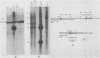Abstract
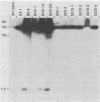
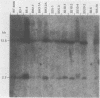
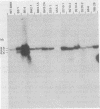
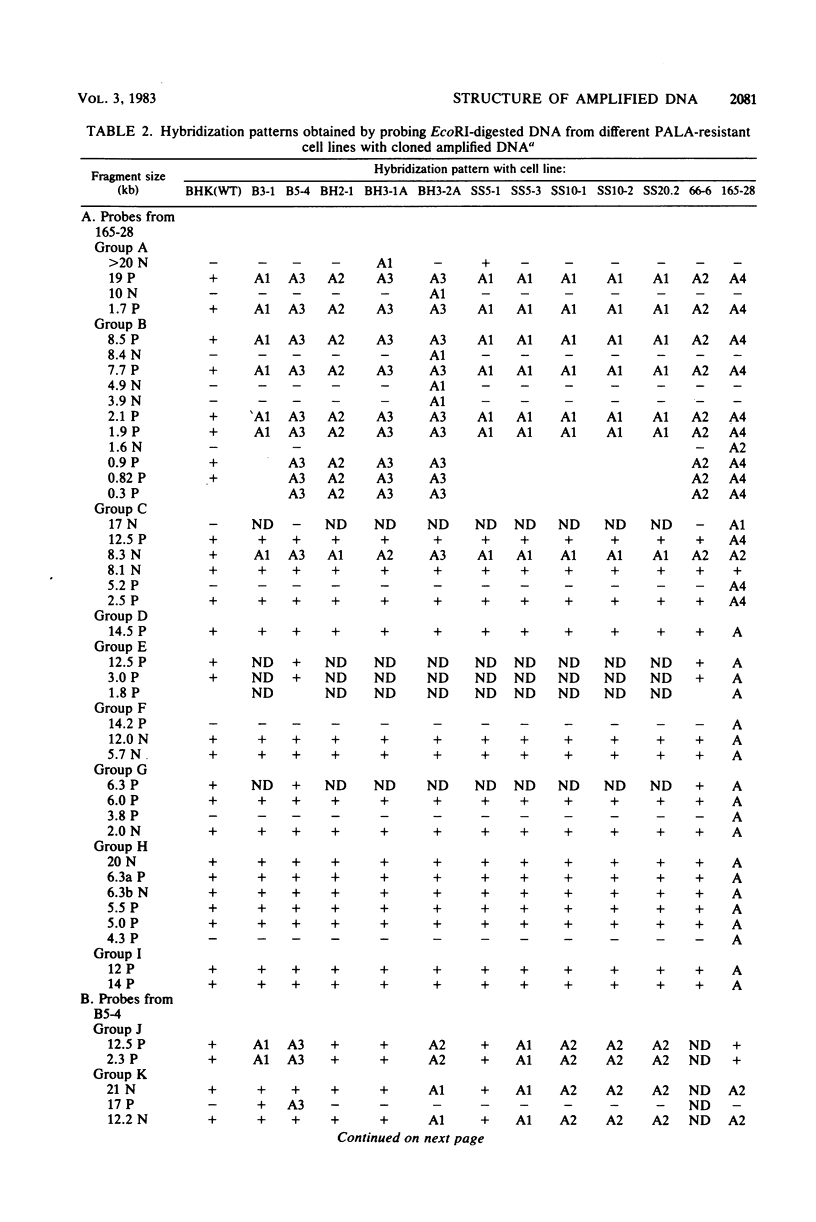
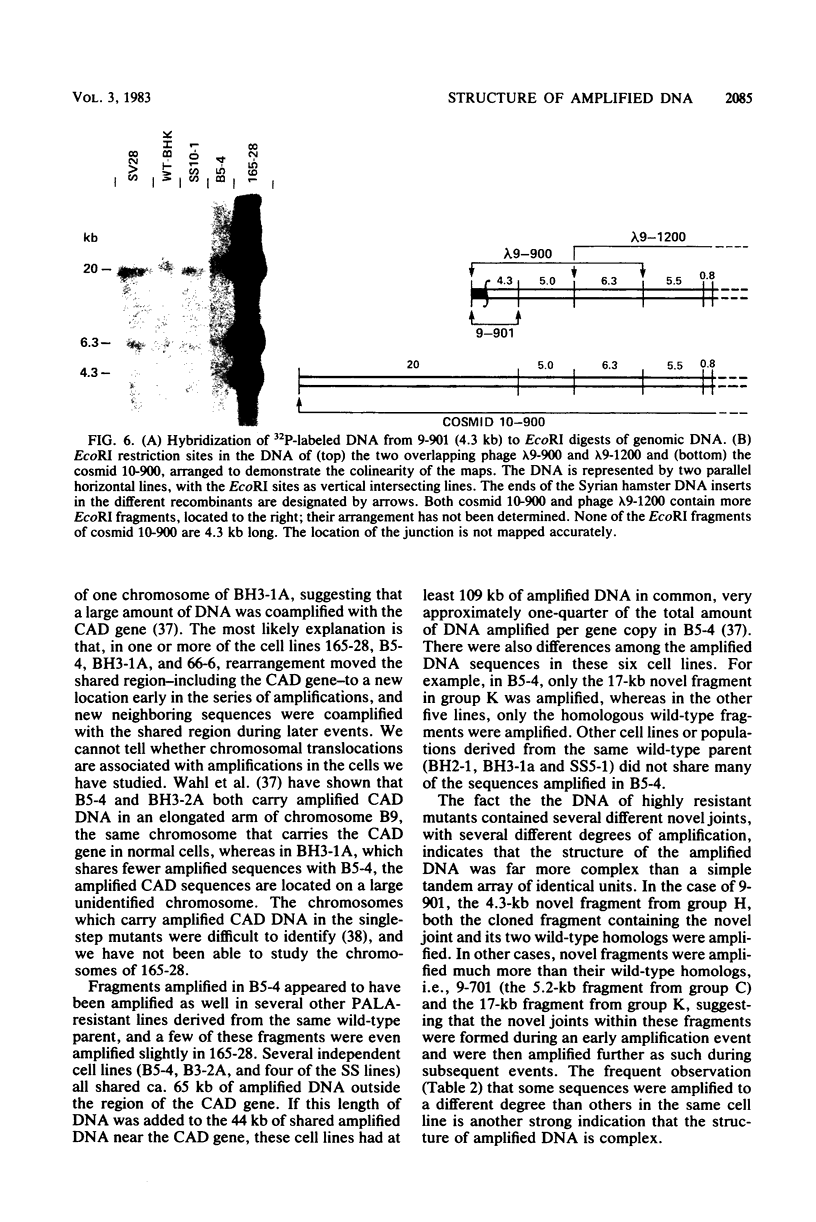
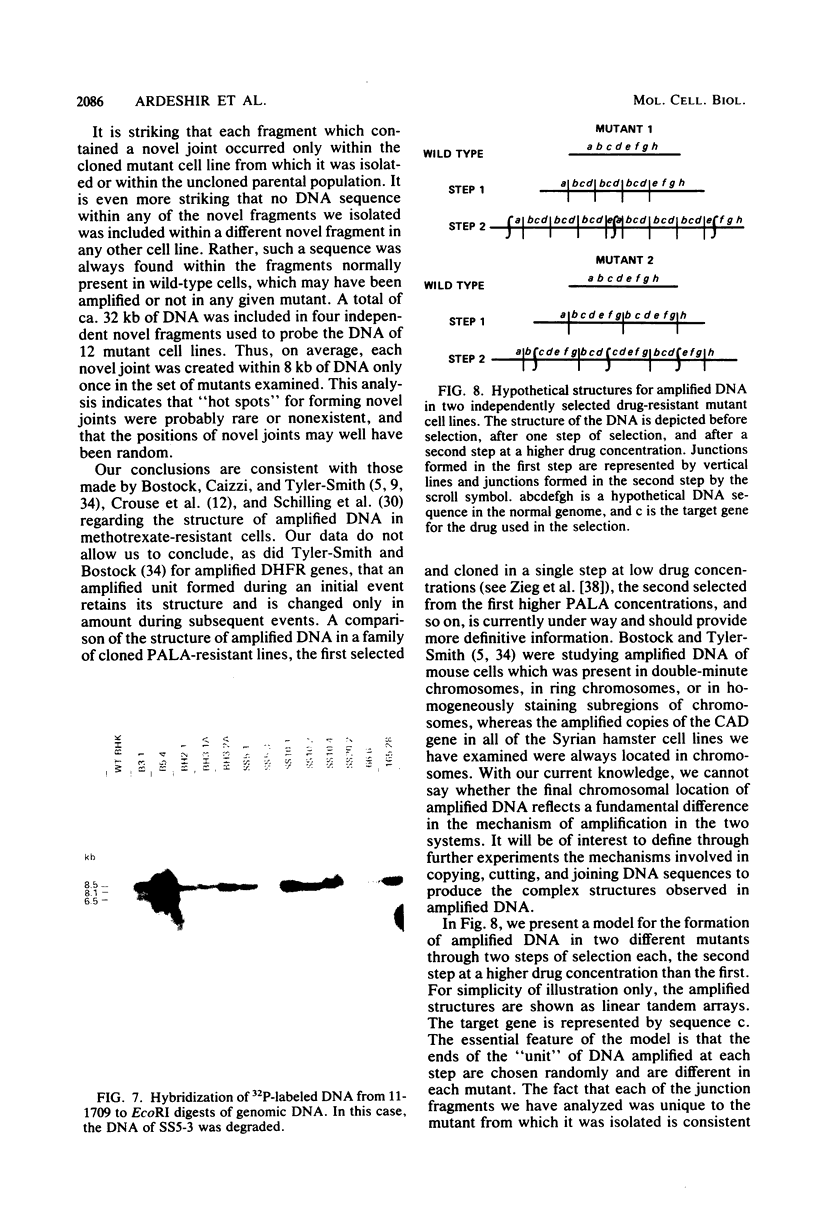
Syrian hamster cell lines selected in multiple steps for resistance to high levels of N-(phosphonacetyl)-L-aspartate (PALA) contain many copies of the gene coding for the pyrimidine pathway enzyme CAD. Approximately 500 kilobases of additional DNA was coamplified with each copy of the CAD gene in several cell lines. To investigate its structure and organization, we cloned ca. 162 kilobases of coamplified DNA from cell line 165-28 and ca. 68 kilobases from cell line B5-4, using a screening method based solely on the greater abundance of amplified sequences in the resistant cells. Individual cloned fragments were then used to probe Southern transfers of genomic DNA from 12 different PALA-resistant mutants and the wild-type parents. A contiguous region of DNA ca. 44 kilobases long which included the CAD gene was amplified in all 12 mutants. However, the fragments cloned from 165-28 which were external to this region were not amplified in any other mutant, and the external fragments cloned from B5-4 were not amplified in two of the mutants. These results suggest that movement or major rearrangement of DNA may have accompanied some of the amplification events. We also found that different fragments were amplified to different degrees within a single mutant cell line. We conclude that the amplified DNA was not comprised of identical, tandemly arranged units. Its structure was much more complex and was different in different mutants. Several restriction fragments containing amplified sequences were found only in the DNA of the mutant cell line from which they were isolated and were not detected in DNA from wild-type cells or from any other mutant cells. These fragments contained novel joints created by rearrangement of the DNA during amplification. The cloned novel fragments hybridized only to normal fragments in every cell line examined, except for the line from which each novel fragment was isolated or the parental population for that line. This result argues that "hot spots" for forming novel joints are rare or nonexistent.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Beach L. R., Palmiter R. D. Amplification of the metallothionein-I gene in cadmium-resistant mouse cells. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2110–2114. doi: 10.1073/pnas.78.4.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Bostock C. J., Clark E. M. Satellite DNA in large marker chromosomes of methotrexate-resistant mouse cells. Cell. 1980 Mar;19(3):709–715. doi: 10.1016/s0092-8674(80)80047-x. [DOI] [PubMed] [Google Scholar]
- Bostock C. J., Tyler-Smith C. Gene amplification in methotrexate-resistant mouse cells. II. Rearrangement and amplification of non-dihydrofolate reductase gene sequences accompany chromosomal changes. J Mol Biol. 1981 Dec 5;153(2):219–236. doi: 10.1016/0022-2836(81)90275-8. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brennand J., Chinault A. C., Konecki D. S., Melton D. W., Caskey C. T. Cloned cDNA sequences of the hypoxanthine/guanine phosphoribosyltransferase gene from a mouse neuroblastoma cell line found to have amplified genomic sequences. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1950–1954. doi: 10.1073/pnas.79.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brison O., Ardeshir F., Stark G. R. General method for cloning amplified DNA by differential screening with genomic probes. Mol Cell Biol. 1982 May;2(5):578–587. doi: 10.1128/mcb.2.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caizzi R., Bostock C. J. Gene amplification in methotrexate-resistant mouse cells. IV. Different DNA sequences are amplified in different resistant lines. Nucleic Acids Res. 1982 Nov 11;10(21):6597–6618. doi: 10.1093/nar/10.21.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S., Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982 Aug 12;298(5875):679–681. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- Cowell J. K. Double minutes and homogeneously staining regions: gene amplification in mammalian cells. Annu Rev Genet. 1982;16:21–59. doi: 10.1146/annurev.ge.16.120182.000321. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Simonsen C. C., McEwan R. N., Schimke R. T. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982 Jul 10;257(13):7887–7897. [PubMed] [Google Scholar]
- Dalla-Favera R., Wong-Staal F., Gallo R. C. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature. 1982 Sep 2;299(5878):61–63. doi: 10.1038/299061a0. [DOI] [PubMed] [Google Scholar]
- Dolnick B. J., Berenson R. J., Bertino J. R., Kaufman R. J., Nunberg J. H., Schimke R. T. Correlation of dihydrofolate reductase elevation with gene amplification in a homogeneously staining chromosomal region in L5178Y cells. J Cell Biol. 1979 Nov;83(2 Pt 1):394–402. doi: 10.1083/jcb.83.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L., Sternberg N. In vitro packaging of lambda Dam vectors and their use in cloning DNA fragments. Methods Enzymol. 1979;68:281–298. doi: 10.1016/0076-6879(79)68020-5. [DOI] [PubMed] [Google Scholar]
- George D. L., Powers V. E. Cloning of DNA from double minutes of Y1 mouse adrenocortical tumor cells: evidence for gene amplification. Cell. 1981 Apr;24(1):117–123. doi: 10.1016/0092-8674(81)90507-9. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J., Brown P. C., Schimke R. T. Amplified dihydrofolate reductase genes in unstably methotrexate-resistant cells are associated with double minute chromosomes. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5669–5673. doi: 10.1073/pnas.76.11.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe T. D., Swyryd E. A., Bruist M., Stark G. R. Stable mutants of mammalian cells that overproduce the first three enzymes of pyrimidine nucleotide biosynthesis. Cell. 1976 Dec;9(4 Pt 1):541–550. doi: 10.1016/0092-8674(76)90036-2. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M., Guild G. M., Prestidge L. S., Hogness D. S. A new high-capacity cosmid vector and its use. Gene. 1980 Nov;11(3-4):271–282. doi: 10.1016/0378-1119(80)90067-0. [DOI] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Chang A. C., Cohen S. N., Schimke R. T. Structure and genomic organization of the mouse dihydrofolate reductase gene. Cell. 1980 Feb;19(2):355–364. doi: 10.1016/0092-8674(80)90510-3. [DOI] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Schimke R. T., Urlaub G., Chasin L. A. Amplified dihydrofolate reductase genes are localized to a homogeneously staining region of a single chromosome in a methotrexate-resistant Chinese hamster ovary cell line. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5553–5556. doi: 10.1073/pnas.75.11.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Wahl G. M., Stark G. R. Structure of the gene for CAD, the multifunctional protein that initiates UMP synthesis in Syrian hamster cells. Mol Cell Biol. 1982 Mar;2(3):293–301. doi: 10.1128/mcb.2.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts J. M., Buck L. B., Axel R. A structure for amplified DNA. Cell. 1983 May;33(1):53–63. doi: 10.1016/0092-8674(83)90334-3. [DOI] [PubMed] [Google Scholar]
- Seed B. Diazotizable arylamine cellulose papers for the coupling and hybridization of nucleic acids. Nucleic Acids Res. 1982 Mar 11;10(5):1799–1810. doi: 10.1093/nar/10.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swyryd E. A., Seaver S. S., Stark G. R. N-(phosphonacetyl)-L-aspartate, a potent transition state analog inhibitor of aspartate transcarbamylase, blocks proliferation of mammalian cells in culture. J Biol Chem. 1974 Nov 10;249(21):6945–6950. [PubMed] [Google Scholar]
- Tyler-Smith C., Alderson T. Gene amplification in methotrexate-resistant mouse cells. I. DNA rearrangement accompanies dihydrofolate reductase gene amplification in a T-cell lymphoma. J Mol Biol. 1981 Dec 5;153(2):203–218. doi: 10.1016/0022-2836(81)90274-6. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C., Bostock C. J. Gene amplification in methotrexate-resistant mouse cells. III. Interrelationships between chromosome changes and DNA sequence amplification or loss. J Mol Biol. 1981 Dec 5;153(2):237–256. doi: 10.1016/0022-2836(81)90276-x. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Vitto L., Padgett R. A., Stark G. R. Single-copy and amplified CAD genes in Syrian hamster chromosomes localized by a highly sensitive method for in situ hybridization. Mol Cell Biol. 1982 Mar;2(3):308–319. doi: 10.1128/mcb.2.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Vincent B. R., Delbrück S., Eckhart W., Meinkoth J., Vitto L., Wahl G. The cloning and reintroduction into animal cells of a functional CAD gene, a dominant amplifiable genetic marker. Cell. 1981 Dec;27(2 Pt 1):267–277. doi: 10.1016/0092-8674(81)90410-4. [DOI] [PubMed] [Google Scholar]