Abstract
Borrelia burgdorferi, the spirochete that causes Lyme disease, differentially regulates synthesis of the outer membrane lipoprotein OspC to infect its host. OspC is required to establish infection but then repressed in the mammal to avoid clearance by the adaptive immune response. Inverted repeats (IR) upstream of the promoter have been implicated as an operator to regulate ospC expression. We molecularly dissected the distal inverted repeat (dIR) of the ospC operator by site-directed mutagenesis at its endogenous location on the circular plasmid cp26. We found that disrupting the dIR but maintaining the proximal IR prevented induction of OspC synthesis by DNA supercoiling, temperature, and pH. Moreover, the base-pairing potential of the two halves of the dIR was more important than the nucleotide sequence in controlling OspC levels. These results describe a cis-acting element essential for the expression of the virulence factor OspC.
Introduction
Lyme disease is caused by the spirochete Borrelia burgdorferi, which is transmitted via an Ixodes tick [1]–[3]. B. burgdorferi is maintained in an enzootic cycle between its tick vector and a vertebrate host reservoir [4]–[6]. Naïve larvae acquire B. burgdorferi when feeding on an infected animal and can then transmit the bacterium to uninfected hosts as nymphs during feeding, completing the cycle. B. burgdorferi encounters disparate, and hostile, environments as it transitions through the enzootic cycle; the spirochete has evolved a repertoire of strategies, which involve the regulation of a variety of genes, to respond and adapt to these changes during its lifecycle [6]–[8].
The differential syntheses of outer surface lipoproteins (Osp), which are the interface between B. burgdorferi and its hosts, is paramount to the ability to infect, survive and replicate in both the tick and the vertebrate [6]–[10]. These regulated genes encode wide-ranging functions; for example, VlsE mediates evasion from the vertebrate immune system [11]–[13], while OspA binds to a tick midgut protein and protects the spirochetes from the incoming blood meal [14]–[17]. One of the best-studied lipoproteins is OspC [18], which is required for transmission and the early stages of mammalian infection [19]–[22]. The ospC gene is carried on the conserved 26-kb circular plasmid cp26 [23], [24]. The precise function of OspC remains elusive, but the outer membrane lipoprotein appears to have several activities [25], including providing initial protection from the innate immune system [26] and facilitating dissemination [27]. Additionally, OspC has a ligand-binding domain essential for its function [28] and binds the tick salivary protein Salp15 [29] as well as mammalian plasminogen [30], [31], which may assist in transmission and dissemination, respectively. OspC is highly immunogenic, so its synthesis must be repressed for the spirochete to persist in the mammal [32]–[34]. B. burgdorferi that continue to produce OspC during infection of immunocompetent mice are cleared [34] and ospC expressed in trans from a shuttle vector is selected against during mammalian infection [35]. While ospC is present in all B. burgdorferi strains examined, the sequence is variable, with only strains carrying certain alleles capable of disseminating and establishing infection in humans [13], [36]–[39].
OspC synthesis is induced in vitro in response to increased temperature, which presumably mimics a signal that occurs during tick feeding [40]–[42]. Subsequent studies showed that numerous factors such as pH [43], [44], DNA supercoiling [42], oxygen [45], carbon dioxide [46], acetate [47], and transition metals [48] also control ospC expression. External signals are transmitted through a unique signaling pathway involving the sequential action of two alternative sigma factors, RpoN (σ54) and RpoS (σs) [49], [50]. RpoN, in collaboration with the response regulator Rrp2 [51]–[54] and the transcription factor BosR [55]–[58], activates transcription of rpoS; RpoS, in turn, activates transcription of ospC [49] and other genes [54], [59], [60]. Another level of control of ospC expression is exerted during translation of rpoS. The small RNA DsrABb [61], [62] and the RNA chaperone Hfq [63] control rpoS translation and subsequently ospC transcription. However, there is no evidence for post-transcriptional control of the ospC gene [42], [44]–[49], [52], [61], [64].
Previous studies identified an ospC operator consisting of two overlapping inverted repeats (IRs) 42 bp upstream of the major transcriptional start site [23], [65]–[67]. The operator is highly conserved in B. burgdorferi sensu stricto [67], but there is some controversy about its role in ospC gene regulation. Eggers et al. [64] showed that the operator is required for full ospC expression in vitro, while Yang et al. [68] found the operator is dispensable for induction and repression. In addition, mutants lacking the operator were unable to persist in immunocompetent mice and were also cleared from SCID mice injected with transferred anti-OspC antibodies, suggesting that the operator is required to repress ospC expression during infection [69]. One caveat to these previous studies is that ospC was expressed in trans in an ospC null background strain. In the current study, we molecularly dissect the ospC operator in its native locus on cp26 and show that the distal IR is an important cis-acting element controlling OspC expression.
Materials and Methods
Bacterial Strains and Culture Conditions
Low-passage B. burgdorferi strain 297 (BbAH130) [49] and all mutant strains were maintained in Barbour-Stoenner-Kelly II (BSK-II) liquid medium, pH 7.6, containing 6% rabbit serum [70]. In temperature shift experiments, cultures were passaged twice at 23°C, before inoculating cultures at 1×105 cells ml−1 and growing at 23°C to late log phase (5 to 9×107 cells ml−1) or inoculating cultures at 1×103 cells ml−1 and growing at 34°C to late log phase. For temperature downshift experiments, cultures were grown to log phase at 34°C before inoculating cultures at 1×105 cells ml−1 and growing at 23°C to late log phase. In experiments examining the effect of pH, cultures at 34°C in BSK II at pH 8.0 were passaged into BSK II at pH 7.0. To determine the effect of DNA supercoiling, cultures were grown at 23°C (inoculated at 1×105 cells ml−1) in the presence of 10 ng ml−1 coumermycin A1 (50 mg ml−1 stock in DMSO) or DMSO (solvent control) until late log phase [42]. Cell density was determined using a Petroff-Hausser counting chamber [71].
Construction of ospC Promoter Mutants
Mutations in the ospC operator were generated by allelic exchange on cp26 [72]. Constructs containing the operator mutants were made by overlap extension PCR [73]. The 5′ portion of the upstream construct was amplified by PCR of genomic DNA using KOD polymerase (Novagen) with the primers ospC U866F and ospCp mutHup(297)R or ospCp mutDH(297)R2 (Table 1). The 3′ portion of the upstream construct, which also includes the ospC gene, was amplified by PCR using the primers ospC D697R+AatII+AgeI and ospCp mutHup(297)F (Table 1). PCR products were separated and purified in a 1% agarose gel. Paired 5′ and 3′ portions of the upstream construct were then combined and extended for six cycles in a thermal cycler. Next, the primers ospC U866F and ospC D697R+AatII+AgeI (Table 1) were added and the combined upstream construct was amplified by PCR. To generate the construct for the wild-type control strain with the antibiotic resistance cassette, PCR was done using primers ospC U866F and ospC D697R+AatII+AgeI with genomic DNA as a template. PCR products were separated in a 1% agarose gel, gel purified, polyadenylated, and cloned into pCR®2.1-TOPO. PCR of genomic DNA with the primers ospC D673F+AatII and ospC D1572R+AgeI (Table 1) was used to amplify the downstream construct, which was cloned into pCR®2.1-TOPO as described above. The accuracy of all DNA constructs was confirmed by sequencing. The downstream sequence was inserted into the upstream ospC operator mutation constructs using the synthetic AatII and AgeI restriction sites. Lastly, the kanamycin resistance cassette flgBp-aphI [74] was inserted downstream of the ospC gene into the engineered AatII site. The orientation of flgBp-aphI was determined by PCR using the primers kanR 488R and ospC D1572R+AgeI (primers a and b, respectively). The plasmid was linearized by digestion with AhdI and electroporated into competent B. burgdorferi [61], [71], [72]. Transformants were cloned in liquid BSK-II by diluting the electroporated cells to less than one cell per well of a 96-well plate in medium containing kanamycin (200 µg ml−1) at 34°C and a 1.5% CO2 atmosphere [16]. Total genomic DNA was isolated from positive colonies and sequenced by the Murdock DNA Sequencing Facility at The University of Montana using the primer ospC U291F to confirm the site-directed ospC operator mutations.
Table 1. Oligonucleotides used in this studya.
| Name | Sequence (5′-3′) |
| ospC U866F | AGCTTAATTTTTTCCACAATGG |
| ospC D697R+AatII+AgeI | ACCGGT AATGACGTCTGACTTATATTGACTTTATTTTTCCAG |
| ospC D673F+AatII | GACGTC GGAAAAATAAAGTCAATATAAGTCAAG |
| ospC D1572R+AgeI | ACCGGT AATGGAAAAATTCCTAATGTCG |
| ospC U291F | ATTAGTTGGCTATATTGGG |
| kanR 488R | TCACTCGCATCAACCAAACC |
| ospCp mutHup(297)F | TAAGACAATATTGAAAAAATTCTTCAAT |
| ospCp mutHup(297)R | ATTGAAGAATTTTTTCAATATTGTCTTA |
| ospCp mutDH(297)R2 | TTTCAATTTTTTATTTTTTCAAATATTTGAAAAAATTCTTCAATATTT |
| flaB 423F | TTCTCAAAATGTAAGAACAGCTGAAGA |
| flaB 542R | TGGTTTGTCCAACATGAACTC |
| flaB probe | 6-FAM-TCACTTTCAGGGTCTCAAGCGTCTTGGAC-TAMRA |
| ospC F | CATGGGCAACTTGGAATTGA |
| ospC R | TTGCCAAGTTTTCTACTGCTTTAAATAG |
| ospC probe | 6-FAM-TAAAGATAAGGGCGCTGCAGAGC-TAMRA |
Synthetic restriction sites are underlined.
RNA Isolation and qRT-PCR Analysis
RNA was isolated from 40-ml cultures of B. burgdorferi grown at 23°C containing 10 ng ml−1 coumermycin A1 or DMSO as a solvent control using TRIzol (Invitrogen) as previously described [61], [75]. Samples were treated with Turbo DNase (Ambion) to remove contaminating DNA. Samples were screened by PCR to ensure that contaminating DNA had been removed using the primers flaB 423F and flaB 542R. cDNA was synthesized using 1 µg RNA with SuperScript® III for qRT-PCR (Invitrogen). flaB and ospC primers and probes were designed using Primer Express® version 3.0 (Applied Biosystems). TaqMan absolute qRT-PCR was performed in 96-well plates using an Applied Biosystems 7300 Real-Time PCR System and standard curves for flaB and ospC were generated using a portion of the flaB ORF (nucleotides 278–551 of the ORF) cloned into pCR®2.1-TOPO and B. burgdorferi strain 297 (BbAH130) genomic DNA, respectively. Values represent the mean (±SE) from three independent experiments.
SDS-PAGE and Immunoblotting
B. burgdorferi cultures were grown to late log phase at 23°C or 34°C and total cell lysates collected as previously described [61], [72]. Equal amounts of protein were separated on pre-cast Novex 4–20% Tris-Glycine polyacrylamide gels (Invitrogen). Proteins were transferred by electroblotting to PVDF Immobilon™ membranes (Millipore) and membranes blocked in Blocking Buffer (138 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, 0.05% Tween 20, 4% dry milk, and 1% goat serum) overnight at 4°C. Membranes were incubated with rabbit anti-OspC antibodies (1∶1000) [68], [76] or anti-FlaB antibodies (1∶1000) (a kind gift from Tom Schwan) in Blocking Buffer for 1 h at room temperature. Rabbit antibodies were detected by incubating membranes with goat anti-rabbit HRP-linked antibodies (Bio-Rad Laboratories) (1∶20,000) in Blocking Buffer for 1 h at room temperature. HRP-linked antibodies were visualized by chemiluminescence using Amersham™ ECL Plus (GE Healthcare) and images were collected using an LAS-3000 Intelligent Dark Box (Fujifilm Medical Systems USA). Images were processed using ImageJ (US National Institutes of Health and available at http://rsbweb.nih.gov/ij/) and Pixelmator (Pixelmator Team, Ltd).
Results
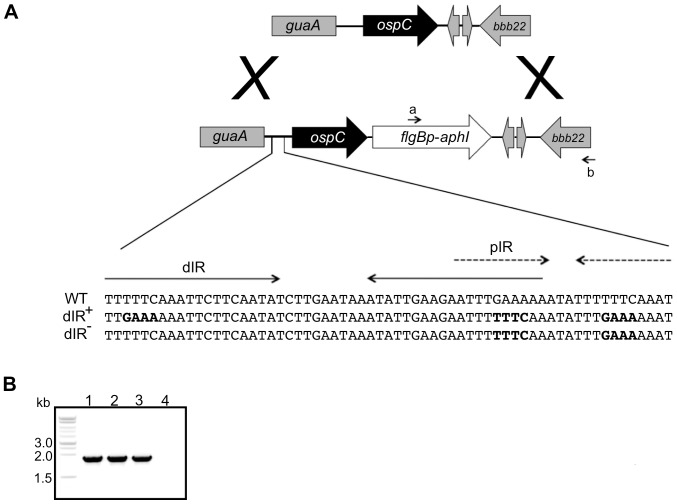
The ospC operator consists of a set of overlapping inverted repeats that are highly conserved in B. burgdorferi sensu stricto strains (Fig. 1) [65]–[67]. We hypothesized that the dIR has a specific role in responding to changes in DNA supercoiling, which is known to regulate ospC expression [42]. Increased negative DNA supercoiling can alter DNA structure, including extrusion of cruciforms from IRs, which can have a regulatory effect on transcription [77]–[79]. We generated site-directed mutations in the native ospC operator on cp26 that specifically disrupted the dIR (dIR–) and that changed the dIR sequence but retained complementarity (dIR+) (Fig. 2A; sequence changes are in bold). The orientation of the kanamycin resistance cassette, flgBp-aphI, downstream of the ospC gene was determined by PCR using the primer sets a+b (Fig. 2B). Changes in the operator sequence of the mutant strains were confirmed by DNA sequencing; about one-third of the kanamycin-resistant clones contained the site-directed mutations.
Figure 1. The sequence of the B. burgdorferi strain 297 ospC operator.
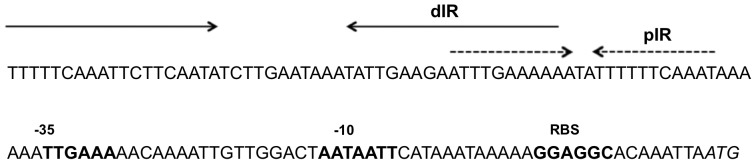
Two overlapping inverted repeats (IR): proximal (pIR, dashed arrows) and distal (dIR, solid arrows) of the ospC operator. The promoter (−10 and −35) and ribosome-binding site (RBS) are in bold and the translational start site is in italics.
Figure 2. The ospC operator and mutagenesis strategy.
(A) ospC operator mutations are linked to the kanamycin resistance cassette (flgBp-aphI). The sequence upstream of the ospC gene in B. burgdorferi strain 297 is shown (WT) with the dIR (solid arrows) and the overlapping pIR (dashed arrows). The nucleotides that have been changed are marked in bold. The strain nomenclature is as follows: dIR+ has the nucleotide sequence changed but the complementarity of the inverted repeats maintained, and dIR− has the distal inverted repeat disrupted but the complementarity of the proximal IR intact. (B) PCR of genomic DNA from ospC operator mutants (lane 1, WT with flgBp-aphI cassette; lane 2, dIR+; lane 3, dIR−; and lane 4, no template control) using primers primers kanR 488R (a) and ospC D1572R+AgeI (b) to determine the orientation of the flgBp-aphI antibiotic resistance cassette.
The Role of the dIR in ospC Expression Induced by Relaxation of DNA Supercoiling
We have previously shown that a decrease in negative DNA supercoiling causes an increase in ospC transcription [42]. The antibiotic coumermycin A1 relaxes DNA supercoiling by inhibiting DNA gyrase [80]–[83]. Wild-type and mutant strains were grown at 23°C in the presence of coumermycin A1 or DMSO (solvent only control) until late log phase to examine if the increase in ospC expression by the relaxation of supercoiling is mediated by the dIR of the operator. Transcript levels of ospC and flaB were measured by qRT-PCR. The fold increase in ospC transcript in the dIR+ strain grown in coumermycin A1, compared to the solvent control, was significantly greater than that seen in the dIR− strain (about thirteenfold compared to less than twofold) (Fig. 3). The change in flaB transcript levels was about twofold or less for both the dIR+ and dIR− strains (Fig. 3). These data suggest that the ability to form the dIR is an important regulator of coumermycin A1-mediated ospC expression.
Figure 3. ospC expression induced by relaxation of supercoiling is dependent on the dIR of the operator.
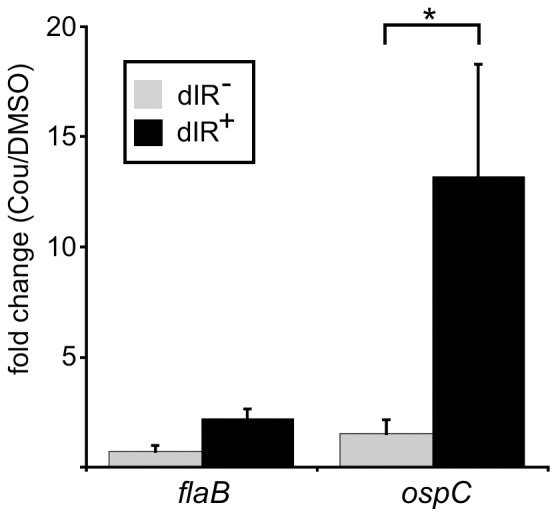
qRT-PCR analyses of flaB (gray bars) and ospC (black bars) mRNA levels from strains grown to late log phase at 23°C in 10 ng ml−1 coumermycin A1 (Cou) or in DMSO as a solvent control. Values represent the mean and error bars the SE of the mean from three independent experiments. * = P<0.05 by an unpaired t-test.
To examine if the changes in ospC transcript levels in response to coumermycin A1 treatment were reflected in OspC protein levels, total cell lysates were analyzed by Western blot using polyclonal antibodies to OspC to determine the levels of OspC synthesis. OspC levels increased in the wild-type 297 strain in response to relaxation of supercoiling when grown at 23°C in the presence of coumermycin A1 compared to cultures grown in the DMSO control (Fig. 4). Very little OspC was produced when the dIR− strain was grown in coumermycin A1 compared with the wild-type strain and the dIR+ strain (Fig. 4). We have confirmed these results with a second independently constructed clone of each of the mutant strains (data not shown). Taken together, these results suggest that the ability to form the dIR is more important than the actual sequence, at least in regard to the nucleotides that we mutated. These data imply that the increase in OspC levels stimulated by relaxation of DNA supercoiling is mediated via the dIR of the operator and that the secondary structure of the operator, rather than its sequence, plays a dominant role in regulating OspC levels.
Figure 4. The role of the dIR in OspC synthesis mediated by relaxation of DNA supercoiling.
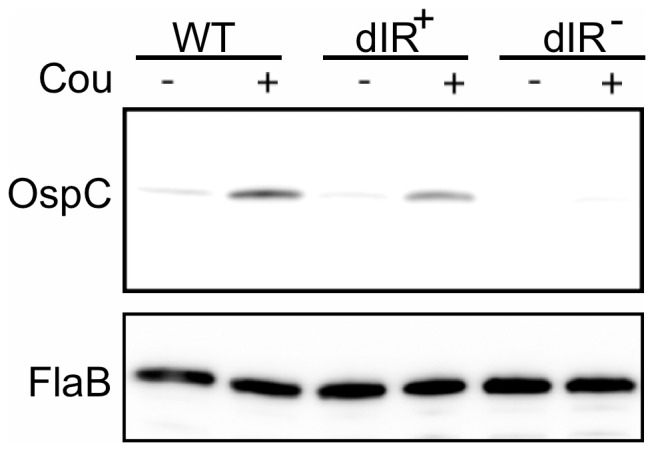
Immunoblot analysis of whole-cell lysates from strains grown to late log phase at 23°C in 10 ng ml−1 coumermycin A1 (Cou) (+) or in DMSO as a solvent control (−). Membranes were probed with antibodies against OspC (upper panel) or FlaB (lower panel).
The Role of the dIR in Temperature-regulated OspC Synthesis
A temperature shift from 23°C to 34°C is commonly used in vitro to mimic an environmental signal during tick feeding that induces virulence gene expression [8], [40]. To assess the role of the dIR in temperature-regulated OspC expression, cultures were grown at 23°C, shifted to 34°C and grown to late log phase. Total cell extracts were analyzed by Western blot. The wild-type 297 strain increased OspC synthesis upon a temperature shift (Fig. 5A). Similar to the coumermycin A1 treatment, OspC was not induced in response to temperature shift when the dIR was disrupted, while regenerating the complementarity of the dIR restored the ability to respond to increased temperature with increased OspC levels in the dIR+ strain (Fig. 5A). A second clone of each mutant, from an independent transformation, showed the same pattern of OspC levels during temperature shift (data not shown).
Figure 5. The dIR is required for OspC synthesis regulated by temperature.
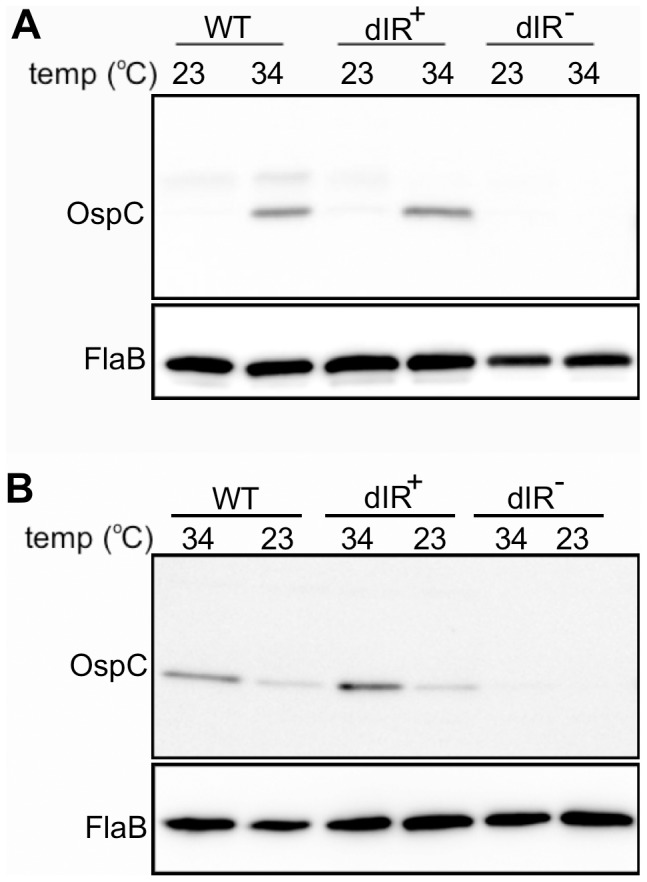
(A) Immunoblot analysis of whole-cell lysates from strains grown at 23°C and then temperature shifted to 34°C and grown to late log phase. The wild-type parental strain (297 WT) and the strain with a wild-type ospC operator linked to the antibiotic resistance cassette (WT) are controls. (B) Immunoblot analysis of whole-cell lysates from strains grown at 34°C and then temperature shifted to 23°C and grown to late log phase. Membranes were probed with antibodies against OspC (upper panels) or FlaB (lower panels).
We next assayed if the dIR was also involved in reducing OspC levels at 23°C. Cultures were grown to late log phase at 34°C and then passaged and grown at 23°C to late log phase. Again, changing the dIR sequence but maintaining complementarity in the dIR+ strain allowed for the reduction in OspC levels similar to the wild-type strain shifted to 23°C (Fig. 5B).
The Role of the dIR in pH-regulated OspC Synthesis
OspC levels have also been shown to be regulated by pH [43], [44], which is considered an environmental signal that changes during tick feeding: reducing the pH to 7.0 increases and raising the pH to 8.0 decreases OspC levels. To examine if pH-regulated OspC expression is mediated through the dIR, cultures were grown at 34°C in medium at pH 7.0 and then passaged into medium at pH 8.0. Total cell extracts were collected at late log phase and OspC levels were analyzed by Western blot. The dIR– strain did not show increased OspC levels at pH 7.0 and the dIR+ strain behaved like the wild-type strain in response to changing the pH (Fig. 6). Thus, all the signals examined, DNA supercoiling, temperature, and pH, control OspC levels through the dIR of the operator in vitro, suggesting a common mechanism, and the complementarity of the dIR may be more important than the specific sequence.
Figure 6. The role of the dIR in OspC synthesis mediated by pH increase.
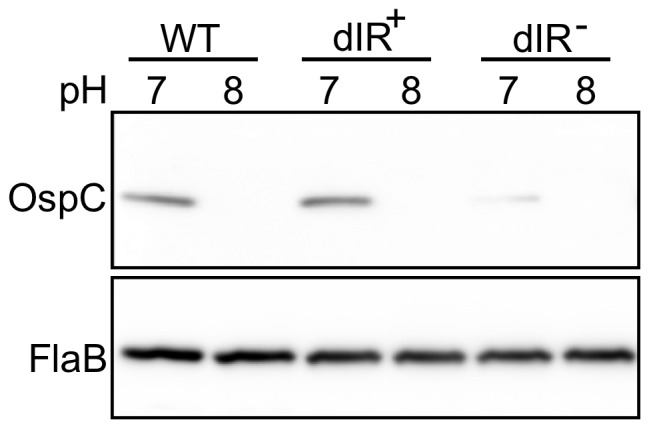
Immunoblot analysis of whole-cell lysates from strains grown to late log phase at 34°C at pH 7.0 or shifted to pH 8.0. Membranes were analyzed by probing with antibodies against OspC (upper panel) or FlaB (lower panel).
Discussion
Induction and repression of ospC transcription are crucial for B. burgdorferi to establish and maintain, respectively, an infection in mammals. A number of external factors, including temperature, pH, oxygen, carbon dioxide, acetate, and transition metals, regulate ospC expression [40]–[48], [72], [84]. Induction of ospC expression is generally accepted to be dependent on the RpoN-RpoS sigma factor cascade [49], which includes the regulatory proteins Rrp2 [51]–[54] and BosR [55]–[58]. Considerably less is known concerning the repression of ospC transcription, including the signals and accessory proteins involved. Here we show that the cis-acting dIR of the ospC operator functions to control expression and our data indicate that the base-pairing potential of the two halves of the inverted repeat, rather than the specific sequence, is essential for induction, thus providing a level of ospC-specific regulation downstream of RpoS.
The large intergenic region upstream of the ospC gene contains the operator and is highly conserved among B. burgdorferi sensu stricto strains, much more so than the ospC gene itself, likely indicating selective pressure on the regulation of ospC expression [65], [67]. Certain features of the operator, including the proximal IR (pIR), are broadly conserved throughout B. burgdorferi sensu lato genospecies, but, inexplicably, the dIR does not overlap the pIR in B. afzelii strains and is not even obviously present in B. garinii strains [67], suggesting alternate modes of gene regulation. Although we and others previously had shown that deleting the operator has little effect on the regulation of ospC transcription [68], [69], our current results more closely agree with Eggers et al. [64], who showed that the entire operator was required for full ospC expression in response to a temperature shift. These data suggest that the dIR plays a role in ospC regulation. This discrepancy may be explained by the differences in the experimental approaches between the studies. In the present work, we have utilized a more precise method to dissect the operator: site-directed mutations were generated in cis in the endogenous operator on cp26, while the other studies utilized truncated operator mutants in trans on a shuttle vector in an ospC null background. Thus, ospC expression in trans from a plasmid much smaller than cp26 (7 kb compared to 26 kb), albeit still circular, with a strong promoter fused to a selectable marker, may affect operator function, especially when DNA topology is likely involved [42], [68], [85]. In fact, OspC levels expressed in trans were elevated at 23°C compared to wild type, even though the plasmid-borne ospC contained the entire operator region [68].
Mutations that disrupt the dIR, but maintain the pIR (dIR− strain) prevent an increase in OspC levels in response to temperature, pH or relaxation of supercoiling, suggesting that all these signals function through a similar mechanism. Thus, the dIR is required for an increase in the amount of OspC. The finding that the ospC induction by relaxation of supercoiling at 23°C with coumermycin A1 (Fig. 3) depends on the dIR suggests that DNA topology has a regulatory role. These data imply that the regulatory element may be the DNA structure rather than the sequence, although we may not have mutated the nucleotides in the dIR+ strain that are important in regulation. We propose that the inverted repeats mediate the effect of DNA supercoiling, possibly by extruding a cruciform, or bind a trans-acting factor that recognizes an alternative DNA secondary structure. This provides a molecular mechanism for our previous observation that relaxation of supercoiling induces ospC expression [42].
Finally, we are aware of only a handful of studies in which site-directed mutations were introduced in cis into the genome of B. burgdorferi [51], [73], [86], [87] and this work provides an important caveat for interpreting genetic experiments involving introduction of DNA in trans, albeit the methodology is considerably more convenient. Our results add another level of complexity to ospC regulation suggesting that the DNA structure of the operator serves to mediate the external signals affecting expression.
Acknowledgments
We thank Christian Eggers for thoughtful and critical reading of the manuscript; Meghan Lybecker, Steve Lodmell, Aaron Bestor, Kit Tilly, Paul Policastro, Melissa Hargreaves, Amanda Brinkworth, and Cassie Abel Simonich for helpful discussions; and Tom Schwan for anti-FlaB antibodies and Mike Minnick for assistance generating anti-OspC antibodies.
Funding Statement
This work was supported by Public Health Service grants AI051486 to D.S.S. and P20 GM103546 to the Center for Biomolecular Structure and Dynamics from the National Institutes of Health. L.L.H.-H. was supported by a Watkins Scholarship from The University of Montana, an Undergraduate Research Internship through the National Science Foundation EPSCoR program under Grant EPS-0701906, an Undergraduate Research Award from the Davidson Honors College, and an Honors Fellowship through the Montana Integrative Learning Experience for Students (MILES) program under Grant 52005905 from the Howard Hughes Medical Institute-Undergraduate Science Education Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, et al. (1982) Lyme disease–a tick-borne spirochetosis? Science 216: 1317–1319. [DOI] [PubMed] [Google Scholar]
- 2. Benach JL, Bosler EM, Hanrahan JP, Coleman JL, Bast TF, et al. (1983) Spirochetes isolated from the blood of two patients with Lyme disease. N Engl J Med 308: 740–742. [DOI] [PubMed] [Google Scholar]
- 3. Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, et al. (1983) The spirochetal etiology of Lyme disease. N Engl J Med 308: 733–740. [DOI] [PubMed] [Google Scholar]
- 4. Lane RS, Piesman J, Burgdorfer W (1991) Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol 36: 587–609. [DOI] [PubMed] [Google Scholar]
- 5.Piesman J, Schwan TG (2010) Ecology of borreliae and their arthropod vectors. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norfolk, UK: Caister Academic Press. 251–278.
- 6. Radolf JD, Caimano MJ, Stevenson B, Hu LT (2012) Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh SK, Girschick HJ (2004) Molecular survival strategies of the Lyme disease spirochete Borrelia burgdorferi . Lancet Infect Dis 4: 575–583. [DOI] [PubMed] [Google Scholar]
- 8. Samuels DS (2011) Gene regulation in Borrelia burgdorferi . Annu Rev Microbiol 65: 479–499. [DOI] [PubMed] [Google Scholar]
- 9. Schwan TG (2003) Temporal regulation of outer surface proteins of the Lyme-disease spirochaete Borrelia burgdorferi . Biochem Soc Trans 31: 108–112. [DOI] [PubMed] [Google Scholar]
- 10. Kenedy MR, Lenhart TR, Akins DR (2012) The role of Borrelia burgdorferi outer surface proteins. FEMS Immunol Med Microbiol 66: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J-R, Hardham JM, Barbour AG, Norris SJ (1997) Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89: 275–285. [DOI] [PubMed] [Google Scholar]
- 12. McDowell JV, Sung S-Y, Hu LT, Marconi RT (2002) Evidence that the variable regions of the central domain of VlsE are antigenic during infection with Lyme disease spirochetes. Infect Immun 70: 4196–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brisson D, Drecktrah D, Eggers CH, Samuels DS (2012) Genetics of Borrelia burgdorferi . Annu Rev Genet 46: 515–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pal U, de Silva AM, Montgomery RR, Fish D, Anguita J, et al. (2000) Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J Clin Invest. 106: 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pal U, Li X, Wang T, Montgomery RR, Ramamoorthi N, et al. (2004) TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi . Cell 119: 457–468. [DOI] [PubMed] [Google Scholar]
- 16. Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV (2004) Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med 199: 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Battisti JM, Bono JL, Rosa PA, Schrumpf ME, Schwan TG, et al. (2008) Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect Immun 76: 5228–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuchs R, Jauris S, Lottspeich F, Preac-Mursic V, Wilske B, et al. (1992) Molecular analysis and expression of a Borrelia burgdorferi gene encoding a 22 kDa protein (pC) in Escherichia coli . Mol Microbiol 6: 503–509. [DOI] [PubMed] [Google Scholar]
- 19. Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, et al. (2004) Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci USA 101: 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, et al. (2004) OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest 113: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tilly K, Bestor A, Jewett MW, Rosa P (2007) Rapid clearance of Lyme disease spirochetes lacking OspC from skin. Infect Immun 75: 1517–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilly K, Bestor A, Rosa PA (2013) Lipoprotein succession in Borrelia burgdorferi: similar but distinct roles for OspC and VlsE at different stages of mammalian infection. Mol Microbiol: In press. [DOI] [PMC free article] [PubMed]
- 23. Marconi RT, Samuels DS, Garon CF (1993) Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol 175: 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Šadžiene A, Wilske B, Ferdows MS, Barbour AG (1993) The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun 61: 2192–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Radolf JD, Caimano MJ (2008) The long strange trip of Borrelia burgdorferi outer-surface protein C. Mol Microbiol. 69: 1–4. [DOI] [PubMed] [Google Scholar]
- 26. Stewart PE, Wang X, Bueschel DM, Clifton DR, Grimm D, et al. (2006) Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect Immun 74: 3547–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Q, McShan K, Liang FT (2008) Essential protective role attributed to the surface lipoproteins of Borrelia burgdorferi against innate defences. Mol Microbiol 69: 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Earnhart CG, LeBlanc DV, Alix KE, Desrosiers DC, Radolf JD, et al. (2010) Identification of residues within ligand-binding domain 1 (LBD1) of the Borrelia burgdorferi OspC protein required for function in the mammalian environment. Mol Microbiol 76: 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, et al. (2005) The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature 436: 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lagal V, Portnoï D, Faure G, Postic D, Baranton G (2006) Borrelia burgdorferi sensu stricto invasiveness is correlated with OspC-plasminogen affinity. Microbes Infect 8: 645–652. [DOI] [PubMed] [Google Scholar]
- 31. Önder Ö, Humphrey PT, McOmber B, Korobova F, Francella N, et al. (2012) OspC is potent plasminogen receptor on surface of Borrelia burgdorferi . J Biol Chem 287: 16860–16868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang FT, Nelson FK, Fikrig E (2002) Molecular adaptation of Borrelia burgdorferi in the murine host. J Exp Med 196: 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, et al. (2004) Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect Immun 72: 5759–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu Q, Seemanapalli SV, McShan K, Liang FT (2006) Constitutive expression of outer surface protein C diminishes the ability of Borrelia burgdorferi to evade specific humoral immunity. Infect Immun 74: 5177–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, et al. (2006) Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun 74: 3554–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, et al. (1999) Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun 67: 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Earnhart CG, Buckles EL, Dumler JS, Marconi RT (2005) Demonstration of OspC type diversity in invasive human Lyme disease isolates and identification of previously uncharacterized epitopes that define the specificity of the OspC murine antibody response. Infect Immun 73: 7869–7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dykhuizen DE, Brisson D, Sandigursky S, Wormser GP, Nowakowski J, et al. (2008) The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am J Trop Med Hyg 78: 806–810. [PMC free article] [PubMed] [Google Scholar]
- 39. Wormser G, Brisson D, Liveris D, Hanincová K, Sandigursky S, et al. (2008) Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis 198: 1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA (1995) Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA 92: 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stevenson B, Schwan TG, Rosa PA (1995) Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi . Infect Immun 63: 4535–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alverson J, Bundle SF, Sohaskey CD, Lybecker MC, Samuels DS (2003) Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi . Mol Microbiol 48: 1665–1677. [DOI] [PubMed] [Google Scholar]
- 43. Carroll JA, Garon CF, Schwan TG (1999) Effects of environmental pH on membrane proteins in Borrelia burgdorferi . Infect Immun 67: 3181–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, et al. (2000) Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi . Mol Microbiol 37: 1470–1479. [DOI] [PubMed] [Google Scholar]
- 45. Seshu J, Boylan JA, Gherardini FC, Skare JT (2004) Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi . Infect Immun 72: 1580–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hyde JA, Trzeciakowski JP, Skare JT (2007) Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J Bacteriol 189: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu H, Caimano MJ, Lin T, He M, Radolf JD, et al. (2010) Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi . PLoS Pathog 6: e1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Troxell B, Ye M, Yang Y, Carrasco SE, Lou Y, et al. (2013) Manganese and zinc regulate virulence determinants in Borrelia burgdorferi. Infect Immun: In press. [DOI] [PMC free article] [PubMed]
- 49. Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, et al. (2001) Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci USA 98: 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Caimano MJ, Eggers CH, Hazlett KR, Radolf JD (2004) RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect Immun 72: 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang XF, Alani SM, Norgard MV (2003) The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi . Proc Natl Acad Sci USA 100: 11001–11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, et al. (2007) Insights into the complex regulation of rpoS in Borrelia burgdorferi . Mol Microbiol 65: 277–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boardman BK, He M, Ouyang Z, Xu H, Pang X, et al. (2008) Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi . Infect Immun 76: 3844–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ouyang Z, Blevins JS, Norgard MV (2008) Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi . Microbiology 154: 2641–2658. [DOI] [PubMed] [Google Scholar]
- 55. Hyde JA, Shaw DK, Smith R III, Trzeciakowski JP, Skare JT (2009) The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both homeostatic and pathogenic properties of the Lyme disease spirochete. Mol Microbiol 74: 1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, et al. (2009) BosR (BB0647) governs virulence expression in Borrelia burgdorferi . Mol Microbiol 74: 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hyde JA, Shaw DK, Smith R III, Trzeciakowski JP, Skare JT (2010) Characterization of a conditional bosR mutant in Borrelia burgdorferi . Infect Immun 78: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ouyang Z, Deka RK, Norgard MV (2011) BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog 7: e1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, et al. (2005) Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci USA 102: 5162–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, et al. (2007) Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol 65: 1193–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lybecker MC, Samuels DS (2007) Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi . Mol Microbiol 64: 1075–1089. [DOI] [PubMed] [Google Scholar]
- 62. Archambault L, Linscott J, Swerdlow N, Boyland K, Riley E, et al. (2013) Translational efficiency of rpoS mRNA from Borrelia burgdorferi: effects of the length and sequence of the mRNA leader region. Biochem Biophys Res Commun 433: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lybecker MC, Abel CA, Feig AL, Samuels DS (2010) Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi . Mol Microbiol 78: 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eggers CH, Caimano MJ, Radolf JD (2004) Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J Bacteriol 186: 7390–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Margolis N, Hogan D, Tilly K, Rosa PA (1994) Plasmid location of Borrelia purine biosynthesis gene homologs. J Bacteriol 176: 6427–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Margolis N, Hogan D, Cieplak W Jr, Schwan TG, Rosa PA (1994) Homology between Borrelia burgdorferi OspC and members of the family of Borrelia hermsii variable major proteins. Gene 143: 105–110. [DOI] [PubMed] [Google Scholar]
- 67. Tilly K, Casjens S, Stevenson B, Bono M, Samuels DS, et al. (1997) The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol 23: 361–373. [DOI] [PubMed] [Google Scholar]
- 68. Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, et al. (2005) Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi . J Bacteriol 187: 4822–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xu Q, McShan K, Liang FT (2007) Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi . Mol Microbiol 64: 220–231. [DOI] [PubMed] [Google Scholar]
- 70. Barbour AG (1984) Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57: 521–525. [PMC free article] [PubMed] [Google Scholar]
- 71.Samuels DS (1995) Electrotransformation of the spirochete Borrelia burgdorferi. In: Nickoloff JA, editor. Electroporation Protocols for Microorganisms. Totowa, New Jersey: Humana Press. 253–259. [DOI] [PMC free article] [PubMed]
- 72. Gilbert MA, Morton EA, Bundle SF, Samuels DS (2007) Artificial regulation of ospC expression in Borrelia burgdorferi . Mol Microbiol 63: 1259–1273. [DOI] [PubMed] [Google Scholar]
- 73. Samuels DS, Mach KE, Garon CF (1994) Genetic transformation of the Lyme disease agent Borrelia burgdorferi with coumarin-resistant gyrB . J Bacteriol 176: 6045–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bono JL, Elias AF, Kupko III JJ, Stevenson B, Tilly K, et al. (2000) Efficient targeted mutagenesis in Borrelia burgdorferi . J Bacteriol 182: 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hoon-Hanks LL, Morton EA, Lybecker MC, Battisti JM, Samuels DS, et al. (2012) Borrelia burgdorferi malQ mutants utilize disaccharides and traverse the enzootic cycle. FEMS Immunol Med Microbiol 66: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Damman CJ, Eggers CH, Samuels DS, Oliver DB (2000) Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system. J Bacteriol 182: 6791–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hatfield GW, Benham CJ (2002) DNA topology-mediated control of global gene expression in Escherichia coli . Annu Rev Genet 36: 175–203. [DOI] [PubMed] [Google Scholar]
- 78. Saier MH Jr (2008) The bacterial chromosome. Crit Rev Biochem Mol Biol 43: 89–134. [DOI] [PubMed] [Google Scholar]
- 79. Dorman CJ (2013) Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat Rev Microbiol 11: 349–355. [DOI] [PubMed] [Google Scholar]
- 80. Drlica K, Snyder M (1978) Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol 120: 145–154. [DOI] [PubMed] [Google Scholar]
- 81. Gellert M, O'Dea MH, Itoh T, Tomizawa J (1976) Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci USA 73: 4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Maxwell A (1993) The interaction between coumarin drugs and DNA gyrase. Mol Microbiol 9: 681–686. [DOI] [PubMed] [Google Scholar]
- 83. Samuels DS, Garon CF (1993) Coumermycin A1 inhibits growth and induces relaxation of supercoiled plasmids in Borrelia burgdorferi, the Lyme disease agent. Antimicrob Agents Chemother 37: 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Montgomery RR, Malawista SE, Feen KJ, Bockenstedt LK (1996) Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J Exp Med 183: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Beaurepaire C, Chaconas G (2007) Topology-dependent transcription in linear and circular plasmids of the segmented genome of Borrelia burgdorferi . Mol Microbiol 63: 443–453. [DOI] [PubMed] [Google Scholar]
- 86. Knight SW, Kimmel BJ, Eggers CH, Samuels DS (2000) Disruption of the Borrelia burgdorferi gac gene, encoding the naturally synthesized GyrA C-terminal domain. J Bacteriol 182: 2048–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Earnhart CG, Rhodes DV, Marconi RT (2011) Disulfide-mediated oligomer formation in Borrelia burgdorferi outer surface protein C, a critical virulence factor and potential Lyme disease vaccine candidate. Clin Vaccine Immunol 18: 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]