Bivalent histone modification signatures combining activating and repressive histone marks are thought to poise the expression of developmental genes and influence cell fate and differentiation. However, the role of bivalent domains during development is controversial. In this review, Voigt et al. discuss recent studies describing bivalent histone domains and propose that bivalent domains and associated chromatin-modifying complexes safeguard proper and robust differentiation.
Keywords: Polycomb, Trithorax, bivalent domains, chromatin, embryonic stem cells
Abstract
Histone modifications and chromatin-associated protein complexes are crucially involved in the control of gene expression, supervising cell fate decisions and differentiation. Many promoters in embryonic stem (ES) cells harbor a distinctive histone modification signature that combines the activating histone H3 Lys 4 trimethylation (H3K4me3) mark and the repressive H3K27me3 mark. These bivalent domains are considered to poise expression of developmental genes, allowing timely activation while maintaining repression in the absence of differentiation signals. Recent advances shed light on the establishment and function of bivalent domains; however, their role in development remains controversial, not least because suitable genetic models to probe their function in developing organisms are missing. Here, we explore avenues to and from bivalency and propose that bivalent domains and associated chromatin-modifying complexes safeguard proper and robust differentiation.
Histone proteins and their post-translational modifications have emerged as important players in the regulation of gene expression and other chromatin-associated processes. The four core histones—H2A, H2B, H3, and H4—are subject to a host of covalent modifications, including methylation, acetylation, phosphorylation, and ubiquitination, among others (Vaquero et al. 2003; Campos and Reinberg 2009; Bannister and Kouzarides 2011). These marks are thought to exert their function through direct modulation of chromatin structure and through effector proteins that feature modification-specific binding domains (Taverna et al. 2007; Voigt and Reinberg 2011). Moreover, several histone modifications have been implicated as carriers of epigenetic information that can be transmitted through cell division, instructing gene expression patterns in the daughter cells (Probst et al. 2009; Margueron and Reinberg 2010).
Two prominent systems of chromatin-modifying activities are the Trithorax group (trxG) and Polycomb group (PcG) proteins. trxG proteins were discovered as activators of Hox genes in Drosophila (Schuettengruber et al. 2007). A subset of trxG protein complexes in flies, yeast, and mammals catalyze the trimethylation of histone H3 Lys 4 (H3K4me3), a mark generally associated with active transcription. In mammals, the responsible enzymes are SET1A, SET1B, and mixed lineage leukemia (MLL) proteins 1–4 (Shilatifard 2012). These proteins require additional subunits for activity, forming the multisubunit SET1A/B and MLL1–4 complexes. The PcG proteins were identified as silencers of Hox genes in Drosophila (Schuettengruber et al. 2007; Simon and Kingston 2009). Mutations in PcG genes lead to ectopic expression of key developmental regulators in flies, giving rise to characteristic body patterning defects. In vertebrates and flies, PcG proteins form the multisubunit Polycomb-repressive complexes (PRCs) 1 and 2 (Margueron and Reinberg 2011; Simon and Kingston 2013). PRC2 catalyzes H3K27me3, a pivotal mark in the establishment of repressive chromatin in both early development and adult organisms. PRC1 subsumes a diverse range of complexes that all contain the RING1A/B ubiquitin ligase and several additional subunits (Gao et al. 2012; Tavares et al. 2012; Simon and Kingston 2013). Some PRC1 complexes catalyze ubiquitination of H2A Lys 119 (H2AK119ub), whereas others likely act by directly compacting chromatin.
Even though the molecular decoding of histone marks such as H3K4me3 and H3K27me3 is far from being understood, genome-wide studies have provided intriguing clues as to how some of these marks might function in vivo. In particular, extensive efforts have been exerted toward mapping and understanding the chromatin landscape of embryonic stem (ES) cells, in part owing to the great promise of these cells for biological research and medical application. ES cells are derived from the inner cell mass of mammalian preimplantation blastocysts. They are capable of self-renewal, yielding offspring of equivalent developmental potential. ES cells are also pluripotent and can differentiate into all lineages of the developing and adult organism. Unraveling the mechanisms that govern ES cell self-renewal and pluripotency is crucial to our understanding of development. The ES cell state is controlled through a network of core transcription factors (TFs), most notably Nanog, Sox2, and Oct4 (Orkin and Hochedlinger 2011; Young 2011). Even though the action of such master regulator TFs is primarily guided by DNA sequences, it has become increasingly clear that chromatin and its associated factors provide additional layers of regulation in gene expression.
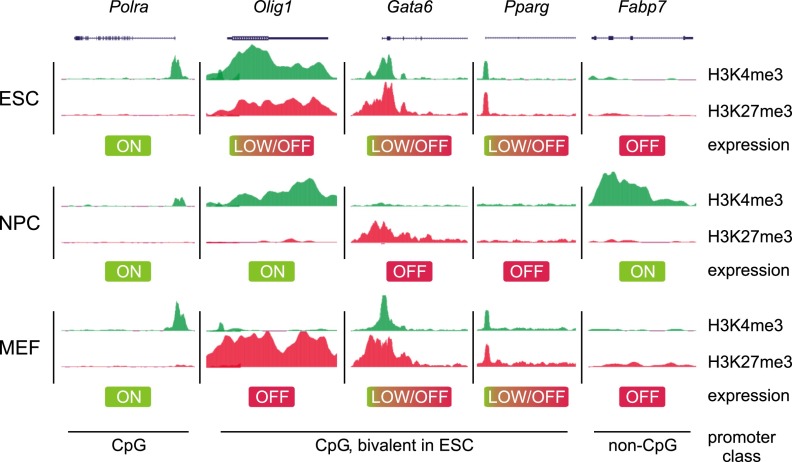
Genome-wide mapping studies of chromatin modifications in ES cells have revealed the presence of distinct histone marks at certain genomic domains, such as H3K4me1 and acetylation of H3K27 (H3K27ac) within active enhancers as well as H3K4me3 and H3K27me3 within active and repressed promoters, respectively (Zhou et al. 2011; Calo and Wysocka 2013). Although these correlations hold true for virtually all mammalian cell types, certain chromatin signatures do appear to be more specific to ES cells. For example, there is a large cohort of developmental gene promoters that are simultaneously marked by both activating H3K4me3 and repressive H3K27me3 modifications (Fig. 1). These patterns of seemingly opposing histone marks on the same promoter are referred to as “bivalent” domains, a term first coined by Bernstein et al. (2006). Although first described for ES cells, where they are most prevalent, later observations also detected bivalent domains in cell types of restricted potency. By exhibiting both active and repressive features, bivalent genes are posited as being in a poised state, enabling them to be rapidly activated upon suitable developmental cues and/or environmental stimuli.
Figure 1.
Bivalent domains mark CpG-rich promoters of developmental genes in ES cells. Whereas promoters of housekeeping genes such as Polm are marked solely with H3K4me3, bivalent promoters carry both H3K4me3 and H3K27me3. The latter genes either are expressed at low levels or are silent in ES cells but may become activated upon differentiation to neural precursor cells (NPCs) or be expressed in cells of reduced differentiation potential, such as MEFs. Conversely, other genes may become silent upon differentiation. Promoters with low CpG content may be activated during differentiation by other means, as they lack H3K4me3 and H3K27me3 in ES cells, and their regulation is likely distinct from CpG-rich promoters. The ChIP-seq traces are based on data from Mikkelsen et al. (2007).
In this review, we first summarize the studies describing bivalent domains in different systems and explore the role of PcG and trxG group proteins in their establishment. We also discuss the controversy surrounding the general importance of bivalency in development. Based on recent advances in the field, we propose a model for the formation and interpretation of bivalent domains in ES cells and finally discuss the functional implications of bivalency as a means to fine-tune gene expression and license proper differentiation.
Discovery of bivalent domains in ES cells and beyond
By combining chromatin immunoprecipitation (ChIP) and DNA tiling arrays, Bernstein et al. (2006) analyzed H3K4me3 and H3K27me3 modification patterns in regions containing highly conserved noncoding elements in mouse ES cells. These regions are enriched for developmental genes such as the Hox genes. As observed in previous studies (Bernstein et al. 2005; Kim et al. 2005), the vast majority of transcriptional start sites (TSSs) were found as being marked by H3K4me3. The distribution of H3K27me3 was generally broader, and ∼75% of the H3K27me3 sites spanned TSSs that were also marked by H3K4me3 (Bernstein et al. 2006). Sequential ChIP demonstrated that TSSs of select genes were simultaneously marked by both modifications (Bernstein et al. 2006). Strikingly, most such bivalent genes encode TFs of developmental importance. Despite the presence of H3K4me3, the bivalent genes under study were expressed only at low levels (Bernstein et al. 2006). Upon differentiation toward the neuronal lineage, some bivalent genes became expressed and lost the H3K27me3 mark, whereas those that were silenced lost H3K4me3 but retained H3K27me3 (Bernstein et al. 2006). These observations led to the intriguing model that bivalent domains maintain developmental genes in a silent state in ES cells while keeping them poised for subsequent expression upon differentiation.
A different approach to study the specifics of ES cell chromatin was taken by the Fisher laboratory (Azuara et al. 2006). Replication timing was assessed as a surrogate for chromatin accessibility and the transcriptional status of genes, with early replicating regions being typically more transcriptionally active and accessible. Some neural genes replicated early despite not being expressed in ES cells. In differentiated, nonneuronal cells, these genes remained unexpressed but replicated at the expected later stages (Azuara et al. 2006). Early replicating but inactive genes in ES cells were marked by active marks such as H3K9ac and H3K4me3 but also with the repressive H3K27me3 (Azuara et al. 2006). To explore the role of H3K27me3 at those genes, Azuara et al. (2006) analyzed ES cells lacking the Eed subunit of PRC2. This deficiency results in an almost complete absence of H3K27me3. Several bivalent genes were significantly up-regulated in Eed−/− cells, indicating their premature expression (Azuara et al. 2006). This study thus provided independent evidence for the existence of bivalent domains.
Subsequent reports extended these observations to a genome-wide scale. Mikkelsen et al. (2007) employed ChIP combined with next-generation sequencing (ChIP-seq) to analyze the genome-wide distribution of various trimethylation marks. Virtually all promoters with high CpG density (CpG islands), were marked by H3K4me3 in mouse ES cells. About 22% of these promoters (∼2500) additionally exhibited H3K27me3 and were expressed only at low levels (Fig. 1; Mikkelsen et al. 2007). Genes carrying this bivalent signature included developmental TFs, morphogens, and cell surface molecules. To assess whether bivalent domains are a unique feature of pluripotent cells, differentiated cells were analyzed as well. After differentiation of mouse ES cells to neural progenitor cells, most bivalent genes lost one of the marks, whereas 8% remained bivalent (see Fig. 1 for examples). In mouse embryonic fibroblasts (MEFs), 4% of CpG-rich promoters were bivalent (Mikkelsen et al. 2007). Genes associated with unrelated lineages resolved to monovalent markings, whereas some genes of related lineages retained bivalency. These studies were the first to indicate that nonpluripotent cells also contain bivalent domains.
The Schübeler group (Mohn et al. 2008) monitored bivalent domains of mouse ES cells during terminal differentiation to glutamatergic pyramidal neurons via radial glial neuronal progenitor cells, steps resembling the differentiation process in vivo. Interestingly, even though 675 bivalent domains were resolved, ∼550 genes gained H3K27me3 and formed novel bivalent domains in the neural progenitors (Mohn et al. 2008). Upon terminal differentiation, ∼1000 bivalent domains were lost, and ∼340 reformed. Nonpluripotent cells may thus contain bivalent genes that are not present in ES cells.
Bivalent genes were further identified in human ES cells (Pan et al. 2007; Zhao et al. 2007). They largely overlap with those found in mouse ES cells, and 2157 genes were classified as bivalent in at least two of the three studies (Mikkelsen et al. 2007; Pan et al. 2007; Sharov and Ko 2007; Zhao et al. 2007). As in mice, bivalent genes in human ES cells are highly enriched for TFs and other developmental genes, and genes induced upon ES cell differentiation lose H3K27me3 (Pan et al. 2007; Zhao et al. 2007). Somatic cells such as fibroblasts can be reprogrammed to so-called induced pluripotent stem cells (iPSCs) by expression of select pluripotency factors (Takahashi and Yamanaka 2006; Stadtfeld and Hochedlinger 2010). The chromatin state of iPSCs closely resembles that of ES cells, and the vast majority of bivalent domains are recapitulated in iPSCs (Maherali et al. 2007; Mikkelsen et al. 2008; Guenther et al. 2010). Human T cells retain restricted differentiation potential and feature many genes that are only weakly expressed despite the presence of H3K4me3 and H3K9/H3K14 acetylation (Roh et al. 2006; Barski et al. 2007). These promoters were marked additionally with H3K27me3 (Roh et al. 2006). Moreover, multipotent human hematopoietic stem cells contain ∼3000 bivalently marked promoters (Cui et al. 2009). Taken together, these studies show that bivalent domains also exist in human stem cells.
Several reports suggested both the existence of bivalent domains in cancer cells and a link between bivalent domains in ES cells and genes deregulated in cancer. Various cancer cells exhibit bivalent genes partially overlapping the set of bivalent genes in ES cells (e.g., see Rodriguez et al. 2008; Bapat et al. 2010). However, these genes often exhibit DNA methylation in cancer cells and thus are completely silenced (Rodriguez et al. 2008), and some CpG island promoters only establish bivalency after drug-induced DNA demethylation (McGarvey et al. 2008). Loci bivalent in ES cells are often found to be DNA-hypermethylated in adult cancers, and it was speculated that bivalency or the presence of PcG proteins might predispose those genes for later DNA hypermethylation (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007).
Existence of bivalent domains in developing embryos and other organisms
Studies initially reporting bivalent domains were based on cultured ES cells, which are kept in an artificial state of permanent pluripotency. Stem cells in developing embryos are only transiently pluripotent, raising the question as to whether bivalency and other characteristics of ES cell chromatin are present in developing organisms as well. Using a ChIP protocol suitable for low numbers of cells, Rugg-Gunn et al. (2010) demonstrated that bivalent domains do exist in pluripotent epiblast cells of early post-implantation embryos in mice. Trophoblast and extraembryonic endoderm stem cells, however, contain few bivalent domains, owing to low levels of PRC2 that catalyzes H3K27me3 (Rugg-Gunn et al. 2010). Strikingly, H3K9me3 appears to functionally replace H3K27me3 in these stem cells, with H3K9me3 bivalent genes being similarly repressed in stem cells and resolved upon differentiation (Rugg-Gunn et al. 2010). The simultaneous presence of H3K9me3 and H3K4me3 was confirmed by sequential ChIP. Notably, H3K9me3 is present along with H3K4me3 and H3K27me3 at a subset of bivalent genes in cultured ES cells as well (Bilodeau et al. 2009; Yeap et al. 2009; Yuan et al. 2009). Bivalent domains were further detected in the pluripotent inner cell mass of preimplantation embryos and, to a lesser extent, in the extraembryonic tissue and trophectoderm of mouse blastocysts (Alder et al. 2010; Dahl et al. 2010). Pluripotent cells in the mouse embryo thus contain bivalent domains, demonstrating their existence beyond ES cells cultured in vitro.
Additional evidence for bivalent domains in developing embryos came from studies with zebrafish. Similar to other vertebrates, zebrafish embryos initiate transcription during the maternal–zygotic transition. H3K4me3 and H3K27me3 patterns emerge during that process, and bivalent domains detectable by sequential ChIP are formed at inactive genes, including the Hox clusters and other developmental TFs (Vastenhouw et al. 2010). According to another study, H3K4me3, H3K27me3, and bivalent domains are present even before the onset of zygotic transcription, albeit to a lesser extent (Lindeman et al. 2011). Although differing with regard to when bivalency is established during embryogenesis, both studies further affirm its existence in vivo.
While there is ample evidence supporting the existence of bivalent marks in mammals and zebrafish, there appear to be exceptions in other organisms. Using gastrula stage Xenopus embryos undergoing the midblastula transition, Akkers et al. (2009) detected very few bivalent domains. Moreover, genes with signals for H3K4me3 and H3K27me3 originated largely from distinct areas of the embryo, often being expressed in parts of the embryo, and only a minority of them corresponding to bivalent genes in mouse ES cells (Akkers et al. 2009). Although the use of late stage Xenopus embryos that have already undergone substantial lineage specification may partially explain this discrepancy with the zebrafish studies, it nevertheless seems plausible that modes of gene regulation differ between Xenopus, zebrafish, and higher vertebrates.
This notion is supported by the comparatively late appearance of repressive histone marks during lineage specification in Xenopus development (Schneider et al. 2011), suggesting that bivalent domains might be restricted to certain organisms. Indeed, while Drosophila features a repertoire of PcG and trxG complexes similar to that in mammals, bivalent domains appear to be absent. Analysis of Drosophila embryos and testis-derived stem cells did not yield evidence for significant coexistence of both marks (Schuettengruber et al. 2009; Gan et al. 2010). Inherent differences in gene regulation between arthropods, lower vertebrates, and mammals may account for this apparent discrepancy. For example, CpG island promoters, the sites of bivalent domains, are overwhelmingly more common in mammals. Instead, regulation of RNA polymerase II (Pol II) pausing may constitute an alternative means to coordinate the expression of early developmental genes in Drosophila (Muse et al. 2007; Zeitlinger et al. 2007; Boettiger and Levine 2009).
Molecular conformation of bivalent domains
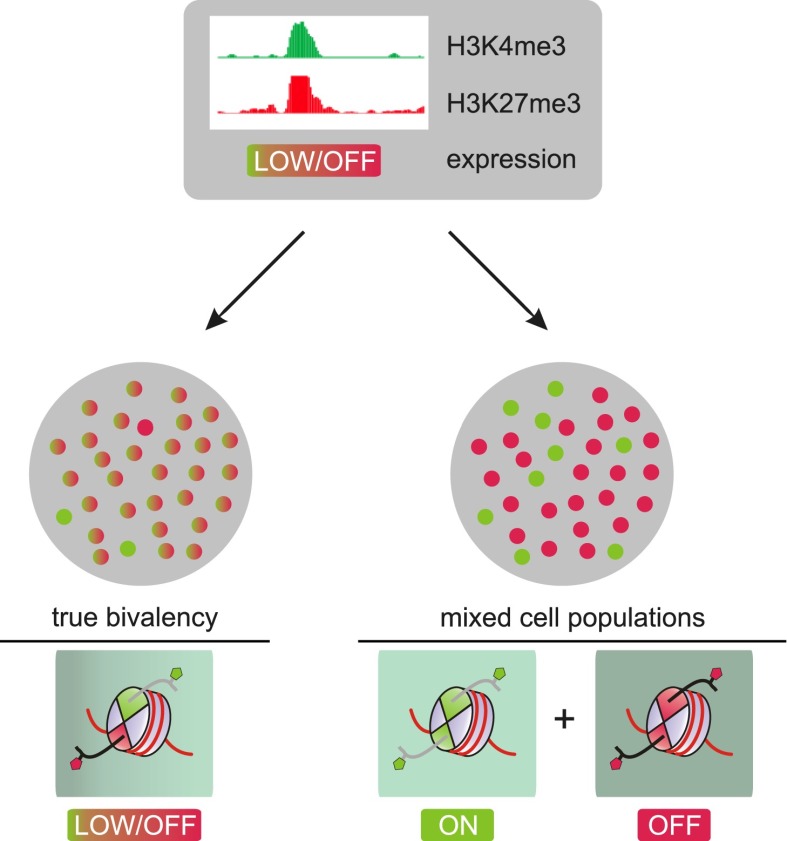
In accordance with their posited function in gene regulation, bivalent promoters must contain H3K27me3 and H3K4me3 simultaneously. ChIP assays performed individually for each mark are often drawn upon to establish bivalency at a given locus. However, these assays are unable to unequivocally establish the coexistence of both marks on the same allele in a given cell. Thus, it has been argued that the observed bivalency simply reflects cellular heterogeneity arising from the averaging of cells that carry either, but not both, marks at a given locus (Fig. 2). However, given that bivalent domains can still be observed—albeit in lower proportion—in unipotent cells such as T cells and MEFs, an admixture of cell populations appears to be an unlikely explanation for the observed coexistence of these marks. Experiments on sorted populations of T cells, ES cells, and embryonic tissue likewise argue against an admixture of cells with active expression or gene silencing feigning bivalency (Roh et al. 2006; Pan et al. 2007; Alder et al. 2010; Dahl et al. 2010).
Figure 2.
Bivalent domains and heterogeneity. Two scenarios could potentially explain the co-occurrence of H3K4me3 and H3K27me3 observed by ChIP-seq on bivalent promoters. As ChIP-seq cannot establish physical co-occurrence of two marks on the same allele, admixture of cells that either express (green) or do not express (red) the gene in focus could explain the occurrence of both marks as well as the low expression level in the overall population. In contrast, in the case of “true” bivalency, virtually all cells in the population carry both marks simultaneously at the promoter in question, leading to low, if any, expression for that gene in all cells.
In further addressing cellular heterogeneity as a cause of apparent bivalency, a recent study assessed the epigenetic landscape of naïve pluripotent mouse ES cells. Conventional mouse ES cell culture requires the presence of serum, and the heterogeneous expression of pluripotency factors and developmental regulators has been partly attributed to serum components (Chambers et al. 2007; Hayashi et al. 2008; Toyooka et al. 2008). Interestingly, the requirement for serum can be bypassed through the use of two small-molecule inhibitors (2i conditions) (Ying et al. 2008; Wray et al. 2011). The 2i inhibitors PD0325901 and CHIR99021 target mitogen-activated protein kinase kinase (Mek) and glycogen synthase kinase-3 (Gsk3), respectively. ES cells cultured under this “2i” regime are more homogeneous and exhibit tighter control of developmental gene expression. Interestingly, these naïve pluripotent ES cells display reduced levels of H3K27me3 at developmental promoters, and, consequently, fewer genes were classified as bivalent (Marks et al. 2012) because they fell below an arbitrary threshold previously used to identify H3K27me3 peaks. Nevertheless, it should be noted that the averaged H3K27me3 signals over these weakly expressing/inactive promoters were still appreciably higher compared with active promoters, which are typically devoid of H3K27me3. While it is clear that the relative abundance of bivalent loci (as defined by a certain computational signal cutoff) may differ in a context-dependent manner, unambiguous bivalent promoters are still present in highly homogeneous ES cells. Thus, we argue that bivalency cannot be solely a consequence of an admixture of cell populations.
Brookes et al. (2012) took an interesting approach to address the issue of ES cell heterogeneity by combining single-cell approaches with genome-wide analyses. First, they determined that of the ∼3600 genes classified as bivalent in this study based on the presence of H3K27me3 and H3K4me3 in ChIP-seq, ∼2400 were bound by RNA Pol II phosphorylated at Ser 5 (S5P) within its C-terminal domain (CTD), the form of RNA Pol II that initiates transcription. However, the levels of RNA Pol II phosphorylated at Ser 2 (S2P) within its CTD, a hallmark of productive mRNA elongation, were insignificant. Accordingly, these genes undergo transcription initiation but not elongation. Considering the incompatibility of H3K27me3 with transcription elongation (e.g., see Schmitges et al. 2011; see also below) and the association of RNA Pol II (S5P) with H3K4 trimethylases (Ng et al. 2003; Milne et al. 2005; see also below), this observation is supportive of a simultaneous rather than a separate occurrence of H3K27me3 and H3K4me3. Indeed, using sequential ChIP, the co-occurrence of PRC1 and PRC2 with the initiating S5P form of RNA Pol II was further confirmed on select bivalent promoters (Brookes et al. 2012). In contrast, the remaining ∼1200 genes initially classified as bivalent also contained the elongating form of RNA Pol II (S2P) with detectable transcript levels and thus likely exist in distinct active and repressed states in different subpopulations (Brookes et al. 2012). Further supporting this notion, FISH analysis revealed that the Lefty locus was marked by RNA Pol II S2P in some cells, indicating productive transcription, but by the catalytic PRC2 subunit Ezh2 in others, indicating repression in those cells (Brookes et al. 2012). Therefore, by integrating the transcriptional status of bivalent promoters with H3K27me3 and H3K4me3, this study clarified that while heterogeneity can lead to apparent bivalency at some loci, “true” bivalency exists at the majority of loci. However, it will be interesting to address the extent to which cell-intrinsic heterogeneity such as allelic differences may contribute to this overall abundance in bivalency. This will entail the use of more comprehensive single-cell-based approaches, possibly in combination with allele-specific ChIP.
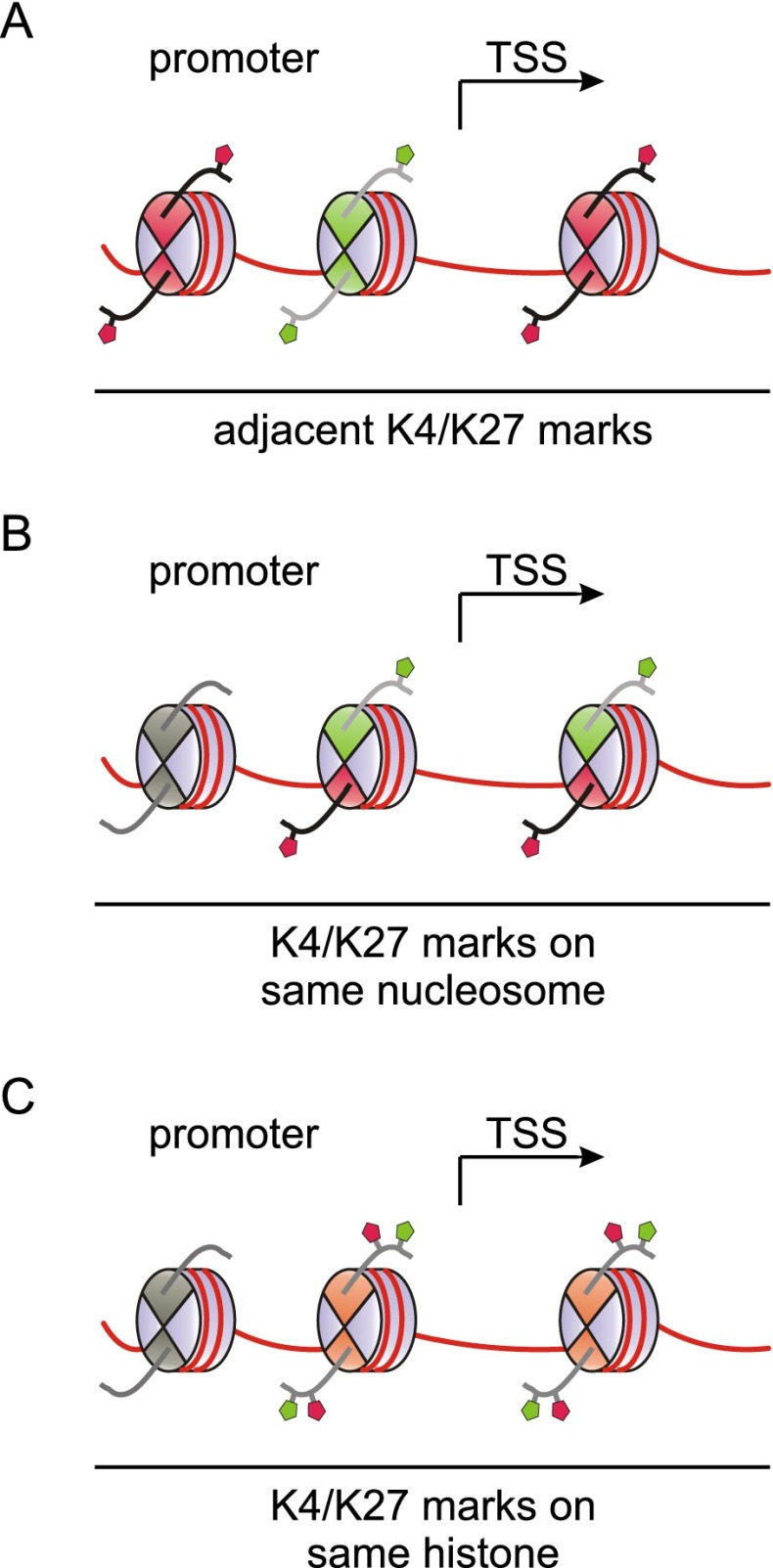
To accommodate both H3K4me3 and H3K27me3 in a bivalent configuration, promoters may principally adopt one of several conformations. The active and repressive histone modifications may be present on adjacent nucleosomes, the same nucleosome, or even the same copy of H3 within a nucleosome (Fig. 3). Sequential ChIP has been instrumental in showing that H3K4me3 and H3K27me3 are present on the same nucleosome or on neighboring ones at or around the TSS of bivalent genes. Bernstein et al. (2006) performed re-ChIP on three genes, and subsequent studies on T cells (Roh et al. 2006), human ES cells (Pan et al. 2007; De Gobbi et al. 2011), mouse trophoblast stem cells (Alder et al. 2010), zebrafish embryos (Vastenhouw et al. 2010), and 3T3 cells (Xie et al. 2012) provided re-ChIP-based evidence for the coexistence of these marks at genes in various systems. These studies employed sonicated chromatin fragments with one to three nucleosomes and thus indicate mark coexistence on either neighboring or the same nucleosomes. These conformations may confer equivalent bivalent functionality; however, potential differences between them have not been analyzed yet. Sequential ChIP on mononucleosomes generated by micrococcal nuclease digestion resolves both conformations and demonstrates mark coexistence on single nucleosomes. Such assays have been performed for select genes in C2C12 (Seenundun et al. 2010) and mouse ES cells (Voigt et al. 2012). However, to date, genome-wide sequential ChIP analysis has not been reported, and the results obtained on single genes may not extend to all genes that were classified as bivalent based on conventional ChIP-seq.
Figure 3.
Potential conformations of bivalent domains. At bivalent promoters, H3K4me3 and H3K27me3 marks may be present in different conformations. As illustrated in A, H3K4me3 and H3K27me3 may occupy neighboring nucleosomes in the vicinity of the TSS. Alternatively, both modifications may co-occupy the same nucleosome in either an asymmetric fashion featuring differentially modified copies of H3 within a single nucleosome (B) or a symmetric conformation involving H3 molecules that carry both modifications simultaneously (C). Recent data indicate that the latter conformation is unlikely to be present in cells, whereas an asymmetric conformation can be observed. See the text for details.
With these issues in mind, Voigt et al. (2012) sought to develop alternative strategies to directly probe for bivalency. Assessing the overall extent of H3K4me3/H3K27me3 co-occurrence on single nucleosomes in a quantitative fashion could provide strong support for the widespread existence of bivalent domains. With an approach combining mononucleosome ChIP with mass spectrometry (MS)-based quantitative profiling of histone modifications, Voigt et al. (2012) recently showed that ∼15% of all H3 histones within H3K4me3-carrying mononucleosomes were marked with H3K27me3. These MS-based findings indicate that both marks coexist at a sizable number of nucleosomes in ES cells, supporting widespread co-occurrence of H3K27me3 and H3K4me3 at thousands rather than a few select promoters. For MEFs, a lower amount of H3K27me3 was found on H3K4me3-containing nucleosomes (Voigt et al. 2012), in line with MEFs exhibiting fewer bivalent genes.
It has been argued that H3K4me3 and H3K27me3 cannot coexist on nucleosomes because PRC2 is inhibited by the active marks H3K4me3 and H3K36me3 (Schmitges et al. 2011). Moreover, MS-based studies found that H3K4me3 and H3K27me3 do not coexist on individual histones in HeLa cells (Young et al. 2009). Given the recent observation that sister histones within a nucleosome are often unequally modified (Voigt et al. 2012), bivalent domains could feature asymmetrically modified nucleosomes. Indeed, nucleosomes with only one H3K4me3 mark could still be methylated by PRC2, presumably on the unmodified H3 tail, whereas inhibition of PRC2 required the presence of H3K4me3 on both copies of H3 (Voigt et al. 2012). MS analysis of ES cell-derived histones confirmed the presence of these marks on distinct copies of H3 in vivo. In conclusion, these data suggest that bivalent domains feature nucleosomes that carry H3K4me3 and H3K27me3 on opposite H3 copies. Of note, this observed asymmetry in H3K27me3 and H3K4me3 is compatible with the reported reduction in H3K27me3 signals at some H3K4me3-marked nucleosomes relative to their neighbors that do not carry H3K4me3 (Pan et al. 2007; Marks et al. 2012).
Generation of bivalent domains
Controlling their access to genomic loci is thought to be a major way of regulating the activity of trxG and PcG proteins, the central players in setting up and maintaining bivalency. Several recruitment mechanisms have been proposed, including specific DNA sequence elements, DNA methylation status, particular histone modifications, TFs, and noncoding RNAs (ncRNAs), among others. Not surprisingly, many of these elements have been implicated in the generation of bivalent domains as well. One of the key clues as to how bivalent domains might be generated came from analyses of their underlying DNA sequences. It was uncovered early on that bivalent domains strongly correlate with CpG islands in ES cells (Bernstein et al. 2006). CpG islands are a prominent feature of promoters in vertebrate genomes and are present at ∼70% of all promoters (Saxonov et al. 2006; Deaton and Bird 2011). Virtually all CpG-rich promoters in ES cells are devoid of DNA methylation (Weber et al. 2007; Fouse et al. 2008; Meissner et al. 2008; Mohn et al. 2008) while being trimethylated at H3K4 (Guenther et al. 2007; Mikkelsen et al. 2007). Conversely, essentially all H3K4me3 sites map to CpG islands (Mikkelsen et al. 2007; Pan et al. 2007), which consequently holds true for all bivalent domains as well. CpG islands thus appear to play a major role in the establishment of bivalent domains. Indeed, artificially introduced CpG islands obtain both modifications, indicating that they suffice to recruit activities placing these marks (Mendenhall et al. 2010; Thomson et al. 2010; Woo et al. 2010; Lynch et al. 2012). Here we summarize how CpG islands together with associated factors may guide H3K4me3 and H3K27me3.
Establishment of H3K4me3 at bivalent loci
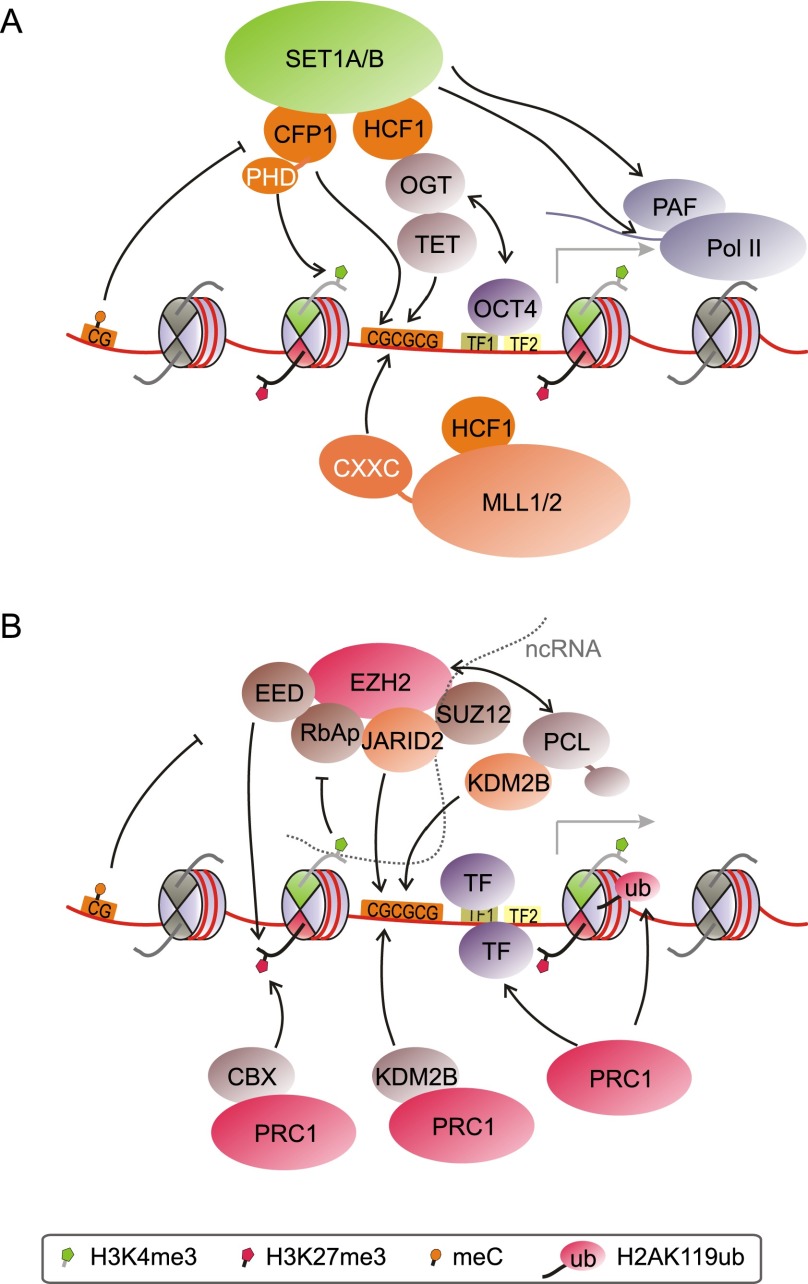
Unmethylated CpG islands are key factors in controlling H3K4me3 levels through recruitment of H3K4 methyltransferases (Fig. 4A). The SET1A/B complexes are responsible for the bulk of H3K4me3, while the MLL complexes likely assume more gene-specific roles (Shilatifard 2012). MLL1 and MLL2 contain so-called CXXC or zinc finger CXXC (ZF-CXXC) DNA-binding domains that specifically recognize unmethylated CpG islands (Birke et al. 2002; Bach et al. 2009), likely helping in the recruitment of these MLL complexes. Conversely, the specificity of CXXC domains for unmethylated CpG islands excludes them from sites of DNA methylation. Similarly, the CXXC finger protein 1 (Cfp1) subunit of SET1A/B complexes possesses a DNA-binding domain that is specific for unmethylated CpG (Lee et al. 2001). Knockdown of Cfp1 strongly diminishes H3K4me3 levels at CpG islands in 3T3 cells (Thomson et al. 2010). Cfp1 is recruited to engineered insertions of nonpromoter CpG islands, which gain H3K4me3 (Thomson et al. 2010). This establishment of H3K4me3 at ectopic CpG islands was confirmed in mouse ES cells (Mendenhall et al. 2010). A recent study found that loss of Cfp1 most strongly affects H3K4 methylation at promoters of highly expressed genes in ES cells (Clouaire et al. 2012). Correspondingly, H3K4me3 was not significantly altered at bivalent genes (Clouaire et al. 2012). A recent study revealed that Cfp1—like its yeast ortholog, SPP1 (Shi et al. 2007)—contains a plant homeodomain (PHD) finger that binds to H3K4me3 (Eberl et al. 2013), indicating a potential feed-forward mechanism for the deposition of H3K4me3 at active genes. These findings indicate a less crucial role for Cfp1 at weakly transcribing bivalent promoters, where CXXC-mediated recruitment of MLL1/2 might be more prevalent.
Figure 4.
Modes of H3K4 methyltransferase and PRC recruitment to CpG-rich promoters. (A) SET1A/B and MLL complexes are recruited in both transcription-independent and transcription-dependent ways. Mechanisms involve recognition of unmethylated CpG islands by CXXC domain proteins, interactions with the OGT/TET system, TFs, H3K4me3 itself, and the transcription machinery. (B) Recruitment of PRC2 likely relies on interactions with DNA, histones, histone modifications, auxiliary proteins, and ncRNAs. PRC1 is targeted by recognition of unmethylated CpG islands through CXXC proteins such as KDM2B (by binding to H3K27me3) and through interaction with TFs. The box denotes the histone and DNA modifications present in this figure as well as in Figures 5 and 6.
Host cell factor 1 (HCF1) is an integral component of the SET1A/B and MLL1/2 complexes. HCF1 interacts with and is modified by O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) (Capotosti et al. 2011). OGT modifies serine and threonine residues by the addition of O-GlcNAc (Hanover et al. 2012). Recent studies found that OGT also interacts with the ten-eleven translocation (TET) family of proteins (Chen et al. 2013; Deplus et al. 2013; Vella et al. 2013). TET proteins are viewed as potential mediators of active DNA demethylation, as their hydroxylase function can convert 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine (Wu and Zhang 2011; Williams et al. 2012). Interestingly, TET proteins seem to recruit OGT to CpG-rich promoters in ES cells (Fig. 4A; Chen et al. 2013; Deplus et al. 2013; Vella et al. 2013). TET1 and TET3 contain CXXC domains, whereas localization of TET2 to CpG islands is mediated by the CXXC domain protein IDAX (Ko et al. 2013). Knockdown of TET1 or TET2 led to loss of OGT from chromatin (Chen et al. 2013; Vella et al. 2013), while knockdown of TET2 also reduced O-GlcNAc modification of histones (Chen et al. 2013). Moreover, interaction with TET2/3 stimulates OGT activity toward HCF1 (Deplus et al. 2013). Given this connection between OGT, TET proteins, and HCF1, it appears likely that TET proteins are key factors in recruiting SET1A/B and/or MLL1/2 complexes to CpG islands (Fig. 4A). In line with this notion, OGT- and TET-binding sites overlap to a large extent with H3K4me3 sites at promoters (Deplus et al. 2013; Vella et al. 2013). Both OGT activity and its interaction with TET2/3 promoted chromatin association of SET1A complexes and H3K4 methylation in HEK293 cells (Deplus et al. 2013). Taken together, TET proteins and OGT may play a central role in establishing H3K4me3 at CpG-rich promoters in ES cells.
Recent studies also suggested a CpG-dependent role of certain histone variants in bivalency. Unlike canonical histones, which are synthesized and deposited in a strictly replication-coupled fashion during S phase, histone variants are expressed throughout the cell cycle and can be deposited independently of DNA replication (Sarma and Reinberg 2005; Talbert and Henikoff 2010). Genome-wide studies have shown that both H2A.Z and H3.3 are highly enriched on CpG-rich promoters, correlating with H3K4 methylation states at active and bivalent promoters as well as enhancers (Barski et al. 2007; Goldberg et al. 2010; Ku et al. 2012; Hu et al. 2013). Interestingly, depletion of H2A.Z impairs recruitment of MLL complexes primarily to enhancers but also to active and bivalent promoters (Hu et al. 2013). In addition, OCT4 levels are significantly decreased at one-third of its binding sites upon H2A.Z depletion, indicating that H2A.Z further plays a role in directing OCT4 to promoters and active enhancers (Hu et al. 2013). Taken together with studies showing interaction of OCT4 with MLL complexes (Ang et al. 2011) and with OGT/HCF1 (Pardo et al. 2010; van den Berg et al. 2010), these findings may indicate a role for pluripotency factors such as OCT4 and for the histone variant H2A.Z in targeting MLL complexes. In addition to pluripotency factors, other TFs such as the estrogen receptor and E2F6 also have been shown to interact with MLL complexes, adding further potential avenues for their recruitment (Ruthenburg et al. 2007).
Several ncRNA species, especially long ncRNAs, have been implicated in the recruitment and function of many chromatin-modifying enzymes (for review, see Wang and Chang 2011; Rinn and Chang 2012; Mercer and Mattick 2013). Expression profiles of several types of ncRNAs suggest that they are involved in cell fate decisions (Jacquier 2009; Guttman et al. 2011; Pauli et al. 2011). Even though ncRNAs are mostly implicated in targeting of repressive complexes, at least one study suggests a role for ncRNAs in the recruitment of MLL complexes. The long ncRNA HOTTIP is transcribed from the 5′ end of the HoxA locus and is bound by the WDR5 protein, which is a subunit of SET1/MLL complexes (Wang et al. 2011), ATAC (Wang et al. 2008) and NSL (Cai et al. 2010) acetyltransferase complexes, and the CHD8 chromatin remodeling factor (Thompson et al. 2008). Knockdown of HOTTIP diminished the presence of MLL1 complexes at several TSSs within the 5′ end of the HoxA locus, suggesting that HOTTIP mediates targeting of the MLL1 complex in cis to neighboring loci (Wang et al. 2011). Further studies are required to explore whether as yet unidentified long ncRNAs may be involved in targeting SET1 and MLL complexes to additional loci and whether RNA-mediated targeting is a common theme for these complexes.
In addition to the recruitment mechanisms described above, the process of transcription itself likely plays important roles in the establishment and reinforcement of H3K4me3 (Fig. 4A). The underlying mechanisms have been studied mostly in yeast, but analogous events likely take place during transcription in metazoans as well (for review, see Ruthenburg et al. 2007; Smith and Shilatifard 2010). In yeast, SET1 is recruited to the 5′ end of actively transcribed loci through direct interaction with the RNA Pol II CTD in its S5P form (Ng et al. 2003). Human MLL1/2 complexes were shown to similarly interact with S5P (Milne et al. 2005). Moreover, yeast SET1 also interacts with polymerase-associated factor 1 (Paf1), a protein complex that is associated with elongating RNA Pol II, mediating recruitment of SET1 to transcribed loci during early elongation (Krogan et al. 2003). A similar mechanism might recruit SET1 and MLL complexes in mammals, leading to additional deposition of H3K4me3 during transcription. Active transcription might therefore reinforce H3K4me3 deposition at actively transcribed genes and, to a lesser extent, at minimally transcribed bivalent loci.
CpG islands and PRCs
CpG islands likewise play an important role in establishing and maintaining H3K27me3 at bivalent domains (Fig. 4B). In contrast to H3K4me3, however, not all CpG islands are marked with H3K27me3. Moreover, whereas H3K4me3 is highly localized at promoters and thus marks only a minute fraction of nucleosomes, the distribution patterns of H3K27me3 are more complex. H3K27me3 marks ∼10%–15% of all H3 histones in ES cells as assessed by quantitative MS (Peters et al. 2003; Voigt et al. 2012). If considering H3K27me2 as well, ∼50% of all nucleosomes in ES cells are modified by PRC2 (Voigt et al. 2012). Many ChIP-seq studies revealed “lawns” of H3K27me3 mostly spanning intergenic regions and inactive genes (e.g., see Pauler et al. 2009; Young et al. 2011; Marks et al. 2012). H3K27me3 is also enriched in subtelomeric regions (Rosenfeld et al. 2009) and at long terminal repeat retrotransposons (Leeb et al. 2010). These regions likely account for the bulk of H3K27me2/3 present in the ES cell genome. In addition, a relatively smaller amount of H3K27me3 also exhibits more localized patterns around the TSS, sometimes extending into the promoter (e.g., see Mikkelsen et al. 2007; Young et al. 2011). In ES cells, these TSSs are almost exclusively bivalent (Mikkelsen et al. 2007; Ku et al. 2008). Interestingly, when analyzing the genomic localization of components of the PRC2 complex, defined peaks are predominantly found around gene promoters (Boyer et al. 2006; Bracken et al. 2006; Ku et al. 2008), indicating more efficient recruitment or retention at promoters. PRC2 appears to be more spread out over inactive regions and repeats, giving rise to a flat distribution rather than defined peaks. Here, we focus on modes of H3K27me3 establishment at bivalent loci; however, aspects of PRC2 recruitment to these loci may hold true for more extensive areas of PRC2 activity, such as inactive regions and transposons, as well.
The correlation between promoter-associated H3K27me3 and CpG islands was noted early on (Bernstein et al. 2006; Boyer et al. 2006; Mikkelsen et al. 2007). Subsequent studies aimed to distinguish CpG islands that feature H3K4me3 alone from those carrying H3K27me3 as well. A key difference appears to be the transcriptional status of the associated genes. In human and mouse ES cells, >97% of all promoter-associated Ezh2 recruitment sites corresponded to CpG islands (Ku et al. 2008). Interestingly, further analysis revealed that Ezh2-positive CpG islands contain binding sites for TFs that were either absent or inactive in ES cells or corresponded to transcriptional repressors. Conversely, Ezh2-free CpG islands featured binding sites for transcriptional activators that are highly expressed and active in ES cells, such as Myc, NFY, and Ets1 (Ku et al. 2008). CpG island promoters may thus assume and maintain a bivalent conformation by default if transcriptional activators are absent (Ku et al. 2008).
This concept implies that productive transcription, through proper assembly of TFs, coactivators, and the splicing machinery, is likely to preclude stable PRC2 recruitment or binding around the TSS, whereas either the absence of transcription or transcription at levels below a certain threshold is sufficient to allow PRC2 recruitment to promoter CpG islands. In support of this, introduction of ectopic CpG islands is sufficient to recruit PRC2 as long as these CpG islands are devoid of activating sequences (Mendenhall et al. 2010). A CpG island-containing region of the HoxD locus was similarly sufficient to recruit PRC1 and PRC2; however, this recruitment was chiefly attributed to other sequence elements within the region (Woo et al. 2010). Moreover, replacement of a fragment of the murine α-globin locus with its CpG island-containing human counterpart sufficed to induce H3K4me3 and H3K27me3 modification and recruit PcG proteins, establishing bivalency at the normally nonbivalent mouse locus (Lynch et al. 2012). By modulating the associated promoter sequences, Lynch et al. (2012) further demonstrated that the presence of H3K27me3 correlated inversely with the degree of activation. First, these data indicate that unmethylated CpG islands mediate recruitment of both SET1/MLL and PRC2 activities, as both H3K4me3 and H3K27me3 are established at ectopically introduced CpG islands. Moreover, higher levels of transcription appear to displace PRC2 and thus H3K27me3, whereas low levels permit default recruitment of PRC2 to CpG islands.
These data suggest that PRC recruitment in mammals likely employs mechanisms distinct from that in Drosophila, where CpG islands are absent and Polycomb-repressive elements (PREs) and associated TF binding partially account for PRC recruitment (Schuettengruber et al. 2007; Simon and Kingston 2009). Nonetheless, certain TFs that chiefly act as repressors may augment targeting of PRCs to CpG islands in the case of mammals. Candidates include REST and SNAIL, whose target sequences are found at a subset of PRC targets (Dietrich et al. 2012; Arnold et al. 2013) and might fulfill, at least in part, the role of Drosophila PREs.
Targeting PRC1 and PRC2 to bivalent loci
As the core PRC2 complex does not contain any DNA-binding domains itself, the mechanisms of its recruitment to gene loci have remained elusive. Even though PRC2 localizes to unmethylated CpG islands, no CXXC domain-containing proteins are known to interact with PRC2. Genomic binding sites of Jarid2, an interactor of PRC2, exhibit significant overlap with PRC2 target sites in ES cells (Peng et al. 2009; Shen et al. 2009; Landeira et al. 2010; Li et al. 2010; Pasini et al. 2010). Jarid2 lacks a CXXC domain but exhibits unspecific DNA binding with a slight bias toward GC-rich sequences (Fig. 4B; Li et al. 2010). Recruitment of Jarid2 and the PRC2 core component Ezh2 appear to be codependent, but the exact role of Jarid2 in recruiting PRC2 remains unclear. Similarly, AEBP2, a zinc finger protein that binds DNA with low specificity, interacts and colocalizes with PRC2 at some promoters (Kim et al. 2009). PHF1 (PCL1), MTF2 (PCL2), and PHF19 (PCL3), orthologs of Drosophila Polycomb-like (PCL), also interact with PRC2 and have been implicated in its recruitment (Margueron and Reinberg 2011; Simon and Kingston 2013). These and other proteins shown to transiently interact with PRC2 may mediate its recruitment to specific loci, but it remains unclear whether any of these proteins can completely account for its preference for CpG islands in ES cells.
Targeting of PRC2 complexes to specific genomic sites in mammals likely occurs through multiple means. Given the paucity of sequence-specific factors identified to date, other modes of interaction may explain PRC2 recruitment to CpG islands. PRC2 forms multiple contacts with nucleosomes that generate affinity for chromatin in a sequence-independent fashion (Fig. 4B; see also Margueron and Reinberg 2011). Although each such interaction is of low affinity, the combination of these interactions may allow for a consolidated and spatially accurate recruitment of PRC2 based on local chromatin features, akin to coincidence detection (Margueron and Reinberg 2011; Voigt and Reinberg 2011). Specifically, Jarid2 and AEBP2 each interact with DNA and with PRC2, and the PRC2 core components RbAp46/48 and Eed bind to histones H3 and H4. Whereas Eed also binds to H3K27me3 and might function in perpetuating the mark (Margueron et al. 2009), H3K4me3 abrogates RbAp46/48 recognition of H3 and inhibits PRC2 activity (Schmitges et al. 2011). Similarly, H3K36me3 inhibits PRC2 activity (Schmitges et al. 2011; Yuan et al. 2011) and may serve to exclude PRC2 from active genes. Besides histone modifications, contact with H3 in neighboring nucleosomes stimulates PRC2, leading to more efficient methylation in compact chromatin regions (Yuan et al. 2012). On the other hand, CpG methylation abrogates PRC2 interaction with nucleosomes in vitro (Fig. 4B; Bartke et al. 2010).
Additional interactions with nucleosomes may stem from PCL proteins, which contain two PHD fingers and a tudor domain. Unexpectedly, the tudor domains of PHF1 and PHF19 recognize H3K36me3 as well as H3K27me3, although weakly in the latter case (Ballaré et al. 2012; Brien et al. 2012; Musselman et al. 2012; Cai et al. 2013). Corecruitment of the H3K36 demethylases NO66 and possibly KDM2B (see below) resolves PRC2 inhibition by H3K36me3, suggesting a pathway to repress previously active genes (Ballaré et al. 2012; Brien et al. 2012). All three PCL proteins are expressed in ES cells, and each co-occupies subsets of PRC2-bound loci. Knockdown of PHF19 diminished overall H3K27me3 levels and significantly decreased binding of the PRC2 core component Suz12 at ∼65% of overall target loci and >80% of bivalent loci in ES cells (Hunkapiller et al. 2012). It is notable that some bivalent promoters exhibit appreciable H3K36me3 in mouse ES cells (Cai et al. 2013). In this case, PHF19 may help target PRC2 to these bivalent domains in part via H3K36me3 recognition to constrain further productive transcription.
Taken together, these findings might explain how PRC2 and H3K27me3 are largely excluded from actively transcribed genes and regions silenced by DNA methylation. Instead of active recruitment to target loci, exclusion of PRC2 from inappropriate regions might be a predominant mode of regulating its genomic localization. Bivalent domains form a special case, as H3K4me3 would be expected to prevent PRC2 activity. However, as mentioned above, the presence of H3K4me3 on only one, but not both, copies of H3 per nucleosome is permissive to PRC2 activity (Voigt et al. 2012). Thus, the resulting asymmetric conformation with H3K4me3 and H3K27me3 occupying opposite H3 tails allows the coexistence of active and repressive marks within single nucleosomes at bivalent loci (Voigt et al. 2012).
We discussed how DNA elements, TFs, nucleosomes, and their associated modifications may be highly instructive to PRC2 recruitment. However, they may not fully explain the observed H3K27me3 patterns. In exploring additional means for sequence-directed recruitment, many groups have assessed a potential role for RNA molecules. These endeavors were motivated by the well-established action of the Xist ncRNA in X inactivation and studies showing a direct interaction between a region of Xist and Ezh2 (for review, see Brockdorff 2013). Ezh2 and other PRC2 subunits interact with a wide range of long ncRNAs (Wang and Chang 2011; Rinn and Chang 2012), suggesting that these proteins recognize secondary structures rather than primary sequences. Indeed, instead of providing locus specificity based on sequence complementarity, secondary structures may direct recruitment in a co- or post-transcriptional fashion to loci in cis or in trans.
Transcripts of 50–200 nucleotides (nt) originating from several PRC2 target loci in ES cells have been implicated in PRC2 recruitment (Kanhere et al. 2010). These short ncRNAs span CpG-rich sequences and are predicted to form stem–loop structures similar to those of long ncRNAs. The Suz12 subunit of PRC2 interacts with these short ncRNAs, potentially recruiting PRC2 in cis (Kanhere et al. 2010). Notably, activation of bivalent genes abrogates production of their short ncRNAs (Kanhere et al. 2010). However, it remains unclear whether these short transcripts directly elicit PRC2 recruitment in vivo. Interestingly, the local abundance of CpG islands may predict whether a locus is marked purely by H3K4me3 or also by H3K27me3, as the likelihood of H3K27me3 modification increases with CpG island density (Orlando et al. 2012). Short transcripts from promoters with multiple CpG islands are highly likely to form the stem–loop structures implicated in ncRNA-mediated PRC2 recruitment (Orlando et al. 2012). Nonetheless, the underlying mechanisms remain unclear, rendering the role of ncRNAs in recruitment as controversial at present.
A subset of bivalent genes is further occupied by PRC1. All PRC1 complexes contain the RING1A/B ubiquitin ligase and a member of the PCGF protein family, bridging RING1B and its interaction partners. PRC1 forms several subcomplexes with unique subunit composition (Gao et al. 2012; Tavares et al. 2012; Simon and Kingston 2013). RING1B occupies ∼40%–50% of all bivalent domains in ES cells (Ku et al. 2008; Brookes et al. 2012). RING1B-bound bivalent genes are highly enriched for developmental factors and are well conserved between mice and humans. Moreover, they exhibit larger regions of H3K27me3 and are more likely to remain repressed upon differentiation (Ku et al. 2008). PRC1 complexes that contain CBX proteins may be recruited, at least in part, by binding to H3K27me3. In mouse ES cells, CBX7 is likely the predominant CBX protein that helps recruit PRC1 to H3K27me3-containing sites (Morey et al. 2012, 2013). However, other H3K27me3-independent determinants control PRC1 targeting and depend on the subunit composition of each particular PRC1 complex (Fig. 4B). Candidates include TFs such as E2F6, YY1, and REST as well as ncRNAs (Simon and Kingston 2013).
Notably, Fbxl10/KDM2B was recently shown to recruit some PRC1 complexes to unmethylated CpG islands via its CXXC domain, rendering it an intriguing candidate for targeting some PRC1 complexes to bivalent promoters (Farcas et al. 2012; He et al. 2013; Wu et al. 2013). KDM2B is present at low levels at virtually all unmethylated CpG islands in ES cells but is excluded from sites of DNA methylation. However, substantial KDM2B-PRC1 binding is observed only at a fraction of all unmethylated CpG islands. This suggests a “sampling” mechanism whereby KDM2B-PRC1 complexes continually probe unmethylated CpG loci for their susceptibility to repression, and stable recruitment may further depend on pre-existing repressive determinants (Farcas et al. 2012). Likewise, factors involved in transcription may prevent accumulation of high levels of these PRC1 complexes at active loci.
Several studies have connected PRCs to H2A.Z. As mentioned above, H2A.Z is enriched at both active and bivalent promoters but not at loci marked exclusively by H3K27me3 (Ku et al. 2012; Hu et al. 2013). Loss of H2A.Z reduces PRC2 occupancy at both bivalent promoters and enhancers (Creyghton et al. 2008; Hu et al. 2013). Initial reports suggested that targeting of H2A.Z to bivalent promoters may depend on PRC1/2 complexes and vice versa (Creyghton et al. 2008). However, recent studies indicate that H2A.Z deposition is independent of the PRCs (Illingworth et al. 2012). Nevertheless, understanding H2A.Z recruitment may shed light on how PRCs are targeted to specific loci. This question is of special interest given that OCT4 targeting is dependent on H2A.Z in ES cells (Hu et al. 2013). In addition to interacting with the MLL complexes, OCT4 has also been shown to interact with PRC1 subcomplexes as well as with the histone deacetylase-containing NuRD complex (Pardo et al. 2010; van den Berg et al. 2010). The NuRD complex facilitates PRC2 recruitment through its deacetylation of H3K27 (Reynolds et al. 2012). Notably, depletion of NuRD leads to deregulation of several bivalent genes and is accompanied by increased H3K27ac and reduced H3K27me3 (Reynolds et al. 2012). Taken together, these studies underscore the importance of H2A.Z as a central player orchestrating the deposition of pluripotency factors and epigenetic regulators at bivalent loci.
A model for the generation and maintenance of bivalent domains
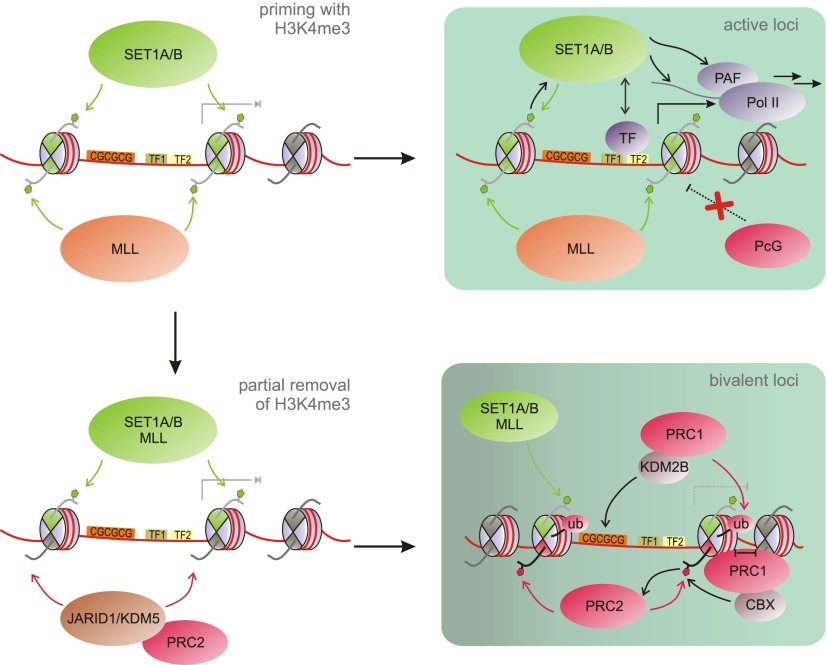
Based on the work described in the preceding sections, we propose the following model for the establishment of bivalent domains (Fig. 5). All promoters with a high CpG content, including bivalent promoters, are “primed” with basal levels of H3K4me3 in ES cells. Recruitment of SET1A/B/MLL complexes is mediated at least in part by CXXC domain-containing proteins or through the action of TET enzymes, OGT, and histone variants. In the presence of activating signals and TFs such as OCT4, H3K4me3 at these promoters is reinforced and sustained by cotranscriptional deposition. The act of productive transcription as well as the ensemble of TFs and coactivators may suffice to exclude PcG proteins from active genes through competition for binding to the underlying GC-rich DNA sequences or through repulsion of PRC2 by nucleosomes symmetrically modified with H3K4me3 or H3K36me3. Spurious H3K27me3 may be removed from active genes by demethylases such as UTX, a component of MLL3/4 complexes (Agger et al. 2007; Lee et al. 2007).
Figure 5.
A step-wise model for the generation of bivalent domains. MLL and SET1A/B complexes “prime” CpG-rich promoters with basal levels of H3K4me3. For recruitment mechanisms, see Figure 4A. Loci where suitable TFs and activators are present will experience reinforcement of H3K4me3 deposition through cotranscriptional means, leading to sustained activation (active loci). In the absence of activating TFs, H3K4 demethylases can counterbalance the activity of MLL/SET1 complexes, allowing PRC2 to methylate one copy of H3 at the promoter to generate a bivalent set of marks. At some bivalent loci, PRC2 is joined by PRC1, reinforcing gene repression as well as PRC recruitment in a feed-forward loop. See Figure 4 for a key of the histone modifications present in this figure.
In the case of bivalent genes, activating signals or corresponding TFs are absent, and the associated genes are either silent or transcribed at low levels. Under these circumstances, H3K4me3 demethylases of the Jarid1/KDM5 class (Mosammaparast and Shi 2010) can compete with H3K4me3 deposition, leading to removal of H3K4me3 from at least one copy of H3 at the TSS-associated nucleosome. An interesting candidate in this context is RBBP2 (KDM5A), which interacts and colocalizes with PRC2 (Pasini et al. 2008). PRC2 would then be able to overcome H3K4me3-mediated exclusion, methylating the available H3 to establish an asymmetric, bivalent nucleosome. Again, demethylases may counterbalance H3K27me3 deposition, leading to equilibrium between H3K4me3 and H3K27me3. Directly or indirectly, PRC2 is recruited to the bivalent promoters through their unmethylated CpG islands, for which it competes with SET1A/B and MLL complexes. Interestingly, a higher density of CpG islands correlates with more efficient PRC2 recruitment and may enable PRC2 to successfully compete with the machinery that establishes basal H3K4me3 levels but may not suffice to overcome transcription-associated reinforcement of H3K4me3. The degree of transcription may thus play a major role in controlling bivalency. PRC1 cooperates with PRC2 in setting up bivalency at many loci, and each may reinforce the other's recruitment. PRC1—targeted to CpG islands by, e.g., KDM2B—locally compacts chromatin, which stimulates PRC2. Enhanced deposition of H3K27me3 in turn supports CBX-mediated recruitment of PRC1, resulting in a feed-forward loop.
Notably, the correlation between CpG density and both H3K4me3 and H3K27me3 appears weaker in differentiated cell types, indicating that different or additional mechanisms may control recruitment in those systems. The factors mediating the establishment of bivalent domains in ES cells likely control their maintenance as well. As long as developmental signals are absent, the equilibrium between activating and repressive signals remains unchanged. The methyltransferase and demethylase activities involved remain targeted to these loci, and their continuous interplay maintains the bivalent modification status. As discussed in the next section, the presence of the PRC1 and PRC2 complexes and their associated marks may ensure repression of the appropriate genes in ES cells while still allowing for subsequent activation when activating TFs tip the balance toward expression of the bivalent gene.
Function of bivalency—fine-tuning gene expression and safeguarding differentiation
The initial observations uncovering bivalency (Azuara et al. 2006; Bernstein et al. 2006) led to the elegant and inherently appealing concept that bivalent domains keep genes in a poised or primed state that allows for either rapid activation or stable silencing upon differentiation while maintaining low expression levels and a reversible, silenced state in ES cells. Subsequent studies confirmed this concept on a genome-wide scale, showing widespread resolution of bivalent genes and associated expression changes upon differentiation (Mikkelsen et al. 2007; Pan et al. 2007; Zhao et al. 2007). This striking correlation has been seen as indicative of a causal role of bivalency in the timely activation of genes during differentiation. However, it was not evident from these studies whether bivalent features indeed facilitate gene activation as well as silencing and whether they are required for developmental plasticity.
In this section, we summarize studies aimed at uncovering the relevance of bivalent domains for development, explore how reversible silencing of bivalent genes is achieved in ES cells at the molecular level, and discuss how bivalency may be resolved upon differentiation. We suggest that bivalency acts as a means to fine-tune expression of crucial factors during development and defend against unscheduled gene activation, collectively contributing to the robustness and reduced noise in these processes.
Probing the function of bivalency in development
It is beyond doubt that bivalently marked genes do change expression during differentiation. Moreover, as outlined in the preceding sections, strong evidence for their existence both in vivo and in vitro have been provided by ChIP and other approaches. Nevertheless, it remains contested whether bivalent features indeed facilitate gene activation as well as silencing during development and whether they are required for developmental plasticity in a physiological context. The most direct and convincing way to clarify the relevance of bivalent domains during development would entail specifically ablating bivalent domains to analyze potential developmental defects. Such an approach is nontrivial and requires a better understanding of factors directing trxG and PcG proteins to bivalent domains. To directly address the role of bivalency in poising gene expression would require specifically ablating PcG and trxG protein targeting to bivalent loci without affecting the expression of other genomic features normally bound by these complexes. Such studies have yet to be performed but are certainly tenable with the advent of programmable genomic editing tools.
Nevertheless, some insights into the relevance of bivalent domains for development can still be gained from analysis of PcG and trxG gene knockouts. Although loss of H3K4me3 or H3K27me3 is not limited to bivalent loci in these models, valuable information can be obtained especially for PcG proteins, as the vast majority of their genic targets in ES cells correspond to bivalent genes. Nonetheless, the absence of H3K27me3 and PRCs at repetitive elements and other targets must be considered when interpreting PcG mutant phenotypes. Different groups have observed a general propensity of PcG mutant ES cells to up-regulate developmental genes, supporting a crucial role for PcG proteins and thus bivalent domains in development. For instance, several bivalent genes are prematurely expressed in Eed−/− ES cells (Azuara et al. 2006; Boyer et al. 2006). Likewise, Suz12−/− ES cells show higher expression of lineage-specific genes (Pasini et al. 2007). However, despite the misexpression of lineage genes, cell viability and self-renewal are not compromised in PRC2-deficient ES cells (Pasini et al. 2007; Chamberlain et al. 2008; Shen et al. 2008; Leeb et al. 2010). The overall mild defects of PRC2-deficient ES cells in self-renewal may be partially explained by PRC1-mediated compensatory effects and the absence of TFs that could robustly activate the affected genes in the undifferentiated state. Indeed, simultaneous depletion of RING1B and EED in ES cells provokes an even stronger inclination toward differentiation, although self-renewal can still be preserved under careful culture conditions (Leeb et al. 2010). In contrast to the relatively mild effects on self-renewal, all PRC2-deficient ES cells exhibit aberrant differentiation potential (Pasini et al. 2007; Chamberlain et al. 2008; Shen et al. 2008; Leeb et al. 2010), which parallels the post-implantation lethality phenotypes observed in PRC2 knockout mouse models (Faust et al. 1995; O'Carroll et al. 2001; Pasini et al. 2004). Taken together, these knockout models demonstrate that PRCs—presumably to a large degree through control of bivalent target genes encoding developmental factors—are vital for proper differentiation.
Notably, the recent discovery that FBXL10/KDM2B is key in targeting a subset of PRC1 complexes to CpG-rich promoters may allow for the specific modulation of PRCs at bivalent loci (Farcas et al. 2012; He et al. 2013; Wu et al. 2013). Its depletion in ES cells causes derepression of PcG target genes comparable with RING1B knockout cells and leads to premature and defective differentiation (He et al. 2013; Wu et al. 2013), underscoring the importance of PcG repression as a safeguard mechanism at bivalent loci for proper development, especially in the context of lineage specification.
Histone modifications, binding proteins, and PRCs in reversible silencing at bivalent promoters
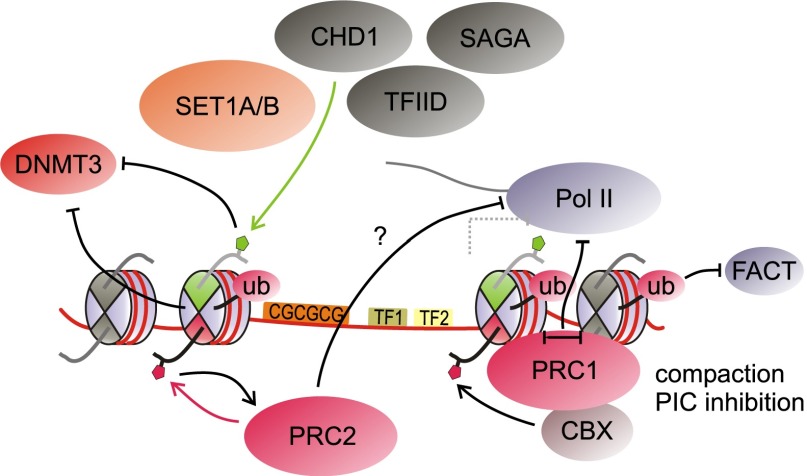
Both the histone modifications and the protein complexes present at bivalent promoters likely mediate the impact of bivalency on transcription. Many proteins that bind H3K4me3 and function as effectors have been described, most of which are associated with active transcription (Fig. 6). The PHD finger of the TAF3 subunit of TFIID recognizes H3K4me3 (Vermeulen et al. 2007), whereas the TAF1 subunit binds to acetylated lysines on histones H3 and H4 via its bromodomains (Jacobson et al. 2000). These interactions likely contribute to recruitment of this general TF. Indeed, a recent study demonstrated that the TAF3–H3K4me3 interaction directly contributes to preinitiation complex formation in a reconstituted transcription system and to TFIID recruitment in vivo (Lauberth et al. 2013). As mentioned above, H3K4me3 binding by the PHD finger of Cfp1 may be instrumental in mediating a feed-forward loop for H3K4me3 methylation (Clouaire et al. 2012; Eberl et al. 2013). Other factors bound by H3K4me3 include the chromatin remodeler CHD1, the SAGA complex, and the H3K9me2-, H3K27me2-, and H4K20me1-specific demethylase PHF8 (Horton et al. 2010; Vermeulen et al. 2010). In combination with other proteins involved in transcription, these factors may also serve to exclude PcG proteins from actively transcribed loci, contributing to the equilibrium between transcription and repression found at bivalent domains.
Figure 6.
Histone mark-binding proteins at bivalent promoters. Several binding proteins recognizing H3K4me3 and H3K27me3 may be present at bivalent genes. TFIID, SAGA, and CHD1 complexes, among others, are recruited at least in part by H3K4me3. Cfp1 binds to H3K4me3 as a subunit of SET1A/B complexes, supporting further H3K4me3 methylation. H3K4me3, together with other factors, prevents DNMT3-mediated DNA methylation. On the other hand, H3K27me3 recruits certain PRC1 complexes but also reinforces binding of PRC2 itself through interaction with its EED subunit. PRC1 complexes compact chromatin and interfere with preinitiation complex (PIC) assembly. Some PRC1 complexes further catalyze H2.Aub1, which may impair FACT recruitment, among other effects. See Figure 4 for a key of the histone modifications present in this figure.
To ensure proper governance of potency, ES cells employ elaborate schemes to prevent permanent DNA silencing of developmental promoters; for example, through the concerted actions of H3K4me3, TET enzymes, and H2A.Z. Promoters with high CpG content contain H3K4me3 but are virtually devoid of DNA methylation (Weber et al. 2007; Fouse et al. 2008; Meissner et al. 2008). H3K4me3 interferes with the recruitment of the de novo DNA methyltransferases DNMT3A and DNMT3B (Ooi et al. 2007; Zhang et al. 2010), enforcing the mutual exclusion of these marks. In addition to affecting recruitment, H3K4me3 diminishes activity of these enzymes in vitro (Zhang et al. 2010). The presence of the TET enzymes at CpG islands may further ensure the absence of DNA methylation at those sites (Wu and Zhang 2011; Williams et al. 2012). Moreover, H2A.Z at CpG-rich promoters may further antagonize DNA methylation (Zilberman et al. 2008). H3K4me3, possibly along with other factors such as H2A.Z, may thus function to a large degree by keeping genes in a state permissive for activation by precluding irreversible repression through DNA methylation (Fig. 6). Avoidance of DNA methylation is essential for bivalent genes as well, as they are required to retain plasticity for subsequent activation or repression.
However, an inevitable consequence of such a permissive chromatin state may be a resultant low level of transcription emanating from these bivalent promoters. Indeed, as previously mentioned, most promoters of protein-coding genes (Guenther et al. 2007) and essentially all bivalent promoters (Brookes et al. 2012) harbor the initiating (S5P) form of RNA Pol II in ES cells, indicative of transcriptional competence. Furthermore, several groups have ascertained the presence of low but appreciable levels of regulated transcription arising from PRC-bound bivalent loci (Kanhere et al. 2010; Walker et al. 2010; Min et al. 2011; Brookes et al. 2012). Interestingly, the extent of RNA Pol II engagement and elongation is modulated by the presence of PRC1 and PRC2 (Stock et al. 2007; Chopra et al. 2011; Min et al. 2011).
Thus, we propose that bivalency constitutes an apparent “catch-22” situation but nevertheless a necessary one. A key role of H3K27me3 as well as of the PcG machinery on bivalent domains is to precisely curtail overt productive transcription and fine-tune gene expression. In this context, understanding the molecular underpinnings of PcG protein-mediated gene repression is pertinent. In the case of PRC2, the initial deposition of H3K27me3 may serve to impede subsequent deposition of H3K36me3, given the noncompatibility of both modifications to coexist on the same histone tail (Schmitges et al. 2011; Voigt et al. 2012). Conversely, in the event that developmental genes are inappropriately expressed, the ability of PHF19 to recognize H3K36me3 may be a primary trigger for increased PRC2 loading at these loci to dampen transcription.
To bring about robust silencing, H3K27me3 further serves to recruit PRC1–CBX complexes, fortifying gene repression. Mechanistically, this may be achieved through enforcing a blockage to RNA Pol II engagement (Fig. 6), a role that can be fulfilled by PRC1 through chromatin compaction (for example, see Francis et al. 2004; Grau et al. 2011) and interference with the preinitiation transcription complex (Min et al. 2011; Lehmann et al. 2012). The PRC1-associated H2AK119 monoubiquitination (H2AK119ub1) modification may also interfere with FACT recruitment (Zhou et al. 2008). However, considering that PRC1 occupies only a subset of all bivalent promoters, additional buffering mechanisms are likely in place to keep these PRC2-only target genes in a state of low activity or inactivity. On this note, global run-on sequencing (GRO-seq) analysis in mouse ES cells shows that in the absence of PRC1, RNA Pol II remains engaged but largely confined to the 5′ proximal regions of bivalent promoters containing only PRC2 (Min et al. 2011). It is of great importance to identify what additional determinants are in place to limit RNA Pol II progression on these genes and, in particular, how H3K27me3-modified nucleosomes directly contribute to this process in a manner that is independent of PRC1. It remains theoretically possible that bivalency and its associated marks are a consequence of the transcriptional status at such genes rather than being instructive to their harboring initiating, not elongating, RNA Pol II. In vitro transcription assays will help clarify how H3K27me3-modified nucleosomes may affect the dynamics of RNA Pol II transcription along chromatin templates and, of special interest, whether (and how) different conformations of bivalent H3K4me3–H3K27me3 nucleosomes regulate transcription.
Expanding bivalency—players beyond trxG and PcG proteins
Thus far, we discussed how PcG and trxG complexes and their associated cofactors help establish the bivalent chromatin state. Many of these components are ubiquitously expressed in all tissues. However, given the overwhelming prevalence of bivalent domains in ES cells, it is conceivable that the pluripotency factors themselves may also play a regulatory role in landscaping this state. Indeed, one such example is Utf1. Utf1 is a transcriptional regulator whose expression is largely restricted to pluripotent tissues as well as germ cells. Interestingly, Utf1 was recently shown to bind to bivalent promoters, where it fulfills disparate roles in both limiting PRC2 recruitment and promoting mRNA degradation of spurious mRNAs that are inappropriately transcribed from these loci (Jia et al. 2012). In line with the facultative transcriptional state of bivalent promoters, Utf1 provides yet another layer of defense to limit spurious and unscheduled expression of lineage genes in ES cells. Taken together, an interplay between PRCs and the (post-)transcriptional apparatus is important in regulating expression of these genes in anticipation of developmental cues.
As our understanding of bivalent chromatin states increases, more novel players are likely to emerge. For example, a recent assessment of H3K27me3-containing mononucleosomes revealed an enrichment of H2A.Zub1, likely imposed by RING1B (Ku et al. 2012). Interestingly, a proportion of H2A.Zub1 can be further acetylated, constituting a dual-modified form of H2A.Z. The coexistence of these two seemingly opposing post-translational modifications on H2A.Z expands our current concept of bivalency beyond the classical H3K4me3–H3K27me3 definition, highlighting how different layers of regulation are encoded within developmental gene promoters. Again, how these disparate patterns of histone modifications impinge on the transcriptional machinery awaits investigation.
Resolution of bivalent domains during differentiation
During ES cell differentiation, a significant share of bivalent loci undergoes either activation or silencing. For activation to occur, a bivalent promoter needs to be cleared of repressive factors—most importantly, the PRCs and their associated marks. Moreover, appropriate TFs need to be recruited to the promoter and H3K4me3 to be installed. Conversely, stable repression of the gene requires removal of the H3K4me3 mark, exclusion of cognate TFs, and efficient silencing through PcG proteins, DNA methylation, and other means. These processes are initiated by signaling events that communicate developmental cues to the cell nucleus. These pathways often culminate in activation of specific TFs, leading to transcription of target genes. However, signals from the cell surface may also modulate chromatin-modifying activities, changing chromatin structure to facilitate or preclude transcription (Badeaux and Shi 2013). Despite advances toward unraveling both chromatin regulation and signal transduction in development, it remains largely unclear how these processes connect to guide lineage commitment.
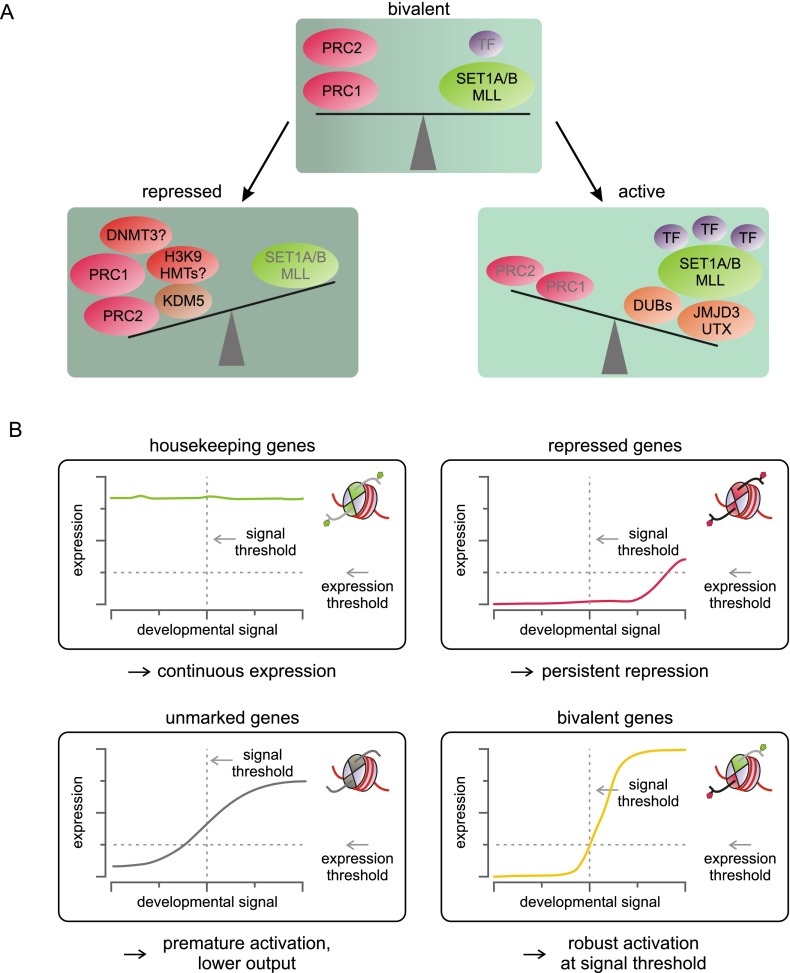
Compared with the establishment of bivalent domains, less is known about the factors controlling their resolution. In line with our model for the establishment of bivalent domains (Fig. 5), activation of bivalent genes can be conceived of as shifting the metastable equilibrium of the bivalent state toward activation (Fig. 7A). For many bivalent genes, the cognate TFs are either absent or inactive in ES cells (Ku et al. 2008), and their recruitment during differentiation likely plays a key role in activating transcription from these loci (Fig. 7A). In support of this view, ectopic expression of developmental TFs in ES cells leads to changes in transcriptional programs, strongly affecting bivalent genes (Nishiyama et al. 2009; Sharov et al. 2011). The presence of H3K27me3 thus not only represses gene expression, but may also confer responsiveness to environmental changes (Sharov et al. 2011). In contrast, expression of housekeeping genes remains largely unaffected, likely due to the continuous presence of active transcription complexes. To allow robust activation of bivalent genes, repressive marks and associated complexes must be displaced from bivalent promoters and gene bodies. We propose that this step introduces an important barrier against spurious activation of developmental genes, reducing noise and increasing activation thresholds, thereby conferring robustness to the system (Fig. 7B).
Figure 7.
Bivalency represents a dynamic equilibrium between activation and repression that keeps genes in a plastic, inducible state and at the same time increases robustness. (A) In the bivalent state, activating stimuli and repressive complexes counterbalance each other in a metastable equilibrium. Activating TFs in concert with H3K27 demethylases and H2.A-deubiquitinating enzymes (DUBs) shift the equilibrium toward activation, ultimately displacing repressive factors and converting bivalent loci to active ones. In contrast, removal of the activating stimuli that are present at low levels at bivalent domains shifts genes to a repressed state, a process that requires robust H3K4 demethylase activity. H3K9 methylation and DNA methylation may buttress repression. (B) For proper differentiation, genes need to be activated once a developmental signal reaches a certain threshold, while ensuring that significant expression of these genes does not occur prior to reaching that signal threshold. Only H3K4me3-marked genes such as housekeeping genes are constitutively active, regardless of signal levels. In contrast, fully repressed genes may require extremely high levels of signal for sufficient activation. Genes carrying neither active nor repressive marks may be induced, but the absence of repressive signals renders them vulnerable to noise and may lead to subthreshold activation. Bivalent genes exhibit the correct activation thresholds because higher levels of signal are required to displace the counterbalancing repressive factors.
Several studies have shown that PcG proteins are removed from specific loci following developmental signals through the action of TFs, histone demethylases, and enhancers and through the introduction of modifications that counteract binding of PcG proteins (Delest et al. 2012). Both UTX and JMJD3 are capable of demethylating H3K27me3, and both proteins are required for proper differentiation (Agger et al. 2007; Lee et al. 2007). UTX is part of the MLL3/4 complexes (Lee et al. 2007), whereas JMJD3 interacts with proteins involved in transcriptional elongation, such as the FACT subunit SPT16, and with KIAA1718, a demethylase for H3K9me2, H3K27me2, and H4K20me1 (Chen et al. 2012). Intriguingly, KIAA1718 recognizes H3K4me3 via its PHD finger (Horton et al. 2010), rendering it an ideal candidate for removing H3K27 methylation in the context of bivalency. Both UTX and JMJD3 rarely localize to bivalent promoters in ES cells but are recruited upon differentiation and are necessary for gene activation (Agger et al. 2007; Lan et al. 2007; Lee et al. 2007; Burgold et al. 2008). JMJD3 has also been shown to interact with Smad2 and Smad3, leading to its recruitment to Smad target genes in Nodal signaling (Dahle et al. 2010; Kim et al. 2011). Correspondingly, knockdown of JMJD3 diminishes expression of Nodal target genes (Dahle et al. 2010). Alternative signaling-dependent targeting mechanisms may exist that couple demethylation to additional developmental signals. Notably, enhancers appear to be critically involved in clearance of PcG proteins from bivalent promoters at least in part by mediating demethylase recruitment (Seenundun et al. 2010; Taberlay et al. 2011; Vernimmen et al. 2011). Interestingly, many enhancers likewise exist in a poised state featuring H3K27me3, and activation of these enhancers entails acetylation of H3K27 (Creyghton et al. 2010; Rada-Iglesias et al. 2011), which must be preceded by demethylation of H3K27me3.
Removal of H3K27me3 is expected to render bivalent promoters more amenable for transcription at least in part due to the dissociation of CBX-containing PRC1 complexes and reduced compaction. Moreover, removal of H3K27me3, an EED-binding target, will destabilize binding of PRC2 itself, potentially initiating a negative feedback loop augmented by the incurrence of symmetric H3K4me3 when transcription commences. Notably, JMJD3 has also been implicated in regulating the release of paused RNA Pol II into productive elongation, traveling along the gene body to clear H3K27me3 (Chen et al. 2012; Estarás et al. 2013). With respect to removal of PRC1 and deubiquitinylation of H2A, multiple proteins have been identified that possess deubiquitinylation activity toward H2A in mammals, including MYSM1, USP3, USP7, USP16, USP21, and USP22 (Weake and Workman 2008; Atanassov et al. 2011). Their regulation and targeting, however, remains largely elusive. Besides demethylation of H3K27me3 and loss of CBX binding, triggers that displace PRC1 are likewise mostly uncharacterized at present. The issue of PRC1 displacement is especially crucial in light of findings that key developmental genes are more likely to feature a bivalent state involving both PRC2 and PRC1 complexes (Ku et al. 2008). The combined action of both repressive complexes may further increase signaling thresholds, as displacement of both silencing systems is required for activation of those key genes.
After removal of H3K27me3 and other PcG signals, robust H3K4me3 needs to be established at bivalent promoters that become activated, presumably involving recruitment of SET1A/B and MLL complexes and feed-forward loops with active transcription as described above. Importantly, whereas PRC2 cannot modify H3 copies that carry H3K4me3 (Schmitges et al. 2011; Voigt et al. 2012), it is yet to be analyzed whether H3K4 trimethylases can act on the H3 copies that carry H3K27me3. Given the potential role of bivalent domains as a means to increase the activation threshold for developmental genes, it appears likely that at least certain H3K4 trimethylases are sensitive to H3K27me3. Notably, recent evidence suggests that the Cfp1 subunit of SET1A/B complexes is excluded from CpG islands harboring PcG proteins (Vernimmen et al. 2011), indicating the requirement for H3K27me3 removal and PcG clearance prior to H3K4 methylation by these complexes.
During differentiation, a sizable number of bivalent domains also undergo silencing. In this case, the equilibrium established at these bivalent promoters shifts to repression, implying a loss of activating input (Fig. 7A). In ES cells, bivalent promoters are often bound by the core pluripotency factors such as OCT4 (Boyer et al. 2005; Loh et al. 2006), presumably contributing to a basal level of activation at those promoters. Upon differentiation, these factors are down-regulated and may disappear from bivalent loci, tipping the balance toward repression. This reduction in activating signals may allow the PRCs to establish a stronger foothold around those promoters, forging repression, a process that is counterbalanced by H3K4me3 and its associated factors in ES cells. Similar to the presence of H3K27me3 as a barrier for activation, H3K4me3 may function to raise the threshold required to completely silence a gene upon differentiation, again conferring robustness to the system.
Through the loss of activating signals and perhaps the compromised recruitment of H3K4 trimethylases to bivalent domains that are being silenced, JARID1/KDM5 H3K4me3 demethylases may overcome counterbalancing activities and remove the singular, asymmetric H3K4me3 from the bivalent nucleosomes, allowing those H3 tails to be modified by PRC2. Both PRC2 and PRC1 may then establish a fully repressed environment. Moreover, other factors such as DNA methylation and H3K9 methylation may contribute to silencing at those loci. Even though most bivalent promoters remain unmethylated, loss of H3K4me3 correlates with increased DNA methylation at some loci (Meissner et al. 2008; Mohn et al. 2008). Interestingly, promoters that lose both H3K4me3 and H3K27me3 have a high probability of becoming DNA-hypermethylated (Meissner et al. 2008), supporting a role for both marks in preventing DNA methylation. The extent of H3K9me2 methylation markedly increases during differentiation, spanning large regions of silenced chromatin (Wen et al. 2009). Formerly bivalent genes may likely be included in those regions as well, being subjected to additional means to stabilize their silencing.
Conclusion
In the short history since their discovery, bivalent domains have garnered great attention as a means to poise gene expression in ES cells and beyond. Current evidence suggests that bivalent domains function in the fine-tuning of gene expression during development. The simultaneous presence of active and repressive modifications and associated complexes helps to maintain bivalent loci in a state that is both responsive to developmental cues and at the same time refractory to subthreshold noise. Despite tremendous progress toward understanding the establishment of bivalency as well as the action of marks and complexes in poising transcription, future work is clearly required to directly probe the importance of bivalency in developing organisms and further our knowledge of exactly how PcG proteins regulate transcription. The bivalency field is still in development.
Acknowledgments
We thank Dr. Lynne Vales for critical reading of this manuscript. P.V. is supported by fellowships from the Deutsche Akademie der Naturforscher Leopoldina (LPDS 2009-5) and the Empire State Training Program in Stem Cell Research (NYSTEM, contract no. C026880). W.W.T. is supported by a Druckenmiller post-doctoral fellowship from The New York Stem Cell Foundation. Work in the Reinberg laboratory was supported by grants from the National Institute of Health (GM064844 and R37GM037120) and the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.219626.113.
References
- Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs K-J, Stunnenberg HG, Veenstra GJC 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell 17: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O'Neill L, Azuara V 2010. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development 137: 2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Y-S, Tsai S-Y, Lee D-F, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold P, Schöler A, Pachkov M, Balwierz PJ, Jørgensen H, Stadler MB, van Nimwegen E, Schübeler D 2013. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res 23: 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Koutelou E, Dent SY 2011. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett 585: 2016–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK 2009. Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene 28: 815–823 [DOI] [PubMed] [Google Scholar]
- Badeaux AI, Shi Y 2013. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol 14: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, et al. 2012. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19: 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat SA, Jin V, Berry N, Balch C, Sharma N, Kurrey N, Zhang S, Fang F, Lan X, Li M et al. 2010. Multivalent epigenetic marks confer microenvironment-responsive epigenetic plasticity to ovarian cancer cells. Epigenetics 5: 716–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23: 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke M, Schreiner S, García-Cuéllar M-P, Mahr K, Titgemeyer F, Slany RK 2002. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res 30: 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M 2009. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien GL, Gambero G, O'Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al. 2012. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 19: 1273–1281 [DOI] [PubMed] [Google Scholar]
- Brockdorff N 2013. Noncoding RNA and Polycomb recruitment. RNA 19: 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. 2012. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G 2008. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 3: e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 285: 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Rothbart SB, Lu R, Xu B, Chen W-Y, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, et al. 2013. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 49: 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J 2013. Modification of enhancer chromatin: What, how, and why? Mol Cell 49: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D 2009. Histones: Annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, Conaway RC, Herr W 2011. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144: 376–388 [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T 2008. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234 [DOI] [PubMed] [Google Scholar]
- Chen S, Ma J, Wu F, Xiong L-J, Ma H, Xu W, Lv R, Li X, Villen J, Gygi SP, et al. 2012. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev 26: 1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X 2013. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hendrix DA, Core LJ, Tsui C, Lis JT, Levine M 2011. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell 42: 837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, Lee J-H, Skalnik D, Bird A 2012. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev 26: 1714–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA 2008. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh T-Y, Schones DE, Childs RW, Peng W, Zhao K 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P 2010. Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS ONE 5: e9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle Ø, Kumar A, Kuehn MR 2010. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 3: ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobbi M, Garrick D, Lynch M, Vernimmen D, Hughes JR, Goardon N, Luc S, Lower KM, Sloane-Stanley JA, Pina C, et al. 2011. Generation of bivalent chromatin domains during cell fate decisions. Epigenetics Chromatin 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delest A, Sexton T, Cavalli G 2012. Polycomb: A paradigm for genome organization from one to three dimensions. Curr Opin Cell Biol 24: 405–414 [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. 2013. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J 32: 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N, Rappsilber J, Södersten E, Hansen K 2012. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet 8: e1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M 2013. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell 49: 368–378 [DOI] [PubMed] [Google Scholar]
- Estarás C, Fueyo R, Akizu N, Beltrán S, Martínez-Balbás MA 2013. RNA polymerase II progression through H3K27me3-enriched gene bodies requires JMJD3 histone demethylase. Mol Biol Cell 24: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW et al. 2012. KDM2B links the Polycomb repressive complex 1 (PRC1) to recognition of CpG islands. Elife 1: e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T 1995. The eed mutation disrupts anterior mesoderm production in mice. Development 121: 273–285 [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, Chen X 2010. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol 11: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE 2011. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev 25: 2210–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA 2010. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC 2012. Bittersweet memories: Linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol 13: 312–321 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Lopes SM, Tang F, Surani MA 2008. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3: 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y 2013. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X 2010. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17: 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K 2013. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12: 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M, Krogan N, Ren B, Song JS, Reiter JF 2012. Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet 8: e1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Botting CH, Grimes GR, Bickmore WA, Eskeland R 2012. PRC1 and PRC2 are not required for targeting of H2A.Z to developmental genes in embryonic stem cells. PLoS ONE 7: e34848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422–1425 [DOI] [PubMed] [Google Scholar]
- Jacquier A 2009. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844 [DOI] [PubMed] [Google Scholar]
- Jia J, Zheng X, Hu G, Cui K, Zhang J, Zhang A, Jiang H, Lu B, Yates J, Liu C, et al. 2012. Regulation of pluripotency and self-renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell 151: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J 2009. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res 37: 2940–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Yoon S-J, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J 2011. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol 357: 492–504 [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al. 2013. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497: 122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, Bernstein BE 2012. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol 13: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jørgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. 2010. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12: 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG 2013. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152: 1021–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Voo KS, Skalnik DG 2001. Identification and characterization of the DNA binding domain of CpG-binding protein. J Biol Chem 276: 44669–44676 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan K-P, Reinberg D, Di Croce L, Shiekhattar R 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A 2010. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann L, Ferrari R, Vashisht AA, Wohlschlegel JA, Kurdistani SK, Carey M 2012. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J Biol Chem 287: 35784–35794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata C, Mathavan S, Müller F, Aleström P, et al. 2011. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell 21: 993–1004 [DOI] [PubMed] [Google Scholar]
- Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Lynch MD, Smith AJH, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. 2012. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. 2007. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2010. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149: 590–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB 2008. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res 68: 5753–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS 2013. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20: 300–307 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL 2005. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci 102: 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT 2011. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 25: 742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10: 47–62 [DOI] [PubMed] [Google Scholar]
- Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L 2013. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep 3: 60–69 [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y 2010. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 79: 155–179 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde M-E, Hong Z, Allen C, Roy S, Nuñez JK, Nickoloff J, et al. 2012. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19: 1266–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. 2009. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5: 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin S-P, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K 2011. Chromatin connections to pluripotency and cellular reprogramming. Cell 145: 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Guenther MG, Frampton GM, Young RA 2012. CpG island structure and trithorax/polycomb chromatin domains in human cells. Genomics 100: 320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA 2007. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1: 299–312 [DOI] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J 2010. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell 6: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PAC, Helin K 2008. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-repressive complex 2. Genes Dev 22: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP 2009. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF 2011. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AHFM, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G 2009. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B 2012. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J 31: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Muñoz M, Vives L, Frangou CG, Groudine M, Peinado MA 2008. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc Natl Acad Sci 105: 19809–19814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh T-Y, Cuddapah S, Cui K, Zhao K 2006. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci 103: 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ 2009. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics 10: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J 2010. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci 107: 10783–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J 2007. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D 2005. Histone variants meet their match. Nat Rev Mol Cell Biol 6: 139–149 [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci 103: 1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Arteaga-Salas JM, Mentele E, David R, Nicetto D, Imhof A, Rupp RAW 2011. Stage-specific histone modification profiles reveal global transitions in the Xenopus embryonic epigenome. PLoS ONE 6: e22548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu Q-C, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ 2010. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J 29: 1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Ko MSH 2007. Human ES cell profiling broadens the reach of bivalent domains. Cell Stem Cell 1: 237–238 [DOI] [PubMed] [Google Scholar]
- Sharov AA, Nishiyama A, Piao Y, Correa-Cerro LS, Amano T, Thomas M, Mehta S, Ko MSH 2011. Responsiveness of genes to manipulation of transcription factors in ES cells is associated with histone modifications and tissue specificity. BMC Genomics 12: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu Y-J, Fujiwara Y, Kim J, Mao X, Yuan G-C, Orkin SH 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan G-C, Lee Y, Orkin SH 2009. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo J-HA, Lake A, Davrazou F, Chan SM, Martin DGE, Fingerman IM, Briggs SD, et al. 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem 282: 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2012. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A 2010. The chromatin signaling pathway: Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K 2010. Induced pluripotency: History, mechanisms, and applications. Genes Dev 24: 2239–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu C-C, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA 2011. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell 147: 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S 2010. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11: 264–275 [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ 2007. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Tremblay V, Lin G, Bochar DA 2008. CHD8 is an ATP-dependent chromatin remodeling factor that regulates β-catenin target genes. Mol Cell Biol 28: 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr ARW, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H 2008. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135: 909–918 [DOI] [PubMed] [Google Scholar]
- van den Berg DLC, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Loyola A, Reinberg D 2003. The constantly changing face of chromatin. Sci Aging Knowledge Environ 2003: RE4. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, et al. 2013. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell 49: 645–656 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, Lynch MD, De Gobbi M, Garrick D, Sharpe JA, Sloane-Stanley JA, Smith AJH, Higgs DR 2011. Polycomb eviction as a new distant enhancer function. Genes Dev 25: 1583–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Reinberg D 2011. Histone tails: Ideal motifs for probing epigenetics through chemical biology approaches. Chembiochem 12: 236–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Leroy G, Drury WJ 3rd, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D 2012. Asymmetrically modified nucleosomes. Cell 151: 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL 2010. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 6: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-L, Faiola F, Xu M, Pan S, Martinez E 2008. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem 283: 33808–33815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL 2008. Histone ubiquitination: Triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP 2009. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet 41: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. 2007. Epigenetic stem cell signature in cancer. Nat Genet 39: 157–158 [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K 2012. DNA methylation: TET proteins—guardians of CpG islands? EMBO Rep 13: 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE 2010. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13: 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y 2011. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K 2013. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49: 1134–1146 [DOI] [PubMed] [Google Scholar]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W 2012. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci 109: 8161–8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap L-S, Hayashi K, Surani MA 2009. ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q-L, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA 2011. Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA 2009. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics 8: 2266–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME, Oshlack A, Majewski IJ 2011. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res 39: 7415–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh Y-H, Yaw L-P, Robson P, Lim B, Ng H-H 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23: 2507–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. 2012. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337: 971–975 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. 2010. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 38: 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung W-K, Shahab A, Kuznetsov VA, et al. 2007. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1: 286–298 [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]