X-rays [70,71]. Follow-up studies on females with scoliosis exposed to multiple diagnostic X-rays have reported a standardised incidence ratio of 1.8, which increased with the number of X-rays performed [70]. However, the number of breast cancer cases in the study was small (n=11), and therefore the study was later expanded to 5573 patients and again found an increasing risk of breast cancer with the increasing number of radiographic examinations. Similar trends were seen in females examined by X-rays for the treatment of tuberculosis, with 147 breast cancer cases reported in contrast to 113.6 expected among 4940 patients [71]. These results should be taken with caution owing to the recall bias in the questionnaire data collection and low statistical power in some of the studies, but they nevertheless raise an important point to consider in relation to nationwide screening programmes. Altering protocols, such as reducing the number of views performed in screening [70], have been suggested to lower the risk.
The main long-term studies available are from radiology workers or the Hiroshima and Nagasaki atomic bomb survivors. Translocations are the aberration of choice in assessing the long-term radiation damage as discussed previously. Retrospective studies in radiology technologists have shown increased translocations after exposure to low doses [72]; however, uncertainties remain for retrospective dose estimates based on biological markers of exposure.
Biomarkers for radiotherapy
Nowadays, the use of radiation is very prominent in cancer diagnosis and treatment. Although vital for a patient’s survival, radiation therapy (RT) carries with it acute side effects, such as dermatitis, cystitis and bone marrow suppression, and late effects, such as cardiovascular disease, infertility, fibrosis or second malignancies. Patient response to radiation treatment can vary greatly from normal to severe with the late stage toxicity seen in 5–10% of patients, which can be irreversible [73]. Extreme radiosensitivity usually results from a mutation in a vital repair pathway gene, such as ATM or NBS1, seen in the diseases Ataxia telangiectasia (AT) and Nijmegen breakage syndrome (NBS) or in genes, such as DNA-PKcs, Ku70, Ku86 and DNA ligase IV [74,75] observed in immune-deficient individuals. In patients without these diseases, adverse side effects are still evident, which are thought to result from several variants, each contributing to the sensitivity of the individual [76]. This severity of adverse effects in some patients limits the dose used during treatments, as the individual response is not known. Establishing an assay to predict a patient’s response to radiation could provide an individualised treatment plan with a higher probability of cancer cure. Assays being developed at the moment involve a sample of blood or cells being irradiated ex-vivo and changes before and after irradiation being analysed. For radioresistant patients, higher doses could be used to treat the cancer, whereas for radiosensitive patients, lower doses or alternative treatment options may be employed, such as surgery.
Tissue response
RT is used to treat cancer, as the cancer cells are more sensitive to IR than surrounding normal healthy tissue. The reason for this is not fully understood. Intense research is being carried out to investigate the reason why tissues differ in their response, as it could lead to a more tailored treatment of tumour control and alleviate normal tissue damage. This difference is particularly evident in experimental studies of microbeam radiotherapy (MRT) that uses an array of beams that are micrometres in width, where spatially fractionated high doses kill tumour cells but cause little damage to normal tissue. In a study by Crosbie et al [77], γ-H2AX foci revealed cell migration in a xenograft tumour model and a limited repair response after MRT treatment, whereas MRT-treated normal skin showed little migration but an efficient repair response. Recent work has utilised γ-H2AX foci to allow the microbeam tracks to be traced within tissues, providing an in situ map of DNA damage and estimating the dose received between these tracks [78]. This factor is important in optimising RT and further understanding the tissue response after MRT. Differences in the DNA repair response between cell types, as determined by foci levels, were far less significant than differences between individuals [79] or mouse strains with different genetic background [80]. The tissue environment also greatly affects its response to irradiation and can also be altered to improve successful tumour removal. Hypoxic conditions in cancer cells develop when a tumour outgrows its own blood supply, resulting in the cancer cells being resistant to radiotherapy. An oxygen enhancement ratio of 2.6 is observed in anoxic cells for DSB induction [81]. Nitric oxide has long been known as a radiosensitiser and increases the yield of radiation-induced DSBs in hypoxic cells [82]. However, its role is not clear-cut, as it reportedly has both antitumour and protumour roles, often depending on its concentration [83]. Many chemicals and drugs such as efaproxiral [84], cisplastin [85] and 5-fluorouracil [86] are being investigated in clinical trials for use as radiosensitisers in radiotherapy treatment (reviewed in [87]). The levels of biomarkers in the tissue environment may also be used to optimise radiotherapy. Radiotherapy is delivered in a series of fractions based on normal tissue fraction sensitivity. An increased expression of biomarkers of DSBs and homologous recombination, 53BP1 and RAD51, have been observed in epidermal basal cells after a course of breast radiotherapy when there is a decrease in cell sensitivity to the fraction size [88]. Such studies may enable effective fraction size determination during radiotherapy and therefore improve treatment outcome.
The development of radiotherapy equipment has led to higher precision and also higher doses being used, such as for intensity-modulated radiotherapy (IMRT). With greater control over the shape and intensity of the radiation beam, IMRT improves tumour control while minimising exposure to the sensitive surrounding tissues. Although this treatment is still only around 10 years in use, studies are in favour of IMRT over the conventional three-dimensional conformal radiotherapy (3D-CRT) [89]. Owing to the higher doses being used and the complex treatment regimens, requiring longer beam-on times and more scatter radiation, recent studies have focused on determining the treatment-associated risk and trying to quantify the exposure of normal tissues to radiation. A 7-year follow-up study of prostate cancer patients treated with IMRT by Spratt et al[90] found survival outcomes of 98.8% in low-risk groups and 67.9% in high-risk groups. In comparison with 3D-CRT, step-and-shoot IMRT has shown similar levels of high-dose exposure of peripheral lymphocytes in prostate patients as determined by γ-H2AX foci [91], but it has also shown greater levels of low-dose exposure. This increase in low-dose exposure has raised concerns for second malignancies and highlighted the need for further long-term studies.
Radiosensitivity
The fibroblast clonogenic assay was previously used to assess the normal tissue radiosensitivity but has shown to be variable owing to differences in protocols [92], and it sometimes showed little correlation with fibrosis and, therefore, morbidity [93,94]. One overwhelming limitation was that it took 2–3 months to produce results, too long for a patient awaiting radiotherapy, in contrast with the micronucleus assay that takes <2 weeks. The micronucleus assay performed on ex vivo irradiated peripheral blood lymphocytes from 38 prostate cancer patients has shown a correlation of micronuclei with RT-related morbidity. Lee et al [95] reported a significant difference in micronuclei yields between average and overreactors after doses ≥2 Gy. It has been shown to be a reliable technique that correlates with morbidity and patient response [96]. It has previously been shown that the apoptotic response to IR is dependent on the genetic factors and therefore is a source of variation in radiosensitivity [97,98]. Assays such as the leukocyte apoptosis assay or tunel assay have therefore been used to try to establish a link between apoptotic response and cancer susceptibility or radiosensitivity. A decreased apoptotic response to radiation has been observed in AT patients and in RT patients displaying toxicity in comparison with normal donors [99,100]. This correlation between apoptosis and toxicity has also been identified in gene sets [73]. This is because of the impaired genes involved in the repair process in immune-comprised patients or radiosensitive patients and their inability to remove DNA damage after irradiation. The T-lymphocyte apoptosis assay used by Ozsahin et al [101] can predict Grade 2 or Grade 3 late-stage toxicity with a 70% level of accuracy. Sources of variation using the leukocyte apoptosis assay include hereditary factors, cell type and age [100]. Most importantly, for this assay from a clinical point of view, it can produce results in 48 h. Despite these promising results, no large-scale study has yet confirmed a technique capable of identifying radiosensitive patients in a clinical setting.
Variation in radiosensitivity has a genetic basis and may account for around 70% of variation in normal tissue toxicity [102]. Therefore, gene expression arrays and genome-wide association studies could therefore be useful in identifying the variants that are associated with this radiosensitivity. So far, gene expression studies have been used to successfully classify tumours and predict tumour response to treatment [103] but are still not routinely used in the clinics. Recent work in gene expression focuses on either predicting toxicity in patients before treatment or monitoring gene expression changes during treatment. Gene sets have been developed to identify late radiation toxicity in patients from a population of ex vivo irradiated lymphocytes, with a 64% [104] and 55% [73] success rate of predicting severe acute and late toxicity in individual patients. Expression of the gene P21 by QPCR has been shown to predict adverse sensitivity 2 h after irradiation [45]. The gene BCL-X has been shown to exhibit protective properties in cells exposed to IR and chemotherapy and to be a determinant of the rate of apoptosis and therefore clinical outcome [105]. Also, a clinical study investigating the link between radiotherapy toxicity and gene expression in the peripheral blood of cancer patients has identified genes and pathways that could be involved [106]. Since loss of γ-H2AX foci is a measure of the DNA repair capacity, the technique has also been used to try to identify individuals with DNA repair deficiencies. The γ-H2AX technique can distinguish between ATM−/− homozygote and ATM± heterozygote individuals 8 h after exposure of a blood sample to 1 Gy and has also been used to identify patients with solid tumours at a risk of adverse side effects because of impaired DNA repair [107]. γ-H2AX measurement by flow cytometry has been shown by Bourton et al [108] to be a possible marker capable of predicting the risk of radiotherapy patients developing excessive normal tissue toxicity and to a lesser degree that may be used for the prediction of oral mucositis [109]. However, other studies show no correlation between γ-H2AX foci and acute [110] or late tissue damage [111]. Scoring of chromosome aberrations in ex vivo-irradiated lymphocytes has also been correlated with acute and late normal tissue damage in radiotherapy patients [112,113]. Chua et al [114] also showed a correlation between dicentrics, acentrics and rings and γ-H2AX and late normal tissue damage in ex vivo-irradiated lymphocytes from breast cancer radiotherapy patients.
For years, it has been known that human response to irradiation varies greatly. It is now thought that a person’s degree of radiosensitivity is genetically defined and is generally not caused by one mutation in one gene, like in AT patients, but is caused by a number of variations in multiple genes. These variations can be analysed using single nucleotide polymorphisms (SNPs) and are of huge interest in establishing interindividual differences in response to irradiation. A role for non-genetic factors, such as smoking, is also observed [115]. Recruiting patients for large population studies, however, is a re-occurring problem for researchers. In the USA, only 9.3% of patients undergoing radiotherapy take part in clinical trials, which, together with the fragmented nature of the healthcare system, results in smaller trials with little statistical power [116]. In the UK and Europe, large studies involving thousands of donors have been established to investigate the genetic sources that contribute to the variation among radiotherapy patients. These studies include the Genetic Predictors of Adverse Radiotherapy Effects Project (Gene-PARE), GENEtic pathways for the Prediction of the effects of Irradiation (GENEPI), Radiation Genomics (RadGenomics) and Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy (RAPPER), which attempt to link genomic data to the toxicity observed in cancer patients. These studies all aim to identify sources of radiosensitivity in patients, which could be useful to doctors in developing treatment plans tailored to each individual patient. However, recent results from the RAPPER study, involving a large study group of 1613 patients with breast and prostate cancer, failed to find a correlation between SNPs and radiation toxicity [117]. The RAPPER study is still ongoing, with plans to include larger studies across the Radiogenomics Consortium into the analysis.
CONCLUSION
The future of biomarker development has enormous promise for the improvement of medical care of individuals receiving both planned and accidental radiation exposures. Variation among the human population is now being researched extensively through functional assays, gene expression and detection of SNPs. This genomic analysis of the response to radiation exposure has the potential to individualise treatment for each patient. Overall, future biomarker research is focused on developing strategies capable of monitoring exposure, treatment success and individual patient response. Biomarker research therefore has an important role to help improve clinical outcomes and patient safety.
REFERENCES
- 1.International Organisation for Standardisation Radiation protection-performance criteria for service laboratories performing biological dosimetry by cytogenetics. ISO 19238. Geneva, Switzerland. International Organization for Standardization; 2004 [Google Scholar]
- 2.International Organisation for Standardisation Radiation protection—performance criteria for laboratories performing cytogenetic triage for assessment of mass casualties in radiological or nuclear emergencies—general principles and application to dicentric assay. ISO 19238. Geneva, Switzerland: International Organization for Standardization; 2008 [Google Scholar]
- 3.International Atomic Energy Agency Cytogenetic dosimetry: applications in preparedness for and response to radiation emergencies. Vienna, Austria: International Atomic Energy Agency; 2011 [Google Scholar]
- 4.Szluinska M, Edwards AA, Lloyd DC. Statistical methods for biological dosimetry. Chilton, UK: Health Protection Agency; 2005 [Google Scholar]
- 5.Lloyd DC, Edwards AA, Moquet JE, Guerrero-Carbajal YC. The role of cytogenetics in early triage of radiation casualities. Appl Radiat Isot 2000;52:1107–12 [DOI] [PubMed] [Google Scholar]
- 6.Vaurijoux A, Gruel G, Pouzoulet F, Grégoire E, Martin C, Roch-Lefèvre S, et al. Strategy for population triage based on dicentric analysis. Radiat Res 2009;171:541–8 10.1667/RR1664.1 [DOI] [PubMed] [Google Scholar]
- 7.Thierens H, Vral A, Barbé M, Aousalah B, De Riddler L. A cytogenetic study of nuclear power plant workers using the micronucleus-centromere assay. Mutat Res 1999;445:105–11 [DOI] [PubMed] [Google Scholar]
- 8.Thierens H, Vral A, Morthier R, Aousalah B, De Riddler L. Cytogenetic monitoring of hospital workers occupationally exposed to ionizing radiation using the micronucleus centromere assay. Mutagenesis 2000;15:245–9 [DOI] [PubMed] [Google Scholar]
- 9.Thomas P, Umegaki K, Fenech M. Nucleoplasmic bridges are a sensitive measure of chromosome rearrangement in the cytokinesis-block micronucleus assay. Mutagenesis 2003;18:187–94 [DOI] [PubMed] [Google Scholar]
- 10.International Atomic Energy Agency The radiological accident in Istanbul. Vienna, Austria: International Atomic Energy Agency; 2000 [Google Scholar]
- 11.Wolff HA, Hennies S, Herrmann M, Rave-Fränk M, Eickelmann D, Virsik P, et al. Comparison of the micronucleus and chromosome aberration techniques for the documentation of cytogenetic damage in radiochemotherapy-treated patients with rectal cancer. Strahlenther Onkol 2011;187:52–8 [DOI] [PubMed] [Google Scholar]
- 12.Willems P, August L, Slabbert J, Romm H, Oestreicher U, Thierens H, et al. Automated micronucleus (MN) scoring for population triage in case of large scale radiation events. Int J Radiat Biol 2010;86:2–11 10.3109/09553000903264481 [DOI] [PubMed] [Google Scholar]
- 13.Rossnerova A, Spatova M, Schunck C, Sram RJ. Automated scoring of lymphocyte micronuclei by the MetaSystems Metafer image cytometry system and its application in studies of human mutagen sensitivity and biodosimetry of genotoxin exposure. Mutagenesis 2011;26:169–75 10.1093/mutage/geq057 [DOI] [PubMed] [Google Scholar]
- 14.Fenech M. The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys 2010;98:234–43 10.1097/HP.0b013e3181b85044 [DOI] [PubMed] [Google Scholar]
- 15.Johnson RT, Rao PN. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature 1970;226:717–22 [DOI] [PubMed] [Google Scholar]
- 16.Pantelias GE, Maillie HD. The use of peripheral blood mononuclear cell prematurely condensed chromosomes for biological dosimetry. Radiat Res 1984;99:140–50 [PubMed] [Google Scholar]
- 17.Hatzi VI, Terzoudi GI, Paraskevopoulou C, Makropoulos V, Matthopoulos DP, Pantelias GE. The use of premature chromosome condensation to study the influence of environmental factors on human genetic material in interphase cells. ScientificWorldJournal 2006;6:1174–90 10.1100/tsw.2006.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasanna PG, Blakely WF. Premature chromosome condensation in human resting peripheral blood lymphocytes for chromosome aberration analysis using specific whole-chromosome DNA hybridisation probes. Methods Mol Biol 2005;291:49–57 [DOI] [PubMed] [Google Scholar]
- 19.Hayata I, Kanda R, Minamihisamatsu M, Furukawa M, Sasaki MS. Cytogenetical dose estimation for 3 severely exposed patients in the JCO criticality accident in Tokai-mura. J Radiat Res 2001;42:S149–55 [DOI] [PubMed] [Google Scholar]
- 20.Lindholm C, Stricklin D, Jaworska A, Koivistoinen A, Paile W, Arvidsson E, et al. Premature chromosome condensation (PCC) assay for dose assessment in mass casualty accidents. Radiat Res 2010;173:71–8 10.1667/RR1843.1 [DOI] [PubMed] [Google Scholar]
- 21.Darroudi F, Natarajan AT, Bentvelzen PA, Heidt PJ, Van Rotterdam A, Zoetelief J. Detection of total- and partial-body irradiation in a monkey model: a comparative study of chromosomal aberration, micronucleus and premature chromosome condensation assays. Int J Radiat Biol 1998;74:207–15 [DOI] [PubMed] [Google Scholar]
- 22.Lindholm C, Edwards A. Long-term persistence of translocations in stable lymphocytes from victims of a radiological accident. Int J Radiat Biol 2004;80:559–66 10.1080/09553000412331283498 [DOI] [PubMed] [Google Scholar]
- 23.Tawn EJ, Whitehouse CA. Persistence of translocation frequencies in blood lymphocytes following radiotherapy: implications for retrospective radiation biodosimetry. J Radiol Prot 2003;23:423–30 [DOI] [PubMed] [Google Scholar]
- 24.Bauchinger M, Schmid E, Zitzelsberger H, Braselmann H, Nahrstedt U. Radiation-induced chromosome aberrations analysed by two-color fluorescence in situ hybridization with composite whole chromosome-specific DNA probes and a pancentromeric DNA probe. Int J Radiat Biol 1993;64:179–84 [DOI] [PubMed] [Google Scholar]
- 25.Lucas JN, Awa A, Straume T, Poggensee M, Kodama Y, Nakano M, et al. Rapid translocation frequency analysis in humans decades after exposure to ionising radiation. Int J Radiat Biol 1992;62:53–63 [DOI] [PubMed] [Google Scholar]
- 26.Edwards AA, Lindholm C, Darroudi F, Stephan G, Romm H, Barquinero J, et al. Review of translocations detected by FISH for retrospective biological dosimetry applications. Radiat Prot Dosimetry 2005;113:396–402 10.1093/rpd/nch452 [DOI] [PubMed] [Google Scholar]
- 27.Sigurdson AJ, Ha M, Hauptmann M, Bhatti P, Sram RJ, Beskid O, et al. International study of factors affecting human chromosome translocations. Mutat Res 2008;652:112–21 10.1016/j.mrgentox.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucker JD, Luckinbill LS. Estimating the lowest detectable dose of ionising radiation by FISH whole-chromosome painting. Radiat Res 2011;175:631–7 10.1667/RR2506.1 [DOI] [PubMed] [Google Scholar]
- 29.Bhatti P, Yong LC, Doody MM, Preston DL, Kampa DM, Ramsey MJ, et al. Diagnostic X-ray examinations and increased chromosome translocations: evidence from three studies. Radiat Environ Biophys 2010;49:685–92 10.1007/s00411-010-0307-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-strand breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858–68 [DOI] [PubMed] [Google Scholar]
- 31.Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A 2003;100:5057–62 10.1073/pnas.0830918100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothkamm K, Balroop S, Shekdar J, Fernie P, Goh V. Leukocyte DNA damage after multi-detector row CT: a quantitative biomarker of low-level radiation exposure. Radiology 2007;242:244–51 10.1148/radiol.2421060171 [DOI] [PubMed] [Google Scholar]
- 33.Sak A, Grehl S, Erichsen P, Engelhard M, Grannass A, Levegrün S, et al. Gamma-H2AX foci formation in peripheral blood lymphocytes of tumour patients after local radiotherapy to different sites of the body: dependence on the dose-distribution, irradiated site and time from start of treatment. Int J Radiat Biol 2007;83:639–52 [DOI] [PubMed] [Google Scholar]
- 34.Horn S, Bernard S, Rothkamm K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS One 2011;6:e25113 10.1371/journal.pone.0025113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner HC, Brenner DJ, Chen Y, Bertucci A, Zhang J, Wang H, et al. Adapting the γ-H2AX for automated processing in human lymphocytes. 1. Technological aspects. Radiat Res 2011;175:282–90 10.1667/RR2125.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pope IA, Barber PR, Horn S, Ainsbury E, Rothkamm K, Vojnovic B. A portable microfluidic fluorescence spectrometer device for H2AX-based biological dosimetry. Radiation Measurements 2011;46:907–11 10.1016/j.radmeas.2011.02.004 [DOI] [Google Scholar]
- 37.Ainsbury EA, Bakhanova E, Barquinero JF, Brai M, Chumak V, Correcher V, et al. Review of retrospective dosimetry techniques for external ionising radiation exposures. Radiat Prot Dosimetry 2011;147:573–92 10.1093/rpd/ncq499 [DOI] [PubMed] [Google Scholar]
- 38.Kabacik S, Mackay A, Tamber N, Manning G, Finnon P, Paillier F, et al. Gene expression following ionising radiation: identification of biomarkers for dose estimation and prediction of individual response. Int J Radiat Biol 2011;87:115–29 10.3109/09553002.2010.519424 [DOI] [PubMed] [Google Scholar]
- 39.Amundson SA, Do KT, Fornace AJ., Jr Induction of stress genes by low doses of gamma rays. Radiat Res 1999;152:225–31 [PubMed] [Google Scholar]
- 40.Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, et al. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLoS One 2008;3:e1912 10.1371/journal.pone.0001912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fält S, Holmberg K, Lambert B, Wennborg A. Long-term global gene expression patterns in irradiated human lymphocytes. Carcinogenesis 2003;24:1837–45 10.1093/carcin/bgg134 [DOI] [PubMed] [Google Scholar]
- 42.Morandi E, Severini C, Quercioli D, Perdichizzi S, Mascolo M, Horn W, et al. Gene expression changes in medical workers exposed to radiation. Radiat Res 2009;172:500–8 10.1667/RR1545.1 [DOI] [PubMed] [Google Scholar]
- 43.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med 2007;4:e106 10.1371/journal.pmed.0040106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q. Human in vivo radiation-induced biomarkers: gene expression changes in radiotherapy patients. Cancer Res 2004;64:6368–71 10.1158/0008-5472.CAN-04-1883 [DOI] [PubMed] [Google Scholar]
- 45.Badie C, Dziwura S, Raffy C, Tsigani T, Alsbeih G, Moody J, et al. Aberrant CDKNIA transcriptional response associates with abnormal sensitivity to radiation treatment. Br J Cancer 2008;98:1845–51 10.1038/sj.bjc.6604381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabacik S, Ortega-Molina A, Efeyan A, Finnon P, Bouffler S, Serrano M, et al. A minimally invasive assay for individual assessment of the ATM/CHEK2/p53 pathway activity. Cell Cycle 2011;10:1152–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grace MB, McLeland CB, Gagliardi SJ, Smith JM, Jackson WE, 3rd, Blakely WF. Development and assessment of a quantitative reverse transcription-PCR assay for simultaneous measurement of four amplicons. Clin Chem 2003;49:1467–75 [DOI] [PubMed] [Google Scholar]
- 48.Kang CM, Park KP, Song JE, Jeoung DI, Cho CK, Kim TH, et al. Possible biomarkers for ionising radiation exposure in human peripheral blood lymphocytes. Radiat Res 2003;159:312–19 [DOI] [PubMed] [Google Scholar]
- 49.Manning G, Kabacik S, Finnon P, Bouffler S, Badie C. High and low dose responses of transcriptional biomarkers in ex vivo X-irradiated human blood. Int J Radiat Biol Feb. 2013 doi: 10.3109/09553002.2013.769694. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 50.Brengues M, Paap B, Bittner M, Amundson S, Seligmann B, Korn R, et al. Biodosimetry on small blood volume using gene expression assay. Health Phys 2010;98:179–85 10.1097/01.HP.0000346706.44253.5c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meadows SK, Dressman HK, Daher P, Himburg H, Russell JL, Doan P, et al. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PLoS One 2010;5:e11535, 2010. 10.1371/journal.pone.0011535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gruel G, Voisin P, Vaurijoux A, Roch-Lefevre S, Grégoire E, Maltere P, et al. Broad modulation of gene expression in CD4+ lymphocyte subpopulations in response to low doses of ionising radiation. Radiat Res 2008;170:335–44 10.1667/RR1147.1 [DOI] [PubMed] [Google Scholar]
- 53.Mori M, Benotmane MA, Tirone I, Hooghe-Peters EL, Desaintes C. Transcriptional response to ionising radiation in lymphocyte subsets. Cell Mol Life Sci 2005;62:1489–501 10.1007/s00018-005-5086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bustin SA. Why the need for QPCR publication guidelines? The case for MIQE. Methods 2010;50:217–26 10.1016/j.ymeth.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 55.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009;55:611–22 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 56.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 2001;29:365–71 10.1038/ng1201-365 [DOI] [PubMed] [Google Scholar]
- 57.Brenner DJ, Doll R, Goodhead DT, Hall EJ, Land CE, Little JB, et al. Cancer risks attributable to low doses of ionising radiation: assessing what we really know. Proc Natl Acad Sci U S A 2003;100:13761–6 10.1073/pnas.2235592100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenner DJ, Hall EJ. Computed tomography—an increasing source of radiation exposure. N Engl J Med 2007;357:2277–84 [DOI] [PubMed] [Google Scholar]
- 59.Stephen AE, Segev DL, Ryan DP, Mullins ME, Kim SH, Schnitzer JJ, et al. The diagnosis of acute appendicitis in a pediatric population: to CT or not to CT. J Pediatr Surg 2003;38:367–71 10.1053/jpsu.2003.50110 [DOI] [PubMed] [Google Scholar]
- 60.Preston DL, Cullings H, Suyama A, Funamoto S, Nishi N, Soda M, et al. Solid cancer incidence in atomic bomb survivors exposed in utero or as young children. J Natl Cancer Inst 2008;100:428–36 10.1093/jnci/djn045 [DOI] [PubMed] [Google Scholar]
- 61.Pearce MS, Salotti JA, Little MP, McHugh K, Lee C, Kim KP, et al. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet 2012;380:499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beels L, Bacher K, De Wolf D, Werbrouck J, Thierens H. Gamma-H2AX foci as a biomarker for patient X-ray exposure in pediatric cardiac catheterization: are we understanding radiation risks? Circulation 2009;10:1903–9 [DOI] [PubMed] [Google Scholar]
- 63.Löbrich M, Rief N, Kühne M, Heckmann M, Fleckenstein J, Rübe C, et al. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci U S A 2005;102:8984–9 10.1073/pnas.0501895102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jost G, Golfier S, Pietsch H, Lengsfeld P, Voth M, Schmid TE, et al. The influence of x-ray contrast agents in computed tomography on the induction of dicentrics and gamma-H2AX foci in lymphocytes of human blood samples. Phys Med Biol 2009;54:6029–39 10.1088/0031-9155/54/20/001 [DOI] [PubMed] [Google Scholar]
- 65.Frankenberg D, Kelnhofer K, Bär K, Frankenberg-Schwager M. Enhanced neoplastic transformation by mammography X rays relative to 200 kVp X rays: indication for a strong dependence on photon energy of the RBE(M) for various end points. Radiat Res 2002;157:99–105 [DOI] [PubMed] [Google Scholar]
- 66.Göggelmann W, Jacobsen C, Panzer W, Walsh L, Roos H, Schmid E. Re-evaluation of the RBE of 29 kV x-rays (mammography x-rays) relative to 220 kV x-rays using neoplastic transformation of human CGL1-hybrid cells. Radiat Environ Biophys 2003;42:175–82 [DOI] [PubMed] [Google Scholar]
- 67.Andrieu N, Easton DF, Chang-Claude J, Rookus MA, Brohet R, Cardis E, et al. Effect of chest x-rays on the risk of breast cancer among BRCA1/2 mutation carriers in the international BRCA1/2 carrier cohort study: a report from the EMBRACE, GENEPSO, GEO-HEBON and IBCCS collaborators group. J Clin Oncol 2006;24:3361–6 [DOI] [PubMed] [Google Scholar]
- 68.Colin C, Devic C, Noël A, Rabilloud M, Zabot MT, Pinet-Isaac S, et al. DNA double-strand breaks induced by mammographic screening procedures in human mammary epithelial cells. Int J Radiat Biol 2011;87:1103–12 10.3109/09553002.2011.608410 [DOI] [PubMed] [Google Scholar]
- 69.Office for National Statistics Breast cancer: incidence, mortality and survival. London, UK: Office for National Statistics; 2010 [Google Scholar]
- 70.Hoffman DA, Lonstein JE, Morin MM, Visscher W, Harris BS, 3rd, Boice JD., 3rd Breast cancer in women with scoliosis exposed to multiple diagnostic x-rays. J Natl Cancer Inst 1989;81:1307–12 [DOI] [PubMed] [Google Scholar]
- 71.Boice JD, Jr, Preston D, Davis FG, Monson RR. Frequent chest X-ray fluoroscopy and breast cancer incidence among tuberculosis patients in Massachusetts. Radiat Res 1991;125:214–22 [PubMed] [Google Scholar]
- 72.Bhatti P, Preston DL, Doody MM, Hauptmann M, Kampa D, Alexander BH, et al. Retrospective biodosimetry among United States radiologic technologists. Radiat Res 2007;167:727–34 10.1667/RR0894.1 [DOI] [PubMed] [Google Scholar]
- 73.Svensson JP, Stalpers LJ, Esveldt-van Lange RE, Franken NA, Haveman J, Klein B, et al. Analysis of gene expression using gene sets discriminates cancer patients with and without late radiation toxicity. PLoS Med 2006;3:e422 10.1371/journal.pmed.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badie C, Goodhardt M, Waugh A, Doyen N, Foray N, Calsou P, et al. A DNA double-strand break defective fibroblast cell line (180BR) derived from a radiosensitive patient represents a new mutant phenotype. Cancer Res 1997;15:4600–7 [PubMed] [Google Scholar]
- 75.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol 1999;1:699–702 [DOI] [PubMed] [Google Scholar]
- 76.Barnett GC, West CM, Dunning AM, Elliott RM, Coles CE, Pharoah PD, et al. Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer 2009;9:134–42 10.1038/nrc2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crosbie JC, Anderson RL, Rothkamm K, Restall CM, Cann L, Ruwanpura S, et al. Tumor cell response to synchrotron microbeam radiation therapy differs markedly from cells in normal tissues. Int J Radiat Oncol Biol Phys 2010;77:886–94 10.1016/j.ijrobp.2010.01.035 [DOI] [PubMed] [Google Scholar]
- 78.Rothkamm K, Crosbie JC, Daley F, Bourne S, Barber PR, Vojnovic B, et al. In situ biological dose mapping estimates the radiation burden delivered to “spared” tissue between synchrotron x-ray microbeam radiotherapy tracks. PLoS One 2012;7:e29853 10.1371/journal.pone.0029853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chua ML, Somaiah N, Bourne S, Daley F, A'Hern R, Nuta O, et al. Inter-individual and inter-cell type variation in residual DNA damage after in vivo irradiation of human skin. Radiother Oncol 2011;99:225–30 10.1016/j.radonc.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 80.Rübe CE, Grudzenski S, Kühne M, Dong X, Rief N, Löbrich M, et al. DNA double-strand break repair of blood lymphocytes and normal tissues analysed in a preclinical mouse model: implications for radiosensitivity testing. Clin Cancer Res 2008;14:6546–55 10.1158/1078-0432.CCR-07-5147 [DOI] [PubMed] [Google Scholar]
- 81.Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res 2008;68:605–14 10.1158/0008-5472.CAN-07-5472 [DOI] [PubMed] [Google Scholar]
- 82.Wardman P, Rothkamm K, Folkes LK, Woodcock M, Johnston PJ. Radiosensitization by nitric oxide at low radiation doses. Radiat Res 2007;167:475–84 10.1667/RR0827.1 [DOI] [PubMed] [Google Scholar]
- 83.Lancaster JR, Jr, Xie K. Tumors face NO problems? Cancer Res 2006;66:6459–62 10.1158/0008-5472.CAN-05-2900 [DOI] [PubMed] [Google Scholar]
- 84.Stea B, Suh JH, Boyd AP, Cagnoni PJ, Shaw E, REACH Whole-brain radiotherapy with or without efaproxiral for the treatment of brain metastases: determinants of response and its prognostic value for subsequent survival. Int J Radiat Oncol Biol Phys 2006;64:1023–30 10.1016/j.ijrobp.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 85.Keys HM, Bundy BN, Stehman FB, Muderspach LI, Chafe WE, Suggs CL, et al. Cisplatin, radiation and adjuvant hysterectomy compared with radiation and adjuvant hysterectomy for bulky stage IB cervical carcinoma. N Engl J Med 1999;340:1154–61 10.1056/NEJM199904153401503 [DOI] [PubMed] [Google Scholar]
- 86.Whitney CW, Sause W, Bundy BN, Malfetano JH, Hannigan EV, Fowler WC, Jr, et al. Randomized comparison of fluorouracil plus cisplatin versus hydroxyurea as an adjunct to radiation therapy in stage IIB-IVA carcinoma of the cervix with negative para-aortic lymph nodes: a gynecologic oncology group and southwest oncology group study. J Clin Oncol 1999;17:1339–48 [DOI] [PubMed] [Google Scholar]
- 87.Wardman P. Chemical radiosensitizers for use in radiotherapy. Clin Oncol 2007;19:397–417 10.1016/j.clon.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 88.Somaiah N, Yarnold J, Daley F, Pearson A, Gothard L, Rothkamm K, et al. The relationship between homologous recombination repair and the sensitivity of human epidermis to the size of daily doses over a 5-week course of breast radiotherapy. Clin Cancer Res 2012;18:5479–88 10.1158/1078-0432.CCR-10-3297 [DOI] [PubMed] [Google Scholar]
- 89.Lin SH, Wang L, Myles B, Thall PF, Hofstetter WL, Swisher SG, et al. Propensity score-based comparison of long-term outcomes with 3-dimensional conformal radiotherapy vs intensity-modulated radiotherapy for esophageal cancer. Int J Radiat Oncol Biol Phys 2012;84:1078–85 10.1016/j.ijrobp.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spratt DE, Pei X, Yamada J, Kollmeier MA, Cox B, Zelefsky MJ. Long-term survival and toxicity in patients treated with high-dose intensity modulated radiation therapy for localized prostate cancer. Int J Radiat Oncol Biol Phys 2013;85:686–92 10.1016/j.ijrobp.2012.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zwicker F, Swartman B, Sterzing F, Major G, Weber K, Huber PE, et al. Biological in-vivo measurement of dose distribution in patients' lymphocytes by gamma-H2AX immunofluorescence staining: 3D conformal- vs. step-and-shoot IMRT of the prostate gland. Radiat Oncol 2011;6:62 10.1186/1748-717X-6-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bentzen SM, Hendry JH. Variability in the radiosensitivity of normal cells and tissues. Report from a workshop organised by the European society for therapeutic radiology and oncology in Edinburgh, UK, 19 September 1998. Intl J Radiat Biol 1999;75:513–17 [DOI] [PubMed] [Google Scholar]
- 93.Peacock J, Ashton A, Bliss J, Bush C, Eady J, Jackson C, et al. Cellular radiosensitivity and complication risk after curative radiotherapy. Radiother Oncol 2000;55:173–8 [DOI] [PubMed] [Google Scholar]
- 94.Russell N, Grummels A, Hart AA, Smolders IJ, Borger J, Bartelink H, et al. Low predictive value of intrinsic fibroblast radiosensitivity for fibrosis development following radiotherapy for breast cancer. Int J Radiat Biol 1998;73:661–70 [DOI] [PubMed] [Google Scholar]
- 95.Lee TK, Allison RR, O'Brien KF, Johnke RM, Christie KI, Naves JL, et al. Lymphocyte radiosensitivity correlated with pelvic radiotherapy morbidity. Int J Radiat Oncol Biol Phys 2003;57:222–9 [DOI] [PubMed] [Google Scholar]
- 96.Cantena C, Conti D, Parasacchi P, Marenco P, Bortolato B, Botturi M, et al. Micronuclei in cytokinesis-blocked lymphocytes may predict patient response to radiotherapy. Intl J Radiat Biol 1996;70:301–8 [DOI] [PubMed] [Google Scholar]
- 97.Finnon P, Robertson N, Dziwura S, Raffy C, Zhang W, Ainsbury L, et al. Evidence for significant heritability of apoptotic and cell cycle responses to ionising radiation. Hum Genet 2008;123:485–93 10.1007/s00439-008-0500-1 [DOI] [PubMed] [Google Scholar]
- 98.Camplejohn RS, Hodgson S, Carter N, Kato BS, Spector TD. Heritability of DNA-damage-induced apoptosis and its relationship with age in lymphocytes from female twins. Br J Cancer 2006;95:520–4 10.1038/sj.bjc.6603257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crompton NE, Miralbell R, Rutz HP, Ersoy F, Sanal O, Wellmann D, et al. Altered apoptotic profiles in irradiated patients with increased toxicity. Int J Radiat Oncol Biol Phys 1999;45:707–14 [DOI] [PubMed] [Google Scholar]
- 100.Crompton NE, Shi YQ, Emery GC, Wisser L, Blattmann H, Maier A, et al. Sources of variation in patient response to radiation treatment. Int J Radiat Oncol Biol Phys 2001;49:547–54 [DOI] [PubMed] [Google Scholar]
- 101.Ozsahin M, Crompton NE, Gourgou S, Kramar A, Li L, Shi Y, et al. CD4 and CD8 T-lymphocyte assay can predict radiation-induces late toxicity: a prospective study in 399 patients. Clin Cancer Res 2005;11:7426–33 10.1158/1078-0432.CCR-04-2634 [DOI] [PubMed] [Google Scholar]
- 102.Turesson I, Nyman J, Holmberg E, Odén A. Prognostic factors for acute and late skin reactions in radiotherapy patients. Int J Radiat Oncol Biol Phys 1996;36:1065–75 [DOI] [PubMed] [Google Scholar]
- 103.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov J, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999;286:531–7 [DOI] [PubMed] [Google Scholar]
- 104.Rieger KE, Hong WJ, Tusher VG, Tang J, Tibshirani R, Chu G. Toxicity from a radiation therapy associated with abnormal transcriptional responses to DNA damage. Proc Natl Acad Sci U S A 2004;101:6635–40 10.1073/pnas.0307761101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhan Q, Alamo I, Yu K, Boise LH, Cherney B, Tosato G, et al. The apoptosis-associated gamma-ray response of BCL-X(L) depends on normal p53 function. Oncogene 1996;13:2287–93 [PubMed] [Google Scholar]
- 106.Sonis S, Haddad R, Posner M, Watkins B, Fey E, Morgan TV, et al. Gene expression changes in peripheral blood cells provide insight into the biological mechanisms associated with regimen-related toxicities in patients being treated for head and neck cancers. Oral Oncol 2007;43:289–300 [DOI] [PubMed] [Google Scholar]
- 107.Rübe CE, Fricke A, Schneider R, Simon K, Kühne M, Fleckenstein J, et al. DNA repair alterations in children with pediatric malignancies: novel opportunities to identify patients at risk for high-grade toxicities. Int J Radiat Oncol Biol Phys 2010;78:359–69 10.1016/j.ijrobp.2009.08.052 [DOI] [PubMed] [Google Scholar]
- 108.Bourton EC, Plowman PN, Smith D, Arlett CF, Parris CN. Prolonged expression of the γ-H2AX DNA repair biomarker correlates with excess acute and chronic toxicity from radiotherapy treatment. Int J Cancer 2011;129:2928–34 10.1002/ijc.25953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fleckenstein J, Kühne M, Seegmüller K, Derschang S, Melchior P, Gräber S. The impact of individual in vivo repair of DNA double-strand breaks on oral mucositis in adjuvant radiotherapy of head-and-neck cancer. Int J Radiat Oncol Biol Phys 2011;81:1465–72 10.1016/j.ijrobp.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 110.Werbrouck J, Duprez F, De Neve W, Thierens H. Lack of a correlation between H2AX foci kinetics in lymphocytes and the severity of acute normal tissue reactions during IMRT treatment for head and neck cancer. Int J Radiat Biol 2011;87:46–56 10.3109/09553002.2010.518213 [DOI] [PubMed] [Google Scholar]
- 111.Werbrouck J, De Ruyck K, Beels L, Vral A, Van Eijkeren M, De Neve W, et al. Prediction of late normal tissue complications in RT treated gynaecological cancer patients: potential of the gamma-H2AX foci assay and association with chromosomal radiosensitivity. Oncol Rep 2010;23:571–8 [PubMed] [Google Scholar]
- 112.Hoeller U, Borgmann K, Bonacker M, Kuhlmey A, Bajrovic A, Jung H, et al. Individual radiosensitivity measured with lymphocytes may be used to predict the risk of fibrosis after radiotherapy for breast cancer. Radiother Oncol 2003;69:137–44 [DOI] [PubMed] [Google Scholar]
- 113.Borgmann K, Hoeller U, Nowack S, Bernhard M, Röper B, Brackrock S, et al. Individual radiosensitivity measured with lymphocytes may predict the risk of acute reaction after radiotherapy. Int J Radiat Oncol Biol Phys 2008;71:256–64 10.1016/j.ijrobp.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 114.Chua ML, Somaiah N, A’Hern R, Davies S, Gothard L, Yarnold J, et al. Residual DNA and chromosomal damage in ex vivo irradiated blood lymphocytes correlated with late normal tissue response to breast radiotherapy. Radiother Oncol 2011;99:362–6 10.1016/j.radonc.2011.05.071 [DOI] [PubMed] [Google Scholar]
- 115.Health Protection Agency Human radiosensitivity. Report of the independent advisory group on ionising radiation. Chilton, UK: Health Protection Agency; 2013 [Google Scholar]
- 116.Movsas B, Moughan J, Owen J, Coia LR, Zelefsky MJ, Hanks G, et al. Who enrolls onto clinical oncology trials? A radiation patterns of care study analysis. Int J Radiat Oncol Biol Phys 2007;68:1145–50 10.1016/j.ijrobp.2007.01.051 [DOI] [PubMed] [Google Scholar]
- 117.Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol 2012;13:65–77 10.1016/S1470-2045(11)70302-3 [DOI] [PubMed] [Google Scholar]