Abstract
Inflammatory macrophages are abundant in kidney disease, stimulating repair, or driving chronic inflammation and fibrosis. Damage associated molecules (DAMPs), released from injured cells engage pattern recognition receptors (PRRs) on macrophages, contributing to activation. Understanding mechanisms of macrophage activation during kidney injury may lead to strategies to alleviate chronic disease. We identified Triggering-Receptor-in-Myeloid-cells (TREM)-1, a regulator of TLR signaling, as highly upregulated in kidney inflammatory macrophages and tested the roles of these receptors in macrophage activation and kidney disease. Kidney DAMPs activated macrophages in vitro, independently of TREM-1, but partially dependent on TLR-2/−4, MyD88. In two models of progressive interstitial kidney disease, TREM-1 blockade had no impact on disease or macrophage activation in vivo, but TLR-2/−4, or MyD88 deficiency was anti-inflammatory and anti-fibrotic. When MyD88 was mutated only in the myeloid lineage, however, there was no bearing on macrophage activation or disease progression. Instead, TLR-2/−4 or MyD88 deficiency reduced activation of mesenchyme lineage cells resulting in reduced inflammation and fibrosis, indicating that these pathways play dominant roles in activation of myofibroblasts but not macrophages. To conclude, TREM-1, TLR2/4 and MyD88 signaling pathways are redundant in myeloid cell activation in kidney injury, but the latter appear to regulate activation of mesenchymal cells.
Introduction
Inflammatory monocytes which are recruited to sites of tissue injury and differentiate into tissue effector macrophages have been shown to play important roles in the progression of chronic kidney diseases and the resolution of acute kidney injury [1], [2]. We previously showed that in mouse models of progressive kidney injury, three subpopulations of macrophages can be discerned in the kidney parenchyma defined by the cell surface marker Ly6C [3]. The Ly6Chigh population was activated similarly to the M1 activation defined in vitro, and the Ly6Clow subpopulation bore some similarities to M2a macrophages with the hallmarks of a profibrotic population. We also showed that Ly6Chigh macrophages differentiate into Ly6Clow macrophages [3]. Overall macrophages in this model promote injury and fibrosis. Since all populations of macrophages were activated we hypothesized that danger associated molecular patterns (DAMPs) may play important roles in their activation and that specific pattern recognition receptors (PRRs) may regulate the response of macrophages to DAMPs.
Identification of injury molecules and receptors that activate monocytes when they enter the injured kidney is critical to the development of new treatments focused on macrophages. Increasing evidence points to DAMPs, released from injured parenchymal cells, as critical factors that contribute to the pro-inflammatory phenotype in the injured kidney via PRR-binding and subsequent activation of NF-κB, MAPK and inflammasome signaling [4], [5]. The PRRs that have been most intensely investigated as receptors for DAMPs are the Toll-Like Receptors (TLRs). TLRs are germline encoded transmembrane receptors that recognize pathogen-associated molecular patterns and initiate an intracellular signaling cascade leading ultimately to an inflammatory response. All of the TLRs, with the exception of TLR-3, require the MyD88 adapter protein for maximal response. A number of DAMPs have been identified as ligands for different TLRs, including mitochondrial DNA (TLR-9), histones (TLR-4), hyaluronan fragments (TLR-2 and 4), high mobility group box -1 (TLR-4), and several heat shock proteins (TLR-2 and 4) [6]–[12]. One hypothesis is that the array of DAMPs and associated bound molecules to which monocytes are exposed, determines the state of activation of the myeloid cells.
Triggering receptors expressed on myeloid cell (TREM) family are a cell surface Immunoglobulin domain receptor family restricted to myeloid lineage cells. The TREM family function as modulators of cellular response, regulating positively and negatively the activation of myeloid cells during inflammation. The majority of TREM family members lack cytoplasm signaling motif but associate with an ITAM containing signaling adaptor protein, DAP12 which can recruit activating kinases including Syk. TREM-1 was first characterized in infections, was highly upregulated and has been implicated as an amplifier of inflammation [13] [14]–[17], functioning as an important co-activator of the TLR [13], [17], [18] and NOD Like Receptor (NLR) [19], [20] signaling pathways. Several recent studies suggested that TREM-1 may be an important and targetable effector molecule not only in infections but also in sterile inflammation [21] in pancreas [22], [23], joints [24], gut [25], [26] and eyes [4], [27]. In addition, TREM-1 is cleaved, and soluble TREM-1 is readily detected in biological fluids of patients suffering from a variety of diseases [28], suggesting a possible role as a decoy receptor that competes for putative ligands and negatively regulates TREM-1 pathway activation.
In these studies we identified TREM-1 as a highly expressed receptor in macrophages during sterile kidney injury. Furthermore, we investigated the role of TREM-1 and the related TLR receptor signaling pathways in macrophage activation and disease progression in the kidney.
Materials and Methods
Animals
CsfR1-iCre [29] (FVB) mice were bred with Myd88fl/fl mice (C57BL/6) (Jackson Laboratories) and the F2 generation was used for the experiments. Tlr2−/−, Tlr4−/−, Tlr2-4−/− and Myd88−/− (C57BL/6) mice were previously reported [30]. Dap12−/− mice were previously reported [31], [32]. All experiments were performed under a protocol approved by the Department of Comparative Medicine, University of Washington (Permit number 4244-01). All surgery was performed under ketamine and xylazine anesthesia and all efforts were made to minimize suffering.
Mouse Model of Kidney Injury with Fibrosis
Mice were anesthetized with ketamine/xylazine (100/10 mg/kg i.p.) and Unilateral Ureteral Obstruction (UUO) or unilateral Ischemia and Reperfusion Injury (U-IRI) were performed in adult (8–12wk) mice as previously described [33]. In the U-IRI model, left kidney was clamped for 40 minutes (females) at 36.8–37.3°C core temperature. For the TREM-1 experiments, mice received daily i.p. injections of 40 µg of purified TREM1-Fc or hIgG1, as control, diluted in PBS, starting at the day of surgery until sacrifice at day 5.
Tissue Preparation and Histology
Mouse tissues were prepared and stained as previously described [33], [34]. Primary antibodies against the following proteins were used for immunolabeling: CD11b (e-bioscience 1∶200), Ly6C (e-bioscience 1∶200), anti-TREM-1 (R&D 1∶200), F4/80 (Invitrogen 1∶200) and αSMA (Sigma 1∶400). Slides were incubated with Fluorescence (Cy3 or FITC)-conjugated secondary antibodies (1∶400–1∶800, Jackson ImmunoResearch), mounted with Vectashield/DAPI, and images were captured using a Nikon TiE Inverted Widefield Fluorescence microscope at the Lynn and Mike Garvey Cell Imaging Core at Institute for Stem Cell and Regenerative Medicine of University of Washington. For morphometric analysis of collagen fibril staining, deparaffinized sections (3 µm) were stained with 0.1% picrosirius red [35], [36]. Area of positive fluorescence/stain in 200× magnification of 10 randomly selected images per mouse were quantified using Image J software (http://rsbweb.nih.gov/ij/) [34], [37].
Q-PCR
RNA was isolated from kidney tissue samples using TRIzol (Invitrogen) according to standard protocol. First-strand cDNA was synthesized using the iScript kit (Bio-rad). Real-time PCR was performed using iTaq SYBR green supermix with ROX (Bio-rad) and 7900HT ABI detection system (Applied Biosystems). Target genes were normalized by Hypoxanthine phosphoribosyltransferase (HPRT) expression. The mRNA expression was calculated using the 2-ΔΔCt method and expressed as an n-fold difference relative to the control group.
Kidney Danger Associated Molecular Pattern Preparation
Kidneys were collected from normal mice (control), or day 5 after UUO. Under sterile conditions kidney vasculature was flushed with ice cold PBS. Under sterile conditions kidneys were decapsulated, minced and incubated with 2ml of LIBERASE TL (0.2mg/ml in DMEM/F12, Roche) and digested by shaking vigorously in a water-bath (37°C, 30min). Five ml of PBS was added, then the single cell preparation filtered through a 40 µm Cell Strainer. The filtrate was centrifuged (2000rpm, 5min at 4°C) to pellet any cellular debris and the cell-free supernatant was filtered using 0.2 µm low protein binding syringe filter. Polymyxin B beads (Sigma P1411), 20 µl, were added to the supernatant and incubated at 4°C for 30 minutes to remove any contamination by endotoxin. Supernatant containing DAMPs centrifuged (2000rpm, 5min, 4°C) to remove beads, and aliquots were stored at −80°C until use.
Bone Marrow Macrophage and Pericyte Isolation
BMDMφ were generated by flushing femurs with DMEM/F12 as previously described [29]. BMDMφ was cultured for 7 days in macrophage medium (DMEM/F12 medium (Cellgro), containing 10% FBS (Invitrogen), 1% Penicillin/Streptomycin (Cellgro), 20% L929 conditioned media containing M-CSF and stimulated on day 8. Pericytes from normal kidney from C57BL/6 wild-type, Tlr2–4−/− and Myd88−/− mice were isolated using MACS (Miltenyi Biotech) and rabbit polyclonal anti-PDGFRβ antibody as detailed previously [37].
Bone Marrow Macrophage and Pericyte Stimulation
BMDMφ were incubated in 24well/plate (0.5×106 cells/well) with 1ml of DMEM/F12 serum-free media per well, containing 100 µl of crude DAMPS from injured or normal kidney, or 100 µl of Liberase mix as control. To activate TREM-1 in BMDMφ, anti-TREM-1 antibodies (R&D) at 1 or 2 µg/ml or isotype IgG control (R&D) were applied. To block TREM-1 activity, purified TREM1-Fc or human IgG (Sigma) was applied at 1 or 3 µg/ml. Pericytes between passages 2 and 5 were stimulated in 12-well plates pre-coated with gelatin when 60–80% confluent. The concentrations of IL-6 and MCP-1 in the supernatants of primary pericytes were determined using enzyme-linked immunosorbent assay kits (BioLegend) according to the vendor’s protocols.
TREM1-Fc Generation and Purification
Trem1 ORF was cloned from cDNA from LPS activated BMDMφs. To generate a soluble TREM1-Fc fusion protein, the cDNA encoding the extracellular region of Trem1 (position 57 to position 659 in the gene; NCBI Reference NM_021406.5) was amplified by PCR, digested and subcloned, in frame, into the multiple cloning site of (pFUSE-hIgG1-Fc1, InvivoGen) containing the Fc region of human IgG1 (hinge, CH2, CH3), and confirmed by sequencing. Plasmid DNA was obtained by Maxiprep (Qiagen) and transfected using lipofectamine LTX reagent (Invitrogen) to approximately 60% confluent 293T cells in 75 flasks that were seeded the day before. After 24h of transfection, cells were washed with PBS, and incubated with DMEM/F12 serum-free media for 4 days. Supernatant was collected, centrifuged, filtered and TREM1-Fc protein was purified by affinity to Protein A - Sepharose beads column (Invitrogen) using standard methods described [38]. After elution, TREM1-Fc was dialyzed for 16h at 4°C, using 20.000 MWCO cassettes (Thermo Scientific) to exchange the current elution buffer to PBS. Presence and purity of TREM1-Fc protein was confirmed by SDS PAGE gel stained with Gel Code Blue Safe Protein Stain (Thermo Scientific) and western blot using anti-TREM-1 antibody (R&D).
SDS PAGE and Western Blotting
Kidneys or BMDMφs lysates were separated on 10% SDS-PAGE gel (Bio-rad) then semi-dry transferred to an Immobilon PVDF membrane as described [37], [39]. After blocking, membranes were incubated overnight with primary antibodies, anti-TREM-1 (R&D, 1∶1000), anti-MyD88 (ProSci, 1∶500), β-Actin (Santa Cruz, 1∶1000), anti-HMGB1 (BioLegend 1∶1000), anti-mouse IgG (Jackson ImmunoResearch, 1∶2500). Horseradish peroxidase-conjugated secondary antibodies (Pierce) were applied and enhanced chemiluminescence (Thermo Scientific) was used to detect proteins, and images collected by FluorChemQ machine (Alpha Innotech Corporation).
Statistical Analysis
Statistical evaluation was carried out using the One Way Analysis of Variance (ANOVA) followed by Tukey post-test using GraphPad Prism (GraphPad Software). A p value <0.05 was considered to be significant. Error bars indicate standard error of mean.
Results
TREMs are Highly Upregulated in Mouse Models of Chronic Kidney Injury
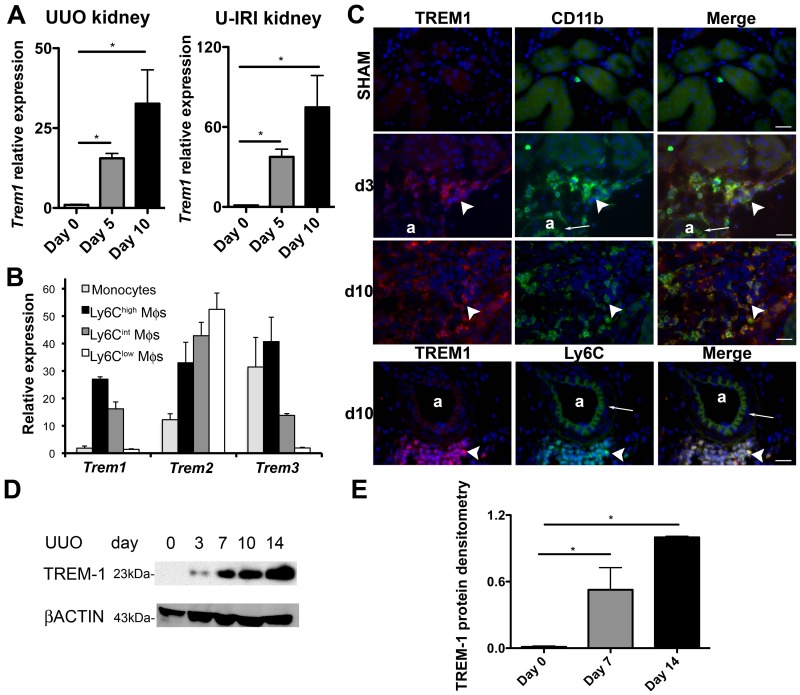
We purified macrophage subpopulations, discriminated by the subpopulation marker, Ly6C, from kidneys 5 days after inducing progressive interstitial kidney disease by unilateral ureteral obstruction (UUO) using flow sorting (see Methods S1). Neutrophils and NK cells were excluded (Fig. S1). The transcriptome of those macrophage subpopulations was interrogated by microarray to identify regulated genes that separate M1 (Ly6Chigh) from M2a (Ly6Clow) type macrophages in vivo. Using a highly stringent algorithm to identify regulated genes (Fig. S2), we discovered that the majority of genes were down regulated from comparing Ly6C+ to Ly6Clow cells and that the genes could be clustered in terms of biological processes, including immune response, response to stimulus, migration and chemotaxis. Among the immune response genes were many associated with either pattern recognition or activation (Trem1, Trem3, Pglyrp1, Clec4d, Clec4e, Tsg6 and Schlafen4, S100a8 and S100a9), strongly suggesting that Ly6Clow macrophages down-regulate activation pathways compared with Ly6C+ macrophages. Since TREM-1 has been shown to be an important co-activator of macrophages in inflammatory diseases, we investigated the function of TREM-1 further in sterile kidney injury. To validate the transcriptional analysis we quantified transcripts for the TREM family by quantitative RT-PCR (Q-PCR) of whole kidney or purified macrophage subpopulations or autologous blood monocytes during the evolution of the UUO model of progressive kidney injury ( Fig. 1A–B ). Trem1 was highly upregulated in whole kidney, 5 and 10 days after UUO, as well as in a second model of kidney injury with chronic inflammation and fibrosis, unilateral ischemia and reperfusion injury (U-IRI) ( Fig. 1A ). In the UUO model, Trem1 was particularly upregulated in Ly6Chigh and intermediate macrophages purified from the kidney ( Fig. 1B ). Trem3 expression mirrored the regulation pattern of Trem1 except that Trem3 was highly expressed by blood monocytes ( Fig. 1B ). Trem2 was also expressed by monocytes, upregulated in Ly6Chigh macrophages, but unlike Trem1 and Trem3, was further upregulated in Ly6Clow macrophages ( Fig. 1B ). Similarly to transcript levels, TREM-1 protein was not detected in normal kidney but highly upregulated during the progression of the UUO model ( Fig. 1D–E ), and its expression was restricted to CD11b+ and Ly6C+ cells ( Fig. 1C ).
Figure 1. TREMs are highly expressed in macrophages during kidney injury.
(A) Q-PCR for Trem1 expression in whole kidney 0, 5 and 10 days after UUO and U-IRI. (B) Q-PCR for TREM family transcript expression in blood monocytes, and different sub-populations of kidney macrophages purified at day 5 after UUO. (C) Fluorescence images showing CD11b (green), Ly6C (green) and TREM-1 (red) expression in tissue sections from control kidney (sham), day 3 and 10 after UUO (a, arteriole; Bar = 25 µm; arrowhead shows interstitial CD11b+ and TREM-1+ cells; arrow shows autofluorescent arteriole internal elastic lamina). (D) Western blot of whole kidney lysates detecting TREM-1 (23kD) and β-Actin (43kD), 0, 3, 7, 10 and 14 days after UUO. (E) TREM-1 protein densitometry normalized to endogenous control β-Actin. (n = 3–5/group, 3 independent experiments; *P<0.05).
TLR-2 and TLR-4 but not TREM-1, Regulate Activation of Macrophages in vitro by Kidney DAMPs
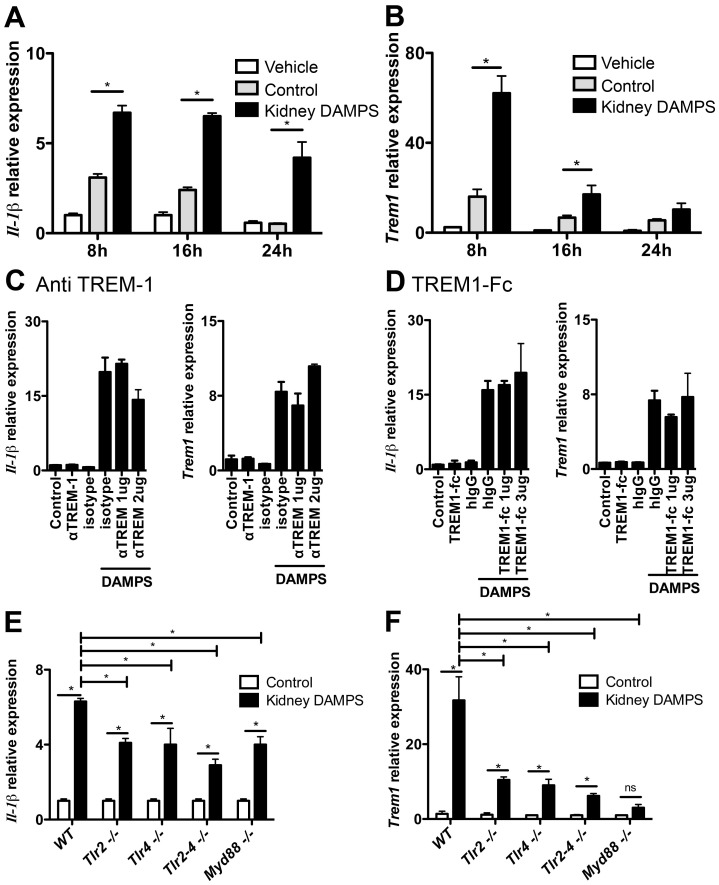
To study the signaling pathways in DAMP-mediated activation of macrophages in kidney injury and the role of TREM-1 and TLRs in this activation, we separated extracellular soluble factors from injured kidney (kidney DAMPs) or normal kidney (control) and applied these factors to primary cultures of quiescent bone marrow derived macrophages (BMDMφs) as a model of macrophage activation in vitro. Kidney DAMPs specifically stimulated Il-1β expression, an effect lasting 24h ( Fig. 2A ). Kidney DAMPs also activated Trem1 expression highly at 8h, but this response returned to baseline at 24h, a finding suggestive that TREM-1 may play a role in enhancing DAMP responses in macrophages and consistent with the findings that kidney macrophages are activated and produce TREM-1 ( Fig. 2B ). Kidney DAMPs did not induce Tnf-a production (not shown), similarly to our previous studies which showed kidney macrophages did not show Tnf-a activation [3]. Pre-incubation of BMDMφs with IFNγ for 8h markedly augmented subsequent kidney DAMP responses (Fig. S3A) suggesting either that cells have been primed by IFNγ to respond more strongly to DAMPs, similar to the enhancement during TLR agonist stimulation, or, alternatively that IFNγ upregulates DAMP receptors. To explore the nature of kidney DAMPs further, the crude kidney DAMPs were separated by SDS PAGE and proteins detected by Coomassie blue stain (Fig. S3B) (see Methods S1). Although, multiple protein bands were visualized in the normal kidney preparation (control), several bands appeared in the DAMPs preparation only suggesting these might be candidate protein DAMP molecules in the soluble preparation (Fig. S3B). High-mobility group protein B1 (HMGB1), which has been previously described to be a ligand for TREM-1 [40], [41], was one such DAMP molecule found in abundance in kidney DAMPs (Fig. S3C). Boiling almost completely attenuated biological activity of kidney DAMPs (Fig. S3D). When kidney DAMPs were exposed to trypsin or pronase, however, there was no impact on DAMPs activity, suggesting these DAMP factors specific to macrophages are either resistant to degradation or are non-proteinaceous (Fig. S3E).
Figure 2. TLR-2 and TLR-4 but not TREM-1, regulate activation of macrophages in vitro by kidney DAMPs.
(A–B) Q-PCR showing Il-1β and Trem1 expression in BMDMφ stimulated for 8, 16 or 24h with soluble factors prepared from normal kidney (control) or disease kidney (kidney DAMPs). (C–D) Graphs showing Il-1β and Trem1 expression by Q–PCR, 16h after BMDMφ were stimulated with kidney DAMPs and (C) activating anti-TREM-1 antibodies, or (D) TREM1-Fc, which blocks TREM-1 receptor by competing for ligands. (E–F) Q-PCR showing (E) Il-1β and (F) Trem1 expression in BMDMφ isolated from WT, Tlr2−/−, Tlr4−/−, Tlr2–4−/−, and Myd88−/− mice stimulated with kidney DAMPs for 16h. Q-PCR results were normalized to their respective control group. (*P<0.05, n = 5–7/group, 3 independent experiments; ns, p is not significant).
We hypothesized that cell surface TREM-1 may function as a co-activator of macrophages during activation by kidney DAMPs. We cloned mouse Trem1, and generated an Fc-fusion protein with the Trem1 ectodomain and the human IgG1 Fc domain to use as a decoy receptor to block TREM-1 activation (Fig. S4A), a method used successfully by others, such as in attenuating macrophage activation by LPS [13], [17]. To validate our protein, we stimulated BMDMφs with LPS and treated with TREM1-Fc or anti-TREM-1 antibodies, which triggers TREM-1 cell surface clustering and activates TREM-1 signaling. TREM1-Fc abrogated macrophage activation by LPS, and anti-TREM-1 amplified this activation (Fig. S4B–C). We then investigated the role of TREM-1 in BMDMφ-activation by kidney DAMPs. In contrast to the results observed with LPS, activation of surface TREM-1 by anti-TREM-1 antibodies did not augment DAMP-mediated activation of macrophages in vitro ( Fig. 2C ), and purified TREM1-Fc did not significantly inhibit DAMP-mediated activation of macrophages in vitro ( Fig. 2D ). Because TREM-1 is only expressed after exposure to DAMPs, we also pre-activated macrophages with kidney DAMPs, and subsequently blocked DAMP-mediated activation with TREM1-Fc or activated with anti-TREM-1, but this experiment also provided no evidence of inhibition or amplification of activation (Fig. S4D). In addition, we pre-incubated kidney DAMPs overnight with TREM1-Fc conjugated beads. TREM-1 bound DAMPs were then separated by centrifugation and the remaining supernatant was applied to macrophages. Compared with controls, TREM1-Fc adsorption did not attenuate DAMP activity (Fig. S4E). Furthermore, we tested whether DAMPs could activate DAP12 by signaling through membrane bound TREM-1. We stably expressed a fusion protein of TREM-1 with DAP12 in NFAT-Lacz reporter cells (BWZ TREM1/DAP12) (see Methods S1). Signaling via DAP12 activates the NFAT promoter driving β-galactosidase production, which can be detected by a colorimetric assay. Using this cell line, anti-TREM-1 antibodies in suspension or coated to a plate robustly activated DAP12 signaling and production of β-galactosidase (Fig. S4F–G). However, kidney DAMPs either coated to plates or in suspension did not activate LacZ, and therefore DAP12, in these reporter cells, suggesting TREM-1 is not a major target for endogenous activators of innate immune responses. Finally, to test whether TREM-1 may have ligands on dying epithelial cells, the binding capacity of TREM1-Fc to apoptotic kidney proximal epithelial cells (LLC-PK1) was evaluated, but no differences were seen compared to healthy epithelial cells (data not shown) [42].
Because TREM-1 is a regulator of TLR signaling, we next investigated whether TLR-2, TLR-4 and MyD88 signaling pathways played any role in DAMP-mediated activation. Using macrophages deficient in TLR-2, TLR-4, TLR-2 and 4 or MyD88, we assessed their responsiveness to kidney DAMPs. Both Toll like single receptor deficiency in macrophages attenuated the production of Il-1β in response to DAMPs, but this inhibition was not enhanced significantly in macrophages lacking both receptors, suggesting they have overlapping specificities for DAMP activity ( Fig. 2E ). MyD88 deficient macrophages were also hypo-responsive to kidney DAMPs ( Fig. 2E ). The hypo-responsiveness was most strikingly seen in terms of Trem1 transcript induction ( Fig. 2F ). Collectively these findings suggest TREM-1 does not play a role in macrophage activation in vitro by DAMPs, and TLR-2, TLR-4 and the MyD88 signaling pathway are activated by kidney DAMPs, but that other signaling pathways are also responsible for activation.
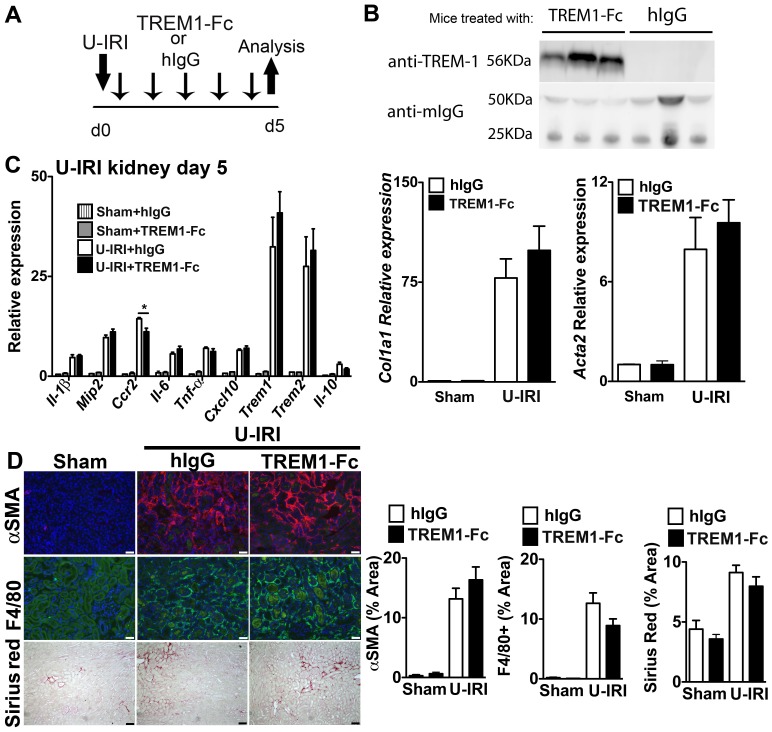
Circulating TREM1-Fc does not Prevent Macrophage Activation, Injury and Fibrosis in Models of Kidney Disease
Our studies indicated that TREM-1 did not play a functional role in BMDMφ activation in vitro. To test whether this translated to in vivo kidney disease, we induced two different models of kidney injury in mice, UUO and U-IRI, which are characterized by progressive interstitial inflammation and fibrosis [35], and administered TREM1-Fc daily by i.p. injections at a dose of 40 µg/mouse, to give a predicted ECF volume concentration of 4 µg/ml or control human IgG1 ( Fig. 3A ). On d2 of the experiment, 2 µl of venous blood was assessed for the presence of TREM1-Fc, which was abundant in mice receiving TREM1-Fc injections but not in mice receiving hIgG injections ( Fig. 3B ). In both models, whole kidney analysis of macrophage activation genes indicated that TREM1-Fc had no clear impact on macrophage activation ( Fig. 3C, S 5A). Because both of these models result in a fibrogenic process, we also evaluated Col1a1 and Acta2 transcripts, as indicators of myofibroblast activation and fibrogenesis finding no differences ( Fig. 3C, S 5A). TREM1-Fc administration had no impact on the extent of macrophage recruitment to the kidney, as well as the extent of αSMA+ myofibroblasts and collagen deposition ( Fig. 3D, S 5B). Since TREM-1 signals via the co-receptor DAP12 we evaluated the effect of DAP12 deficiency on the extent of U-IRI kidney disease. Consistent with our observations with administration of TREM1-Fc in vivo, Dap12−/− mice had similar disease severity compared with strain-matched controls (Table S1).
Figure 3. Treatment with soluble TREM1-Fc does not prevent macrophage activation, injury and fibrosis in sterile kidney injury.
(A) Schema showing experimental design. Mice were subjected to unilateral ischemia and reperfusion injury (U-IRI) and treated daily with 40 µg/mouse of TREM1-Fc or hIgG, as control. (B) Western blot showing presence of TREM1-Fc (approximately 56kD) in 2 µl of plasma collected at day 2 from mice treated daily with 40 µg of TREM1-Fc. Anti-mouse IgG was used as endogenous control. (C) Q-PCR for different inflammatory transcripts (left) or pro-fibrotic transcripts, Collagen1a1 (Col1a1) and alpha smooth muscle actin (Acta2), from whole kidney day 5 after U-IRI. (D) Representative images (left) and quantitative graphs (right) showing+F4/80 cells (green),+αSMA (red) or collagen deposition (Sirius Red staining) day 5 after U-IRI. (*P<0.05, n = 5–7/group, 3 independent experiments; Bar marker = 50 µm; Q-PCR results were normalized to sham +hIgG).
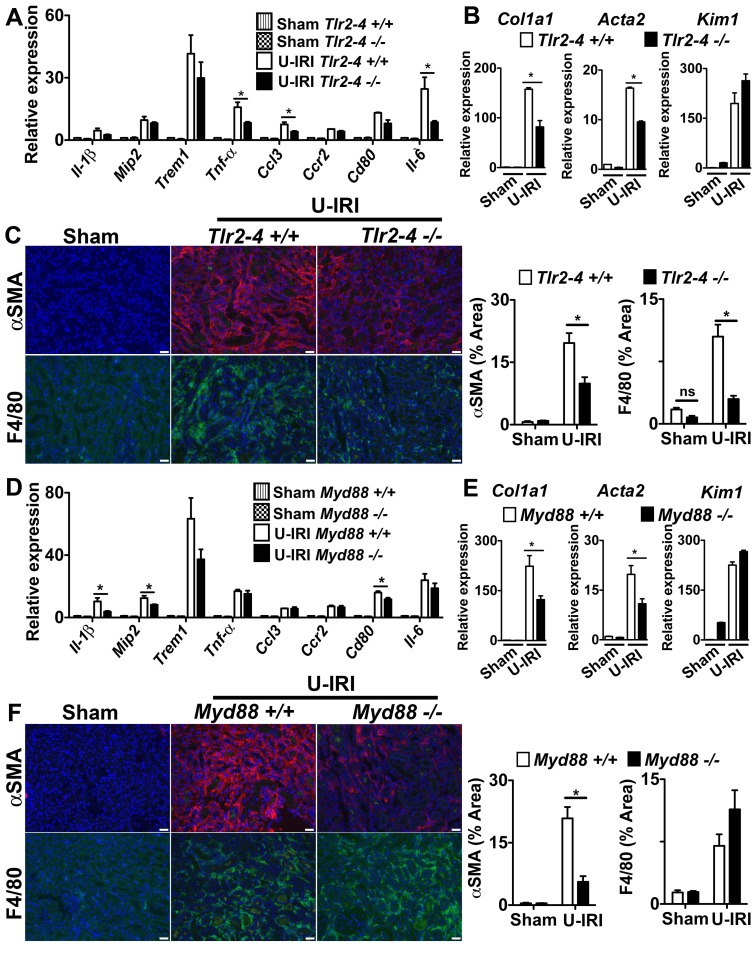
The TLR-2, TLR-4 and MyD88 Pathways Play a Role in Inflammation and Fibrosis in the U-IRI Model of Sterile Kidney Injury
The observations from in vitro studies ( Fig. 2 ) suggested that TLR-2, TLR-4 and MyD88 might be partially responsible for macrophage activation in vivo. To explore this possibility further, we studied the progression of two models of kidney disease in mice deficient in TLR-2, TLR-4 and MyD88 ( Fig. 4 ; S6). Mice lacking MyD88 or both TLRs showed evidence of significant reduction in pro-inflammatory cytokine production in the U-IRI model ( Fig. 4A,D ), but the pattern of reduction was distinct, suggesting that MyD88 deficiency may disrupt signaling from TLRs other than TLR-2 and TLR-4 or disrupt signaling from MyD88-dependent cytokine receptors such as the IL-1 or IL-18 receptors. In both experiments, the Trem1 transcript level, which is an indicator of macrophage activation, was similar to the wild type group, suggesting that macrophage activation was not different. More strikingly was the observation that both Col1a1 and Acta2 gene transcripts were reduced by approximately 50% in mice following kidney U-IRI and this was similar between MyD88 and TLR-2/TLR-4 deficiency ( Fig. 4B,E ). These gene transcripts are restricted to the pericyte/fibroblast/myofibroblast lineage, suggesting that deficiency of MyD88 and TLR-2/TLR-4 was impacting mesenchyme cell activation in the U-IRI model predominantly. Furthermore, the epithelial injury marker Kim-1 was elevated similarly in MyD88 or TLR-2/TLR-4 deficiency, suggesting that epithelial injury was not different between the mutant and wild type mice and that these activating receptors may not be critical in epithelial cells ( Fig. 4B,E ). In addition, the extent of αSMA+ myofibroblasts and the extent of macrophage recruitment to the kidneys was reduced when MyD88 or TLR-2/TLR-4 were deleted ( Fig. 4C,F ). Although deficiency of TLR-2, TLR-4 and MyD88 all had anti-inflammatory and anti-fibrotic effects in the U-IRI model, the UUO model was completely insensitive to deletion of these genes, indicating that the factors stimulating activation and injury in the UUO model did not involve the TLR-2, TLR-4 or MyD88 pathways (Fig. S6).
Figure 4. The TLR-2/TLR-4/MyD88 pathways play a role in fibrosis in the U-IRI model of sterile kidney injury.
(A–C) Tlr2–4−/− (D–F) Myd88−/− mice or respective controls were subjected to U-IRI and kidney harvested for tissue analysis 5 days later. Q-PCR (A,D) for different inflammatory transcripts, (B,E) pro-fibrotic transcripts, collagen1a1 (col1a1) and alpha smooth muscle actin (Acta2), and the tubule injury marker, kidney injury molecule-1 (Kim-1) from whole kidney day 5 after U-IRI. (C,F) Representative fluorescent images (left) and quantitative graphs (right) showing+αSMA (red) cells and+F4/80 cells (green). (*P<0.05, n = 5–7/group, 3 independent experiments; ns, p is not significant; Bar = 50 µm; Q-PCR results were normalized to wild type control).
The TLR-2/TLR-4/MyD88 Pathway is Dispensable in Macrophage Activation in Kidney Fibrosis, but Important in Mesenchymal Cell Activation
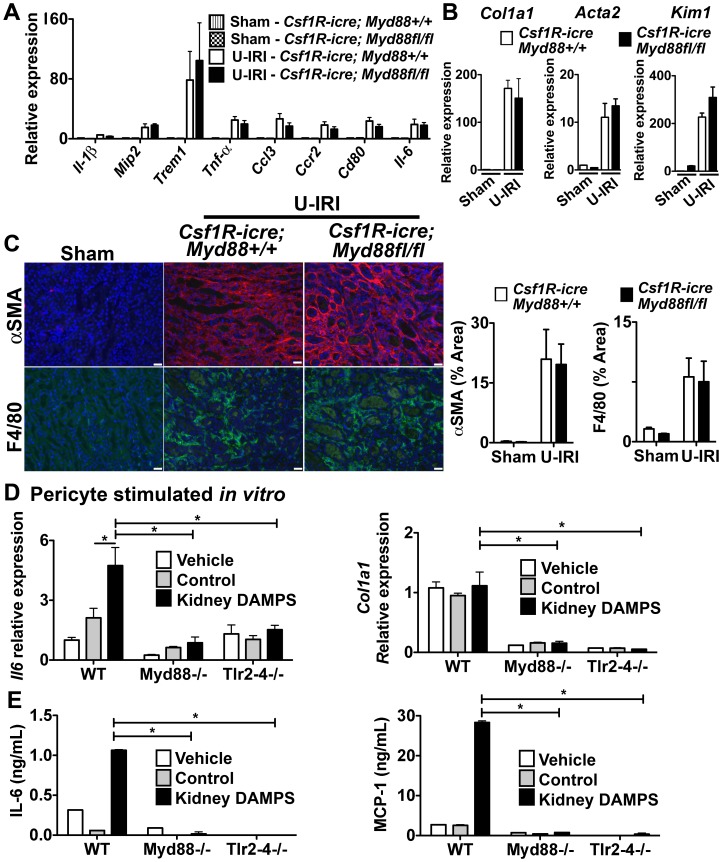
To test the importance of these activating receptors in myeloid cells further, we generated mice lacking MyD88 somatically in myeloid lineage cells (macrophages, dendritic cells and neutrophils) Csf1R-iCre; Myd88fl/fl [29]. To validate the deletion of MyD88 in myeloid lineage cells, we generated BMDMφs from these mice and Csf1R-iCre; Myd88+/+ littermates. As expected the WT mice expressed high levels of MyD88, which was further induced by LPS (Fig. S7A), but Csf1R-iCre; Myd88fl/fl showed no detectable MyD88 protein. Similarly, BMDMφs from Csf1R-iCre; Myd88fl/fl mice were completely insensitive to LPS (Fig. S7B), a result phenocopied using BMDMφs from mice with germline MyD88 deficiency. These results indicate that MyD88 is completely deleted in myeloid lineage cells in the Csf1R-iCre; Myd88fl/fl mice. Unexpectedly, deletion of MyD88 only in myeloid cells resulted in no difference in pro-inflammatory cytokines or chemokines in the U-IRI kidney injury model ( Fig. 5A ). In addition, whereas the MyD88 deficient mice showed protection from myofibroblast activation ( Fig. 4E ), Csf1R-iCre; Myd88fl/fl mice showed no evidence of a reduction in myofibroblast activation ( Fig. 5B ) and the extent of αSMA+ myofibroblasts ( Fig. 5C ). Although the Csf1R-iCre; Myd88fl/fl mice were an F2 generation of FVB crossed with C57bl/6 mouse strain, the extent of disease observed in the Csf1R-iCre; Myd88+/+ was identical to C57bl/6 wild type (Fig. 4), therefore these findings suggest that in the U-IRI model, these pathways are dispensable for macrophage activation.
Figure 5. The TLR-2/TLR-4/MyD88 pathway is dispensable in macrophage activation in kidney fibrosis, but important in mesenchyme cell activation.
(A–C) Csf1R-icre; Myd88fl/fl mice or respective controls were subjected to U-IRI and kidney harvested for tissue analysis 5 days later. Q-PCR (A) for different inflammatory transcripts, (B) pro-fibrotic transcripts, collagen1a1 (col1a1) and alpha smooth muscle actin (Acta2), and the tubule injury marker, kidney injury molecule-1 (Kim-1) from whole kidney day 5 after U-IRI. (C) Representative fluorescent images (left) and quantitative graphs (right) showing+αSMA (red) cells and+F4/80 cells (green). (D–E) Primary pericytes were isolated from Myd88−/− and Tlr2–4−/− mice and stimulated in vitro for 8h with kidney DAMPs. (D) Graph showing Il-6 and Col1a1 transcript expression by Q-PCR. (E) Graph showing IL-6 and MCP-1 concentration in supernatant by ELISA. (*P<0.05, n = 5–7/group, 3 independent experiments; ns, p is not significant; Bar = 50 µm; Q-PCR results were normalized to wild type control).
Because TLR-2, TLR-4 and Myd88 germline deficiency had marked affects on myofibroblast activation and fibrogenesis, we hypothesized that these receptors may be important mesenchymal cell (myofibroblast progenitor) activation. We therefore generated primary cultures of kidney pericytes (myofibroblast progenitors) and stimulated them with kidney DAMPs. Pericytes robustly activated Il-6 and MCP-1 ( Fig. 5D–E ) but not Il-1β in response to DAMPs (Data not shown). Our studies predicted that MyD88, TLR-2 and TLR-4 may play a role in DAMP- mediated activation in pericytes. To test these we cultured MyD88, and TLR-2/TLR-4 deficient pericytes. Pericytes deficient in MyD88 or TLR-2/TLR-4 were not activated when stimulated with kidney DAMPs ( Fig. 5D–E ). Pericytes in culture express high-baseline levels of Col1a1 transcript, and DAMPs did not further increase Col1a1 production. However, pericytes deficient in MyD88 and TLR-2/TLR-4 expressed very low baseline levels of Col1a1 transcript when compared to wild type pericytes, and stimulation with kidney DAMPs did not induce Col1a1 ( Fig. 5D ), indicating that TLR-2/TLR-4 and MyD88 pathways are important for mesenchyme cell activation.
Discussion
These studies show that although TREM-1 is highly upregulated in two models of chronic kidney disease in mice, and can regulate TLR sensitivity to LPS, it plays no role in sterile activation of monocytes/macrophages that are recruited to the injured kidney. These studies also show that, although the TLR-2, TLR-4 and MyD88 signaling pathways may play a small role in activation of macrophages by kidney DAMPs in vitro, they are largely dispensable in vivo. However, MyD88-dependent TLR-2, TLR-4 signaling appears to be critical in activation of mesenchymal cells of the kidney.
TREM genes were highly upregulated in the kidney in Ly6C+ macrophages in response to injury. Several studies report a similar response in sterile inflammation in other organs, and report that TREM-1 blockade either by TREM1-Fc or by a peptide, attenuates the disease process [23]–[26]. Our studies in the kidney do not support a functional role for TREM-1 despite the similarities between the studies. This may reflect differences in DAMPs released in different organs, or the differing roles of macrophages in each tissue. The fact that the mouse TREM-3 gene lies adjacent to the TREM-1, and studies have shown that both receptors have redundant functions in the mouse [43], [44], suggests that TREM-3 could be compensating for TREM-1 in our model. However, like TREM-1, TREM-3 is reported to signal through DAP12 to function as an amplifier of inflammation. Our findings indicate that Dap12−/− mice (Table S1) and Trem-2−/− mice (not shown) had similar disease severity compared to strain-matched controls, suggesting that these pathways are dispensable in these models of kidney injury.
The studies presented here show a novel method for studying kidney DAMPs. The preparation method although crude, robustly and reliably activates leukocytes and can be used to dissect individual DAMP factors. The fact that DAMPs are heat sensitive, but not sensitive to trypsin and pronase, indicates that they may be non-proteinaceous factors, including nucleic acids, lipids or products of extracellular matrix. However, they also could be protein complexes resistant to proteolytic digestion. Consistent with this a known DAMP, HMGB1 was readily identified in the preparation. Further studies beyond the scope of the current studies should identify these DAMPs.
Because TLRs have been implicated as effectors in kidney diseases [45], [46] and TREM-1 has been previously linked to TLR signaling, we investigated TLR-2 and TLR-4 as putative receptors for kidney DAMPs and macrophage activation. The studies presented here show a significant role for TLR-2, TLR-4 and MyD88 in vivo, particularly in the U-IRI model of chronic inflammation, but indicate they are not important in macrophage activation, even though macrophages during chronic kidney injury contribute to disease progression and fibrogenesis [2] [3]. Both the in vitro studies and in vivo studies implicate other signaling pathways that are more important in macrophage activation in kidney disease. Strikingly, we also identified C-type lectin (Clec) 4d and Clec4e as kidney macrophage PRRs in the transcriptional profiling experiments. Clec4e, also known as MINCLE, was reported to be a critical PRR in macrophages responding to the mitochrondrial spliceosomal protein SAP130, and signals through FcRγ [47]. We previously published that FcRγ deficient mice exhibit a significant reduction in injury and fibrosis in these models of kidney injury. However the interacting receptors involved in this activation were not determined [35]. Since these current studies have ruled out a role for TREM-1 in macrophage activation in the kidney future studies should determine whether the Clec receptors play dominant roles via FcRγ in kidney macrophage activation.
Although TLR signaling has been implicated in kidney diseases in other studies [45], the role of these receptors in different cell compartments in the kidney has been controversial and has been thought to depend on the model of injury. Using a bone marrow chimeric approach, it has been suggested that TLRs may play a more important role in parenchymal cells rather than myeloid cells during acute kidney injury [48], [49]. Our findings would support that. However our studies do not indicate that epithelial cell injury is reduced when MyD88 or TLR-2 and TLR-4 are absent, implicating other kidney cells. Until recently, the existence of the mesenchymal cells in the kidney, known as pericytes and resident fibroblasts, has been underappreciated but with new genetic tools their roles in fibrogenesis and innate immunity as well as vascular biology have recently become established [33], [37], [50]. Pericytes are embedded in the peritubular capillaries of the kidney and, like dendritic cells, form a barrier at the vascular interface [51]–[53]. Using a microarray approach, we have recently published that during a progressive kidney disease, genes involved in the immune response were highly upregulated in the pericyte to myofibroblast transition in the kidney [37]. Although surprising, our studies suggest that pericytes may form an important early immune defense to injury by release of pro-inflammatory cytokines and chemokines and that this response is highly dependent on TLR-2, TLR-4 and MyD88. Further studies are required.
We conclude that TREM-1 and TLR-2, TLR-4 and MyD88 are dispensable in macrophage activation in sterile kidney injury but that these pathways are important in activation of kidney pericytes.
Supporting Information
Ly6C Macrophage subpopulations purified from UUO kidney. (A) Representative plots of total kidney cells from single cell preparation 5 days after UUO were selected for viability and singularity by initial forward and side scatter gates. (B) Ly6G+ and NK1.1+ cells were negative gated to exclude neutrophils and NK cells. The different macrophage subpopulation were sorted by gating populations of CD11b+ cells with three levels of Ly6C expression: Ly6Chigh, Ly6Cint, and Ly6low.
(TIF)
Transcriptional analysis of activated macrophages in sterile kidney injury. Clustered profiles of 63 differentially expressed genes between Ly6C+ (Ly6Chigh and Ly6Cint, n = 3/group) and Ly6Clow (n = 2) macrophages depicted using a heatmap. Note the progressive decline in Trem1 expression levels across Ly6Chigh, Ly6Cint, and Ly6Clow sub-populations. Gene Ontology relational representation of highly enriched functional modules corresponding to differentially expressed genes between Ly6C+ and Ly6Clow macrophages. Prominent processes include immune response, migration, chemotaxis, and cytokine binding and activity.
(TIF)
Temperature sensitive kidney DAMPs activate macrophages ex vivo. (A) Q-PCR for Il-1β in BMDMφ primed with IFNγ (0, 250 or 500 U/ml) for 8 hours, washed, and further stimulated with kidney DAMPs for 12 hours. (B) Coomassie blue stained SDS PAGE of crude preparation of soluble extracellular factors from normal (control) and disease kidney (kidney DAMPs). (C) Western blotting showing HMGB1 expression in soluble extracellular factors from control and kidney DAMPs. (D) Q-PCRs from BMDMφs treated with DAMPs for 16 h showing the effect of temperature changes on kidney DAMP activity. (E) Q-PCR showing the effect of kidney DAMPs digestion for 16 h with Trypsin (1∶20 w/w ratio) or Pronase (1∶50 w/w ratio) prior application to BMDMφ for 16 h. (*P<0.05, n = 5–7/group, 3 independent experiments; ns, p is not significant).
(TIF)
TREM-1 pathway is important for BMDMφ activation by LPS in vitro , but dispensable for activation by kidney DAMPs. (A) Schema of TREM1-Fc fusion protein and Western blot of purified TREM1-Fc, detected by anti-TREM-1 antibodies. (B–C) Q-PCR for Il-1β in BMDMφs stimulated with LPS and treated with (B) TREM1-Fc or (C) anti-TREM-1 antibodies. (D) Q-PCR for Il-1β in BMDMφ pre-incubated with kidney DAMPs for 8 h to induce TREM-1 expression, followed by kidney DAMPs in the presence of anti-TREM-1 antibodies or TREM1-Fc for 16 h further. (E) Q-PCR showing BMDMφ response to DAMPs for 16 h that were pre-adsorbed by hIgG or TREM1-Fc coated protein-A beads. (F–G) Colorimetric assay reporting Lacz activity in BWZ-Lacz reporter cells expressing TREM1-DAP12 chimera protein stimulated with kidney DAMPs for 16 h in wells (F) pre-coated with kidney DAMPs or (G) in suspension (anti-TREM-1 antibodies are positive control). (n = 3–5/group, 3 independent experiments; *P<0.05).
(TIF)
Treatment with soluble TREM1-Fc does not prevent macrophage activation, injury and fibrosis in UUO model of sterile kidney injury. Mice were subjected to unilateral ureter obstruction (UUO) and treated daily with 40 µg/mouse of TREM1-Fc or hIgG, as control. (A) Q-PCR for different inflammatory transcripts (left) or pro-fibrotic transcripts, Collagen1a1 (Col1a1) and alpha smooth muscle actin (Acta2), from whole kidney day 5 after UUO. (B) Representative images (left) and quantitative graphs (right) showing+F4/80 cells (green),+αSMA (red) or collagen deposition (Sirius Red staining) day 5 after UUO. (*P<0.05, n = 5–7/group, 3 independent experiments; Bar marker = 50 µm; Q-PCR data were normalized to sham+higG control).
(TIF)
The TLR2/4/MyD88 pathway is dispensable in the UUO model of sterile kidney injury. (A,C,E) Q-PCR for different inflammatory molecules, pro-fibrotic transcripts, collagen1a1 (col1a1) and alpha smooth muscle actin (Acta2), and the tubule injury marker, kidney injury molecule-1 (Kim-1) from whole kidney day 5 after UUO in (A) Myd88−/−, (C) Tlr2–4−/−, and mice lacking MyD88 only in myeloid cells lineage, (E) Csf1R-icre; MyD88fl/fl. (B,D,F) Graphs showing quantification of fluorescent images for+αSMA cells and+F4/80 cells. (*P<0.05, n = 5–7/group; Q-PCR data were normalized to wild type sham).
(TIF)
Validation of MyD88 conditional ablation in myeloid cells expressing Csf1R. Csf1R-iCre mice were crossed with Myd88fl/fl to generate Csf1R-icre; Myd88fl/fl mice, which selectively ablates MyD88 expression in myeloid cells expressing Csf1R. (A) Western blot showing basal or LPS-induced MyD88 expression of BMDMφ isolated from Csf1R-icre; Myd88+/+ or Csf1R-icre; MyD88fl/fl. (B) Q-PCR for Il-1β expression of BMDMφ from Csf1R-icre; Myd88+/+, Csf1R-icre; MyD88fl/fl, Myd88+/+ and Myd88−/− mice stimulated with LPS for 16h. (*P<0.05, n = 3–5/group; Q-PCR data were normalized to wild type control).
(TIF)
Quantitative PCR from kidney tissue day 5 after U-IRI injury.
(DOCX)
(DOCX)
Acknowledgments
We wish to thank Dr. Chien Liang Chen (Kaohsiung Veterans General Hospital, Kaohsiung, Taiwan) for generating TREM1-Fc. Dr. Li-Li Hsiao (Harvard Medical School, MA), Dr. Roderick Jensen (Virginia Tech, VA), Michael J Lombardi from the Microarray Biotechnology Center, Harvard Medical School for assistance with microarrays, and Naoki Nakagawa (University of Washington).
Funding Statement
The Duffield Lab is funded by National Institutes of Health (NIH) Grants (DK84077, DK87389, DK93493, DK94768) Genzyme GRIP Award, University of Washington. This work was also supported by NIH Grants HL086883 (to W.A.A.) and AI 073441 (to J.A.H.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nelson PJ, Rees AJ, Griffin MD, Hughes J, Kurts C, et al. (2012) The renal mononuclear phagocytic system. Journal of the American Society of Nephrology 23: 194–203 doi:10.1681/ASN.2011070680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duffield JS (2010) Macrophages and Immunologic Inflammation of the Kidney. Seminars in Nephrology 30: 234–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin SL, Castano AP, Nowlin BT, Lupher MLJ, Duffield JS (2009) Bone Marrow Ly6Chigh Monocytes Are Selectively Recruited to Injured Kidney and Differentiate into Functionally Distinct Populations. The Journal of Immunology 183: 6733–6743. [DOI] [PubMed] [Google Scholar]
- 4. Rosin DL, Okusa MD (2011) Dangers Within: DAMP Responses to Damage and Cell Death in Kidney Disease. J Am Soc Nephrol 22: 416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anders HJ (2010) Toll-like receptors and danger signaling in kidney injury. Journal of the American Society of Nephrology 21: 1270–1274 doi:10.1681/ASN.2010030233 [DOI] [PubMed] [Google Scholar]
- 6. Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, et al. (2010) Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464: 104–107 doi:10.1038/nature08780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang H, Evankovich J, Yan W, Nace G, Zhang L, et al. (2011) Endogenous histones function as alarmins in sterile inflammatory liver injury through Toll-like receptor 9 in mice. Hepatology 54: 999–1008 doi:10.1002/hep.24501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang D, Liang J, Fan J, Yu S, Chen S, et al. (2005) Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nature Medicine 11: 1173–1179 doi:10.1038/nm1315 [DOI] [PubMed] [Google Scholar]
- 9. Termeer C, Benedix F, Sleeman J, Fieber C, Voith U, et al. (2002) Oligosaccharides of Hyaluronan activate dendritic cells via toll-like receptor 4. J Exp Med 195: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsung A, Sahai R, Tanaka H, Nakao A, Fink MP, et al. (2005) The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med 201: 1135–1143 doi:10.1084/jem.20042614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Asea A, Rehli M, Kabingu E, Boch JA, Bare O, et al. (2002) Novel signal transduction pathway utilized by extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. J Biol Chem 277: 15028–15034 doi:10.1074/jbc.M200497200 [DOI] [PubMed] [Google Scholar]
- 12. Ohashi K, Burkart V, Flohé S, Kolb H (2000) Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J Immunol 164: 558–561. [DOI] [PubMed] [Google Scholar]
- 13. Bouchon A, Dietrich J, Colonna M (2000) Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol 164: 4991–4995. [DOI] [PubMed] [Google Scholar]
- 14. Sharif O, Knapp S (2008) From expression to signaling: Roles of TREM-1 and TREM-2 in innate immunity and bacterial infection. Immunobiology 213: 701–713. [DOI] [PubMed] [Google Scholar]
- 15. Gibot S, Alauzet C, Massin F, Sennoune N, Faure GC, et al. (2006) Modulation of the triggering receptor expressed on myeloid cells-1 pathway during pneumonia in rats. J Infect Dis 194: 975–983 doi:10.1086/506950 [DOI] [PubMed] [Google Scholar]
- 16. Gibot S, Massin F, Marcou M, Taylor V, Stidwill R, et al. (2007) TREM-1 promotes survival during septic shock in mice. Eur J Immunol 37: 456–466 doi:10.1002/eji.200636387 [DOI] [PubMed] [Google Scholar]
- 17. Bouchon A, Facchetti F, Weigand MA, Colonna M (2001) TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 410: 1103–1107 doi:10.1038/35074114 [DOI] [PubMed] [Google Scholar]
- 18. Bleharski JR, Kiessler V, Buonsanti C, Sieling PA, Stenger S, et al. (2003) A role for triggering receptor expressed on myeloid cells-1 in host defense during the early-induced and adaptive phases of the immune response. J Immunol 170: 3812–3818. [DOI] [PubMed] [Google Scholar]
- 19. Netea MG, Azam T, Ferwerda G, Girardin SE, Kim SH, et al. (2006) Triggering receptor expressed on myeloid cells-1 (TREM-1) amplifies the signals induced by the NACHT-LRR (NLR) pattern recognition receptors. Journal of Leukocyte Biology 80: 1454–1461 doi:10.1189/jlb.1205758 [DOI] [PubMed] [Google Scholar]
- 20. Klesney-Tait J, Turnbull IR, Colonna M (2006) The TREM receptor family and signal integration. Nat Immunol 7: 1266–1273 doi:10.1038/ni1411 [DOI] [PubMed] [Google Scholar]
- 21. Derive M, Massin F, Gibot S (2010) Triggering receptor expressed on myeloid cells-1 as a new therapeutic target during inflammatory diseases. Self Nonself 1: 225–230 doi:10.4161/self.1.3.12891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yasuda T, Takeyama Y, Ueda T, Shinzeki M, Sawa H, et al. (2008) Increased levels of soluble triggering receptor expressed on myeloid cells-1 in patients with acute pancreatitis. Crit Care Med 36: 2048–2053 doi:10.1097/CCM.0b013e31817b8824 [DOI] [PubMed] [Google Scholar]
- 23. Kamei K, Yasuda T, Ueda T, Qiang F, Takeyama Y, et al. (2009) Role of triggering receptor expressed on myeloid cells-1 in experimental severe acute pancreatitis. J Hepatobiliary Pancreat Sci 17: 305–312. [DOI] [PubMed] [Google Scholar]
- 24. Murakami Y, Akahoshi T, Aoki N, Toyomoto M, Miyasaka N, et al. (2009) Intervention of an inflammation amplifier, triggering receptor expressed on myeloid cells 1, for treatment of autoimmune arthritis. Arthritis Rheum 60: 1615–1623. [DOI] [PubMed] [Google Scholar]
- 25. Schenk M, Bouchon A, Seibold F, Mueller C (2007) TREM-1–expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest 117: 3097–3106 doi:10.1172/JCI30602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gibot S, Massin F, Alauzet C, Montemont C, Lozniewski A, et al. (2008) Effects of the TREM-1 pathway modulation during mesenteric ischemia-reperfusion in rats. Critical Care Medicine 36: 504–510. [DOI] [PubMed] [Google Scholar]
- 27. Wu M, Peng A, Sun M, Deng Q, Hazlett LD, et al. (2011) TREM-1 Amplifies Corneal Inflammation after Pseudomonas aeruginosa Infection by Modulating Toll-Like Receptor Signaling and Th1/Th2-Type Immune Responses. Infection and Immunity 79: 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ford JW, McVicar DW (2009) TREM and TREM-like receptors in inflammation and disease. Current Opinion in Immunology 21: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin SL, Li B, Rao S, Yeo EJ, Hudson TE, et al. (2010) Macrophage Wnt7b is critical for kidney repair and regeneration. Proceedings of the National Academy of Sciences 107: 4194–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chun CD, Liles WC, Frevert CW, Glenny RW, Altemeier WA (2010) Mechanical ventilation modulates Toll-like receptor-3-induced lung inflammation via a MyD88-dependent, TLR4-independent pathway: a controlled animal study. BMC Pulm Med 10: 57 doi:–––10.1186/1471–2466–10–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, et al. (2006) Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol 177: 2051–2055. [DOI] [PubMed] [Google Scholar]
- 32. Bakker AB, Hoek RM, Cerwenka A, Blom B, Lucian L, et al. (2000) DAP12-deficient mice fail to develop autoimmunity due to impaired antigen priming. Immunity 13: 345–353. [DOI] [PubMed] [Google Scholar]
- 33. Lin S-L, Kisseleva T, Brenner DA, Duffield JS (2008) Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. The American Journal of Pathology 173: 1617–1627 doi:10.2353/ajpath.2008.080433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grgic I, Campanholle G, Bijol V, Wang C, Sabbisetti VS, et al.. (2012) Targeted proximal tubule injury triggers interstitial fibrosis and glomerulosclerosis. Kidney Int. doi:10.1038/ki.2012.20. [DOI] [PMC free article] [PubMed]
- 35. Castano AP, Lin SL, Surowy T, Nowlin BT, Turlapati SA, et al. (2009) Serum amyloid P inhibits fibrosis through Fc gamma R-dependent monocyte-macrophage regulation in vivo. Science Translational Medicine 1: 5ra13–5ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, et al. (2005) Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest 115: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schrimpf C, Xin C, Campanholle G, Gill SE, Stallcup W, et al. (2012) Pericyte TIMP3 and ADAMTS1 modulate vascular stability after kidney injury. J Am Soc Nephrol 23: 868–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harlow E, Lane D (1999) Using antibodies : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. 496 pp.
- 39. Chau BN, Xin C, Hartner J, Ren S, Castano AP, et al. (2012) MicroRNA-21 Promotes Fibrosis of the Kidney by Silencing Metabolic Pathways. Science Translational Medicine 4: 121ra18–121ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu J, Li J, Salcedo R, Mivechi NF, Trinchieri G, et al. (2012) The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Research 72: 3977–3986 doi:––10.1158/0008–5472.CAN-12–0938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mezayen El R, Gazzar El M, Seeds MC, McCall CE, Dreskin SC, et al. (2007) Endogenous signals released from necrotic cells augment inflammatory responses to bacterial endotoxin. Immunology Letters 111: 36–44 doi:10.1016/j.imlet.2007.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, et al. (2008) Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest 118: 1657–1668 doi:10.1172/JCI34487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klesney-Tait J, Keck K, Li X, Gilfillan S, Otero K, et al. (2013) Transepithelial migration of neutrophils into the lung requires TREM-1. J Clin Invest 123: 138–149 doi:10.1172/JCI64181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chung D-H, Seaman WE, Daws MR (2002) Characterization of TREM-3, an activating receptor on mouse macrophages: definition of a family of single Ig domain receptors on mouse chromosome 17. Eur J Immunol 32: 59–66. [DOI] [PubMed] [Google Scholar]
- 45. Gonçalves GM, Castoldi A, Braga TT, Camara NOS (2011) New roles for innate immune response in acute and chronic kidney injuries. Scand J Immunol 73: 428–435 doi:–10.1111/j.1365–3083.2011.02523.x [DOI] [PubMed] [Google Scholar]
- 46. Anders HJ, Banas B, Schlöndorff D (2004) Signaling Danger: Toll-Like Receptors and their Potential Roles in Kidney Disease. J Am Soc Nephrol 15: 854–867. [DOI] [PubMed] [Google Scholar]
- 47. Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, et al. (2008) Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol 9: 1179–1188. [DOI] [PubMed] [Google Scholar]
- 48. Pulskens WP, Teske GJ, Butter LM, Roelofs JJ, van der Poll T, et al. (2008) Toll-Like Receptor-4 Coordinates the Innate Immune Response of the Kidney to Renal Ischemia/Reperfusion Injury. PLoS ONE 3: e3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leemans JC, Stokman G, Claessen N, Rouschop KM, Teske GJD, et al. (2005) Renal-associated TLR2 mediates ischemia/reperfusion injury in the kidney. J Clin Invest 115: 2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Humphreys BD, Lin S-L, Kobayashi A, Hudson TE, Nowlin BT, et al. (2010) Fate Tracing Reveals the Pericyte and Not Epithelial Origin of Myofibroblasts in Kidney Fibrosis. The American Journal of Pathology 176: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Armulik A, Abramsson A, Betsholtz C (2005) Endothelial/pericyte interactions. Circ Res 97: 512–523 doi:10.1161/01.RES.0000182903.16652.d7 [DOI] [PubMed] [Google Scholar]
- 52. Armulik A, Genove G, Mae M, Nisancioglu MH, Wallgard E, et al. (2010) Pericytes regulate the blood-brain barrier. Nature 468: 557–561 doi:10.1038/nature09522 [DOI] [PubMed] [Google Scholar]
- 53. Majesky MW, Dong XR, Hoglund V, Mahoney WMJ, Daum G (2011) The adventitia: a dynamic interface containing resident progenitor cells. Arteriosclerosis, Thrombosis, and Vascular Biology 31: 1530–1539 doi:10.1161/ATVBAHA.110.221549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ly6C Macrophage subpopulations purified from UUO kidney. (A) Representative plots of total kidney cells from single cell preparation 5 days after UUO were selected for viability and singularity by initial forward and side scatter gates. (B) Ly6G+ and NK1.1+ cells were negative gated to exclude neutrophils and NK cells. The different macrophage subpopulation were sorted by gating populations of CD11b+ cells with three levels of Ly6C expression: Ly6Chigh, Ly6Cint, and Ly6low.
(TIF)
Transcriptional analysis of activated macrophages in sterile kidney injury. Clustered profiles of 63 differentially expressed genes between Ly6C+ (Ly6Chigh and Ly6Cint, n = 3/group) and Ly6Clow (n = 2) macrophages depicted using a heatmap. Note the progressive decline in Trem1 expression levels across Ly6Chigh, Ly6Cint, and Ly6Clow sub-populations. Gene Ontology relational representation of highly enriched functional modules corresponding to differentially expressed genes between Ly6C+ and Ly6Clow macrophages. Prominent processes include immune response, migration, chemotaxis, and cytokine binding and activity.
(TIF)
Temperature sensitive kidney DAMPs activate macrophages ex vivo. (A) Q-PCR for Il-1β in BMDMφ primed with IFNγ (0, 250 or 500 U/ml) for 8 hours, washed, and further stimulated with kidney DAMPs for 12 hours. (B) Coomassie blue stained SDS PAGE of crude preparation of soluble extracellular factors from normal (control) and disease kidney (kidney DAMPs). (C) Western blotting showing HMGB1 expression in soluble extracellular factors from control and kidney DAMPs. (D) Q-PCRs from BMDMφs treated with DAMPs for 16 h showing the effect of temperature changes on kidney DAMP activity. (E) Q-PCR showing the effect of kidney DAMPs digestion for 16 h with Trypsin (1∶20 w/w ratio) or Pronase (1∶50 w/w ratio) prior application to BMDMφ for 16 h. (*P<0.05, n = 5–7/group, 3 independent experiments; ns, p is not significant).
(TIF)
TREM-1 pathway is important for BMDMφ activation by LPS in vitro , but dispensable for activation by kidney DAMPs. (A) Schema of TREM1-Fc fusion protein and Western blot of purified TREM1-Fc, detected by anti-TREM-1 antibodies. (B–C) Q-PCR for Il-1β in BMDMφs stimulated with LPS and treated with (B) TREM1-Fc or (C) anti-TREM-1 antibodies. (D) Q-PCR for Il-1β in BMDMφ pre-incubated with kidney DAMPs for 8 h to induce TREM-1 expression, followed by kidney DAMPs in the presence of anti-TREM-1 antibodies or TREM1-Fc for 16 h further. (E) Q-PCR showing BMDMφ response to DAMPs for 16 h that were pre-adsorbed by hIgG or TREM1-Fc coated protein-A beads. (F–G) Colorimetric assay reporting Lacz activity in BWZ-Lacz reporter cells expressing TREM1-DAP12 chimera protein stimulated with kidney DAMPs for 16 h in wells (F) pre-coated with kidney DAMPs or (G) in suspension (anti-TREM-1 antibodies are positive control). (n = 3–5/group, 3 independent experiments; *P<0.05).
(TIF)
Treatment with soluble TREM1-Fc does not prevent macrophage activation, injury and fibrosis in UUO model of sterile kidney injury. Mice were subjected to unilateral ureter obstruction (UUO) and treated daily with 40 µg/mouse of TREM1-Fc or hIgG, as control. (A) Q-PCR for different inflammatory transcripts (left) or pro-fibrotic transcripts, Collagen1a1 (Col1a1) and alpha smooth muscle actin (Acta2), from whole kidney day 5 after UUO. (B) Representative images (left) and quantitative graphs (right) showing+F4/80 cells (green),+αSMA (red) or collagen deposition (Sirius Red staining) day 5 after UUO. (*P<0.05, n = 5–7/group, 3 independent experiments; Bar marker = 50 µm; Q-PCR data were normalized to sham+higG control).
(TIF)
The TLR2/4/MyD88 pathway is dispensable in the UUO model of sterile kidney injury. (A,C,E) Q-PCR for different inflammatory molecules, pro-fibrotic transcripts, collagen1a1 (col1a1) and alpha smooth muscle actin (Acta2), and the tubule injury marker, kidney injury molecule-1 (Kim-1) from whole kidney day 5 after UUO in (A) Myd88−/−, (C) Tlr2–4−/−, and mice lacking MyD88 only in myeloid cells lineage, (E) Csf1R-icre; MyD88fl/fl. (B,D,F) Graphs showing quantification of fluorescent images for+αSMA cells and+F4/80 cells. (*P<0.05, n = 5–7/group; Q-PCR data were normalized to wild type sham).
(TIF)
Validation of MyD88 conditional ablation in myeloid cells expressing Csf1R. Csf1R-iCre mice were crossed with Myd88fl/fl to generate Csf1R-icre; Myd88fl/fl mice, which selectively ablates MyD88 expression in myeloid cells expressing Csf1R. (A) Western blot showing basal or LPS-induced MyD88 expression of BMDMφ isolated from Csf1R-icre; Myd88+/+ or Csf1R-icre; MyD88fl/fl. (B) Q-PCR for Il-1β expression of BMDMφ from Csf1R-icre; Myd88+/+, Csf1R-icre; MyD88fl/fl, Myd88+/+ and Myd88−/− mice stimulated with LPS for 16h. (*P<0.05, n = 3–5/group; Q-PCR data were normalized to wild type control).
(TIF)
Quantitative PCR from kidney tissue day 5 after U-IRI injury.
(DOCX)
(DOCX)